This study describes the harmful potential of commensal E. coli in ANP, which has not been demonstrated in previous studies. Our work provides new insights into gut bacterium-ANP cross talk, suggesting that nonpathogenic commensals could also exhibit adverse effects in the context of diseases.
KEYWORDS: acute necrotizing pancreatitis, commensal E. coli, gut microbiota, intestinal barrier dysfunction, intestinal epithelial cells
ABSTRACT
An increase of Escherichia-Shigella was previously reported in acute necrotizing pancreatitis (ANP). We investigated whether Escherichia coli MG1655, an Escherichia commensal organism, increased intestinal injury and aggravated ANP in rats. ANP was induced by retrograde injection of 3.5% sodium taurocholate into the biliopancreatic duct. Using gut microbiota-depleted rats, we demonstrated that gut microbiota was involved in the pancreatic injury and intestinal barrier dysfunction in ANP. Using 16S rRNA gene sequencing and quantitative PCR, we found intestinal dysbiosis and a significant increase of E. coli MG1655 in ANP. Afterward, administration of E. coli MG1655 by gavage to gut microbiota-depleted rats with ANP was performed. We observed that after ANP induction, E. coli MG1655-monocolonized rats presented more severe injury in the pancreas and intestinal barrier function than gut microbiota-depleted rats. Furthermore, Toll-like receptor 4 (TLR4)/MyD88/p38 mitogen-activated protein (MAPK) and endoplasmic reticulum stress (ERS) activation in intestinal epithelial cells were also increased more significantly in the MG1655-monocolonized ANP rats. In vitro, the rat ileal epithelial cell line IEC-18 displayed aggravated tumor necrosis factor alpha-induced inflammation and loss of tight-junction proteins in coculture with E. coli MG1655, as well as TLR4, MyD88, and Bip upregulation. In conclusion, our study shows that commensal E. coli MG1655 increases TLR4/MyD88/p38 MAPK and ERS signaling-induced intestinal epithelial injury and aggravates ANP in rats. Our study also describes the harmful potential of commensal E. coli in ANP.
IMPORTANCE This study describes the harmful potential of commensal E. coli in ANP, which has not been demonstrated in previous studies. Our work provides new insights into gut bacterium-ANP cross talk, suggesting that nonpathogenic commensals could also exhibit adverse effects in the context of diseases.
INTRODUCTION
Acute necrotizing pancreatitis (ANP) is a critical and life-threatening disease, usually accompanied by serious complications such as systemic inflammatory response syndrome, multiple-organ dysfunction syndrome, pancreatic infections, and sepsis. Intestinal barrier dysfunction and inflammatory injury have been shown to play a vital role in the initiation and propagation of these complications (1). Gut microbiota dysbiosis is known to be an important contributor to intestinal injury (2). Zhu et al. (3) collected fecal samples from acute pancreatitis (AP) patients and healthy controls and then performed 16S rRNA gene sequencing. These researchers found that AP significantly induced dysbiosis, including an increased abundance of Escherichia-Shigella, and the disturbed microbiota was closely correlated with systemic inflammation and gut barrier dysfunction. Further, these researchers demonstrated that the antibiotic-treated mice and germfree mice exhibited alleviated pancreatic injury in cerulein-induced AP, whereas subsequent fecal microbiota transplantation in turn exacerbated the disease. Zhu et al. concluded that gut microbiota dysbiosis worsened the severity of AP in patients and mice. However, which kinds of bacteria participate in the pathogenesis of AP and their mechanisms remain unclear.
Our previous research showed that ANP induced ileal inflammation and increased intestinal permeability in rats, as well as an alteration of gut microbiota and a significantly higher abundance of Escherichia-Shigella by 16S rRNA sequencing (4). In the genus Escherichia, Escherichia coli MG1655 is a commensal, nonpathogenic strain. Becker et al. (5) reported that E. coli MG1655 triggered interleukin-18 (IL-18) secretion in the human intestinal epithelial cell lines Caco-2 and SW837 in vitro. Bambou et al. (6) also showed that E. coli MG1655 was capable of activating NF-κB and inducing CCL-20 and IL-8 in HT29-19A and Caco-2 cells. In addition, an in vivo study (7) found that in Toxoplasma gondii-induced acute ileitis mice, E. coli MG1655 exacerbated ileal histological damages and increased T lymphocyte infiltration of the ileal mucosa and lamina propria. However, whether E. coli MG1655 plays a part in ANP has not been studied.
It is commonly acknowledged that the response of mammalian cells to Gram-negative bacteria is mainly orchestrated through Toll-like receptor 4 (TLR4), a kind of pattern recognition receptor. Upon recognition of lipopolysaccharide (LPS), TLR4 elicits complex downstream signaling events, among which the TLR4/MyD88/p38 mitogen-activated protein (MAPK) pathway is a classical proinflammatory pathway. This pathway is suggested to be involved in the regulation of intestinal barrier function and tight-junction proteins (TJPs) (8, 9). Moreover, Awla et al. (10) demonstrated that TLR4-deficient mice showed less pancreatic injury and inflammation after AP induction, supporting a role of TLR4 in the pathogenesis of AP. On the other hand, endoplasmic reticulum stress (ERS) signaling, including three principal branches (IRE1α/sXBP1, PERK/eIF2α, and ATF6) of the unfolded protein response, has also been associated with inflammation and TJPs (11, 12). We wondered whether bacterium-induced TLR4 activates ERS in the intestinal epithelia, leading to more severe intestinal barrier dysfunction in ANP. In this study, we sought to investigate whether E. coli MG1655 increases intestinal injury and aggravates ANP and to explore its underlying mechanism.
RESULTS
Gut microbiota participated in the pathogenesis of ANP.
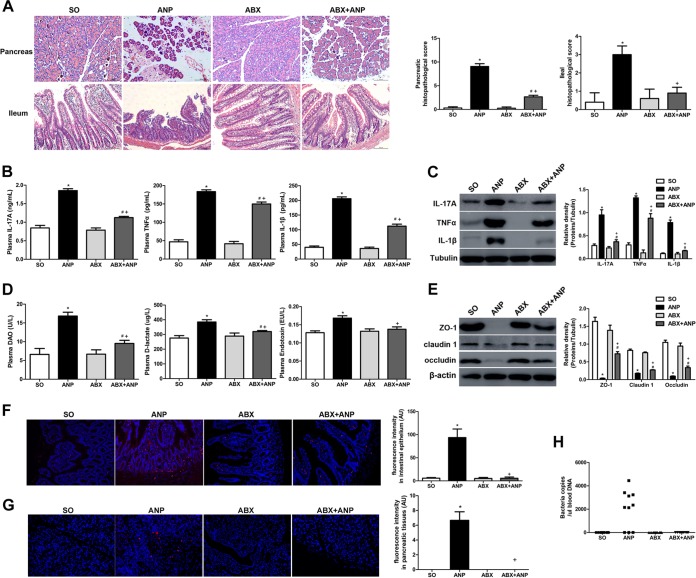
After ANP induction, conventional rats showed pancreatic injury and systemic inflammation. Meanwhile, the ileum presented barrier dysfunction, manifested by increased inflammatory cytokines (IL-17A, tumor necrosis factor alpha [TNF-α], and IL-1β) and intestinal permeability (diamine oxidase [DAO], d-lactate, and endotoxin) and decreased TJPs (ZO-1, claudin 1, and occludin), especially at 24 and 48 h (P < 0.05) (see Fig. S1 in the supplemental material).
