Abstract
Histone deacetylase 5 (HDAC5) and HDAC9 are class IIa HDACs that function as signal-responsive repressors of the epigenetic program for pathological cardiomyocyte hypertrophy. The conserved deacetylase domains of HDAC5 and HDAC9 are not required for inhibition of cardiac hypertrophy. Thus, the biological function of class IIa HDAC catalytic activity in the heart remains unknown. Here we demonstrate that catalytic activity of HDAC5, but not HDAC9, suppresses mitochondrial reactive oxygen species generation and subsequent induction of NF-E2–related factor 2 (NRF2)–dependent antioxidant gene expression in cardiomyocytes. Treatment of cardiomyocytes with TMP195 or TMP269, which are selective class IIa HDAC inhibitors, or shRNA-mediated knockdown of HDAC5 but not HDAC9 leads to stimulation of NRF2-mediated transcription in a reactive oxygen species–dependent manner. Conversely, ectopic expression of catalytically active HDAC5 decreases cardiomyocyte oxidative stress and represses NRF2 activation. These findings establish a role of the catalytic domain of HDAC5 in the control of cardiomyocyte redox homeostasis and define TMP195 and TMP269 as a novel class of NRF2 activators that function by suppressing the enzymatic activity of an epigenetic regulator.
Keywords: histone deacetylase (HDAC); cardiac hypertrophy; oxidative stress; mitochondria; nuclear factor 2 (erythroid-derived 2–like factor, NFE2L2, Nrf2); cardiomyocyte; histone deacetylase; mitochondria; NRF2; oxidative stress
Introduction
Heart failure (HF)4 is estimated to afflict ∼7 million adults in the United States alone (1). Despite recent advances in the treatment of HF, patients with this condition continue to have poor long-term outcomes, highlighting a large unmet medical need (2). A hallmark of HF is cardiomyocyte hypertrophy, a process whereby cardiac muscle cells grow but do not proliferate. Cardiac hypertrophy has traditionally been viewed as a compensatory mechanism that normalizes wall stress and enhances cardiac performance in the face of a pathogenic insult. However, suppression of left ventricular hypertrophy reduces morbidity and mortality in patients with cardiovascular disease (3, 4); thus, cardiac hypertrophy is now recognized as a target for novel therapeutic intervention (5).
In the early 2000s, histone deacetylases (HDACs), which are epigenetic regulatory enzymes, were shown to serve critical roles in the control of cardiac hypertrophy (6–8). The 18 mammalian HDACs are grouped into four classes: class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 6, 7, 9, and 10), class III (SirT1–SirT7), and class IV (HDAC11). Class II HDACs are further divided into two subclasses: IIa (HDACs 4, 5, 7, and 9) and IIb (HDACs 6 and 10) (9). Class IIa HDAC5 and HDAC9 associate with pro-hypertrophic transcription factors such as myocyte enhancer factor 2 (MEF2) and block transcription of genes that are required for stress-induced growth of the heart (8, 10). In response to signals for cardiac hypertrophy, HDAC5 and HDAC9 undergo phosphorylation-dependent nuclear export, which leads to derepression of downstream target genes (11, 12).
The biological functions of class IIa HDAC catalytic domains have remained elusive. Class IIa HDACs are unable to deacetylate histones (13), and bona fide endogenous substrates of these enzymes have yet to be identified; class IIa HDAC catalytic activity can only be monitored using an artificial substrate (14). Furthermore, with regard to the heart, the deacetylase domains of HDAC5 and HDAC9 are dispensable for inhibition of cardiomyocyte hypertrophy.
In this study, we employed chemical biology and genetic approaches to elucidate roles for class IIa HDAC catalytic domains in cardiomyocytes. The findings reveal a previously unrecognized function for HDAC5 catalytic activity in the suppression of oxidative stress and antioxidant gene expression governed by NF-E2–related factor 2 (NRF2).
Results
Class IIa HDAC catalytic domain inhibition has no effect on hypertrophy of cardiomyocytes
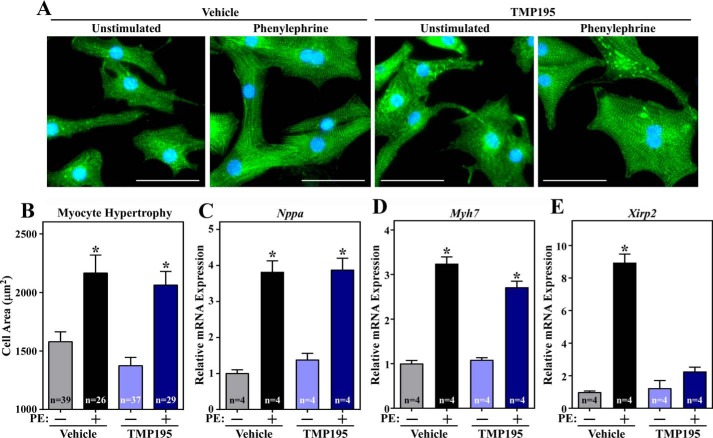
Class IIa HDACs function as signal-responsive repressors of cardiac hypertrophy. Prior studies demonstrated that ectopic expression of truncated versions of HDAC5 and HDAC9 lacking catalytic domains leads to inhibition of cardiomyocyte hypertrophy (8). To rule out possible confounding issues related to HDAC overexpression and to corroborate these findings at the level of endogenous class IIa HDACs, cultured neonatal rat ventricular myocytes (NRVMs) were treated with a recently discovered small molecule, TMP195, which selectively inhibits class IIa catalytic activity but does not suppress other HDAC isoforms (15). The cell area of NRVMs was quantified based on α-actinin immunostaining (Fig. 1A). The results revealed that TMP195 treatment did not reduce phenylephrine (PE)-mediated hypertrophic growth of NRVMs, nor did it alter cell size when administered in the absence of PE (Fig. 1B). Consistent with this, quantitative PCR analysis demonstrated that TMP195 treatment failed to impact agonist-dependent induction of expression of classical hypertrophic marker genes, including Nppa and Myh7 (Fig. 1, C and D). In contrast, the class IIa HDAC inhibitor did repress expression of Xirp2, which is a direct target of the MEF2 transcription factor (Fig. 1E). Thus, although pharmacological inhibition of class IIa HDAC catalytic activity is capable of influencing cardiac gene expression, it is not sufficient to suppress agonist-dependent hypertrophy of cardiomyocytes.
Figure 1.
Class IIa HDAC catalytic domain inhibition does not suppress agonist-dependent hypertrophy of cardiomyocytes. A, NRVMs were treated with vehicle control (Veh; DMSO, 0.1% final concentration) or TMP195 (3 μm) in the absence or presence of phenylephrine (PE, 10 μm) for 48 h. Cells were fixed and analyzed by indirect immunofluorescence with an antibody against sarcomeric α-actinin. Scale bars = 50 μm. B, the cell area was quantified. C–E, quantitative RT-PCR analysis of Nppa, Myh7, and Xirp2 mRNA expression with RNA prepared from parallel plates of NRVMs. For B–E, values represent means + S.E.; *, p < 0.05 versus corresponding conditions but without PE. For B, n = the number of cells quantified. For C–E, n = plates of NRVMs per condition.
