Abstract
Phosphodiesterase-6 (PDE6) plays a central role in both rod and cone phototransduction pathways. In the dark, PDE6 activity is suppressed by its inhibitory γ-subunit (Pγ). Rhodopsin-catalyzed activation of the G protein transducin relieves this inhibition and enhances PDE6 catalysis. We hypothesized that amino acid sequence differences between rod- and cone-specific Pγs underlie transducin's ability to more effectively activate cone-specific PDE6 than rod PDE6. To test this, we analyzed rod and cone Pγ sequences from all major vertebrate and cyclostome lineages and found that rod Pγ loci are far more conserved than cone Pγ sequences and that most of the sequence differences are located in the N-terminal region. Next we reconstituted rod PDE6 catalytic dimer (Pαβ) with various rod or cone Pγ variants and analyzed PDE6 activation upon addition of the activated transducin α-subunit (Gtα*-GTPγS). This analysis revealed a rod-specific Pγ motif (amino acids 9–18) that reduces the ability of Gtα*-GTPγS to activate the reconstituted PDE6. In cone Pγ, Asn-13 and Gln-14 significantly enhanced Gtα*-GTPγS activation of cone Pγ truncation variants. Moreover, we observed that the first four amino acids of either rod or cone Pγ contribute to Gtα*-GTPγS–mediated activation of PDE6. We conclude that physiological differences between rod and cone photoreceptor light responsiveness can be partially ascribed to ancient, highly conserved amino acid differences in the N-terminal regions of Pγ isoforms, demonstrating for the first time a functional role for this region of Pγ in the differential activation of rod and cone PDE6 by transducin.
Keywords: phototransduction, G protein, photoreceptor, vision, protein structure, inhibition mechanism, γ-subunit, PDE6, phosphodiesterase (PDE), retinal protein, transducin
Introduction
Although the speed and sensitivity of the photoresponses of rod and cone photoreceptors are quite different, the underlying molecular components of the visual excitation pathway are believed to be homologous in both classes of photoreceptors (1, 2). In rod photoreceptors, the cGMP signaling cascade is initiated when light-activated rhodopsin binds the heterotrimeric G protein transducin (Gt).2 This leads to guanine nucleotide exchange of GDP to GTP and dissociation of the activated transducin α-subunit (Gtα*-GTP). Gtα*-GTP then associates with and activates the rod PDE6 holoenzyme (consisting of two catalytic subunits, α and β, and two inhibitory rod-specific γ-subunits, rPγ). Cone photoreceptors express homologous forms of the visual pigment (cone opsins), cone transducin, and cone PDE6 holoenzyme (comprised of two identical α' catalytic subunits and two cone-specific Pγ subunits, cPγ; for a review, see Ref. 3).
One approach to identify the biochemical differences in the rod and cone visual transduction pathways has been to genetically introduce a cone-specific isoform of a phototransduction gene into rod photoreceptor cells. As recently summarized (2), transgenic incorporation of various cone opsin genes, the cone transducin α-subunit, or the cone PDE6 α'-subunit into mouse rod photoreceptor cells had a very modest or no effect on the light sensitivity of the rod photoresponse. Biochemical and molecular approaches have identified several intrinsic differences between rod and cone opsins (e.g. spontaneous activation, chromophore dissociation, and regeneration), but evidence is lacking that differences in photoactivated rod and cone opsins can account for the different light sensitivity and photoresponse kinetics of rods and cones (4). Characterization of the biochemical properties of rod and cone transducin α-subunits have failed to reveal significant functional differences (5–7). Likewise, the PDE6 catalytic dimers in rods and cones have equivalent hydrolytic activities (8–10).
The photoreceptor PDE6 is the only member of the 11-member phosphodiesterase superfamily that is known to be regulated by a distinct regulatory protein, the Pγ subunit (3, 11). The mechanism of rod PDE6 activation involves binding of Gtα*-GTP to the PDE6 holoenzyme, causing displacement of the C-terminal domain of rPγ from the entrance of the active site to accelerate hydrolysis of cGMP. The lifetime of Gtα*-GTP and activated rod PDE6 is determined by the GTPase activity of the transducin α-subunit, which is regulated by a complex of three GTPase-accelerating proteins (RGS9-1, Gβ5L, and R9AP (12)). Importantly, the rPγ subunit also serves as feedback regulator of this GTPase accelerating proteins complex in rods (13). The cone cPγ sequence is highly homologous to rPγ, and both Pγ isoforms bind to and inhibit the rod PDE6 catalytic dimer with similar affinity (10). However, the cone PDE6 holoenzyme is more easily activated by rod Gtα*-GTPγS than rod the PDE6 holoenzyme (8).
The rod and cone Pγ genes (PDE6G and PDE6H) appear to have evolved with our earliest vertebrate ancestors at the same time when PDE6 catalytic subunit genes arose (14, 15) with their unique catalytic and regulatory properties (3). It is noteworthy that the sea lamprey (Petromyzon marinus), a cyclostome that diverged from other vertebrate lineages about 500 million years ago, has a duplex retina in which rod and cone photoreceptors show similar physiological differences to light, as observed with mammalian photoreceptors (16, 17). Interestingly, lamprey rods and cones express the same PDE6 catalytic subunit along with distinct rod- and cone-specific Pγ isoforms (18).
To test the hypothesis that differences in rPγ and cPγ subunits may contribute to the physiological differences in rod and cone light responsiveness, we examined whether amino acid sequence differences between rPγ and cPγ underlie the ability of transducin to more effectively activate cone PDE6 relative to rod PDE6. We first carried out a phylogenetic analysis of available Pγ subunit sequences to identify rod- and cone-specific sequence differences that might underly their different biochemical properties. We then reconstituted the purified rod PDE6 catalytic dimer (Pαβ) with different rPγ or cPγ mutants and tested the relative affinity and extent of catalytic activation upon addition of the persistently activated transducin α-subunit (Gtα*-GTPγS). Our results demonstrate that the N-terminal amino acids of rPγ and cPγ are responsible for the differential activation of PDE6 by transducin, from which we conclude that the evolution of separate genes for rod and cone Pγ represents one of the regulatory mechanisms distinguishing the rod and cone phototransduction pathways in vertebrate photoreceptors.
Results
Evolutionary analysis reveals that rod Pγ is far more conserved than cone Pγ
To identify differences in the sequences of the rod (PDE6G) and cone (PDE6H) Pγ genes, we sampled the largest phylogenetic diversity possible from public databases. The sequences we obtained include representation from PDE6G and PDE6H sequences from all major lineages of craniates, including cyclostomes (hagfish and lamprey), chondrichthyans (sharks and rays), neopterygian fishes (including teleosts), squamates (lizards and snakes), archosaurs (crocodilians and birds), and mammals. At present, PDE6G and PDE6H sequence data are lacking only for sturgeons, paddlefishes, and bichirs.
Phylogenetic analyses revealed strong support for two clades representing PDE6G and PDE6H for all vertebrate classes; however, because of strong sequence conservation, we found little support for shallow nodes across the tree (Fig. 1). Overall, we found that PDE6G sequences are highly conserved throughout craniates. In contrast, the PDE6H clade has undergone greater amino acid sequence diversification, especially outside of the mammalian class. In particular, PDE6H sequences for neopterygian fishes show radical alterations compared with other PDE6H sequences. In addition, we identified eight N terminus residues that are highly conserved in PDE6G but divergent or missing in PDE6H. We also uncovered two residues that are differentiated, but highly conserved, within PDE6G and PDE6H (Fig. 1).
