Abstract
The first domesticated companion animal, the dog, is currently represented by over 190 unique breeds. Across these numerous breeds, dogs have exceptional variation in lifespan (inversely correlated with body size), presenting an opportunity to discover longevity-determining traits. We performed a genome-wide association study on 4169 canines representing 110 breeds and identified novel candidate regulators of longevity. Interestingly, known functions within the identified genes included control of coat phenotypes such as hair length, as well as mitochondrial properties, suggesting that thermoregulation and mitochondrial bioenergetics play a role in lifespan variation. Using primary dermal fibroblasts, we investigated mitochondrial properties of short-lived (large) and long-lived (small) dog breeds. We found that cells from long-lived breeds have more uncoupled mitochondria, less electron escape, greater respiration, and capacity for respiration. Moreover, our data suggest that long-lived breeds have higher rates of catabolism and β-oxidation, likely to meet elevated respiration and electron demand of their uncoupled mitochondria. Conversely, cells of short-lived (large) breeds may accumulate amino acids and fatty acid derivatives, which are likely used for biosynthesis and growth. We hypothesize that the uncoupled metabolic profile of long-lived breeds likely stems from their smaller size, reduced volume-to-surface area ratio, and therefore a greater need for thermogenesis. The uncoupled energetics of long-lived breeds lowers reactive oxygen species levels, promotes cellular stress tolerance, and may even prevent stiffening of the actin cytoskeleton. We propose that these cellular characteristics delay tissue dysfunction, disease, and death in long-lived dog breeds, contributing to canine aging diversity.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00062-6) contains supplementary material, which is available to authorized users.
Keywords: Aging, Dogs, Mitochondria, Uncoupling, GWAS, Primary cells
Introduction
The domestic dog, Canis lupus familiaris, is an emerging model for aging biology and age-related disorders. Genomic studies suggest that canine domestication began between 20,000 and 40,000 years ago (Botigue et al. 2017). Throughout this period and up to the present, humans and dogs have shared intimate proximity with common environmental exposures (air, water, food, and pathogens). With age, dogs undergo a similar functional decline as humans (Hoffman et al. 2018); they develop many of the same age-associated diseases, including cancers, metabolic syndromes, and neurodegeneration (Kaeberlein et al. 2016). Moreover, genetic analysis has demonstrated strong parallel co-evolution between our species, particularly in metabolic and neurological processes (Wang et al. 2013). Together, these factors suggest scientific findings in dogs may be informative to human biology and aging.
Dogs have immense genetic and phenotypic diversity across individual breeds. Extensive human-led breeding efforts have created significant variation in size, disease prevalence, and longevity, among many other parameters. For example, there is a 30-fold difference in weight and 2.5-fold difference in lifespan between the Chihuahua (~ 5 lb, ~ 16 years) and Great Dane (~ 150 lb, ~ 7 years). Interestingly, body size is a strong predictor of lifespan in dogs, with smaller breeds living longer than larger breeds (Greer et al. 2007). This holds true with the hundreds of breeds registered with the American Kennel Club (AKC) which fall along this size-lifespan continuum (Fig. 1a). Importantly, mortality-data modeling suggests that the dominant reason small dogs live longer is by aging more slowly, where the rate of mortality increases with age slower than their larger counterparts (Kraus et al. 2013). Furthermore, although larger breeds tend to have higher levels of inbreeding, variation in longevity cannot be explained by variation in the strength of inbreeding (Yordy et al., Body size, inbreeding, and lifespan in domestic dogs, in preparation).
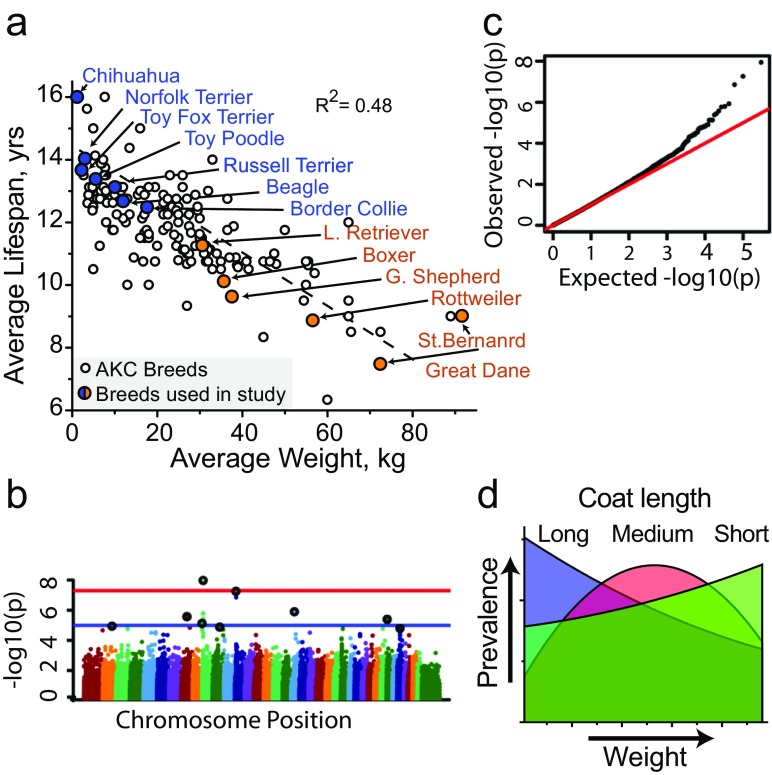
Fig. 1.
Thermoregulatory and mitochondrial genes associated with canine breed longevity. a Scatter plot of breeds registered with the American Kennel Club plotted average lifespan by average weight. Cells from breeds used in this study are highlighted in blue (long-lived) or orange (short-lived). b Manhattan (blue and red lines correspond to p value cutoffs of 10−5 and 10−8) and (c) quantile-quantile plots from the GWAS of breed-average life expectancy (n = 4169 individuals, 110 breeds, see Table 1). d FGF5 and RSPO2 govern variation in coat length and phenotype; this plot shows the relative distribution of coat length versus increasing breed weight (see methods). Note that the smaller the breed, the more likely it is to have longer hair and vice versa
Dog breed size has been associated with SNPs in the pro-growth hormone insulin-like growth factor-1 (IGF-1) (Sutter et al. 2007; Hayward et al. 2016). Suppressed IGF-1 signaling can increase longevity, and small dogs are known to have reduced circulating IGF-1 (Greer et al. 2011). Growth signaling from IGF-1 certainly plays a significant role in controlling breed size, but it is not clear if (or to what degree or mechanism) it might regulate lifespan in dogs. Variation in IGF-1 and the rate of aging would suggest that differences in cellular and tissue physiology underlie canine longevity. The only known cellular property that correlates with breed longevity is telomere length, where long-lived breeds display longer telomeres (Fick et al. 2012). However, the involvement of telomeres in the aging process is not fully understood and even disputed. To elucidate what governs the variation in aging rate between dog breeds, additional unbiased studies are needed. Dogs provide an excellent model to identify novel longevity pathways, which will not only help us understand the true nature of aging but can also be exploited for human and canine health.