We used gut microbiota-depleted rats to investigate the role of gut microbiota in the pathogenesis of ANP (Fig. S2 in the supplemental material). Gut microbiota-depleted rats (the ABX group) showed no differences in the pancreas and ileum compared to conventional rats (the SO group) (Fig. 1). After ANP induction in gut microbiota-depleted rats, the ABX+ANP group displayed less pancreatic injury and lower levels of IL-17A, TNF-α, and IL-1β in the plasma than the ANP group (P < 0.05) (Fig. 1A and B), which was consistent with the results of gut microbiota-depleted mice and germfree mice in the study by Zhu et al. (3). Further, compared to the ANP group, the ABX+ANP group also exhibited significantly alleviated ileal histological damages and intestinal barrier dysfunction, including decreased levels of inflammatory cytokines (IL-17A, TNF-α, and IL-1β) and intestinal permeability (DAO, d-lactate, and endotoxin) and increased expression of TJPs (ZO-1, claudin 1, and occludin) (P < 0.05) (Fig. 1C to E). As determined by fluorescence in situ hybridization (FISH) with a universal bacterial probe, the ABX+ANP group showed less bacterial invasion into the intestinal epithelia than did the ANP group (P < 0.05) (Fig. 1F). Bacterial translocation to the pancreas determined by FISH and bacterial DNA in peripheral blood determined by quantitative PCR (qPCR) were detected in 7 of 10 rats in the ANP group, while none was detected in the ABX+AP group (Fig. 1G and H). Altogether, these data suggest that gut microbiota may participate in the pathogenesis of ANP.
FIG 1.
Gut microbiota participated in the pathogenesis of ANP. (A) Histological analysis of the pancreas and ileum (H&E, ×200). (B) IL-17A, TNF-α, and IL-1β levels in the plasma as determined by ELISA. (C) IL-17A, TNF-α, and IL-1β expression in the ileum as determined by Western blotting. (D) DAO, d-lactate, and endotoxin levels in the plasma. (E) ZO-1, claudin 1, and occludin expression in the ileum as determined by Western blotting. (F and G) Gut bacterial translocation to the intestinal epithelia (F) and pancreas (G) as determined by FISH using a universal bacterial probe (red represents bacteria, and blue represents nuclei). (H) Bacterial DNA detection in peripheral blood as determined by qPCR with universal primers. n = 10 per group. Three independent experiments were performed. Data were analyzed by using one-way ANOVA, followed by Bonferroni’s posttest. *, P < 0.05 versus SO group; #, P < 0.05 versus ABX group; +, P < 0.05 versus ANP group. SO, sham operated.
E. coli MG1655 was associated with the pathogenesis of ANP.
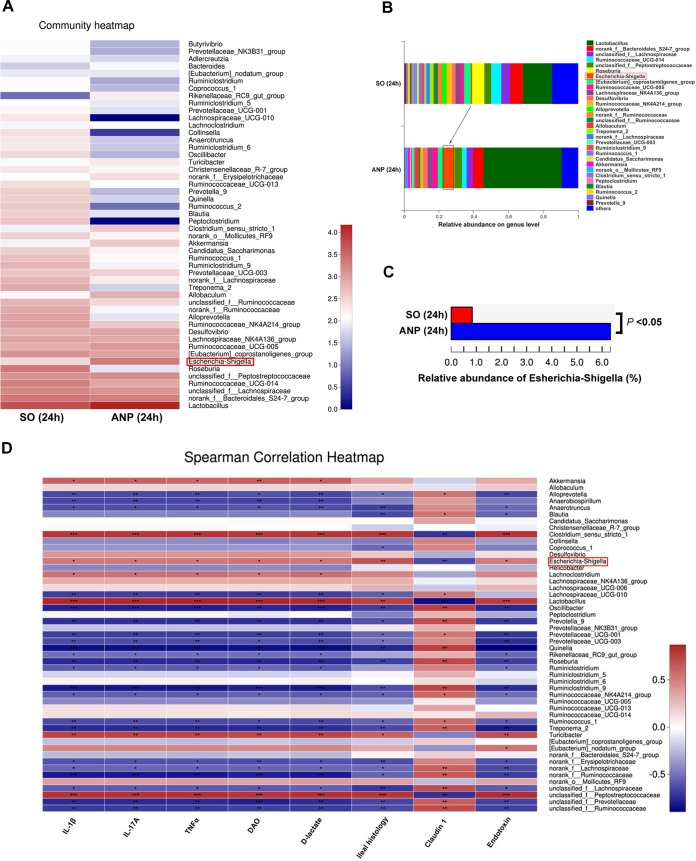
To clearly define which kind of bacteria was involved in ANP pathogenesis, we performed 16S rRNA gene sequencing and found that ANP (24 h) altered the composition of gut microbiota (Fig. 2A and B). Of note, the ANP rats had a higher abundance of Escherichia and Shigella spp. (P < 0.05) at the genus level (Fig. 2A to C), a finding in accordance with the results for AP patients in the study by Zhu et al. (3). Using Spearman correlation analysis, we discovered that Escherichia and Shigella spp. were positively related to inflammatory cytokines in the ileum (IL-1β, IL-17A, and TNF-α), intestinal permeability (DAO, d-lactate, and endotoxin), and ileal histology and negatively related to the tight-junction protein (claudin 1) (Fig. 2D). The results indicated that Escherichia and Shigella may be associated with the pathogenesis of ANP.
FIG 2.
ANP induced an alteration of gut microbiota and a significant increase of Escherichia and Shigella levels. (A) Heatmap of bacterial genera in SO and ANP (24 h) groups as determined by 16S rRNA gene sequencing. (B) Relative abundance of genera in the gut microbiota of the SO and ANP (24 h) groups. (C) Comparison of Escherichia and Shigella levels between the SO and ANP (24 h) groups. (D) Heatmap of Spearman correlation between indicators of intestinal injury and different genera. 16S rRNA sequencing was performed once, with 10 rats in each group. Comparison between two groups (SO versus ANP) was performed using a Student t test, and the correlation between indicators of intestinal injury and different genera was analyzed by using a Spearman test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. SO, sham operated.
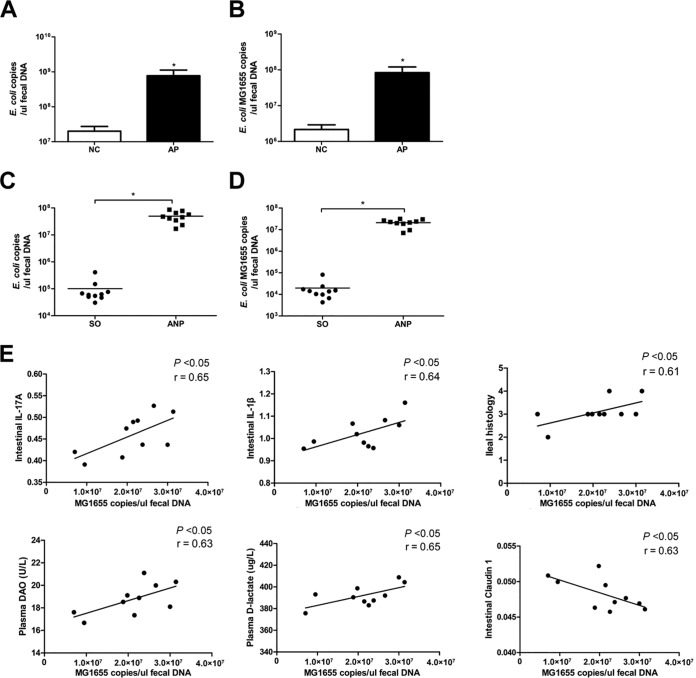
Since the genus Shigella is pathogenic, we focused on Escherichia. There are at least five species in the genus Escherichia, including E. coli, Escherichia blattae, Escherichia fergusonii, Escherichia hermannii, and Escherichia vulneris. The latter three species are pathogenic, and E. blattae was isolated from the Blatta orientalis cockroach . By quantitative PCR (qPCR), we verified that E. coli was significantly increased in AP patients (P < 0.05) (Fig. 3A). We also found that E. coli MG1655, which is a commensal E. coli K-12-derived strain, may be associated with intestinal pathological situations (4–6). By qPCR, we demonstrated a marked increase of E. coli MG1655 in AP patients (P < 0.05) (Fig. 3B). Similarly, we observed larger amounts of E. coli (species) and E. coli MG1655 (strain) in ANP rats than in SO rats (P < 0.05) (Fig. 3C and D). To examine the potential relationship between E. coli MG1655 and intestinal injury in ANP, a Spearman test was performed. The results showed that E. coli MG1655 was positively related to inflammatory cytokines in the ileum (IL-1β and IL-17A), intestinal permeability (DAO and d-lactate), and ileal histology and negatively related to claudin 1 protein (Fig. 3E). Altogether, the data indicated that E. coli MG1655 may be associated with the pathogenesis of ANP.
