Abstract
The C-terminal domain (CTD) of RNA polymerase II contains a repetitive heptad sequence (YSPTSPS) whose phosphorylation states coordinate eukaryotic transcription by recruiting protein regulators. The precise placement and removal of phosphate groups on specific residues of the CTD are critical for the fidelity and effectiveness of RNA polymerase II–mediated transcription. During transcriptional elongation, phosphoryl-Ser5 (pSer5) is gradually dephosphorylated by CTD phosphatases, whereas Ser2 phosphorylation accumulates. Using MS, X-ray crystallography, protein engineering, and immunoblotting analyses, here we investigated the structure and function of SSU72 homolog, RNA polymerase II CTD phosphatase (Ssu72, from Drosophila melanogaster), an essential CTD phosphatase that dephosphorylates pSer5 at the transition from elongation to termination, to determine the mechanism by which Ssu72 distinguishes the highly similar pSer2 and pSer5 CTDs. We found that Ssu72 dephosphorylates pSer5 effectively but only has low activities toward pSer7 and pSer2. The structural analysis revealed that Ssu72 requires that the proline residue in the substrate's SP motif is in the cis configuration, forming a tight β-turn for recognition by Ssu72. We also noted that residues flanking the SP motif, such as the bulky Tyr1 next to Ser2, prevent the formation of such configuration and enable Ssu72 to distinguish among the different SP motifs. The phosphorylation of Tyr1 further prohibited Ssu72 binding to pSer2 and thereby prevented untimely Ser2 dephosphorylation. Our results reveal critical roles for Tyr1 in differentiating the phosphorylation states of Ser2/Ser5 of CTD in RNA polymerase II that occur at different stages of transcription.
Keywords: RNA polymerase II, phosphatase, mass spectrometry (MS), X-ray crystallography, DNA transcription, intrinsically disordered protein, CTD code, phosphoryl motif, proline isomerization, tyrosine phosphorylation
Introduction
The CTD2 is an intrinsically disordered domain found in eukaryotic RNA polymerase II, the polymerase responsible for transcribing all protein-coding mRNAs as well as some small nuclear RNAs and microRNAs in eukaryotes (1, 2). Although the CTD sequence seems surprisingly simple with the consensus heptad (Y1S2P3T4S5P6S7) repeated 17–52 times in various species, this domain undergoes extensive post-translational modification (PTM) during transcription that predominantly involves phosphorylation (3). Different PTMs or combinations of PTMs orchestrate the process of transcription by recruiting various regulatory proteins at specific stages of the eukaryotic transcriptional cycle (2, 4). The coordination of CTD phosphorylation/dephosphorylation by CTD-modifying enzymes is thus essential for the accurate transcription of genetic information (5, 6).
Phosphorylations at Ser2 and Ser5 of the CTD are the primary modifications in each round of transcription (7). pSer5 is detected predominantly during initiation of transcription and required for capping enzyme recruitment (8, 9). During the transition from initiation to productive elongation, Ser5 sites gradually become dephosphorylated, whereas Ser2 sites become phosphorylated and eventually become the major phosphorylated species in the later stages of transcription (7, 10). At the end of transcription, all remaining phosphate groups on the CTD are removed because nonphosphorylated CTD is required for RNA polymerase II to recycle and bind the next promoter (11). Other than phosphorylation at Ser2 and Ser5, the other three sites (namely Tyr1, Thr4, and Ser7) are also phosphorylated in vivo (1). Among the three alternative phosphorylation sites, Tyr1 phosphorylation is particularly interesting because Tyr1 is highly conserved in all species, and mutation in this residue leads to defects in RNA polymerase II stability (12), antisense transcription (13), and elongation/termination (14, 15). However, the molecular mechanism of how Tyr1 and its phosphorylation regulate transcription is still unclear.
Just as proper placement of phosphorylation marks on the CTD is essential for engaging in transcription, so too is specific and accurately timed removal of CTD phosphorylation marks. For example, deficiency in pSer5 removal causes termination defects leading to transcriptional read-through (16). Failure in pSer2 dephosphorylation leads to cell death because the promoter binding of RNA polymerase II requires a hypophosphorylated CTD (17). However, it is currently puzzling how CTD phosphatases distinguish the highly similar phosphoryl motifs once they are recruited to RNA polymerase II. It is particularly challenging for dephosphorylating Ser2 and Ser5 when both are phosphorylated during elongation, and their flanking residues are very similar (Y1S2P3 and T4S5P6). Nature accomplishes this goal with precision as observed during transcription elongation and termination when pSer5 levels diminish rapidly but pSer2 levels steadily increase (18). This task is mainly accomplished by Ssu72, a conserved essential CTD phosphatase that dephosphorylates pSer5 at the transition from elongation to termination (19, 20).
To understand the molecular mechanism of this high specificity of pSer5 dephosphorylation, we herein investigate the structural characteristics of Ssu72 specificity. Using synthetically phosphorylated CTD peptides and recombinant CTD mutants, we found that Ssu72 has weak phosphatase activity against pSer2 in addition to its previously reported strong activity against pSer5. The unique binding mode of Ssu72 toward its substrate peptides defines the structural basis for the pSer5 preference (Protein Data Bank (PDB) code 6NPW), which requires the proline residue in the SP motif in the cis configuration to form a tight β-turn. The bulky side chain of Tyr1 makes it energetically unfavorable for pSer2-Pro3 to adopt this configuration due to steric hindrance. We further reveal the role of Tyr1 in Ssu72's ability to distinguish pSer5 from pSer2 using CTD substrates with Tyr1 mutated to other residues, which alter Ssu72's inherent specificity. Additionally, structural analysis reveals that the cis-proline configuration of Ser2 is even more unfavorable when Tyr1 phosphorylation occurs, which may help prevent inappropriate pSer2 dephosphorylation by Ssu72 during the transcription cycle. Our results provide a structural explanation for the accurate removal of CTD phosphate groups and reveal a new role of Tyr1 in preventing inappropriate removal of Ser2 phosphorylation during transcription.
Results
Dephosphorylation activity toward other heptad residues of the CTD by Ssu72
As shown from previous kinetic and crystallographic studies from our and other laboratories, Ssu72 predominantly dephosphorylates pSer5 on CTD heptad repeats (21–23). However, Ssu72 has also been reported to dephosphorylate pSer7, albeit with much lower activity than that for pSer5 (24). Ssu72 is part of the cleavage and polyadenylation factor complex (25) forming stable interactions with the N-terminal domain of the scaffolding protein symplekin (26, 27). This complex formation enhances Ssu72 phosphatase activity by increasing protein stability in vitro (21). We hence used the Ssu72–symplekin complex in our biochemical and structural study. For all experiments below, when we show results of Ssu72 reactions, the material includes symplekin complexed with Ssu72 for better stability in vitro and higher experimental reproducibility.
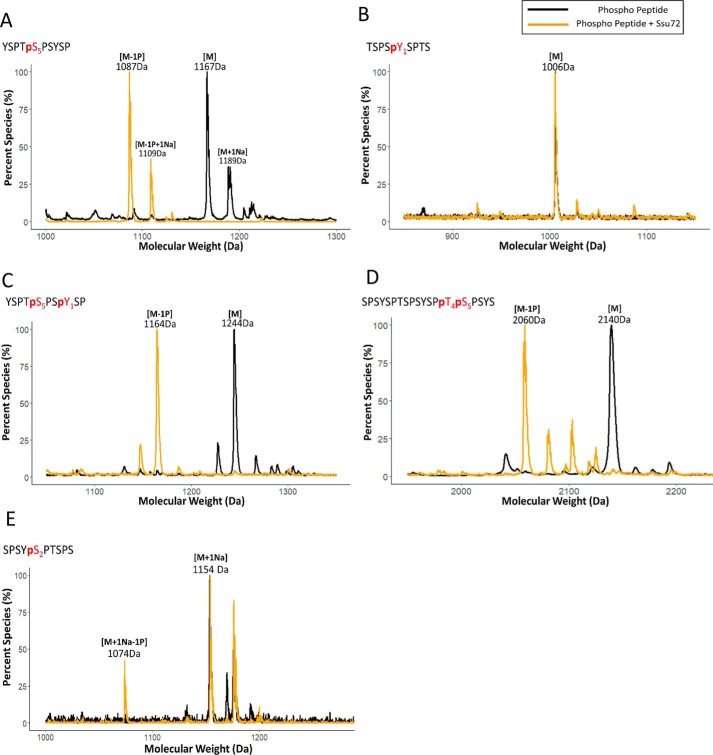
To test whether Ssu72 can dephosphorylate other sites of the CTD in addition to the reported pSer5 and pSer7, we needed a highly sensitive assay to monitor CTD dephosphorylation products even if their abundance is low. Instead of using Malachite green assay, which suffers from a narrow window of detection and a poor signal/background ratio (28), we utilized matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS, which can detect low-abundance products in the picomolar range. Using a short 10-mer pSer5 CTD peptide treated with Ssu72 as a proof of concept, we found that adding 2,5-dihydroxybenzoic acid as a matrix allows for effective ionization of both phosphorylated and dephosphorylated CTD species (Fig. 1A). We observed an 80-Da downshift in our MALDI spectra when the phosphoryl peptide was incubated with Ssu72 (Fig. 1A, gold trace), consistent with the removal of a single phosphate group from pSer5. Therefore, our MALDI-based assay is suitable to detect phosphatase activity for Ssu72 against suboptimal substrates.
