SUMMARY
Systematic measurement biases make normalization an essential step in single-cell RNA sequencing (scRNA-seq) analysis. There may be multiple competing considerations behind the assessment of normalization performance, of which some may be study specific. We have developed “scone”— a flexible framework for assessing performance based on a comprehensive panel of data-driven metrics. Through graphical summaries and quantitative reports, “scone” summarizes trade-offs and ranks large numbers of normalization methods by panel performance. The method is implemented in the open-source Bioconductor R software package SCONE. We show that top-performing normalization methods lead to better agreement with independent validation data for a collection of scRNA-seq datasets. SCONE can be downloaded at http://bioconductor.org/packages/scone/.
Graphical Abstract

In Brief
We have developed an approach for exploratory analysis and normalization of scRNA-seq data that enables execution of a wide array of normalization procedures and provides principled assessment of their performance based on a comprehensive set of data-driven performance metrics.
INTRODUCTION
Normalization is a common preprocessing step in the analysis of -omic data, such as high-throughput transcriptome microarray and sequencing (RNA sequencing [RNA-seq]) data. The goal of normalization is to account for observed differences in measurements between samples and/or features (e.g., genes) resulting from technical artifacts or unwanted biological effects (e.g., batch effects) rather than biological effects of interest. Accordingly, two types of normalization are often considered: between-sample and within-sample. This article focuses on the former. To derive gene expression measures from single-cell RNA sequencing (scRNA-seq) data and subsequently compare these measures between cells, analysts must normalize read counts (or other expression measures) to adjust for obvious differences in sequencing depths. When there are other significant biases in expression quantification, it may be necessary to further adjust expression measures for more complex unwanted technical factors related to sample and library preparation.
As previously discussed (Bacher and Kendziorski, 2016; Vallejos et al., 2017), normalization of scRNA-seq data is often accomplished via methods developed for bulk RNA-seq or even micro-array data. These methods tend to neglect prominent features of scRNA-seq data such as “zero inflation,” i.e., an artifactual excess of zero read counts observed in some single-cell protocols (e.g., SMART-seq) (Finak et al., 2015; Kharchenko et al., 2014); transcriptome-wide nuisance effects (e.g., batch) comparable in magnitude to the biological effects of interest (Hicks et al., 2018); and uneven sample quality, e.g., in terms of alignment rates and nucleotide composition (Ilicic et al., 2016). In particular, widely used global-scaling methods, such as reads per million (RPM) (Mortazavi et al., 2008), trimmed mean of M values (TMM) (Robinson and Oshlack, 2010), and DESeq (Anders and Huber, 2010), are not well suited to handle large or complex batch effects and may be biased by low counts and zero inflation (Vallejos et al., 2017). Other more flexible methods, such as remove unwanted variation (RUV) (Gagnon-Bartsch and Speed, 2012; Risso et al., 2014) and surrogate variable analysis (SVA) (Leek and Storey, 2007; Leek, 2014), depend on tuning parameters (e.g., the number of unknown factors of unwanted variation).
A handful of normalization methods specifically designed for scRNA-seq data have been proposed. These include scaling methods (Lun et al., 2016a, 2016b; Qiu et al., 2017), regression-based methods for known nuisance factors (Buettner et al., 2015; Bacher et al., 2017), and methods that rely on spike-in sequences from the External RNA Controls Consortium (ERCC) (Ding et al., 2015; Vallejos et al., 2015). While these methods address some of the problems affecting bulk normalization methods, each suffers from limitations with respect to their applicability across diverse study designs and experimental protocols. Global-scaling methods define a single normalization factor per cell and thus are unable to account for complex batch effects. Explicit regression on known nuisance factors (e.g., batch, number of reads in a library) may miss unknown, yet unwanted variation, which may still confound the data (Risso et al., 2014). Unsupervised normalization methods that regress gene expression measures on unknown unwanted factors may perform poorly with default parameters (e.g., number of factors adjusted for) and require tuning, while ERCC-based methods suffer from differences between endogenous and spiked-in transcripts (Risso et al., 2014; Vallejos et al., 2017). Protocols using unique molecular identifiers (UMI) still require normalization; while UMIs remove amplification biases, they are often sensitive to sequencing depth and differences in capture efficiency before reverse transcription (Vallejos et al., 2017).
Because of the prevalence of confounding in single-cell experiments, the lack of a uniformly optimal normalization across datasets, and the ambiguity in tuning parameter guidelines for commonly used normalization methods, we recommend the inspection and evaluation of many approaches and the use of multiple data-driven metrics to guide the selection of suitable approaches for a given dataset. We have developed the “scone” framework for implementing and assessing the performance of a range of normalization procedures, each consisting of defined normalization steps, such as scaling and supervised or unsupervised regression-based adjustments. “scone” evaluates the performance of each procedure and ranks them by aggregating over a panel of performance metrics that consider different aspects of a desired normalization outcome, including both removal of unwanted variation and preservation of wanted variation.
We demonstrate that the “scone” methodology is generally applicable to different scRNA-seq protocols and study designs. The modularity of the Bioconductor R software package SCONE allows researchers to tune and compare a set of default normalizations as well as to include user-defined methods, providing a useful framework for both practitioners and method developers.
RESULTS
scRNA-Seq Data Are Affected by Batch Effects and Other Unwanted Variation
We briefly illustrate the difficulties of normalizing scRNA-seq data with a published SMART-seq dataset, processed on Fluidigm C1 (Gaublomme et al., 2015). We focus on a subset of 420 mouse Th17 T cells harvested after in vitro differentiation of CD4+ naive T cells under 48 h of pathogenic (IL-1β+IL-6+IL-23) or non-pathogenic (TGF-β1+IL-6) conditioning. Prior to conditioning, these cells had been extracted from two strains of mice: wild-type (WT) B6 and transgenic B6 mice with an IL17a GFP reporter. We have aligned the publicly available reads to the mouse genome, counting over RefSeq gene intervals to generate a cells-by-genes count matrix. After sample and gene filtering, 337 libraries and 7,590 genes are retained for normalization and downstream analysis, preserving ~80% of all reads and cells (Figure S1B; see STAR Methods).
Even after scaling the gene-level read counts by total counts (TC), mouse-specific effects are prominently featured in principal-component analysis (PCA; Figure 1A). In the space defined by the first two “principal components” (PC), the distances between cells from mouse 7 and mouse 8 are larger than the distances between pathogenic and non-pathogenic cells collected from the same mouse. Because of the partially confounded design, these mouse effects may result from multiple sources, including (1) true biological differences between mice or (2) mouse-specific technical biases. The study design prevents us from teasing apart these two effects, but we can account for technical contributions to the read counts by examining the association of the expression PCs with RNA-seq library quality control (QC) metrics (Figure 1B; see Tables S1 and S2; STAR Methods).
Figure 1. Exploratory Data Analysis of Mouse Th17 Dataset (Gaublomme et al., 2015) and “scone” Workflow.
(A)Principal-component analysis (PCA) of the log-transformed, total-count-normalized (TC) read count data for all genes and cells passing quality filtering (see STAR Methods). Cells are color-coded by biological condition; shape represents the donor mouse (batch). For two of the three conditions, samples were extracted from only one mouse (IL1B_IL6_IL23–48h-IL-17A/GFP+ and TGFB1_IL6–48h-IL-17A/GFP+ from mice 7 and 8), while samples from the third condition (TGFB1_IL6–48h) came from two distinct mice (mice 5 and 6). Cells cluster by both biological condition and batch, the latter representing unwanted variation.
(B) Absolute Spearman correlation coefficient between the first three principal components (PCs) of the expression measures (as computed in A) and a set of quality control (QC) measures (Table S1).
(C) Heatmap of pairwise Pearson correlation coefficients between QC measures.
(D) PCA of the QC measures for all cells in (A). PCs of QC measures are labeled “qPCs” to distinguish them from expression PCs. Single-cell QC profiles cluster by batch, representing aspects of batch covariation.
(E) Boxplot of the first qPC, stratified by both biological condition and batch. Note that there are different numbers of cells in each stratum.
(F) Schematic view of the “scone” workflow. The yellow box summarizes the five main steps of the “scone” workflow. (i) QC measures are obtained for each sample, using Picard tools (Table S1), Cell Ranger (Table S2), or other tools such as scater. (ii) (Optional) Sample-level QC measures are used to filter out low-quality samples. Subsequently, lowly expressed genes are identified and filtered out to reduce the impact of noisy features on downstream analysis. (iii) Data are normalized via many combinations of scaling procedures and regression-based procedures, modeling both known and unknown variation as indicated in the regression model diagram. (iv) Normalized data are evaluated and ranked according to a panel of eight performance metrics, spanning three categories: (1) Clustering properties (e.g., removing batch effects and preserving biological heterogeneity), (2) association with control genes and QC metrics (e.g., preserving association with positive controls and removing association with QC measures), and (3) global distributional properties (e.g., reducing global expression variability). (v) One or more highly ranked normalization procedures are analyzed in parallel, and downstream conclusions are compared.
The first three expression PCs exhibit large correlations with measures of genomic alignment rate, primer contamination, intronic alignment rate, and 5′ bias (see STAR Methods). The correlation structure between these QC measures reflects constraints on library quality in the study (Figure 1C). While some of these pairwise associations represent natural dependencies between similar QC measures (e.g., total number of reads and total number of aligned reads are positively correlated), others reflect mouse-specific technical biases in the study. Applying PCA to the matrix of QC measures, we can see how these metrics provide a candidate basis for representing batch (i.e., mouse) effects (Figure 1D). Inter-batch QC differences are relatively large as in Figure 1A, while intra-batch differences between pathogenic and non-pathogenic cells are noticeably smaller (Figure 1E). Mouse 6 libraries are technically similar to mouse 7 libraries, whereas cells from mice 5 and 8 are technically distinct; these relationships are similar to those observed in the PCA of gene expression measures, suggesting that the corresponding structure in the expression data is artifactual. It is surely possible that some of the observed associations between read counts and QC measures result from biological confounding rather than direct technical bias, i.e., a cell’s biological state may impact transcriptome integrity and sequencing viability (Vallejos et al., 2017; Wagner et al., 2016). However, unlike factors such as mouse of origin, there exist simple interpretations for correlations between quantified expression measures and library alignment statistics.
We have focused on a specific dataset in this section, but we note that many of these observations are not unique to this example and are general features of scRNA-seq. To highlight this, we have performed similar exploratory data analyses on a set of developing human cortical neurons assayed using a 2014 Fluidigm protocol (Pollen et al., 2014) and a set of human peripheral blood mononuclear cells (PBMC) assayed using the 10× chromium platform (Zheng et al., 2017) (Figures S2 and S3).
scone: An Exploratory Framework for the Implementation and Evaluation of scRNA-Seq Normalization
As illustrated in Figure 1, simple global scaling alone is insufficient for normalizing scRNA-seq data and more flexible and aggressive procedures aimed at removing unwanted variation (e.g., batch effects) may be generally beneficial. Here, we present a general framework for implementing and evaluating normalization procedures for scRNA-seq data (Figure 1F).
The full “scone” pipeline consists of two steps prior to normalization, namely (1) QC assessment and (2) optional sample and gene filtering, resulting in the selection of high-quality cell gene expression profiles.