FIG 3.
E. coli MG1655 was associated with the pathogenesis of ANP. (A and B) Quantity of E. coli (species) (A) and E. coli MG1655 (strain) (B) as determined by qPCR in normal controls (NC) and AP patients. (C and D) Quantity of E. coli (species) (C) and E. coli MG1655 (strain) (D) as determined by qPCR in SO and ANP rats. (E) Correlation analysis of E. coli MG1655 and IL-17A and IL-1β in the intestine, DAO and d-lactate in the plasma, and ileal histopathological scores as determined by using a Spearman test. n = 10 per group. Three independent experiments were performed. Comparison between two groups (NC versus AP and SO versus ANP) was performed by Student t test, and the correlation between E. coli MG1655 and indicators of intestinal injury was analyzed by using a Spearman test. *, P < 0.05 versus the NC or SO group. SO, sham operated.
E. coli MG1655 aggravated pancreatic and intestinal injury in ANP.
First, we administered E. coli MG1655 by gavage to conventional rats (Fig. 4A). As shown in Fig. 4A, E. coli MG1655 could not stably colonize the guts of conventional rats. Compared to SO rats, rats given E. coli MG1655 by gavage (MG1655 group) did not show any changes in the pancreas and ileum (Fig. 4B to I). Compared to the ANP group, the MG1655+ANP group displayed no statistical differences in pancreatic injury and systemic inflammation (IL-17A, TNF-α, and IL-1β in the plasma) (Fig. 4B and C) or ileal histological damages and intestinal barrier dysfunction (Fig. 4B and D to I). In the present study, these data appeared to indicate that E. coli MG1655 given via gavage to conventional rats did not have any significant effects on pancreatic and intestinal injury in ANP.
FIG 4.
Administration of E. coli MG1655 by gavage to conventional rats showed no effects on the pancreas and ileum. (A) Time axis of E. coli MG1655 administration by gavage and ANP induction in conventional rats and quantity of fecal E. coli MG1655 as determined by qPCR. *, P < 0.05. (B) Histological analysis of the pancreas and ileum (H&E, ×200). (C) IL-17A, TNF-α, and IL-1β levels in the plasma as determined by ELISA. (D) IL-17A, TNF-α, and IL-1β expression in the ileum as determined by Western blotting. (E) DAO, d-lactate, and endotoxin levels in the plasma. (F) ZO-1, claudin 1, and occludin expression in the ileum as determined by Western blotting. (G and H) Gut bacterial translocation to the intestinal epithelia (G) and the pancreas (H) as determined by FISH using a probe of E. coli (red represents E. coli, and blue represents nuclei). (I) E. coli MG1655 DNA detection in peripheral blood as determined by qPCR. n = 10 per group. Three independent experiments were performed. Data were analyzed using one-way ANOVA, followed by Bonferroni’s posttest. *, P < 0.05 versus the SO group; #, P < 0.05 versus the ANP group. SO, sham operated.
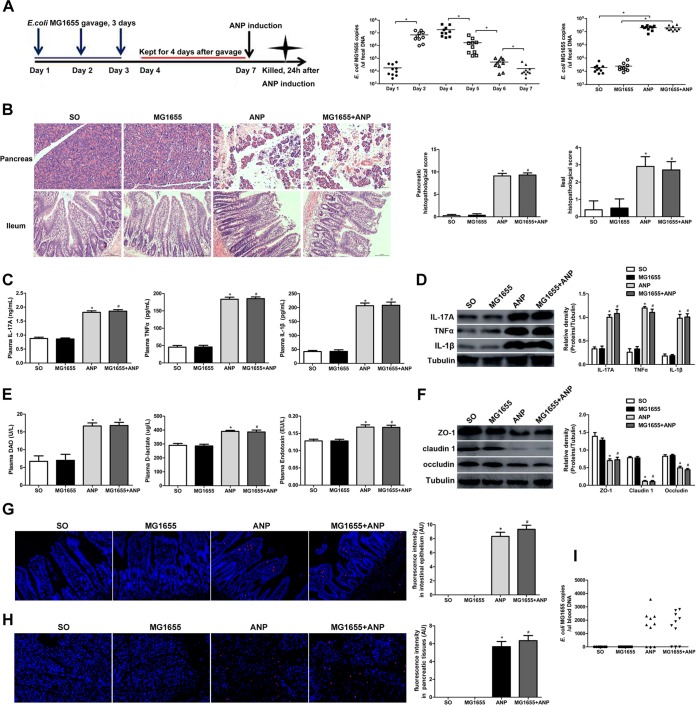
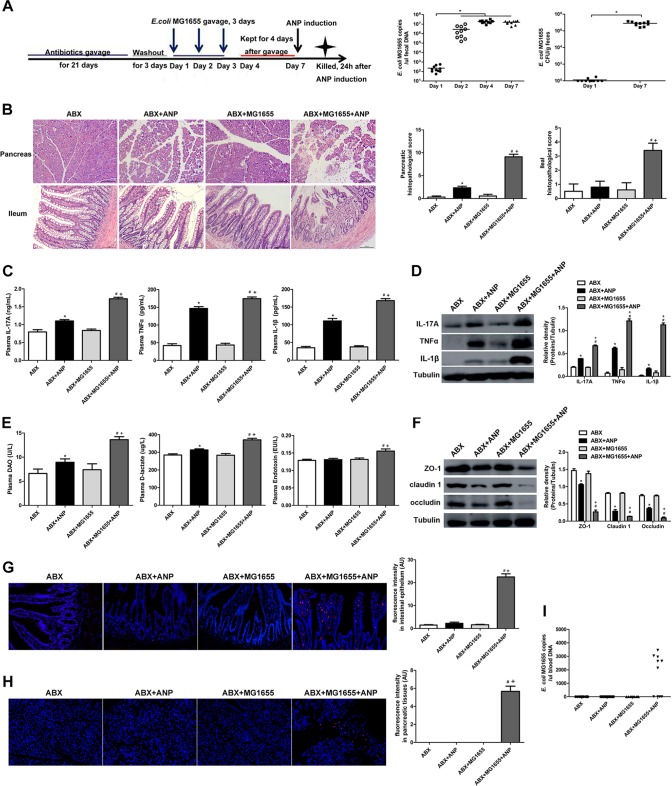
Next, we administered E. coli MG1655 by gavage to gut microbiota-depleted rats and verified its stable colonization in the gut by a fecal culture and qPCR (Fig. 5A). Compared to the ABX group, the ABX+MG1655 group showed no differences in the pancreas and ileum (Fig. 5B to I). After ANP induction, the ABX+MG1655+ANP group had more severe pancreatic injury and significantly elevated levels of IL-17A, TNF-α, and IL-1β in the plasma compared to the ABX+ANP group (P < 0.05) (Fig. 5B and C). Meanwhile, compared to the ABX+ANP group, the ABX+MG1655+ANP group presented exacerbated ileal histological damages and intestinal barrier dysfunction, manifested by increased inflammatory cytokines (IL-17A, TNF-α, and IL-1β) and intestinal permeability (DAO, d-lactate, and endotoxin) and by decreased TJPs (ZO-1, claudin 1, and occludin) (P < 0.05) (Fig. 5B and D to F). In addition, FISH, applying a probe of E. coli, showed that an enormous amount of E. coli invaded the intestinal epithelia in the ABX+MG1655+ANP group (Fig. 5G). E. coli translocation to the pancreas as determined by FISH and E. coli DNA in peripheral blood as determined by qPCR were detected in 6 of 10 rats in the ABX+MG1655+ANP group, while none was detected in the other groups (Fig. 5H and I). These results suggest that E. coli MG1655 administered via gavage to gut microbiota-depleted rats could exacerbate pancreatic and intestinal injury in ANP.
