Abstract
Prostate cancer is the second leading cause of cancer death among men in the United States. The androgen receptor (AR) antagonist enzalutamide is a Food and Drug Administration–approved drug for treatment of patients with late-stage prostate cancer and is currently under clinical study for early-stage prostate cancer treatment. After a short positive response period, tumors will develop drug resistance. In this study using RNA-Seq and bioinformatics analyses, we observed that NOTCH signaling is a deregulated pathway in enzalutamide-resistant cells. NOTCH2 and c-MYC gene expression positively correlated with AR expression in samples from patient with hormone refractory disease in which AR expression levels correspond to those typically observed in enzalutamide resistance. Cleaved NOTCH1, HES1 (Hes family BHLH transcription factor 1), and c-MYC protein expression levels are elevated in two enzalutamide-resistant cell lines, MR49F and C4-2R, indicating NOTCH signaling activation. Moreover, inhibition of the overexpressed ADAM metallopeptidase domain 10 (ADAM10) in the resistant cells induces an exclusive reduction in cleaved NOTCH1 expression. Furthermore, exposure of enzalutamide-resistant cells to both PF-03084014 and enzalutamide increased cell death, decreased colony formation ability, and resensitized cells to enzalutamide. Knockdown of NOTCH1 in C4-2R increased enzalutamide sensitivity by decreasing cell proliferation and increasing cleaved PARP expression. In a 22RV1 xenograft model, PF-03084014 and enzalutamide decreased tumor growth through reducing cell proliferation and increasing apoptosis. These results indicate that NOTCH1 signaling may contribute to enzalutamide resistance in prostate cancer, and inhibition of NOTCH signaling can resensitize resistant cells to enzalutamide.
Keywords: drug resistance, prostate cancer, Notch pathway, androgen receptor, anticancer drug, ADAM, androgen receptor, enzalutamide, Notch1, PF-03084014
Introduction
Prostate cancer is the most diagnosed cancer in men in the United States (1). Despite the high survival rate of prostate cancer patients, most develop resistance to therapies, leading to a more aggressive form of the disease and ultimately death. Enzalutamide, initially approved as a second-line treatment of metastatic prostate cancer patients, has shown significant beneficial results when used as first-line therapy for patients with metastatic prostate cancer or even early stage prostate cancer in combination with hormonal therapy (2, 3). Shortly after a brief response period to enzalutamide, patients will develop resistance leading to death (4). With enzalutamide in ongoing clinical trial on early stage prostate cancer patients, it is of high importance to understand the mechanism by which prostate cancer patients develop resistance to enzalutamide and identify novel approaches to overcome resistance. By understanding and identifying these mechanisms, targeted combination therapies can be used to treat patients who develop drug resistance.
In general, mechanisms of androgen deprivation therapy (ADT)2 resistance and specifically enzalutamide resistance mainly highlight the role of the androgen receptor (AR) in resistance development, including AR amplification, activating mutations, and AR variants (5, 6). In this study, we hypothesized that genes and pathways outside of the AR signaling pathway contribute to enzalutamide resistance in prostate cancer cells. To test our hypothesis, we used RNA-Seq to compare previously developed enzalutamide-resistant cell lines to their parental cell lines to identify potential non-AR pathways that contribute to resistance. Our results show that the Notch signaling pathway is linked to enzalutamide resistance. Abrogation of this pathway in vitro and in vivo restores sensitivity of prostate cancer cells to enzalutamide.