Following these initial steps, “scone” uses a two-part normalization template to define an ensemble of normalization procedures: (1) scaling of the counts to account for between-sample differences in sequencing depth and other parameters of the read count distributions; (2) regression-based adjustment for known unwanted factors, such as processing batches, and unknown unwanted factors (Risso et al., 2014). Figure 1F displays some of the methods that can be employed in our normalization template. For instance, one can apply either TC scaling or more robust scaling procedures designed to reduce the effect of outliers (e.g., TMM [Robinson and Oshlack, 2010] or DESeq [Anders and Huber, 2010]). Additionally, confounding factors can be adjusted for by regressing scaled gene expression measures on quantities known to influence them (e.g., batches or the QC measures shown in Figure 1). Alternatively, unsupervised procedures can estimate hidden unwanted factors and regress them out of the data (e.g., RUV of Risso et al. [2014]).
The evaluation step of “scone” involves the comparison and ranking of all normalization procedures to identify sets of top-performing procedures. To achieve this, “scone” calculates a set of eight performance metrics, aimed at capturing different aspects of successfully normalized data. We classify these metrics into three broad categories: (1) clustering of samples according to factors of wanted and unwanted variation; (2) association of expression principal components with factors of wanted and unwanted variation; and (3) between-sample distributional properties of the expression measures (see STAR Methods). Overall, these metrics capture the trade-offs between the ability of a normalization procedure to remove unwanted variation, preserve biological variation of interest, and maintain minimum global technical expression variability. These trade-offs are rooted in the confounding commonly encountered in single-cell assays; “scone” provides a reproducible basis for managing these trade-offs via normalization. A simple ranking of normalization procedures can be obtained by averaging ranks based on the eight individual metrics, although the multidimensional aspect of normalization performance is lost in this one-dimensional representation. To account for this, we advocate the inspection of the full space of normalization performance measures, through specialized plots (Figures 2A–2C) and the “scone” report browser (Figure 7). Notably, in order to avoid an evaluation dominated by how normalization procedures handle zeros, we force to zero all values that are initially zeros as well as any negative values produced by a normalization procedure (see STAR Methods).
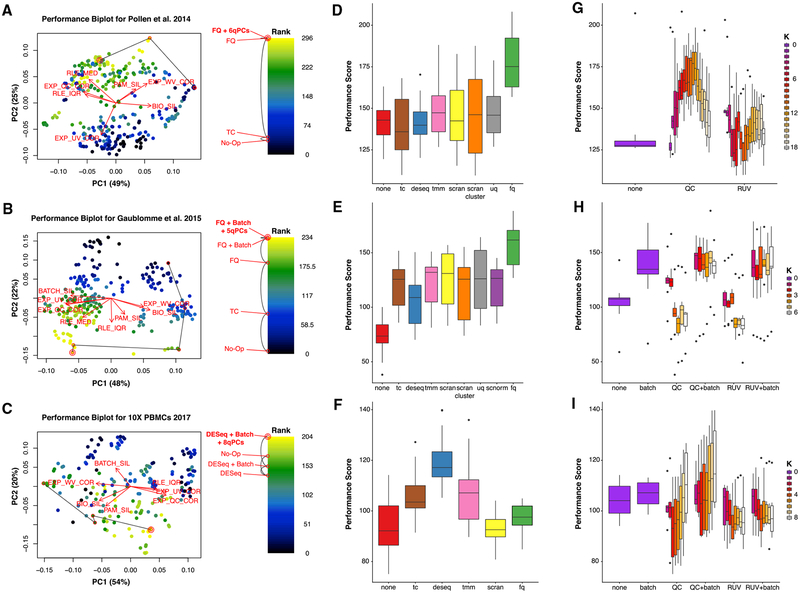
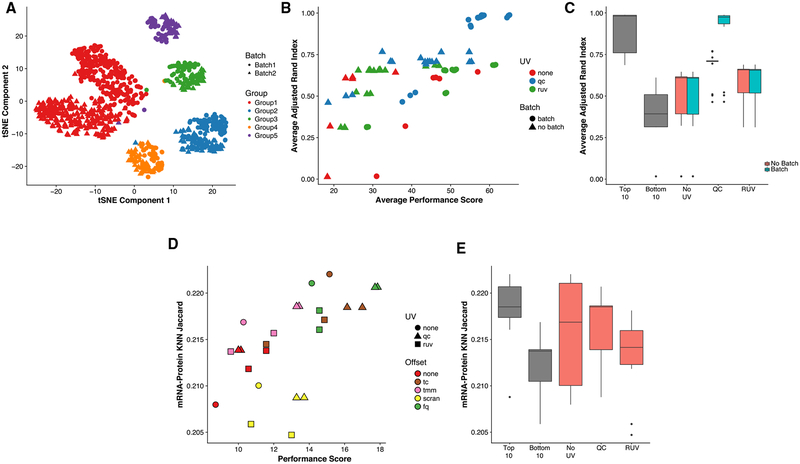
Figure 2. Normalization Performance Assessment for Three scRNA-Seq Datasets (Pollen et al., 2014; Gaublomme et al., 2015; Zheng et al., 2017).
(A–C) Biplot (Gabriel, 1971) showing the first two PCs of eight rank-transformed “scone” performance metrics, or fewer if some are undefined or invariant: preservation of biological clustering (“BIO_SIL”), batch effect removal (“BATCH_SIL”), cluster heterogeneity (“PAM_SIL”), preservation of association with positive control genes (“EXP_WV_COR”), removal of unwanted associations (negative control genes, “EXP_UV_COR,” or sample-level QC measures, “EXP_WC_COR”), and global distributional uniformity (“RLE_MED” and “RLE_IQR”). Each point corresponds to a normalization procedure and is color coded by the rank of the “scone” performance score (mean of eight “scone” performance metric ranks). The red arrows correspond to the PCA loadings for the eight performance metric ranks. The direction and length of a red arrow can be interpreted as a measure of how much each metric contributes to the first two PCs. Red circles mark the best normalization (w/double circle), no normalization, and other normalization procedures relating the two (see labels). Abbreviations are as follows: No-Op, no normalization; TC, total-count normalization; FQ, full-quantile normalization; DESeq, relative log-expression scaling (Anders and Huber, 2010); Batch, regression-based batch normalization; and kqPCs, regression-based adjustment for first k qPCs.
(D–F) Boxplot of “scone” performance score, stratified by scaling normalization method, for the three scRNA-seq datasets presented in the same order as in (A)–(C).
(G–I) Boxplot of “scone” performance score, stratified by regression-based normalization method (batch, QC, and RUV), for the three scRNA-seq datasets presented in the same order as in (A)–(C).
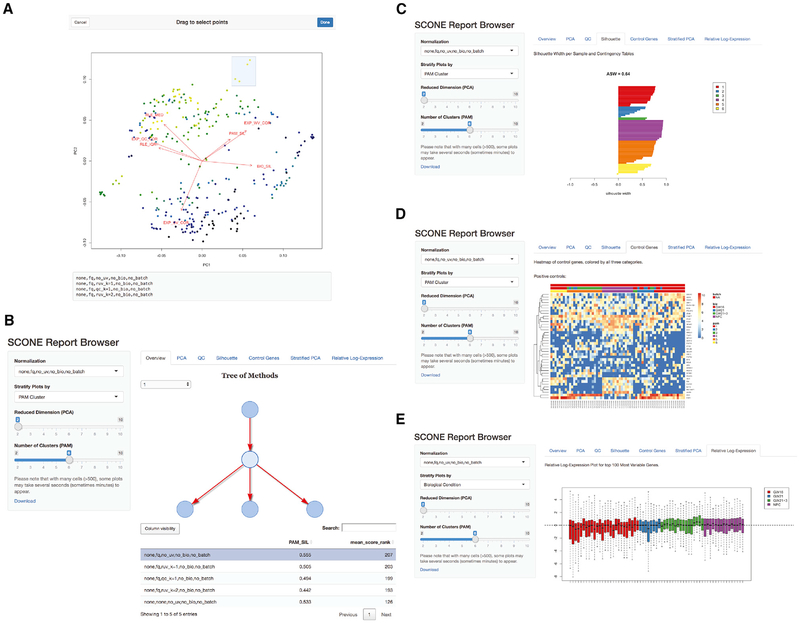
Figure 7. Report Browser Shiny Interface.
(A) Selecting normalization procedures of interest using the interactive biplot function biplot_interactive and its drag-and-drop window selection tool. This tool is useful for exploring performance clusters and selecting procedures that perform similarly across the eight performance metrics.
(B) Browsing normalized products. The SCONE Report Browser presents an interactive tree representation (top-right panel) of selected procedures. Procedures may be further selected via a sortable performance table (bottom-right panel) or a drop-down menu (side panel). The report will then produce plots corresponding to various analyses of the normalized data.
(C) Report Browser “Silhouette” tab: for the selected procedure, the silhouette width of each normalized sample is computed, grouping samples by biological condition, batch, or PAM clustering. The drop-down menu in the left bar allows the user to switch between the three categorical labels; the slider in the left panel allows the user to select the number of clusters for PAM, recomputed for each normalization procedure.
(D) Report Browser “Control Gene” tab: if the user provides positive and negative control genes, the gene-level expression measures for these genes are visualized using silhouette-sorted heatmaps, including annotations for biological condition, batch, and PAM clustering.
(E) Report Browser “Relative Log-Expression” tab: a boxplot of relative log-expression (RLE) measures is shown for the selected normalization procedure. Boxes (per-cell) are color coded by biological condition, batch, or PAM clustering (drop-down selection in the left panel). If the majority of genes are not expected to be differentially expressed, the RLE distributions of the samples should be similar and centered around zero.
Below we draw on evidence from multiple single-cell datasets, generated from various technological platforms, to show how no single normalization method is uniformly optimal: insights from the “scone” framework will highlight how performance depends on the design of the experiment and other characteristics of the data.
“scone” Removes Unwanted Variation while Preserving Biological Variation of Interest
A useful representation of the normalization performance landscape is the “biplot” (Gabriel, 1971), in which each point corresponds to a normalization procedure, and the dimensions of variation, represented by red arrows, correspond to “scone” performance metrics (Figures 2A–2C).
The “scone” biplot naturally represents trade-offs between these metrics, as illustrated in Figure 2 for three datasets. For the cortical neuron dataset of Pollen et al. (2014) (Figure 2A), there are two major bundles of red arrows, representing (1) preservation of biological heterogeneity and (2) distributional uniformity irrespective of library quality. The existence of this trade-off suggests that wanted variation is confounded with measurement artifacts. The top-ranked procedure according to “scone” involved full-quantile (FQ) normalization followed by adjustment for 6 QC PCs. Tracing a path from the performance coordinates of unnormalized data to those of the top-ranked normalization, we notice that FQ without adjustment also performs very well according to “scone,” occupying a middle position between these two trade-offs. Both procedures may reasonably be carried to downstream analysis, but the “scone” biplot highlights a tension in this decision. Despite the large number of normalization procedures represented in the analysis, the SCONE run time for this dataset is just over a minute with 1 CPU.
The “scone” biplot for the SMART-seq dataset of Gaublomme et al. (2015) demonstrates a more complex “fan” of trade-offs between batch effect removal and preservation of wanted variation (Figure 2B). Compared to no normalization, global-scaling and FQ normalization primarily improve distributional properties of the data, reducing the amount of global expression variability between samples (captured by the RLE metrics; see STAR Methods). Regression-based normalization, including batch regression and RUV, remove unwanted variation at the expense of wanted variation; the biplot can help identify those normalizations that balance the trade-off between removing too much biological variation and too little technical variation. Unlike the biplot for Pollen et al. (2014), the arrows corresponding to associations with QC metrics or negative control genes are closely aligned. However, the factors of unwanted variation computed from negative control genes (via RUVg) and the QC measures are not always correlated (Figure S4), suggesting that regression normalizations based on these factors are complementary approaches. Because of the greater number of cells in this dataset, the SCONE run time is longer, lasting 7 min (when SCnorm was precomputed).