Figure 1.

Phylogenetic analysis of PDE6G (rod) and PDE6H (cone) sequences. Branch colors on the gene tree (left) correspond to the species tree (inset). The tree is rooted between putative rod and cone Pγ subunits. Multiple sequence alignment is plotted on the right. In the N-terminal region, several residues (Rod Conserved, gray boxes) are more than 90% conserved in the PDE6G alignment but divergent or missing in the PDE6H alignment. Two Rod/Cone Differentiated sequences (black boxes) are more than 80% conserved in either PDE6G and PDE6H alignments but differentiated between the two genes.
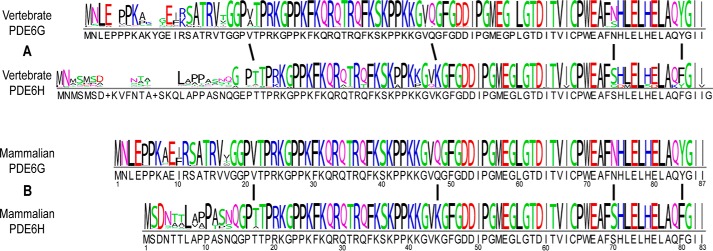
According to the consensus logo (Fig. 2), vertebrate PDE6G and PDE6H residues show high sequence similarity except for the N-terminal region of the protein sequence (Fig. 2A). Within this N-terminal region, PDE6G sequences are significantly more highly conserved than PDE6H sequences, as judged by comparison of the consensus logo for all vertebrates (Fig. 2A). This pattern of greater conservation of rod versus cone residues in the N-terminal region is even more evident when the analysis is confined to mammalian sequences (Fig. 2B).
Figure 2.
Consensus logo of PDE6G and PDE6H subunits. A, the vertebrate consensus logo for PDE6G and PDE6H was generated based on the multiple sequence alignment of 101 rod and 103 cone sequences shown in Fig. 1. B, the mammalian consensus logo is based on a multiple sequence alignment of a subset of the entire set of sequences and consisting of 50 mammalian PDE6G sequences and 51 mammalian PDE6H sequences.
Our analyses also reveal four highly conserved rod–cone differences among inhibitory subunit sequences that are virtually invariant since the last common ancestor of PDE6G and PDE6H (Fig. 2, A and B, indicated by vertical lines). With vertebrate PDE6G as a reference, these include residues Val-21, Gln-48, Asn-74, and Tyr-84 (Fig. 2A). The mutually exclusive nature of these residues is even more strongly pronounced when considering mammalian sequences alone (Fig. 2B), perhaps indicating purifying selection during the radiation of mammals. (Note that the insertion after Gly-59 of PDE6G (Fig. 2A) was found in only a single species and is unlikely to have biological significance.) In summary, PDE6H sequences have diversified much more than their rod sisters in the evolutionary history of craniates, and much of this sequence diversity is found within the first ∼15 N-terminal amino acid residues.
Gtα*-GTPγS activation of PDE6 is more effective with cone Pγ than rod Pγ
It has been reported that the rod and cone PDE6 holoenzymes are very similar in their catalytic properties and their interactions with their inhibitory Pγ subunits but differ in how rod and cone PDE6 are activated by transducin (10). We hypothesized that structural differences in rod and cone Pγ isoforms may be responsible for these different interactions between transducin and rod and cone PDE6.
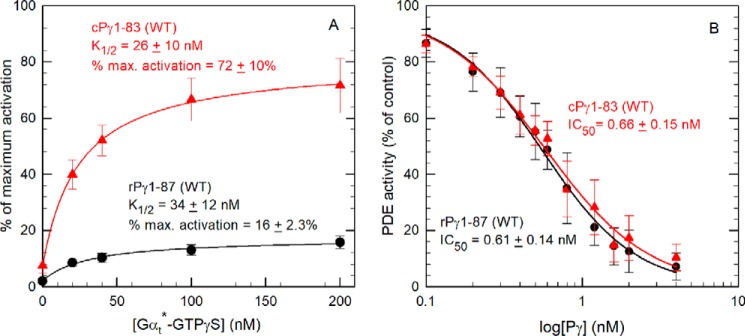
To test this, we first compared full-length recombinant rPγ and cPγ reconstituted with purified rod Pαβ catalytic dimers and tested the ability of rod Gtα*-GTPγS to activate PDE6 and accelerate cGMP hydrolysis. Fig. 3A shows that PDE6 containing cPγ is activated to a much greater extent (72% of the maximum catalytic activity of Pαβ lacking Pγ) than rPγ (16% maximum activation) when tested under identical experimental conditions. In contrast to the 4.5-fold difference in the extent of activation, the concentration dependence of Gtα*-GTPγS activation (K½) did not significantly differ for the two PDE6 enzyme preparations (Table 1).
Figure 3.
PDE6 reconstituted with cPγ is more effectively activated by Gtα*-GTPγS than PDE6 containing rPγ. A, Gtα*-GTPγS activation assay. 1 nm Pαβ was preincubated with a 10-fold molar excess of WT rPγ or cPγ to reconstitute the PDE6 holoenzyme. Gtα*-GTPγS was then added at the indicated concentrations and incubated for 1 h prior to measuring PDE activity with 2 mm cGMP as substrate. PDE activity is reported relative to the activity of Pαβ in the absence of Pγ. The data are the mean (± S.D.) for 19 or 20 separate determinations of PDE6 reconstituted with WT rPγ or cPγ, respectively. The data were fit by nonlinear regression analysis using a three-parameter hyperbolic equation and are reported in Table 1; basal activities for PDE6 reconstituted with rPγ and cPγ were 2% ± 0.8% and 8% ± 3%, respectively. B, Pγ inhibition assay. Pαβ (0.2 nm) was incubated with the indicated concentrations of WT rPγ or cPγ for 10 min, and then the catalytic activity was measured using 2 mm cGMP as substrate. The data are the mean (± S.D.) of eight experiments with rPγ and cPγ. A three-parameter logistic equation was used to estimate the IC50: rPγ, IC50 = 0.56 nm; cPγ, IC50 = 0.58 nm.
Table 1.
