Abstract
Pseudokinases play key roles in many biological processes but they are poorly understood compared to active kinases. Eight putative pseudokinases have been predicted in Plasmodium species. We selected the unique pseudokinase belonging to tyrosine kinase like (TKL) family for detailed structural and functional analysis in P. falciparum and P. berghei. The primary structure of PfpTKL lacks residues critical for kinase activity, supporting its annotation as a pseudokinase. The recombinant pTKL pseudokinase domain was able to bind ATP, but lacked catalytic activity as predicted. The sterile alpha motif (SAM) and RVxF motifs of PfpTKL were found to interact with the P. falciparum proteins serine repeat antigen 5 (SERA5) and protein phosphatase type 1 (PP1) respectively, suggesting that pTKL has a scaffolding role. Furthermore, we found that PP1c activity in a heterologous model was modulated in an RVxF-dependent manner. During the trophozoite stages, PbpTKL was exported to infected erythrocytes where it formed complexes with proteins involved in cytoskeletal organization or host cell maturation and homeostasis. Finally, genetic analysis demonstrated that viable strains obtained by genomic deletion or knocking down PbpTKL did not affect the course of parasite intra-erythrocytic development or gametocyte emergence, indicating functional redundancy during these parasite stages.
Subject terms: Phosphorylation, Parasite biology
Introduction
Malaria remains one of the most life-threatening parasitic diseases in the world, with 219 million cases leading to an estimated 435,000 deaths worldwide in 20171. Among the Plasmodium species that infect humans, P. falciparum is the most virulent and is responsible for more than 90% of deaths, mostly in African children less than 5 years old. Thanks to preventative measures such as bed nets and treatments mainly involving artemisinin-based combination therapy (ACT), the global incidence of malaria fell by 38% between 2010 and 20162,3. However, the rapid spread of ACT-resistant Plasmodium strains in South-East Asia is alarming, and innovative antimalarial strategies are therefore needed4–6.
The life cycle of Plasmodium species is complex because the parasite infects two hosts and progresses through various stages of development that are replicative, invasive or involved in sexual reproduction7,8. To sustain development and respond to environmental changes, the parasite uses post-translational modifications (PTMs) for the dynamic modulation of protein function, in many cases phosphorylation9. This reversible modification is catalyzed by protein kinases and the phosphate groups are removed by protein phosphatases. Phosphoproteomics approaches have revealed that kinases and phosphatases play key roles in cell growth, division, motility, invasion and communication with host cells during the Plasmodium life cycle10–13. The essential role of these enzymes has also been demonstrated using phosphatase and kinase inhibitors14–16, most of which are deleterious to the parasite17. Reverse genetics has helped to decipher the role of several of these enzymes at each developmental stage12,18,19. These studies, as well as the more detailed characterization of specific enzymes, have improved our understanding of Plasmodium signal transduction pathways20.
Although phosphorylated tyrosine residues can be detected in the late Plasmodium development stages13, the parasite kinome is mainly composed of serine/threonine (S/T) kinases21. Hanks-type S/T kinases are membrane-bound or cytoplasmic enzymes that share common features22, including a 12-subdomain signature in the catalytic domain required for kinase activity23,24. Active kinases take part in regulatory networks that allow the integration of signals by controlling protein structure and activity. Enzymes that lack one of the catalytic residues required for kinase activity are classified as pseudokinases25. They constitute 10% of the mammalian kinome and fulfil a plethora of biological roles, depending on their subdomain composition and their ability to bind ATP and/or divalent ions26,27. They can introduce PTMs, either as active kinases if they have evolved compensatory mechanisms to overcome the lack of one or more key subdomains (e.g., CASK28, Haspin29, WNK130, and other examples that have been reviewed31,32) or as AMPylation catalyzers33, demonstrating the functional versatility of kinase domains. Some pseudokinases also play a role in cell signaling. They can notably function as protein–protein interaction platforms or allosteric regulators of various enzymes, including kinases34, or of protein trafficking and subcellular localization35. Their importance in metazoan cell homeostasis is highlighted by their role in diseases, including cancer36.
In apicomplexan parasites, most pseudokinases are poorly characterized, but the highly polymorphic ROP5 family in Toxoplasma gondii has been studied in detail37. TgROP5 binds ATP but lacks kinase activity in vitro, probably due to the absence of a catalytic aspartate residue in subdomain VI. However, the conservation of this non-canonical motif is necessary for parasitic virulence37. TgROP5 forms a complex with the kinases TgROP17 and TgROP18, causing the synergistic neutralization of host immunity-related GTPases (IRGs), favoring infection by protecting the parasite from cell-mediated clearance38,39. Reverse genetics has been used to confirm that TgROP5 is required for parasite virulence in genetically divergent strains40, revealing that parasite pseudoenzymes can play essential roles at the host–pathogen interface.
The Plasmodium kinome was initially predicted to include eight pseudokinases41, five of which are probably essential for the intra-erythrocytic development of the parasite19. Here, we report the characterization of the unique Plasmodium pseudo-tyrosine kinase-like (pTKL) protein, previously named TKL542. We evaluated its potential for kinase activity, characterized its interactions to establish its molecular function, and investigated its biological role using different knockout and knock-down approaches.
Results
PfpTKL transcript sequencing and protein annotation
To confirm PfpTKL transcription, we performed overlapping PCR on reverse transcribed Pf3D7 total RNA using primer pairs derived from the predicted PF3D7_1106800 sequence (Supplementary Fig. S1 and Table S1). We analyzed 6–12 sequences amplified from two independent reverse transcribed RNA preparations for each of the eight transcript fragments (length 600–1200 bp). The deduced amino acid sequence (1483 residues) was compared to the PlasmoDB43 predicted sequence. Both sets of data were identical with the exception of minor substitutions that did not affect the overall annotation (Supplementary Fig. S1).
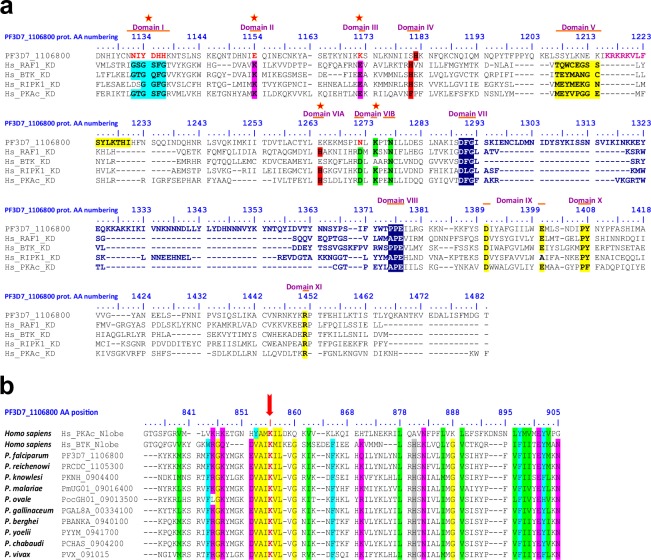
More detailed analysis revealed the presence of multiple domains (Figs 1 and 2; summarized in Table 1 and showed in Fig. 3a). From the N-terminus to the C-terminus, we identified two membrane occupation recognition nexus (MORN) motifs (Fig. 2a), one classical sterile alpha motif (SAM) predicted to bind proteins (Fig. 2c,d; Supplementary Fig. S5), one Bruton’s tyrosine kinase (BTK)-like N-terminal lobe, designated as BTK-like domain (Fig. 1b; Supplementary Fig. S3) and one kinase domain (KD; Fig. 1a). We found that the KD lacks key residues required for ATP stabilization and phosphoryl transfer, confirming that PfpTKL is a pseudokinase (Fig. 1a). Two putative RVxF motifs, described as protein phosphatase 1 (PP1) catalytic subunit PP1c binding motifs44, were also identified (Fig. 2b). RVxF1 was localized to the C-terminal side of the SAM whereas RVxF2 was localized within the KD. Low-complexity regions enriched in asparagine, lysine or serine were identified between the predicted folded domains.
Figure 1.
In silico annotation of PF3D7_1106800 (PfpTKL) catalytic domains. (a) PfpTKL has a pseudo-kinase domain. Sequence alignment between human PKAc (used as the kinase domain reference), RAF1 (used as a TKL reference), RIPK1 (used as a pseudo-TKL reference), BTK (used as a tyrosine kinase reference) and PfpTKL kinase domains. The kinase subdomains (I to XI on the sequence alignment) were delineated according to Hanks & Hunter23 and are shaded. Non canonical S/T kinase subdomain amino acids are shown in red and marked with a red star. The activation loop residues are shown in dark blue. (b) PfpTKL possesses a Bruton’s tyrosine kinase (BTK) N-terminal lobe-like domain upstream of its pseudo-kinase domain. Protein sequence alignment of BTK-like domains of PfpTKL and its Plasmodium homologs with the N-terminal lobe sequences of human BTK and PKAc. The alignment was created with MAFFT89 and conserved residues were shaded using BioEdit v7.2.5116. The ATP-binding lysine conserved across all the aligned sequences is indicated with a red arrow.
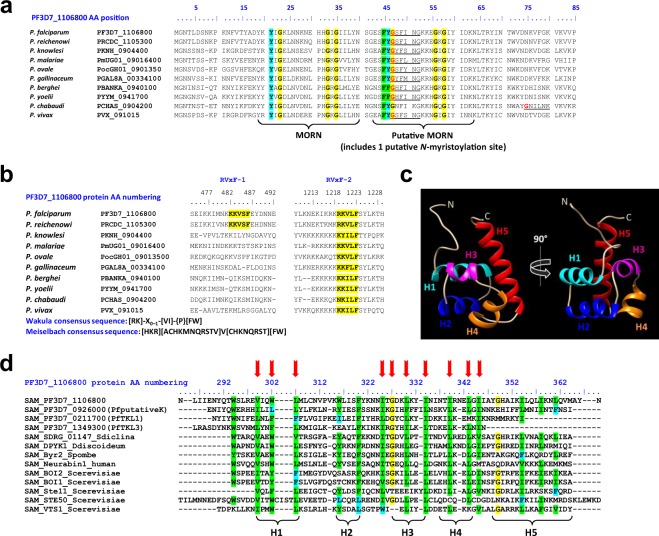
Figure 2.
In silico annotation of PfpTKL interaction motifs and domains. (a) Plasmodium PfpTKL homologs share common N-terminal features beside the MORN motif(s). The alignment was created with MAFFT89 and residues 100% conserved over all of the sequences were shaded using BioEdit v7.2.5116. The putative N-myristoylation site is shown in red. (b) Plasmodium PfpTKL homologs possess one or two RVxF motifs. The alignment was created with MAFFT89. RVxF motif consensus sequences used in silico to detect RVxF1 and RVxF2 are displayed below the alignment86,87. (c) The PfpTKL SAM undergoes classic folding. Tertiary structure PfpTKL SAM predicted with Phyre281 (75 residues (97%) modeled at >90% accuracy). Structure annotation and shading were achieved with Chimera v1.10.1117. (d) Plasmodium SAMs showing classic composition compared with SAM references. Sequence alignment created with MAFFT89 between the four P. falciparum SAMs and reference SAMs (Sdiclina = Saprolegnia diclina, Scerevisiae = Saccharomyces cerevisiae, Spombe = Schizosaccharomyces pombe, Ddiscoideum = Dictyostelium discoideum). Conserved residues were shaded using BioEdit v7.2.5116. Arrows indicate residues conserved in SAMs. Below the alignment are delineated the five α-helices (H1-H5) of the SAM core118.
Table 1.