Figure 1.
Analysis of the specificity of Ssu72 toward phosphorylated CTD peptides using MALDI. A–E, sequences of CTD peptides have been indicated in the top left-hand corner of each respective graph with phosphoryl residues highlighted in red color and a “p” indicating phosphorylation. The color scheme of the trace is depicted at the top right-hand corner, showing that the black trace is the controlled experiment with no phosphatase treatment, and the gold trace is with Ssu72 treatment. Three independent biological duplicates have been sampled for each assay with consistent results. One representative of the samples is shown in each panel. “M” notation indicates the substrate peptide, “−#P” notation indicates an approximate number of phosphates removed based on mass shift, and “+Na” indicates the formation of sodium adduct.
Ssu72 exhibits high structural similarity to low-molecular-weight tyrosine phosphatases and has an almost identical reaction mechanism (23). To test whether Ssu72 has phosphatase activity against pTyr1 in CTD, we first evaluated phosphatase activity against a 9-mer pTyr1 CTD peptide by MALDI-TOF in which no dephosphorylation was detected (Fig. 1B). To ensure that the lack of activity is not due to the failure of targeting Ssu72 to the substrate, we treated a doubly Tyr1-Ser5 phosphorylated 10-mer peptide with Ssu72 as pSer5 recruits Ssu72 to the substrate (Fig. 1C). We observed one phosphate being rapidly removed in the reaction, which we expect to be the dephosphorylation of pSer5 as shown previously (Fig. 1A), but no further dephosphorylation of the peptide was detected (Fig. 1C). Our results indicate that Ssu72 cannot dephosphorylate pTyr1 of the RNA polymerase II CTD.
We next asked whether Ssu72 could dephosphorylate other serine/threonine residues of the CTD. Previously, it was reported that Ssu72 could dephosphorylate pSer7 with activity about 4000 times less than that against pSer5 (24), but no activity against pThr4 or pSer2 has been reported. To test whether Ssu72 can dephosphorylate pThr4, we used a doubly phosphorylated Thr4-Ser5 CTD peptide as a substrate to ensure the recruitment of the phosphatase. When the peptide was analyzed for its molecular weight by MALDI, only one phosphate was removed even after prolonged incubation (Fig. 1D, gold trace). We have previously shown crystallographically that Ssu72 maintains strong pSer5 specificity even in the context of pThr4, suggesting the removed phosphate is derived from pSer5 and not pThr4 (29). Therefore, Ssu72 shows no detectable phosphatase activity against pThr4.
In contrast, pSer2 can potentially act as a substrate for Ssu72 as it is flanked by a proline to form an SP motif for recognition, similar to pSer5. To determine whether pSer2 is subject to dephosphorylation by Ssu72, we used a 10-mer synthetic peptide containing pSer2 as substrate. Incubation of this peptide with Ssu72 resulted in a peak appearing with 80 Da removed, consistent with at least partial removal of the pSer2 mark from the phosphoryl peptide (Fig. 1E, gold trace). Longer incubation increases the intensity of the peak, but full dephosphorylation is never reached as observed with pSer5 peptide (Fig. 1A). This result is surprising because there have been no previous reports of Ssu72 activity against pSer2. Our sensitive MALDI-TOF data suggest that Ssu72 can dephosphorylate pSer2 in vitro but with less activity compared with pSer5.
Ssu72 activity against full-length CTD
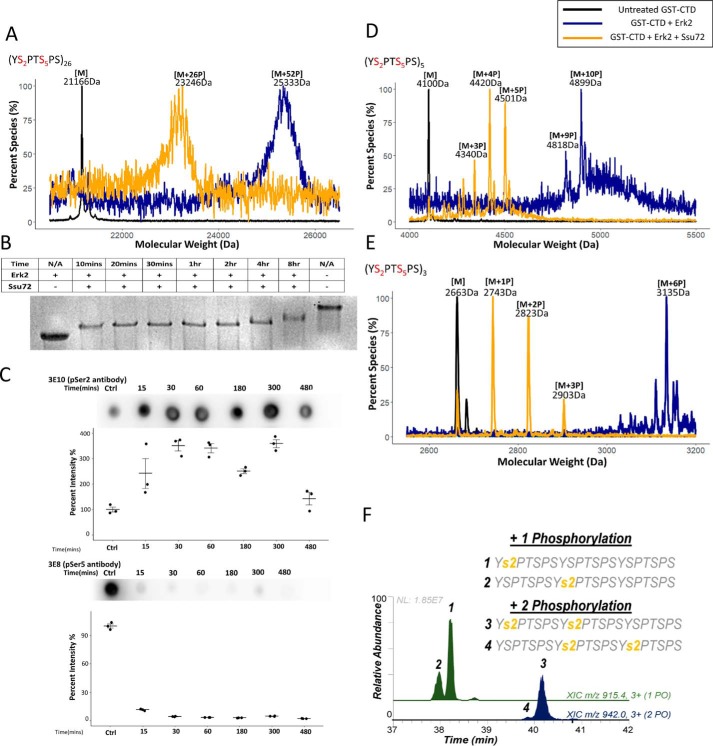
Although Ssu72 only shows weak phosphatase activity against pSer2 in CTD peptides, we considered the possibility that the dephosphorylation can be substantially more prevalent when full-length CTD is used as a substrate due to the high local concentration of pSer2. To test this, we used full-length yeast CTD (26 repeats with two SP motifs in each repeat, Ser2-Pro3 and Ser5-Pro6; Fig. S1) as the substrate for Ssu72 (Fig. S1). To achieve the saturated phosphorylation at all SP motifs, we incubated CTD with Erk2 under the condition of excess ATP and extended reaction time. The number of phosphates added was quantified using MALDI-TOF (Fig. 2A, blue trace). WT full-length yeast CTD was almost completely phosphorylated on all SP motifs (∼52 phosphorylation sites detected for the 52 SP motifs from 26 CTD repeats) (Fig. 2A, blue trace). Using this fully phosphorylated CTD as a substrate with all Ser2 and Ser5 phosphorylated, we characterized the pattern of dephosphorylation of CTD by Ssu72 using MALDI-TOF, electrophoretic mobility shift assay (EMSA) and immunoblotting assays (Fig. 2, A–C). A time-course EMSA indicated that the Ssu72-mediated CTD dephosphorylation occurs in a two-phase mode, a fast phase is reached within the first ∼10 min and then followed by a slow phase where gradual dephosphorylation is occurring with prolonged incubation (Fig. 2B). A sample taken after 90 min of the reaction was analyzed by MALDI-TOF and contained ∼26 phosphates on the CTD (Fig. 2A, gold trace). To determine the identities of the phosphoryl species, we monitored the dephosphorylation using immunoblotting with phosphoryl-specific antibodies (Fig. 2C). Incubation of fully phosphorylated yeast CTD with Ssu72 shows the rapid loss of pSer5 signal as detected by the pSer5-specific antibody (3E8) over time (Fig. 2C). In contrast, the pSer2 signal during the dephosphorylation increased first because the removal of pSer5 allows better pSer2 antibody (3E10) binding (2). The level of pSer2 eventually starts to drop after 5 h of incubation. Overall, when Ssu72 dephosphorylates full-length CTD with both pSer2 and pSer5 present, it dephosphorylates pSer5 rapidly, leaving pSer2 as an intermediate species that undergoes very slow dephosphorylation.