Results
Long-term enzalutamide treatment of prostate cancer cell lines induces global gene expression changes after acquiring resistance
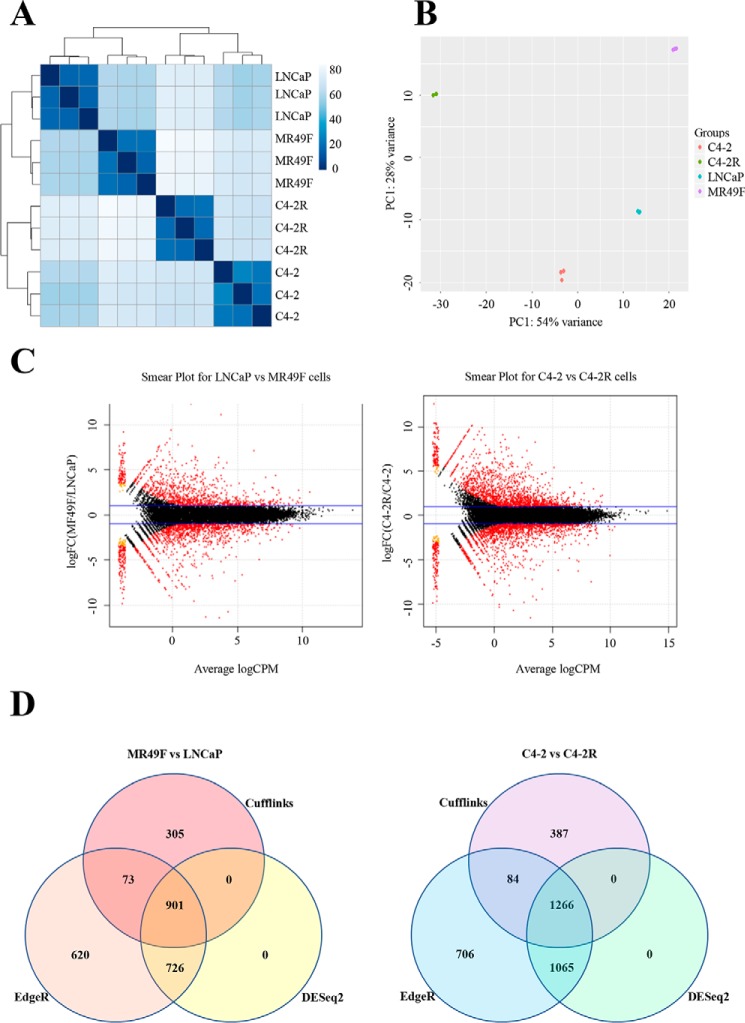
Kuruma et al. (5) and Liu et al. (6) developed enzalutamide-resistant cell lines MR49F and C4-2R derived from the parental cell lines LNCaP and C4-2, respectively. These cell lines were developed by extensive treatment with high concentrations of enzalutamide in a tissue culture setting or in an in vivo xenograft mouse model to serve as a model to study enzalutamide resistance (5, 6). To compare enzalutamide-sensitive and enzalutamide-resistant cell lines, RNA was isolated from LNCaP, C4-2, MR49F, and C4-2R after a 4-h treatment with enzalutamide. Approximately 30,000 features were detected in all cell lines after RNA-Seq, excluding genes with zero counts. Sample-to-sample matrix heat map reveals that the biological replicates are consistent within each sample group (Fig. 1A). In addition, principle component analysis (PCA) shows that our replicates cluster together within each group (Fig. 1B). Fig. 1, A and B shows that LNCaP is the closest to MR49F. On the other hand, C4-2 has the most similarity with C4-2R. This validates our choice for pairing LNCaP with MR49F and C4-2 with C4-2R. For differentially expressed genes analysis, we used three different algorithms: DESeq2, edgeR, and Cufflinks. Using edgeR with a 1% FDR and 2-fold change cutoff, we were able to identify a significant number of genes that are differentially expressed in LNCaP versus MR49F and C4-2 versus C4-2R (Fig. 1C). Overall, the intersection of the three algorithms showed 901 genes that have been differentially expressed in MR49F compared with LNCaP and 1266 genes differentially expressed in C4-2R compared with C4-2 (Fig. 1D). Finally, Tables 1 and 2 show the number of genes that are up- or down-regulated in LNCaP versus MR49F and C4-2 versus C4-2R discovered by the independent algorithms. Our data indicate that long-term enzalutamide treatment leads to alteration of global gene expression.
Figure 1.