Class IIa HDAC catalytic domain inhibition alters gene expression in cardiomyocytes
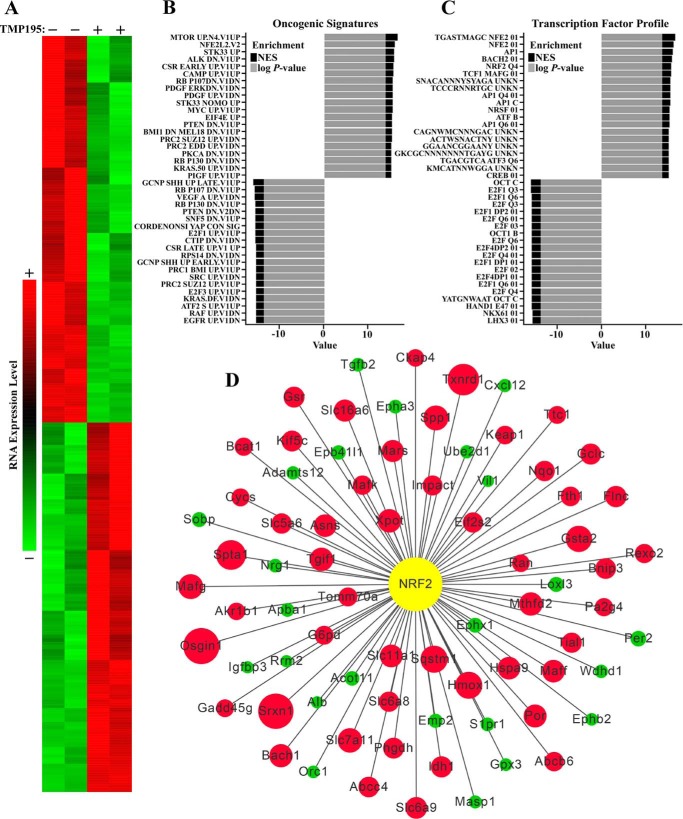
To further explore the role of class IIa HDAC catalytic activity in cardiomyocytes, whole-transcriptome RNA-Seq analysis was performed using RNA from NRVMs treated with TMP195 or vehicle control for 48 h. TMP195 significantly down-regulated expression of 623 mRNA transcripts and up-regulated expression of 594 mRNA transcripts, as illustrated by the heatmap (Fig. 2A). Pathway enrichments were computed using gene set enrichment analysis (GSEA) and databases available through the Molecular Signatures Database. Any enrichment with a normalized p value of less than 0.05 was considered to be significant. Across multiple databases (e.g. Oncogenic Signatures, Kyoto Encyclopedia of Genes and Genomes, and Reactome), there was a significant enrichment of metabolic and oxidative pathways (Fig. 2B and data not shown); pathways enriched in TMP195-treated NRVMs are displayed with a positive log p value and normalized enrichment score (NES), whereas pathways that are down-regulated upon exposure of cells to the class IIa HDAC inhibitor are shown as a negative log p value and NES.
Figure 2.
Class IIa HDAC catalytic domain inhibition alters gene expression in cardiomyocytes. A, NRVMs were treated with vehicle control (DMSO, 0.1% final concentration) or TMP195 (3 μm) for 48 h. Shown is a heatmap representing significantly up-regulated or down-regulated transcripts determined by RNA-Seq analysis. B, pathway enrichments were computed using GSEA and databases available through the Molecular Signatures Database. The top 20 increased and decreased pathways in the Oncogenic Signature Database are displayed as a waterfall plot; UNKN, unknown. C, conserved cis-regulatory elements for genes enriched (positive log p value and NES) or decreased (negative log p value and NES) in TMP195-treated NRVMs. D, a network interaction model highlights the impact of TMP195 treatment on NRF2 target gene expression in NRVMs. The color of the node indicates the direction of the expression change with TMP195 treatment (green, down-regulation; red, up-regulation). The magnitude of change is indicated by the size of the node.
Further evaluation revealed that gene sets enriched in TMP195-treated NRVMs showed striking conservation of cis-regulatory elements predicted to bind to the Cap′N′Collar transcription factors NF-E2 and NF-E2–related factor 2 (NRF2) (Fig. 2C). Network interaction analysis of this subset indicated that antioxidant genes that are known to be stimulated by NRF2, including Hmox1, Srxn1, and Txnrd1, were dramatically up-regulated in TMP195-treated NRVMs compared with controls (Fig. 2D). Therefore, we focused the remainder of this study on elucidating the mechanisms and consequences of class IIa HDAC–mediated control of NRF2-dependent transcription.
Inhibiting class IIa HDACs triggers NRF2-dependent gene expression
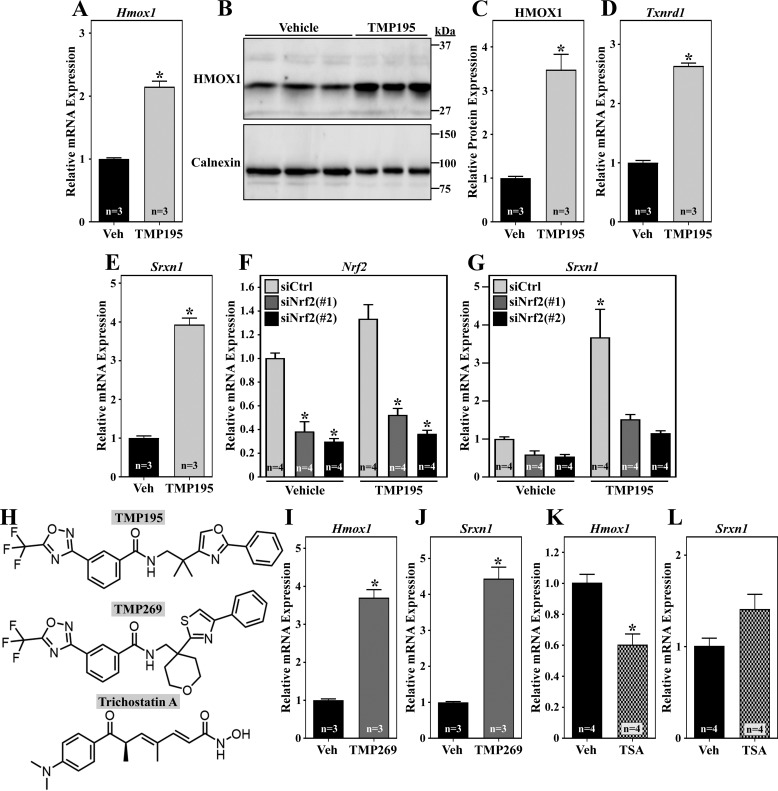
Quantitative PCR and immunoblot analyses of independent samples confirmed that TMP195 stimulates Hmox1 mRNA and HMOX1 protein expression in NRVMs (Fig. 3, A–C). Induction of Txnrd1 and Srxn1 mRNA transcripts by TMP195 was also validated (Fig. 3, D and E). TMP195-mediated antioxidant gene expression was attenuated by knockdown on endogenous NRF2, further suggesting that the class IIa HDAC inhibitor stimulates the activity of this transcription factor (Fig. 3, F and G).
Figure 3.
Class IIa HDAC catalytic domain inhibition stimulates NRF2 target gene expression in cardiomyocytes. A–C, NRVMs were treated with vehicle control (Veh; DMSO, 0.1% final concentration) or TMP195 (3 μm) for 48 h. Hmox1 mRNA expression was assessed by qRT-PCR (A), and HMOX1 protein levels were determined by immunoblotting (B and C); calnexin served as a loading control. (D and E) NRVMs were treated as described in A, and Txnrd1 and Srxn1 mRNA was quantified by qRT-PCR. F and G, NRVMs were transfected with control siRNA or two distinct siRNAs targeting rat Nrf2; 24 h post-transfection, cells were treated with TMP195 for another 48 h. Nrf2 and Srxn1 mRNA was quantified by qRT-PCR. H, structures of HDAC inhibitors. I and J, NRVMs were treated with TMP269 (3 μm) or DMSO control for 36 h prior to qRT-PCR analysis. K and L, NRVMs were treated with TSA (200 nm) or DMSO for 36 h prior to qRT-PCR analysis. For all graphs, values represent means + S.E. For C–E and I–L, *, p < 0.05 versus vehicle. For F, *, p < 0.05 versus siControl from the same treatment group. For G, *p < 0.05 versus siControl + vehicle.
To begin to address whether TMP195 stimulates NRF2-mediated gene expression via inhibition of class IIa HDAC catalytic activity as opposed to an off-target action, NRVMs were treated with a structurally distinct class IIa HDAC inhibitor, TMP269 (15) (Fig. 3H). Similar to TMP195, TMP269 induced expression of the NRF2 target genes Hmox1 and Srxn1 (Fig. 3, I and J).
Using left ventricular lysates from adult rats as a source of cardiac HDACs for enzymatic assays, we confirmed that TMP195 is a selective class IIa HDAC inhibitor (Fig. S1). Nonetheless, at higher concentrations, the compound is capable of suppressing class IIb HDAC activity. To rule out the possibility that TMP195 triggers NRF2-dependent antioxidant gene expression by targeting other HDAC isoforms, NRVMs were treated with the hydroxamic acid HDAC inhibitor trichostatin A (TSA), which potently suppresses class I and IIb HDAC activity but has minimal effects on class IIa HDACs (16). In contrast to TMP195 and TMP269, TSA failed to stimulate Hmox1 or Srxn1 mRNA expression (Fig. 3, K and L). Taken together, the results suggest that selective inhibition of class IIa HDAC catalytic activity stimulates NRF2-dependent gene expression in cardiomyocytes.