We observe similar trade-offs between removal of technical variation and preservation of biological variation in the 10× Genomics dataset (Figure 2C). Unique to this case is the relatively good performance of no normalization: most normalization procedures scored worse than doing nothing. Nevertheless, “scone” identifies a procedure that balances the trade-offs between the different metrics, involving the DESeq scaling factor and regression-based adjustment for all 8 PCs of the QC matrix. Given the profound heterogeneity of PBMCs measured by 10×, we were interested in evaluating performance scores by stratifying cells according to both their batch and cell-type cluster. This approach avoids some of the confounding effects that muddle our interpretation of performance metrics (e.g., library quality differing between known biological clusters). The SCONE package supports this stratified evaluation (see STAR Methods), though we found that this option does not significantly change performance ranking for this dataset (Figure S5A). The SCONE run time for this large dataset is much longer than the other two datasets, at 60 h.
“scone’s” Normalization Performance Ranking Is Data Adaptive
Global-scaling normalization methods are ranked similarly for both C1 datasets (Figures 2D and 2E), underperforming the more aggressive FQ normalization. For the dataset of Pollen et al. (2014), globally scaled data do not show improved performance when compared to unscaled data (Figure 2B). Conversely, scaling by DESeq size factors outperforms other scaling normalizations as well as FQ normalization for the 10× Genomics PBMC dataset (Figure 2F). For all of these datasets, single-cell-specific methods, such as those implemented in the R packages scran and SCnorm, do not outperform methods developed for bulk RNA-seq.
The inclusion of a batch regression step in the normalization strategy is desirable for the SMART-seq dataset; procedures including QC or RUV factors without batch normalization perform poorly (Figure 2H). This result indicates that, for this study, preexisting batch classifications are better proxies of inter-batch effects than QC or RUVg factors, despite their problematic associations with biological condition. In contrast, QC-based regression normalization outperforms RUVg for the Pollen et al. (2014) dataset (Figure 2B), as well as in the 10× dataset when paired with batch adjustment (Figure 2I). Taken together, these observations suggest that there is no single normalization method that uniformly outperforms the others and that “scone” is able to identify appropriate normalization procedures in a data-dependent fashion.
“scone’s” Subsample-Based Normalization Performance Ranking Approximates Full-Data Ranking
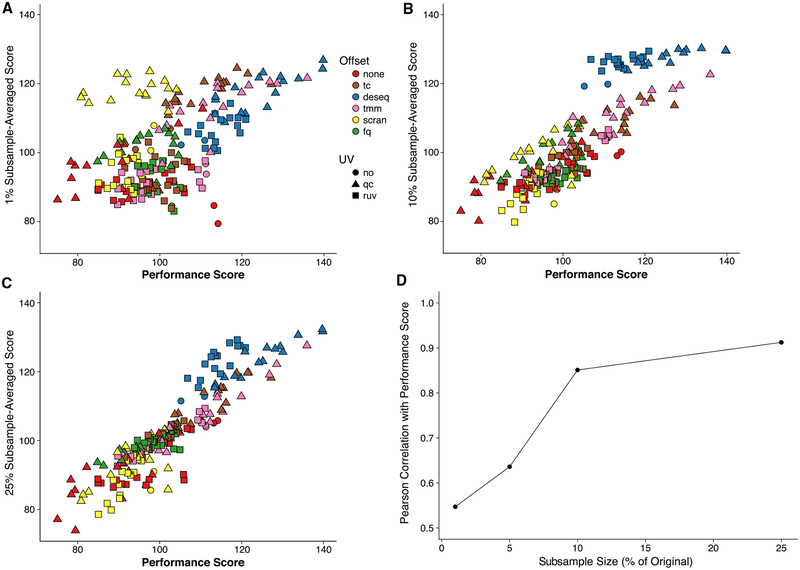
One potential drawback of “scone” is its computational complexity, implementing and ranking hundreds of normalization procedures per dataset. This can be especially problematic when applying “scone” to large datasets. In such cases, an efficient strategy is to use a random subset of the cells for the purpose of ranking normalizations, applying only the best-performing normalization procedure to the full dataset. For the 10× PBMC dataset, this subsampling strategy leads to a ranking that is highly consistent with the ranking based on the full data, as illustrated in Figure 3. As little as 10% of the cells is enough to yield more than 80% correlation with the full ranking (Figure 3D).
Figure 3. “scone” Analyses for Subsamples of 10× PBMC Dataset (Zheng et al., 2017).
(A–C) Average subsample performance score versus full-sample performance score. We randomly extracted 10 subsamples from the full dataset corresponding to a fixed percentage of the original sample size, applied “scone” independently for each subsample, and averaged the 10 performance scores to obtain a final performance score per procedure. Plots are shown for subsamples comprising (A) 1%, (B) 10%, and (C) 25% of the original sample.
(D) Pearson correlation coefficient between average subsample performance score and full-sample performance score for different subsample percentages. When sampling at least 10% of the cells, we observed correlations greater than 0.8 with scores for the full data.
External Measures of Differential Expression Validate “scone” Performance
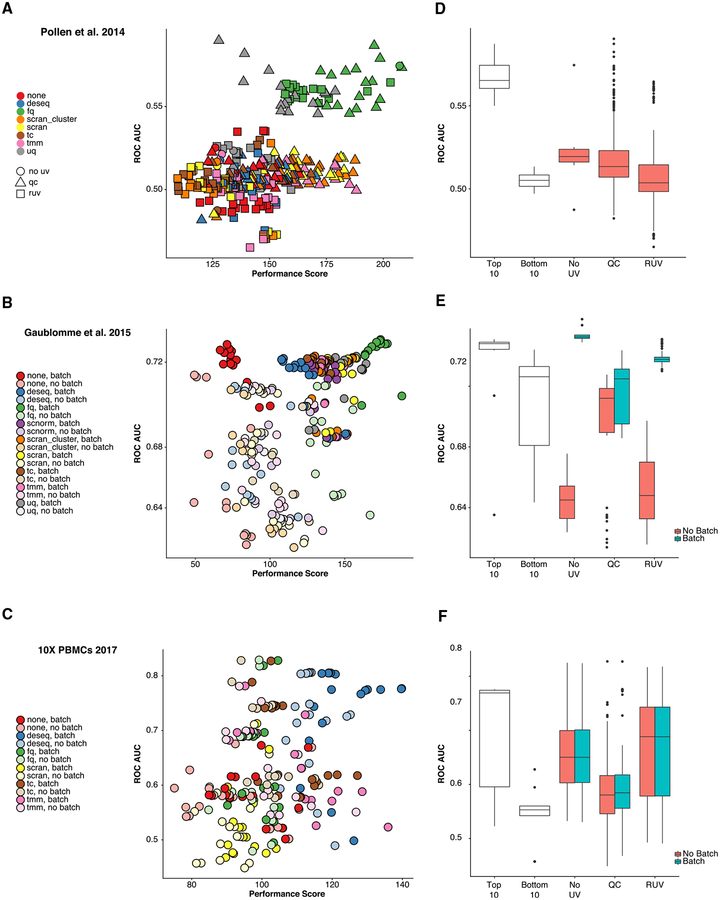
We validate “scone’s” performance assessment by relating normalized expression measures to controls derived from external differential expression studies. For the Pollen et al. (2014) dataset, we consider a set of positive (DE) and negative (non-DE) control genes for differential expression between CP+SP (cortical plate and subplate) and SZ+VZ (subventricular zone and ventricular zone) tissues from an independent bulk microarray dataset Miller et al. (2014). As positive controls, we select the 1,000 most significantly differentially expressed genes from that study, as ranked by limma p values (Ritchie et al., 2015) (all 1,000 also had p values < 0.01). As negative controls, we take the 1,000 least significantly differentially expressed genes. We assess our ability to discriminate these two sets of genes in a comparison of GW16 (gestational week 16) and GW21+3 (gestational week 21, cultured for 3 weeks) cells based on the normalized Pollen et al. (2014) dataset (using “limma” with “voom” weights [Law et al., 2014]), generating receiver operating characteristic (ROC) curves (see STAR Methods). Our approach identifies two clusters of procedures, including one cluster with low ROC area under the curve (AUC) and low-to-moderate “scone” performance and another cluster with high ROC AUC and moderate-to-high “scone” performance (Figure 4A). The latter includes all FQ procedures as well as a subset of upper-quartile (UQ) procedures paired with QC adjustment. This example highlights the advantage in considering method classes rather than individual procedures, as in Figure 2D: while there is a spread in “scone” performance scores for any one scaling method, FQ performs well on average, and this performance is validated by external comparisons.
Figure 4. Relationship between “scone” Performance Scores and External Differential Expression Validation in Three scRNA-Seq Datasets (Pollen et al., 2014; Gaublomme et al., 2015; Zheng et al., 2017).
(A–C) ROC AUC versus “scone” performance score. Normalization procedures in the top-right corner are deemed best both by “scone” and by independent differential expression (DE) validation. (A) Comparing GW16 (gestational week 16) and GW21+3 (gestational week 21, cultured for 3 weeks) cells in Pollen et al. (2014), highlighting performance differences between scaling methods and the type of regression-based adjustment. (B) Comparing pathogenic and non-pathogenic cells in Gaublomme et al. (2015), performance differs between scaling methods and regression-based batch adjustment. (C) Comparing B cells and dendritic cells in 10× dataset (Zheng et al., 2017); performance differs between scaling methods but not by batch adjustment.
(D–F) Boxplots of ROC AUC for the bottom 10 (bot10) and top 10 (top10) procedures as ranked by “scone” and for procedures with RUV, QC adjustment, and neither (RUV, QC, or No_UV respectively). Boxplots are further stratified by batch adjustment, when appropriate. Datasets are presented in the same order as in (A)–(C).
For the SMART-seq dataset of Gaublomme et al. (2015), we utilize a separate study of bulk differential expression between pathogenic and non-pathogenic Th17 cells (Lee et al., 2012). For each normalization procedure, we test for differences in expression between Th17-positive pathogenic cells and unsorted non-pathogenic cells, generating ROC curves. We observe a relatively low correlation between the ROC AUC and the “scone” performance scores (Figure 4B; Spearman correlation coefficient of 0.4). However, the improved performance of both the FQ method (Figure 2E) and our batch adjustment (Figure 2H) are both validated by external differential expression data.
For the PBMC dataset (Zheng et al., 2017), we process an independent bulk microarray dataset of Nakaya et al. (2011). We compute sets of positive and negative control genes by comparing baseline B cell and baseline dendritic cell (DC) micro-array samples. For each normalization procedure, we use these sets to evaluate differential expression between the single-cell clusters of B cells and dendritic cells, as defined by Seurat’s clustering procedure (Butler et al., 2018) (see STAR Methods). DESeq scaling performs well on average, as suggested by the “scone” performance score (Figure 2F). Stratified evaluation does not significantly change the correlation of the performance ranking with external validation performance (Figure S5B).
While the “scone” ranking is not necessarily correlated with the AUC-based ranking across the whole performance range, we find an overall high level of agreement between the two rankings at the level of method classes, with the exception of RUV and QC methods. When considering many normalization procedures, users may also rely on top-ranking procedures to provide a basis for further exploration and downstream analysis. We found that the top ten normalizations as ranked by “scone” consistently performed well in terms of ROC AUC and better than procedures that consisted of scaling only (Figures 4D–4F). Taken together, these results indicate that the “scone” performance ranking is a good way of identifying suitable normalization procedures for a given dataset.