FIG 5.
E. coli MG1655 aggravated pancreatic and intestinal injury in ANP. (A) Time axis of E. coli MG1655 administration by gavage and ANP induction in gut microbiota-depleted rats and quantity of fecal E. coli MG1655 as determined by qPCR and bacterial cultivation. *, P < 0.05. (B) Histological analysis of the pancreas and ileum (H&E, ×200). (C) IL-17A, TNF-α, and IL-1β levels in the plasma as determined by ELISA. (D) IL-17A, TNF-α, and IL-1β expression in the ileum as determined by Western blotting. (E) DAO, d-lactate, and endotoxin levels in the plasma. (F) ZO-1, claudin 1, and occludin expression in the ileum as determined by Western blotting. (G and H) Gut bacterial translocation to the intestinal epithelia (G) and the pancreas (H) as determined by FISH using a probe of E. coli (red represents E. coli, and blue represents nuclei). (I) E. coli MG1655 DNA detection in peripheral blood as determined by qPCR. n = 10 per group. Three independent experiments were performed. Data were analyzed using one-way ANOVA, followed by Bonferroni’s posttest. *, P < 0.05 versus the ABX group; #, P < 0.05 versus the ABX+MG1655 group; +, P < 0.05 versus the ABX+ANP group.
E. coli MG1655-mediated ANP aggravation involved TLR4 and ERS signaling.
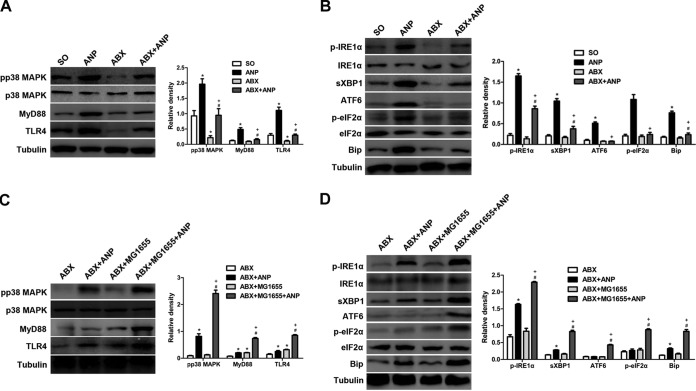
To further examine the molecular mechanism of E. coli MG1655 aggravating ANP, we measured TLR4 and ERS signaling changes in the ileum. The ANP group showed activation of TLR4/MyD88/p38 MAPK and ERS signaling; the latter was indicated by an increase in Bip, IRE1α phosphorylation, sXBP1, eIF2α phosphorylation, and ATF6 induction (P < 0.05) (see Fig. S3A and B in the supplemental material). However, the two signaling pathways were downregulated in the ABX+ANP group compared to the ANP group (P < 0.05) (Fig. 6A and B). After administration of E. coli MG1655 by gavage to conventional rats with ANP, we observed no statistical differences in TLR4 and ERS signaling between the MG1655+ANP group and the ANP group (see Fig. S3C and D in the supplemental material). Interestingly, after E. coli MG1655 was administered via gavage to gut microbiota-depleted rats with ANP, the levels of TLR4 and ERS signaling molecules were significantly higher in the ABX+MG1655+ANP group than in the ABX+ANP group (P < 0.05) (Fig. 6C and D). Furthermore, immunohistochemistry demonstrated that TLR4, phosphorylated IRE1α (p-IRE1α), p-eIF2α, and ATF6 were abundantly expressed in intestinal epithelial cells (see Fig. S4 in the supplemental material). The data collectively demonstrated that increased TLR4 and ERS activation in the intestinal epithelia, to a certain extent, was involved in E. coli MG1655-mediated ANP aggravation.
FIG 6.
E. coli MG1655-mediated ANP aggravation involved TLR4 and ERS signaling. (A and B) Comparison of TLR4 (A) and ERS (B) signaling molecules after ANP induction between gut microbiota-depleted rats and conventional rats. *, P < 0.05 versus the SO group; #, P < 0.05 versus the ABX group; +, P < 0.05 versus the ANP group. (C and D) Comparison of TLR4 (C) and ERS (D) signaling molecules after ANP induction between E. coli MG1655-monocolonized rats and gut microbiota-depleted rats. n = 10 per group. Three independent experiments were performed. Data were analyzed using one-way ANOVA, followed by Bonferroni’s posttest. *, P < 0.05 versus the ABX group; #, P < 0.05 versus the ABX+MG1655 group; +, P < 0.05 versus the ABX+ANP group. SO, sham operated.
E. coli MG1655 increased TNF-α-induced intestinal epithelial cell injury in vitro.
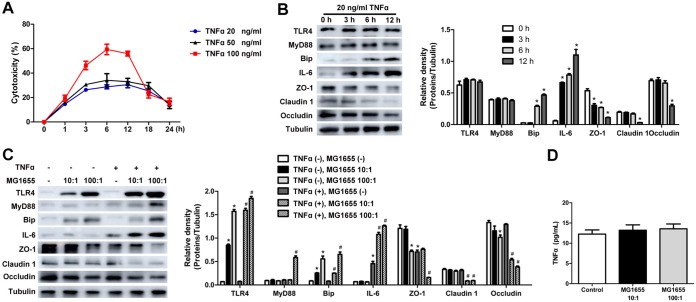
TNF-α plays a crucial role in ANP and is also a classical cytokine to induce intestinal epithelial cell injury (1). In vivo, we demonstrated that the TNF-α level in the plasma and the TNF-α protein expression in the intestine were significantly increased in MG1655-monocolonized ANP rats (Fig. 5C and D). Thus, TNF-α was administered to stimulate IEC-18 cells in vitro. A cell counting kit-8 (CCK-8) assay demonstrated that TNF-α damaged cell viability in a dose-dependent manner (Fig. 7A). After 20-ng/ml TNF-α stimulation, the cytotoxicity reached the highest level at 12 h, with no statistical differences among samples examined at 3, 6, and 12 h. IL-6 production and loss of TJPs (ZO-1, claudin 1, and occludin) were induced in a time-dependent manner (Fig. 7B). Taking these findings into consideration, we chose 20 ng/ml and 3 h, respectively, as the optimal concentration and the optimal preincubation time prior to E. coli MG1655 intervention.
FIG 7.
E. coli MG1655 increased TNF-α-induced intestinal epithelial cell injury in vitro. (A) Cytotoxicity of 20-, 50-, and 100-ng/ml TNF-α treatments of IEC-18 cells. (B) TLR4, MyD88, Bip, IL-6, ZO-1, claudin 1, and occludin levels as determined by Western blotting in IEC-18 cells incubated with 20 ng/ml TNF-α for 3, 6, and 12 h. *, P < 0.05 versus the 0-h group. (C) TLR4, MyD88, Bip, IL-6, ZO-1, claudin 1, and occludin levels as determined by Western blotting in IEC-18 cells challenged with TNF-α and E. coli MG1655 in combination. (D) TNF-α level in the culture supernatant as determined by ELISA. The control represents IEC-18 cells without MG1655 addition. Three independent experiments were performed. Data were analyzed using one-way ANOVA, followed by Bonferroni’s posttest. *, P < 0.05 versus the control group without TNF-α and MG1655 intervention; #, P < 0.05 versus the TNF-α group.