Figure 2.
Fast–slow two-phase dephosphorylation of CTD by Ssu72. Three independent biological duplicates have been sampled for each assay with consistent results. One representative of the samples is shown in each panel. The color scheme of the trace is depicted at the top right-hand corner. Samples with no kinase/phosphatase treatment are used as a control and shown as a black trace. Samples treated in saturating condition with the kinase Erk2 are shown as a blue trace, and samples further treated by Ssu72 are shown as a gold trace. Mass labels indicate m/z at peak intensity. “#P” notation indicates an approximate number of phosphates added based on mass shift. A general figure legend has been depicted at the top right-hand corner. A, a portion of MALDI-MS spectra of WT GST-yCTD variant treated with Erk2 (blue) followed by dephosphorylation with Ssu72 (gold) with the control shown in black. B, electromobility shift assay using a 10% native gel for Erk2-treated GST-yCTD after the indicated time of Ssu72 treatment. C, immunoblotting of Ssu72-mediated dephosphorylation of GST-yeast CTD blotted by pSer5 (3E8) and pSer2 (3E10) antibodies. The error bars represent the S.E. D, portion of MALDI-MS of 3C protease–digested GST-5CTD treated with Erk2 (blue) followed by dephosphorylation with Ssu72 (gold) with the control reaction shown in black. E, portion of MALDI-MS of 3C protease–digested GST-3CTD treated with Erk2 (blue) followed by dephosphorylation with Ssu72 (gold) with the no-kinase control reaction shown in black. F, phosphorylation localized to residues highlighted in yellow from UVPD-MS of product in E. Extracted ion chromatograms of 3C protease–digested GST-3CTD with one (green; m/z 915.4) and two phosphates (blue; m/z 942.0) are shown.
To identify the phosphorylation site with single-residue resolution, we intended to use tandem MS. However, the lack of basic residues on CTD prevents the trypsin proteolysis of CTD to smaller fragments for identification. Thus, we investigated whether the same fast–slow two-phase dephosphorylation by Ssu72 can also be observed on shorter recombinant CTDs so that we can map the shorter CTD without the necessity of trypsinolysis. Indeed, when we used Erk2 to phosphorylate a CTD variant with recombinant GST followed by five CTD repeats at the C terminus, a maximum of 10 phosphates were added, accounting for all Ser2 and Ser5 motifs (Fig. 2D, blue trace). The treatment of Ssu72 results in the removal of approximately half of the phosphoryl groups from the serine residues (Fig. 2D, gold trace). Another recombinant CTD was generated with three CTD repeats cloned C-terminal to the GST recombinant protein. The phosphorylation under saturated conditions resulted in the addition of six phosphates (Fig. 2E, blue trace), up to half of which were removed with Ssu72 treatment (Fig. 2E, gold trace). To identify the position of phosphorylation with single-residue resolution, we applied UV photodissociation (UVPD) using 193-nm photons as an alternative to existing collision- and electron-based activation methods for tandem MS analysis (30). UVPD provides several compelling advantages in the context of MS/MS analysis of the CTD (31). First, because the UVPD process is not modulated by mobile protons (like collision-induced dissociation), UVPD generates rich fragmentation patterns for both positively and negatively charged peptides. This makes UVPD especially suitable for highly phosphorylated peptides like those found in the CTD. Second, UVPD is a fast high-energy activation method that does not dislodge labile modifications, making it well-suited for analysis of phosphopeptides (unlike collision-induced dissociation). Both of these advantages make UVPD a natural choice for CTD analysis. Using UVPD-MS on the product of Ssu72 dephosphorylation, we identified that the major species of dephosphorylation are consistent with a mixture of CTD peptides with pSer2 at different heptads (Fig. 2F). Overall, our results on CTD peptides and recombinant CTD proteins support a model in which Ssu72 preferentially dephosphorylates pSer5 rapidly but can also dephosphorylate pSer2 with much lower activity.
Structural determinants for Ssu72 recognition of pSer2
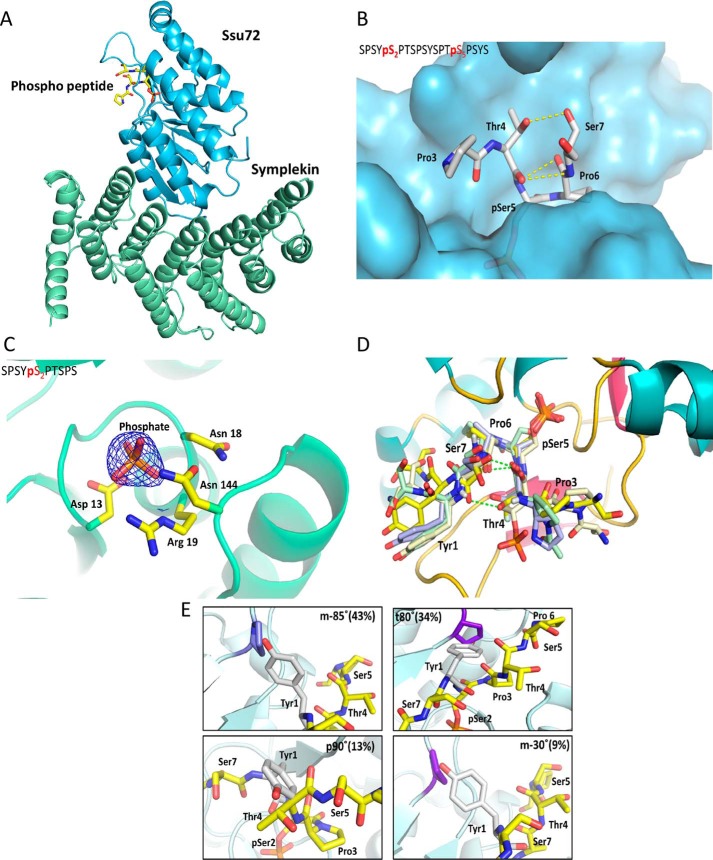
Because both Ser2 and Ser5 from the CTD have similar flanking residues but their phosphorylation states are linked to different biological functions, we wondered why Ssu72 favors pSer5 over pSer2 as substrate and used crystallography to identify structural elements that distinguish the two SP motifs. We incubated a catalytically inactivated Ssu72 (C13D/D144N) complex with a 19-mer CTD peptide with Ser5 and Ser2 phosphorylated on two separate heptad repeats (Table S1) (Fig. 3A). When both phosphoryl sites are available, the crystal structure reveals high positive density at the active site best fitted with pSer5 bound, whereas pSer2 extends outward into the bulk solvent (Figs. 3, A and B, and S2A). We next crystallized Ssu72 with a CTD peptide containing only pSer2. A much smaller positive density with a tetrahedral shape was observed in the active site, consistent with a phosphate or sulfate group (Fig. 3C). From our previous experience working with Ssu72, such electron density has often been observed in the active site even when no phosphoryl substrate is included in crystallization. The lack of electron density at the active site for the CTD peptide containing pSer2 and the weak kinetic activity of pSer2 peptide both suggest the lack of stronger interaction between the pSer2 peptide and Ssu72. Thus, consistent with the biochemical assays, our crystal structures also indicate that Ssu72 recognizes pSer5 as a substrate much better than pSer2.
Figure 3.
Structural rationale for preference to dephosphorylate Ser5 position by Ssu72. A, overall structure of the Ssu72 C13D/D144N (shown in blue)–symplekin (shown in green) complex with the phosphoryl-CTD peptide (shown in yellow) B, surface representation of the Ssu72–symplekin C13D/D144N complex (shown in blue) soaked with the peptide Ser2-Ser5 (sequence details in Table S1) represented as sticks with carbon atoms colored white and intramolecular hydrogen bond holding the tight β-turn conformation of the peptide highlighted as yellow dashes. C, 2Fo − Fc map (contoured at 1.8σ) shown as a blue mesh for the Ser2 peptide soaked in Ssu72–symplekin C13D/D144N complex. D, superimposition of Ssu72–peptide complex structures of PDB codes 3P9Y (green), 3O2Q (blue), and 4IMI (light yellow) onto the structure of Ssu72 bound to the doubly phosphorylated Ser2-Ser5 peptide (obtained in this study; shown in yellow). E, the four different rotamers of the tyrosine most frequently observed modeled in the active site of Ssu72 assuming pSer2 is bound to the active site.