Using genome-wide association (GWAS), we identify novel longevity-associated genes that are known to regulate thermogenic and bioenergetic processes. We investigate these processes with a comparative metabolic and biophysical analysis of primary dermal fibroblasts derived from short- and long-lived dog breeds. Previous studies have indicated that primary dermal fibroblasts can recapitulate organismal metabolism and have demonstrated their utility in uncovering physiological differences between short- and long-lived animals (Murakami et al. 2003; Wang and Miller 2012). We analyzed fibroblasts for bioenergetic-metabolite (organic acids, amino acids, and acylcarnitines) profiles, mitochondrial and electron transport chain (ETC) properties, stress tolerance, and cytoskeletal dynamics. We report that, even after isolation and culturing, canine fibroblasts retain breed-specific metabolism that correlates with donor breed longevity. We show that cells from long-lived breeds have more mitochondrial uncoupling, greater respiration with less ROS formation, and potentially more flexible cytoskeletons. Our data suggests that long-lived breeds utilize greater amino and fatty acid catabolism, which is required to sustain their uncoupled-energetic demand. We also demonstrate greater stress tolerance in cells from long-lived breeds and posit that these breeds age more slowly due to superior tissue homeostasis conferred by these properties.
Results
The functionality and properties of cells are known to contribute to the robustness and longevity of whole organisms. To narrow the range of cellular processes to analyze, all of which could potentially contribute to aging variation in dogs, we performed a GWAS for breed-average lifespan. The analysis included 150,000 loci in 4169 individuals representing 110 breeds. We used a linear mixed model as described previously (Hayward et al. 2016), and identified top candidate genes that associate with canine longevity (Fig. 1b, c, Table 1, Supplemental File 1). Interestingly, our GWAS identified FGF5 and RSPO2 to associate with breed longevity (Table 1), which are two out of the three known genes that control canine coat phenotypes such as hair length (Housley and Venta 2006; Cadieu et al. 2009). We next checked whether coat length in dogs correlated with the size and longevity of breeds. We found that on average, coat length was inversely related to breed size, where larger breeds tend to have shorter hair and smaller breeds are likelier to have longer hair (Fig. 1d). The principal purpose of hair is thermoregulation, and it is logical that smaller breeds would have longer hair to retain heat, and for larger dogs to have shorter hair to keep cool. This not only indicates that canines possess thermoregulatory strategies but that similar mechanisms may influence breed longevity.
Table 1.
GWAS of breed-average life expectancy
| Across breed life expectancy GWAS results with p values below or near 1E-05. | ||
|---|---|---|
| Top marker(s) in region (chr: position) | p value | Candidate gene(s) |
| 10: 8183593 | 1.15 × 10−8 | MSRB3, HMGA2 |
| 13: 8654384 | 5.46 × 10−8 | RSPO2, ANGPT1 |
| 19: 19107981 | 1.19 × 10−6 | PRDM5 |
| 8: 36016977 | 2.67 × 10−6 | SLC38A6, TRMT5 |
| 32: 4503644 | 4.12 × 10−6 | FGF5 |
| 10: 2185035 | 7.44 × 10−6 | ATP23 |
| 10: 2201298 | ||
| 10: 2224239 | ||
| 2: 59036686 | 1.24 × 10−5 | |
| 11: 47400651 | 1.28 × 10−5 | |
| 34: 18341784 | 1.54 × 10−5 | IGF2BP2 |
Other top GWAS hits were loci within or near mitochondrial and bioenergetic regulators including IGFBP2, MSRB3, ATP23, and TRMT5 (Table 1). IGFBP2 inhibits IGF signaling and its overexpression increases lifespan in mice (Hoeflich et al. 2016). Other identified genes—MSRB3, ATP23, and TRMT5—are known to regulate various mitochondrial properties, such as ROS production, respiration, and ATP synthesis (Supplemental File 1). Importantly, and in addition to the aforementioned properties, mitochondria also have a prominent role in thermogenesis via proton uncoupling. Overall, the candidate genes identified by GWAS suggested mitochondrial-bioenergetic and thermoregulatory mechanisms as major factors driving canine aging diversity.
To investigate differences in mitochondrial bioenergetics in cells derived from different dog breeds, we moved to ex vivo primary cell culture. We used a targeted mass spectrometry approach to profile major bioenergetic metabolites. These included organic acids, amino acids, and acylcarnitine species (fatty acid derivatives primed for β-oxidation) in cells from a subset of breeds: Boxers (short-lived), Chihuahuas (long-lived), and Labradors (in between Boxer and Chihuahua) (Supplemental File 2). Three independently prepared samples from each of five to seven donors per breed were used. Unsupervised hierarchical clustering of measured metabolites clearly segregated by donor breed (Fig. 2a), suggesting that each breed has its own unique metabolite signature. Principal component analysis (PCA) of the separate metabolite classes revealed that acylcarnitines distinguished the breeds with good resolution (Fig. 2b). Moreover, acylcarnitine clustering and PCA positioned Labradors as an intermediate between Boxers and Chihuahuas (Fig. 2b, Fig. S1, S2), which mirrors the average lifespan of these breeds. These data indicate that bioenergetic metabolites (acylcarnitines in particular) derived from primary canine fibroblasts can predict the donor’s breed, and importantly for our study, that there are breed-distinct cellular metabolic profiles that are retained in vitro.
Fig. 2.