Effectiveness of Pγ constructs in promoting Gtα*-GTPγS activation of PDE6
Pαβ (1 nm) was preincubated with one of the rPγ or cPγ constructs (10 nm) to reconstitute the PDE6 holoenzyme. Various concentrations of Gtα*-GTPγS were then added and incubated for 1 h at room temperature. The PDE activity was measured using 2 mm cGMP as substrate and expressed as the percent activity referenced to fully activated Pαβ. The K½ and maximum percent activation were calculated by fitting the data to a three-parameter hyperbola. The IC50 of all Pγ constructs was determined using 0.2 nm Pαβ and 2 mm cGMP as substrates. The values represent the mean ± S.D. for n individual experiments.
| Pγ construct | K1/2 | Maximum activation | IC50 |
|---|---|---|---|
| nm | % | nm | |
| rPγ1–87 (WT) | 34 ± 12 | 16 ± 2.3 (19) | 0.61 ± 0.14 (12) |
| rPγ5–87 | 35 ± 6.5 | 15 ± 3.8 (3) | 0.81 |
| rPγ9–87 | 32 ± 4.1 | 15 ± 2.6 (3) | 0.51 |
| rPγ19–87 | 37 ± 9.9 | 28 ± 3.6 (9) | 0.74 ± 0.16 (5) |
| rPγ19–87V21T | 37 ± 14 | 55 ± 5.5 (5) | 0.86 ± 0.13 (3) |
| rPγ1–4_9–87 (Δ5–8) | 30 ± 2.1 | 25 ± 4.1 (3) | 0.58 |
| rPγ1–4_19–87 (Δ5–18) | 58 ± 9.0 | 55 ± 5.9 (4) | 0.40 (2) |
| rPγ1–8_19–87 (Δ9–18) | 78 ± 16 | 64 ± 2.6 (8) | 0.64 (2) |
| rPγ1–8_19–87V21T | 66 ± 12 | 81 ± 11 (3) | 0.56 |
| rPγV21T | 49 ± 1.4 | 27 ± 6.3 (3) | 0.42 |
| rPγ1–18_cPγ15–83 | 66 ± 18 | 30 ± 1.3 (4) | 0.46 |
| cPγ1–83 (WT) | 26 ± 10 | 72 ± 10 (20) | 0.66 ± 0.15 (13) |
| cPγ5–83 | 35 ± 15 | 66 ± 11 (3) | 0.68 |
| cPγ9–83 | 40 ± 18 | 62 ± 12 (3) | 0.62 |
| cPγ13–83 | 33 ± 17 | 72 ± 14 (5) | 0.83 |
| cPγ15–83 | 44 ± 18 | 45 ± 8.3 (11) | 0.79 ± 0.18 (5) |
| cPγ1–4_9–83 (Δ5–8) | 40 ± 8.6 | 80 ± 1.2 (3) | 0.69 |
| cPγ1–4_15–83 (Δ5–14) | 25 ± 6.5 | 83 ± 11 (3) | 0.70 (2) |
| cPγ1–8_15–83 (Δ9–14) | 52 ± 4.6 | 83 ± 11 (3) | 0.67 |
| cPγT17V | 32 ± 6.4 | 74 ± 7.8 (3) | 0.69 |
| cPγK44QS70NF80Y | 25 ± 2.0 | 72 ± 5.4 (4) | 0.59 ± 0.04 (3) |
| cPγ1–14_rPγ19–87 | 40 ± 20 | 67 ± 8.5 (4) | 0.54 |
To evaluate whether this 4.5-fold difference in maximum transducin activation of PDE6 could be accounted for by a lower intrinsic binding affinity of cPγ for Pαβ, we measured the concentration dependence of rPγ and cPγ to inhibit cGMP hydrolysis of Pαβ. As shown in Fig. 3B, no significant difference between IC50 values of rPγ and cPγ was observed. We conclude that the differences in overall binding affinity of Pγ to Pαβ cannot account for the greater efficacy of cPγ to promote transducin activation of PDE6.
The N-terminal region of cone Pγ is the locus for the differences in transducin activation of PDE6
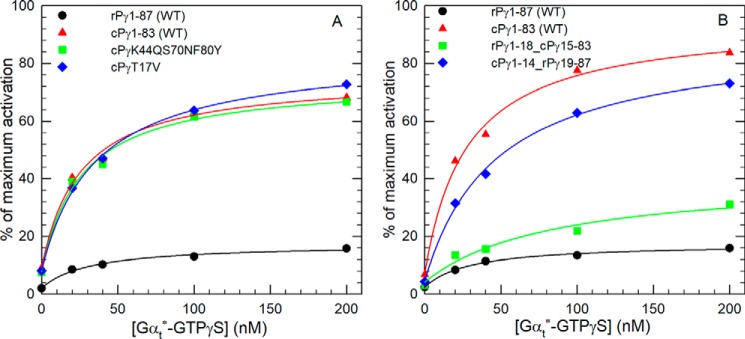
To identify the structural basis for the differences in the ability of rPγ and cPγ to permit efficient activation of PDE6 by transducin, we first hypothesized that one or more of the four highly conserved amino acid differences identified in the Pγ sequence alignment (vertical lines in Fig. 2B) were responsible. To test this, we generated site-directed mutants of cPγ to see whether converting the cone residue to its rod counterpart would suppress the extent of transducin activation of reconstituted PDE6. As seen in Fig. 4A, the cPγ triple mutant (K44Q,S70N,F80Y) was as effective as WT cPγ in supporting Gtα*-GTPγS activation of PDE6; a similar result was observed for the cPγT17V construct (Fig. 4A). From this result, we concluded that the variable N-terminal region of rPγ and cPγ was most likely responsible for the observed differences in the ability of transducin to activate PDE6.
Figure 4.
Four highly conserved sites that differ between rPγ and cPγ do not alter transducin activation efficacy, whereas the N-terminal region plays a primary role in regulating transducin activation. A, an rPγ amino acid was substituted for the cPγ residue at three sites (K44Q,S70N,F80Y) or at one site (T17V), and the site-directed mutants were then reconstituted with Pαβ prior to addition of varying amounts of Gtα*-GTPγS. B, chimeric Pγ mutants consisting of the N-terminal region of one Pγ fused to the remaining C-terminal sequence of the other Pγ were generated and assayed for the ability of PDE6 to be activated by increasing concentrations of Gtα*-GTPγS. Experiments were performed and analyzed as described in the legend for Fig. 3A, with values for K½ and percent of maximum activation provided in Table 1.
To evaluate the role of the N-terminal region of Pγ, we created two rod–cone Pγ chimeras consisting of the N-terminal region of one Pγ with the remaining C-terminal portion of the other Pγ: rPγ1–18–cPγ15–83 and cPγ1–14–rPγ19–87. We first verified that the two chimeric Pγ constructs inhibited Pαβ with affinities similar to that of the WT Pγ proteins (Table 1). As seen in Fig. 4B, the chimera containing the cPγ N-terminal region behaved essentially the same as WT cPγ, whereas the N-terminal region of rPγ enhanced Gtα*-GTPγS activation of PDE6 less than 2-fold (30% maximum activation) compared with WT rPγ (16% maximum activation). We conclude that differences in the N-terminal region of rPγ and cPγ, thought previously to lack functional significance, are responsible for regulating the ability of transducin to bind to and efficiently activate catalysis of the PDE6 holoenzyme.