To understand why Ssu72 strongly prefers Ser5 dephosphorylation even though the flanking residues of Ser2 are quite similar (Y1S2P3 versus T4S5P6), we analyzed all published X-ray crystal structures of Ssu72 bound with various CTD peptides and noticed that all substrates adopt the same configuration of a sharp β-turn (Fig. 3D). In this configuration, the proline residue next to the phosphoryl-serine is recognized by a hydrophobic pocket of Ssu72 in cis-proline configuration, regardless of flanking residue PTM states, peptide length, or the presence of scaffold protein symplekin (Fig. 3D). Although cis-proline only accounts for 10–20% of the overall proline population, the flanking cis-proline to substrate appears to be a prerequisite for Ssu72 recognition as peptidomimetics with a nonrotatable trans-proline cannot be bound to the active site (32). To retain the same substrate-bound configuration at the active site with cis-proline bound in the hydrophobic pocket when pSer7 is recognized at the active site, the pSer7 peptide reverses the orientation of the N and C termini of the peptide (Fig. S2B) (24). The 180° flip of the peptide directionality allows the same cis-proline placed in the hydrophobic pocket but loses favorable interactions that stabilize pSer5 recognition, resulting in much weaker activity against pSer7.
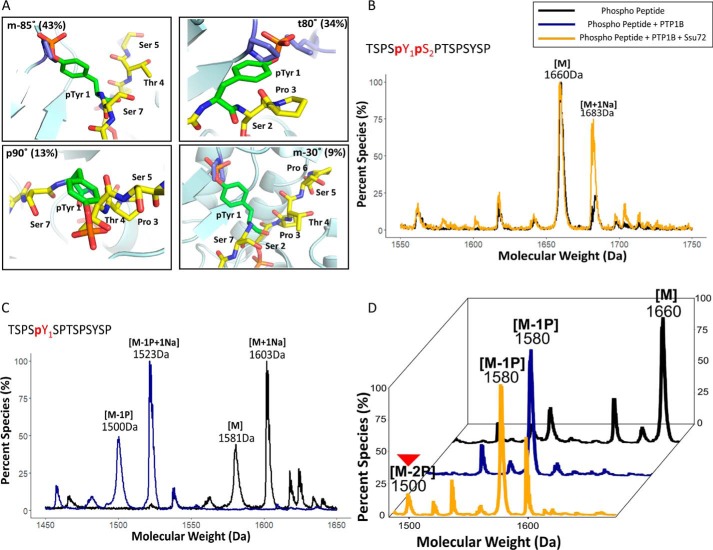
Because Ssu72 appears to bind each of its substrates in an identical configuration (Fig. 3D), the recognition of the pSer2 CTD peptide is likely retained with Pro3 in the cis-proline conformation and bound in the hydrophobic pocket close to the active site. However, such an arrangement would place the Tyr1 with its bulky side chain in the elongated tunnel of the Ssu72 active site (Fig. 3E). When we modeled the flanking residues into the active site next to pSer2, we noticed that Tyr1 could barely be fitted into the tight β-turn (Fig. 3E). The bulky side chain of tyrosine causes unfavorable van der Waals contacts or even steric clashes in all four of its most frequently used rotamer states (33) (Fig. 3E). The most common tyrosine rotamer observed in folded proteins (called m-85° based on its torsion angle with 44% occurrence frequency) would place the phenol ring of Tyr1 rather close to Pro53 of Ssu72 (1.2 Å). If Tyr1 adopts a t80° configuration instead (∼34% frequency), the hydroxyl group of the Tyr1 side chain would directly collide with Pro46 of Ssu72. The p90° rotamer (observed in 19% of protein structures) would clash into the ligand itself, and the m-30° rotamer (9% appearance) also places the Tyr1 side chain in an unfavorable van der Waals contact with Pro53 and Lys44 (Fig. 3E). Considering the dynamic states of the protein, the Tyr1 side chain would have to adopt a rotamer configuration not energetically favored to maintain the substrate-binding mode for Ssu72 recognition. Thus, the bulky side chain of Tyr1 places pSer2 substrate at a disadvantage to form the substrate-recognition mode, distinguishing it from the more favorable pSer5 substrate.
Tyr1 differentiates the two SP motifs for Ssu72 dephosphorylation
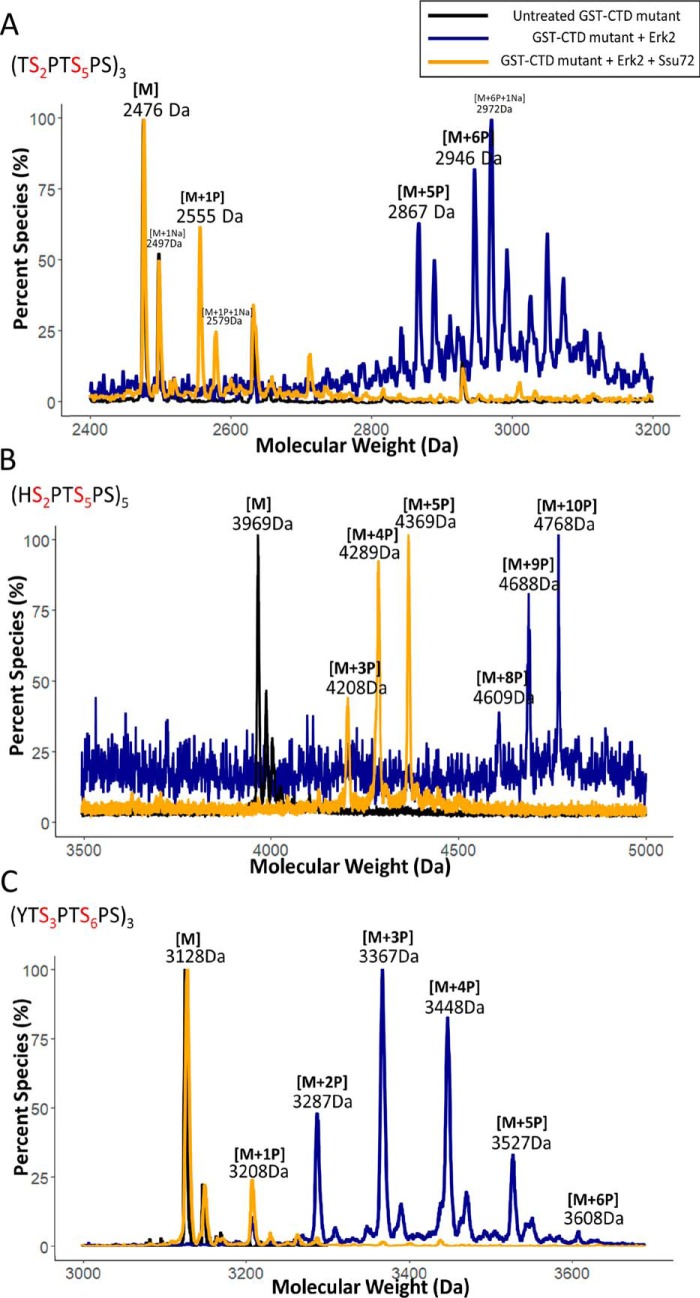
Our structural studies suggest that the residue preceding the phosphorylated CTD SP motif allows Ssu72 to differentiate the two different phosphoserine sites (pSer5 versus pSer2). To evaluate whether the identity of the residue preceding the phosphorylated SP motif is critical to Ssu72 specificity, we engineered CTDs with various mutations adjacent to the SP motif (Fig. S1) and analyzed the dephosphorylation process by Ssu72 when kinases phosphorylate all SP motifs. We speculate that the bulkiness of the flanking Tyr1 sterically hinders the cis-proline configuration for substrate recognition at pSer2 compared with pSer5 by Ssu72. Thus, we replaced Tyr1 with a small residue to test whether the removal of the bulky side chain is sufficient to eliminate the substrate differentiation. We generated a three-repeat CTD with each Tyr1 mutated to threonine (Fig. 4A), thus making both Ser2 and Ser5 sites preceded by a small residue with no potential steric hindrance for the cis-proline configuration formation. We then treated these TSPTSPS repeats with a saturating amount of Erk2, resulting in the phosphorylation of each SP motif (Fig. 4A, blue trace). When using these hyperphosphorylated CTDs as a substrate for Ssu72, all phosphorylations from the hyperphosphorylated substrate were removed rapidly with no discrimination as detected by MALDI-TOF (Fig. 4A, gold trace). This is dramatically different from WT CTD where Ssu72 shows fast–slow two-phase dephosphorylation (Fig. 2) where pSer5 is removed rapidly and pSer2 is removed much slower (Fig. 2, A and F). The result of this mutant strongly supports our hypothesis that bulkiness of the residue in front of the SP motif protects pSer2 from Ssu72 dephosphorylation.