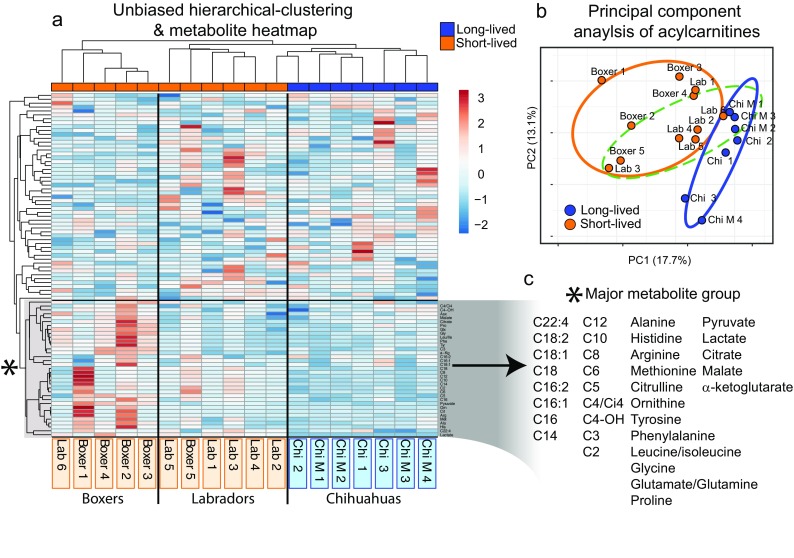
Primary canine fibroblasts retain breed-specific bioenergetic-metabolite profiles in vitro. Multiple donors were utilized from: Chihuahuas, Chihuahua-mixed, Labrador Retrievers, and Boxers for targeted mass spectrometry of organic acids, amino acids, and acylcarnitines. Raw metabolite values were normalized to protein concentrations and averaged across three replicates for every donor cell line. a Unbiased hierarchical clustering of donor metabolite profiles, note the clustering of donors by breed. b Principal component analysis of acylcarnitine metabolites. Circles denote 90% confidence interval (orange—short-lived breeds, blue—long-lived, dashed green—Labradors). c Magnification of major metabolite branch driving distinctions between breeds (see Supplemental File 2 for raw data, Fibroblast donor information)
Next, we utilized the Seahorse analyzer to measure rates of oxygen consumption and extracellular acidification, which represent oxidative phosphorylation/respiration and anaerobic glycolysis. We assessed metabolic rates before and after co-treating cells with oligomycin (which stimulates glycolysis) and FCCP (which maximizes respiration). In doing so, we could measure the baseline and maximum metabolic rates of cells. From these rates, we calculated the “metabolic potential or capacity,” which is the degree of change in respiration or glycolysis from baseline to maximum possible levels. We observed no trends in respiration or glycolysis at baseline levels. However, the respiration capacity clustered strongly by breed longevity, where long-lived breeds had on average 65% greater potential to upregulate their respiration (**p = 0.008), when stimulated (Fig. 3a, b). Furthermore, the respiration capacity significantly correlated with breed longevity (R = 0.74, *p = 0.024). Greater respiration capacity in long-lived breed cells could arise from two possible mechanisms: more abundant mitochondria or less electron escape from electron transport chain (ETC) complexes within mitochondria. To examine these possibilities, we utilized mitochondria-specific dyes. MitoTracker Green staining showed that the abundance of mitochondria was not significantly different between breeds (Fig. 3c, e). This suggests that the greater respiration capacity of long-lived breeds derives from innate properties of the mitochondria and ETC.
Fig. 3.
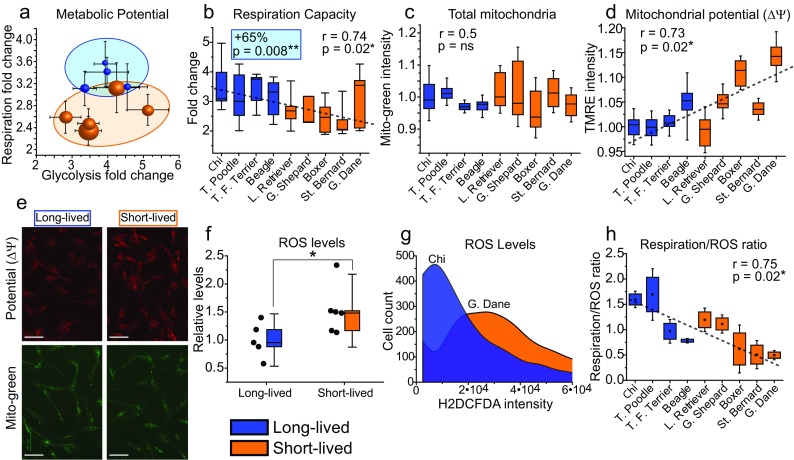
Fibroblasts from long-lived breeds have greater respiration capacity, lower mitochondrial potential, and less electron escape from electron transport chains than short-lived breeds. The Seahorse assay results are from 3 to 5 (dependent on breed) independent experiments. a Plot depicting the Seahorse assay results for metabolic potential with fold change of respiration (y-axis) and glycolysis (x-axis), standard error shown. Each point corresponds to an individual breed, and the size of the point approximately correlates to the average size of the breed. Note the clustering of breeds by longevity on the respiration y-axis. b Box plot representations of the fold change in respiration depicted in a. Note the trend of decreasing respiration fold change with decreasing breed longevity. c Box plots of the relative MitoTracker Green intensity (mitochondrial mass) in primary canine fibroblasts. d Box plots of relative TMRE (mitochondrial membrane potential—ΔΨ intensity). e Representative fluorescent microscopy images of fibroblasts stained with the MitoTracker Green and TMRE as quantified in c and d. Results are pooled from three independent experiments with standard deviations shown. Note the equivalent mitochondrial abundance between breeds but increasing potential with decreasing breed longevity. f Box plots depicting the average and relative levels of ROS in fibroblasts from long- and short-lived breeds, measured by H2DCFDA staining. g Example of H2DCFDA staining in fibroblasts from the Chihuahua and Great Dane. h Electron transport chain efficiency calculated from the ratio of oxygen consumption under maximum ETC flux (measured by the Seahorse) to ROS H2DCFDA intensity. Note short-lived breeds have and produce more ROS relative to oxygen consumed
Next, to examine ETC properties, we stained the fibroblasts with tetramethylrhodamine ethyl ester (TMRE), which measures the mitochondrial membrane potential (ΔΨ). The ΔΨ represents the proton electrochemical gradient that is responsible for ATP production via oxidative phosphorylation. The movement of protons across the electrochemical gradient can be disconnected or “uncoupled” from ATP synthesis (notably for thermogenesis). Furthermore, the degree of uncoupling inversely correlates with the ΔΨ, where increased uncoupling results in lower ΔΨ and lower uncoupling results in a higher ΔΨ. We found that the ΔΨ correlated with breed longevity (R = 0.73, *p = 0.027), where long-lived breeds had lower ΔΨ, with the difference being as high as 15% between breeds (Fig. 3d, e). The lower ΔΨ in long-lived breeds suggests more proton uncoupling and further shows innate variation in mitochondrial and ETC properties between long- and short-lived breeds.
Differences in mitochondrial membrane potential (ΔΨ) and the degree of uncoupling influence ROS production. More specifically, the higher the ΔΨ, the higher the rate of mitochondrial ROS production (Mookerjee et al. 2010). The main source of ROS is electrons that escape from the ETC, which generate superoxides (precursors to harsher ROS) instead of oxygen reduction by respiration. Because we observed differences in the ΔΨ between long- and short-lived breeds, we next measured cellular ROS using H2DCFDA dye. We found that cells from long-lived breeds had reduced levels of ROS compared to short-lived breeds (Fig. 3f, g). This suggests that long-lived breed fibroblasts do have less electron escape in their ETCs, as their baseline respiration is equivalent to short-lived breeds but ROS levels are lower. We also measured the ratio of oxygen consumed to ROS produced. A higher ratio indicates less electron escape, as more oxygen is converted to water versus ROS. We found that the respiration/ROS ratio correlated with breed longevity (R = 0.75, *p = 0.02), particularly under maximum ETC flux and respiration, with long-lived breeds having a higher ratio (Fig. 3h). These data suggest that longer-lived breeds have a greater respiration capacity due to less electron escape from their ETCs. Overall, the data show that mitochondria of longer-lived breeds are more uncoupled, have lower ΔΨ, and have less electron escape to ROS. Furthermore, these data suggest that long-lived breed cells would more readily handle bioenergetic stress.