PfpTKL domain/motif annotation results.
| Domain/motif name | PfpTKL region (AA) | Data from the literature | PfpTKL annotation data (this study) |
|---|---|---|---|
| Kinase domain (KD) |
1080–1483 |
Kinase domains are composed of 12 subdomains23, six of which are critical for kinase activity25. PfpTKL lacks some canonical KD subdomains41. PbpTKL is a pseudokinase19. |
PfpTKL KD (Fig. 1a): —belongs to the tyrosine kinase-like (TKL) family42 (Supplementary Fig. S1). —lacks the following subdomains: -I (glycine triad) -II (ATP-binding lysine) although there might be an alignment offset because the PfpTKL KD N-terminal lobe is longer than in reference kinases -III (αC helix glutamate can interact with subdomain II lysine) —includes some specificities: -VIB aspartate (ATP γ-phosphate transfer catalyzer) is mutated to asparagine -a ~50-residue insertion in its activation loop (shown in red in Fig. 1a). |
| Bruton’s tyrosine kinase-like (BTK-like) domain |
830–905 (Fig. 1b) |
BTK is named after Dr Ogden Carr Bruton, who discovered X-linked agammaglobulinemia (XLA) in boys in 1952. This cytoplasmic tyrosine kinase (mostly in B lymphocytes) needs to bind lipids via its PH-TH module to adopt an active conformation104. BTK is a promising target in tumors105. |
BTK-like domain: —was predicted by most of the prediction tools we used. —alignment with HsPKA and HsBTK N-terminal lobe sequences shows the conservation of canonical KD subdomains II (this lysine is shaded in red in Fig. 1b), IV (highlighted in gray) and V. —tertiary structure prediction superimposes well with HsBTK N-terminal lobe (Supplementary Fig. S3). |
| Membrane occupation recognition nexus (MORN) domain |
MORN1: 19–40 MORN2: 43–63 (Fig. 2a) |
First identified in Junctophilins106, MORN motifs have been shown to bind: -lipids and regulate kinase activity in plant PIPKs107,108 -the cytoskeleton as in TgMORN1109 -membranes and is required for the localization of retinophilin in the Drosophila melanogaster rhabdomere110 |
There is no signal peptide at the N-terminus of PfpTKL. MORN1 follows the MORN motif signature: a 14–16 residue core domain starting with Y/FxG and ending with GxG (Supplementary Fig. S4). MORN2 does not follow a canonical MORN signature but contains a putative N-myristoylation site that is conserved across all Plasmodium species (Fig. 2a). |
| RVxF motifs |
RVxF1: 482–486 RVxF2: 1219–1223 (Fig. 2b) |
RVxF motifs: - so-called because of their binding signature111 - are the most frequent binding motifs used by PP1 partners to interact with PP1c112 - can be predicted with consensus sequences (Fig. 2b) |
RVxF1 follows the Meiselbach consensus sequence87. It is located in a low-complexity region, which may increase its interaction potential with PP1c50. It is conserved among PfpTKL homologs in P. praefalciparum, P. reichenowi, P. billcollinsi and P. blacklocki (Fig. 3c). RVXF2 follows the Wakula consensus sequence86. It is located in a basic charged region, which could reinforce its binding potential75. It is conserved across all PfpTKL homologs in the genus Plasmodium (Figs 2b and 3c). |
| Sterile alpha motif (SAM) |
299–364 (Fig. 2d) |
SAMs belong to the alpha protein family (289 folds – SCOP database113). They typically fold into 4–5 orthogonal α helices. They are involved in interactions with protein or RNA71,72. In the Plasmodium proteome: - there are four SAMs (PfpTKL, PfTKL1, PfTKL3, PF3D7_0926000)66 - there are two SAM-like domains involved in interactions with RNA (prototypical HTH motifs) in Pfg27114,115. |
The PfpTKL SAM domain is predicted to fold classically (Fig. 2c). The PfpTKL SAM core domain includes all SAM canonical residues (Fig. 2d). The PfpTKL SAM domain is close to SAMs involved in protein–protein interactions (Supplementary Fig. S5). This phylogenetic analysis includes Plasmodium SAM and SAM-like sequences as well as reference SAM and SAM-like sequences across the tree of life. |
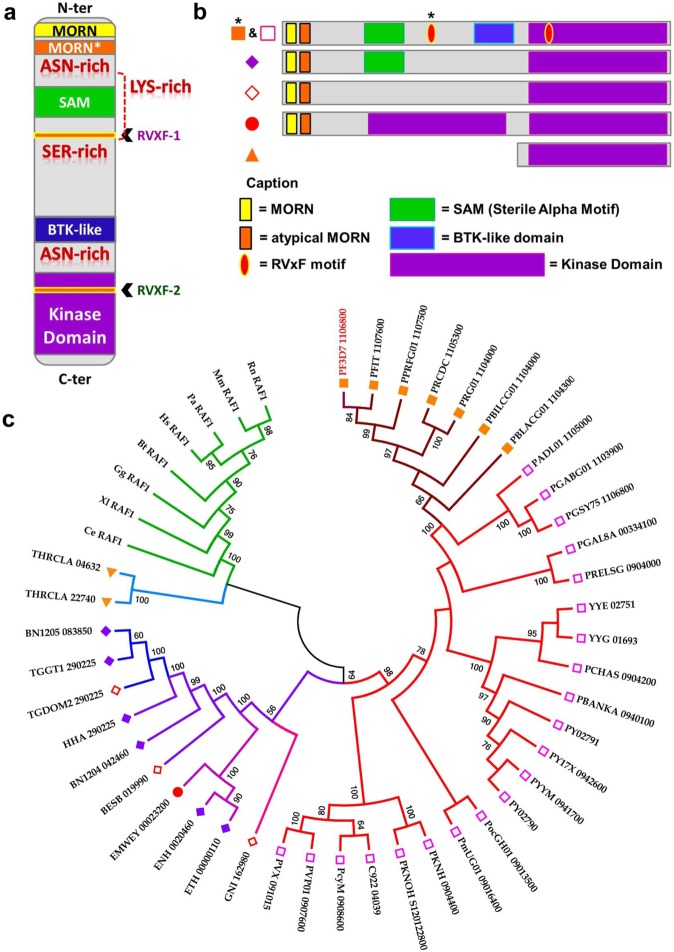
Figure 3.
Molecular structure and phylogenetic analysis of PfpTKL and its homologs in the Plasmodium genus. (a) PfpTKL molecular structure. All protein domains/motifs (MORN, SAM, RVxF, BTK-like, kinase) identified in silico are shown, as well as amino acid repeat regions (Asn = asparagine, Lys = lysine and Ser = serine, delineated using ScanProsite80). (b) Comparison of PfpTKL and its homologs. Sequence alignment was created in MAFFT89 and was used to assess conserved domains. Symbols on the left are linked to the proteins included in the phylogenetic analysis displayed in (c). (c) Phylogenetic analysis of PfpTKL and its homologs. Maximum likelihood phylogenetic analysis of PfpTKL homologs from: Alveolata: PF3D7 = Plasmodium falciparum 3D7, PFIT = P. falciparum IT, PPRFG01 = P. praefalciparum G01, PRCDC = P. reichenowi CDC, PRG01 = P. reichenowi G01, PBILCG01 = P. billcollinsi G01, PBLACG01 = P. blacklocki G01, PADL01 = P. adleri G01, PGAB01 = P. gaboni G01, PGSY75 = P. gaboni SY75, PYYM = P. yoelii yoelii YM, PY = P. yoelii 17XNL, PBANKA = P. berghei ANKA, PCHAS = P. chabaudi, YYE = P. vinckei vinckei VINCKEI, YYG = P. vinckei petteri CR, PGAL8A = P. gallinaceum 8A, PRELSG = P. relictum SGS1-like, PocGH01 = P. ovale curtisi GH01, PmUG01 = P. malariae UG01, PKNOH = P. knowlesi Malayan PK1, PKNH = P. knowlesi H, C922 = P. inui San Antonio 1, PcyM = P. cynomolgi M, PVP01 = P. vivax 01, PVX = P. vivax Sal 01, GNI = Gregarina niphandrodes, ETH = Eimeria tenella, ENH = Eimeria necatrix, EMWEY = Eimeria maxima, BESB = Besnoitia besnoiti, BN1204 = Neospora caninum, HHA = Hammondia hammondi, TGDOM2 = Toxoplasma gondii DOM2, TGGT1 = T. gondii GT1, BN1205 = T. gondii VEG; Stramenopile: THRCLA = Thraustotheca clavata. Eight RAF1 protein sequences from Opisthokonta: Mammals (Rn = Rattus norvegicus, Mm = Mus musculus, Pa = Pongo abelii, Hs = Homo sapiens, Bt = Bos taurus), Aves (Gg = Gallus gallus), Amphibia (Xl = Xenopus laevis), Chromaderea (Ce = Caenorhabditis elegans) were included in the analysis as an outgroup.
Phylogenetic classification of PfpTKL
We constructed a phylogenetic tree comprising PfpTKL homologs and a RAF1 outgroup. The tree with the highest log likelihood is presented in Fig. 3c. The topology of the tree reflects the general classification of eukaryotes45, with the apicomplexa clustering together in one clade (Alveolata), as well as the Stramenopile Thraustotheca clavata and the Metazoa (RAF1 outgroup). Within the alveolates, Toxoplasma, Eimeria, Neospora, Hammondia and Besnoitia (Eucoccidia) cluster in one clade, as well as Plasmodium (Haemosporida). Gregarina niphandrodes forms a monophyletic clade within the alveolates. This classification reflects the molecular structure of all of the proteins displayed in Fig. 3b. Stramenopile proteins contain only one KD. All alveolate proteins contain two MORN motifs at their N-terminus (one atypical) and at least one KD at their extreme C-terminus. Eucoccidia proteins occasionally contain a SAM domain in addition to MORN motifs and a KD. Eimeria maxima stands out with two KDs, but nevertheless clusters with other Eimeria proteins. The Plasmodium clade members are the most complex in terms of molecular composition. In addition to the domains listed for Eucoccidia, they also contain at least one RVxF motif in their KD and a BTK-like domain upstream of the C-terminal KD. Interestingly, the P. falciparum, P. praefalciparum, P. reichenowi, P. billcollinsi and P. blacklocki sequences clustered together as previously described46 and stand out among all Plasmodium sequences given the presence of a second RVxF motif located downstream of the SAM domain. Importantly, this analysis revealed that pTKL is specific to certain apicomplexan species.
The PfpTKL KD binds ATP but is catalytically inactive
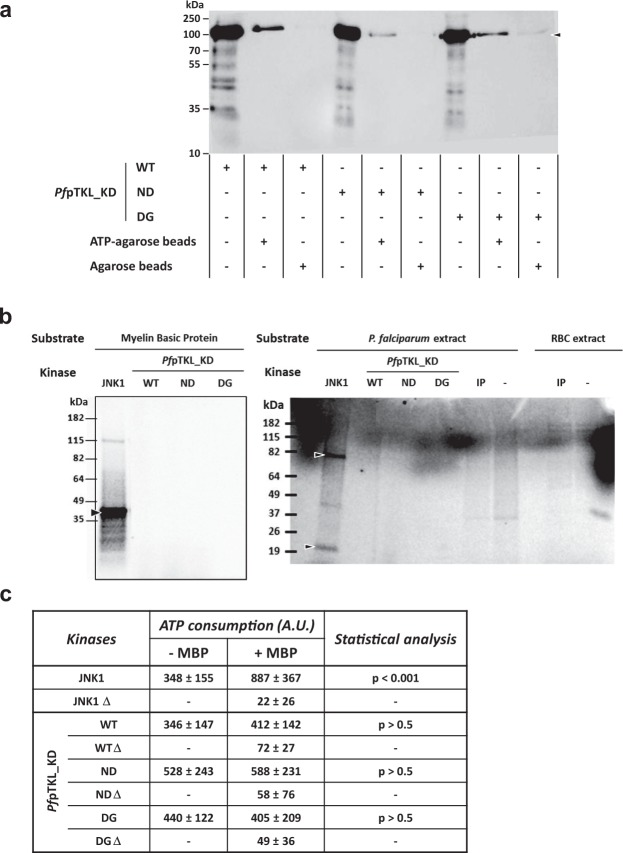
The PfpTKL KD was produced as a recombinant protein named pTKL_KD_WT (Supplementary Fig. S6) and its ability to bind ATP was evaluated using a pull-down assay based on ATP-agarose beads (Fig. 4a). To determine whether the ATP-binding activity of PfpTKL KD is cation dependent, the amino acid triplet 1290DFG1292 in subdomain VII was mutated to 1290GFE1292 (pTKL_KD_DG) because this motif is required for cation binding to stabilize ATP and favor phosphate group transfer. The pTKL_KD_DG mutant retained its ability to bind ATP (Fig. 4a) indicating that PfpTKL belongs to the class 2 (nucleotide-only binding) pseudokinases26.
Figure 4.
PfpTKL kinase domain (pTKL_KD_WT) and its mutants (pTKL_KD_ND and pTKL_KD_DG) bind ATP but do not display any kinase activity in vitro. (a) Plasmodium pTKL kinase domain binds ATP. Wild-type and mutated recombinant pTKL kinase domains were incubated with ATP-agarose or agarose beads. Eluates were separated by SDS-PAGE and His6-tagged proteins bound to the beads were detected by western blot. (b) Plasmodium pTKL kinase domain does not catalyze kinase reactions. Kinase reactions performed with γ33P-ATP were stopped by the addition of loading buffer and separated by SDS-PAGE. Gels are displayed according to the substrate: myelin basic protein (left panel), P. falciparum and erythrocyte extracts (right panel). WT = wild-type recombinant kinase domain, ND = N1272D mutant and DG = 1290DFG1292 to 1290GFE1292 mutant. IP = immunoprecipitated PbpTKL-AID-HA. JNK1 was used as an active kinase control. (c) Plasmodium pTKL kinase domain binds ATP but does not display any kinase activity. WT and mutant recombinant pTKL kinase domains were distributed in plates in the presence or absence of myelin basic protein used here as a substrate. JNK1 was used as an active kinase control and all kinases were also heated (∆) to generate negative controls. After incubation at 37 °C for 2 h, the quantity of free ATP remaining in wells was evaluated by luminometry. The ATP consumption displayed in the table corresponds to the difference between the luminometry value obtained without kinase and the luminometry value obtained with the maximum amount of kinase (80 ng/well for JNK1, and 8 µg/well for PfpTKL) (mean ± SD; three biological replicates and three technical replicates per data point – raw data are available in Supplementary Table S4).