Figure 4.
Differentiation of two SP motifs of CTD by Ssu72. Three independent biological duplicates have been sampled for each assay with consistent results. One representative of the samples is shown in each panel. The color scheme of the trace is depicted at the top right-hand corner. A, portion of MALDI-MS of GST-3CTD variant with every Tyr replaced by Thr in each heptad repeat. B, portion of MALDI-MS of GST-5CTD variant with every Tyr replaced by His in each heptad repeat. C, an engineered octamer repeat of three with a Thr residue inserted after Tyr. For sample C, the variants were first treated with Erk2 (blue) followed by dephosphorylation with Ssu72 (gold) with nonphosphorylated control shown in black. Mass labels indicate m/z at peak intensity. “M” notation indicates the substrate peptide, and “+#P” notation indicates an approximate number of phosphates added based on mass shift.
To further establish that the steric hindrance of the residue in front of the SP motif disfavors the dephosphorylation by Ssu72, we replaced Tyr1 with another bulky residue to see whether it also allows distinction of Ser5/Ser2. When we generated a five-heptad repeat with Tyr1 replaced with histidine at each repeat (Fig. 4B), Erk2 phosphorylated all 10 SP motifs (Fig. 4B, blue trace), but Ssu72 only removed approximately half of these phosphorylations when further dephosphorylation ground to a halt (Fig. 4B, gold trace). This is similar to the consensus sequence with tyrosine in the first position when the dephosphorylation by Ssu72 exhibits fast–slow two-phase dynamics (Fig. 2C). Careful inspection of the structure of Ssu72 reveals that, when modeled into the Tyr1 position, the histidine side chain is placed in the active site with unfavorable van der Waals interactions with Ssu72, similar to Tyr1 (Fig. S3). Thus, the presence of any bulky residue in front of a CTD SP motif appears to attenuate Ssu72 dephosphorylation activity.
We also tested whether the steric hindrance can be relieved by inserting a small residue in between a bulky residue and the SP motif to allow Ssu72 to dephosphorylate SP motifs more efficiently. We generated a three-CTD octamer containing the repeating sequence YTSPTSPS (Fig. 4C), which has Thr inserted between Tyr1 and the Ser2-Pro3 SP motif. The insertion of a residue with a small side chain allows all SP motifs to be dephosphorylated rapidly by Ssu72 (Fig. 4C, gold trace). Thus, the bulkiness of the residue in the position immediately preceding the SP motif differentiates the dephosphorylation of CTD by Ssu72.
Tyr1 phosphorylation prevents the dephosphorylation of Ser2 by Ssu72
Our results for the full CTD suggest that the activity of Ssu72 against pSer2 is low with Tyr1 hindering the effective formation of substrate-recognition configuration such that the pSer5 is preferably dephosphorylated. Because the accuracy of dephosphorylation of pSer5/Ser2 is important during transcription, we wondered whether PTM on flanking residues will impact Ssu72 specificity. Previously, we have shown that Thr4 phosphorylation reduced the dephosphorylation of pSer5 by Ssu72 by 3–4-fold (29). With the weak activity of pSer2 by Ssu72, we wanted to quantify how Tyr1 phosphorylation impacts the Ssu72 phosphatase activity. Both Tyr1 and Ser2 phosphorylation species are observed using ChIP-Seq in the transcription elongation stage (1). Recent reports using MS to identify CTD phosphorylation of engineered RNA polymerase II identified the existence of a Tyr1-Ser2 double phosphorylation motif (50 and 20% of all diphosphorylated heptads in yeast and human, respectively), even though the abundance of double phosphorylation is substantially lower than that of single phosphorylation (34). If Ssu72 binds substrate in the cis-proline conformation as found in all Ssu72 structures, phosphorylation of Tyr1 should further hinder the formation of the tight β-turn required for substrate recognition of pSer2 by Ssu72 (Fig. 5A). To test whether pTyr1 indeed prevents dephosphorylation of pSer2 in vitro, we used a synthetic peptide containing Tyr1 and Ser2, both phosphorylated on the same heptad as substrate, and incubated it with Ssu72. Little dephosphorylation can be detected in the sensitive MALDI-TOF assay (Fig. 5B, gold trace). Thus, phosphorylation of the preceding Tyr1 prevents pSer2 from being dephosphorylated by Ssu72.
Figure 5.
Phosphorylation of Tyr1 protects the dephosphorylation of Ser2 by Ssu72. A, modeling phosphorylated Tyr1 with CTD pSer2 in the active site for Ssu72 binding. The isomeric states of Tyr1 are the four most frequently observed rotamers. B, mass spectrometry analysis using MALDI-TOF for the synthetic phosphopeptide Tyr1-Ser2 before Ssu72 treatment (black) and after Ssu72 treatment (gold). Mass labels indicate m/z at peak intensity. C, mass spectrometry analysis using MALDI-TOF for the synthetic phosphopeptide control shown in black and PTP1B treatment shown in blue. D, mass spectrometry analysis using MALDI-TOF for the synthetic phosphopeptide after treatment with PTP1B and then followed by Ssu72. Three independent biological duplicates have been sampled for each assay with consistent results. One representative of the samples is shown in each panel. The trace after PTP1b treatment is in blue, and the trace after Ssu72 treatment is in gold. Mass labels indicate m/z at peak intensity. “M” notation indicates the substrate peptide, and “−#P” notation indicates an approximate number of phosphates removed based on mass shift.
To establish that the phosphorylation of Tyr1 inhibits pSer2 from being dephosphorylated by Ssu72, we tested whether this protection from dephosphorylation could be reversed by removing the Tyr1 phosphorylation mark. PTP1b is a potent tyrosine phosphatase (35). Although PTP1b is not a physiological phosphatase for RNA polymerase II in vivo, its activity against pTyr1 allows us to use it as a tool to interrogate the effect of pTyr1 on the dephosphorylation of pSer2. We first validated the in vitro specificity of PTP1b on every single phosphorylable residue of the CTD heptad repeat and found that pTyr1 is the only residue it dephosphorylates (Fig. 5C), whereas other residues (Ser2,5,7 and Thr4) are not subject to dephosphorylation by PTP1b (Fig. S4). When we treated Tyr1-Ser2 doubly phosphorylated CTD peptide with PTP1b, one phosphate was removed with a loss of 80 Da, consistent with the phosphate on pTyr1 (Fig. 5D, blue trace). Following PTP1b treatment, we next added Ssu72 to the reaction, which resulted in a product appearing at 1500 Da, which corresponded to a peptide missing both phosphate groups (Fig. 5D, gold trace). This is in contrast to the double phosphorylated peptide treated by Ssu72 alone where no phosphate was removed (Fig. 5B). Thus, once Tyr1 phosphorylation is removed, pSer2 is subject to dephosphorylation by Ssu72. This suggests that the timing of Tyr1 phosphorylation/dephosphorylation can be important for ensuring proper Ser2 phosphorylation state near the end of transcription.