Residues in the N-terminal region of rod Pγ impair the activation efficiency of Gtα*-GTPγS
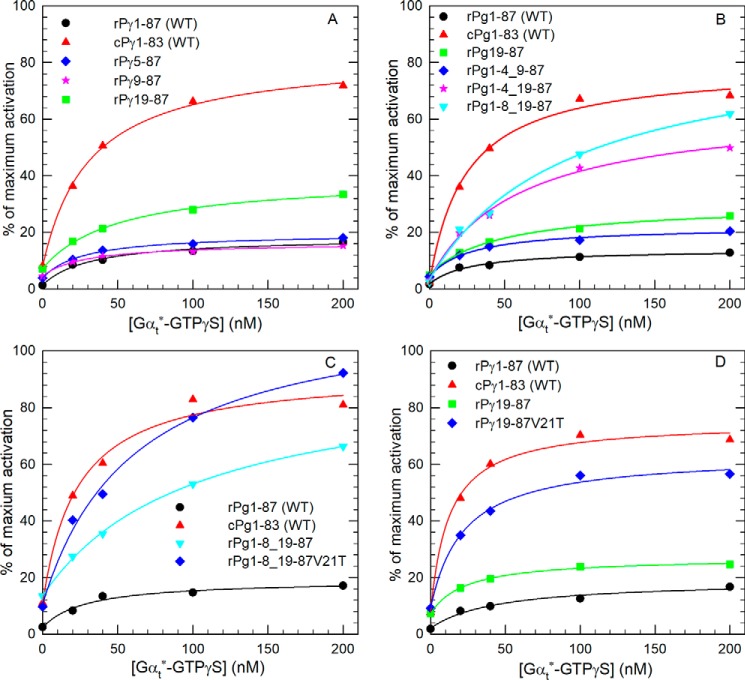
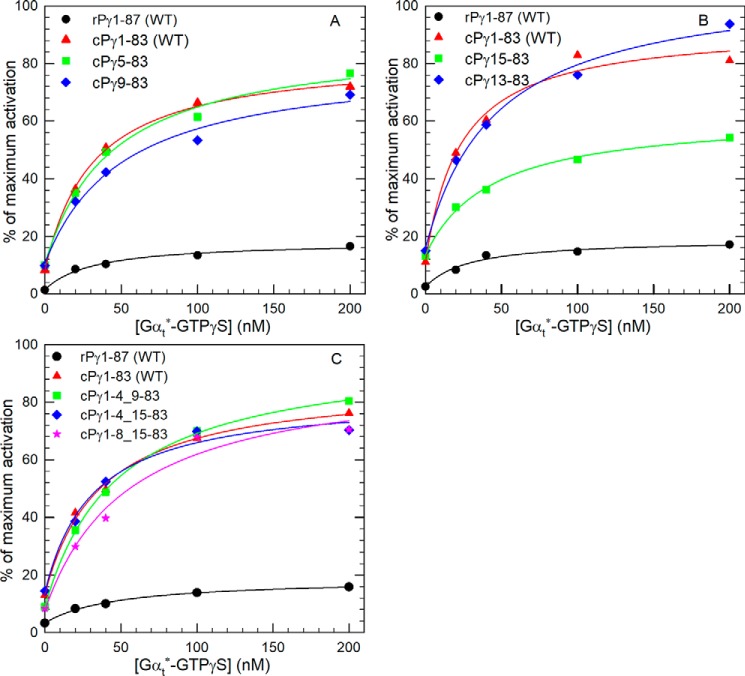
Because of sequence dissimilarity in the N-terminal region of rPγ and cPγ sequences (Fig. 2), it was difficult to predict potential functional sites responsible for suppressing the ability of transducin to activate Pαβ reconstituted with rPγ. Instead, we generated several N-terminal truncation mutants of rPγ to test whether removing a portion of the N terminus would enhance Gtα*-GTPγS activation of PDE6. As shown in Fig. 5A, removal of the first four or eight amino acids from the rPγ N terminus did not alter Gtα*-GTPγS activation efficiency, whereas removal of the first 18 amino acids resulted in less than a 2-fold increase in transducin-activated PDE catalysis.
Figure 5.
Residues in the N-terminal region of rPγ affect the Gtα*-GTPγS activation efficiency. A, three N-terminal truncated rPγ mutants (rPγ5–87, rPγ9–87, and rPγ19–87) were reconstituted with Pαβ to test Gtα*-GTPγS activation efficiency. B, three rPγ mutants with internal deletions were tested to evaluate the role of the N terminus in promoting PDE6 activation by Gtα*-GTPγS. C and D, Gtα*-GTPγS activation assays of PDE6, in which rPγ mutants were tested with and without the site-directed mutant V21T. Gtα*-GTPγS activation assays were performed and analyzed as described in Fig. 3A, and the K½ and percent of maximum activation are summarized in Table 1.
We next constructed several internal deletion mutants of rPγ to search for possible “inhibitory” elements within the N-terminal region that could suppress the ability of transducin to activate rod PDE6. Deletion of amino acid residues 5–8 (i.e. rPγ1–4_9–87) resulted in a modest increase in the maximum extent of Gtα*-GTPγS activation of PDE6 (25% of Pαβ activity; Fig. 5B and Table 1). Further enhancement of Gtα*-GTPγS activation of PDE6 was observed when amino acids 5–18 were deleted (55% of maximum Pαβ activity; Table 1). A positive role of the first four amino acids of the rPγ sequence in promoting transducin activation can be inferred by comparing the rPγ19–87 truncation mutant (28% of maximum Pαβ activity) with the internal deletion mutant (rPγ1–4_19–87; 55% of maximum activity). We further localized the region of rPγ that suppresses transducin activation of rod PDE6 by testing another internal deletion mutant of rPγ lacking only amino acids 9–18; in this instance, Pαβ reconstituted with rPγ1–8_19–87 achieved close to the same level of transducin activation (64%) as we observed for WT cPγ (72%; Fig. 5B and Table 1). These results demonstrate that amino acids 9–18 of mammalian rPγ represent a highly conserved sequence (E(I/F)RSATRV*G) that is absent in mammalian cPγ and that significantly reduces the ability of transducin to maximally activate PDE6. These results also indicate that the first eight amino acids of rPγ can enhance the ability of Gtα*-GTPγS to activate PDE6, although this effect is only observed when residues 9–18 are deleted.
Having identified a rPγ-specific element that decreased the ability of transducin to fully activate PDE6, we sought to determine whether substituting the nearby highly conserved rod–cone Pγ difference (Val-21 in rPγ and Thr-17 in cPγ) would enhance transducin activation when the rod inhibitory region was deleted. Fig. 5C shows that the rPγ construct (Pγ1–8_19–87V21T) achieved a slightly greater maximum extent of activation (80%) compared with WT cPγ (72%); a similar enhancing effect of this V21T substitution was also observed when the N-terminal 18 amino acids were deleted (rPγ18–87V21T; Fig. 5D). When tested as a single site substitution, the rPγV21T construct caused a 10% increase in Gtα*-GTPγS activation of PDE6 (Table 1). These results indicate that the valine at position 21 of the rPγ sequence contributes to the decreased ability of Gtα*-GTPγS to bind to and effectively activate PDE6.
The cone Pγ residues Asn-13 and Gln-14 contribute to efficient Gtα*-GTPγS activation of PDE6
To determine which N-terminal residues of cPγ modulate the ability of transducin to effectively activate PDE6, several cPγ N-terminal mutants were created. Fig. 6A shows that removal of the first four (cPγ5–83) or eight (cPγ9–83) residues of cPγ has an insignificant effect on the ability of Gtα*-GTPγS to activate PDE6 containing these mutant cPγ constructs. Although truncation of the first 14 amino acids from cPγ (cPγ15–83) significantly reduced the maximum extent of transducin activation, inclusion of amino acids Asn-13 and Gln-14 (cPγ13–83) was sufficient to restore transducin activation to levels observed for WT cPγ (Fig. 6B and Table 1).
Figure 6.