“scone’s” Normalization Performance Ranking Is Associated with Improved Representation of Cell-Cell Similarity
Our validation of the “scone” performance scoring in the previous section assumes that there were different cell populations to compare. In many scRNA-seq studies, however, the goal is to identify novel cell sub-populations via cluster analysis. Here, we aim to assess the ability of “scone” to identify normalization procedures or classes thereof that will lead to the best clustering of a given dataset, using some notion of ground truth for cell clusters.
We simulate 10 datasets mimicking typical characteristics of a dataset comprising multiple cell populations using the Bio-conductor R package “splatter” (Zappia et al., 2017), with simulation parameters estimated from our 10× Genomics dataset (Figure 5A; see STAR Methods). The “scone” performance score is highly correlated with the adjusted Rand index (ARI) calculated between the simulated clusters and the clusters identified by k-means on the normalized data (Figures 5B and 5C; see STAR Methods). Because these are simulated data, we can also evaluate the different normalization methods by their ability to recover the true fold-change between cell types (Figure S6A). Notably, this is only weakly correlated with the ARI (Figure S6B), suggesting that even if, on average, a method leads to less biased fold-change estimation, it may produce data that are too noisy to yield reliable clustering results.
Figure 5. Validating “scone” Performance with Simulated Data and External Cell-Level Data.
(A) t-distributed stochastic neighbor embedding (tSNE) of the first 10 PCs of the log-transformed, TC-normalized UMI counts for a dataset simulated using “splatter,” with parameters inferred from the 10× PBMC dataset (Zheng et al., 2017).
(B) Average adjusted Rand index (ARI) between the true simulated clusters and k-means clusters (k = 5) for normalized data versus “scone” performance score (without BIO_SIL score) across 10 “splatter” simulations (see STAR Methods). A Pearson correlation of 0.73 between the two metrics highlights the ability of “scone” to select procedures that optimize aspects of clustering that are not explicitly accounted for in the performance panel. The top-performing procedure was FQ with adjustment for batch and 1 qPC.
(C) Boxplot of average ARI for the bottom 10 (bot10) and top 10 (top10) procedures as ranked by “scone” and for procedures with RUV, QC adjustment, and neither (“RUV,” “QC,” and “No_UV,” respectively). The boxplot is stratified by batch adjustment for the latter 3 categories.
(D) Jaccard score between kNN graph of protein abundance measures and kNN graph of normalized expression measures (k = 792, 10% of samples; see STAR Methods) versus “scone” performance score. A Pearson correlation of 0.60 between these metrics demonstrates how “scone” selects procedures that improve local representations of cell-cell similarity.
(E) Boxplot of Jaccard score for the bottom 10 (bot10) and top 10 (top10) procedures as ranked by “scone,” procedures with no non-batch unwanted variation normalization (No_UV) and procedures with RUV or QC adjustment (QC or RUV).
We also apply “scone” to the recent cellular indexing of transcriptome and epitopes by sequencing (CITE-seq) the dataset of Stoeckius et al. (2017), in which gene expression and antibody levels for 13 cell-surface proteins had been jointly measured for the same cells. Specifically, we use “scone” to normalize the transcription measures and then examined the extent to which their consistency with the protein measures varies according to normalization. Computing k-nearest-neighbor graphs (k = 792, 10% of cells) for the two spaces, namely, protein abundance and transcript abundance (10 PCs, see STAR Methods), we observe an increase in overlap between the two graphs as the “scone” performance score increased (Figures 5D and 5E), reflecting how procedures ranked highly by “scone” are better at representing surface marker expression similarity.
Using Contrasts to Adjust for Batch Effects in Nested Designs
The “scone” analysis of the Th17 dataset (Figure 2) demonstrates the importance of correcting for batch effects in scRNA-seq data. However, depending on the experimental design, simply including a batch variable in the model is not always a viable option. As an extreme case, imagine a completely confounded design, in which each biological condition is assayed in a distinct batch. In such a case, regressing out the batch indicator from the expression measures will result in the removal of biological effects; conversely, not accounting for batch will make it impossible to attribute the observed differences in expression measures to biological differences between conditions or technical differences between batches. Note that this is not just a thought experiment; several examples of datasets with suboptimal designs are discussed in Hicks et al. (2018).
On the opposite end of the spectrum are experiments designed in such a way that each batch contains cells from each biological condition. Such factorial designs are the optimal choice, when possible. Hicks et al. (2018) and Tung et al. (2017) discuss practical aspects of designing factorial experiments in the context of scRNA-seq.
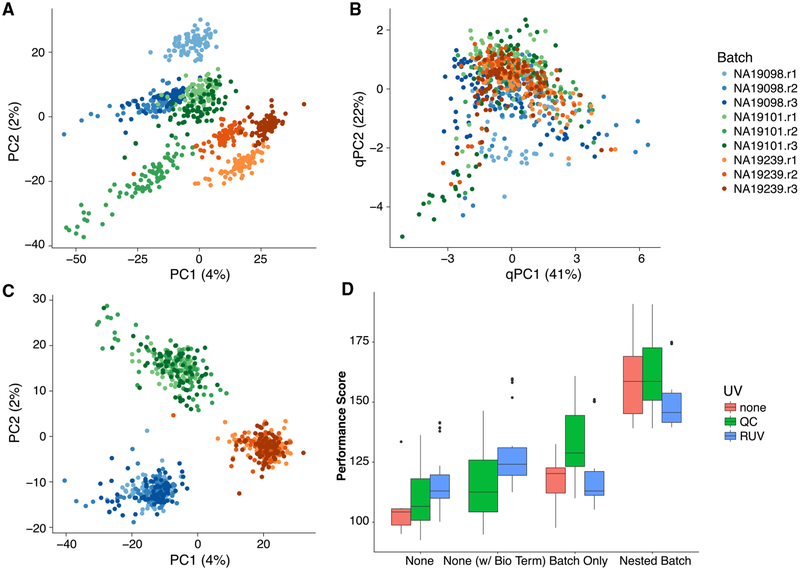
Although optimal, factorial experiments are not always possible or practical. An alternative strategy is to collect multiple batches of cells from each biological condition of interest, in what we refer to as a nested design. A good example of nested design is given by the iPSC dataset analyzed in Tung et al. (2017) (Figure 6). After scaling normalization, the cells clearly cluster by individual, but the cells for each individual are further clustered by batch (Figure 6A). Blindly removing these batch effects with a standard batch correction method, such as ComBat (Leek et al., 2012), removes the biological effects of interest along with the batch effects (Figure S7). Moreover, the QC measures collected as part of the “scone” pipeline are not able to completely capture the batch effects (Figure 6B), as the space of the first two PCs of the QC measures is dominated by the difference between a subset of low-quality cells and the rest of the cells. Explicitly accounting for the nested nature of the design while adjusting for batch effects is the only strategy that effectively removes the unwanted technical variation and preserves the biological signal of interest (Figure 6C; see STAR Methods for details on our nested batch effect correction).
Figure 6. “scone” Results for Human Induced Pluripotent Stem Cells (iPSC) Dataset with Nested Study Design (Tung et al., 2017).
(A) PCA of the log-transformed, TC-normalized UMI counts for all genes and cells passing quality filtering, with points coded by donor (color) and batch (shade). The cells cluster by batch, indicating substantial batch effects.
(B) PCA of QC measures, with points coded by donor and batch. The QC measures do not appear to capture batch effects, but rather intra-batch technical variation.
(C) PCA of log-transformed expression measures after FQ normalization followed by normalization for nested batch effects (top-performing procedure in “scone”), with points coded by donor and batch. As desired, cells cluster by donor but not by batch.
(D) Boxplot of “scone” performance score, stratified by regression-based normalization. Normalization procedures including a nested batch correction per-formed better than those without that step.
The SCONE package is able to detect nested designs by examining the cross-tabulation of the biological and batch factors. Given a nested design, the nested batch effect adjustment based on Equations 3 and 4 in the STAR Methods is automatically applied as one of the different normalization strategies to be compared. Nested designs are common in single-cell studies because of various practical constraints (e.g., processing material from different tissues separately). The “scone” performance scores (Figure 6D) show how only procedures that remove the nested batch effects rank high in the evaluation step. This holds even when removing the two performance metrics that directly involve the batch indicator (BIO_SIL and BATCH_SIL; see STAR Methods), suggesting that the result is not driven by these two metrics alone (Figure S7).
The SCONE Package’s User Interface Facilitates the Exploration of Normalized Data
As part of the Bioconductor R package SCONE, we have developed a Shiny app (Chang et al., 2018) that allows users to interactively explore the data at various stages of the “scone” workflow. In Figure 7, we use the cortical neurons dataset of Pollen et al. (2014) to illustrate the app’s functionality.
The SCONE package provides a function to display an interactive version of the biplot, allowing the user to select a group of normalizations for further exploration (Figure 7A).
The app also provides a hierarchical overview of all the compared normalization strategies (Figure 7B). The hierarchy is based on the series of algorithmic choices that constitute a given normalization strategy. In the example, the first level of the hierarchy represents scaling (e.g., FQ and TMM), while the second represents regression-based methods (QC or RUVg). In general, additional levels are present for the optional imputation and batch correction steps. Alternatively, the user can select a normalization strategy using the drop-down menu in the left panel of the app or using the interactive table at the bottom of the screen; this table can be sorted by the “scone” performance score or by any individual performance metric, making it easy to select, for instance, the procedure that maximizes the preservation of wanted variation (as measured by the EXP_WV_COR metric; see STAR Methods).
Once a normalization approach has been selected for inspection, the Shiny app provides six exploratory tabs for an extended view of the normalized data that should guide the selection of the final procedure. Here, we focus on three examples (see STAR Methods for details). The “Silhouette” tab (Figure 7C) shows the silhouette width for each sample, for clustering based on partitioning around medoids (PAM), clustering by batch, or clustering by biological condition (if available). If the user provides a set of positive and negative control genes, these are visualized in the “Control Genes” panel (Figure 7D) in a heatmap that includes batch, biology, and PAM clustering information. Similarly, the “Relative Log-Expression” tab displays boxplots of the relative log-expression (RLE) measures for the normalized data (Figure 7E).
DISCUSSION
Many different normalization schemes are available, either specifically designed for scRNA-seq or borrowed from the bulk RNA-seq and microarray literatures. Here, we have demonstrated that simple global-scaling normalization is not always sufficient to correctly normalize the data and that more sophisticated strategies may be needed. However, different normalization strategies may perform differently across datasets, depending on the experimental design, protocol, and platform.
Given the small amount of RNA present in a single cell, scRNA-seq data are characterized by a large fraction of dropouts, i.e., genes that are expressed but not detected. To account for the resulting false negatives, “scone” estimates false negative rates that can be used to filter out low-quality samples and, optionally, to impute the expression measure of dropout genes. Zero imputation is still in its infancy; only a handful of methods exist, (Van Dijk et al., 2018; Li and Li, 2018) and it is unclear whether their promised advantages outweigh their limitations. Although we focused on normalization, “scone” can be used to compare different filtering and imputation methods (including none). An alternative approach to imputation is to model the zeros as part of dimensionality reduction. An example of such a method, ZINB-WaVE (Risso et al., 2018), has the ability to include additional covariates to produce a low-dimensional representation of the data that is not influenced by unwanted variation. In principle, the covariates (e.g., QC or RUV factors) selected by “scone” as important for normalization can be included in ZINB-WaVE to provide a more robust projection of the data.