Discussion
As the primary mechanism for PTM, phosphorylation/dephosphorylation require high precision for the accurate transmission of information. Phosphatases, which usually have a pan-spectrum enzymatic activity against phosphorylated substrates, rely heavily on the cellular location of substrate specificity (35). In hyperphosphorylated proteins where different phosphorylation sites encode various functions, however, it is not yet known how these phosphatases distinguish different phosphoryl sites once they are recruited to their targets. The CTD of RNA polymerase II is highly phosphorylated with five phosphorylable residues in each of its repetitive heptad repeats, which begs the question of how CTD phosphatases are capable of removing the correct phosphate marks at the right stage in transcription. During elongation, for example, pSer5 of the CTD is dephosphorylated rapidly, whereas Ser2 phosphorylation is still steadily increasing (4). Herein, we show that Tyr1 residues of the substrate play a critical role in distinguishing the almost identical SP motifs. The major phosphatase involved in elongation, Ssu72, has a unique active site that requires the proline residue in the substrate motif to be in cis configuration. This requirement disfavors pSer2 whose neighboring bulky Tyr1 side chain hinders the formation of an energetically favorable CTD configuration recognized by Ssu72. Using CTD mutants, we proved that the bulky side chain of Tyr1 is responsible for the differentiation between the two SP motifs in CTD. Intriguingly, pSer2 activity by Ssu72 is abolished when the preceding Tyr1 is phosphorylated, the combination of which has been detected in cells (34).
Tyr1 is highly conserved across all species, even in the highly divergent sequence of Drosophila melanogaster CTD where only three of 46 heptads show consensus (36). Furthermore, even conservative mutations (i.e. to phenylalanine) of all Tyr1 positions in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and vertebrates result in either lethal or significant growth-deficient phenotypes (12). Tyr1 phosphorylation is critical for the stability of RNA polymerase II, termination factor recruitment, and antisense transcription (12–15). Our research provides a plausible mechanism for the observed diversified biological implication of Tyr1 conservation and its phosphorylation by regulating the activity of CTD modifiers such as Ssu72 to impact the phosphorylation state of other CTD residues. In human cells, Tyr1 phosphorylation occurs at the beginning of transcription, placing it at the critical point to influence the subsequent events of CTD modification. Tyr1 and Ser2 phosphorylations overlap during transcriptional elongation in vivo as shown in ChIP-Seq analysis (37), and tandemly phosphorylated Tyr1-Ser2 have been detected by MS both in human and yeast RNA polymerase II (34, 38). Our results show that not only does Tyr1 greatly reduce the activity of Ssu72 against pSer2 in vitro, but as a further safety proof, Tyr1-Ser2 double phosphorylation can prevent inappropriate dephosphorylation of pSer2 by Ssu72. This strategy can potentially provide timely control of the phosphorylation state of specific residues in the CTD for the accurate regulation of transcription.
Conclusion
In this investigation, we identified that Tyr1 of the CTD consensus heptad repeat is critical in the differentiation of the two SP motifs for dephosphorylation by CTD phosphatase Ssu72. This allows the removal of Ser5 phosphorylation marks while allowing phosphorylation of Ser2 to accumulate. Phosphorylation of Tyr1 further prevents any inappropriate dephosphorylation of pSer2 by Ssu72. This study reveals a new biological function of Tyr1 by determining the accurate removal of the phosphorylation on other CTD residues.
Experimental procedures
Phosphorylated CTD peptides used in this study were custom-ordered from Anaspec and CPC BioSciences, and their sequences have been listed in Table S1. The gene blocks and oligos for the 3CTD, 5CTD, and its variants are described in Fig. S1 were ordered from Integrated DNA Technologies, Inc. and GenScript. All other reagents and chemicals were purchased from Sigma-Aldrich unless specified otherwise.
Protein expression and purification
Purification of Ssu72
D. melanogaster Ssu72 and symplekin constructs and expression plasmids used in this study are the same as those described previously (29). Ssu72–symplekin was also purified as detailed before (23, 29). Briefly, the Ssu72 and symplekin plasmids were separately transformed into Escherichia coli BL21 (DE3) cells, and these cells were subsequently grown in Luria-Bertani (LB) medium with 50 μg/ml kanamycin at 37 °C. When the OD600 of the culture reached around 0.4–0.6, expression was induced by adding isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mm, after which the temperature was turned down to 16 °C, and the cultures were grown for another 18 h. The cells were pelleted by centrifugation, resuspended in lysis buffer, and lysed by sonication. High-speed centrifugation was used to separate the cellular debris from the aqueous fraction, which was passed through a nickel-nitrilotriacetic acid column (Qiagen); the His-tagged protein bound to the nickel beads and was eluted with a buffer containing a high concentration of imidazole. Thrombin protease was used to cleave off the N-terminal His tag during an overnight dialysis at 4 °C, after which the proteins were passed through ion-exchange and size-exclusion chromatography columns for further purification. The Ssu72 and symplekin purified separately by the above means were then mixed together and incubated at 4 °C overnight to allow for complex formation. The complex was specifically separated from any uncomplexed excess protein by passing this mixture through a gel-filtration column. Homogeneity of the complex was confirmed by running the gel-filtration fractions on an SDS-polyacrylamide gel. The inactive Ssu72 C13D/D144N–symplekin, which was used in all the crystallization experiments, was purified using the same method as described above.
Purification of the kinase Erk2
Human Erk2 used to phosphorylate the GST-CTD and its variants was expressed from pET-His6-ERK2-MEK1_R4F_coexpression vector, which was a gift from Melanie Cobb (Addgene plasmid number 39212). E. coli BL21 (DE3) cells, transformed with the above vector, were grown in LB medium at 37 °C in the presence of 50 μg/ml ampicillin antibiotic to maintain the plasmid selection. When the OD600 of this culture reached 04–0.6, 0.5 mm isopropyl β-d-thiogalactopyranoside (final concentration) was added to induce expression. Erk2 was coexpressed with MEK1 by growing this culture after induction at 37 °C for another 4 h before pelleting the cells by centrifugation at 5000 × g for 20 min. The cells were lysed by sonicating in a buffer containing 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 15 mm imidazole, 10% glycerol, 0.1% Triton X-100, and 10 mm β-mercaptoethanol (BME). The lysate was cleared by centrifugation at 15,000 rpm for 45 min at 4 °C. The supernatant was initially purified by affinity chromatography using nickel-nitrilotriacetic acid (Qiagen) beads, and the target protein bound to the beads via its His6 tag was eluted with buffer containing 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 200 mm imidazole, and 10 mm BME. The eluted protein sample was dialyzed against buffer containing 20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 10 mm BME, and 20% glycerol, after which it was diluted further to a final concentration of 10% glycerol and purified by anion-exchange chromatography with Mono Q resin (GE Healthcare). An NaCl gradient (50–1000 mm) was used to elute the protein from the column into fractions that were analyzed by SDS-PAGE. Fractions containing phosphorylated protein were pooled and concentrated, if necessary, in Vivaspin columns (Sartorius).
Purification of engineered GST-CTD variants
The CTD (three repeats and five repeats) and its variants (sequences are described in Fig. S1) were amplified from synthetic DNA templates generated by Integrated DNA Technologies, Inc. These were then cloned into a pET28a (Novagen) derivative vector encoding an N-terminal His tag followed by GST tag and a 3C protease site to generate the GST-CTD constructs.
GST-CTD proteins were expressed and purified as described in established protocols published previously (30). Concisely, the cell pellet was lysed by sonication in a buffer at pH 8.0 containing 500 mm NaCl, 0.1% Triton X-100, and 10% glycerol. The cell debris was then separated by high-speed centrifugation at 16,000 × g for 45 min at 4 °C, and the supernatant fraction was passed through a nickel-affinity column to purify the His-tagged GST-CTD variants, which were eluted by using a high concentration of imidazole. The eluate was purified further by passing it through a gel-filtration column in the buffer at pH 8.0 with 50 mm NaCl and 10 mm BME. Fractions containing protein were run on an SDS-polyacrylamide gel for size verification and ultimately concentrated and aliquoted before flash freezing in liquid nitrogen.
Purification of PTP1B phosphatase
The expression plasmid containing PTP1b was kindly provided by Dr. Zhong Yin Zhang from Purdue University. E. coli BL21 (DE3) cells containing overexpressed His-tagged PTP1B were lysed, and the tagged protein was purified using the standard protocols described in earlier sections. The nickel column eluate was further purified by gel filtration in a buffer containing 100 mm MES, pH 6.5, 1 mm EDTA, 50 mm NaCl, and 10 mm BME. The fractions containing protein were concentrated before flash freezing in liquid nitrogen.