Identification of residues in the N-terminal region of cPγ that enhance Gtα*-GTPγS activation efficiency. A and B, four N-terminal truncated cPγ mutants (cPγ5–83, cPγ9–83, cPγ13–83, and cPγ15–83) were reconstituted with Pαβ to test the Gtα*-GTPγS activation efficiency of PDE6. C, three internal deletion mutants of cPγ (cPγ1–4_9–83, cPγ1–4_15–83, and cPγ 1–8_15–83) were also tested under the same experimental conditions. The Gtα*-GTPγS activation assay was carried out and analyzed as described in the legend for Fig. 3A, and the results are summarized in Table 1.
Because the first four or eight N-terminal amino acids of rPγ were able to enhance the ability of Gtα*-GTPγS to bind to and activate PDE6 when the rPγ inhibitory region was deleted (Fig. 5B), we also investigated whether the N-terminal amino acids of cPγ contributed to transducin activation efficacy. Comparison of cPγ9–83 (62% of Pαβ activity; Fig. 6A and Table 1) with cPγ1–4_9–83 (80% maximal activation; Fig. 6C and Table 1) revealed a significant enhancing effect of the first four amino acids of cPγ. More significantly, the impaired activation efficiency of cPγ15–83 (Fig. 6B) could be completely reversed by inclusion of the first four or eight N-terminal amino acids (Fig. 6C), rising from 45% for cPγ15–83 to 83% of the Pαβ activity for either cPγ1–4_15–83 or cPγ1–8_15–83 (Table 1). We conclude from Fig. 6 that either residues Asn-13 and Asn-14 or the first several N-terminal amino acids of cPγ are sufficient for Gtα*-GTPγS to maximally activate PDE6 containing these cPγ constructs.
Discussion
Using a phylogenetic analysis to identify functionally important differences between the rod and cone isoforms of Pγ in conjunction with biochemical assays of transducin activation of PDE6 reconstituted with different rPγ and cPγ mutants, we determined that some of the well-established physiological differences in light responsiveness of rod and cone photoreceptors (1, 2) can be ascribed to structural differences in the N-terminal region of rod and cone Pγ. This work also identifies, for the first time, a regulatory role of the N terminus of Pγ in modulating the ability of transducin to bind to the PDE6 holoenzyme and relieve the inhibitory constraint on cGMP hydrolysis imposed by the last 10 C-terminal residues of Pγ (19).
Current evidence suggests that the physiological differences in the response to light exhibited by rods and cones had already evolved prior to the last common ancestor of living vertebrates (16, 18, 20). Our analyses of Pγ sequence evolution provide additional context to this suggestion, as we demonstrate that these physiological differences are explained in part by ancient divergences in the N terminus region of Pγ. A recent paper by Lagman et al. (15) analyzed Pγ sequences but did not attempt a phylogenetic analysis because of high sequence conservation and a proposed lack of phylogenetic signal. However, our phylogenetic analyses of Pγ demonstrate two well-supported clades consisting of PDE6G (rod) and PDE6H (cone) sequences. We do observe generally low support for internal nodes by either bootstrapping or approximate likelihood ratio indices, but the delineation between the major clades is clear. Lagman et al. (15) also proposed a new clade of Pγ called PDE6I. In our analyses, PDE6I sequences are found within the PDE6H clade. This finding does not dispute the existence of PDE6I genes, but our phylogenetic results indicate that they are best interpreted as duplicate PDE6H genes. We chose to root our phylogeny in the most evolutionarily parsimonious manner, which places Pγ sequences from the hagfish Eptatretus stoutii at the base of both the rod and cone clades. However, we note that electrophysiological evidence for cyclostome photoreceptor light responsiveness exists only for the lamprey (17, 21, 22), and other rootings for the Pγ phylogenetic tree could change the affinity of the hagfish Pγ sequences.
On the amino acid sequence level, we find that PDE6G sequences are generally far more conserved than PDE6H sequences (Figs. 1 and 2). Because rod phototransduction shows single-photon sensitivity and has been considered a paragon of the efficacy of natural selection (23, 24), this strong conservation among PDE6G sequences could be interpreted as evidence for purifying selection. Alternatively, sequence divergence among PDE6H sequences might be an indication of diversifying selection, as the kinetics of cone phototransduction and its role in color vision might facilitate selection for adaptive but alternative regulatory properties. These two scenarios are not mutually exclusive, and functional studies from the cyclostomes will be needed to provide further resolution.
In addition to highly reliable single photon detection, rod photoreceptors differ from cone photoreceptors in having a lower level of continuous “dark noise” in their dark-adapted state (25, 26) as well as a longer latency period for the initial “rising” phase of the response to flash stimuli (27). Recently, it has been reported that rod PDE6 activation by transducin requires binding of two activated transducins to the rod PDE6 holoenzyme (28); furthermore, computer simulations of this dimeric activation model for rod PDE6 (29) support the idea that this dimeric transducin activation mechanism serves to reduce rod PDE6 spontaneous activation (i.e. reduced dark noise) and accounts for the longer delay in the rising phase of visual excitation compared with the cone photoresponse in vivo. Because cone PDE6 consists of a catalytic homodimer, Lamb et al. (29) postulated that the symmetrical cone catalytic dimer may permit each cone catalytic subunit to be activated by transducin binding in an independent, noncooperative manner. Our results support an alternative hypothesis in which differences between rod and cone Pγ isoforms in the N-terminal region of the protein and not differences between rod and cone PDE6 catalytic subunits determine whether transducin can activate each catalytic subunit independently (in the presence of cPγ) or, alternatively, require binding of two transducins to the rod PDE6 holoenzyme (containing rPγ) for maximal activation. However, it should be pointed out that our in vitro biochemical data were obtained with protein concentrations several orders of magnitude lower than in the photoreceptor outer segment, and it will be important to evaluate the importance of the N-terminal region of Pγ under physiological conditions.
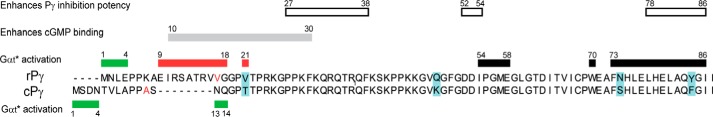
Previous work delineating the sites of interaction of the intrinsically disordered rPγ subunit (11) with the rod PDE6 catalytic dimer have identified several functionally important regions (Fig. 7): the approximately 10 C-terminal amino acid residues of rPγ are known to directly interact with the catalytic domain to regulate access of substrate to the active site and directly control catalytic activity (19, 30); a region in the N-terminal half of rPγ (amino acids 10–30) enhances the affinity of cGMP to noncatalytic binding sites in the regulatory domain of the catalytic dimer (31), most likely by direct binding of this region of rPγ to the cGMP binding pocket (32, 33); and multiple binding sites of rPγ with Pαβ have been identified along practically the entire length of the rPγ sequence, with its central region making a major contribution to the high overall affinity of rPγ for Pαβ (31). In contrast to these functionally important regions of rPγ, the N-terminal region of rPγ (specifically its first 17 residues) was not observed to contribute to the ability of Pγ to inhibit cGMP hydrolysis or to stimulate cGMP binding (31). Because of the difficulty of isolating biochemical quantities of purified cone PDE6 holoenzyme, comparable characterizations of the interactions of cPγ with cone catalytic homodimer are lacking.
Figure 7.