As shown in Fig. 7C, compared to the TNF-α group, the TNF-α+MG1655 (100:1) group showed significantly higher levels of IL-6 and lower levels of ZO-1, claudin 1, and occludin (P < 0.05). On the other hand, TNF-α (20 ng/ml) stimulation induced no changes in TLR4 and MyD88 in IEC-18 cells, whereas coculture with E. coli MG1655 significantly increased TLR4 and MyD88 expression in the TNF-α+MG1655 (100:1) group (P < 0.05). Bip, as the ERS marker, was increased time dependently in response to TNF-α (20 ng/ml) at 3, 6, and 12 h. Furthermore, its level was much higher in the TNF-α+MG1655 (100:1) group than in the TNF-α group (P < 0.05) (Fig. 7B and C). It is worth noting that MG1655 intervention had no significant influence on the TNF-α level in the IEC-18 cell culture supernatant (Fig. 7D), indicating that the results presented above probably cannot be simply explained by E. coli MG1655-induced TNF-α production.
DISCUSSION
In our study, we found that gut microbiota depletion attenuated pancreatic and intestinal injury in ANP; however, the pancreatic injury, systemic inflammation, and intestinal barrier dysfunction were aggravated in ANP after administration of E. coli MG1655 by gavage. In vitro, we observed that E. coli MG1655 increased TNF-α-induced inflammatory cytokine production and loss of TJPs in IEC-18 cells. It was further indicated that TLR4 and ERS signaling may be involved in E. coli MG1655-mediated ANP aggravation.
After the onset of ANP, disease progression can be divided into two phases: an early phase characterized by local inflammation of the pancreas and a late phase dominated by a systemic inflammatory response. The systemic inflammatory response, which greatly affects the prognosis of ANP, is usually amplified by intestinal injury (13). The damaged intestinal barrier promotes gut bacterial translocation and excessive release of inflammatory cytokines, induces gut-derived infection, and eventually leads to high morbidity and mortality rates in ANP.
Gut microbiota is known to be a crucial and potential target in ANP (14), while its cross talk with intestinal injury during ANP remains inexplicit. Zhu et al. (3) and Tsuji et al. (15) reported that the pancreatic inflammation was barely detectable and that serum amylase was marginally elevated in gut microbiota-depleted mice and germfree mice after AP induction by cerulein injection. Although these researchers concluded that gut commensals are essential for the development of cerulein-induced AP, neither the intestinal injury nor a particular bacterial strain was identified. A sodium taurocholate-induced ANP rat model was chosen in our study, and we found that in addition to the pancreatic injury, the intestinal barrier dysfunction (the increase in ileal inflammatory cytokines, intestinal permeability, and the decrease of TJPs) was also ameliorated in gut microbiota-depleted rats after ANP induction. Studies reported that selective decontamination of the digestive tract (SDD) by oral administration of antibiotics could remarkably cut down the amount of bacteria in the intestine and lower the rate of pancreatic secondary infection in ANP (14). The underlying theory of applying SDD was in accord with our findings.
E. coli MG1655 is a commensal nonpathogenic strain derived from E. coli K-12, while some studies have shown that it had harmful potential in some pathological situations. As Bereswill et al. reported (7), E. coli MG1655 given via gavage to gut microbiota-depleted mice caused more severe ileal pathological damages and more CD3+ T lymphocyte infiltration of the ileal mucosa and lamina propria in T. gondii-induced acute ileitis, indicating the proinflammatory potential of commensal E. coli MG1655 in ileitis. We demonstrated a higher abundance of E. coli MG1655 in the feces samples of AP patients and rats. Through in vivo experiments, we found that E. coli MG1655 aggravated pancreatic injury, systemic inflammation, and intestinal barrier dysfunction in ANP rats. In vitro, Bambou et al. and Becker et al. (5, 6) administered E. coli MG1655 to the intestinal epithelial cell (IEC) lines HT29-19A, Caco-2, and SW837 and observed that NF-κB activation and CCL-20, IL-8, and IL-18 production were triggered in IECs. In the present study, we cocultured E. coli MG1655 with TNF-α-preincubated IEC-18 cells and then found increased TNF-α-induced IL-6 production and loss of TJPs in IEC-18 cells. Combining data from both in vivo and in vitro experiments, we provide evidence that E. coli MG1655 aggravates ANP through targeting intestinal epithelial cells. In the healthy gut harboring normal microbiota, commensal E. coli MG1655 grows in the intestinal mucus layer and is prevented from contacting the intestinal epithelia. However, during ANP, intestinal barrier dysfunction and immune disorder allow a large quantity of E. coli MG1655 to contact the intestinal epithelia directly and exert proinflammatory effects on the intestine, eventually leading to ANP deterioration.
In general, the gut microbiota depends on pathogen recognition receptors, such as TLR4, to interact with the host. TLR4 recognizes the LPS of Gram-negative bacteria and activates downstream proinflammatory signaling, including the TLR4/MyD88/p38 MAPK pathway, which is also suggested to be involved in the regulation of TJPs and intestinal permeability (8, 9, 16). It has been reported that TLR4−/− mice exhibit less pancreatic injury and inflammation in pancreatitis compared to wild-type mice (10), supporting the significance of TLR4 in the pathogenesis of pancreatitis. TLR4 has also been demonstrated to be responsible for intestinal injury. Heimesaat et al. (17) demonstrated that TLR4 signaling increased ileal immunopathology in TLR4−/− mice. Administration of E. coli M, an isolate from a diseased mouse, via gavage aggravated T. gondii-induced ileitis in gut microbiota-depleted wild-type mice, whereas TLR4−/− mice displayed less ileal histopathological injury. Lipid A, an ancient component of LPS, showed effects similar to those of TLR4 in that study. Through coculture of E. coli MG1655 and RAW 264.7 cells, Eder et al. (18) revealed that LPS, especially lipid A, is crucial for E. coli MG1655 to evoke TNF release and NF-κB activation in macrophages. TLR4 responses against LPS are conserved within commensal Enterobacteriaceae, including E. coli. As Heimesaat et al. (17) reported, TLR4-mediated sensing of LPS from commensal E. coli exacerbated murine ileitis. Here, we show that E. coli MG1655 increased TLR4 signaling in the ilea of E. coli MG1655-monocolonized ANP rats and in TNF-α-preincubated IEC-18 cells. We propose that E. coli MG1655 might utilize LPS to activate TLR4 signaling in intestinal epithelial cells and eventually cause inflammatory injury. As a paradigm of commensal enterobacteria, E. coli MG1655 might also recapitulate the mechanisms of action (e.g., LPS/TLR4 signaling) of other closely related bacterial species in this context.
ERS is implicated as a cause of inflammation in many disorders and reported to be associated with intestinal barrier dysfunction in inflammatory bowel diseases (19). Accumulating evidence supports that ERS in the pancreas is a driving force behind the pathogenesis of ANP (20), but how ERS in the intestine works in ANP is rarely explored. One study (21) suggested that ERS was induced in the ileal epithelia during 3% sodium taurocholate-induced pancreatitis, indicated by an irregular, dilated endoplasmic reticulum and Bip upregulation. On the other hand, ERS closely interacts with TLR4 (22). The TLR4-ERS axis has been confirmed in several reports. For example, TLR4-induced ERS in intestinal stem cell (ISC) led to ISC apoptosis and necrotizing enterocolitis (23). Saturated fatty acids activate the TLR4-ERS axis in endothelial cells and impair the vasodilator action of insulin, contributing to insulin resistance (24). In our study, we observed not only ERS activation in the ileum but also consistent changes in TLR4 and ERS signaling in ANP with E. coli MG1655 intervention. Therefore, we propose that E. coli MG1655 is first sensed by TLR4 in intestinal epithelial cells during ANP. TLR4 then induces ERS activation. The signaling causes more severe intestinal barrier dysfunction, including increased intestinal inflammation, intestinal permeability, and loss of TJPs. Finally, the injured intestine in turn aggravates pancreatic injury and systemic inflammation. However, it is possible that TLR4 and ERS are two independent signaling pathways in ANP. Further research is necessary to fully understand the cross talk between TLR4 and ERS signaling.