More generally, this manuscript discusses normalization as a modular preprocessing step, transforming read counts into normalized expression measures. Normalization can alternatively be discussed as a parameterization in model-based approaches that combine normalization with other downstream analyses, such as dimensionality reduction and differential expression (Vallejos et al., 2015; Risso et al., 2018; Finak et al., 2015). Performance analyses of normalization as a preprocessing step can also aid such methods, for instance, by informing them about which covariates to include to adjust for unwanted technical effects. While these and other methods do not find a natural place in our SCONE implementation, SCONE’s scoring panel function may be applied to any normalized or processed expression matrix. This generalized scoring functionality facilitates performance comparisons beyond the normalization methods currently supported by SCONE.
We wish to emphasize that although “scone” has demonstrated its usefulness in published studies (e.g., Fletcher et al., 2017; Afik et al., 2017; Gadye et al., 2017; Martin-Gayo et al., 2018), and normalization can help remove unwanted variation from the data, a careful experimental design is a critical aspect of successful use of scRNA-seq. In fact, if the biological effects of interest are completely confounded with unwanted technical effects, no statistical method will be able to extract meaningful signal from the data (Hicks et al., 2018).
Our discussion here surrounds normalization, but the “scone” framework is more general, facilitating the comparison of imputation methods (Van Dijk et al., 2018; Li and Li, 2018), dimensionality reduction techniques (Pierson and Yau, 2015; Risso et al., 2018; Townes et al., 2017), and additional preprocessing steps such as gene and sample filtering.
The present article also focuses on scRNA-seq, but the methodology and software are general and applicable to other types of assays, including microarray, bulk RNA-seq, and adductomic and metabolomic assays. In particular, the user could extend the package by adding different metrics for scRNA-seq, as well as metrics specific to other assays. The SCONE package implementation leverages core Bioconductor packages for efficient parallel computation and on-disk data representation, both essential when analyzing large datasets (Huber et al., 2015).
The computational complexity of “scone” is directly related to the complexity of the normalization methods included in the comparisons. In particular, all scaling methods, RUV and regression-based methods are very efficient, leading to reasonable computing time. With parallelization, computation on a larger dataset can be sped up considerably (e.g., 11 h with 10 processors for the 10× Genomics PBMC dataset of 12,039 cells). For very large datasets, subsampling can be used to decrease computation. One or more random subsamples can be used for evaluating a set of normalization procedures using the “scone” metrics. Following normalization selection, the best procedures can be applied to the full dataset.
The main idea behind “scone” is to use a data-driven approach to select an appropriate normalization strategy for the data at hand. Although it may be infeasible to select the “best” normalization because this would depend on a somewhat subjective definition of optimality, “scone” provides a set of performance metrics (and clustering via the biplot) that can be used to reduce the number of normalization procedures to further explore in the selection of a suitable strategy. One advantage of a panel-based normalization selection framework is that it can be communicated and reproduced by other investigators. We have shown using real data spanning different labs and technologies that “scone” is able to reliably rank normalizations by summarizing multiple dimensions of data quality.
Overall, “scone” provides a flexible and modular framework for the preprocessing of scRNA-seq data that can be used by practitioners to evaluate the impact of the statistical design of a given study and select an appropriate normalization and by method developers to systematically compare a proposed strategy to state-of-the art approaches.
STAR★METHODS
METHOD DETAILS
The “scone” workflow (Figure 1F) consists of five steps: (i) Quality control; (ii) Filtering; (iii) Normalization procedures; (iv) Normalization performance assessment; (v) Exploratory analysis of normalized data. We next provide specific details for these steps. By “log” transformation, we generally refer to the ln(x+1) function, so that zero counts are assigned a value of zero (R function log1p).
Quality Control
For assessing scRNA-seq data quality at the sample level, we rely on over a dozen quality control (QC) metrics evaluated by software packages such as FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), Picard (http://broadinstitute.github.io/picard), and Cell Ranger (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger). These QC measures summarize various aspects of genome alignment, nucleotide composition, and, for the 10x Genomics platform, unique molecular identifiers (UMI) and bar code processing. Summaries of these QC measures are presented in Tables S1 and S2.
QC measures can vary substantially between batches (e.g., C1 run) and also flag low-quality outlying samples (e.g., libraries with low percentages of mapped reads). Including these “bad” samples in downstream analyses may introduce bias in results (e.g., clustering driven by library quality). Even after sample filtering, QC measures can be strongly associated with read counts (Figure 1). It is therefore advisable to both filter samples based on QC measures as well as consider how these measures can be used in the normalization process.
In some cases, it is difficult to obtain low-level (e.g., alignment-based) QC measures. This may occur if sequence data are filtered or omitted when uploading datasets to a public repository. A similar issue arises when expression measures are derived from simulations of count data, rather than sequence-level simulations. In either case, we may consider QC measures derived from count matrices such as those computed by the scater package (McCarthy et al., 2017); we have taken this approach for our simulated data (see Datasets).
Filtering
The goal of data filtering is to remove problematic or noisy observations from downstream analysis. This can simplify many aspects of such analyses, including normalization. In our study, we use gene and sample filtering to combat zero inflation and poor data quality. Our filtering step has three sub-steps, the latter two reducing the size of the dataset:
Define common genes based on read counts: Genes with more than nr reads in at least fs of samples, where nr is the upper-quantile of the non-zero elements of the samples-by-genes count matrix and fs is a user-specified percentage, with default value 25%.
-
Filter samples based on QC measures: Remove samples with low numbers of reads, low proportions of mapped reads (not applied to the 10x Genomics dataset), low numbers of detected common genes, or high areas under the false negative rate (FNR) curve as defined below.
The threshold for each measure is defined data-adaptively: A sample may fail any criterion if the associated metric under-performs by zcut standard deviations from the mean metric value or by zcut median absolute deviations from the median metric value. For all sample-filtered datasets, we have used zcut=2. This sub-step was not applied to the Pollen et al. (2014) dataset due to the small number of samples.
Filter genes based on read counts: Remove genes with fewer than nr reads in at least ns samples, where nr is the upper-quantile of the non-zero elements of the samples-by-genes count matrix, for samples that passed the filtering described in the previous step. We have set a default of ns=5 to accommodate markers of rare populations.
This sub-step ensures that included genes are detected in a sufficient number of samples after sample filtering. Due to the small number of negative control genes in the dataset from Gaublomme et al. (2015) (see below), we force inclusion of all detected (i.e., non-zero count) negative controls.
False Negative Rate Analysis
Single-cell RNA-seq data have many more genes with zero read counts than bulk RNA-seq data. In some cases, the number of zeros far exceeds predictions from standard low-mean count distributions; this zero inflation (ZI) can occur for biological reasons (i.e., the gene is simply not expressed) or technical reasons (e.g., low capture efficiency and amplification).
It is informative to examine the distribution of zero counts, as this might reveal problematic cells or genes. For instance, cells with an unusually high proportion of zero counts for highly-expressed genes might have encountered PCR or read alignment problems. Additionally, in an attempt to determine whether zeros are technical, one can employ housekeeping genes which are expected to be expressed both highly and uniformly across cells.
Specifically, let Yij denote the log-read count of gene jϵ{1,…,J} in cell/sample iϵ{1,…,n} and let j0 ⊆ {1,…,J}denote a set of housekeeping genes. For each cell i, fit the following logistic regression model to the housekeeping genes only:
| (Equation 1) |
where is an indicator variable equal to 1 if gene j is “detected” and zero otherwise (as determined by a detection threshold ϵ≥0), is the median log-read count of gene j across all non-zero samples, βi = (βi0,βi1)ϵR2 are cell-specific regression parameters, and the logit function is defined as logit(x)=log(x/(1−x)) for xϵ[0,1].
The motivation behind this model is that the detection rate of gene j in cell i should be related to the “baseline” expression measure of the gene in a cell-specific manner (i.e., cell-specific regression parameters βi) and that undetected housekeeping genes should constitute “false negatives”. In practice, we set ϵ=0.
For a given detection threshold ϵ, Equation (1) yields FNR curves for each cell, where each point on the curve corresponds to a gene’s “failure probability” at a baseline expression . The higher the curve, the lower the quality of the sample. Samples can then be compared based on the area under the curve (AUC) for their respective FNR curves.
Note that we assume that the housekeeping genes are both highly and uniformly expressed across all samples. These assumptions allow us to treat the zeros as “dropouts” rather than as biologically meaningful zeros, hence the interpretation of as failure probability.
Normalization Procedures
A variety of gene- and sample-specific unwanted effects can bias gene expression measures and thus require normalization. Accordingly, we distinguish between two types of normalization: Within-sample normalization (Risso et al., 2011), which adjusts for gene-specific (and possibly sample-specific) effects, e.g., related to gene length and GC-content, and between-sample normalization, which adjusts for effects related to distributional differences in read counts between samples, e.g., sequencing depth, C1 run, library preparation. Here, we focus on between-sample normalization.
Most between-sample normalization methods proposed to date are adaptations of methods for bulk RNA-seq and microarrays and range, as described next, from simple global scaling to regression on gene- and sample-level covariates.
Global-Scaling Normalization
For the simplest linear global-scaling normalization procedures, gene-level read counts are scaled by a single factor per sample.
Total-Count (TC)
The scaling factor is the sum of the read counts across all genes, as in the widely-used reads per million (RPM), counts per million (CPM), and reads per kilobase of exon model per million mapped reads (RPKM) (Mortazavi et al., 2008).
Single-Quantile
The scaling factor is a quantile of the gene-level count distribution, e.g., upper-quartile (UQ) (Bullard et al., 2010).
Trimmed Mean of M Values (TMM) (Robinson and Oshlack, 2010)
The scaling factor is based on a robust estimate of the overall expression fold-change between the sample and a reference sample.
TMM is implemented in the Bioconductor R package edgeR (Robinson et al., 2010). The default behavior used here is that the selected reference sample has an upper quartile closest to the mean upper quartile of all samples.
DESeq (Anders and Huber, 2010)
The scaling factor for a given sample is defined as the median fold-change between that sample and a synthetic reference sample whose counts are defined as the geometric means of the counts across samples. The method is implemented in the Bioconductor R packages DESeq and edgeR (as “RLE”). Note that the method discards any gene having zero count in at least one sample; as zeros are common in single-cell data, the scaling factors are often based on only a handful of genes.
scran (Lun et al., 2016a, 2016b)
To reduce the effect of single-cell noise on normalization, the scaling factors are computed on pooled expression measures and then deconvolved to obtain cell-specific factors. The method is implemented in the Bioconductor R package scran (Lun et al., 2016a, 2016b). Pool sizes from 20 to the 100 (intervals of five) were considered. For the dataset of Pollen et al. (2014), we limit pool size to the total number of cells.
Optionally, cells can be clustered prior to normalization to relax the assumption that the majority of genes are not differentially expressed across groups of cells (“scran_cluster” scaling). In these cases, we used the quick clustering utility in scran, setting a minimum cluster size at 50. Minimum cluster size was then used as the pool size.
While not strictly necessary, our implementation of global-scaling procedures typically preserves the original measurement scale by further rescaling the normalized measures by the inverse mean scaling factor.
Non-linear Scaling Normalization
In some cases, a single scaling factor per sample may not be sufficient to capture the non-linear effects that affect gene expression measures. Hence, some authors have proposed non-linear normalization methods. Although, these are not technically “scaling” methods, they belong to the first step of the “scone” normalization template, in that they are aimed at making the between-sample distributions of expression measures more similar, rather than explicitly correcting for batch or other confounding factors.
Full-Quantile Normalization (FQ)
All quantiles of the read count distributions are matched between samples (Bullard et al., 2010). Specifically, for each sample, the distribution of sorted read counts is matched to a reference distribution defined in terms of a function of the sorted counts (e.g., median) across samples. This approach, inspired from the microarray literature (Irizarry et al., 2003), is implemented in the Bioconductor R package EDASeq (Risso et al., 2011).