The stress tolerance of cells has been shown to correlate with organismal and species longevity (Murakami et al. 2003; Harper et al. 2011), including variation in metabolic response to oxidative and nutrient stress (Wang and Miller 2012). Furthermore, mild mitochondrial uncoupling is known to promote cellular survival, particularly under stress (Baffy et al. 2011). We hypothesized that long-lived breed cells would tolerate oxidative and nutrient stress more effectively due to being more uncoupled and producing less ROS. To test this, we stressed cells with nutrient deprivation or rotenone exposure. Survival of cells was measured by staining with the apoptosis markers annexin-V and propidium iodide followed by flow cytometry, as described previously (Domanskyi et al. 2017). Fibroblasts from long-lived breeds survived on average 8% more after rotenone challenge (*p = 0.01) (Fig. 4b). Moreover, there was a significant correlation between breed longevity and survival after nutrient deprivation (R = 0.58, *p = 0.03), where long-lived breed fibroblasts had up to 25% greater survival (Fig. 4a, c). These data demonstrate that long-lived breed cells possess greater stress tolerance than those from short-lived breeds, especially in low nutrient conditions.
Fig. 4.

Fibroblasts from long-lived breeds have greater stress tolerance and consume more oxygen in low-nutrient conditions. Box plots showing relative survival of fibroblasts after (a) nutrient deprivation and (b) rotenone exposure. c Example of flow cytometry plots (annexin-V by PI staining) used to quantify survival of fibroblasts as depicted in a and b. d Baseline respiration rates measured by the Seahorse under low-nutrient conditions. e Respiration capacity measured by the Seahorse assay under low-nutrient conditions, also shown is the capacity under nutrient-replete conditions. Cells from long-lived donors have elevated baseline and respiration capacity under low-nutrient conditions
Next, we performed the Seahorse analysis on the fibroblasts with low nutrients to assess if mitochondrial response also varied between breeds in this condition. Interestingly, we found a stronger correlation of baseline respiration and breed longevity with low nutrients, where respiration was 52% (*p = 0.05) higher on average in long-lived breeds (Fig. 4d). The respiration capacity was also significantly higher for long-lived breeds and correlated with breed longevity (Fig. 4e). An argument can be made that these low in vitro nutrient conditions more accurately reflect a physiologically relevant state. Cells in tissues (especially in skin) are not saturated with glucose and growth factors, as they are in vitro. Overall, these data suggest that long-lived breed fibroblasts are more uncoupled, produce less ROS, and have a greater respiratory capacity (due to less electron escape in ETCs). These differences are apparent under normal physiological stress and contribute to the cells’ greater stress tolerance and survival.
In addition to metabolic and survival dynamics, the functionality of tissues also depends on physical properties of cells. Here, we applied atomic force microscopy (AFM) combined with a brush computational model to measure the elastic modulus (rigidity) on a subset of canine fibroblasts. There was a correlation of rigidity with breed longevity, despite only four breeds being represented (Beagle, Labrador, St. Bernard, and Great Dane). The short-lived Great Dane had significantly stiffer cells at both early and late passages, while the Beagle (longest-lived breed used for AFM) had the most flexible overall (Fig. S3). These data suggest that shorter lived-breed fibroblasts are stiffer than their longer-lived counterparts, due to an altered cytoskeleton. More flexible cells may be better able to mitigate ROS and other damage because they are more deformable in response to everyday mechanical stresses. This would help maintain cellular function and homeostasis through age.
Discussion
Dogs are an emerging research model with great promise for future biomedical studies. To date, however, there are few resources available for manipulation and mechanistic experiments in the canine model, which imposed a few key limitations on our study. First, our study focuses on primary cell culture, and as such, the physiological relevance of our results may be open to question. Second, for many of the breeds in our study, cells were available from only one donor. Rather than relying on a small number of breeds with many donors, for most of our experiments, we chose to use many breeds with one donor, the exception being mass spectrometry (please see Supplemental File 1). Including a larger number of breeds helps to ensure that the differences we observe are not the result of breed-specific differences unrelated to size or lifespan, but it also raises the possibility of individual differences coming into play (the data in Figs. 2 and 5 have the opposite problem). Despite these limitations, we still observed significant correlations and differences in culture between breeds. Third, our study is descriptive in nature and lacks a validated mechanistic element, although we propose a mechanism in Fig. 6. Finally, we hypothesize that many of our metabolic findings arise from an underlying thermogenic process, but the relevance of dermal fibroblasts to thermogenic processes is unknown. It is possible that these mitochondrial differences may arise by mechanisms unrelated to thermogenesis, though we believe our data is most synonymous with an uncoupling/thermogenic process ultimately driven by size differences. With these caveats in mind, we believe the data and discussion presented here will aid aging and dog model system researchers going forward.
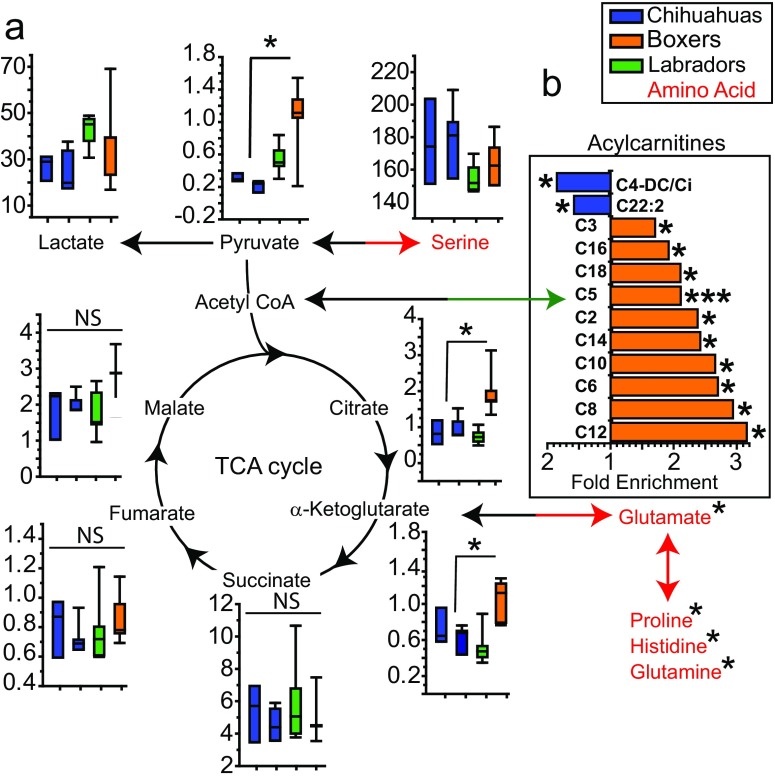
Fig. 5.