Long-term enzalutamide treatment of prostate cancer cell lines induces global gene expression changes after acquiring resistance. A, heat map showing the Euclidian distances between samples, made with the DESeq2 transformed data after a regularized log transformation was performed. B, PCA plot of libraries. Salmon-colored points represent C4-2 samples, green points represent C4-2R samples, teal points represent LNCaP samples, and purple points represent MR49F samples. Overall 54% of the variance is captured by the first principle component (shown on the x axis), and the second principle component (shown on the y axis) accounts for 28% of the variation. The data were normalized in DESeq2, and a regularized log transformation as performed prior to doing the PCA. C, smear plots from edgeR showing, in red, transcripts differentially expressed (false discovery rate < 0.01) between, respectively, enzalutamide-resistant/enzalutamide-sensitive cells. Blue lines indicate 2-fold-change cutoff. The x axis shows the average log (count per million, CPM), and the y axis shows the log2 (fold change). D, Venn diagrams of overlap between differential expression results among the three statistical packages Cufflinks, edgeR, and DESeq2 for both comparisons.
Table 1.
MR49F versus LNCaP, 1% FDR, 2FC cutoff
Total, up-regulated, and down-regulated transcripts differentially expressed (FDR < 0.01 and 2-fold cutoff) between, respectively, enzalutamide-resistant/enzalutamide-sensitive cells using the three different packages Cufflinks, edgeR, and DESeq2.
| Total | Up-regulated in resistant cells | Down-regulated in resistant cells | |
|---|---|---|---|
| DESeq2 | 1627 | 738 | 889 |
| edgeR | 2320 | 1026 | 1294 |
| Cufflinks | 1279 | 719 | 560 |
Table 2.
C4–2R versus C4–2, 1% FDR, 2FC cutoff
Total, up-regulated, and down-regulated transcripts differentially expressed (FDR < 0.01 and 2-fold cutoff) between, respectively, enzalutamide-resistant/enzalutamide-sensitive cells using the three different packages Cufflinks, edgeR, and DESeq2.
| Total | Up-regulated in resistant cells | Down-regulated in resistant cells | |
|---|---|---|---|
| DESeq2 | 2331 | 1206 | 1125 |
| edgeR | 3121 | 1554 | 1567 |
| Cufflinks | 1737 | 871 | 866 |
Notch signaling pathway is enriched in patient samples that mimic enzalutamide-resistant cell lines
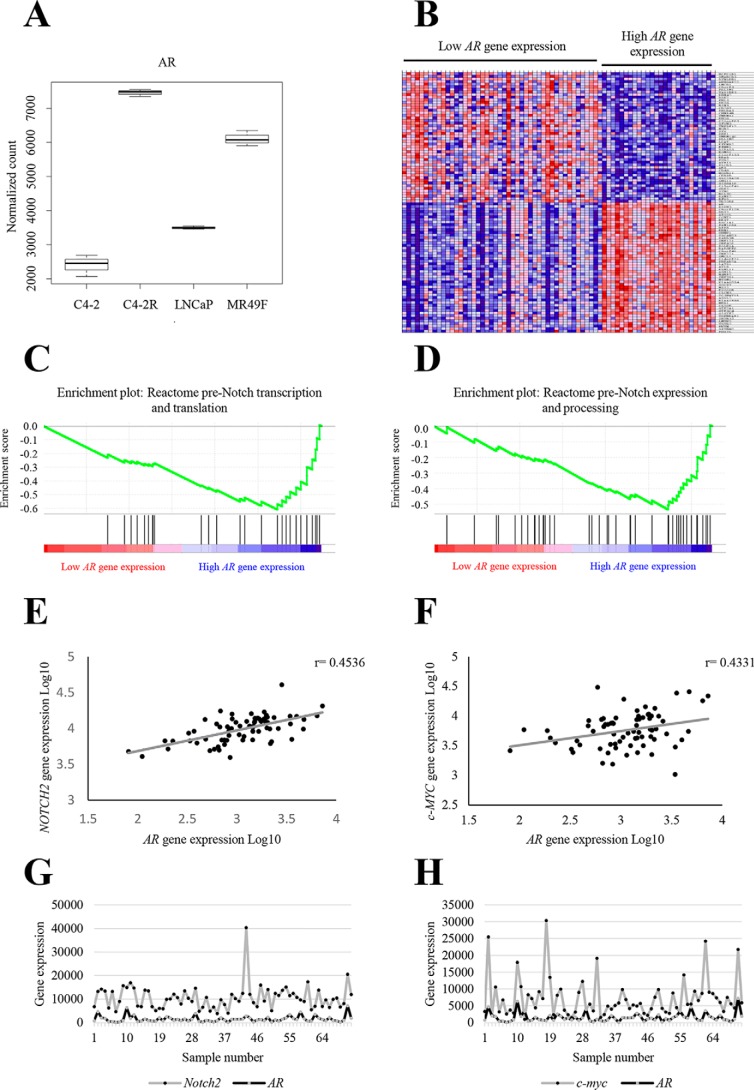