SCnorm (Bacher et al., 2017)
Bacher et al. (2017) noted that a single scaling factor per sample is not enough to account for the systematic variation in the relationship between gene-specific expression measures and sequencing depth. To address this problem, they use quantile regression to estimate the dependence of gene expression measures on sequencing depth, group genes with similar dependence, and use a second quantile regression to estimate scaling factors within each group. In this way, gene expression measures are normalized differently across the range of expression (i.e., highly-expressed genes are scaled differently than lowly-expressed genes). The method is implemented in the Bioconductor R package SCnorm (Bacher et al., 2017).
Regression-Based Normalization
We consider the following generalized linear model (GLM; Figure 1F), which allows adjustment for known and unknown factors of “unwanted variation”:
| (Equation 2) |
where Y is the n×J matrix of gene-level read counts, X is an n×M design matrix corresponding to the M covariates of interest/factors of “wanted variation” (e.g., treatment) and β its associated M×J matrix of parameters of interest, U is an n×H matrix corresponding to known factors of unwanted variation (e.g., batch, sample QC measures) and γ its associated H×J matrix of nuisance parameters, W is an n×K matrix corresponding to unknown factors of unwanted variation and α its associated K×J matrix of nuisance parameters, and g is a link function, such as the logarithm in Poisson/log-linear regression.
The Uγ and Wα terms correspond, respectively, to supervised and unsupervised removal of unwanted variation. A fully supervised version of Equation (2), without W, reduces to a simple GLM fit. The remove unwanted variation (RUV) model of Risso et al. (2014) arises as a special case of Equation (2), when one omits the known unwanted factors U. In many cases, the data-driven unsupervised version of Equation (2), without U, captures effects related to U; for instance, W is often associated with QC measures (Figure S4). However, in many cases, U should still include known batches (e.g., set of samples processed at the same time), as W could capture effects related to sample quality within batches that are targeted for removal in addition to the batch effects captured by U. We also find that, in practice, the computationally simpler approach of fitting a linear model to log-counts Y yields good results. Hence, “scone” relies on a linear model version of Equation (2) with identity link function (g(x)=x).
As detailed in Risso et al. (2014) and implemented in the Bioconductor R package RUVSeq, the unknown, unwanted factors W can be estimated by singular value decomposition (SVD) using three main approaches. Here, we only use RUVg, which estimates the factors of unwanted variation based on negative control genes, assumed to have constant expression across all samples (β=0).
In the current implementation of SCONE, the only covariates that can be included in U are those related to known experimental batches and to sample-level QC. Because QC measures are often highly correlated (Figure 1), SCONE transforms the metrics using principal component analysis prior to fitting the model. The user can choose the maximum number of components to include in the model.
Single-cell RNA-seq data can exhibit severe zero inflation (especially in the case of the 10x Genomics platform, see Figure S1C): anywhere from 10–90% of the read counts are zero (Figure S1I). Regression-based normalization may modify these zeros, rendering their interpretation problematic (cf. technical or biological contributions to zero inflation). To avoid these issues, we restore all values that were initially zero back to zero following normalization. We also make sure that any negative values produced by a normalization procedure are set to zero. As a result, the performance metrics do not reflect differences in the effects of normalization on zero values.
Adjustment for Nested Experimental Designs
The model of Equation (2) should be applied with caution and only after a careful examination of the experimental design. In particular, a common limitation of scRNA-seq datasets is the nesting of unwanted technical effects within the biological effects of interest. For instance, the iPSC dataset of Tung et al. (2017) contains samples derived from three individuals, each processed in three batches. Regressing read counts on the batch covariate in U without adjusting for the covariate of interest in X would then remove the effect of interest (effect of individual donor). Additionally, to avoid collinearity issues between columns of U and X due to nesting, one could either specify suitable contrasts or use a mixed effect model where technical effects are viewed as random. The SCONE implementation of the model in Equation (2) automatically detects nested designs and specifies the right contrasts to avoid removing the effects of interest and to avoid identifiability issues in the estimation procedure. See Tung et al. (2017) for an alternative method that uses random effects.
Specifically, to account for nested batch effects, we consider the following model for each gene (cf. ANOVA). For illustration purposes and ease of notation, we do not include the gene subscript and the additional known or unknown covariates allowed in Equation (2), although the latter can be included in the SCONE package implementation. Let Yijk denote the expression measure of sample k in batch j of condition i, with i=1,.,a conditions, j=1,.,bi batches for condition i, and k=1,.,nj(i) samples for batch j of condition i. We fit the following model gene by gene
| (Equation 3) |
where α is an intercept, βi a biological effect of interest, and γj(i) a nested batch effect. Given the a+1 constraints
| (Equation 4) |
the model is identifiable and can be fit using standard R functions such as lm. The batch-corrected gene expression measures are then given by the residuals . When additional factors of unwanted variation are included in the model, their effects are similarly subtracted from the original matrix to produce the normalized matrix.
Note that although “scone” is able to handle the special case of nested designs, no adjustment method is able to remove batch effects while preserving biological effects of interest if the experimental design is completely confounded. For instance, if only one batch had been processed per individual in the iPSC dataset of Tung et al. (2017), it would have been impossible to determine if the differences between samples were due to batch effects or biology (Hicks et al., 2018).
Normalization Performance Assessment
Seurat Clustering Analyses
It is common to see clear biological clustering at early stages of a single-cell analysis (e.g., major blood cell types). However, an asset of single-cell approaches is their ability to resolve deeper and more subtle biological heterogeneities. Thus, one might wish to maintain the large-scale clustering evident in loosely normalized data, by passing this clustering to SCONE as a biological classification to be preserved after normalization.
We took this approach for our two largest datasets, namely, the 10x PBMC and CITE-seq CBMC datasets. After sample filtering, we loaded the UMI matrices for these datasets into the widely-used Seurat analysis pipeline (Butler et al., 2018). Following TC normalization, log-transformation, scaling, and principal component analysis (PCA), we clustered the cells in the first 10 principal components (PCs) at a “resolution” of 0.6. The resulting clusters were treated as biological conditions for evaluating the biological cluster tightness (BIO_SIL, see below).
For the PBMC dataset, we manually collapsed clusters based on expression of the following marker genes: IL7R (CD4 T-cells), CD14 and LYZ (CD14+ Monocytes), MS4A1 (B cells), CD8A (CD8 T-cells), FCGR3A and MS4A7 (FCGR3A+ Monocytes), GNLY and NKG7 (NK-cells), FCER1A and CST3 (Dendritic cells), and PPBP (Megakaryocytes). These RNA markers were discussed as part of the Seurat vignette.
Normalization Performance Metrics
Different normalization procedures can lead to vastly different distributions of gene expression measures. We have previously shown, in the context of bulk RNA-seq, that the choice of normalization procedure had a greater impact on differential expression results than the choice of DE test statistic (Bullard et al., 2010). A natural and essential question is therefore whether normalization is beneficial and, if so, which method is most appropriate for a given dataset. In order to address this question, we have developed eight normalization performance metrics that relate to three aspects of the distribution of gene expression measures.
Clustering Properties
The following three metrics evaluate normalization procedures based on how well the samples can be grouped according to factors of wanted and unwanted variation: Clustering by wanted factors is desirable, while clustering by unwanted factors is undesirable.
As clustering quality measures, we use silhouette widths (Rousseeuw, 1987). For any clustering of n samples, the silhouette width of sample i is defined as
| (Equation 5) |
where a(i) denotes the average distance (by default, Euclidean distance over first three PCs of expression measures) between the ith sample and all other samples in the cluster to which i was assigned and b(i) denotes the minimum average distance between the ith sample and samples in other clusters. Intuitively, the larger the silhouette widths, the better the clustering. Thus, the average silhouette width across all n samples provides an overall quality measure for a given clustering.
BIO_SIL: Group the n samples according to the value of a categorical covariate of interest (e.g., known cell type, genotype) and compute the average silhouette width for the resulting clustering.
BATCH_SIL: Group the n samples according to the value of a nuisance categorical covariate (e.g., batch) and compute the average silhouette width for the resulting clustering.
PAM_SIL: Cluster the n samples using partitioning around medoids (PAM) for a range of user-supplied numbers of clusters and compute the maximum average silhouette width for these clusterings. “scone” provides an option for stratified scoring, computing the PAM_SIL metric in all distinct strata defined jointly by biological and batch classification. The reported PAM_SIL metric is a weighted average across all strata, weighting by the total number of cells in each. This option is useful when prior biological classifications are poor proxies for single-cell states, i.e., when additional heterogeneity is expected. We used this option for all datasets but the 10x Genomics PBMC and CITE-seq CBMC datasets for which the biological classification were data-derived clusters (see above Seurat clustering analyses).
Large values of BIO_SIL and PAM_SIL and low values of BATCH_SIL are desirable.
Association with Controlgenes and QC Metrics
The next three metrics concern the association of log-count principal components (default 3) with “evaluation” principal components of wanted or unwanted variation.
EXP_QC_COR: The weighted coefficient of determination (details below) for the regression of log-count principal components on all principal components of user-supplied QC measures (using prcomp with scale=TRUE; QC measures described in Tables S1 or S2).
EXP_UV_COR: The weighted coefficient of determination for the regression of log-count principal components on factors of unwanted variation UV (default 3) derived from negative control genes (preferably different from those used in RUV). The sub-matrix of log-transformed unnormalized counts for negative control genes is row-centered and scaled (i.e., for each row/gene, expression measures are transformed to have mean zero and variance one across columns/cells) and factors of unwanted variation are defined as the right-singular vectors as computed by the svds function from the rARPACK package.
EXP_WV_COR: The weighted coefficient of determination for the regression of log-count principal components on factors of wanted variation WV (default 3) derived from positive control genes. The WV factors are computed in the same way as the UV factors above, but with positive instead of negative control genes.
Large values of EXP_WV_COR and low values of EXP_QC_COR and EXP_UV_COR are desirable.
The weighted coefficients of determination are computed as follows. For each type of evaluation criterion (i.e., QC, UV, or WV), regress each expression PC on all supplied evaluation PCs. Let SSTk, SSRk, and SSEk denote, respectively, the total sum of squares, the regression sum of squares, and the residual sum of squares for the regression for the kth expression PC. The coefficient of determination is defined as usual as
and our weighted average coefficient of determination as
| (Equation 6) |
“scone” provides an option for stratified scoring for these metrics, computing them in all distinct strata defined jointly by biological and batch classification. As for the stratified PAM_SIL metric, the reported metric is a weighted average across all strata, weighting by the total number of cells in each. This option was turned off for all analyses expect the one presented in S5.
Global Distributional Properties
When comparing distributions of expression measures between samples, gene-level relative log-expression (RLE) measures, defined as log-ratios of read counts to median read counts across samples, are more informative than log-counts (Gandolfo and Speed, 2018):
| (Equation 7) |
for gene j in cell i. For similar distributions, the RLE should be centered around zero and have have similar spread across samples.
RLE_MED: Mean squared median relative log-expression.
RLE_IQR: Variance of inter-quartile range (IQR) of RLE.
Low values of RLE_MED and RLE_IQR are desirable.
As for PAM_SIL and all correlation metrics above, “scone” provides an option for stratified scoring for these metrics. This option was turned off for all analyses expect the one presented in S5.
Ranking and Selecting Normalization Procedures
In the “scone” framework, the expression measures are normalized according to a user-specified set of methods (including no normalization) and the eight metrics above are computed for each normalized dataset.