Functionally important sites of the rPγ and cPγ inhibitory subunits. Blue-shaded residues represent highly conserved rod-cone Pγ differences. Solid green boxes represent regions that enhance Gtα* activation. The solid red boxes represent the regions that impair Gtα* activation. The red letters identify two amino acids in the bovine Pγ sequences that differ from the mammalian consensus sequences: M17V (rPγ) and T11A (cPγ). Also shown are previously identified regions that enhance rPγ inhibition potency (white boxes), enhance noncatalytic cGMP binding to the GAFa domain (gray box), or are critical for activation of PDE6 by activated transducin (black boxes) (31).
Upon rod PDE6 activation by transducin, Gtα*-GTP interacts with multiple sites within the C-terminal half of rPγ (Fig. 7, black boxes) to displace rPγ from the catalytic domain, allowing diffusion of cGMP into the active site (34–36). Studies of the interactions of rPγ with the activated transducin α-subunit have also identified a second region of interaction in the polycationic central region of rPγ (37, 38) whose function is uncertain. This work is the first report showing that the N-terminal region of Pγ (preceding its polycationic region) plays an important role in Gtα*-GTP activation of PDE6. For both rPγ and cPγ, the first four amino acids significantly contribute to enhancing the maximum extent of transducin activation under conditions where neighboring amino acids have been deleted (Δ5–8; Figs. 5 and 6). Of greater significance are the inhibitory (rPγ residues 9–18 and Val-21; Fig. 5) and stimulatory loci (cPγ Asn-13 and Gln-14; Figs. 4 and 6) we discovered that modulate the efficacy with which transducin can bind to Pγ and activate PDE6 catalysis.
The fact that these sites near the N terminus of Pγ overlap with the region of rPγ shown previously to enhance cGMP binding affinity to the GAFa domain (Fig. 7) is consistent with the following allosteric mechanism. We hypothesize that rPγ residues 9–21 may be allosterically coupled to the central polycationic region of rPγ (residues ∼22–45) and reduce the affinity of transducin binding to this region of rPγ, which, in turn, reduces the ability of transducin to bind to and displace the C-terminal residues of rPγ required for disinhibition of PDE6 catalysis. Conversely, residues 13–17 in cPγ may serve to allosterically enhance transducin binding to the central region of cPγ, facilitating full activation of PDE6 catalysis. The lower affinity of cGMP for the GAFa domains of cone PDE6 than for rod PDE6 (8, 39) and the well-established reciprocal regulation of rPγ and cGMP binding affinity to the PDE6 catalytic dimer (reviewed in Ref. 3) also likely contributes to differences in the allosteric regulation of rPγ and cPγ interactions with activated transducin.
In conclusion, this work demonstrates that the N-terminal region of the rod and cone inhibitory Pγ subunits allosterically regulate the efficiency with which activated transducin is able to displace Pγ from the active site of the PDE6 holoenzyme. This supports the hypothesis that the activation mechanism of PDE6 differs in rods and cones and can account for at least some of the physiological differences in rod and cone light responsiveness. These results will also contribute to a better understanding of the molecular etiology of disease-causing mutations in the catalytic and inhibitory subunits of photoreceptor PDE6, particularly those that occur in the regulatory domains of these proteins (Ref. 40 and references cited therein).
Experimental procedures
Materials
Bovine retinas were purchased from W. L. Lawson, Inc. Synthetic peptides (rPγ19–87 and cPγ15–83) were purchased from New England Peptide. All synthetic DNAs of rPγ and cPγ mutants were purchased from Thermo Fisher–Invitrogen. Site-directed mutants of Pγ were generated using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). Filtration membranes and chemicals were from Millipore–Sigma.
Sequence alignments and phylogenetic analyses
PDE6G and PDE6H sequences from a diversity of vertebrate species were obtained from Uniprot or from the literature (14, 18). Additional sequences were obtained from the gene models of 56 additional craniate species with whole-genome sequences using a custom phylogenomics pipeline. This pipeline uses BLAST (e value = 0.0001) to search individual datasets using PDE6G and PDE6H query sequences (CNRG_HUMAN, P18545.1; CNCG_BOVIN, P22571.1) (41). Redundant sequences were then removed using cdhit (−c = 1.0 (42)), and the resulting dataset was aligned using the progressive algorithm implemented in PASTA with default parameters (43). Following the initial PASTA alignment, we removed four sequences that were either shorter than 50 amino acid residues or had internal deletions longer than 30 amino acids. Sequences were then realigned using PASTA under default parameters; we confirmed that additional iterations of progressive alignment did not produce improvements. The resulting Pγ amino acid sequences from different vertebrate species are listed in Table S1.
The phylogeny of PDE6G and PDE6H was then estimated using IQ-tree under the best fit model (44); see Fig. S1 for the resulting tree with species labels. Node support was ascertained using both bootstrap and approximate likelihood ratio scores (45). The final tree with tip label and approximate likelihood ratio scores is available as a .tre file in the Github repository associated with this paper. All scripts and command lines used in sequence analyses are located at https://github.com/plachetzki/PDE6_GAMMA.3
Construction and purification of Pγ mutants
Codon-optimized synthetic DNA fragments coding for various bovine rPγ and cPγ constructs used in this study (Fig. S1) were inserted into the NdeI and BamHI sites of the pET11a vector, followed by transformation into the Escherichia coli BL21(DE3) strain. The sequence of all Pγ mutants was confirmed by DNA sequencing (Functional Biosciences).
Following expression of recombinant Pγ mutants in E. coli BL21(DE3), the bacterial extract was purified by HiTrap SP FF column from GE Healthcare. The Pγ mutants were further purified by C18 reverse-phase HPLC following standard procedures (46). The purity of these proteins was determined to be >90%, as judged by SDS-PAGE. Protein concentrations were determined by the bicinchoninic acid protein assay using bovine γ-globulin as a standard.
PDE6 and Pαβ purification and activity assays
The bovine rod PDE6 holoenzyme was purified from bovine retinas as described previously (47). Pαβ catalytic dimers were prepared by limited trypsin proteolysis and repurified by Mono Q anion exchange chromatography prior to use (47). PDE6 catalytic activity was measured in 20 mm HEPES, 10 mm MgCl2, and 0.5 mg/ml BSA using a colorimetric assay (48). The PDE6 concentration was estimated based on the rate of cGMP hydrolysis of trypsin-activated PDE6 and knowledge of the kcat of the enzyme (5600 mol cGMP hydrolyzed per 1 mol Pαβ per second (39). The IC50 values of all Pγ constructs reported in Table 1 were determined using 0.2 nm Pαβ and 2 mm cGMP as substrates (39, 49); in all cases, the IC50 values did not differ significantly from those observed for WT rPγ and cPγ (Fig. 3B). Note that the difficulty in obtaining biochemical quantities of purified bovine cone photoreceptor PDE6 holoenzyme and bovine cone transducin precluded carrying out complementary experiments with these cone isoforms.
Purification of Gtα*-GTPγS and measurements of transducin activation of PDE6
Transducin α-subunits were extracted from the PDE6-depleted rod outer segment membranes by addition of 50 μm GTPγS. The extracted Gtα*-GTPγS was purified on a Blue Sepharose column as described previously (50, 51), followed by gel filtration chromatography to remove residual PDE6. The concentration of Gtα*-GTPγS was determined by a colorimetric protein assay. Purified Gtα*-GTPγS (in 50% glycerol) was stored at −20 °C.