Our bioinformatics data from comparing enzalutamide-resistant and enzalutamide-sensitive cell lines revealed that the normalized counts of AR were significantly and consistently increased in MR49F and C4-2R compared with LNCaP and C4-2, respectively (Fig. 2A). This indicates that cell lines with acquired resistance exhibit a higher expression of AR compared with cells that are still sensitive the drug. Because of the lack of clinical samples from patients treated with enzalutamide, we analyzed a cohort of 72 patients with hormone-refractory prostate cancer (Fig. 2B). Of the 72 patients, 26 had high AR gene expression mimicking cell lines that are resistant to enzalutamide, and 46 had low AR gene expression mimicking cell lines that are sensitive. After separating the samples into AR low versus AR high, the data set was subjected to gene set enrichment analysis (GSEA). Genes from the pre-Notch transcription and translation signaling pathway and the pre-Notch expression and processing signaling pathway are significantly enriched in the AR high group (Fig. 2, C and D). These pathways are subgroups of the Notch signaling pathway from Reactome. Furthermore, we used Pearson correlation analysis to identify genes that correlate with AR expression in the data set. NOTCH2 expression positively correlated with AR expression, with a correlation coefficient of 0.4331 (Fig. 2, E and G). In addition, one of the downstream targets of Notch signaling c-MYC positively correlated with AR expression, with a correlation coefficient of 0.4536 (Fig. 2, F and H). These data indicate that Notch signaling pathway may play a role in resistance to enzalutamide in prostate cancer.
Figure 2.
Notch signaling pathway is enriched in patient samples that mimic enzalutamide-resistant cell lines. A, box plot of normalized AR counts for all samples using DESeq2. B, heat map representing a cohort of patients from the TCGA database, separated into samples with high AR expression (26 samples) and samples with low AR expression (46 samples). C, GSEA enrichment plot of the Reactome pre-Notch transcription and translation gene set in the cohort of patient with AR low versus AR high. The plot indicates a negative enrichment, meaning that the queried gene set correlates with AR high samples. D, GSEA enrichment plot of the Reactome pre-Notch expression and processing gene set in the cohort of patient with AR low versus AR high. The plot indicates a negative enrichment, meaning that the queried gene set correlates with AR high samples. E, NOTCH2 gene expression analysis in the cohort of patients and its correlation with AR gene expression indicates a strong positive correlation with a correlation coefficient of r = 0.4331. F, c-MYC gene expression analysis in the cohort of patients and its correlation with AR gene expression indicates a strong positive correlation with a correlation coefficient of r = 0.4536. G and H, NOTCH2 and c-MYC counts matched to the sample number from the cohort of patients.