In conclusion, our findings demonstrated that commensal E. coli MG1655 increases TLR4/MyD88/p38 MAPK and ERS signaling-induced intestinal injury and aggravates ANP in rats. However, it remains unclear whether other commensals should be blamed for this aggravation. Further research might clarify this question in the future. Our study describes harmful potential of the commensal E. coli in ANP. Our work provides new insights into bacterium-ANP cross talk, suggesting that nonpathogenic commensals could have negative effects in the context of diseases.
MATERIALS AND METHODS
Patients with AP.
Of the patients admitted to the Gastroenterology Department of Shanghai General Hospital from September 2018 to November 2018, only those who met the diagnostic criteria of AP were included in the study (25). Patients with chronic or other acute inflammatory diseases were excluded. Healthy volunteers were assessed as the normal control (NC) group. Informed written consent was obtained from all subjects. The study protocol was approved by the medical ethics committee of Shanghai General Hospital.
Feces samples from the NC group were collected immediately after a standardized physical examination, whereas feces from AP patients were collected within the first week of AP onset. Feces were collected in sterile tubes, transferred to the laboratory immediately and stored at –80°C for qPCR analysis.
Study design.
All animal experiments were conducted according to the guidelines of the Animal Care and Use Committee of Shanghai Jiaotong University. Male Sprague-Dawley (SD) rats were obtained from SLAC Laboratory Animal Co., Ltd., Shanghai, China, and maintained under specific-pathogen-free conditions.
SD rats were randomly divided into a sham-operated (SO) group and an ANP group. ANP was induced by retrograde injection of 3.5% sodium taurocholate (Sigma-Aldrich, St. Louis, MO) at a volume of 0.1 ml/100 g into the biliopancreatic duct, as previously described (4), while in the SO group normal saline was used as a control. Rats were sacrificed at 6, 12, 24, and 48 h after ANP induction. The time point of 24 h was chosen in the following experiments according to the course of ANP and the pancreatic and ileal injuries.
Gut microbiota-depleted rats were used in order to investigate the role of gut microbiota in ANP. According to the previously published protocol (26), rats were administered a combination of four antibiotics (ABX), i.e., ampicillin, neomycin, metronidazole, and vancomycin (Sangon Biotech, Shanghai, China), by gavage (10 mg/ml, 1 ml/100 g of body weight, twice a day) for 21 consecutive days. Afterward, ANP was induced on day 22. Fecal samples were harvested before and after antibiotic treatment. Validation of the gut bacterial depletion was performed by a fecal culture, in which brain heart infusion agar (Oxoid, Ltd., Basingstoke, Hampshire, UK) plates were incubated anaerobically at 37°C for 48 h (26).
We then studied the role of E. coli MG1655 in the pathogenesis of ANP. E. coli MG1655 suspensions (109 to 1010 CFU/ml) were administered by gavage (3 ml, once a day) for three consecutive days (7). ANP was induced on day 7. Fresh feces were harvested before and after MG1655 was given via gavage. For MG1655 cultivation, Luria-Bertani (LB) agar plates (Oxoid) were incubated aerobically at 37°C for 48 h.
Blood samples were collected from the abdominal aorta. Pancreas and distal ileum samples were fixed in 10% neutral-buffered formaldehyde solution for later histological evaluation. Segments of the pancreas and distal ileum were harvested, snap-frozen in liquid nitrogen, and stored at –80°C for further analysis.
Histology.
Fixed pancreatic and ileal tissues were embedded in paraffin, cut into 4 μm sections, stained with hematoxylin and eosin (H&E), and examined using a light microscope. Pancreatic and ileal histological changes were evaluated, respectively, according to the Schmidt (27) and Chiu (28) standards.
Plasma cytokine analysis.
The levels of TNF-α, IL-1β, and IL-17A in plasma were measured by enzyme-linked immunosorbent assay (ELISA; eBioscience, San Diego, CA) according to the manufacturer’s instructions.
Assessment of intestinal barrier dysfunction.
Intestinal inflammatory cytokines (IL-17A, TNF-α, and IL-1β), intestinal permeability (DAO, d-lactate, and endotoxin), TJPs (occludin, claudin 1, and ZO-1), and bacterial translocation were measured to assess intestinal barrier dysfunction.
TNF-α, IL-1β, IL-17A, occludin, claudin 1, and ZO-1 in the ileum were measured by Western blotting. Ileal tissues were lysed in freshly prepared radioimmunoprecipitation assay buffer containing complete protease inhibitor cocktail (Yeasen, Shanghai, China). Concentrations of protein extracts were determined by using a BCA assay kit (Beyotime, Shanghai, China). After heating at 100°C for 10 min, proteins (40 μg per lane) were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% fat-free milk for 2 h at room temperature and then incubated with primary antibodies overnight at 4°C, followed by horseradish peroxidase-conjugated secondary antibodies. All primary antibodies were as follows (1:1,000): anti-tubulin (Abcam, Cambridge, MA), anti-occludin and anti-claudin 1 (Abcam), anti-ZO-1 (Fisher Scientific, Waltham, MA), and anti-TNF-α, anti-IL-1β, and anti-IL-17A (ABclonal, Wuhan, Hubei, China). Protein bands were visualized using an enhanced chemiluminescence kit (Pierce, Rockford, IL). The density of each band was measured using ImageJ software and normalized to tubulin levels.
DAO activity in the plasma was measured by a commercial kit (Jiancheng Bioengineering Institute, Nanjing, China), d-lactate was measured by ELISA (Abcam), and endotoxin was measured by a chromogenic limulus amebocyte lysate kit (Usen Biotech, Shanghai, China). All procedures were performed according to the manufacturers’ protocols.
Bacterial translocation was examined by FISH according to previous publications (29). Briefly, ileal and pancreatic sections were deparaffinized (60 min at 60°C, 2 × 10 min in 100% xylene, and 5 min in 100% ethanol), air dried, and then incubated with specific probes at 52°C in a humid chamber for 18 h. The following two probes were applied: a universal bacterial probe (EUB338, 5′-GCTGCCTCCCGTAGGAGT-3′) and a probe specific to E. coli (EC1531, 5′-CACCGTAGTGCCTCGTCATCA-3′) (30). The sections were washed and counterstained by DAPI (4′,6′-diamidino-2-phenylindole). Images were acquired using a confocal microscope (Olympus, Tokyo, Japan).
Cell culture and CCK-8 assay.
IEC-18 rat ileal epithelial cells (ATCC CRL-1589) were grown in Dulbecco modified Eagle medium (Thermo Fisher Scientific) containing 0.1 U/ml bovine insulin (Sigma-Aldrich), 5% fetal bovine serum, and 1× penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2. The medium was replaced every 3 days, and cultures were passaged before confluence.
A total of 4,000 IEC-18 cells were plated into each well of a 96-well plate and cultured overnight. Then, 20, 50, or 100 ng/ml TNF-α (Peprotech, Rocky Hill, NJ) was added to the culture medium, followed by coculture with cells for 1, 3, 6, 12, 18, and 24 h. Subsequently, 10 µl of CCK-8 solution (Dojindo Co., Ltd., Kumamoto, Japan) was added to each well, and the cells were incubated for 3 h. A microplate reader was used to measure the absorbance at 450 nm.
E. coli MG1655 stimulation of IEC-18 cells.
E. coli MG1655 was grown aerobically in LB medium at 37°C and 220 rpm. Bacteria were harvested by centrifugation (2,500 × g, 10 min), washed two times with phosphate-buffered saline, and subsequently diluted to obtain final multiplicities of infection of 100 and 10 bacteria per cell. Then, 2 × 105 cells were seeded in a 6-cm dish. IEC-18 cells were preincubated with 20 ng/ml TNF-α (Peprotech) for 3 h, and then E. coli MG1655 was added at ratios of 100:1 and 10:1 for 6 h.