To measure transducin activation of PDE6, 1 nm Pαβ was preincubated with 10 nm Pγ mutant for 10 min prior to addition of the indicated concentrations of Gtα*-GTPγS for 1 h at room temperature. The PDE6 activity was then assayed at a final concentration of 0.2 nm using 2 mm cGMP as substrate. Note that these assay conditions were chosen to maximize differences between Pαβ reconstituted with rPγ and cPγ; however, even with a 10-fold higher concentration of purified rod PDE6 holoenzyme (2 nm) and a 1000-fold excess of Gtα*-GTPγS, we failed to observe more than 50% maximal activation by Gtα*-GTPγS (similar to literature values; see Refs. 8, 10, 31).
Data analysis
All experiments were repeated at least three times. The transducin concentration dependence of PDE6 activation (Figs. 3A and 4–6) was analyzed by nonlinear regression (Sigmaplot v.12.5, SPSS, Inc.) using a three-parameter hyperbolic equation: y = y0 + a × x/(b + x), where a is the maximum percent activation, b is the K½, and y0 is the basal PDE6 activity in the absence of Gtα*-GTPγS. Pγ inhibition potency (Fig. 3B) was determined using a three-parameter logistic equation: y = a/(1 + (x/x0)b), where a is the amplitude, b is the slope, and x0 is the IC50 value.
Author contributions
X. W., D. C. P., and R. H. C. data curation; X. W., D. C. P., and R. H. C. formal analysis; X. W. and D. C. P. investigation; X. W., D. C. P., and R. H. C. methodology; X. W. and R. H. C. writing-original draft; X. W., D. C. P., and R. H. C. writing-review and editing; R. H. C. conceptualization; R. H. C. resources; R. H. C. supervision; R. H. C. funding acquisition; R. H. C. validation; R. H. C. project administration.
Supplementary Material
Acknowledgment
We thank Karyn B. Cahill for construction of some of the Pγ constructs used in this paper.
This work was supported by NEI, National Institutes of Health Grant 1R01EY05798-29 and NIGMS, National Institutes of Health Grant P20GM113131 (to R. H. C.) and National Science Foundation IOS Award 1755337 and United States Department of Agriculture–National Institute of Food and Agriculture Hatch Project 00654 (to D. C. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- Gt
- heterotrimeric G protein transducin
- PDE
- phosphodiesterase.
References
- 1. Korenbrot J. I. (2012) Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: facts and models. Prog. Retin. Eye Res. 31, 442–466 10.1016/j.preteyeres.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingram N. T., Sampath A. P., and Fain G. L. (2016) Why are rods more sensitive than cones? J. Physiol. 594, 5415–5426 10.1113/JP272556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cote R. H. (2006) In Cyclic Nucleotide Phosphodiesterases in Health and Disease (Beavo J. A., Francis S. H., and Houslay M. D., eds.) pp. 165–193, CRC Press, Boca Raton, FL [Google Scholar]
- 4. Kefalov V. J. (2012) Rod and cone visual pigments and phototransduction through pharmacological, genetic, and physiological approaches. J. Biol. Chem. 287, 1635–1641 10.1074/jbc.R111.303008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deng W. T., Sakurai K., Liu J., Dinculescu A., Li J., Pang J., Min S. H., Chiodo V. A., Boye S. L., Chang B., Kefalov V. J., and Hauswirth W. W. (2009) Functional interchangeability of rod and cone transducin α-subunits. Proc. Natl. Acad. Sci. U.S.A. 106, 17681–17686 10.1073/pnas.0901382106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gopalakrishna K. N., Boyd K. K., and Artemyev N. O. (2012) Comparative analysis of cone and rod transducins using chimeric Ga subunits. Biochemistry 51, 1617–1624 10.1021/bi3000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mao W., Miyagishima K. J., Yao Y., Soreghan B., Sampath A. P., and Chen J. (2013) Functional comparison of rod and cone Gαt on the regulation of light sensitivity. J. Biol. Chem. 288, 5257–5267 10.1074/jbc.M112.430058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gillespie P. G., and Beavo J. A. (1988) Characterization of a bovine cone photoreceptor phosphodiesterase purified by cyclic GMP-Sepharose chromatography. J. Biol. Chem. 263, 8133–8141 [PubMed] [Google Scholar]
- 9. Mou H., and Cote R. H. (2001) The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory γ subunit. J. Biol. Chem. 276, 27527–27534 10.1074/jbc.M103316200 [DOI] [PubMed] [Google Scholar]
- 10. Muradov H., Boyd K. K., and Artemyev N. O. (2010) Rod phosphodiesterase-6 PDE6A and PDE6B subunits are enzymatically equivalent. J. Biol. Chem. 285, 39828–39834 10.1074/jbc.M110.170068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo L. W., and Ruoho A. E. (2008) The retinal cGMP phosphodiesterase γ-subunit: a chameleon. Curr. Protein Pept. Sci. 9, 611–625 10.2174/138920308786733930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arshavsky V. Y., and Wensel T. G. (2013) Timing is everything: GTPase regulation in phototransduction. Invest. Ophthalmol. Vis. Sci. 54, 7725–7733 10.1167/iovs.13-13281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skiba N. P., Hopp J. A., and Arshavsky V. Y. (2000) The effector enzyme regulates the duration of G protein signaling in vertebrate photoreceptors by increasing the affinity between transducin and RGS protein. J. Biol. Chem. 275, 32716–32720 10.1074/jbc.C000413200 [DOI] [PubMed] [Google Scholar]
- 14. Lamb T. D., Patel H., Chuah A., Natoli R. C., Davies W. I., Hart N. S., Collin S. P., and Hunt D. M. (2016) Evolution of vertebrate phototransduction: cascade activation. Mol. Biol. Evol. 33, 2064–2087 10.1093/molbev/msw095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lagman D., Franzén I. E., Eggert J., Larhammar D., and Abalo X. M. (2016) Evolution and expression of the phosphodiesterase 6 genes unveils vertebrate novelty to control photosensitivity. BMC Evol. Biol. 16, 124 10.1186/s12862-016-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamb T. D. (2013) Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119 10.1016/j.preteyeres.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 17. Morshedian A., and Fain G. L. (2017) Light adaptation and the evolution of vertebrate photoreceptors. J. Physiol. 595, 4947–4960 10.1113/JP274211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muradov H., Boyd K. K., Kerov V., and Artemyev N. O. (2007) PDE6 in lamprey Petromyzon marinus: implications for the evolution of the visual effector in vertebrates. Biochemistry 46, 9992–10000 10.1021/bi700535s [DOI] [PubMed] [Google Scholar]
- 19. Granovsky A. E., Natochin M., and Artemyev N. O. (1997) The γ subunit of rod cGMP-phosphodiesterase blocks the enzyme catalytic site. J. Biol. Chem. 272, 11686–11689 10.1074/jbc.272.18.11686 [DOI] [PubMed] [Google Scholar]