HDAC5 suppresses cardiomyocyte NRF2 activity
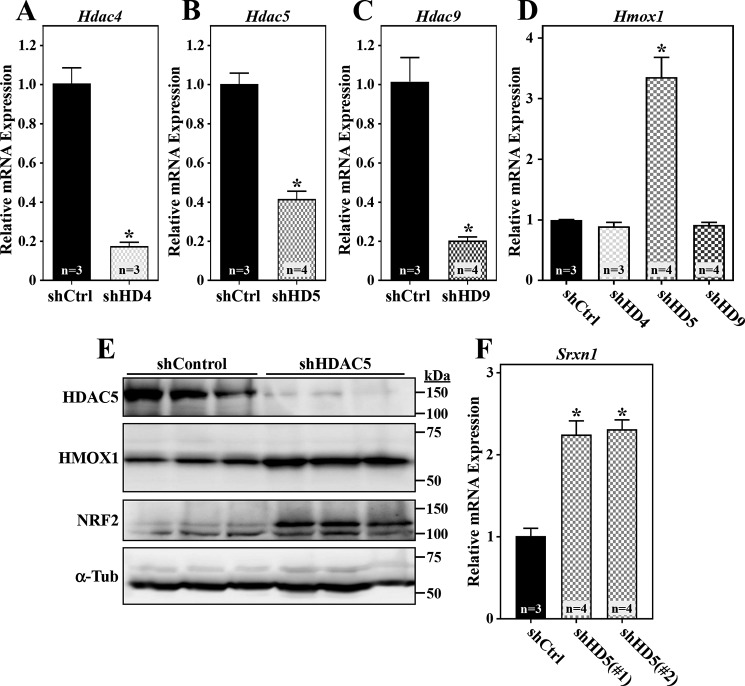
Class IIa HDACs consist of HDACs 4, 5, 7, and 9. HDACs 4, 5, and 9 are well-established regulators of cardiomyocyte gene expression, whereas HDAC7 appears to be more relevant in endothelial cells (17, 18). To address the contributions of the three cardiomyocyte class IIa HDACs to the control of NRF2 function, shRNA-encoding adenoviruses were employed to knock down expression of endogenous HDAC4, HDAC5, and HDAC9 in NRVMs (Fig. 4, A–C). Reduced expression of HDAC5, but not HDAC4 or HDAC9, led to dramatic induction of Hmox1 mRNA and HMOX1 protein expression, which correlated with increased NRF2 abundance (Fig. 4, D and E). An adenovirus encoding shRNA that targets a distinct region of the HDAC5 mRNA transcript also triggered NRF2 target gene expression, confirming an on-target mechanism of action (Fig. 4F).
Figure 4.
HDAC5 knockdown stimulates NRF2 target gene expression in cardiomyocytes. A–D, NRVMs were infected with adenoviruses (m.o.i. = 10) encoding nontargeting control shRNA (shCtrl) or shRNAs targeting HDAC4 (shHD4), HDAC5 (shHD5), and HDAC9 (shHD9). After 72 h of infection, total RNA was isolated, and Hdac4 (A), Hdac5 (B), Hdac9 (C), and Hmox1 (D) mRNA levels were determined by qRT-PCR. E, protein homogenates were prepared from NRVMs infected with adenoviruses encoding shCtrl or shHD5 for 72 h. Immunoblotting was performed with antibodies against the indicated proteins. F, NRVMs were infected with adenoviruses encoding shCtrl or shRNAs targeting two distinct regions of the Hdac5 mRNA transcript (shHD5(#1) and shHD5(#2)) for 72 h prior to preparing total RNA for qRT-PCR analysis of Srxn1 mRNA expression. For all graphs, values represent means + S.E.; *, p < 0.05 versus shCtrl.
Class IIa HDAC inhibitors do not activate NRF2 via a common electrophile-mediated pathway
Most small-molecule NRF2 activators function by covalently modifying cysteine residues in KEAP1 to disrupt the interaction between KEAP1 and NRF2, thereby preventing ubiquitin-dependent NRF2 degradation (19). Although the findings described above demonstrate that HDAC5 inhibition is sufficient to stimulate NRF2 target gene expression in cardiomyocytes, it remained possible that class IIa HDAC inhibitors also function via direct KEAP1 inhibition. Thus, to address the potential of TMP195 to stimulate NRF2 activity via KEAP1, a series of experiments was performed to compare the action of this class IIa HDAC inhibitor with a prototypical thiol-reactive NRF2 activator, AI-1.
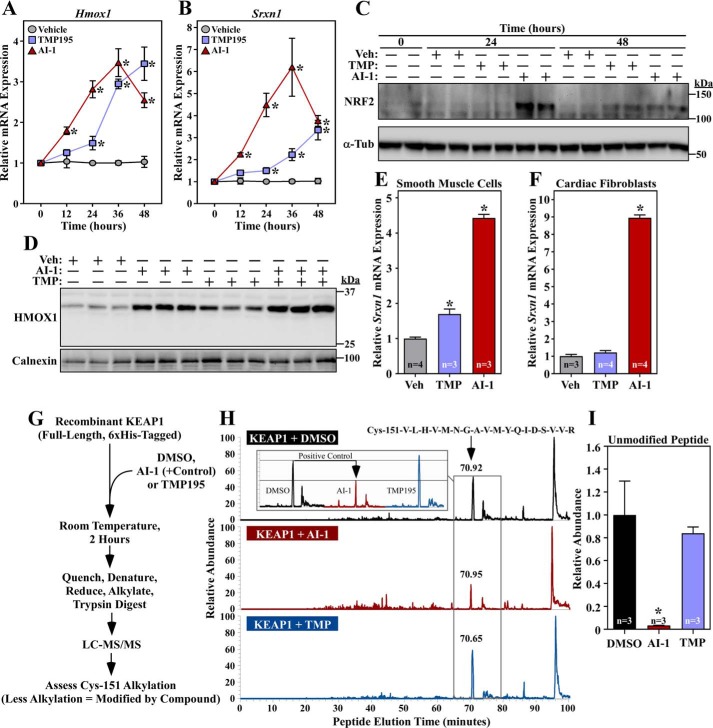
Quantitative PCR analysis revealed distinct kinetics of NRF2 target gene expression in NRVMs treated with TMP195 or AI-1. AI-1 rapidly induced expression of Hmox1 and Srxn1, whereas the response to TMP195 was delayed and more prolonged (Fig. 5, A and B). The differential kinetics of target gene induction by AI-1 and TMP195 correlated with the rate of induction of NRF2 protein abundance (Fig. 5C), suggesting that the compounds stimulate NRF2 activity through different mechanisms. In further support of this notion, combined treatment of NRVMs with AI-1 and TMP195 led to an additive increase in HMOX1 protein expression (Fig. 5D). Additionally, AI-1 stimulated NRF2 target gene expression in all cell types tested (NRVMs, rat aortic smooth muscle cells, and cardiac fibroblasts), whereas TMP195 was only able to increase NRF2 activity in muscle cells (Fig. 5, E and F). Finally, MS analysis of recombinant KEAP1 failed to reveal evidence of covalent modification by TMP195 but confirmed that AI-1 is capable of coupling to cysteine 151, which is a residue in KEAP1 that is commonly targeted by electrophile activators of NRF2 (Fig. 5, G–I) (20, 21). Together, the data suggest that the primary mechanism by which TMP195 stimulates NRF2 in cardiomyocytes is through HDAC5 inhibition rather than by electrophilic adduction to KEAP1.
Figure 5.
Class IIa HDAC catalytic domain inhibition does not stimulate NRF2 through KEAP1 inactivation. A and B, NRVMs were treated with vehicle control (DMSO, 0.1% final concentration), TMP195 (3 μm), or AI-1 (10 μm) for the indicated times. Hmox1 (A) and Srxn1 (B) mRNA levels were determined by qRT-PCR. Values represent means + S.E.; n = plates of cells/condition. *, p < 0.05 versus vehicle at the equivalent time. C, protein homogenates were prepared from NRVMs treated as described in A and subjected to immunoblotting with antibodies specific for NRF2 or α-tubulin (α-Tub). Veh, vehicle. D, NRVMs were treated for 36 h with compounds and vehicle control as described above. Protein homogenates were immunoblotted with antibodies against HMOX1; calnexin served as a loading control. E and F, primary rat aortic smooth muscle cells (E) and neonatal rat cardiac fibroblasts (F) were treated for 48 h with compounds and vehicle control as described above; Srxn1 mRNA expression was assessed by qRT-PCR. Values represent means + S.E. *, p < 0.05 versus vehicle. G, schematic of the in vitro assay to determine whether TMP195 covalently couples to KEAP1. H, the signals of unmodified tryptic peptides that contain KEAP1 Cys-151, which is a common site of electrophilic addition, were measured on an LTQ IonTrap following in vitro incubation of KEAP1 with vehicle control, TMP195, or AI-1; AI-1 served as a positive control. The LC peaks for the Cys-151–containing peptide are indicated by a black arrow; a reduced signal indicates that a compound conjugated to the peptide. The inset shows a side-by-side comparison of the peak for the Cys-151–containing peptide in the different treatment groups. I, the MS experiment in H was repeated to include three additional samples per treatment group. Relative abundance reflects the area under the curve normalized to the DMSO group. Values represent means + S.E. *, p < 0.05 versus DMSO-treated peptide.