Bioenergetic metabolites differ significantly between the Boxers and Chihuahuas. Boxplots from left to right: Chihuahuas, Chihuahua-mixed, Labradors, and Boxers. Raw metabolite values were normalized to protein concentrations and averaged across three replicates for every donor cell line. Asterisk indicates significant difference, in the respective metabolite, between the Boxers and Chihuahuas. a Glycolysis and citric acid cycle metabolite pathway schematic. b Fold enrichment of significantly different acylcarnitines between the Boxers and Chihuahuas (Labradors had intermediate concentrations for all acylcarnitines, see Supplemental File 2 for raw data and fibroblast donor information)
Fig. 6.
Model. We hypothesize that smaller dogs, due to extra demand for thermogenesis, evolved or responded to have more uncoupled mitochondria, which in turn results in lowered electrical potential (due to proton leak) and elevated β-oxidation and catabolism (in order to fuel uncoupled mitochondria). We suggest that the secondary changes associated with these adaptations happen to be beneficial for longevity; these include decreased production of reactive oxygen species, metabolic profiles which are less favorable for tumorigenesis, increased cytoskeletal flexibility, and resistance to stress (affording superior tissue homeostasis and longevity). Conversely, larger breeds might preferentially use energy for biosynthesis to maintain their greater mass. Overall, our data suggest that lifespan variation across canine breeds is partially regulated by innate cellular properties
In this study, we demonstrate innate variation in mitochondrial bioenergetics between cells of short- and long-lived dog breeds. We show that primary canine fibroblasts have equivalent mitochondrial mass but that long-lived (smaller) breeds have a lower ΔΨ (Fig. 3c, d), suggesting more uncoupling. When the fibroblasts are not saturated with nutrients, which is more representative of the in vivo tissue microenvironment, we observe higher baseline respiration in long-lived breeds (Fig. 4c), which also supports a greater metabolic rate and uncoupling. Mild mitochondrial uncoupling lowers ΔΨ and ROS levels and increases ETC flux and respiration (Brand et al. 2004). In fact, mild uncoupling in primary human dermal fibroblasts suppresses ROS (Cho et al. 2014). We find that fibroblasts from long-lived breeds have greater respiration capacity (consume more oxygen with maximized ETC flux) and produce less ROS (Fig. 3b, f). This suggests that longer-lived breeds have less electrons escaping their ETC to form ROS. Regardless of nutrient conditions, long-lived breed fibroblasts have greater respiration capacity (Figs. 3b and 4e), further supporting lower electron escape in their ETCs.
The innate variation in uncoupling and electron escape has functional consequences on stress tolerance. Specifically, short-lived breeds display lower respiration, more ROS, and greater cell death under nutrient and oxidative stress (Fig. 4). These properties are also regulated by the distribution and motility of mitochondria in a cell, which is dependent on the cytoskeleton. The fibroblasts from the shortest-lived breed in our study had the stiffest actin-cytoskeleton, while the longest-lived breed had the most flexible (Fig. S3), which potentially contributes to variation in mitochondrial function and stress tolerance. Furthermore, mitochondrial stress and ROS hasten telomere attrition and its associated dysfunctions (Sanderson and Simon 2017). Inferior mitochondrial stress response and ROS management in short-lived breeds may explain their reduced telomere length in comparison to long-lived breeds (Fick et al. 2012).
In addition to dampened ROS, an alternative or parallel mechanism that could promote cellular function downstream of uncoupling would be caloric restriction (CR)–like metabolism. A CR-like metabolic state induces quality control processes, suppresses ROS and oxidative damage, and promotes β-oxidation. Long-lived breeds may have lower concentrations of acylcarnitines due to higher fatty acid oxidation (Fig. 5b). This would explain how long-lived breeds maintain their uncoupled energetics, which requires greater substrate oxidation to supply electrons to the ETC (Fig. 6). CR suppresses glycolysis, and interestingly, a recent study observed lower rates of glycolysis in cells from small dogs in a Seahorse assay (Jimenez et al. 2018). We did not observe glycolysis differences in our Seahorse assay and did not find significant lactate differences in our mass spectrometry data (Fig. 5). We believe that the discrepancy between the findings of Jimenez et al. and our study is the result of specific methodological differences. Greater reliance on glycolysis in larger dogs would predispose them to cancer and other diseases. Furthermore, decreased uncoupling combined with lower β-oxidation with accumulation of acylcarnitines promotes inflammation, oxidative stress, cell death, and disease (McCoin et al. 2015). Together, these metabolic properties reduce cellular stress tolerance and may contribute to the suppressed longevity of large dogs.
Mass spectrometry revealed unique bioenergetic-metabolite signatures that persist in primary culture and distinguish breeds, with acylcarnitines providing the best resolution (Fig. 2b). Studies on plasma metabolites and lipids in dogs also showed strong clustering by breed, with differences in long-chain fatty acids being a significant factor (Lloyd et al. 2016; Lloyd et al. 2017). We observed enrichment of acylcarnitines in the short-lived Boxers versus Chihuahuas, with Labradors having intermediate concentrations (Fig. 5b). Long-chain acylcarnitines (C14 and above) normally undergo β-oxidation. The accumulation of these long-chain acylcarnitines suggests that this process is partially blocked. Such a blockade could result in diminished respiration. Indeed, we observed lower respiration in short-lived breeds, including Labrador and Boxer cells (Figs. 3b and 4d). The Boxers also had higher concentrations of many amino acids, supporting enrichment in biosynthesis (Fig. 5a) and agreeing with results from a study which examined in vivo plasma metabolites in dogs (Middleton et al. 2017).Overall, these data suggest that longer-lived breeds, like the Chihuahua, may utilize amino acid catabolism and β-oxidation to fuel their uncoupled energetic demands, whereas short-lived breeds, like the Boxer, preferentially shuttle metabolites into biosynthetic pathways (Fig. 6). Indeed, mitochondrial uncoupling is associated with increased β-oxidation (Sukumar et al. 2016).