Next, we tested the catalytic activity of pTKL_KD_WT and pTKL_KD_DG using γ33P-ATP and three different substrates: myelin basic protein (MBP), P. falciparum extracts and erythrocyte extracts (Fig. 4b). The positive control JNK1 displayed kinase activity against both MBP and the P. falciparum extracts, confirming the assay was working correctly, but neither pTKL_KD_WT nor pTKL_KD_DG were active against any substrate (Fig. 4b). To determine the reason for inactivity, we mutated N1272 back to a canonical phosphate group-transferring subdomain VIB aspartate (pTKL_KD_ND), but this mutant remained inactive despite retaining the ability to bind ATP (Fig. 4a,b).
Finally, we conducted a luminescence assay in the presence of MBP, which confirmed the ability of JNK1 to hydrolyze ATP whereas pTKL_KD_WT, pTKL_KD_ND and pTKL_KD_DG lacked such activity (Fig. 4c, Supplementary Table S4).
Collectively, these experiments showed that the PfpTKL pseudo-KD was able to bind ATP but did not display any kinase activity.
PfpTKL RVxF1 and RVxF2 bind PfPP1c in vitro but only RVxF2 is functional in Xenopus laevis oocytes
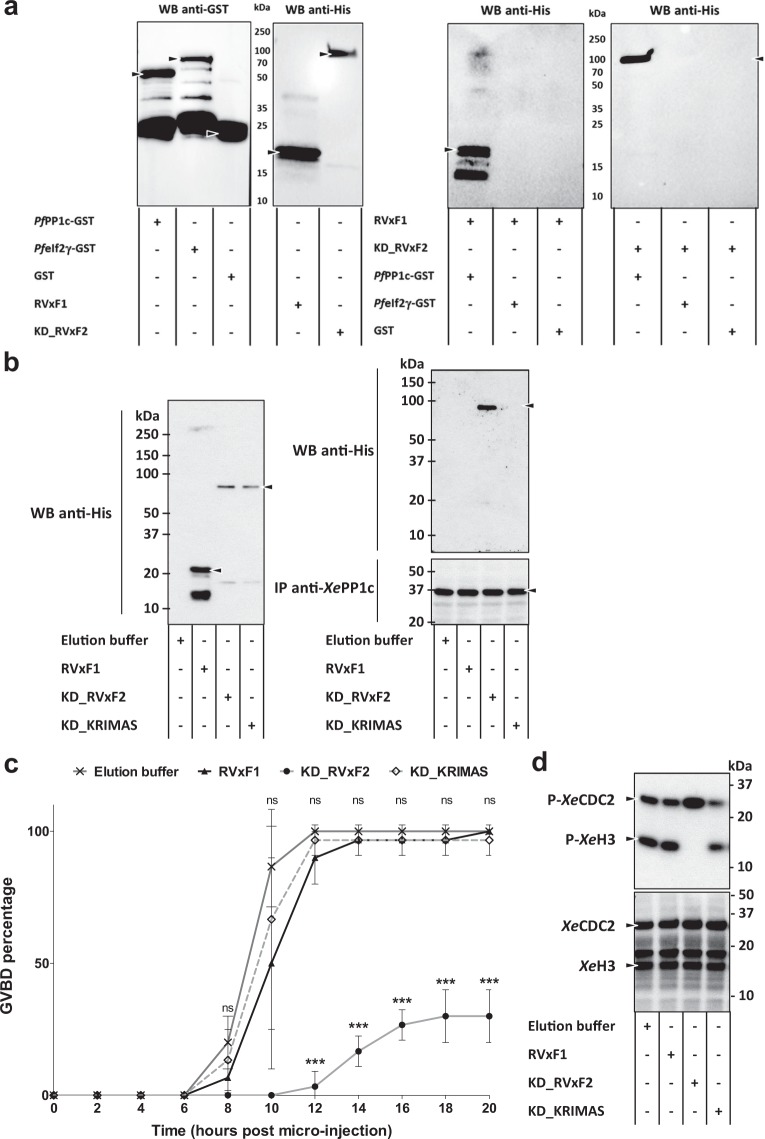
The ability of the PfpTKL RVxF1 and RVxF2 motifs to bind the PfPP1 catalytic subunit (PfPP1c) was determined using glutathione S-transferase (GST) pull-down assays. A portion of PfpTKL containing RVxF1 (residues 371–531) was produced as the recombinant protein His6-pTKL_RVxF1 (Supplementary Fig. S6) to test the first motif, and pTKL_KD_WT (containing RVxF2, as described above) was used to test the second one. PfeIf2γ-GST and GST were used as negative controls47. Both His6-pTKL_RVxF1 and pTKL_KD_WT were only pulled down by PfPP1c-GST beads (Fig. 5a, right panel, lanes 1 and 4).
Figure 5.
Both PfpTKL RVxF1 and RVxF2 motifs bind PfPP1c in vitro but only RVxF2 exerts a functional role in Xenopus laevis oocytes. (a) RVxF1 and RVxF2 both directly bind PfPP1c in vitro. PfPP1c-GST, PfeIf2γ-GST or GST (the last two = negative control) beads were incubated with His6-pTKL_RVxF1 (RVxF1) or pTKL_KD_WT (KD_RVxF2). Following washes, beads were eluted in Laemmli buffer. Proteins were separated by SDS-PAGE and His6-tagged proteins bound to beads were identified by western blot (WB, right panel – both blot first lanes show the specific and direct interaction of RVxF1 and KD_RVxF2 with PfPP1c-GST). All inputs were analyzed by western blot as well (left panel). Uncropped blots are available in the Supplementary Information. (b) Only RVxF2 binds XePP1c in vivo. RVxF1, KD_RVxF2 and KD_KRIMAS (containing RVxF2 inactivated by mutation) were micro-injected into X. laevis oocytes. To check for the presence of RVxF-containing proteins, oocytes were lysed and His6-tagged proteins were separated by SDS-PAGE and detected by western blot (left panel). Oocytes micro-injected with RVxF1, KD_RVxF2 or KD_KRIMAS and incubated with progesterone were lysed 15 h after micro-injection and anti-XePP1c immunoprecipitation assays (IP) were performed. After washes, proteins bound to beads were detected by western blot (right panel). (c,d) RVxF2 inhibits progesterone-induced germinal vesicle breakdown (GVBD) in X. laevis oocytes. (c) Chart showing GVBD percentage over time in progesterone-incubated X. laevis oocytes micro-injected either with proteins or with control buffers (mean ± SD; three biological replicates and two technical replicates with 10 oocytes per data point). Oocyte images (external view and hemisections) are available for each sample in the Supplementary Information. ***, p < 0.001. (d) GVBD of oocytes micro-injected with RVxF-containing proteins was assayed by analyzing XeCDC2 dephosphorylation and XeH3 phosphorylation by western blot. Only KD_RVxF2 (third lane) was able to block progesterone-induced XeH3 phosphorylation. Lower panel, western blot analysis of total XeCDC2 and XeH3. Uncropped blots are available in the Supplementary Information.
To investigate the PfpTKL–PP1c interaction in more detail, we used Xenopus laevis oocytes as a model system given that X. laevis and P. falciparum PP1c share 85% identity at the amino acid level. X. laevis oocytes arrested at G2/M meiotic prophase I can be used to study signaling pathways involved in cell cycle progression because the induction of meiosis with progesterone triggers germinal vesicle breakdown (GVBD). GVBD is easily detected by the rise of the nucleus to the oocyte surface (visible as a white maturation spot in the animal hemisphere), XeCDC2 dephosphorylation (necessary for the activation of downstream pathways) and histone H3 (XeH3) phosphorylation, an indicator of chromosome condensation48. Previously, we showed how X. laevis oocytes could be used to investigate the function of P. falciparum PP1c regulators: XePP1c contributes to inhibit GVBD, so the micro-injection of oocytes with a PP1c inhibitor triggers GVBD whereas the introduction of an activator prevents progesterone-induced GVBD49.
Accordingly, we injected X. laevis oocytes with recombinant His6-pTKL_RVxF1 and pTKL_KD_WT followed by co-immunoprecipitation assays to determine their XePP1c-binding activity. Only pTKL_KD_WT was able to bind XePP1c (Fig. 5b). Interestingly, pTKL_KD_WT did not trigger GVBD in untreated arrested oocytes, showing that it may not be a direct XePP1c inhibitor (Supplementary Fig. S9a), but it was able to block GVBD in the presence of progesterone in 60–70% of the oocytes. The micro-injection of His6-pTKL_RVxF1 had no effect, similar to the protein elution buffer used as a negative control (Fig. 5c). This led us to the conclusion that pTKL_KD_WT is probably a PP1c activator, which was supported by the relatively high phosphorylation of XeCDC2 and the absence of phosphorylation of XeH3 (Fig. 5d). To examine the contribution of RVxF2 in more detail, we produced the recombinant protein pTKL_KD_KRIMAS in which the RVxF2 motif 1216KRKVLF1221 was mutated to 1216KRIMAS1221 to reduce its ability to bind PP1c50 (Supplementary Fig. S6). We found that pTKL_KD_KRIMAS was unable to bind XePP1c and its micro-injection did not affect progesterone-induced GVBD (Fig. 5b–d).
These experiments showed that PfpTKL was able to bind and modulate PP1c via the RVxF2 motif, whereas the RVxF1 motif-containing region bound PP1c in vitro but did not modulate its activity.
The PfpTKL SAM domain interacts with PfSERA5
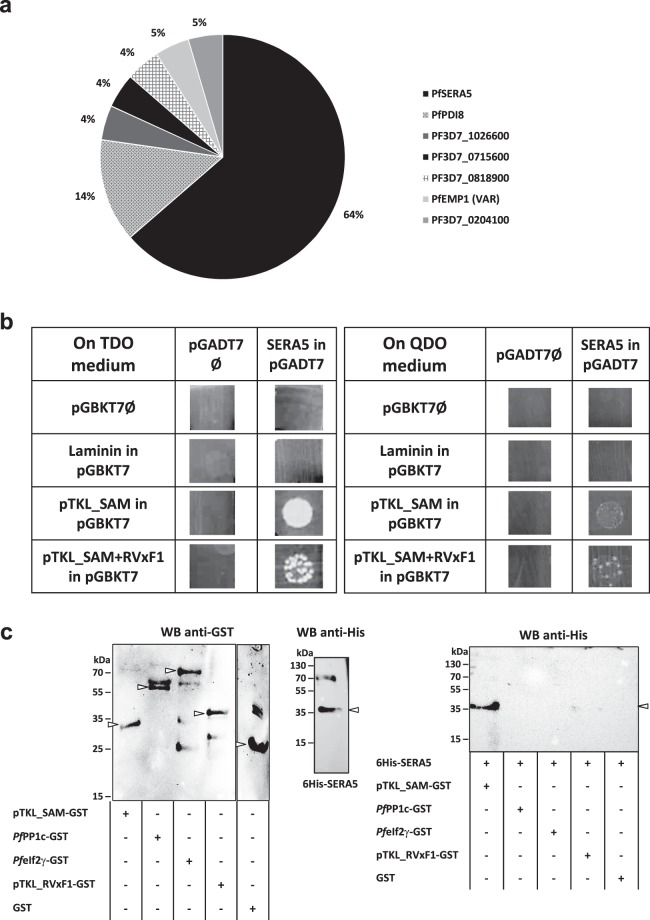
In silico analysis of the PfpTKL SAM domain showed that it might be involved in protein–protein interactions (Fig. 2e). To identify potential interaction partners, we screened a P. falciparum cDNA library using a yeast two-hybrid (Y2H) assay with the 838–1594 bp portion of PfpTKL cDNA as the bait, corresponding to the SAM and RVxF1. We found that 32 clones of the 6000 screened were able to grow on stringent media, indicating strong interactions with the PfpTKL SAM + RVxF1 region. These represented 12 P. falciparum proteins (Supplementary Table S5). Interestingly, PfSERA5 (PF3D7_0207600) was detected in 64% of the in-frame screening hits (Fig. 6a). PfSERA5 is the most abundant pseudo-protease in the parasitophorous vacuole51. To confirm this interaction, a Y2Hgold strain transformed with pGADT7 (either empty or encoding the longest PfSERA5 fragment identified during the Y2H screen) was mated to a Y187 strain transformed with pGBKT7 (empty or encoding the interaction negative control laminin, SAM + RVXF1 or SAM alone). The only diploids that could grow on stringent media were those expressing both PfSERA5 and at least the PfpTKL SAM domain, thus excluding RVxF1 as a requirement for PfSERA5 binding (Fig. 6b). These data confirm the strong and specific interaction between PfSERA5 and the SAM domain of PfpTKL.
Figure 6.