The performance assessment results can be visualized using biplots (Gabriel, 1971) and the normalization procedures ranked based on a function of the performance metrics. In particular, we define a performance score by orienting the metrics (multiplying by ±1) so that large values correspond to good performance, ranking procedures by each metric, and averaging the ranks across metrics.
Note that a careful, global interpretation of the metrics is recommended, as some metrics tend to favor certain methods over others, e.g., EXP_UV_COR naturally favors RUVg, especially when the same set of negative control genes are used for normalization and evaluation. We have used non-overlapping sets of control genes for all of the analyses discussed here.
Performance Assessment on Subsampled Datasets
For the analysis presented in Figure 3, we have randomly and independently drawn 10 subsets of cells, running “scone” separately on each subset, and averaging the “scone” performance scores across the 10 subsets. We present results over a range of subsample sizes (1%, 5%, 10%, and 25% of cells) and correlate subsampled performance scores with performance scores derived from the full data.
Evaluation of “scone” Rankings
To independently evaluate the “scone” normalization performance metrics, we compared the “scone” rankings to rankings based on external data available for our three primary datasets, a CITE-seq dataset, and simulated data.
SMART-seq C1 Th17 Dataset (Gaublomme et al., 2015)
To evaluate “scone” performance on the Th17 dataset, we analyzed independent bulk microarray data from Lee et al. (2012), available on GEO with accession GSE39820. We computed sets of positive (DE) and negative (non DE) control genes by comparing cytokine activation IL-1beta, IL-6, IL-23 and cytokine activation TGF-beta1, IL-6 microarray samples, using the Bio-conductor R package limma (Ritchie et al., 2015). Specifically, we identified as positive controls the 1,000 genes with the smallest p-values and as negative controls the 1,000 genes with the largest p-values on the microarray data, excluding any “scone” controls from these sets. For each normalization procedure, we then applied limma with voom weights (Law et al., 2014) to the scRNA-seq data to perform differential expression analysis between Th17-positive pathogenic cells and unsorted non-pathogenic cells. We used the resulting p-values to infer the positive and negative controls derived from the microarray data and produce a receiver operating characteristic (ROC) curve. The ranking of normalization procedures by “scone” was then compared to the ranking by the ROC area under the curve (AUC).
Fluidigm C1 Cortex Dataset (Pollen et al., 2014)
We similarly processed independent bulk microarray data from Miller et al. (2014), available from the BrainSpan atlas (http://brainspan.org/static/download.html). We computed sets of positive (DE) and negative (non DE) control genes by comparing CP+SP and SZ+VZ tissues, using the limma package (Ritchie et al., 2015) as for the Th17 dataset above. For each normalization procedure, we then applied limma with voom weights (Law et al., 2014) to the scRNA-seq data to perform differential expression analysis between the GW16 and GW21+3 conditions. The ranking of normalization procedures by “scone” was compared to the ranking by the ROC AUC, as detailed for the Th17 dataset.
10x Genomics PBMC Dataset (Zheng et al., 2017)
We processed independent bulk microarray data from Nakaya et al. (2011), available on GEO with accession GSE29618. We computed sets of positive (DE) and negative (non DE) control genes by comparing baseline B cell and baseline dendritic cell (DC) microarray samples, using the limma package (Ritchie et al., 2015) as for the Th17 dataset above. For each normalization procedure, we then applied limma with voom weights (Law et al., 2014) to the scRNA-seq data to perform differential expression analysis between the B cell and dendritic cell clusters, as defined by Seurat (Butler et al., 2018). The ranking of normalization procedures by “scone” was compared to the ranking by the ROC AUC, as detailed for the Th17 dataset.
CITE-seq Dataset (Stoeckius et al., 2017)
Antibody-associated cellular indexing of transcriptome and epitopes by sequencing (CITE-seq) UMI counts were extracted from GEO entry GSE100866 (CBMC_8K_13AB_10X), for a collection of human cord blood mononuclear cells (CBMC) and mouse cells. Cell UMI profiles were transformed using the centered log-ratio. Means and standard deviations for each of the 13 antibody measures were computed across all mouse samples (<0.1 human RNA UMI fraction, see Datasets) and the mean plus the standard deviation was subtracted from all abundances, as in the CITE-seq manuscript. For human samples (see Datasets), we constructed a k-nearest-neighbor (kNN) graph using the Euclidean metric in 13-dimensional antibody space (k=792 or 10% of all human samples). For each normalization procedure, we applied PCA to log-transformed scRNA-seq data, selected the top 10 PCs, and used them to constructed a kNN graph with the same choice of k. The ranking of normalization procedures by “scone” was then compared to the ranking by the Jaccard similarity score of the RNA and protein kNN adjacency matrices. Note that we considered other values for k (e.g., k=8 or 1% of samples; data not shown), but found that the mean and range of Jaccard similarity scores decreased considerably for smaller neighborhood sizes. Mouse cells were not utilized beyond preprocessing.
Simulated Datasets
We simulated 10 independent datasets (20,000 genes by 1,000 cells each) using the Bioconductor R package splatter (Zappia et al., 2017). Simulation parameters were inferred from a subset of 100 cells from the 10x Genomics “pbmc4k” dataset (Zheng et al., 2017), setting the differential expression probability to 0.3 and adding five cell populations (or “groups”) of different sizes: one population comprising 50% of the cells, one comprising 20%, and the remaining three populations comprising 10% of cells each. We switched on dropouts and added a batch effect (two batches of 500 cells) to make normalization more challenging.
We ran “scone” on each simulated UMI dataset, including a sample filtering scheme similar to the one above and additionally requiring at least 1,000 UMIs, greater than 80% of common genes detected, below 0.65 AUC, and using a zcut of 3 for greater data-adaptive leniency. Negative control genes (200 for evaluation) were perfect (log-fold change of zero) and extracted from the simulation. 200 positive control genes were selected based on maximum absolute average fold-change as reported by the simulation.
The scater package (McCarthy et al., 2017) was used to compute the following QC measures per simulated library: log10 total UMIs, log10 total UMI features, percent UMIs in top 50, 100, 200, and 500 features. Batch information – not group information – was extracted from the simulation. We considered procedures with zero to three factors of QC or RUVg. We used PCA to decompose the log-normalized count matrix following each normalization. We then performed k-means clustering on the space of the first 10 principal components, specifying the right number of clusters (k=5). Finally, we computed the adjusted Rand index (ARI) between the true simulated clusters and the clusters inferred by k-means. We reported the average ARI across the 10 simulated datasets.
DATA AND SOFTWARE AVAILABILITY
Bioconductor R Software Package SCONE
Our “scone” framework is implemented in the open-source R package SCONE, freely available through the Bioconductor Project (Gentleman et al., 2004; Huber et al., 2015) at https://bioconductor.org/packages/scone.
The package implements methods for sample and gene filtering, provides wrappers for commonly-used normalization procedures, and allows the user to define custom normalization methods in the form of simple R functions.
The main function in the package, called scone, can be used to run and compare different normalization strategies. Through its integration with the BiocParallel and rhdf5 Bioconductor packages, “scone” can take full advantage of parallel processing and on-disk data representation, to avoid storing all the normalization results in memory and instead store the results in HDF5 files. See the package vignette at https://bioconductor.org/packages/scone for additional details.
Although here we used PCA to evaluate the performance of normalization methods (in particular, regarding the metrics EXP_QC_COR, EXP_UV_COR, and EXP_WV_COR), the SCONE package allows the choice of other dimensionality reduction techniques. SCONE also has an optional imputation step that can be turned on to evaluate the performance of imputation methods and their effect on normalization.
In addition to the ability to retrieve normalized expression measures, SCONE allows the user to retrieve the related design matrix. This is useful for downstream differential expression analysis, where rather than removing the unwanted variation and using the residuals as expression measures, one may want to include the factors of unwanted variation as additional covariates in a regression model.
Datasets
SMART-seq C1 Th17 Dataset (Gaublomme et al., 2015)
Cells were harvested from two C57BL/6J and three IL 17A GFP+ mice. Unsorted non-pathogenic cells were collected from the first two mice and both IL-17A-sorted pathogenic and non-pathogenic cells were collected from the three remaining mice. Cells were sorted and a Fluidigm C1-based SMART-seq protocol was used for single-cell RNA extraction and sequencing. Following sample filtering, 337 cells were retained from four donor mice – one mouse was filtered out due to the small number of acceptable cells. We consider the problem of normalizing 7,590 gene features over these 337 cells. For scone, we provided negative and positive control genes based on Table S6 from Yosef et al. (2013).
Fluidigm C1 Cortex Dataset (Pollen et al., 2014)
65 cells from the developing cortex were assayed using the Fluidigm C1 microfluidics system. Each cell was sequenced at both high and low depths; we focus on the high-coverage data. The data are available as part of the Bioconductor R package scRNAseq (https://bioconductor.org/packages/scRNAseq). No sample filtering was applied to this dataset and 4,706 genes were retained following gene filtering. For scone, we provided default negative control genes from the “housekeeping” list and positive control genes related to neurogenesis as annotated in MSigDB (JEPSEN_SMRT_TARGETS and GO_NEURAL_PRECURSOR_CELL_ PROLIFERATION; http://software.broadinstitute.org/gsea/msigdb/cards/).
Fluidigm C1 iPSC Dataset Dataset (Tung et al., 2017)
Three batches of 96 libraries from each of three YRI iPSC lines were sequenced using the Fluidigm C1 microfluidics system. The full dataset, including UMI counts, read counts, and quality metrics, was obtained from https://github.com/jdblischak/singleCellSeq. Library-level QC measures included: (i) Proportion of reads aligning to ERCC spike-ins (matching the pattern “ÊRCC”); (ii) number of unique molecules; (iii) well number as reported in online metadata (“wel”); (iv) concentration as reported in online metadata (“concentration”); (v) number of detected molecule classes (genes with more than zero UMI). Following gene and sample filtering, we retained 6,818 genes and 731 libraries; retained samples had more than 24,546 reads, more than 80% of common genes detected, and FNR AUC below 0.65. For scone, we provided default negative control genes from the “housekeeping” list, as well as positive control genes from the “cellcycle_genes” default list. ERCC genes were used as negative controls for RUVg normalization. Patient was used as a proxy for biological condition, while batch was defined as an individual C1 run.
10x Genomics PBMC Dataset (Zheng et al., 2017)
We considered scRNA-seq data from two batches of peripheral blood mononuclear cells (PBMC) from a healthy donor (4k PBMCs and 8k PBMCs). The data were downloaded from the 10x Genomics website (https://www.10xgenomics.com/single-cell/) using the cellrangerRkit R package (v. 1.1.0). After filtering, 12,039 cells and 10,310 genes were retained. For scone, we provided default negative control genes from the “housekeeping” list and positive control genes as the top 513 most common genes annotated in the MSigDB C7 immunological signature collection (http://software.broadinstitute.org/gsea/msigdb/collections.jsp). Seurat-derived clusters (Butler et al., 2018) were used as a biological condition, while batch was defined as an individual 10x run.
CITE-seq Dataset (Stoeckius et al., 2017)
One cellular indexing of transcriptome and epitopes by sequencing (CITE-seq) dataset was extracted from GEO entry GSE100866 (CBMC_8K_13AB_10X), for a collection of human cord blood mononuclear cells (CBMC) and mouse cells. 8,005 cells were called as human based on greater than 90% human-mapped UMI fraction. After filtering, 7,978 cells and 7,231 genes were retained. For scone, we provided default negative control genes from the “housekeeping” list and positive control genes as the top 513 most common genes annotated in the MSigDB C7 immunological signature collection (http://software.broadinstitute.org/gsea/msigdb/ collections.jsp). Seurat-derived clusters (Butler et al., 2018) were used as a biological condition. QC features were limited to the fraction of human, mouse, and ERCC UMIs (3 features).