HDAC5 inhibition stimulates cardiac NRF2 activity by triggering oxidative stress
To further address the mechanism by which inhibition of HDAC5 catalytic activity leads to induction NRF2 in NRVMs, experiments were performed to determine whether HDAC5 and NRF2 associate physically. Using a mammalian two-hybrid approach, ectopic expression of HDAC5 failed to block reporter gene expression driven by a hybrid protein containing the complete ORF of NRF2 fused to the GAL4 DNA-binding domain (Fig. S2A). In contrast, HDAC5 potently suppressed GAL4-MEF2–driven reporter gene expression (Fig. S2B), which is consistent with the ability of HDAC5 to directly bind to MEF2 (10). Likewise, endogenous NRF2 failed to co-immunoprecipitate with FLAG-tagged HDAC5 in transiently transfected HEK293 cells, even under conditions where NRF2 abundance was elevated because of treatment of cells with the proteasome inhibitor MG132 (Fig. S2, C and D). Conversely, endogenous MEF2A was readily co-immunoprecipitated with HDAC5 (Fig. S2D). Consistent with this, HDAC5 also failed to co-immunoprecipitate with KEAP1, whereas NRF2 was detected in association with its inhibitor (Fig. S2, E and F). These findings indicate that HDAC5 does not physically interact with NRF2:KEAP1 complexes and suggest that the class IIa HDAC represses NRF2 activity through an indirect mechanism.
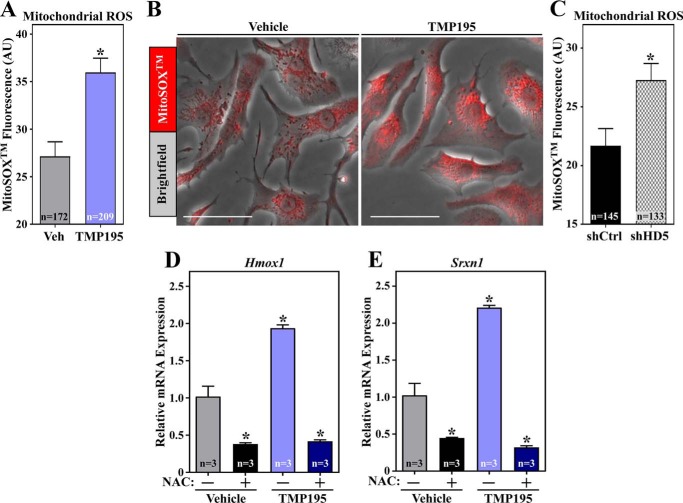
ROS can stimulate NRF2 by modifying cysteine residues in KEAP1 (21). Thus, we addressed the possibility that HDAC5 inhibition promotes NRF2-dependent transcription by triggering cardiomyocyte oxidative stress. Indeed, treatment of NRVMs with TMP195 for 48 h led to a significant increase in mitochondrial ROS levels, as did selective knockdown of HDAC5 using shRNA (Fig. 6, A–C). Furthermore, pretreatment of NRVMs with the antioxidant N-acetylcysteine completely blocked TMP195-induced expression of Hmox1 and Srxn1 mRNA and also reduced the basal levels of these transcripts in unstimulated cardiomyocytes (Fig. 6, D and E). These data suggest that HDAC5 inhibition stimulates NRF2 in cardiomyocytes, at least in part, by triggering oxidative stress.
Figure 6.
Class IIa HDAC catalytic domain inhibition and HDAC5 knockdown elevate cardiomyocyte ROS levels and trigger NRF2 target gene expression. NRVMs were treated with vehicle control (DMSO, 0.1% final concentration) or TMP195 (3 μm) for 48 h. The cells were subsequently washed with PBS and incubated with MitoSOXTM (3 μm) in PBS for 20 min at 37 °C. A, mitochondrial ROS levels were determined by live-cell imaging and quantifying the fluorescence intensity of MitoSOXTM in single cells. Values represent means +S.E., with n = the number of cells quantified. *, p < 0.05 versus vehicle (Veh). AU, artificial unit. B, bright-field images of representative cells with the MitoSOXTM signal overlaid. Scale bars = 50 μm. C, NRVMs were infected with adenoviruses encoding shCtrl or shHD5 for 72 h, and mitochondrial ROS was quantified as in A. NRVMs were pretreated with N-acetylcysteine (NAC, 5 mm) for 1 h and subsequently incubated with DMSO vehicle or TMP195 (3 μm) for 48 h. D and E, Hmox1 (D) and Srxn1 (E) mRNA levels were determined by qRT-PCR. Values represent means + S.E., with n = plates of cells per condition. *, p < 0.05 versus vehicle.
HDAC5 catalytic activity reduces cardiomyocyte oxidative stress
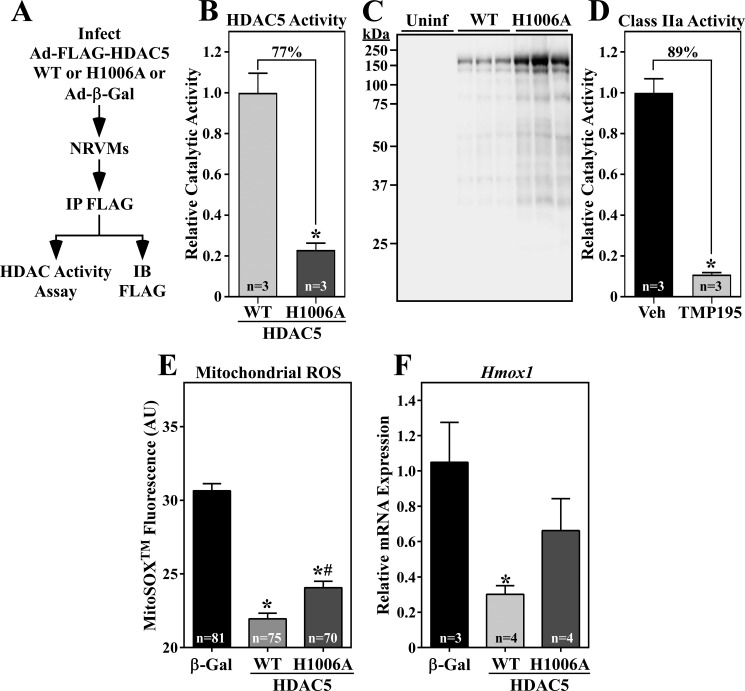
To determine whether increasing HDAC5 catalytic activity is sufficient to alter cardiomyocyte redox signaling, NRVMs were infected with adenoviruses encoding FLAG-tagged versions of WT HDAC5 or HDAC5 harboring a histidine-to-alanine substitution at amino acid 1006 within the catalytic domain (H1006A) (Fig. 7A) (13). In vitro enzymatic assays employing FLAG immunoprecipitates from the cells demonstrated that the catalytic activity of HDAC5 (H1006A) was attenuated (decreased by 77%) but not abolished (Fig. 7, B and C). For comparison, 3 μm TMP195 reduced class IIa HDAC catalytic activity in living NRVMs by 89% (Fig. 7D).
Figure 7.