PfpTKL SAM domain interacts specifically with PfSERA5. (a) PfSERA5 is the main binding partner of the PfpTKL SAM. P. falciparum proteins interacting with the PfpTKL SAM + RVxF1 region were identified by yeast two-hybrid (Y2H) screening. The percentage occurrence of each interacting protein is plotted for clones in frame with GAL4AD. All the hits returned by the screening are available in Supplementary Table S5. (b) PfSERA5 interacts strongly and specifically with the PfpTKL SAM in yeast. The chart shows the results of the mating run between Y2Hgold cells transformed with pGADT7 (either empty (Ø) or cloned with the longest PfSERA5 cDNA sequenced during Y2H screening) and Y187 cells transformed with pGBKT7 (empty (Ø), encoding laminin as a negative interaction control or SAM + RVxF-1 or SAM only). Diploids were either streaked on TDO/A medium (left panel) or QDO/A medium (right panel). (c) PfSERA5 interacts directly with the PfpTKL SAM in vitro. His6-tagged PfSERA5 (His6-SERA5) was incubated with pTKL_SAM-GST, pTKL_RVxF1-GST, PfeIf2γ-GST, PfPP1c-GST or GST (the last three = negative controls). After washes, proteins were eluted in Laemmli buffer, separated by SDS-PAGE and detected by western blot (WB, right panel). Inputs were also detected by western blot (left and middle panels). His6-tagged PfSERA5 binds directly to pTKL_SAM-GST (first lane on right). Uncropped blots are available in the Supplementary Information.
The GST pull-down assay described above was repeated using PfSERA5-His6 as the bait for PfpTKL_SAM-GST and PfpTKL_RVxF1-GST beads, with unrelated GST-tagged proteins as negative controls (Supplementary Fig. S6). PfSERA5-His6 alone remained bound to PfpTKL_SAM-GST beads, confirming the binding specificity between the pseudo-protease and the PfpTKL SAM domain (Fig. 6c, right panel, lane 1).
PbpTKL is exported in infected host erythrocytes
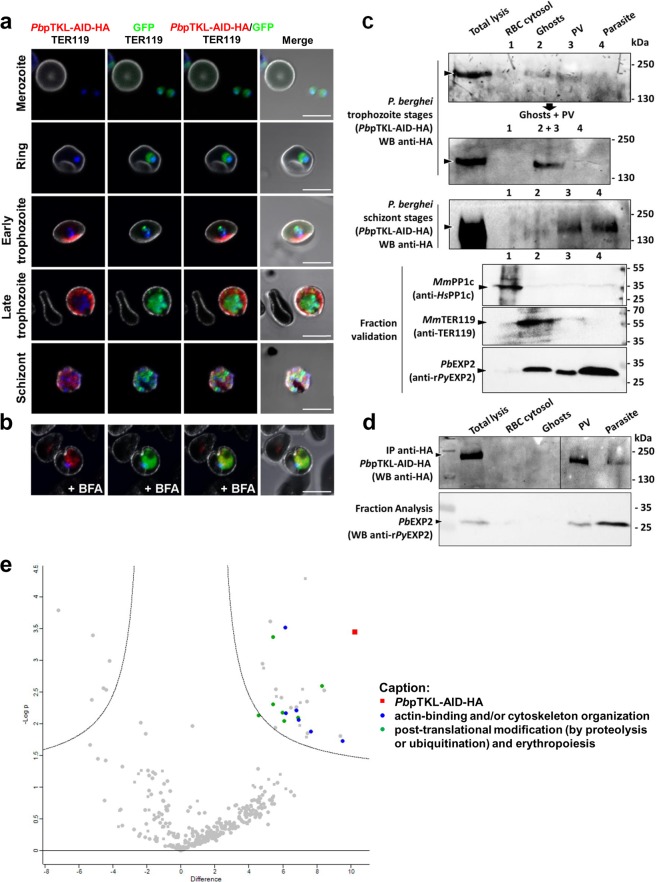
Following unsuccessful attempts to knock-in pTKL in P. falciparum, we generated a P. berghei line expressing an auxin-induced degradation–hemagglutinin (AID-HA)-tagged PbpTKL to study the subcellular localization of pTKL throughout the asexual life stages (Supplementary Fig. S7a). PfpTKL and PbpTKL share a global 36% identity and a protein domain conservation of 50–70% (Supporting Information). The only difference between the two orthologs is the absence of RVxF1 in P. berghei, but as described above this sequence does not appear to be required for PP1c modulation (Fig. 5). Immunofluorescence assays using synchronized intra-erythrocytic stages of the PbpTKL-AID-HA line revealed that PbpTKL was exported within the infected erythrocytes and was not detected in the parasite from the early to late trophozoite stages (Fig. 7a). Small amounts of PbpTKL were secreted during the early trophozoite stages, but from the late trophozoite to schizont stages the protein filled the entire erythrocyte cytosol, and probably also the parasitophorous vacuole (Fig. 7a). The active export of PbpTKL within the infected erythrocytes was further confirmed by treating PbpTKL-AID-HA trophozoites with brefeldin A (BFA), described as an inhibitor of protein transport52. In BFA-treated trophozoites, PbpTKL was solely detected within the parasite cytosol as expected (Fig. 7b,d). Given that pTKL lacks a PEXEL motif and a predicted signal peptide, this also indicates that PbpTKL is a PEXEL-negative protein (Table 1).
Figure 7.
PbpTKL is exported in the erythrocyte during the trophozoite stages and binds host erythrocyte proteins. (a,b) PbpTKL is actively exported from the parasite between the trophozoite and schizont stages. (a) Confocal microscopy images showing the localization of the PbpTKL-AID-HA protein (red) in infected erythrocytes at different stages (top to bottom: merozoite, ring, early trophozoite, late trophozoite and schizonts). Parasite nuclei are stained with Hoechst (blue). The erythrocyte membrane is labeled after detection of TER119 (white). Bar = 5 µm. (b) Confocal microscopy images showing the localization of PbpTKL-AID-HA in brefeldin A (+BFA) treated trophozoites. PbpTKL-AID-HA (red, BFA aggregates) is no longer exported but retained within the parasite cytosol (GFP signal). Bar = 5 µm. (c) PbpTKL associates with ghosts during the trophozoite stages based on sequential lysis results (Supplementary Fig. S8). Fractions were verified by western blot (WB): anti-HsPP1c recognizes mouse PP1c in the erythrocyte cytosol, anti-TER119 recognizes mouse erythrocyte ghosts, and anti-rPyEXP2 recognizes PbEXP2 in the parasite cytosol, parasitophorous vacuole and vesicles in the host erythrocyte cytosol119. Anti-HA immunoprecipitation assays were performed on each fraction and PbpTKL-AID-HA was detected by anti-HA western blot. Uncropped blots are provided in the Supplementary Information. (d) PbpTKL is actively exported to the host erythrocyte cytosol. Sequential lysis on P. berghei trophozoites treated with BFA (verified by fraction analysis using anti-rPyEXP2, as opposed to fraction analysis without treatment shown in (c). Anti-HA IP assays were performed on each fraction and PbpTKL-AID-HA was detected by anti-HA western blot. Uncropped blots can be found in the Supplementary Information. (e) Exported PbpTKL binds erythrocyte proteins. Volcano plot showing proteins identified by mass spectrometry after anti-HA immunoprecipitation assays on PbpTKL-AID-HA versus wild-type pG230 P. berghei-infected erythrocytes (y-axis shows negative log p-value, x-axis shows fold-change between wild-type and PbpTKL-AID-HA samples). Squares = P. berghei proteins; circles = mouse proteins. Proteins absent from wild-type samples and detected in at least two of three PbpTKL-AID-HA ghost samples are shown on the upper right panel of the chart (see caption). Three biological replicates were analyzed per sample type (raw and interactome mouse protein analysis data are available in Supplementary Table S6).
To confirm these observations, the sequential lysis of infected erythrocytes was performed on trophozoites and schizonts, followed by anti-HA immunoprecipitation (Supplementary Fig. S8). During the trophozoite stages, PbpTKL-AID-HA was detected in the parasitophorous vacuole fraction and was also associated with erythrocyte ghosts. However, it was not detected in the infected erythrocyte cytosolic fraction, suggesting that the protein may be associated only with the erythrocyte membrane and cytoskeleton. During the schizont stages, PbpTKL-AID-HA was detected in both the parasite and the parasitophorous vacuole fractions, but was no longer associated with erythrocyte ghosts (Fig. 7c).
PbpTKL interactome in trophozoite-infected erythrocytes
To determine whether the exported PbpTKL interacts with erythrocyte proteins, we screened for interaction partners in trophozoite-infected mouse erythrocytes using anti-HA immunoprecipitation. We screened the total lysates of erythrocytes infected with a wild-type parental P. berghei line and ghost lysates of erythrocytes infected with the P. berghei line expressing PbpTKL-AID-HA. Proteins that remained bound to the anti-HA beads were analyzed by mass spectrometry, and those absent in the wild-type samples but present in at least two ghost samples from three experiments were selected. The resulting list of 23 mouse proteins (Fig. 7e; Supplementary Table S6) was assessed for cellular functions using the Uniprot database53, GO annotation and interactome data available in the STRING database54. Six of the proteins were found to be involved in the organization or dynamic modulation of the actin cytoskeleton, in agreement with the localization data presented above (Fig. 7a,c). Three further proteins were found to be involved in the intracellular trafficking of ions and vesicles. Another seven proteins were members of the proteasome complex or involved in the regulation of protein ubiquitination, such as ISG1555. The list also included two ribosomal proteins involved in translation and two proteins involved in the physiological response to oxidative stress. The last three proteins were not considered relevant due to the subcellular localization data available on Uniprot53 (Supplementary Table S6). Attempts to find PbpTKL interaction partners in schizonts did not lead to any exploitable data given the low expression level of PbpTKL during these stages (PbpTKL-AID-HA was detected in only one of three replicates).
PbpTKL is not required to complete the intra-erythrocytic cycle or gametocyte emergence
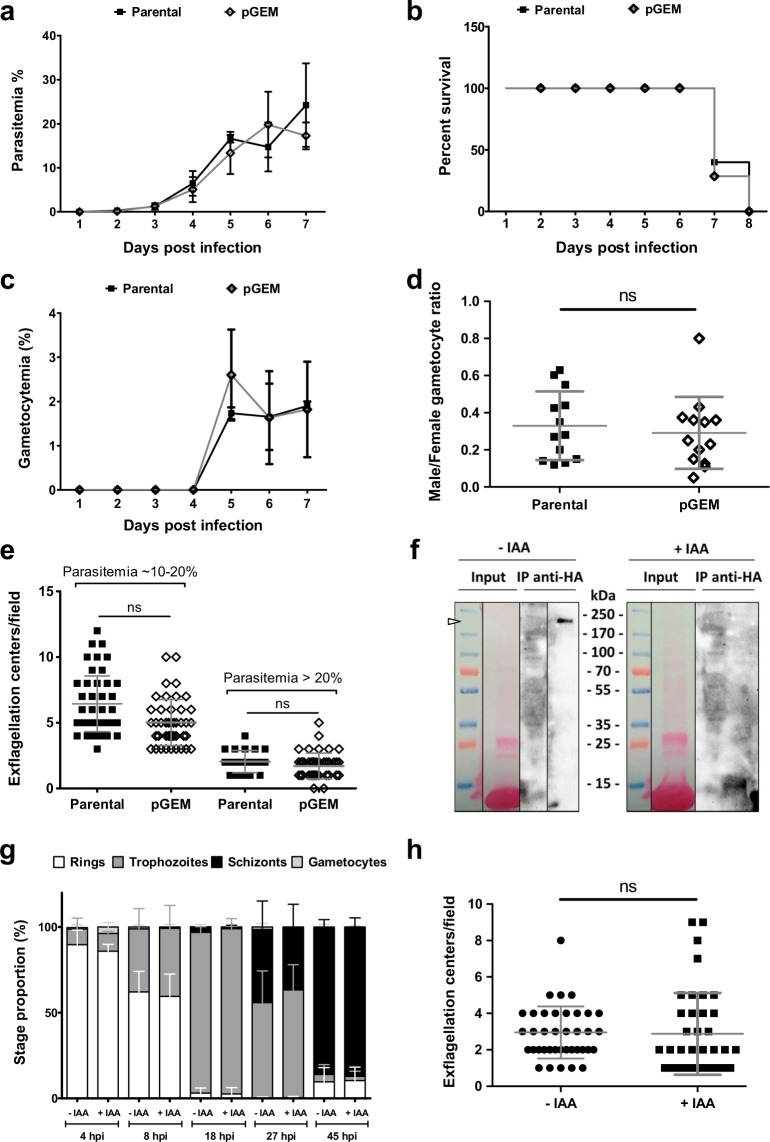
PbpTKL was recently found to be dispensable for the completion of intra-erythrocytic development56. This result was thus in conflict with the systematic functional analysis of the P. berghei kinome19. Both studies were based on similar approaches (i.e., gene disruption by double homologous recombination), but differed mainly in the length of the cloned homology regions, which favor recombination during PlasmoGEM screening56. We therefore attempted to generate a pTKL-knockout P. berghei line (pGEM) by introducing the readily available PlasmoGEM pTKL construct (PbGEM-342364) into the PbGFP ANKA strain (Supplementary Fig. S7b). After selection, pyrimethamine-resistant parasites were cloned and analyzed by genomic PCR to confirm the correct recombination events (Supplementary Fig. S7b). However, both the 5′ end of the PbpTKL transcript (amplified using primers F5′/GW2, including the MORN domains) and its 3′ end (amplified using primers F28/R29, potentially including at least the KD) were detected by RT-PCR (Supplementary Fig. S7b). Furthermore, primers GW1/GT did not detect the dhfr resistance cassette, thus excluding the expression of a chimeric transcript comprising the PbpTKL 5′ region, resistance cassette and PbpTKL 3′ region. Phenotypic analysis of PbpTKL knockout parasites showed identical parasitemia and survival rates compared to the wild-type parental strain (Fig. 8a,b). Furthermore, there was no difference between the lines in terms of gametocyte production, male/female gametocyte ratio and male gametocyte activation (exflagellation) (Fig. 8c–e). Because we could not exclude the possibility that the lack of phenotype reflected the partial expression of PbpTKL, we used the AID-HA knock-in approach57 in a TIR1-expressing P. berghei ANKA pG230 strain. Clones were confirmed by diagnostic PCR (Supplementary Fig. S7a), and the degradation of PbpTKL-AID-HA following the incubation of parasites in vitro with auxin was confirmed by western blot (Fig. 8f). The intra-erythrocytic cycle was completed and male gametocyte exflagellation occurred similarly in the parasite cultures in the presence or absence of auxin (Fig. 8h).