Evaluation of intestinal epithelial cell injury.
The injury of IEC-18 cells was evaluated by inflammatory cytokine (IL-6) and TJPs (occludin, claudin 1, and ZO-1) by Western blotting as described above. Anti-IL-6 was purchased from ABclonal. The TNF-α level in the culture supernatant was measured by ELISA as previously described (6).
Measurement of TLR4 and ERS signaling.
TLR4 and ERS signaling molecules, including TLR4, MyD88, p38 MAPK, pp38 MAPK, Bip, IRE1α, p-IRE1α, sXBP1, ATF6, eIF2α, and p-eIF2α, were measured by Western blotting as previously described. Primary antibodies were as follows (1:1,000): anti-TLR4 (ABclonal); anti-MyD88, anti-p38 MAPK, and anti-pp38 MAPK (Cell Signaling Technology, Denver, MA); anti-IRE1α (Novus, Littleton, CO); anti-p-IRE1α (Cell Signaling Technology); and anti-Bip, anti-sXBP1, anti-ATF6, anti-eIF2α, and anti-p-eIF2α (ABclonal).
Fecal DNA isolation and quantification.
As previously described (4), bacterial DNA was extracted from fecal samples with an E.Z.N.A. soil DNA kit (Omega Bio-Tek, Norcross, GA) according to the manufacturer’s protocols. The details are described in the supplemental material. The concentration of DNA was determined by using a NanoDrop 2000 (Thermo Fisher Scientific).
Blood DNA isolation and quantification.
As previously described (31), bacterial DNA was extracted from plasma with the E.Z.N.A. blood DNA kit (Omega Bio-Tek) according to the manufacturer’s protocols (detailed procedures can be found on the product website). The DNA concentration was determined by using a NanoDrop 2000 (Thermo Fisher Scientific).
16S rRNA sequencing and data analysis.
Bacterial DNA was amplified using 338F (5′-ACTCCACGGGAGGCA-3′) and 806R (5′-GGACTACHVGGGTWTCT-3′) primers covering V3-V4 region of the bacterial 16S rRNA gene. PCRs were performed in triplicate using a 20-µl mixture, including 4 µl of FastPfu buffer, 2 µl of 2.5 mM deoxynucleoside triphosphates, 0.8 μl of each primer (5 μM) (Sangon Biotech), 0.4 μl of FastPfu polymerase (TransStart FastPfu DNA polymerase; TransGenBioTech, Beijing, China), and 10 ng of DNA template. The PCR products were evaluated using 2% (wt/vol) agarose gel electrophoresis, purified using AxyPrep DNA gel extraction, and quantified by using a QuantiFluor-ST (Promega, Madison, WI). Equimolar concentrations of amplification products were pooled and sequenced on an Illumina MiSeq platform according to the manufacturer’s protocol.
The raw pyrosequencing reads were deposited and quality filtered using Trimmomatic and FLASH software. The high-quality sequences were assigned to samples according to barcodes. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (v7.1; http://drive5.com/uparse/). Chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the SILVA 119 16S rRNA database using a confidence threshold of 70%. OTUs that reached 97% similarity were used for α-diversity estimations, including the OTU number and ACE index, and Good’s coverage and rarefaction curve analysis using mothur (v1.30.2; www.mothur.org/). The principal component analysis was conducted in accordance with the Bray-Curtis distance matrix calculated using the OTU information from each sample.
Quantitative PCR analysis of bacteria.
Quantitative PCR was performed using SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan) following the manufacturer’s protocol. The bacterium-specific primer sequences were as follows: universal bacteria, forward (5′-CCTACGGGAGGCAGCAG-3′) and reverse (5′-ATTACCGCGGCTGCTGG-3′); E. coli (species), forward (5′-GTTAATACCTTTGCTCATTGA-3′) and reverse (5′-ACCAGGGTATCTAATCCTGTT-3′) (32); and E. coli MG1655 (strain), forward (5′-TTAGCAGTGATGCCTCGGGA-3′) and reverse (5′-GGCGTCTCCAGAAAGTCGTG-3′). The primer set for E. coli MG1655 (ATCC 700926) was designed based on its genome (GenBank accession no. U00096.3) by Sangon Biotech, Shanghai, China. The copy number of target DNA was determined by comparison with 10-log-fold dilution standards of plasmid DNA analyzed on the same plate (13). The bacterial quantity is presented as a power of 10 bacteria per µl of fecal DNA.
Statistical analysis.
Data are presented as means ± the standard errors of the mean. The distribution of data was first assessed by Kolmogorov-Smirnov test. If the data followed a normal distribution, parametric tests (Student t test for two groups and one-way analysis of variance [ANOVA] for three or more groups) were performed. For ANOVA, Bonferroni’s posttest was carried out for data with an F value at P < 0.05 and no significant variance inhomogeneity. If data were not normally distributed, nonparametric tests (Mann-Whitney test for two groups and Kruskal-Wallis test with Dunn’s posttest for three or more groups) were performed using GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA). Correlations were analyzed by using the Spearman test. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China for Young Scholars (no. 81600500), the Medical-Engineering Cross Project of Shanghai Jiaotong University (YG2015MS29 and YG2014ZD10), the Clinical Research Cultivating Program of Shanghai Hospital Development Center (SHDC12017X09), and the Shanghai General Hospital Clinical Research Innovation Team Project (CTCCR-2016B03).
We thank Baokun He (Shanghai Key Laboratory of Pancreatic Disease, Shanghai JiaoTong University School of Medicine, Shanghai, China) for enlightening ideas and advice regarding our study.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00059-19.