HDAC5 gain-of-function reduces cardiomyocyte oxidative stress and NRF2 target gene expression. A, schematic of the HDAC5 activity assay. IB, immunoblot. B and C, 72 h after infection, NRVM protein homogenates were immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were incorporated into in vitro HDAC activity assays employing a class IIa HDAC–specific substrate, as described under “Experimental procedures.” A portion of each immunoprecipitate was subjected to immunoblotting with anti-FLAG antibody. Values represent means +S.E., with n = plates of cells per condition. The signal from immunoprecipitates of lysates from Ad-β-gal–infected NRVMs was subtracted as background, and the values were normalized to the amount of immunoprecipitated WT or H1006A determined in (C). *, p < 0.05 versus WT. Uninf, uninfected. D, class IIa HDAC activity was quantified in living NRVMs using a cell-permeable substrate. Values were obtained 5.5 h after treatment with TMP195 (3 μm) or DMSO vehicle control. Values represent means + S.E., with n = the number of independent wells of NRVMs quantified. *, p < 0.05 versus vehicle control. E, NRVMs were infected with an adenovirus encoding β-gal, WT HDAC5, or HDAC5 (H1006A) for 72 h. The cells were subsequently washed with PBS and incubated with MitoSOXTM (3 μm) in PBS for 20 min at 37 °C. Mitochondrial ROS levels were determined by live-cell imaging and quantifying the fluorescence intensity of MitoSOXTM in single cells. Values represent means + S.E., with n = the number of cells quantified. *, p < 0.05 versus β-gal; #, p < 0.5 versus WT. F, NRVMs were infected with adenoviruses as described above, and Hmox-1 mRNA expression was determined by qRT-PCR. Values represent means + S.E., with n = the number of plates of cells per condition. *, p < 0.05 versus β-gal.
Next, mitochondrial ROS levels were quantified in NRVMs ectopically expressing WT HDAC5, HDAC5 (H1006A), or a β-gal control via adenoviruses. As shown in Fig. 7E, mitochondrial ROS was significantly decreased in cardiomyocytes expressing WT HDAC5 compared with β-gal. HDAC5 (H1006A) also reduced ROS levels, but to a lesser extent than HDAC5 WT, which is consistent with its impaired catalytic function. Furthermore, ectopic WT HDAC5, but not HDAC5 (H1006A), suppressed Hmox1 mRNA expression in NRVMs (Fig. 7F). These data further suggest that HDAC5 catalytic activity suppresses cardiomyocyte redox signaling.
Discussion
Here we describe a role of HDAC5 catalytic activity in repression of cardiomyocyte oxidative stress and NRF2-dependent antioxidant gene expression. These functions were initially uncovered using a new generation of HDAC inhibitors that selectively target class IIa HDAC catalytic domains and were validated using HDAC5 gain- and loss-of-function approaches in cultured cardiomyocytes. The data suggest that alterations in HDAC5 abundance and/or catalytic activity could contribute to cardiac mitochondrial dysfunction and oxidative stress, which are drivers of HF pathogenesis (22). Thus, HDAC5 action in the heart extends beyond transcriptional repression of pro-hypertrophic gene expression.
Selective inhibitors of class IIa HDAC catalytic activity have only recently become available. Class IIa HDAC catalytic activity is largely resistant to commonly used HDAC inhibitors such as TSA (16). Several years ago, MC1568 was described as the first class IIa HDAC–selective inhibitor (23). However, we found that MC1568 fails to inhibit class IIa HDAC enzymatic activity (24). TMP195 and TMP269 are powerful chemical tools for studying the biological consequences and potential therapeutic benefits of selectively inhibiting class IIa HDAC activity. These compounds were discovered in a high-throughput screening campaign employing recombinant HDAC9 (15). They possess a nonchelating zinc-binding group, trifluoromethyloxadiazole, which is less promiscuous for off-target actions than the hydroxamic acid, zinc-chelating warheads common in many HDAC inhibitors, including TSA. Our data confirmed the selectivity of TMP195 for class IIa HDACs over other HDACs in the heart.
TMP195 did not alter agonist-mediated growth of cardiomyocytes, which is consistent with prior data showing that versions of HDAC5 and HDAC9 lacking deacetylase domains are still capable of inhibiting MEF2 transcriptional activity and cardiac hypertrophy (8). We note, however, that TMP195 did reduce expression of the gene encoding Xirp2, which is a direct target of MEF2A (25, 26). Thus, it is possible that class IIa HDAC catalytic activity promotes expression of a subset of MEF2 target genes.
NRF2 activity has been shown to prevent pathological remodeling of the heart. NRF2-deficient mice develop exaggerated hypertrophy and cardiac dysfunction in response to pressure overload (27), and electrophilic compounds that stimulate NRF2 via covalent attachment to KEAP1 block cardiac hypertrophy and improve ventricular function in mice (28–31). It is likely that direct activation of cardiac NRF2 through KEAP1 targeting will be better tolerated than indirect activation via HDAC5 inhibition because the latter approach triggers oxidative stress in cardiomyocytes. Nonetheless, despite triggering elevated ROS levels, TMP195 did not elicit overt cytotoxic effects in cultured NRVMs, possibly because of increased expression of compensatory NRF2-dependent antioxidant genes.
TMP195 stimulated NRF2 target gene expression in cardiomyocytes and smooth muscle cells but not in cardiac fibroblasts, suggesting that HDAC5 regulates this pathway in a cell type–specific manner. Consistent with this, TMP195 treatment failed to induce NRF2 target gene expression in monocytes, B lymphocytes, or T lymphocytes (15). This differential regulation could be a reflection of the relative abundance and activity of mitochondria in muscle cells over other cell types.
The mechanism by which HDAC5 inhibition promotes cardiomyocyte oxidative stress remains unknown. HDAC5 is detected in mitochondrial fractions from cultured NRVMs (data not shown), suggesting the possibility that this class IIa HDAC directly regulates mitochondrial function through a nontranscriptional mechanism. A recent report demonstrated that HDAC5 depletion with siRNA in HeLa cells results in elevated ROS levels in association with reduced expression of FTH1 and FTHL2, proteins that regulate iron homeostasis (32). Although we cannot rule out the possibility that altered iron abundance contributes to oxidative stress in NRVMs in which HDAC5 has been inhibited or depleted, we note that expression of FTH1 was not reduced in TMP195-treated NRVMs; FTHL2 was not detected in our RNA-Seq dataset. Therefore, the mechanisms by which HDAC5 inhibition leads to oxidative stress in HeLa cells and cardiomyocytes appear to be distinct.
In summary, we describe a novel function for a class IIa HDAC in the control of cardiac redox homeostasis, with HDAC5 catalytic function reducing oxidative stress in cardiomyocytes. Previously, oxidation of conserved cysteine residues in class IIa HDACs, including HDAC5, was shown to lead to nuclear export of these transcriptional repressors through a phosphorylation-independent mechanism (33–35). This process results in derepression of pro-hypertrophic genes and exaggerated cardiomyocyte hypertrophy. Our findings reveal a second level of interplay between ROS signaling and HDAC5 and suggest the possibility of augmenting HDAC5 catalytic activity as a therapeutic strategy to dampen oxidative stress in the context of cardiac disease.
Experimental procedures
Reagents
Vendors and catalog numbers for all of the antibodies, chemicals, and kits employed in this study are provided in Table S2. Oligonucleotide sequences for cloning and quantitative PCR analysis are provided in Table S1.
Cell culture
NRVMs and neonatal rat ventricular fibroblasts (NRVFs) were prepared and cultured from hearts of 1-day-old Sprague-Dawley rats as described previously (36). For adenoviral infection, viruses (m.o.i. 10) were added when seeding the cells. After incubation with adenoviral particles overnight, cells were washed and maintained in DMEM (Corning) supplemented with l-glutamine, penicillin–streptomycin, and 0.1% Nutridoma-SP (Roche Applied Sciences). Infected cells were harvested or analyzed 48 h or 72 h after infection. NRVFs and HEK293A cells were maintained in high glucose DMEM (Corning) containing 1% l-glutamine, penicillin–streptomycin (Corning), and 10% FBS (Gemini). Passage 1 NRVFs were used for experiments. Rat aortic smooth muscle cells (RASMCs, passage 8), were cultured in minimal essential medium (Corning) supplemented with 1% l-glutamine, penicillin–streptomycin, and 10% FBS. NRVFs and rat aortic smooth muscle cells were equilibrated in corresponding medium with 0.1% FBS for 24 h prior to incubation with DMSO, TMP195, or AI-1 for an additional 48 h.
RNA-Seq analysis
NRVMs cultured in serum-free, Nutridoma-supplemented medium were treated with either DMSO or 3 μm TMP195 for 48 h. Total RNA was prepared using the RNeasy Plus Mini Kit (Qiagen). Libraries were prepared using the Ovation® RNA-Seq Systems 1–16 for Model Organisms (NuGEN). Libraries were sequenced by the University of Colorado Denver Genomics and Microarray Core on a HiSeq 2500 (Illumina). All analyses were performed using the rat RN6 genome and RN6 RefSeq gene annotations.