Figure 8.
PbpTKL is dispensable for intra-erythrocytic cycle completion, gametocyte emergence and male gametocyte exflagellation. (a–e) Data collected using mice infected either with the wild-type PbGFP strain (parental) or the PbGEM-342364-transfected strain (pGEM) – 106 parasites/mouse, seven mice per group. (a) Knocking out PbpTKL does not impact parasitemia. Parasitemia follow-up in mice infected either with the parental or pGEM P. berghei parasites (mean ± SD). (b) Knocking out PbpTKL does not improve mouse survival. Percent survival of mice infected with either parental or pGEM P. berghei parasites. (c) Knocking out PbpTKL does not affect gametocyte emergence. Gametocytemia follow-up in mice infected with either with the parental or pGEM P. berghei parasites (mean ± SEM). (d) Knocking out PbpTKL does not affect male/female gametocyte ratio. Male/female ratios were evaluated in mice infected either with the parental or pGEM P. berghei parasites (data points and means ± SD are shown). (e) Knocking out PbpTKL does not affect male gametocyte exflagellation. Exflagellation center numbers were assayed for parental and pGEM parasites at a parasitemia of 10–20% and > 20% (data points and means ± SD are shown). (f–h) Data collected using cloned pG230 P. berghei parasites expressing PbpTKL-AID-HA, after incubation with or without the auxin 3-idoleacetic acid (IAA). (f) PbpTKL-AID-HA is degraded in vitro in the presence of IAA. PbpTKL-AID-HA parasites were incubated either with (+IAA) or without IAA (−IAA). After total lysis of incubated parasites, immunoprecipitation assays (IP) were performed using anti-HA agarose beads. PbpTKL-AID-HA was detected by western blot (WB) using anti-HA antibodies. Uncropped blots are available in the Supplementary Information. (g) Inducible knockdown of PbpTKL does not prevent completion of the intra-erythrocytic cycle. In vitro follow-up of PbpTKL-AID-HA parasite stages after synchronized infection in mice (five per group) – parasites were cultured in the presence (+IAA) or absence (−IAA) of IAA, hpi = hours post-intravenous injection of Nycodenz-enriched schizonts (mean ± SD). (h) Inducible knockdown of PbpTKL does not affect male gametocyte exflagellation. Exflagellation centers were assayed for PbpTKL-AID-HA parasites after incubation with (+IAA) or without IAA (−IAA) (mean ± SD). ns, no significant difference.
Together, these data show that PbpTKL is probably not required to complete the intra-erythrocytic cycle or to support gametocyte production and male gametocyte activation.
Discussion
The Plasmodium kinome is mainly composed of S/T kinases with key roles in parasite development20. Although the multiple roles of pseudokinases are well known in higher eukaryotic cells28–36, little is known about apicomplexan pseudokinases. We therefore characterized pTKL, a pseudokinase specific to certain apicomplexan parasites, including Plasmodium species. Our results demonstrated that in P. berghei, PbpTKL is expressed mostly during the trophozoite stages and the protein is exported to the host cell where it associates with ghosts, i.e. the erythrocyte membrane and cytoskeleton58. During the schizont stages, PbpTKL is retained within the parasite and parasitophorous vacuole. Because PbpTKL is PEXEL-negative and lacks a signal peptide as well as a transmembrane domain, its export mechanism remains unclear. We propose that pTKL crosses the parasite plasma membrane using its hydrophobic MORN motifs. Either both of them or mostly MORN2 which contains a putative N-myristoylation site could also allow the protein to associate with erythrocytic ghosts. This is further supported by the fact that MORN motifs bind to membranes and cytoskeleton (Table 1). Importantly, our interactome analysis revealed that PbpTKL interacts with actin cytoskeleton-binding proteins (Fig. 7e). PbpTKL may therefore be involved in the remodeling of the erythrocyte cytoskeleton or membrane, as shown for other unessential exported proteins such as PfFIKK7.1 and PfFIKK1259, PbSMAC60 and PbRESA61.
When exported to the host erythrocyte cytosol, PbpTKL was found to reside at the crosstalk of two main protein networks. The first is composed of three proteins involved in cellular trafficking, and the second regulates protein expression and degradation (the seven members of this network include ribosomal proteins, regulators of ubiquitination and proteasome components). These proteins are classically found in erythrocyte ghost samples58. They are involved in the last steps of erythropoiesis, during which hemoglobin synthesis increases while other proteins, including ribosomal proteins, are degraded. This process ensures the simplification of the proteome as cells mature from reticulocytes to erythrocytes, in which 98% of all protein molecules are globin62,63. Two proteins involved in oxidative stress regulation were also detected in the PbpTKL ghost interactome. Such proteins play a key role in maintaining erythrocytes by protecting them from the continuous oxidative stress they experience due to their oxygen transport role64. PbpTKL might thus favor the implementation of host–pathogen communication pathways as already described for many classical code-breaking pseudo-kinases and truncated kinases65.
From a biochemical point of view, the PfpTKL catalytic site can bind ATP in a cation-independent manner, confirming that it belongs to the class 2 pseudokinases26. However, the PfpTKL KD was catalytically inactive under our experimental conditions, probably due to the lack of canonical subdomains I-III and VI which usually play key roles in ATP stabilization and γ-phosphate group transfer25. Notably, the native PbpTKL-AID-HA protein also lacked catalytic activity (Fig. 4b). The role of the BTK-like domain and potentially of the SAM domain in terms of regulation of KD activity (as shown for Pf TKL366) remains unclear. However, in addition to interaction data collected from ghost samples, the ability of the pTKL KD to bind ATP could turn the protein into an enzyme cofactor within the erythrocyte cytosol. Furthermore, ATP is an important messenger for erythrocytes67: it is released during hypoxia, oxidative stress or infection by Plasmodium species and may therefore mediate communication between infected cells and uninfected bystander erythrocytes68. The parasite may export an ATP-binding protein and anchor this to the host cell cytoskeleton as a strategy to modulate ATP release during its replication.
In parallel, we showed that PfpTKL interacts with two parasite proteins in vitro and in heterologous models. The RVxF motifs of PfpTKL can bind PfPP1c, the major phosphatase of this parasite69,70. Interaction studies in the X. laevis oocyte model showed that only RVxF2 binds XePP1c in vitro and also influences GVBD. This suggests that RVxF1 may interact transiently with PP1c in vivo to bring pTKL and PP1c closer, probably only during the schizont stages where both pTKL and PP1c are localized in the parasite. RVxF2, which is conserved in all pTKL homologs across the Plasmodium genus, appears to bind PP1c more strongly than RVxF1 and also allows the allosteric modulation of phosphatase activity.
Using a Y2H assay, we found that the PfpTKL SAM domain binds strongly and specifically to PfSERA5, an abundant parasitophorous vacuole pseudo-protease that is expressed during the P. falciparum schizont stages and is involved in the egress of merozoites51. PfSERA5 does not appear to be conserved in P. berghei, so the PbpTKL SAM domain may interact with another member of the SERA family in this species. Although SAM domains feature simple and uniform primary, secondary and tertiary structures, they can bind to different proteins and nucleic acids71,72. The PbpTKL SAM domain may therefore interact with other proteins when it is exported to the host cell cytosol. Although we could not confirm an interaction between PbpTKL and PbPP1c or a PbSERA family member in schizonts due to the low PbpTKL expression level, we found a strong correlation between the interactions identified in vitro and the localization of the protein and its partners in the same compartments during the schizont stages.
Having tested the catalytic and scaffolding roles of pTKL, we next investigated whether the protein is required during the P. berghei blood stages. The phenotypic analysis of PbGEM-342364-transfected P. berghei and the use of the AID knockdown system led us to the conclusion that PbpTKL is probably not required for the completion of the intra-erythrocytic cycle or for gametocyte emergence, in agreement with a recent saturation mutagenesis study in P. falciparum73. Thirteen piggyBac transposons could be inserted within the PfpTKL locus and surrounding intergenic regions without affecting P. falciparum intra-erythrocytic development, suggesting the protein is dispensable for blood stage parasites. However, we cannot exclude the possibility that the PbpTKL MORN motifs or KD may continue to be expressed in the knockout line. Although we also used the in vitro AID system, which is useful to determine whether proteins are essential for invasion, cell cycle completion or gametocyte activation57, it may not be suitable for the analysis of proteins that are rapidly exported to the parasitophorous vacuole or the host cell after their synthesis, even though we observed that PbpTKL degradation was induced in trophozoites in the presence of auxin.
In conclusion, we have shown that the Plasmodium pseudo-kinase pTKL can bind ATP and acts as a protein scaffold at the host–pathogen interface. Although probably unnecessary during the blood stages, it remains to be demonstrated whether this protein is required during the mosquito and/or liver stages of parasite development. Indeed, a recent single-cell transcriptomics study showed that PbpTKL is strongly transcribed during the Plasmodium liver stages (Malaria Cell Atlas74).
Materials and Methods
Materials
Commercial plasmids were sourced from Invitrogen (pCR2.1-TOPO), Novagen (pETDuet-1), GE Life Sciences (pGEX-6P3), Qiagen (pMALpIII), and Clontech (pGBKT7 and pGADT7), whereas pG362 and PbGEM-342364 were kindly provided by Dr N. Philip (The University of Edinburgh, Edinburgh, UK) and the Plasmogem-Wellcome Sanger Institute (Cambridge, UK), respectively. The P. falciparum cDNA bank used for Y2H library preparation was described in an earlier study75.
Primary antibodies were sourced from Invitrogen (anti-His6), Thermo Fisher Scientific (anti-GST), StemCell (TER119) and Roche (rat anti-HA and anti-GFP). Anti-HsPP1c (clone E9) and anti-XePP1c were provided by Santa Cruz Biotechnology. Rabbit anti-HA, anti-Cdc2, anti-phospho-Cdc2 (Tyr15) and anti-phospho-histone H3 (Ser10) polyclonal antibodies were provided by Cell Signaling Technology. Anti-histone H3 was provided by Novus Biological. Secondary highly-cross-adsorbed antibodies were obtained from Invitrogen. Anti-rPyEXP2 was a kind gift from Dr J. Burns (Drexel University College of Medicine, Philadelphia, USA).
Equinatoxin II (EQT) was a kind gift from Dr G. Anderluh (National Institute of Chemistry, Ljubljana, Slovenia)76.
Transcript sequencing and sequence confirmation
Pf3D7 total RNA was extracted from saponin-lysed parasites using TRIzol (Thermo Fisher Scientific) and reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis Super Mix kit (Invitrogen) following the manufacturer’s instructions. The cDNA was used to amplify and clone overlapping regions of the PF3D7_1106800 (PfpTKL) transcript into vector pCR2.1-TOPO using primers F1–R12 (Supplementary Fig. S1 and Table S1). Following the transformation of TOP10 bacteria and clone sequencing, 6–12 sequences per region were compiled into one consensus covering the whole PfpTKL transcript using SeqMan for comparison with PlasmoDB data43.
Protein annotation
The PfpTKL protein sequence was analyzed using SMART77, CATH78, Interpro79 and ScanProsite80. The tertiary structure was determined using the prediction tools Phyre2 Server81, M4T82 and RAPTOR83 (all well ranked by CAMEO84,85). RVxF motifs were identified based on two consensus sequences86,87 (Fig. 2b) and the PlasmoDB Protein Motif Pattern tool. Low-complexity regions and putative post-translational modifications were predicted using ScanProsite80. Signal peptides were predicted using SignalHmmS88.
Phylogenetic analysis
We identified 48 PfpTKL (PF3D7_1106800) homologs via back-and-forth PSI-BLAST analysis, first using the PfpTKL sequence as a query, and then using PSI-BLAST results as queries against the Pf3D7 proteome. Candidates were confirmed as PF3D7_1106800 homologs when pTKL was the highest scoring hit (except for the RAF1 outgroup). Verified sequences were then retrieved from PlasmoDB43 or Uniprot53 for non-Plasmodium proteins before inclusion in the phylogenetic analysis. The 128 SAM amino acid sequences were retrieved from Uniprot53.