Expression of Notch signaling pathway genes is deregulated in enzalutamide-resistant cells
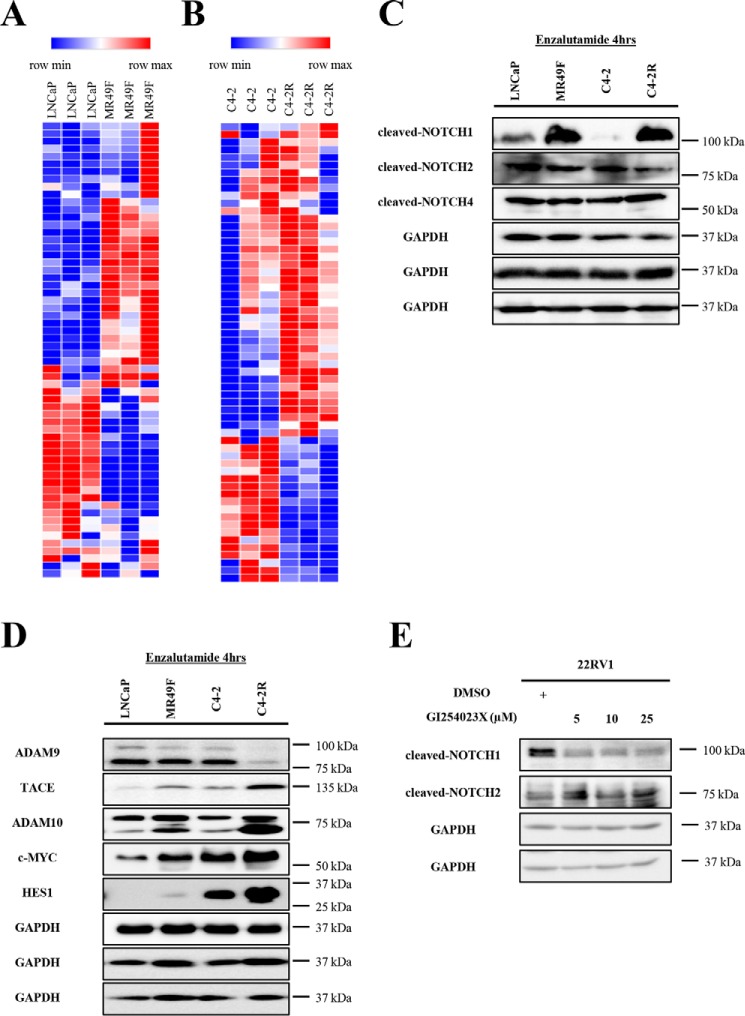
To further explore the implication of Notch signaling in resistance, we compared the expression of genes from this pathway in enzalutamide-resistant versus enzalutamide-sensitive cell lines. As shown in Fig. 3, A and B, more than 30 genes from the Notch signaling pathway are either up- or down-regulated in MR49F and C4-2R compared with LNCaP and C4-2, respectively (Table S1).
Figure 3.
Expression of Notch signaling pathway genes is deregulated in enzalutamide-resistant cells. A, heat map representing gene expression patterns of Notch signaling pathway genes in LNCaP versus MR49F cell lines. B, heat map representing gene expression patterns of Notch signaling pathway genes in C4-2 versus C4-2R cell lines. C and D, LNCaP, MR49F, C4-2, and C4-2R cells were treated with enzalutamide for 4 h and subjected to Western blotting analysis. E, 22RV1 cells were treated with GI254023X (ADAM 10 inhibitor) for 48 h and subjected to Western blotting analysis.
To investigate the functional features of the Notch signaling pathway in enzalutamide-resistant cell lines, we examined the protein expression of important genes from this pathway. The Notch family of proteins is composed of four transmembrane receptors (NOTCH 1, 2, 3, and 4) that are cleaved upon ligand binding on the cell surface rendering their activation and leading to downstream events (7–9). Our results show that cleaved NOTCH1 has a higher expression in MR49F and C4-2R compared with their sensitive counterparts. However, the cleaved forms of NOTCH2 and NOTCH4 were consistently expressed among all cell lines. It is impossible to predict the status of cleaved NOTCH3 expression in these cells because of the lack of a specific antibody to test it. These data suggest that NOTCH1 signaling is overactivated in the resistant cells.
Upon NOTCH-ligand binding, NOTCH proteins exhibit a conformational change leading to S2 cleavage in the NOTCH extracellular domain by ADAM (a disintegrin and metalloprotease) proteins (10). NOTCH S2 cleavage leads to a S3 cleavage by the γ-secretase complex releasing the NOTCH intracellular domain into the cytoplasm (11). To explore the implication of the ADAM family of proteins in the activation of Notch signaling, especially NOTCH1, in enzalutamide-resistant cells, we tested the protein expression of ADAM9, ADAM10, and ADAM17 (TACE). ADAM9 is similarly expressed in LNCaP versus MR49F and down-regulated in C4-2R compared with C4-2 (Fig. 3D). TACE is up-regulated in both resistant cell lines compared with their sensitive counterparts. The active form of ADAM10 (lower band) is highly up-regulated in MR49F and C4-2R compared with LNCaP and C4-2, respectively. These data indicate that upstream proteases responsible for the activation of the Notch signaling pathway are predominantly up-regulated in resistant cells, offering a potential explanation to the increased expression of cleaved NOTCH1.