FastQ files were aligned to RN6 using the TOPHAT2/Cufflinks pipeline. Geometric method was used for normalization. Genes that had an expression greater than 1 were selected for further analysis. Significant differences were computed using a Mann-Whitney U test and corrected for multiple comparisons. Genes that had a p value of less than 0.05 were considered significant.
A heatmap of gene expression was generated using the R Statistical Suite. Pathway analysis was performed using GSEA and databases available through the MSig database via the Broad Institute and the ConsensusPathDB provided by the Max Planck Institute (37, 38). An expression network was created from significantly regulated genes within significantly up-regulated gene sets using Cytoscape (39). The gene sets included to create Fig. 2D were NFE2L2.V2′, NRF2_Q4′, oxidative stress–induced gene expression via nrf2′, transcriptional activation by NRF2′, and NRF2 pathway.
qRT-PCR
RNA was extracted from cells using QIAzol lysis reagent (Qiagen). cDNA was prepared using the Verso cDNA synthesis kit (Thermo Fisher Scientific) with 500 ng of total RNA. qPCR was performed with PowerUpTM SYBR Green Master Mix (Thermo Fisher Scientific) on a StepOnePlus real-time PCR system (Applied Biosystems). Specific primers are listed in Table S1. The relative mRNA expression level was calculated using the 2−ΔΔCT method and normalized to 18S RNA.
Immunoblotting
Total protein was extracted from NRVMs using radioimmune precipitation assay buffer supplemented with a protease and phosphatase inhibitor mixture (Thermo Fisher Scientific). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes (0.45 μm, Life Science Products). Membranes were incubated with primary antibodies overnight at 4 °C. Blots were probed with corresponding HRP-conjugated mouse or rabbit secondary antibodies (Southern Biotech) after primary antibody incubation. Enhanced chemiluminescent HRP substrate (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific) was used for developing blots, and bands were visualized using a FluorChem HD2 Imager (Alpha Innotech).
Indirect immunofluorescence
NRVMs were fixed with 4% paraformaldehyde at room temperature for 10 min, permeabilized with PBS-T (PBS and 0.2% Triton X-100) for 15 min, and blocked with PBS containing 5% BSA for 30 min at room temperature. Cells were subsequently incubated with anti-α-actinin antibody (Sigma) for 1 h. Fluorochrome-conjugated secondary antibody (anti-rabbit Alexa Fluor® 488) was applied for 30 min. Nuclei were counterstained using DAPI (Life Technologies). Images were acquired on an EVOS FL Cell Imaging System (Life Technologies, AMF4300). NRVM cell area was quantified based on α-actinin staining using ImageJ software (National Institutes of Health).
Plasmids
The complementary DNA for human KEAP1 (a gift from Qing Zhong, Addgene plasmid 28023) was PCR-amplified using PFU Turbo DNA Polymerase (Agilent Technologies) and subcloned into pET28a to encode His6-tagged full-length hKEAP1. Site-directed mutagenesis was performed using the QuikChange method (Agilent Technologies). For generation of adenoviral constructs overexpressing FLAG-tagged forms of HDAC5, cDNAs were subcloned into the pENTR/2B entry vector (Invitrogen) and recombined with the pAd/CMV/V5-DESTTM destination vector (Invitrogen). To generate adenovirus constructs for knockdown of HDAC4, HDAC5, and HDAC9, complementary oligonucleotides for shRNAs were designed using BLOCK-iTTM RNAi Designer (Thermo Fisher), synthesized (Integrated DNA Technologies), annealed, and ligated into the pENTR/U6 vector (Invitrogen). Positive clones were recombined with the pAd/BLOCK-iTTM-DEST vector (Invitrogen) to yield shRNA-expressing adenoviral constructs.
siRNA knockdown of NRF2
To knock down endogenous NRF2 expression, NRVMs were transfected with a nonspecific siRNA control or siRNA oligonucleotides targeting Nrf2 (50 nmol/liter, Sigma) using LipofectamineTM RNAiMAX (Thermo Fisher). Twenty-four hours after transfection, cells were treated with DMSO or 3 μm TMP195 for an additional 48 h.
Adenovirus preparation
Adenoviral vectors encoding shRNAs targeting HDAC4, HDAC5, and HDAC9 mRNA transcripts or overexpressing FLAG-HDAC5 (H1006A) were digested by PacI and transfected into HEK293A cells cultured in antibiotic-free medium at 60–70% confluence using Lipofectamine 3000 (Thermo Fisher). Viruses were amplified and recovered from HEK293A cell lysates, and titers were determined using the Sea-Plaque–agarose method (40).
Recombinant KEAP1 protein purification, in vitro labeling, and MS
His6-hKEAP1 was expressed using pET28a in TunerTM (DE3) Escherichia coli (Novagen). Cells were lysed by sonication on ice and pelleted by centrifugation at 27,000 × g in a Sorvall SS-34 fixed-angle rotor for 50 min. Clarified lysates were incubated with 1 mL of prewashed TALON® metal affinity resin (Takara) at 4 °C for 3 h. The resin was pelleted at 3000 rpm for 5 min and washed three times with 13 ml of buffer (200 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10 mm imidazole, and 0.1% β-mercaptoethanol). His6-hKEAP1 was eluted by incubating resin with 2 ml of wash buffer containing 500 mm imidazole at 4 °C for 30 min. Buffer exchange was performed using Amicon ultracentrifugal filter units (Millipore) with 25 mm Tris-HCl (pH 8.0).
His6-hKEAP1 was diluted to 10 μm for in vitro labeling as described previously (20). His6-hKEAP1 was incubated with 5 μl of TMP195 (10 mm), AI-1 (10 mm), or DMSO in 100 μl of buffer at room temperature for 2 h. DTT was added to a final concentration of 10 mm to quench the reaction at room temperature for 15 min. Protein was then denatured and reduced by addition of an equal volume of 8 m urea in 100 mm NH4HCO3 (pH 8.0) and 10 mm DTT solution for 15 min incubation at room temperature. The remaining free cysteine residues were alkylated with 20 mm iodoacetamide for 15 min at room temperature in the dark. Reactions were then diluted to 2 m urea with water, digested with trypsin, and subjected to MS analysis by nano-LC/MS with an LTQ-XL mass spectrometer. Modified sites were manually verified by setting the search range for cysteine to 0–500. Analysis was performed at the University of Colorado School of Medicine Biological Mass Spectrometry Facility.
MitoSOXTM staining
NRVMs were gently rinsed three times with PBS to remove residual medium and incubated with 3 μm MitoSOXTM in prewarmed PBS at 37 °C for 20 min in the dark. Cells were subsequently washed three times with PBS and incubated in PBS during imaging. Images were obtained on an EVOS FL Cell imaging system (Life Technologies, AMF4300). Mean fluorescence intensity quantification based on MitoSOXTM staining within cells was performed using ImageJ software (National Institutes of Health).
HDAC activity assays
To determine the IC50 of TMP195 against class I, IIa, and IIb HDACs, HDAC activity assays were performed as described previously (41). Assays were run simultaneously for class I, IIa, and IIb HDAC activities with parallel sets of sample aliquots. Left ventricular homogenates from Sprague-Dawley rat tissue were prepared as a source of endogenous HDACs using PBS (pH 7.4) containing 0.5% Triton X-100, 300 mm NaCl, and protease/phosphatase inhibitor mixture. Data were plotted using GraphPad Prism and the IC50 of TMP195 was obtained using a nonlinear regression fit.
To assess the catalytic activity of ectopic HDAC5, assays were performed with anti-FLAG immunoprecipitates. NRVMs were infected, at the time of plating, with adenoviruses encoding FLAG-HDAC5 or FLAG-HDAC5 (H1006A) at an m.o.i. of 10. After 16 h of infection, cells were washed and incubated for another 56 h in serum-free medium. Cells were lysed in buffer containing Tris (50 mm (pH 8)), 150 mm NaCl, sodium deoxycholate (0.5%), NP-40 (1%), and SDS (0.1%) and sonicated (4 s at 30% power with the Sonic Dismembrator 500) prior to clarification by centrifugation (12, 000 × g, 5 min at 4 °C). 500 μg of total protein in 500 μl of lysis buffer was immunoprecipitated with 25 μl of prewashed packed anti-FLAG IP resin (GenScript) overnight at 4 °C on a rotator. Beads were washed three times with 500 μl of PBS and resuspended in 200 μl of PBS. Class IIa substrate (10 μl, 1 mm) was added and incubated at 37 °C for 2.5 h. Beads were pelleted at 5000 × g for 30 s, and the supernatant was transferred to a 96-well plate at 100 μl/well. 50 μl of developer solution was then added and incubated at 37 °C for another 20 min. The 7-amino-4-methylcoumarin (AMC) fluorescence signal was measured. Background signals from FLAG-only immunoprecipitates were subtracted. Beads were boiled in sample loading buffer, and proteins were resolved by SDS-PAGE and immunoblotted with anti-FLAG antibody to normalize for the amount of immunoprecipitated HDAC5.