REFERENCES
- 1.Guo ZZ, Wang P, Yi ZH, Huang ZY, Tang CW. 2014. The crosstalk between gut inflammation and gastrointestinal disorders during acute pancreatitis. Curr Pharm Des 20:1051–1062. doi: 10.2174/13816128113199990414. [DOI] [PubMed] [Google Scholar]
- 2.Castoldi A, Favero de Aguiar C, Moraes-Vieira PM, Olsen Saraiva Câmara N. 2015. They must hold tight: junction proteins, microbiota and immunity in intestinal mucosa. Curr Protein Pept Sci 16:655–671. doi: 10.2174/1389203716666150630133141. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, He C, Li X, Cai Y, Hu J, Liao Y, Zhao J, Xia L, He W, Liu L, Luo C, Shu X, Cai Q, Chen Y, Lu N. 2019. Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J Gastroenterol 54:347–358. doi: 10.1007/s00535-018-1529-0. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Huang C, Wang J, Zhou H, Lu Y, Lou L, Zheng J, Tian L, Wang X, Cao Z, Zeng Y. 2017. Dysbiosis of intestinal microbiota and decrease in Paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS One 12:e0176583. doi: 10.1371/journal.pone.0176583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker H, Apladas A, Mertens S, Hausmann M, Fried M, Rogler G. 2010. T2022 probiotic Escherichia coli Nissle 1917 and commensal E. coli Mg1655 trigger IL-18 secretion in intestinal epithelial cell lines. Gastroenterology 138:S-615 10.1016/S0016-5085(10)62835-1. [DOI] [Google Scholar]
- 6.Bambou JC, Giraud A, Menard S, Begue B, Rakotobe S, Heyman M, Taddei F, Cerf-Bensussan N, Gaboriau-Routhiau V. 2004. In vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J Biol Chem 279:42984–42992. doi: 10.1074/jbc.M405410200. [DOI] [PubMed] [Google Scholar]
- 7.Bereswill S, Fischer A, Dunay IR, Kühl AA, Göbel UB, Liesenfeld O, Heimesaat MM. 2013. Proinflammatory potential of Escherichia coli strains K12 and Nissle 1917 in a murine model of acute ileitis. Eur J Microbiol Immunol 3:126–134. doi: 10.1556/EuJMI.3.2013.2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Xia T, Yu X. 2015. Wogonin suppresses inflammatory response and maintains intestinal barrier function via TLR4-MyD88-TAK1-mediated NF-κB pathway in vitro. Inflamm Res 64:423–431. doi: 10.1007/s00011-015-0822-0. [DOI] [PubMed] [Google Scholar]
- 9.Ouyang J, Zhang ZH, Zhou YX, Niu WC, Zhou F, Shen CB, Chen RG, Li X. 2016. Upregulation of tight-junction proteins by p38 mitogen-activated protein kinase/p53 inhibition leads to a reduction of injury to the intestinal mucosal barrier in severe acute pancreatitis. Pancreas 45:1136–1144. doi: 10.1097/MPA.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 10.Awla D, Abdulla A, Regnér S, Thorlacius H. 2011. TLR4 but not TLR2 regulates inflammation and tissue damage in acute pancreatitis induced by retrograde infusion of taurocholate. Inflamm Res 60:1093–1098. doi: 10.1007/s00011-011-0370-1. [DOI] [PubMed] [Google Scholar]
- 11.Ma JH, Wang JJ, Li J, Pfeffer BA, Zhong Y, Zhang SX. 2016. The role of IRE1α-XBP1 pathway in regulation of retinal pigment epithelium tight junction. Invest Ophthalmol Vis Sci 57:5244–5252. doi: 10.1167/iovs.16-19232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qie X, Wen D, Guo H, Xu G, Liu S, Shen Q, Liu Y, Zhang W, Cong B, Ma C. 2017. Endoplasmic reticulum stress mediates methamphetamine-induced blood-brain barrier damage. Front Pharmacol 8:639. doi: 10.3389/fphar.2017.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan C, Ling Z, Huang Y, Cao Y, Liu Q, Cai T, Yuan H, Liu C, Li Y, Xu K. 2015. Dysbiosis of intestinal microbiota associated with inflammation involved in the progression of acute pancreatitis. Pancreas 44:868–875. doi: 10.1097/MPA.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 14.Cen ME, Wang F, Su Y, Zhang WJ, Sun B, Wang G. 2018. Gastrointestinal microecology: a crucial and potential target in acute pancreatitis. Apoptosis 23:377–387. doi: 10.1007/s10495-018-1464-9. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji Y, Watanabe T, Kudo M, Arai H, Strober W, Chiba T. 2012. Sensing of commensal organisms by the intracellular sensor NOD1 mediates experimental pancreatitis. Immunity 37:326–338. doi: 10.1016/j.immuni.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Jiang Y, Steinle JJ. 2017. Toll-like receptor 4 reduces occludin and zonula occludens 1 to increase retinal permeability both in vitro and in vivo. J Vasc Res 54:367–375. doi: 10.1159/000480455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heimesaat MM, Fischer A, Jahn HK, Niebergall J, Freudenberg M, Blaut M, Liesenfeld O, Schumann RR, Göbel UB, Bereswill S. 2007. Exacerbation of murine ileitis by Toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut 56:941–948. doi: 10.1136/gut.2006.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eder K, Vizler C, Kusz E, Karcagi I, Glavinas H, Balogh GE, Vigh L, Duda E, Gyorfy Z. 2009. The role of lipopolysaccharide moieties in macrophage response to Escherichia coli. Biochem Biophys Res Commun 389:46–51. doi: 10.1016/j.bbrc.2009.08.082. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Dai Z, Sun K, Zhang Y, Chen J, Yang Y, Tso P, Wu G, Wu Z. 2017. Intestinal epithelial cell endoplasmic reticulum stress and inflammatory bowel disease pathogenesis: an update review. Front Immunol 8:1271. doi: 10.3389/fimmu.2017.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrera K, Stanek A, Okochi K, Niewiadomska Z, Mueller C, Ou P, John D, Alfonso AE, Tenner S, Huan C. 2018. Acinar cell injury induced by inadequate unfolded protein response in acute pancreatitis. World J Gastrointest Pathophysiol 9:37–46. doi: 10.4291/wjgp.v9.i2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao HX, Fu XS, Zhou XY, Chen X. 2015. Endoplasmic reticulum stress may not be involved in intestinal epithelial cell apoptosis in experimental acute pancreatitis. Dig Dis Sci 60:1690–1698. doi: 10.1007/s10620-015-3542-y. [DOI] [PubMed] [Google Scholar]
- 22.Rath E, Haller D. 2012. Unfolded protein responses in the intestinal epithelium: sensors for the microbial and metabolic environment. J Clin Gastroenterol 46(Suppl):S3–S5. doi: 10.1097/MCG.0b013e318264e632. [DOI] [PubMed] [Google Scholar]
- 23.Afrazi A, Branca MF, Sodhi CP, Good M, Yamaguchi Y, Egan CE, Lu P, Jia H, Shaffiey S, Lin J, Ma C, Vincent G, Prindle T, Weyandt S, Neal MD, Ozolek J, Wiersch J, Tschurtschenthaler M, Shiota C, Gittes G, Billiar TR, Mollen K, Kaser A, Blumberg R, Hackam DJ. 2014. Toll-like receptor 4-mediated endoplasmic reticulum stress in intestinal crypts induces necrotizing enterocolitis. J Biol Chem 289:9584–9599. doi: 10.1074/jbc.M113.526517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JA, Jang HJ, Hwang DH. 2015. Toll-like receptor 4-induced endoplasmic reticulum stress contributes to impairment of vasodilator action of insulin. Am J Physiol Endocrinol Metab 309:E767–E776. doi: 10.1152/ajpendo.00369.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banks PA, Bollen TL, Dervenis C, Gooszen HG, Johnson CD, Sarr MG, Tsiotos GG, Vege SS, Acute Pancreatitis Classification Working Group. 2013. Classification of acute pancreatitis—2012: revision of the Atlanta classification and definitions by international consensus. Gut 62:102–111. doi: 10.1136/gutjnl-2012-302779. [DOI] [PubMed] [Google Scholar]
- 26.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt J, Rattner DW, Lewandrowski K, Compton CC, Mandavilli U, Knoefel WT, Warshaw AL. 1992. A better model of acute pancreatitis for evaluating therapy. Ann Surg 215:44–56. doi: 10.1097/00000658-199201000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu CJ, Scott HJ, Gurd FN. 1970. Intestinal mucosal lesion in low-flow states. II. The protective effect of intraluminal glucose as energy substrate. Arch Surg 101:484–488. doi: 10.1001/archsurg.1970.01340280036010. [DOI] [PubMed] [Google Scholar]
- 29.Braubach P, Lippmann T, Raoult D, Lagier JC, Anagnostopoulos I, Zender S, Länger FP, Kreipe HH, Kühnel MP, Jonigk D. 2017. Fluorescence in situ hybridization for diagnosis of Whipple’s disease in formalin-fixed paraffin-embedded tissue. Front Med (Lausanne) 4:87. doi: 10.3389/fmed.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Møller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, Krogfelt KA, Cohen PS. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun 71:2142–2152. doi: 10.1128/IAI.71.4.2142-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hull N, Miller J, Berry D, Laegreid W, Smith A, Klinghagen C, Schumaker B. 2018. Optimization of Brucella abortus protocols for downstream molecular applications. J Clin Microbiol 56:e01894-17. doi: 10.1128/JCM.01894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santacruz A, Marcos A, Wärnberg J, Martí A, Martin-Matillas M, Campoy C, Moreno LA, Veiga O, Redondo-Figuero C, Garagorri JM, Azcona C, Delgado M, García-Fuentes M, Collado MC, Sanz Y. 2009. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity 17:1906–1915. doi: 10.1038/oby.2009.112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.