Smaller breeds have a higher metabolic rate and utilize more energy per gram of tissue than larger breeds (Speakman et al. 2003; National Research Council (U.S.). Ad Hoc Committee on Dog and Cat Nutrition 2006). Small dogs need to expend more energy via uncoupling and adaptive thermogenesis to maintain homeostatic temperature. The identification of FGF5 and RSPO2, genes that regulate coat phenotypes (Housley and Venta 2006; Cadieu et al. 2009), to associate with breed longevity further supports a connection between thermoregulation and canine longevity (Table 1, Fig. 1d), although esthetic selection on coat length from humans cannot be ruled out. If dogs possess strategies for thermoregulation at the organismal level (through coat length) that would support that cellular mechanisms, such as uncoupling, are present and or evolved. Our data suggest that smaller long-lived dogs have a higher metabolic rate to fuel uncoupled energetics and thermogenesis (Fig. 6). Moreover, naturally higher or induced uncoupling increases mammalian lifespan with elevated respiration and suppressed oxidative damage (Speakman et al. 2004; Caldeira da Silva et al. 2008). Furthermore, our data show a superior response to bioenergetic stress in long-lived breed cells. The increase in uncoupling and superior stress response would confer metabolic and survival advantages for cells and tissues (Fig. 6). This would suppress the functional decline of tissues, staving off dysfunction and disease, and help to explain why small dogs have greater longevity. Overall, our data agree with the “uncoupling to survive” theory of aging, which posits that greater uncoupling promotes longevity (Brand 2000). Interestingly, the “rate of living” theory has garnered recent support in human CR trials (Redman et al. 2018). It may be that increased metabolism is maladaptive to longevity but is beneficial when caused by and paired with uncoupled energetics.
In the future, it will be important to investigate which pathways are responsible for bioenergetic variation between long- and short-lived breeds and how they are regulated. Are cells from small long-lived breeds more uncoupled because of a thermodynamic response (that is retained in vitro epigenetically), or because they are hardwired at the DNA level? IGF-1 may not be responsible for variation in cellular metabolism across breeds, as we observed no differences in its expression (Fig. S4). Additionally, examination of passive versus regulated mitochondrial uncoupling may be informative, as uncoupling proteins associate with human longevity (Rose et al. 2011). Modeling these bioenergetic properties in human populations with variable lifespans is similarly worth pursuing and provides a novel avenue for aging research.
Experimental procedures
Genome-wide association
A GWAS of adult life expectancy was run in GEMMA v. 0.94 using a linear mixed model as described in (Hayward et al. 2016). Four thousand one hundred sixty-nine individuals from 110 breeds were included in the analysis. Dogs were genotyped on a semicustom Illumina SNP array (see Shannon et al. 2015 for detailed methods). Adjusted breed-average life expectancies (Adj. e(2)) were obtained from (Yordy et al., Body size, inbreeding, and lifespan in domestic dogs, in preparation) and are based on data from the Veterinary Medical Database and other sources. Genotype data were pruned in PLINK to exclude SNPs with MAF < 0.05, leaving about 150,000 loci in the analysis. A kinship matrix was calculated in GEMMA, and this matrix was included as a random effect in the association test. Top associations are given in Table 1, more information in Supplemental File 1.
Coat-length classifications for breeds were sourced from Cadieu et al. (2009). Sweeping by weight five breeds at a time and calculating the fraction of short, medium, and long hair within that sweep versus the respective total present, these fractions were then plotted and a three-order polynomial curve was generated with area under the curve (see Supplementary File 2, “Breed coat length distributions” tab).
TMRE and MitoTracker Green
The fibroblasts were plated at a concentration of 25 K in 500 ul media in a 24-well plate and incubated for 18 h. The cells were then stained with 200 nM of either MitoTracker Green (Molecular Probes) or TMRE (Abcam) for 1 h or 30 min at 37 °C, respectively. The cells were then rinsed and immediately imaged with a ZOE Fluorescent Cell Imager (Bio-Rad). Ten representative images were taken for every cell line and dye. The average intensity per image was computed and then averaged across the representative images; this was done on three occasions. Average MitoTracker Green intensity was used to normalize TMRE intensity to the total mitochondrial mass for each breed, respectively.
Mass spectrometry (see Supplementary File 2 for raw data)
Fibroblasts were expanded to fill 6 × 100-mm plates and washed with ice cold PBS, scraped from plates in ice cold PBS, and then spun down in a swinging bucket centrifuge at 500 g for 5 min to pellet cells at 4 °C. Cell pellets were then lysed in 300 μL 0.6% formic acid; 30 μL was then removed for protein quantification, followed by addition of 270 μL acetonitrile to give a final concentration of 0.3% formic acid and 50% acetonitrile. Amino acids, acylcarnitines, and organic acids were analyzed using a stable isotope dilution technique. Amino acids and acylcarnitine measurements were made by flow injection tandem mass spectrometry using sample preparation methods described previously (An et al. 2004; Ferrara et al. 2008). The data were acquired using a Waters TQD mass spectrometer equipped with AcquityTM UPLC system and controlled by MassLynx 4.1 operating system (Waters). Organic acids were quantified using methods described previously (Jensen et al. 2006) employing Trace Ultra GC coupled to ISQ MS operating under Xcalibur 2.2 (Thermo Fisher Scientific).
Metabolite comparisons between breeds were corrected for multiple testing using the Benjamini and Hochberg method.
Principal component analysis and hierarchical clustering of metabolites
PCA was performed using the web-based tool ClustVis (Metsalu and Vilo 2015). PCA plots and heatmaps were exported and modified with Adobe Illustrator.
Seahorse analyzer
We followed the protocol and used the reagents from the Agilent XF Cell Energy Phenotype kit (103325-100) to measure oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), which are readouts of mitochondrial respiration and glycolysis, respectively. Fibroblasts were plated the night prior to assay at 45-K cells in normal cell culture media (DMEM, 10% FBS) in 24-well XF Seahorse plates. One hour prior to assay, media was washed and changed to serum free XF assay media supplemented with 2 mM glutamine and 10 mM glucose (adjusted to a pH of 7.4). Glucose was not added to the assay media in the case of Fig. 4 d and e. Cells were then incubated in a non-CO2 chamber for 1 h before being analyzed in an XF-24 Extracellular Flux Analyzer. FCCP and oligomycin were diluted in XF assay media and used at working concentrations of 1 μm for simultaneous co-treatment.
ROS analysis
Fibroblasts were plated at 50 K in 24-well plates in normal media the day prior to experiment. Media were then removed from wells, washed with warm PBS, and then warm PBS containing 10 μm H2DCFDA (D399, Thermo Fisher Scientific) was added. Cells were incubated for 30 min at 37°, after which, H2DCFDA was removed and trypsin was added for 2 min. Cells were collected, centrifuged, and washed with ice cold PBS. Cells were centrifuged again and resuspended in 500 uL ice cold PBS. We then proceeded to measure H2DCFDA intensity by the FITC channel on a Beckton-Dickinson LSR II. A no stain control for every breed was used for every experiment that was subtracted from the respective stained replicates.
Stress tolerance analysis
Fibroblasts were plated the night prior to stress at 75 K in normal media in 12-well plates. Sixteen hours later, cells were treated with either 20 μM rotenone or replaced with media lacking glucose and serum. Stress conditions lasted 72 h before being collected (including adhered and suspended cells) from wells using Trypsin digestion. Cells were washed in PBS and then suspended in 100 μL of 1× binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4) with 5 μL of annexin-V conjugate and PI. After a 15-min incubation, another 400 μL of binding buffer was added and then cells were analyzed using a 3 laser/8 color Beckton-Dickinson LSR II. A cell was considered to survive if it was negative for both annexin-V and PI stains.