To measure class IIa HDAC activity in living NRVMs, 3 × 105 cells were used for each experiment. NRVMs were incubated with DMSO or 3 μm TMP195 in 100 μl of prewarmed PBS at 37 °C for 3 h. 5 μl of 1 mm class IIa HDAC substrate was then added and incubated at 37 °C for 2.5 h. The AMC fluorescence signal was developed and measured as described above.
Co-immunoprecipitation
HEK293A cells were transfected with an N terminus FLAG-tagged cDNA expression vector encoding human HDAC5 using PEI for 48 h. Cells were lysed in buffer containing Tris (50 mm (pH 7.5)), 150 mm NaCl, and Triton X-100 (0.5%) using a syringe with a 25-gauge needle. Total protein was collected after centrifugation of lysates at 12, 000 × g at 4 °C for 20 min. 500 μg of total protein was diluted in 500 μl of equilibration buffer (50 mm Tris, and 150 mm NaCl (pH 7.4)) and incubated with 25 μl of prewashed packed anti-FLAG IP resin overnight at 4 °C on a rotator. Beads were then washed (three times with equilibration buffer, 0.5 ml per wash), boiled in sample loading buffer, and then loaded on 10% SDS-PAGE gels. Whole-cell lysates were used as input controls.
For immunoprecipitation with anti-KEAP1 antibody, HEK293A cells treated with MG132 (20 μm) for 4 h were lysed in IP lysis buffer as above. 500 μg of total protein homogenate was incubated with 10 μg of anti-KEAP1 antibody (Proteintech) or normal rabbit IgG (Cell Signaling Technologies) overnight at 4 °C on a rotator. The protein complexes were immunoprecipitated using 20 μl of prewashed packed protein A/G Plus–agarose beads (Santa Cruz Biotechnology) for 1 h at room temperature. Beads were then washed (three times with PBS, 0.5 ml per wash), boiled in sample loading buffer, and then loaded on 10% SDS-PAGE gels. In both experiments, proteins were transferred to nitrocellulose membranes (0.45 μm, Life Science Products) and immunoblotted using anti-NRF2, anti-HDAC5 (Cell Signaling Technologies), anti-MEF2 (Santa Cruz Biotechnology), anti-KEAP1 (Proteintech), or anti-FLAG M2-HRP (Sigma) antibodies. For both co-IPs, whole-cell lysates were used as input controls.
Luciferase reporter assays
HEK293A cells (1 × 105) were seeded on 24-well plates and co-transfected with pGB5-Luc, pcDNA3.1-FLAG, or pcDNA3.1-FLAG-HDAC5 and pcDNA3 vectors expressing GAL4, GAL4-NRF2, GAL4-MEF2A, or GAL4-MEF2D using PEI. A Renilla luciferase expression vector (hRLuc/TK) was used as an internal control. Cells were lysed in passive lysis buffer provided with the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured using a BioTek Synergy 2 plate reader.
Statistical analysis
Statistical significance (p < 0.05) was determined using unpaired t test (two groups) or one-way analysis of variance for multiple comparisons via a Tukey post hoc test (GraphPad Prism 7.02).
Author contributions
T. H., F. C. S., R. A. B., M. H., and T. A. M. conceptualization; T. H., F. C. S., and R. A. B. investigation; T. H. and P. D. T. writing-original draft; T. H., F. C. S., R. A. B., P. D. T., M. H., and T. A. M. writing-review and editing; F. C. S., R. A. B., P. D. T., and M. H. formal analysis; M. H. resources; T. A. M. supervision; T. A. M. funding acquisition; T. A. M. project administration.
Supplementary Material
Acknowledgments
We thank Congwu Chi for advice regarding MitoSOXTM staining, Carmen (Kika) Sucharov for providing NRVMs, and Mary Weiser-Evans for providing rat aortic smooth muscle cells.
This work was supported by National Institutes of Health Grants HL116848 and HL127240 and American Heart Association Grant 16SFRN31400013 (to T. A. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1 and S2.
Raw and processed RNA-Seq data were deposited into the GEO online database under accession number GSE130150.
- HF
- heart failure
- HDAC
- histone deacetylase
- NRVM
- neonatal rat ventricular myocyte
- PE
- phenylephrine
- GSEA
- gene set enrichment analysis
- NES
- normalized enrichment score
- TSA
- trichostatin A
- ROS
- reactive oxygen species
- NRVF
- neonatal rat ventricular fibroblast
- m.o.i.
- multiplicity of infection
- cDNA
- complementary DNA
- IP
- immunoprecipitation
- qRT-PCR
- quantitative reverse transcription polymerase chain reaction.
References
- 1. Benjamin E. J., Blaha M. J., Chiuve S. E., Cushman M., Das S. R., Deo R., de Ferranti S. D., Floyd J., Fornage M., Gillespie C., Isasi C. R., Jimenez M. C., Jordan L. C., Judd S. E., Lackland D., et al. (2017) Heart disease and stroke statistics 2017 update: a report from the American Heart Association. Circulation 135, e146–e603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gheorghiade M., Larson C. J., Shah S. J., Greene S. J., Cleland J. G., Colucci W. S., Dunnmon P., Epstein S. E., Kim R. J., Parsey R. V., Stockbridge N., Carr J., Dinh W., Krahn T., Kramer F., et al. (2016) Developing new treatments for heart failure: focus on the heart. Circ. Heart Fail. 9, e002727 [DOI] [PubMed] [Google Scholar]
- 3. Devereux R. B., Wachtell K., Gerdts E., Boman K., Nieminen M. S., Papademetriou V., Rokkedal J., Harris K., Aurup P., and Dahlöf B. (2004) Prognostic significance of left ventricular mass change during treatment of hypertension. JAMA 292, 2350–2356 10.1001/jama.292.19.2350 [DOI] [PubMed] [Google Scholar]
- 4. Gardin J. M., and Lauer M. S. (2004) Left ventricular hypertrophy: the next treatable, silent killer? JAMA 292, 2396–2398 10.1001/jama.292.19.2396 [DOI] [PubMed] [Google Scholar]
- 5. Hill J. A., and Olson E. N. (2008) Cardiac plasticity. N. Engl. J. Med. 358, 1370–1380 10.1056/NEJMra072139 [DOI] [PubMed] [Google Scholar]
- 6. Chang S., McKinsey T. A., Zhang C. L., Richardson J. A., Hill J. A., and Olson E. N. (2004) Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell Biol. 24, 8467–8476 10.1128/MCB.24.19.8467-8476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kook H., Lepore J. J., Gitler A. D., Lu M. M., Wing-Man Yung W., Mackay J., Zhou R., Ferrari V., Gruber P., and Epstein J. A. (2003) Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 112, 863–871 10.1172/JCI19137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., and Olson E. N. (2002) Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gregoretti I. V., Lee Y. M., and Goodson H. V. (2004) Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338, 17–31 10.1016/j.jmb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 10. Lu J., McKinsey T. A., Nicol R. L., and Olson E. N. (2000) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 4070–4075 10.1073/pnas.080064097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harrison B. C., Kim M. S., van Rooij E., Plato C. F., Papst P. J., Vega R. B., McAnally J. A., Richardson J. A., Bassel-Duby R., Olson E. N., and McKinsey T. A. (2006) Regulation of cardiac stress signaling by protein kinase d1. Mol. Cell Biol. 26, 3875–3888 10.1128/MCB.26.10.3875-3888.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKinsey T. A. (2007) Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovasc. Res. 73, 667–677 10.1016/j.cardiores.2006.11.036 [DOI] [PubMed] [Google Scholar]
- 13. Lahm A., Paolini C., Pallaoro M., Nardi M. C., Jones P., Neddermann P., Sambucini S., Bottomley M. J., Lo Surdo P., Carfí A., Koch U., De Francesco R., Steinkühler C., and Gallinari P. (2007) Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 104, 17335–17340 10.1073/pnas.0706487104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heltweg B., Dequiedt F., Marshall B. L., Brauch C., Yoshida M., Nishino N., Verdin E., and Jung M. (2004) Subtype selective substrates for histone deacetylases. J. Med. Chem. 47, 5235–5243 10.1021/jm0497592 [DOI] [PubMed] [Google Scholar]