- 20. Morshedian A., and Fain G. L. (2017) The evolution of rod photoreceptors. Philos. Trans. R Soc. Lond. B Biol. Sci. 372, 20160074 10.1098/rstb.2016.0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asteriti S., Grillner S., and Cangiano L. (2015) A Cambrian origin for vertebrate rods. eLife 10.7554/eLife.07166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morshedian A., and Fain G. L. (2015) Single-photon sensitivity of lamprey rods with cone-like outer segments. Curr. Biol. 25, 484–487 10.1016/j.cub.2014.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hecht S., Shlaer S., and Pirenne M. H. (1942) Energy, quanta, and vision. J. Gen. Physiol. 25, 819–840 10.1085/jgp.25.6.819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pugh E. N., Jr. (2018) The discovery of the ability of rod photoreceptors to signal single photons. J. Gen. Physiol. 150, 383–388 10.1085/jgp.201711970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rieke F., and Baylor D. A. (1996) Molecular origin of continuous dark noise in rod photoreceptors. Biophys. J. 71, 2553–2572 10.1016/S0006-3495(96)79448-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rieke F., and Baylor D. A. (2000) Origin and functional impact of dark noise in retinal cones. Neuron 26, 181–186 10.1016/S0896-6273(00)81148-4 [DOI] [PubMed] [Google Scholar]
- 27. Rotov A. Y., Astakhova L. A., Firsov M. L., and Govardovskii V. I. (2017) Origins of the phototransduction delay as inferred from stochastic and deterministic simulation of the amplification cascade. Mol. Vis. 23, 416–430 [PMC free article] [PubMed] [Google Scholar]
- 28. Qureshi B. M., Behrmann E., Schöneberg J., Loerke J., Bürger J., Mielke T., Giesebrecht J., Noé F., Lamb T. D., Hofmann K. P., Spahn C. M. T., and Heck M. (2018) It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods. Open Biol. 8, 180075 10.1098/rsob.180075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lamb T. D., Heck M., and Kraft T. W. (2018) Implications of dimeric activation of PDE6 for rod phototransduction. Open Biol. 8, 180076 10.1098/rsob.180076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X.-J., Skiba N. P., and Cote R. H. (2010) Structural requirements of the photoreceptor phosphodiesterase gamma-subunit for inhibition of rod PDE6 holoenzyme and for its activation by transducin. J. Biol. Chem. 285, 4455–4463 10.1074/jbc.M109.057406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X. J., Gao X. Z., Yao W., and Cote R. H. (2012) Functional mapping of interacting regions of the photoreceptor phosphodiesterase (PDE6) γ-subunit with PDE6 catalytic dimer, transducin, and regulator of G-protein signaling 9-1 (RGS9-1). J. Biol. Chem. 287, 26312–26320 10.1074/jbc.M112.377333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muradov K. G., Granovsky A. E., Schey K. L., and Artemyev N. O. (2002) Direct interaction of the inhibitory γ-subunit of rod cGMP phosphodiesterase (PDE6) with the PDE6 GAFa domains. Biochemistry 41, 3884–3890 10.1021/bi015935m [DOI] [PubMed] [Google Scholar]
- 33. Zeng-Elmore X., Gao X. Z., Pellarin R., Schneidman-Duhovny D., Zhang X. J., Kozacka K. A., Tang Y., Sali A., Chalkley R. J., Cote R. H., and Chu F. (2014) Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J. Mol. Biol. 426, 3713–3728 10.1016/j.jmb.2014.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Skiba N. P., Artemyev N. O., and Hamm H. E. (1995) The carboxyl terminus of the γ-subunit of rod cGMP phosphodiesterase contains distinct sites of interaction with the enzyme catalytic subunits and the α-subunit of transducin. J. Biol. Chem. 270, 13210–13215 10.1074/jbc.270.22.13210 [DOI] [PubMed] [Google Scholar]
- 35. Liu Y., Arshavsky V. Y., and Ruoho A. E. (1996) Interaction sites of the COOH-terminal region of the γ subunit of cGMP phosphodiesterase with the GTP-bound α subunit of transducin. J. Biol. Chem. 271, 26900–26907 10.1074/jbc.271.43.26900 [DOI] [PubMed] [Google Scholar]
- 36. Guo L. W., Hajipour A. R., and Ruoho A. E. (2010) Complementary interactions of the rod PDE6 inhibitory subunit with the catalytic subunits and transducin. J. Biol. Chem. 285, 15209–15219 10.1074/jbc.M109.086116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Artemyev N. O., Rarick H. M., Mills J. S., Skiba N. P., and Hamm H. E. (1992) Sites of interaction between rod G-protein α-subunit and cGMP-phosphodiesterase γ-subunit: implications for phosphodiesterase activation mechanism. J. Biol. Chem. 267, 25067–25072 [PubMed] [Google Scholar]
- 38. Artemyev N. O. (1997) Binding of transducin to light-activated rhodopsin prevents transducin interaction with the rod cGMP phosphodiesterase γ-subunit. Biochemistry 36, 4188–4193 10.1021/bi963002y [DOI] [PubMed] [Google Scholar]
- 39. Mou H., Grazio H. J. 3rd, Cook T. A., Beavo J. A., and Cote R. H. (1999) cGMP binding to noncatalytic sites on mammalian rod photoreceptor phosphodiesterase is regulated by binding of its γ and δ subunits. J. Biol. Chem. 274, 18813–18820 10.1074/jbc.274.26.18813 [DOI] [PubMed] [Google Scholar]
- 40. Gopalakrishna K. N., Boyd K., and Artemyev N. O. (2017) Mechanisms of mutant PDE6 proteins underlying retinal diseases. Cell Signal. 37, 74–80 10.1016/j.cellsig.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul S. F., and Koonin E. V. (1998) Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem. Sci. 23, 444–447 10.1016/S0968-0004(98)01298-5 [DOI] [PubMed] [Google Scholar]
- 42. Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mirarab S., Nguyen N., Guo S., Wang L. S., Kim J., and Warnow T. (2015) PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J. Comput. Biol. 22, 377–386 10.1089/cmb.2014.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen L. T., Schmidt H. A., von Haeseler A., and Minh B. Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anisimova M., and Gascuel O. (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- 46. Artemyev N. O., Arshavsky V. Y., and Cote R. H. (1998) Photoreceptor phosphodiesterase: interaction of inhibitory γ subunit and cyclic GMP with specific binding sites on catalytic subunits. Methods 14, 93–104 10.1006/meth.1997.0568 [DOI] [PubMed] [Google Scholar]
- 47. Pentia D. C., Hosier S., Collupy R. A., Valeriani B. A., and Cote R. H. (2005) Purification of PDE6 isozymes from mammalian retina. Methods Mol. Biol. 307, 125–140 [DOI] [PubMed] [Google Scholar]
- 48. Cote R. H. (2000) Kinetics and regulation of cGMP binding to noncatalytic binding sites on photoreceptor phosphodiesterase. Methods Enzymol. 315, 646–672 10.1016/S0076-6879(00)15873-2 [DOI] [PubMed] [Google Scholar]
- 49. Hurley J. B., and Stryer L. (1982) Purification and characterization of the γ regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J. Biol. Chem. 257, 11094–11099 [PubMed] [Google Scholar]
- 50. Kleuss C., Pallast M., Brendel S., Rosenthal W., and Schultz G. (1987) Resolution of transducin subunits by chromatography on blue Sepharose. J. Chromatogr. 407, 281–289 10.1016/S0021-9673(01)92625-1 [DOI] [PubMed] [Google Scholar]
- 51. Wensel T. G., He F., and Malinski J. A. (2005) Purification, reconstitution on lipid vesicles, and assays of PDE6 and its activator G protein, transducin. Methods Mol. Biol. 307, 289–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.