Generating phosphorylated GST-CTD substrate
20 μg of GST-3CTD, GST-5CTD, and other GST-CTD variants (described in Fig. S1) were treated with 3 μg of Erk2 prepared as above in a buffer containing 50 mm Tris-Cl, pH 7.5, 50 mm MgCl2, and 2 mm ATP for 16 h at 30 °C.
Phosphatase assay
Ssu72
100 μm synthetic peptide (described in Table S1) was treated with 10–20 μm Ssu72–symplekin complex in 100 mm MES, pH 6.0, at 28 °C for 90 min before being subjected to desalting for MALDI analysis. Similarly, 1 μg of kinase-treated GST-CTD sample was treated with 10–20 μm Ssu72–symplekin in 100 mm MES, pH 6.0. The samples were subsequently run on a 15% native gel for EMSA analysis or were treated with 3C protease before desalting for MALDI.
PTP1B
100 μm peptide (described in Table S1) was treated with 100 μm PTP1B in 100 mm HEPES, pH 7.5, at 28 °C for 60 min before being subjected to desalting for MALDI analysis.
Immunoblotting
60 μg of Erk2-treated GST-yCTD was incubated with 17 μg of Ssu72–symplekin complex for various lengths of time in a buffer containing 100 mm MES, pH 6.5, and 1 mm EDTA. Samples were removed at respective time points, and the reaction was quenched by adding loading dye and heating at 95 °C for 2 min. 0.55 μg of this substrate was then spotted three times (for three technical replicates) on two 0.45-μm nitrocellulose membranes (Bio-Rad) (one for each antibody) and allowed to air dry for 20 min. The membranes were then blocked in 5% BSA in 1× TBST (Tris-buffered saline with 0.2% Tween) for 1 h at room temperature. For probing the CTD Ser2 phosphorylation, the membrane was incubated with the 3E10 antibody (Millipore catalog number 04-1571-1) at a dilution of 1:600 prepared in blocking solution. To detect Ser5 phosphorylation, the second membrane prepared was incubated with the 3E8 antibody (Millipore catalog number 04-1572) prepared at 1:600 dilution in blocking solution. The membranes were incubated with the respective antibodies overnight at 6 °C on a shaker, after which the membranes were washed with a solution of TBST five times for 5 min each. The membranes were then incubated with a 1:30,000 dilution of secondary antibody in blocking buffer for 2 h at room temperature. After another set of washes, the membranes were exposed to SuperSignal West Pico Chemiluminescent Substrate (Pierce catalog number 34079) according to the manufacturer's instructions before imaging using a G:BOX gel doc system (Syngene). The images were analyzed, and the spot intensity was quantified in ImageJ (39). Statistical analysis and p value calculation were done using the t test function in Microsoft Excel. The column scatters plots with mean and S.D. were plotted in RStudio (40). Three independent biological replicates were prepared and imaged as described, all revealing the same trend.
MALDI-TOF MS
Trifluoroacetic acid (TFA) was added to proteins at a concentration at 0.1 mg/ml after 3–4 h of 3C protease treatment or 100 μm peptide. The final concentration of TFA was around 0.1–0.4%, and pHydrion pH test paper (Sigma) was used to verify that the pH was lesser than 4. Samples were subsequently desalted and concentrated using ZipTips with 0.6 μl of C18 resin (Millipore), a wash solution of water/methanol/TFA (95:5:0.1, v/v/v) was used, and samples were finally eluted from the resin in acetonitrile/water/TFA (50:50:0.1, v/v/v). 0.5 μl of 2,5-dihydroxybenzoic acid matrix (saturated in acetonitrile/water, 50:50, v/v) was spotted on a MALDI metal sample plate, and 0.5 μl of the desalted sample prepared above was added to that and thoroughly mixed by pipetting several times. These drops were allowed to air dry at room temperature to form crystals on the sample plate. MALDI-TOF spectra were acquired on a Voyager-DE PRO (Applied Biosystems) instrument; all measurements were made in the positive mode; the preloaded ACTH_linear parameter settings were used for all peptides, GST-3CTD, GST-5CTD, and its variants; the myoglobin_linear parameter settings were used for the GST-yCTD. Laser intensity was adjusted manually to obtain the highest signal to noise. The spectra obtained were postprocessed in Data Explorer (Applied Biosystems) with the noise removal utility, which was used to enhance the visualization of certain spectra when required. All MALDI assays were carried out in three independent biological replicates. The final graphs were plotted in R using the ggplot package (40).
Liquid chromatography–MS for phosphate localization
Following Erk2 and Ssu72 treatment, GST-CTD proteins (∼5 μg) were digested with 3C protease for 5–6 h. Resultant 3CTD peptides were desalted with PepCleanTM C18 spin columns according to the manufacturer's instructions and eluted with water/acetonitrile (30:70, v/v). The solvent was evaporated, and peptides were reconstituted in water/acetonitrile/formic acid (98:2:0.1, v/v/v) for subsequent LC separation. Separations were carried out on a Dionex Ultimate 3000 Nano liquid chromatograph configured for direct injection. PicoFritTM 75-μm-inner-diameter analytical columns (New Objective, Woburn, MA) were packed in-house to 20 cm with 1.8-μm UChrom C18 (NanoLCMS Solutions, Oroville, CA). Mobile phases A and B were water and acetonitrile, respectively, each containing 0.1% formic acid. Separations occurred over a two-step gradient as follows: mobile phase B was increased from 2 to 20% in 15 min and then from 20 to 35% in another 13 min. The flow rate was maintained at 0.300 μl/min during the separation.
An OrbitrapTM Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a Coherent ExciStar XS excimer laser operated at 193 nm and 500 Hz was used in tandem with the liquid chromatograph. Two pulses at 2 mJ were used for UVPD. A targeted analysis was used to select the 3+ charge state of the mono- and diphosphorylated 3CTD peptides, corresponding to m/z 915.4 and 942.0, respectively. Orbitrap resolving power for MS1 and tandem MS spectra was set to 60,000 and 30,000 at m/z 200, respectively.
MS/MS spectra were deconvoluted to neutral forms using the Xtract algorithm and matched to the nine UVPD ion types (a, a•, b, c, x, x•, y, y − 1, and z) within 10-ppm accuracy using ProSight Lite. Phosphate localization was performed by adding a phosphate group (+79.97 Da) to possible serine, threonine, and tyrosine residues to optimize characterization scores in ProSight Lite.
Crystallization, crystal soaking of Ssu72 with CTD peptides, and structure determination
A purified activity-deficient variant of Ssu72 (C13D/D144N) was incubated with symplekin in a 1.2:1 ratio overnight in a cold room and then purified with gel-filtration chromatography. The protein complex was then crystallized in a solution containing 12% PEG 3350 (w/v) and 100 mm HEPES, pH 7.5, at room temperature. Ssu72 C13D/D144N–symplekin crystals were soaked with 0.1 mm pSer2/Ser5 double phosphorylated CTD peptides and 10 mm pSer2 peptide to obtain the phosphatase–CTD peptide complex. The crystals were cryoprotected with mother liquor containing 15% glycerol (v/v) before vitrification in liquid nitrogen.
Crystallographic data for Ssu72–symplekin crystals soaked with CTD peptides were collected at the beamline BL 5.0.3 in the Advanced Light Source (ALS) at National Lawrence Laboratory. All diffraction data were processed with HKL2000 (42). The structures were determined by molecular replacement using the structure of Drosophila Ssu72–symplekin complex (PDB code 4IMI) as a model with PhaserMR in the Phenix Program Suite (41). The ligand was built iteratively in Coot (42), and computational refinements were performed using Phenix Refine (43). The quality of the final refined structures was evaluated by MolProbity (44). The final statistics for data collection and structure determination are summarized in Table S2.
Author contributions
S. I., S. N. S., and Y. Z. conceptualization; S. I., S. N. S., W. Y., N. T. B., B. L., K. S., W. L. M., and J. S. B. data curation; S. I., S. N. S., and Y. Z. formal analysis; S. I. and Y. Z. supervision; S. I., J. S. B., and Y. Z. funding acquisition; S. I., S. N. S., W. Y., J. S. B., and Y. Z. validation; S. I., S. N. S., N. T. B., B. L., and Y. Z. writing-original draft; S. I. and Y. Z. project administration; S. I., S. N. S., and Y. Z. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. J. E. Mayfield for critical comments on the manuscript. Voyager DE-PROT MALDI MS data were collected in the University of Texas (UT) at Austin Proteomics Facility. Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged.