- 15. Lobera M., Madauss K. P., Pohlhaus D. T., Wright Q. G., Trocha M., Schmidt D. R., Baloglu E., Trump R. P., Head M. S., Hofmann G. A., Murray-Thompson M., Schwartz B., Chakravorty S., Wu Z., Mander P. K., et al. (2013) Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat. Chem. Biol. 9, 319–325 10.1038/nchembio.1223 [DOI] [PubMed] [Google Scholar]
- 16. Bradner J. E., West N., Grachan M. L., Greenberg E. F., Haggarty S. J., Warnow T., and Mazitschek R. (2010) Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 6, 238–243 10.1038/nchembio.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang S., Young B. D., Li S., Qi X., Richardson J. A., and Olson E. N. (2006) Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 126, 321–334 10.1016/j.cell.2006.05.040 [DOI] [PubMed] [Google Scholar]
- 18. Haberland M., Montgomery R. L., and Olson E. N. (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kern J. T., Hannink M., and Hess J. F. (2007) Disruption of the Keap1-containing ubiquitination complex as an antioxidant therapy. Curr. Top. Med. Chem. 7, 972–978 10.2174/156802607780906825 [DOI] [PubMed] [Google Scholar]
- 20. Hur W., Sun Z., Jiang T., Mason D. E., Peters E. C., Zhang D. D., Luesch H., Schultz P. G., and Gray N. S. (2010) A small-molecule inducer of the antioxidant response element. Chem. Biol. 17, 537–547 10.1016/j.chembiol.2010.03.013 [DOI] [PubMed] [Google Scholar]
- 21. Zhang D. D., and Hannink M. (2003) Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell Biol. 23, 8137–8151 10.1128/MCB.23.22.8137-8151.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. von Hardenberg A., and Maack C. (2017) Mitochondrial therapies in heart failure. Handb. Exp. Pharmacol. 243, 491–514 [DOI] [PubMed] [Google Scholar]
- 23. Mai A., Massa S., Pezzi R., Simeoni S., Rotili D., Nebbioso A., Scognamiglio A., Altucci L., Loidl P., and Brosch G. (2005) Class II (IIa)-selective histone deacetylase inhibitors: 1: synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J. Med. Chem. 48, 3344–3353 10.1021/jm049002a [DOI] [PubMed] [Google Scholar]
- 24. Lemon D. D., Harrison B. C., Horn T. R., Stratton M. S., Ferguson B. S., Wempe M. F., and McKinsey T. A. (2015) Promiscuous actions of small molecule inhibitors of the protein kinase D-class IIa HDAC axis in striated muscle. FEBS Lett. 589, 1080–1088 10.1016/j.febslet.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang H. T., Brand O. M., Mathew M., Ignatiou C., Ewen E. P., McCalmon S. A., and Naya F. J. (2006) Myomaxin is a novel transcriptional target of MEF2A that encodes a Xin-related α-actinin-interacting protein. J. Biol. Chem. 281, 39370–39379 10.1074/jbc.M603244200 [DOI] [PubMed] [Google Scholar]
- 26. McCalmon S. A., Desjardins D. M., Ahmad S., Davidoff K. S., Snyder C. M., Sato K., Ohashi K., Kielbasa O. M., Mathew M., Ewen E. P., Walsh K., Gavras H., and Naya F. J. (2010) Modulation of angiotensin II-mediated cardiac remodeling by the MEF2A target gene Xirp2. Circ. Res. 106, 952–960 10.1161/CIRCRESAHA.109.209007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J., Ichikawa T., Villacorta L., Janicki J. S., Brower G. L., Yamamoto M., and Cui T. (2009) Nrf2 protects against maladaptive cardiac responses to hemodynamic stress. Arterioscler. Thromb. Vasc. Biol. 29, 1843–1850 10.1161/ATVBAHA.109.189480 [DOI] [PubMed] [Google Scholar]
- 28. Bai Y., Cui W., Xin Y., Miao X., Barati M. T., Zhang C., Chen Q., Tan Y., Cui T., Zheng Y., and Cai L. (2013) Prevention by sulforaphane of diabetic cardiomyopathy is associated with up-regulation of Nrf2 expression and transcription activation. J. Mol. Cell Cardiol. 57, 82–95 10.1016/j.yjmcc.2013.01.008 [DOI] [PubMed] [Google Scholar]
- 29. Eba S., Hoshikawa Y., Moriguchi T., Mitsuishi Y., Satoh H., Ishida K., Watanabe T., Shimizu T., Shimokawa H., Okada Y., Yamamoto M., and Kondo T. (2013) The nuclear factor erythroid 2-related factor 2 activator oltipraz attenuates chronic hypoxia-induced cardiopulmonary alterations in mice. Am. J. Respir. Cell Mol. Biol. 49, 324–333 10.1165/rcmb.2011-0396OC [DOI] [PubMed] [Google Scholar]
- 30. Ichikawa T., Li J., Meyer C. J., Janicki J. S., Hannink M., and Cui T. (2009) Dihydro-CDDO-trifluoroethyl amide (dh404), a novel Nrf2 activator, suppresses oxidative stress in cardiomyocytes. PLoS ONE 4, e8391 10.1371/journal.pone.0008391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xing Y., Niu T., Wang W., Li J., Li S., Janicki J. S., Ruiz S., Meyer C. J., Wang X. L., Tang D., Zhao Y., and Cui T. (2012) Triterpenoid dihydro-CDDO-trifluoroethyl amide protects against maladaptive cardiac remodeling and dysfunction in mice: a critical role of Nrf2. PLoS ONE 7, e44899 10.1371/journal.pone.0044899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hendrick E., Peixoto P., Blomme A., Polese C., Matheus N., Cimino J., Frère A., Mouithys-Mickalad A., Serteyn D., Bettendorff L., Elmoualij B., De Tullio P., Eppe G., Dequiedt F., Castronovo V., and Mottet D. (2017) Metabolic inhibitors accentuate the anti-tumoral effect of HDAC5 inhibition. Oncogene 36, 4859–4874 10.1038/onc.2017.103 [DOI] [PubMed] [Google Scholar]
- 33. Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., and Sadoshima J. (2008) A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978–993 10.1016/j.cell.2008.04.041 [DOI] [PubMed] [Google Scholar]
- 34. Matsushima S., Kuroda J., Ago T., Zhai P., Park J. Y., Xie L. H., Tian B., and Sadoshima J. (2013) Increased oxidative stress in the nucleus caused by Nox4 mediates oxidation of HDAC4 and cardiac hypertrophy. Circ. Res. 112, 651–663 10.1161/CIRCRESAHA.112.279760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oka S., Ago T., Kitazono T., Zablocki D., and Sadoshima J. (2009) The role of redox modulation of class II histone deacetylases in mediating pathological cardiac hypertrophy. J. Mol. Med. 87, 785–791 10.1007/s00109-009-0471-2 [DOI] [PubMed] [Google Scholar]
- 36. Ferguson B. S., Harrison B. C., Jeong M. Y., Reid B. G., Wempe M. F., Wagner F. F., Holson E. B., and McKinsey T. A. (2013) Signal-dependent repression of DUSP5 by class I HDACs controls nuclear ERK activity and cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 110, 9806–9811 10.1073/pnas.1301509110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamburov A., Pentchev K., Galicka H., Wierling C., Lehrach H., and Herwig R. (2011) ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 39, D712–D717 10.1093/nar/gkq1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamburov A., Wierling C., Lehrach H., and Herwig R. (2009) ConsensusPathDB: a database for integrating human functional interaction networks. Nucleic Acids Res. 37, D623–D628 10.1093/nar/gkn698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gonzalez-Hernandez M. B., Bragazzi Cunha J., and Wobus C. E. (2012) Plaque assay for murine norovirus. J. Vis. Exp. e4297 10.3791/4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lemon D. D., Horn T. R., Cavasin M. A., Jeong M. Y., Haubold K. W., Long C. S., Irwin D. C., McCune S. A., Chung E., Leinwand L. A., and McKinsey T. A. (2011) Cardiac HDAC6 catalytic activity is induced in response to chronic hypertension. J. Mol. Cell Cardiol. 51, 41–50 10.1016/j.yjmcc.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.