Statistics
Pearson correlation coefficient (R) and sample size (n) were used to calculate p values for all correlations between experimental results and breed longevity. Spearman correlation coefficients were not significantly different from Pearson in our datasets. When appropriate, we grouped the breeds into long- and short-lived breeds and compared results by the two-tailed t test or single-factor ANOVA. Statistics were run with Microsoft Excel or R statistical programming. For breed lifespan values, we took data from the American Kennel Club: http://akc.com/dog-breeds/ and data available from PetcareRX: https://www.petcarerx.com/article/lifespan-of-a-dog-a-dog-years-chart-by-breed/1223 and averaged the two data sets together.
Primary canine fibroblasts
Fibroblasts acquired from the Miller lab were procured as described previously (Harper et al. 2011). Fibroblasts from the Cornell Veterinary Bank were prepared as follows (briefly): Tissue was minced in MEM and cultured in 15% FBS with 1% l-glutamine and antibiotics in T25 flasks. Fibroblasts incubated in atmospheric oxygen with 5% carbon dioxide at 37°. Once sub-confluent, they were collected via trypsin and centrifugation and frozen (− 70 O/N to liquid nitrogen) in 10% DMSO and 18% FBS media).
In the Libert lab, after thawing and reanimation, fibroblasts were cultured in DMEM (Corning, 10-017) supplemented with 10% FBS on culture plates treated with 0.1% gelatin (including for experiments). To prevent mycoplasma, bacterial and fungal contamination streptomycin, penicillin, and amphotericin b were used in manufacturer-specified concentrations. Fibroblasts were incubated in atmospheric oxygen with 5% carbon dioxide at 37° (see Supplemental File 1 for all canine donors used in this study). Please note we attempted to match fibroblast donor age across breeds, but availability was a limiting factor. Interestingly, donor age within breed appears irrelevant for most metabolite concentrations (see lactate by age graph in Supplemental File 2). While in vitro age (passage) of fibroblasts plays a more apparent role in the rigidity observed by AFM (Fig. S3).
Electronic supplementary material
Supplemental Figs. 1–4 & Methods. Fig. S1 Acylcarnitine profiles have strong predictive power for canine breed. Fig. S2 PCA plot for all metabolites. Fig. S3 Fibroblasts from long-lived dog breeds may have a more flexible cytoskeleton than those from short-lived. Fig. S4 IGF-1 expression in primary fibroblasts is not different across breeds. (DOCX 1285 kb)
Mass spectrometry raw data (XLSX 4537 kb)
Acknowledgements
J.W.N and J.D.Y thank Siri who is a good dog (12 out of 10). We also thank the NVIDIA Corporation for their donation of the Titan Xp GPU used in this work. Lastly, we thank the Cornell Veterinary Bank for primary canine fibroblast donations, as well as William Kohler and Melissa Han of the Miller lab for fibroblast preparations.
Author contributions
Conceptualization, J.W.N. and S.L. Formal analysis, J.W.N. Investigation, J.W.N., M.P., T.M.R., S.V.T., A.B.F., J.D.Y., T.K.Y., D.V., M.W., F.K.H., and O.R.I. Writing—original draft—J.W.N. Writing—review and editing—J.W.N., T.M.R., J.D.Y., T.K.Y., I.S., M.D.H., and S.L. Supervision, S.L. Funding acquisition, J.W.N. and S.L.
Funding information
S.L. and J.W.N. were in part supported by a grant from the American Federation for Aging Research (AFAR, grant no. 2015-030). S.L. received seed grant funding from the Cornell University Center for Vertebrate Genomics. J.W.N. was supported by a Glenn/AFAR Scholarship for Research in the Biology of Aging. R.A.M was supported by the National Institute of Health grant U19-AG023122. I.S. group was supported by the National Science Foundation grant CMMI 1435655.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Justin W. Nicholatos, Phone: (508) 494-1538, Email: jwnichol1@gmail.com
Sergiy Libert, Phone: (281) 435-5687, Email: libert@calicolabs.com.
References
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Baffy G, Derdak Z, Robson SC. Mitochondrial recoupling: a novel therapeutic strategy for cancer? Br J Cancer. 2011;105:469–474. doi: 10.1038/bjc.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botigue LR, Song S, Scheu A, Gopalan S, Pendleton AL, Oetjens M, Taravella AM, Seregely T, Zeeb-Lanz A, Arbogast RM, Bobo D, Daly K, Unterlander M, Burger J, Kidd JM, Veeramah KR. Ancient European dog genomes reveal continuity since the Early Neolithic. Nat Commun. 2017;8:16082. doi: 10.1038/ncomms16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35:811–820. doi: 10.1016/S0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A, Wong A, Mosher DS, Elkahloun AG, Spady TC, Andre C, Lark KG, Cargill M, Bustamante CD, Wayne RK, Ostrander EA. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira da Silva CC, Cerqueira FM, Barbosa LF, Medeiros MH, Kowaltowski AJ. Mild mitochondrial uncoupling in mice affects energy metabolism, redox balance and longevity. Aging Cell. 2008;7:552–560. doi: 10.1111/j.1474-9726.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- Cho SY, Seo DB, Kim WG, Lee SJ. Mild mitochondrial uncoupling prevents premature senescence in human dermal fibroblasts. J Invest Dermatol. 2014;134:540–543. doi: 10.1038/jid.2013.352. [DOI] [PubMed] [Google Scholar]
- Domanskyi S, Nicholatos JW, Schilling JE, Privman V, Libert S. SIRT6 knockout cells resist apoptosis initiation but not progression: a computational method to evaluate the progression of apoptosis. Apoptosis. 2017;22:1336–1343. doi: 10.1007/s10495-017-1412-0. [DOI] [PubMed] [Google Scholar]
- Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, Yandell BS, Newgard CB, Attie AD. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick LJ, Fick GH, Li Z, Cao E, Bao B, Heffelfinger D, Parker HG, Ostrander EA, Riabowol K. Telomere length correlates with life span of dog breeds. Cell Rep. 2012;2:1530–1536. doi: 10.1016/j.celrep.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Greer KA, Canterberry SC, Murphy KE. Statistical analysis regarding the effects of height and weight on life span of the domestic dog. Res Vet Sci. 2007;82:208–214. doi: 10.1016/j.rvsc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Greer KA, Hughes LM, Masternak MM. Connecting serum IGF-1, body size, and age in the domestic dog. Age (Dordr) 2011;33:475–483. doi: 10.1007/s11357-010-9182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Wang M, Galecki AT, Ro J, Williams JB, Miller RA. Fibroblasts from long-lived bird species are resistant to multiple forms of stress. J Exp Biol. 2011;214:1902–1910. doi: 10.1242/jeb.054643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JJ, Castelhano MG, Oliveira KC, Corey E, Balkman C, Baxter TL, Casal ML, Center SA, Fang M, Garrison SJ, Kalla SE, Korniliev P, Kotlikoff MI, Moise NS, Shannon LM, Simpson KW, Sutter NB, Todhunter RJ, Boyko AR. Complex disease and phenotype mapping in the domestic dog. Nat Commun. 2016;7:10460. doi: 10.1038/ncomms10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich A, Reyer A, Ohde D, Schindler N, Brenmoehl J, Spitschak M, Langhammer M, Tuchscherer A, Wirthgen E, Renner-Muller I, Wanke R, Metzger F, Bielohuby M, Wolf E. Dissociation of somatic growth, time of sexual maturity, and life expectancy by overexpression of an RGD-deficient IGFBP-2 variant in female transgenic mice. Aging Cell. 2016;15:111–117. doi: 10.1111/acel.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JM, Creevy KE, Franks A, O'Neill DG, Promislow DEL (2018) The companion dog as a model for human aging and mortality. Aging Cell 17:e12737 [DOI] [PMC free article] [PubMed]