This work was supported by National Institutes of Health Grants R01 GM104896 (to Y. Z.) and R01 GM125882 (to Y. Z. and J. S. B.) and Welch Foundation Grant F-1778 (to Y. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Tables S1 and S2.
The atomic coordinates and structure factors (code 6NPW) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CTD
- C-terminal domain
- PTM
- post-translational modification
- EMSA
- electrophoretic mobility shift assay
- UVPD
- UV photodissociation
- BME
- β-mercaptoethanol
- yCTD
- yeast CTD
- MALDI-TOF
- matrix-assisted laser desorption ionization-time of flight.
References
- 1. Eick D., and Geyer M. (2013) The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- 2. Corden J. L. (2013) RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem. Rev. 113, 8423–8455 10.1021/cr400158h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buratowski S. (2003) The CTD code. Nat. Struct. Biol. 10, 679–680 10.1038/nsb0903-679 [DOI] [PubMed] [Google Scholar]
- 4. Harlen K. M., and Churchman L. S. (2017) The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 10.1038/nrm.2017.10 [DOI] [PubMed] [Google Scholar]
- 5. Jeronimo C., Bataille A. R., and Robert F. (2013) The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem. Rev. 113, 8491–8522 10.1021/cr4001397 [DOI] [PubMed] [Google Scholar]
- 6. Schauer T., Tombácz I., Ciurciu A., Komonyi O., and Boros I. M. (2009) Misregulated RNA Pol II C-terminal domain phosphorylation results in apoptosis. Cell. Mol. Life Sci. 66, 909–918 10.1007/s00018-009-8670-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsin J.-P., and Manley J. L. (2012) The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho E. J., Takagi T., Moore C. R., and Buratowski S. (1997) mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11, 3319–3326 10.1101/gad.11.24.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Komarnitsky P., Cho E.-J., and Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 10.1101/gad.824700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tietjen J. R., Zhang D. W., Rodríguez-Molina J. B., White B. E., Akhtar M. S., Heidemann M., Li X., Chapman R. D., Shokat K., Keles S., Eick D., and Ansari A. Z. (2010) Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17, 1154–1161 10.1038/nsmb.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cho H., Kim T. K., Mancebo H., Lane W. S., Flores O., and Reinberg D. (1999) A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13, 1540–1552 10.1101/gad.13.12.1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsin J.-P., Li W., Hoque M., Tian B., and Manley J. L. (2014) RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife 3, e02112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Descostes N., Heidemann M., Spinelli L., Schüller R., Maqbool M. A., Fenouil R., Koch F., Innocenti C., Gut M., Gut I., Eick D., and Andrau J.-C. (2014) Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife 3, e02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collin P., Jeronimo C., Poitras C., and Robert F. (2019) RNA polymerase II CTD tyrosine 1 is required for efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol. Cell 73, 655–669 10.1016/j.molcel.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 15. Shah N., Maqbool M. A., Yahia Y., El Aabidine A. Z., Esnault C., Forné I., Decker T.-M., Martin D., Schüller R., Krebs S., Blum H., Imhof A., Eick D., and Andrau J.-C. (2018) Tyrosine-1 of RNA polymerase II CTD controls global termination of gene transcription in mammals. Mol. Cell 69, 48–61.e6 10.1016/j.molcel.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 16. Rosado-Lugo J. D., and Hampsey M. (2014) The Ssu72 phosphatase mediates the RNA polymerase II initiation-elongation transition. J. Biol. Chem. 289, 33916–33926 10.1074/jbc.M114.608695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirose Y., and Ohkuma Y. (2007) Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 141, 601–608 10.1093/jb/mvm090 [DOI] [PubMed] [Google Scholar]
- 18. Bartkowiak B., and Greenleaf A. L. (2011) Phosphorylation of RNAPII: to P-TEFb or not to P-TEFb? Transcription 2, 115–119 10.4161/trns.2.3.15004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnamurthy S., He X., Reyes-Reyes M., Moore C., and Hampsey M. (2004) Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 10.1016/S1097-2765(04)00235-7 [DOI] [PubMed] [Google Scholar]
- 20. Sun Z. W., and Hampsey M. (1996) Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 16, 1557–1566 10.1128/MCB.16.4.1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiang K., Nagaike T., Xiang S., Kilic T., Beh M. M., Manley J. L., and Tong L. (2010) Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729–733 10.1038/nature09391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Werner-Allen J. W., Lee C.-J., Liu P., Nicely N. I., Wang S., Greenleaf A. L., and Zhou P. (2011) cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 286, 5717–5726 10.1074/jbc.M110.197129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Zhang M., and Zhang Y. (2011) Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 434, 435–444 10.1042/BJ20101471 [DOI] [PubMed] [Google Scholar]
- 24. Xiang K., Manley J. L., and Tong L. (2012) An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 26, 2265–2270 10.1101/gad.198853.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., and Keller W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10, 1139–1150 10.1016/S1097-2765(02)00707-4 [DOI] [PubMed] [Google Scholar]
- 26. Ghazy M. A., He X., Singh B. N., Hampsey M., and Moore C. (2009) The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol. Cell. Biol. 29, 2296–2307 10.1128/MCB.01514-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He X., Khan A. U., Cheng H., Pappas D. L. Jr., Hampsey M., and Moore C. L. (2003) Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17, 1030–1042 10.1101/gad.1075203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carter S. G., and Karl D. W. (1982) Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods 7, 7–13 10.1016/0165-022X(82)90031-8 [DOI] [PubMed] [Google Scholar]
- 29. Luo Y., Yogesha S. D., Cannon J. R., Yan W., Ellington A. D., Brodbelt J. S., and Zhang Y. (2013) Novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem. Biol. 8, 2042–2052 10.1021/cb400229c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayfield J. E., Robinson M. R., Cotham V. C., Irani S., Matthews W. L., Ram A., Gilmour D. S., Cannon J. R., Zhang Y. J., and Brodbelt J. S. (2017) Mapping the phosphorylation pattern of Drosophila melanogaster RNA polymerase II carboxyl-terminal domain using ultraviolet photodissociation mass spectrometry. ACS Chem. Biol. 12, 153–162 10.1021/acschembio.6b00729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brodbelt J. S. (2014) Photodissociation mass spectrometry: new tools for characterization of biological molecules. Chem. Soc. Rev. 43, 2757–2783 10.1039/C3CS60444F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayfield J. E., Fan S., Wei S., Zhang M., Li B., Ellington A. D., Etzkorn F. A., and Zhang Y. J. (2015) Chemical tools to decipher regulation of phosphatases by proline isomerization on eukaryotic RNA polymerase II. ACS Chem. Biol. 10, 2405–2414 10.1021/acschembio.5b00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lovell S. C., Word J. M., Richardson J. S., and Richardson D. C. (2000) The penultimate rotamer library. Proteins 40, 389–408 [DOI] [PubMed] [Google Scholar]
- 34. Schüller R., Forné I., Straub T., Schreieck A., Texier Y., Shah N., Decker T.-M., Cramer P., Imhof A., and Eick D. (2016) Heptad-specific phosphorylation of RNA polymerase II CTD. Mol. Cell 61, 305–314 10.1016/j.molcel.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 35. Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., and Mustelin T. (2004) Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 10.1016/j.cell.2004.05.018 [DOI] [PubMed] [Google Scholar]
- 36. Chapman R. D., Heidemann M., Hintermair C., and Eick D. (2008) Molecular evolution of the RNA polymerase II CTD. Trends Genet. 24, 289–296 10.1016/j.tig.2008.03.010 [DOI] [PubMed] [Google Scholar]
- 37. Mayer A., Heidemann M., Lidschreiber M., Schreieck A., Sun M., Hintermair C., Kremmer E., Eick D., and Cramer P. (2012) CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336, 1723–1725 10.1126/science.1219651 [DOI] [PubMed] [Google Scholar]
- 38. Suh H., Ficarro S. B., Kang U.-B., Chun Y., Marto J. A., and Buratowski S. (2016) Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. Mol. Cell 61, 297–304 10.1016/j.molcel.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. R Core Team (2017) R: a Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 41. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 43. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.