For each phylogenetic analysis, sequences were aligned using MAFFT89 (Supplementary Information) and evolutionary history was inferred using the maximum likelihood method based on the JTT matrix model in Molecular Evolutionary Genetics Analysis v6 (MEGA6)90,91. All positions with less than 80% site coverage were eliminated, leaving 598 positions in the final PfpTKL dataset and 64 in the final SAM dataset. The initial tree for the heuristic search was obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. For PfpTKL, a discrete gamma distribution was used to model evolutionary rate differences among sites. Evolutionary history was represented by the bootstrap consensus tree inferred from 1000 replicates92. The tree was visualized using MEGA6 and edited in Inkscape v0.92.
Recombinant protein production and purification
GST, PfPP1c-GST, PfPP1c-His6 and PfeIf2γ-GST were produced as previously described47,93. PfpTKL_KD_WT was synthesized (Eurofins) as a re-codonized synthetic gene (synthetic gene 2 in Supplementary Information) and transferred to pMALPIII to stabilize the recombinant protein via an N-terminal fusion to the maltose binding protein94,95 with a C-terminal His6 tag. The resulting construct was introduced into the Escherichia coli strain JM109. The PfpTKL_KD_WT construct was mutated by PCR targeted mutagenesis as previously described96. The N1272D mutant (pTKL_KD_ND) was prepared using primers F22/R23, the 1290GFE1292 mutant (pTKL_KD_DG) was prepared using primers F24/R25, and the 1218KRIMAS1222 mutant (pTKL_KD_KRIMAS) was prepared using primers F26/R27 (Supplementary Table S1).
The pTKL_RVxF1 recombinant proteins with His6 or GST tags were prepared by cloning PfpTKL (cDNA 1114–1594 bp) in vectors pETDuet-1 and pGEX-6P3 using primers F13/R14 (Supplementary Table S1) and introducing them into E. coli strains C41 and AD494, respectively, to produce the proteins His6-pTKL_RVxF1 and GST-pTKL_RVxF1. The SAM domain (cDNA 838–1149 bp) was re-codonized by GenScript (synthetic gene 1 in Supplementary Information), transferred to vector pGEX-6P3 and introduced into E. coli strain AD494 (recombinant protein pTKL_SAM). The His6-tagged SERA5 was obtained by cloning PfSERA5 (PF3D7_0207600) cDNA (432–1089 bp) directly from Y2H screening clone 3457 (Supplementary Table S5) into pETDuet-1 using primers F20/R21 (Supplementary Table S1). The resulting construct was introduced into E. coli strain BL21 (recombinant protein His6-PfSERA5). All recombinant protein constructs were verified by sequencing (Supplementary Information).
The expression of recombinant proteins was induced at 16 °C overnight shaking at 200 rpm in the presence of 0.5 mM IPTG (and 1 mM MgSO4 as a kosmotrope for the pTKL_KD proteins97). Cells were harvested in ice-cold sonication buffer (20 mM Tris pH 7.5 or 8, 300 mM NaCl, 20 mM imidazole, 0.2 U/mL DNase, Roche cOmplete EDTA-free anti-protease cocktail, 0.02 mg/mL lysozyme, 1% Triton X-100, and 1 mM MgSO4 for the pTKL_KD proteins). His6 and GST tagged proteins were purified on Ni-NTA beads (Jena Bioscience) and glutathione beads (Sigma-Aldrich) respectively, according to the manufacturer’s instructions (Supplementary Fig. S6).
Protein concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
Kinase activity assay
Recombinant pTKL_KD_WT, pTKL_KD_ND and pTKL_KD_DG were purified on beads as described above. Beads were suspended in purification washing buffer (20 mM Tris pH 7.5, 300 mM NaCl, 20 mM imidazole, 1 mM MgSO4) and the concentration was determined using the Pierce BCA Protein Assay Kit. 400–800 µg of protein were used per test. Endogenous PbpTKL-AID-HA was immunoprecipitated on HA-agarose beads as described below. Beads were washed and equilibrated in assay washing buffer (20 mM Tris pH 7.5, 1 mM DTT, 200 µM activated sodium orthovanadate, Roche cOmplete EDTA-free anti-protease cocktail). JNK1 (Sigma-Aldrich) was used as a positive control (1 µg per test). We mixed 20 µL of kinase (either beads or free protein) with 5 µg of substrate, 10 µL of 10 μCi ATP [γ-33P] (PerkinElmer Life Sciences) and ATP, final concentration 50 µM, in 40 µL of wash buffer. After incubation for 1 h at 37 °C, kinase reactions were stopped by adding Laemmli buffer. Samples were analyzed by SDS-PAGE. The resulting gel was fixed with 10% acetic acid and 10% methanol before drying. The incorporation of [γ-33P] was visualized and quantified using a PharosFX Plus Molecular Imager.
ATP affinity assay
Recombinant kinases were dialyzed against PBS containing 1 mM DTT and 1 mM EDTA overnight at 4 °C and then assayed for ATP affinity using the ATP Affinity Test Kit (Jena Bioscience) according to the manufacturer’s protocol.
ATP hydrolysis assay
ATP hydrolysis activity was evaluated using the Kinase Glo Luminescent Kinase assay kit (Promega). Beads prepared as described above were distributed in increasing quantities in white 96-well plates in washing buffer (20 mM Tris pH 7.5, 1 mM DTT, 200 µM activated sodium orthovanadate, Roche cOmplete EDTA-free anti-protease cocktail) to 20 µL per well. ATP buffer (washing buffer containing 3.3 µM ATP, 16.7 µg/mL MBP and 3.3 mM MgCl2) was then added to a final volume of 50 µL per well. JNK1 was used as a kinase positive control. After incubation at 37 °C for 2 h shaking at 54 rpm in the dark, Glo reagent equilibrated to room temperature was added (50 µL per well) and the plate was incubated shaking at 100 rpm for 10 min in the dark at room temperature before measuring the luminescence on a TECAN Spark 10 M luminometer (see Supplementary Table S4 for raw data).
GST pull-down assay (RVxF interaction with PfPP1c-GST)
GST pull-down assays were prepared as previously described47. Briefly, 2 μg of recombinant His6-pTKL_RVxF1, pTKL_KD_WT or pTKL_KD_KRIMAS was incubated with PfPP1c-GST, PfeIF2γ-GST (negative interaction control) or GST bound to glutathione-Sepharose beads, and 25 µg of bovine serum albumin (BSA) in binding buffer (20 mM Tris-HCl pH 7.4, 500 mM NaCl, 20 mM HEPES, 0.2 mM EDTA, 0.1% Triton X-100, 1 mM DTT, Roche cOmplete EDTA-free anti-protease cocktail, 1 mM MnCl2, 50 µM ZnCl2) for 2 h at 4 °C on a rotating wheel. After three washes, proteins were analyzed by western blot using an anti-His6 antibody (diluted 1:1000) or an anti-GST antibody (diluted 1:2000).
Ethics statement for animal experimentation
Experiments were carried out in accordance with the principles of the European Community Council recommendations (86/609/EEC) for animal experimentations. All animal procedures were approved and supervised by the local Ethics Animal Committee (CEEA-75 Comité d’Ethique en Expérimentation Animale, Nord - Pas de Calais, France). The ethical approval number for protocols and procedures on mice (male CD1 (22–24 g) from Charles River Laboratories) is 00527.04. Experiments conducted on X. laevis (University of Rennes, UMS 3387) were approved likewise (CEEA 07/2010).
X. laevis oocyte assays
Oocytes were micro-injected 1 h before incubation in 10 µM progesterone as previously described96 (although with recombinant proteins rather than synthetic mRNA). GVBD was assessed by checking for the appearance of a white spot at the surface of activated oocytes and by verifying the absence of any nucleus inside activated oocytes (hemisections of oocytes were prepared). XePP1c was immunoprecipitated 2 or 15 h after micro-injection98.
Yeast two-hybrid assay
A PfpTKL cDNA segment including the SAM domain and RVxF-1 motif (residues 255–371) was transferred to vector pGBKT7 using primers F15/R16 (Supplementary Table S1) and introduced into Y187 competent yeast cells (Clontech) according to the manufacturer’s instructions. The resulting strain was used as a bait to screen protein interactions by mating with Y2Hgold cells transformed with the P. falciparum cDNA library in vector pGADT775. Mating produces diploids that are viable on double dropout/aureobasidine (DDO/A) medium. We streaked 6000 diploids that grew on DDO/A post-mating on triple dropout/A (TDO/A) plates and then on quadruple dropout/A (QDO/A) plates, as increasing stringency allowed for interaction evaluation75. Only 32 diploids were able to develop on QDO/A medium (Supplementary Table S5). Plasmid DNA recovered from 31 of the 32 diploids was introduced into E. coli strain DH5α and checked for the presence of an insert by digestion with SfiI. Finally, the insert sequences were used as BLAST queries against PlasmoDB43 to identify putative PfpTKL SAM interactors. For further Y2H assays, a Y2Hgold strain expressing the SAM alone was generated by mutating construct SAM + RVxF1_pGBKT7 using primers F17/R18.
GST pull-down assay (SAM-GST interaction with PfSERA5–6His)
The pTKL_SAM-GST, PfPP1-GST, PfeIF2γ-GST, pTKL_RVxF1-GST or GST proteins bound to glutathione-Sepharose beads were blocked with 25 μg BSA in binding buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM MnCl2, 50 μM ZnCl2) for 1 h at 4 °C on a rotating wheel. We incubated 1 μg of recombinant PfSERA5-His6 protein with blocked beads for 30 min, and bound proteins were detected as described above.
Purification of stage-specific P. berghei parasites
Parasite stages were isolated as previously described99. Briefly, for schizonts, intra-erythrocytic parasites from mice 3 days post-infection were cultured in RPMI 1640 medium containing 25 mM HEPES, 0.4% Albumax, 0.2 mM hypoxanthine and 20 µg/mL gentamycin at 37 °C with gentle shaking (54 rpm) for 20 h. Infected erythrocytes were separated from non-infected cells on a 60% Nycodenz column (27.6% w/v Nycodenz in 5 mM Tris-HCl pH 7.20, 3 mM KCl, 0.3 mM EDTA).
Generation of transgenic P. berghei parasites
PbpTKL was disrupted by double crossover homologous recombination following the introduction of the plasmoGEM vector PbGEM-342364 (Wellcome Sanger Institute56) into PbGFP ANKA parasites (kind gift from O. Silvie, Université Pierre et Marie Curie, Paris, France) by transfection. The PlasmoGEM construct was linearized using NotI before transfection.
C-terminal AID-HA-tagged PbpTKL strains were generated by single homologous recombination. A 1126-bp region of PbpTKL starting 3664 bp downstream from the start codon and lacking a stop codon was prepared using primers F28/R29 and F31/R32 (Supplementary Table S1) and inserted upstream of the AID-HA sequence in pG362 before the transfection of P. berghei ANKA strain pG230 (both the plasmid and parasite strain were kind gifts from N. Philip (The University of Edinburgh, Edinburgh, UK). The targeting sequence was linearized using BsaBI before transfection.
Transfections were carried out as previously described100. Briefly, Nycodenz-enriched schizonts were electroporated with 5–10 µg of linearized DNA and immediately intravenously injected into naive mice. Transfectants were selected using pyrimethamine because the PbGEM-342364 and pG362 vectors both contain a dhfr/ts expression cassette (Supplementary Fig. S7). From day 1 post-infection, 10 mg/L pyrimethamine (Sigma-Aldrich) was supplied in the drinking water. Drug selection was carried out by repeated passages in mice. Transgenic parasites were cloned by limiting dilution.
Genotypic analysis of mutants
C-terminal AID-HA-tagged PbpTKL parasites were identified by diagnostic PCR using primers F30/Ext and GU533/R3′ (Supplementary Fig. S7a). Immunoprecipitation followed by western blot analysis confirmed the expression of a correctly-sized PbpTKL-AID-HA protein. Knockout parasites were identified by diagnostic PCR as shown in Supplementary Fig. S7b. Primers GT/GW1 were used to confirm integration and primers F5′/R5′ were used to confirm the deletion of PbpTKL. Finally, RT-PCR was used to determine the PbpTKL transcription status of the mutants (Supplementary Table S1 and Fig. S7).
Lysis of infected erythrocytes
P. berghei-infected erythrocytes directly isolated from infected mouse blood or overnight-matured schizonts, treated or not with 5 µg/mL BFA (Sigma-Aldrich) for 6–20 h, were Nycodenz-enriched. Two lysis protocols were used.
For total lysis, a 500-µL dry erythrocyte pellet was lysed in 5 mL ice-cold total lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 1% Triton X-100, 0.5 mM EDTA, 0.2 U/mL DNase, Roche cOmplete EDTA-free anti-protease cocktail) for 5 min. The resulting lysate was fractionated by ultracentrifugation (41,657 g, 4 °C, 15 min) and the supernatant was used for immunoprecipitation.