For the Th17 and cortex datasets, SRA-format files were downloaded from the Sequence Read Archive (SRA) and transformed to FASTQ format using the SRA Toolkit. Reads were aligned with TopHat (v. 2.0.11) (Trapnell et al., 2009) to the appropriate reference genome (GRCh38 for human samples, GRCm38 for mouse). RefSeq mouse gene annotation (GCF_000001635.23_GRCm38.p3) was downloaded from NCBI on Dec. 28, 2014. RefSeq human gene annotation (GCF_000001405.28) was downloaded from NCBI on Jun. 22, 2015. featureCounts (v. 1.4.6-p3) (Liao et al., 2014) was used to compute gene-level read counts.
Supplementary Material
Highlights.
Proper normalization of scRNA-seq datasets is critical for fair interpretation
Different scRNA-seq datasets require different normalization strategies
“scone” assesses and ranks normalization methods according to their performance
High-ranking methods show better agreement with independent validation data
ACKNOWLEDGMENTS
We would like to thank Russell Fletcher and Diya Das for their constant feedback on the “scone” method and R package. We are grateful to Bioconductor core developers for the thorough code review and to all the users who contributed issue reports, bug fixes, and pull requests on GitHub. D.R., A.W., J.N., E.P., S.D., and N.Y. were supported by the National Institutes of Health, BRAIN Initiative (grant U01 MH105979, PI: J.N.). D.R. was supported in part by the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation (grant number 2018–183201) and by Programma per Giovani Ri-cercatori Rita Levi Montalcini granted by the Italian Ministry of Education, University, and Research. M.B.C. was supported by the National Institute of Dental and Craniofacial Research (NIDCR) (F31 DE025176). D.R., J.N., E.P., and S.D. were also supported by the National Institutes of Health, BRAIN Initiative (grant U19 MH114830, PI: Hongkui Zeng).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cels.2019.03.010.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Afik S, Yates KB, Bi K, Darko S, Godec J, Gerdemann U, Swadling L, Douek DC, Klenerman P, Barnes EJ, et al. (2017). Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 45, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Chu L-F, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, and Kendziorski C (2017). SCnorm: robust normalization of single-cell RNA-seq data. Nat. Methods 14, 584–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, and Kendziorski C (2016). Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, and Stegle O (2015). Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol 33, 155–160. [DOI] [PubMed] [Google Scholar]
- Bullard JH, Purdom EA, Hansen KD, and Dudoit S (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinf. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire J, Xie Y, and McPherson J (2018). Shiny: Web Application Framework for R. https://rdrr.io/cran/shiny/.
- Ding B, Zheng L, Zhu Y, Li N, Jia H, Ai R, Wildberg A, and Wang W (2015). Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics 31, 2225–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, et al. (2017). Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell 20, 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel KR (1971). The biplot graphic display of matrices with application to principal component analysis. Biometrika 58, 453–467. [Google Scholar]
- Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, et al. (2017). Injury activates transient olfactory stem cell states with diverse lineage capacities. Cell Stem Cell 21, 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon-Bartsch JA, and Speed TP (2012). Using control genes to correct for unwanted variation in microarray data. Biostatistics 13, 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandolfo LC, and Speed TP (2018). RLE plots: visualizing unwanted variation in high dimensional data. PLoS One 13, e0191629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaublomme JT, Yosef N, Lee Y, Gertner R, Yang L, Wu C, Pandolfi P, Mak T, Satija R, Shalek A, et al. (2015). Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SC, Townes FW, Teng M, and Irizarry RA (2018). Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 19, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, and Teichmann SA (2016). Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, and Speed TP (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol 13, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT (2014). Svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, and Storey JD (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, and Storey JD (2007). Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WV, and Li JJ (2018). An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun 9, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, and Marioni JC (2016a). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Bach K, and Marioni JC (2016b). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gayo E, Cole MB, Kolb KE, Ouyang Z, Cronin J, Kazer SW, Ordovas-Montanes J, Lichterfeld M, Walker BD, Yosef N, et al. (2018). A reproducibility-based computational framework identifies an inducible, enhanced antiviral state in dendritic cells from HIV-1 elite controllers. Genome Biol. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalisation and visualisation of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, and Wold B (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li G, et al. (2011). Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol 12, 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson E, and Yau C (2015). ZIFA: dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 16, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. (2014). Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol 32, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma Y-A, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Schwartz K, Sherlock G, and Dudoit S (2011). GC-content normalization for RNA-seq data. BMC Bioinf. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, and Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Perraudeau F, Gribkova S, Dudoit S, and Vert J-P (2018). A general and flexible method for signal extraction from single-cell RNA-seq data. Nat. Commun 9, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ (1987). Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comp. Appl. Math 20, 53–65. [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, and Smibert P (2017). Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townes FW, Hicks SC, Aryee MJ, and Irizarry RA (2017). Varying-censoring aware matrix factorization for single cell RNA-sequencing. bioRxiv. 10.1101/166736. [DOI] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung P-Y, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, and Gilad Y (2017). Batch effects and the effective design of single-cell gene expression studies. Sci. Rep 7, 39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos CA, Risso D, Scialdone A, Dudoit S, and Marioni JC (2017). Challenges in the normalization of single-cell RNA sequencing datasets. Nat. Methods 14, 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos CA, Marioni JC, and Richardson S (2015). BASiCS: bayesian analysis of single-cell sequencing data. PLoS Comp. Biol 11, e1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Regev A, and Yosef N (2016). Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol 34, 1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. (2013). Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia L, Phipson B, and Oshlack A (2017). Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 18, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bioconductor R Software Package SCONE
Our “scone” framework is implemented in the open-source R package SCONE, freely available through the Bioconductor Project (Gentleman et al., 2004; Huber et al., 2015) at https://bioconductor.org/packages/scone.
The package implements methods for sample and gene filtering, provides wrappers for commonly-used normalization procedures, and allows the user to define custom normalization methods in the form of simple R functions.
The main function in the package, called scone, can be used to run and compare different normalization strategies. Through its integration with the BiocParallel and rhdf5 Bioconductor packages, “scone” can take full advantage of parallel processing and on-disk data representation, to avoid storing all the normalization results in memory and instead store the results in HDF5 files. See the package vignette at https://bioconductor.org/packages/scone for additional details.
Although here we used PCA to evaluate the performance of normalization methods (in particular, regarding the metrics EXP_QC_COR, EXP_UV_COR, and EXP_WV_COR), the SCONE package allows the choice of other dimensionality reduction techniques. SCONE also has an optional imputation step that can be turned on to evaluate the performance of imputation methods and their effect on normalization.
In addition to the ability to retrieve normalized expression measures, SCONE allows the user to retrieve the related design matrix. This is useful for downstream differential expression analysis, where rather than removing the unwanted variation and using the residuals as expression measures, one may want to include the factors of unwanted variation as additional covariates in a regression model.
Datasets
SMART-seq C1 Th17 Dataset (Gaublomme et al., 2015)
Cells were harvested from two C57BL/6J and three IL 17A GFP+ mice. Unsorted non-pathogenic cells were collected from the first two mice and both IL-17A-sorted pathogenic and non-pathogenic cells were collected from the three remaining mice. Cells were sorted and a Fluidigm C1-based SMART-seq protocol was used for single-cell RNA extraction and sequencing. Following sample filtering, 337 cells were retained from four donor mice – one mouse was filtered out due to the small number of acceptable cells. We consider the problem of normalizing 7,590 gene features over these 337 cells. For scone, we provided negative and positive control genes based on Table S6 from Yosef et al. (2013).
Fluidigm C1 Cortex Dataset (Pollen et al., 2014)
65 cells from the developing cortex were assayed using the Fluidigm C1 microfluidics system. Each cell was sequenced at both high and low depths; we focus on the high-coverage data. The data are available as part of the Bioconductor R package scRNAseq (https://bioconductor.org/packages/scRNAseq). No sample filtering was applied to this dataset and 4,706 genes were retained following gene filtering. For scone, we provided default negative control genes from the “housekeeping” list and positive control genes related to neurogenesis as annotated in MSigDB (JEPSEN_SMRT_TARGETS and GO_NEURAL_PRECURSOR_CELL_ PROLIFERATION; http://software.broadinstitute.org/gsea/msigdb/cards/).
Fluidigm C1 iPSC Dataset Dataset (Tung et al., 2017)
Three batches of 96 libraries from each of three YRI iPSC lines were sequenced using the Fluidigm C1 microfluidics system. The full dataset, including UMI counts, read counts, and quality metrics, was obtained from https://github.com/jdblischak/singleCellSeq. Library-level QC measures included: (i) Proportion of reads aligning to ERCC spike-ins (matching the pattern “ÊRCC”); (ii) number of unique molecules; (iii) well number as reported in online metadata (“wel”); (iv) concentration as reported in online metadata (“concentration”); (v) number of detected molecule classes (genes with more than zero UMI). Following gene and sample filtering, we retained 6,818 genes and 731 libraries; retained samples had more than 24,546 reads, more than 80% of common genes detected, and FNR AUC below 0.65. For scone, we provided default negative control genes from the “housekeeping” list, as well as positive control genes from the “cellcycle_genes” default list. ERCC genes were used as negative controls for RUVg normalization. Patient was used as a proxy for biological condition, while batch was defined as an individual C1 run.
10x Genomics PBMC Dataset (Zheng et al., 2017)
We considered scRNA-seq data from two batches of peripheral blood mononuclear cells (PBMC) from a healthy donor (4k PBMCs and 8k PBMCs). The data were downloaded from the 10x Genomics website (https://www.10xgenomics.com/single-cell/) using the cellrangerRkit R package (v. 1.1.0). After filtering, 12,039 cells and 10,310 genes were retained. For scone, we provided default negative control genes from the “housekeeping” list and positive control genes as the top 513 most common genes annotated in the MSigDB C7 immunological signature collection (http://software.broadinstitute.org/gsea/msigdb/collections.jsp). Seurat-derived clusters (Butler et al., 2018) were used as a biological condition, while batch was defined as an individual 10x run.
CITE-seq Dataset (Stoeckius et al., 2017)
One cellular indexing of transcriptome and epitopes by sequencing (CITE-seq) dataset was extracted from GEO entry GSE100866 (CBMC_8K_13AB_10X), for a collection of human cord blood mononuclear cells (CBMC) and mouse cells. 8,005 cells were called as human based on greater than 90% human-mapped UMI fraction. After filtering, 7,978 cells and 7,231 genes were retained. For scone, we provided default negative control genes from the “housekeeping” list and positive control genes as the top 513 most common genes annotated in the MSigDB C7 immunological signature collection (http://software.broadinstitute.org/gsea/msigdb/ collections.jsp). Seurat-derived clusters (Butler et al., 2018) were used as a biological condition. QC features were limited to the fraction of human, mouse, and ERCC UMIs (3 features).
For the Th17 and cortex datasets, SRA-format files were downloaded from the Sequence Read Archive (SRA) and transformed to FASTQ format using the SRA Toolkit. Reads were aligned with TopHat (v. 2.0.11) (Trapnell et al., 2009) to the appropriate reference genome (GRCh38 for human samples, GRCm38 for mouse). RefSeq mouse gene annotation (GCF_000001635.23_GRCm38.p3) was downloaded from NCBI on Dec. 28, 2014. RefSeq human gene annotation (GCF_000001405.28) was downloaded from NCBI on Jun. 22, 2015. featureCounts (v. 1.4.6-p3) (Liao et al., 2014) was used to compute gene-level read counts.