- Housley DJE, Venta PJ. The long and the short of it: evidence that FGF5 is a major determinant of canine ‘hair’-itability. Anim Genet. 2006;37:309–315. doi: 10.1111/j.1365-2052.2006.01448.x. [DOI] [PubMed] [Google Scholar]
- Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB. Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J Biol Chem. 2006;281:22342–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- Jimenez AG, Winward J, Beattie U, Cipolli W (2018) Cellular metabolism and oxidative stress as a possible determinant for longevity in small breed and large breed dogs. PLoS One 13:e0195832 [DOI] [PMC free article] [PubMed]
- Kaeberlein M, Creevy KE, Promislow DE. The dog aging project: translational geroscience in companion animals. Mamm Genome. 2016;27:279–288. doi: 10.1007/s00335-016-9638-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus C, Pavard S, Promislow DE. The size-life span trade-off decomposed: why large dogs die young. Am Nat. 2013;181:492–505. doi: 10.1086/669665. [DOI] [PubMed] [Google Scholar]
- Lloyd AJ, Beckmann M, Tailliart K, Brown WY, Draper J, Allaway D. Characterisation of the main drivers of intra- and inter- breed variability in the plasma metabolome of dogs. Metabolomics. 2016;12:72. doi: 10.1007/s11306-016-0997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AJ, Beckmann M, Wilson T, Tailliart K, Allaway D, Draper J (2017) Ultra high performance liquid chromatography-high resolution mass spectrometry plasma lipidomics can distinguish between canine breeds despite uncontrolled environmental variability and non-standardized diets. Metabolomics 13:15 [DOI] [PMC free article] [PubMed]
- McCoin CS, Knotts TA, Ono-Moore KD, Oort PJ, Adams SH. Long-chain acylcarnitines activate cell stress and myokine release in C2C12 myotubes: calcium-dependent and -independent effects. Am J Physiol Endocrinol Metab. 2015;308:E990–E1000. doi: 10.1152/ajpendo.00602.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton RP, Lacroix S, Scott-Boyer MP, Dordevic N, Kennedy AD, Slusky AR, Carayol J, Petzinger-Germain C, Beloshapka A, Kaput J. Metabolic differences between dogs of different body sizes. J Nutr Metab. 2017;2017:1–11. doi: 10.1155/2017/4535710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookerjee SA, Divakaruni AS, Jastroch M, Brand MD. Mitochondrial uncoupling and lifespan. Mech Ageing Dev. 2010;131:463–472. doi: 10.1016/j.mad.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Salmon A, Miller RA. Multiplex stress resistance in cells from long-lived dwarf mice. FASEB J. 2003;17:1565–1566. doi: 10.1096/fj.02-1092fje. [DOI] [PubMed] [Google Scholar]
- National Research Council (U.S.). Ad Hoc Committee on Dog and Cat Nutrition . Nutrient requirements of dogs and cats. Washington, D.C: National Academies Press; 2006. [Google Scholar]
- Redman LM, Smith SR, Burton JH, Martin CK, Il’yasova D, Ravussin E. Metabolic slowing and reduced oxidative damage with sustained caloric restriction support the rate of living and oxidative damage theories of aging. Cell Metab. 2018;27:805–815 e804. doi: 10.1016/j.cmet.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Crocco P, De Rango F, Montesanto A, Passarino G (2011) Further support to the uncoupling-to-survive theory: the genetic variation of human UCP genes is associated with longevity. PLoS One 6:e29650 [DOI] [PMC free article] [PubMed]
- Sanderson SL, Simon AK. In aged primary T cells, mitochondrial stress contributes to telomere attrition measured by a novel imaging flow cytometry assay. Aging Cell. 2017;16:1234–1243. doi: 10.1111/acel.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon LM, Boyko RH, Castelhano M, Corey E, Hayward JJ, McLean C, White ME, Abi Said M, Anita BA, Bondjengo NI, Calero J, Galov A, Hedimbi M, Imam B, Khalap R, Lally D, Masta A, Oliveira KC, Perez L, Randall J, Tam NM, Trujillo-Cornejo FJ, Valeriano C, Sutter NB, Todhunter RJ, Bustamante CD, Boyko AR. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc Natl Acad Sci U S A. 2015;112:13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, van Acker A, Harper EJ. Age-related changes in the metabolism and body composition of three dog breeds and their relationship to life expectancy. Aging Cell. 2003;2:265–275. doi: 10.1046/j.1474-9728.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Speakman JR, Talbot DA, Selman C, Snart S, McLaren JS, Redman P, Krol E, Jackson DM, Johnson MS, Brand MD. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Mehta GU, Patel SJ, Roychoudhuri R, Crompton JG, Klebanoff CA, Ji Y, Li P, Yu Z, Whitehill GD, Clever D, Eil RL, Palmer DC, Mitra S, Rao M, Keyvanfar K, Schrump DS, Wang E, Marincola FM, Gattinoni L, Leonard WJ, Muranski P, Finkel T, Restifo NP. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 2016;23:63–76. doi: 10.1016/j.cmet.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao KY, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Miller RA. Fibroblasts from long-lived mutant mice exhibit increased autophagy and lower TOR activity after nutrient deprivation or oxidative stress. Aging Cell. 2012;11:668–674. doi: 10.1111/j.1474-9726.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, Zhai W, Yang HC, Fan RX, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng LG, Poyarkov AD, Poyarkov NA, Jr, Tang SS, Zhao WM, Gao Y, Lv XM, Irwin DM, Savolainen P, Wu CI, Zhang YP. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figs. 1–4 & Methods. Fig. S1 Acylcarnitine profiles have strong predictive power for canine breed. Fig. S2 PCA plot for all metabolites. Fig. S3 Fibroblasts from long-lived dog breeds may have a more flexible cytoskeleton than those from short-lived. Fig. S4 IGF-1 expression in primary fibroblasts is not different across breeds. (DOCX 1285 kb)
Mass spectrometry raw data (XLSX 4537 kb)