For sequential lysis, a 500-µL dry pellet was lysed in 3 mL of EQT buffer (20 mM Tris pH 7.4, 130 mM NaCl, 1 mM MgSO4, 6 µg/mL EQT) on wheel for 15 min at room temperature. After centrifugation, the supernatant was fractionated by ultracentrifugation as above to recover ghosts. The EQT lysate ultra-supernatant (corresponding to the erythrocyte cytosol) was kept on ice for further analysis. The ghosts were lysed in ice-cold Triton buffer (20 mM Tris pH 8.0, 50 mM NaCl, 9% Triton X-100, 5 mM EDTA, Roche cOmplete EDTA-free anti-protease cocktail). The pellet, composed of parasites embedded in their parasitophorous vacuole, was further lysed in 3 mL of 0.07% saponin (on ice for 10 min). After centrifugation, the supernatant (the parasitophorous vacuole content) was kept for further analysis while the parasite pellet was lysed in 2 mL of ice-cold Triton buffer. Saponin, parasite and ghost lysates were diluted in total lysis buffer to 5 mL and fractionated by ultracentrifugation as above prior to immunoprecipitation (Supplementary Fig. S8).
Immunoprecipitation assay
Lysates prepared as described above were incubated for 1 h rotating on wheel at 4 °C with 20–30 µL HA-agarose beads (Invitrogen). The beads were washed three times in ice-cold washing buffer (20 mM Tris pH 7.5, 150 mM NaCl, 0.5% Triton X-100) and resuspended in Laemmli sample buffer for western blot analysis and/or mass spectrometry.
Mass spectrometry
PbpTKL-AID-HA was immunoprecipitated from ghosts during the trophozoite stages and from parasite + parasitophorous vacuole fractions during the schizont stages. Total lysis of the P. berghei pG230 strain trophozoites was followed by anti-HA immunoprecipitation to generate wild-type samples. The presence of PbpTKL-AID-HA in the ghost and schizont samples but not the wild-type samples was confirmed by western blot. Three biological replicates of each sample were analyzed by mass spectrometry. The detailed NanoLC-MS/MS protein identification and quantification protocol is provided in the Supplementary Information. Raw mass spectrometry data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository101 with the dataset identifier PXD011986.
The MS data files were processed using MaxQuant v1.5.8.3 and searched using the Andromeda search engine against the Mus musculus database from Swissprot 07/2017 and Plasmodium berghei ANKA from PlasmoDB (v37). The PbpTKL-AID-HA chimeric protein sequence was also integrated. Parent mass and fragment ions were searched with initial mass deviations of 4.5 and 20 ppm, respectively. The minimum peptide length was set to seven amino acids and strict specificity for trypsin cleavage was required, allowing up to two missed cleavage sites. Carbamidomethylation (Cys) was set as a fixed modification, whereas oxidation (Met) and N-terminal acetylation were set as variable modifications. The false discovery rates (FDRs) at the protein and peptide level were set to 1%. Scores were calculated in MaxQuant as previously described102. The reverse and common contaminant hits were removed from the MaxQuant output. Proteins were quantified according to the MaxQuant label-free quantitation (LFQ) algorithm102,103. Matches between runs were not allowed. Wild-type and PbpTKL-AID-HA IP biological replicates were analyzed using Perseus v1.6.2.3. The LFQ data were log2 transformed and proteins identified in at least two of three PbpTKL-AID-HA replicates but never in the wild-type samples were considered “significant”. The data were represented on a volcano plot following the imputation of missing values by creating a Gaussian distribution of random numbers with a standard deviation of 30% relative to the standard deviation of the measured values and a 4 (wild-type) or 1.8 (PbpTKL-AID-HA) standard deviation downshift of the mean.
Phenotypic analysis of transgenic P. berghei parasites
Mice were infected by the intraperitoneal administration of either cryopreserved clones or infected erythrocytes from mice carrying either the wild-type or mutant parasites. Parasitemia and gametocytemia were monitored every day up to 7–8 days using Giemsa-stained blood smears. To follow the intra-erythrocytic cycle, infection was synchronized by intravenously injecting Nycodenz-enriched schizonts into mice99. Blood was collected by cardiac puncture after 3–4 h (typically 0.5–1.2% parasitemia was observed) and cultured at 37 °C shaking at 54 rpm for 48 h. Parasite intra-erythrocytic development was followed using Giemsa-stained smears at precise time points and compared to wild-type cultures. Exflagellation assays were performed 3–4 days after synchronized infection in mice treated with phenylhydrazine 2 days before infection (4 mg/mouse). Following the assessment of gametocytemia, 10 µL of infected blood was added to 50 µL of ookinete medium (RPMI1640 containing 25 mM HEPES and 10% fetal calf serum, pH 8)99 at 21 °C. Exflagellation centers were counted 15 min post-activation by light microscopy (at least 10 fields).
Inducible knockdown of P. berghei pTKL
The degradation of PbpTKL-AID-HA in the presence of 3-indoleacetic acid (IAA, Sigma-Aldrich) was first analyzed by western blot. Nycodenz-enriched infected erythrocytes, with or without exposure to 0.5 µM IAA for 30 min at 37 °C, were lysed (total lysis protocol) and PbpTKL-AID-HA was immunoprecipitated. After confirming protein degradation in the presence of IAA, the intra-erythrocytic cycle was studied in vitro as described above, except that rings were cultured with or without 0.5 µM IAA57.
Immunofluorescence assays and microscopy
P. berghei parasites were synchronized as described above. Infected erythrocytes were collected at each development stage and fixed (4% paraformaldehyde and 0.075% glutaraldehyde in PBS) for 10 min on ice. Following incubation on poly-L-lysine-coated coverslips overnight at 4 °C, infected erythrocytes were blocked and permeabilized (1% BSA and 0.5% Triton X-100 in PBS) for 30 min at room temperature. Coverslips were incubated with rabbit anti-HA antibodies (diluted 1:150 in 1% BSA in PBS) and TER119 (diluted 1:500 as above) for 1 h at 37 °C. Coverslips were then incubated with Hoechst 33342 diluted 1:500 (Sigma-Aldrich) and secondary highly-cross-adsorbed anti-rabbit-AF594 and anti-rat-AF647 (diluted 1:1000 as above) for 30 min at 37 °C. The samples were mounted in Mowiol 4–88 (Sigma-Aldrich). Images were obtained at room temperature with a 63 × Plan Apochromat (1.4 NA) oil objective on the LSM880 confocal microscope (Zeiss) and processed using Zen Software.
Statistical analysis
The Mann-Whitney U test for non-parametric data was used for statistical comparisons of ATP consumption (in vitro Kinase Glo assay), percentages of GVBD observed in Xenopus oocytes and phenotypic analyses performed with P. berghei parental and pGEM lines. p < 0.001 was considered significant.
Supplementary information
Acknowledgements
This work was supported by Université de Lille, CNRS, Inserm and Institut Pasteur de Lille. The authors thank Dr Mathieu Gissot for critical discussion of the experimental design and results collected during the study. The authors also thank Arlette Lescuyer for technical assistance.
Author Contributions
B.G., J.K. and C.P. designed the study. B.G., A.F., K.C., C.L., C.D.W., A.M., V.J., I.C.G. and S.M. performed the experiments. B.G., A.F., K.C., C.L., D.T., A.M., I.C.G., S.M., J.K. and C.P. analyzed the data. B.G., A.F., J.K. and C.P. wrote the paper. All authors read, contributed feedback to, and approved the final manuscript.
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. Raw mass spectrometry data are openly available on PRIDE repository (ProteomeXchange Consortium, PXD011986). Data that support the findings of this study are available from the corresponding author CP upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44542-3.
References
- 1.WHO. World Malaria Report 2018. World Health Organization. Licence: CC BY-NC-SA 3.0 IGO. 2018).
- 2.Pfeffer DA, et al. malariaAtlas: an R interface to global malariometric data hosted by the Malaria Atlas Project. Malar. J. 2018;17:352. doi: 10.1186/s12936-018-2500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. World Malaria Report 2017. World Health Organization. Licence: CC BY-NC-SA3.0 IGO, http://www.who.int/iris/handle/10665/259492 (2017).
- 4.Ouji M, Augereau JM, Paloque L, Benoit-Vical F. Plasmodium falciparum resistance to artemisinin-based combination therapies: A sword of Damocles in the path toward malaria elimination. Parasite. 2018;25:24. doi: 10.1051/parasite/2018021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts L. Drug-resistant malaria advances in Mekong. Science. 2017;358:155–156. doi: 10.1126/science.358.6360.155. [DOI] [PubMed] [Google Scholar]
- 6.WHO. Artemisinin and Artemisinin-based Combination Therapy Resistance: Status Report. World Health Organization. Licence: CC BY-NC-SA 3.0 IGO, http://www.who.int/iris/handle/10665/255213 (2017).
- 7.Hall N, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 8.Cowman AF, Healer J, Marapana D, Marsh K. Malaria: biology and disease. Cell. 2016;167:610–624. doi: 10.1016/j.cell.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 9.Doerig C, Rayner JC, Scherf A, Tobin AB. Post-translational protein modifications in malaria parasites. Nat. Rev. Microbiol. 2015;13:160–172. doi: 10.1038/nrmicro3402. [DOI] [PubMed] [Google Scholar]
- 10.Lasonder E, Green JL, Grainger M, Langsley G, Holder AA. Extensive differential protein phosphorylation as intraerythrocytic Plasmodium falciparum schizonts develop into extracellular invasive merozoites. Proteomics. 2015;15:2716–2729. doi: 10.1002/pmic.201400508. [DOI] [PubMed] [Google Scholar]
- 11.Pease BN, et al. Global analysis of protein expression and phosphorylation of three stages of Plasmodium falciparum intraerythrocytic development. J. Proteome Res. 2013;12:4028–4045. doi: 10.1021/pr400394g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solyakov L, et al. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat. Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- 13.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe. 2011;10:410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derbyshire ER, et al. Chemical interrogation of the malaria kinome. Chembiochem. 2014;15:1920–1930. doi: 10.1002/cbic.201400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doerig C, Billker O, Pratt D, Endicott J. Protein kinases as targets for antimalarial intervention: Kinomics, structure-based design, transmission-blockade, and targeting host cell enzymes. Biochim. Biophys. Acta. 2005;1754:132–150. doi: 10.1016/j.bbapap.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 16.Mitcheson DF, Tobin AB, Alam MM. Applying chemical genetic tools to the study of phospho-signalling pathways in malaria parasites. Biochim. Biophys. Acta. 2015;1854:1650–1656. doi: 10.1016/j.bbapap.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Cabrera DG, et al. Plasmodial kinase inhibitors: license to cure? J. Med. Chem. 2018;61:8061–8077. doi: 10.1021/acs.jmedchem.8b00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttery DS, et al. Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell Host Microbe. 2014;16:128–140. doi: 10.1016/j.chom.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tewari R, et al. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho TG, et al. The ins and outs of phosphosignalling in Plasmodium: Parasite regulation and host cell manipulation. Mol. Biochem. Parasitol. 2016;208:2–15. doi: 10.1016/j.molbiopara.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Anamika, Srinivasan N, Krupa A. A genomic perspective of protein kinases in Plasmodium falciparum. Proteins. 2005;58:180–189. doi: 10.1002/prot.20278. [DOI] [PubMed] [Google Scholar]
- 22.Stancik IA, et al. Serine/Threonine protein kinases from bacteria, archaea and eukarya share a common evolutionary origin deeply rooted in the tree of life. J. Mol. Biol. 2018;430:27–32. doi: 10.1016/j.jmb.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. doi: 10.1096/fasebj.9.8.7768349. [DOI] [PubMed] [Google Scholar]
- 24.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 25.Eyers PA, Murphy JM. Dawn of the dead: protein pseudokinases signal new adventures in cell biology. Biochem. Soc. Trans. 2013;41:969–974. doi: 10.1042/BST20130115. [DOI] [PubMed] [Google Scholar]
- 26.Murphy JM, et al. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 2014;457:323–334. doi: 10.1042/BJ20131174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeqiraj E, van Aalten DM. Pseudokinases-remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol. 2010;20:772–781. doi: 10.1016/j.sbi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee K, et al. CASK functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eswaran J, et al. Structure and functional characterization of the atypical human kinase haspin. Proc. Natl. Acad. Sci. USA. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell. Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Shaw AS, Kornev AP, Hu J, Ahuja LG, Taylor SS. Kinases and pseudokinases: lessons from RAF. Mol. Cell. Biol. 2014;34:1538–1546. doi: 10.1128/MCB.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sreelatha A, et al. Protein AMPylation by an evolutionarily conserved pseudokinase. Cell. 2018;175:809–821 e819. doi: 10.1016/j.cell.2018.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajakulendran T, Sicheri F. Allosteric protein kinase regulation by pseudokinases: insights from STRAD. Sci. Signal. 2010;3:pe8. doi: 10.1126/scisignal.3111pe8. [DOI] [PubMed] [Google Scholar]
- 35.Jacobsen AV, Murphy JM. The secret life of kinases: insights into non-catalytic signalling functions from pseudokinases. Biochem. Soc. Trans. 2017;45:665–681. doi: 10.1042/BST20160331. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Photiou A, Grothey A, Stebbing J, Giamas G. The role of pseudokinases in cancer. Cell. Signal. 2012;24:1173–1184. doi: 10.1016/j.cellsig.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Reese ML, Boothroyd JC. A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J. Biol. Chem. 2011;286:29366–29375. doi: 10.1074/jbc.M111.253435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etheridge RD, et al. The Toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe. 2014;15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reese ML, Shah N, Boothroyd JC. The Toxoplasma pseudokinase ROP5 is an allosteric inhibitor of the immunity-related GTPases. J. Biol. Chem. 2014;289:27849–27858. doi: 10.1074/jbc.M114.567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behnke MS, et al. Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent south american strains of Toxoplasma gondii. PLoS Genet. 2015;11:e1005434. doi: 10.1371/journal.pgen.1005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talevich E, Tobin AB, Kannan N, Doerig C. An evolutionary perspective on the kinome of malaria parasites. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2012;367:2607–2618. doi: 10.1098/rstb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 45.Adl SM, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngoubangoye B, et al. The host specificity of ape malaria parasites can be broken in confined environments. Int. J. Parasitol. 2016;46:737–744. doi: 10.1016/j.ijpara.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Tellier G, et al. Identification of Plasmodium falciparum translation initiation eIF2beta subunit: direct interaction with Protein Phosphatase Type 1. Front. Microbiol. 2016;7:777. doi: 10.3389/fmicb.2016.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slaby S, et al. Maturation of Xenopus laevis oocytes under cadmium and lead exposures: Cell biology investigations. Aquat. Toxicol. 2017;193:105–110. doi: 10.1016/j.aquatox.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Daher W, et al. Regulation of protein phosphatase type 1 and cell cycle progression by PfLRR1, a novel leucine-rich repeat protein of the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2006;60:578–590. doi: 10.1111/j.1365-2958.2006.05119.x. [DOI] [PubMed] [Google Scholar]
- 50.Hendrickx A, et al. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 51.Collins CR, Hackett F, Atid J, Tan MSY, Blackman MJ. The Plasmodium falciparum pseudoprotease SERA5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes. PLoS Pathog. 2017;13:e1006453. doi: 10.1371/journal.ppat.1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marapana DS, et al. Plasmepsin V cleaves malaria effector proteins in a distinct endoplasmic reticulum translocation interactome for export to the erythrocyte. Nat. Microbiol. 2018;3:1010–1022. doi: 10.1038/s41564-018-0219-2. [DOI] [PubMed] [Google Scholar]
- 53.The UniProt, C. UniProt: the universal protein knowledgebase Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maragno AL, et al. ISG15 modulates development of the erythroid lineage. PLoS One. 2011;6:e26068. doi: 10.1371/journal.pone.0026068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bushell E, et al. Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell. 2017;170:260–272 e268. doi: 10.1016/j.cell.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Philip N, Waters AP. Conditional degradation of Plasmodium calcineurin reveals functions in parasite colonization of both host and vector. Cell Host Microbe. 2015;18:122–131. doi: 10.1016/j.chom.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bryk AH, Wisniewski JR. Quantitative Analysis of Human Red Blood Cell Proteome. J. Proteome Res. 2017;16:2752–2761. doi: 10.1021/acs.jproteome.7b00025. [DOI] [PubMed] [Google Scholar]
- 59.Nunes MC, Okada M, Scheidig-Benatar C, Cooke BM, Scherf A. Plasmodium falciparum FIKK kinase members target distinct components of the erythrocyte membrane. PLoS One. 2010;5:e11747. doi: 10.1371/journal.pone.0011747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonager J, et al. Reduced CD36-dependent tissue sequestration of Plasmodium-infected erythrocytes is detrimental to malaria parasite growth in vivo. J. Exp. Med. 2012;209:93–107. doi: 10.1084/jem.20110762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreira CK, et al. The Plasmodium PHIST and RESA-like protein families of human and rodent malaria parasites. PLoS One. 2016;11:e0152510. doi: 10.1371/journal.pone.0152510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mills EW, Wangen J, Green R, Ingolia NT. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 2016;17:1–10. doi: 10.1016/j.celrep.2016.08.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen Anthony T., Prado Miguel A., Schmidt Paul J., Sendamarai Anoop K., Wilson-Grady Joshua T., Min Mingwei, Campagna Dean R., Tian Geng, Shi Yuan, Dederer Verena, Kawan Mona, Kuehnle Nathalie, Paulo Joao A., Yao Yu, Weiss Mitchell J., Justice Monica J., Gygi Steven P., Fleming Mark D., Finley Daniel. UBE2O remodels the proteome during terminal erythroid differentiation. Science. 2017;357(6350):eaan0218. doi: 10.1126/science.aan0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Alessandro A, Righetti PG, Zolla L. The red blood cell proteome and interactome: an update. J. Proteome Res. 2010;9:144–163. doi: 10.1021/pr900831f. [DOI] [PubMed] [Google Scholar]
- 65.Kroiher M, Miller MA, Steele RE. Deceiving appearances: signaling by “dead” and “fractured” receptor protein-tyrosine kinases. Bioessays. 2001;23:69–76. doi: 10.1002/1521-1878(200101)23:1<69::AID-BIES1009>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 66.Abdi A, Eschenlauer S, Reininger L, Doerig C. SAM domain-dependent activity of PfTKL3, an essential tyrosine kinase-like kinase of the human malaria parasite Plasmodium falciparum. Cell. Mol. Life Sci. 2010;67:3355–3369. doi: 10.1007/s00018-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramdani G, Langsley G. ATP, an extracellular signaling molecule in red blood cells: a messenger for malaria? Biomed. J. 2014;37:284–292. doi: 10.4103/2319-4170.132910. [DOI] [PubMed] [Google Scholar]
- 68.Paul A, Pallavi R, Tatu US, Natarajan V. The bystander effect in optically trapped red blood cells due to Plasmodium falciparum infection. Trans. R. Soc. Trop. Med. Hyg. 2013;107:220–223. doi: 10.1093/trstmh/trt010. [DOI] [PubMed] [Google Scholar]
- 69.Bhattacharyya, M. K., Hong, Z., Kongkasuriyachai, D. & Kumar, N. Plasmodium falciparum protein phosphatase type 1 functionally complements a glc7 mutant in Saccharomyces cerevisiae. Int. J. Parasitol. 32, 739-747 (2002). [DOI] [PubMed]
- 70.Kumar R, Adams B, Oldenburg A, Musiyenko A, Barik S. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum: demonstration of its essential role using RNA interference. Malar. J. 2002;1:5. doi: 10.1186/1475-2875-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 72.Qiao F, Bowie JU. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Min, Wang Chengqi, Otto Thomas D., Oberstaller Jenna, Liao Xiangyun, Adapa Swamy R., Udenze Kenneth, Bronner Iraad F., Casandra Deborah, Mayho Matthew, Brown Jacqueline, Li Suzanne, Swanson Justin, Rayner Julian C., Jiang Rays H. Y., Adams John H. Uncovering the essential genes of the human malaria parasitePlasmodium falciparumby saturation mutagenesis. Science. 2018;360(6388):eaap7847. doi: 10.1126/science.aap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reid, A. J. et al. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife7, 10.7554/eLife.33105 (2018). [DOI] [PMC free article] [PubMed]
- 75.Hollin T, De Witte C, Lenne A, Pierrot C, Khalife J. Analysis of the interactome of the Ser/Thr Protein Phosphatase type 1 in Plasmodium falciparum. BMC Genomics. 2016;17:246. doi: 10.1186/s12864-016-2571-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderluh G, Pungercar J, Strukelj B, Macek P, Gubensek F. Cloning, sequencing, and expression of equinatoxin II. Biochem. Biophys. Res. Commun. 1996;220:437–442. doi: 10.1006/bbrc.1996.0391. [DOI] [PubMed] [Google Scholar]
- 77.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dawson NL, et al. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45:D289–D295. doi: 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Castro E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernandez-Fuentes N, Madrid-Aliste CJ, Rai BK, Fajardo JE, Fiser A. M4T: a comparative protein structure modeling server. Nucleic Acids Res. 2007;35:W363–368. doi: 10.1093/nar/gkm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kallberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haas J, et al. Continuous Automated Model EvaluatiOn (CAMEO) complementing the critical assessment of structure prediction in CASP12. Proteins. 2018;86(Suppl 1):387–398. doi: 10.1002/prot.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haas J, et al. The Protein Model Portal–a comprehensive resource for protein structure and model information. Database (Oxford) 2013;2013:bat031. doi: 10.1093/database/bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- 87.Meiselbach H, Sticht H, Enz R. Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem. Biol. 2006;13:49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 88.Burdukiewicz Michał, Sobczyk Piotr, Chilimoniuk Jarosław, Gagat Przemysław, Mackiewicz Paweł. Prediction of Signal Peptides in Proteins from Malaria Parasites. International Journal of Molecular Sciences. 2018;19(12):3709. doi: 10.3390/ijms19123709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 91.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 93.Freville A, et al. Plasmodium falciparum encodes a conserved active inhibitor-2 for Protein Phosphatase type 1: perspectives for novel anti-plasmodial therapy. BMC Biol. 2013;11:80. doi: 10.1186/1741-7007-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Planson AG, Guijarro JI, Goldberg ME, Chaffotte AF. Assistance of maltose binding protein to the in vivo folding of the disulfide-rich C-terminal fragment from Plasmodium falciparum merozoite surface protein 1 expressed in Escherichia coli. Biochemistry. 2003;42:13202–13211. doi: 10.1021/bi035321c. [DOI] [PubMed] [Google Scholar]
- 96.Lenne A, et al. Characterization of a Protein Phosphatase Type-1 and a kinase anchoring protein in Plasmodium falciparum. Front. Microbiol. 2018;9:2617. doi: 10.3389/fmicb.2018.02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bondos SE, Bicknell A. Detection and prevention of protein aggregation before, during, and after purification. Anal. Biochem. 2003;316:223–231. doi: 10.1016/S0003-2697(03)00059-9. [DOI] [PubMed] [Google Scholar]
- 98.Vandomme A, et al. PhosphoTyrosyl phosphatase activator of Plasmodium falciparum: identification of its residues involved in binding to and activation of PP2A. Int. J. Mol. Sci. 2014;15:2431–2453. doi: 10.3390/ijms15022431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Janse CJ, Waters AP. Plasmodium berghei: the application of cultivation and purification techniques to molecular studies of malaria parasites. Parasitol. Today. 1995;11:138–143. doi: 10.1016/0169-4758(95)80133-2. [DOI] [PubMed] [Google Scholar]
- 100.Janse CJ, Ramesar J, Waters AP. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 2006;1:346–356. doi: 10.1038/nprot.2006.53. [DOI] [PubMed] [Google Scholar]
- 101.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 103.Luber CA, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 104.Wang, Q. et al. Autoinhibition of Bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. Elife4, 10.7554/eLife.06074 (2015). [DOI] [PMC free article] [PubMed]
- 105.Molina-Cerrillo J, Alonso-Gordoa T, Gajate P, Grande E. Bruton’s tyrosine kinase (BTK) as a promising target in solid tumors. Cancer Treat. Rev. 2017;58:41–50. doi: 10.1016/j.ctrv.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 106.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 107.Im YJ, et al. The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J. Biol. Chem. 2007;282:5443–5452. doi: 10.1074/jbc.M611342200. [DOI] [PubMed] [Google Scholar]
- 108.Ma H, Lou Y, Lin WH, Xue HW. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 2006;16:466–478. doi: 10.1038/sj.cr.7310058. [DOI] [PubMed] [Google Scholar]
- 109.Lorestani A, et al. Targeted proteomic dissection of Toxoplasma cytoskeleton sub-compartments using MORN1. Cytoskeleton (Hoboken) 2012;69:1069–1085. doi: 10.1002/cm.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mecklenburg KL, et al. Retinophilin is a light-regulated phosphoprotein required to suppress photoreceptor dark noise in. Drosophila. J. Neurosci. 2010;30:1238–1249. doi: 10.1523/JNEUROSCI.4464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Egloff MP, et al. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Camarda G, et al. Regulated oligomerisation and molecular interactions of the early gametocyte protein Pfg27 in Plasmodium falciparum sexual differentiation. Int. J. Parasitol. 2010;40:663–673. doi: 10.1016/j.ijpara.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 115.Sharma A, Sharma I, Kogkasuriyachai D, Kumar N. Structure of a gametocyte protein essential for sexual development in Plasmodium falciparum. Nat. Struct. Biol. 2003;10:197–203. doi: 10.1038/nsb899. [DOI] [PubMed] [Google Scholar]
- 116.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 117.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 118.Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- 119.Meibalan E, et al. Host erythrocyte environment influences the localization of exported protein 2, an essential component of the Plasmodium translocon. Eukaryot. Cell. 2015;14:371–384. doi: 10.1128/EC.00228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. Raw mass spectrometry data are openly available on PRIDE repository (ProteomeXchange Consortium, PXD011986). Data that support the findings of this study are available from the corresponding author CP upon reasonable request.