To explore a potential mechanism involved in the exclusive activation of NOTCH1, the highly activated ADAM10 was inhibited using GI254023X in MR49F and 22RV1 (another enzalutamide-resistant cell line) cells. Upon treatment with GI254023X, cleaved NOTCH1 expression is significantly reduced in 22RV1 and MR49F (data not shown). However, the expression of cleaved NOTCH2 appears to be unaffected by GI254023X treatment. This offers a potential explanation to the exclusive overexpression of cleaved NOTCH1 compared with the expression of cleaved NOTCH2 and cleaved NOTCH4 in resistant cells.
After its internalization, the NOTCH intracellular domain translocates to the nucleus and activates the expression of Notch signaling downstream targets such as HES (hairy and enhancer of split) genes, c-MYC, and others (12). To confirm that Notch signaling is further activated in enzalutamide-resistant cells compared with enzalutamide-sensitive cells, we tested the protein expression of HES1 and c-MYC. Both proteins are up-regulated in MR49F and C4-2R compared with LNCaP and C4-2, respectively (Fig. 3D). This indicates that Notch signaling is activated in enzalutamide-resistant cells.
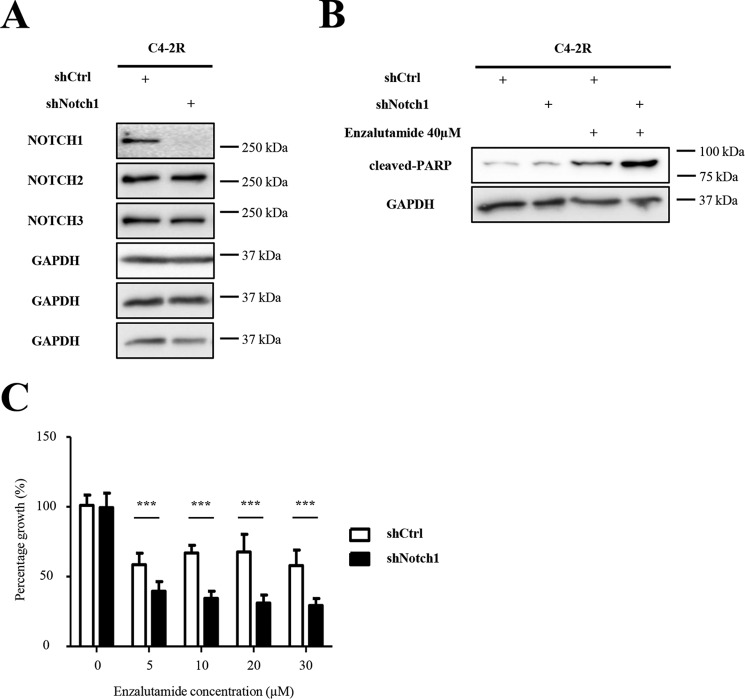
Knockdown of NOTCH1 in C4-2R cells increases sensitivity to enzalutamide
To test whether Notch1 signaling is directly involved in resistance, we designed and generated shRNA vectors targeting NOTCH1 and NOTCH2 in C4-2R cells. Fig. 4A shows successful knockdown of NOTCH1 in C4-2R cells compared with cells infected with shCtrl virus. Exposure of C4-2R–shNOTCH1 cells to enzalutamide induces an increase in apoptosis represented by cleaved PARP expression compared with C4-2R–shCtrl cells (Fig. 4B). Furthermore, exposure of C4-2R–shNOTCH1 cells to increasing concentrations of enzalutamide induces a decrease in cell proliferation (Fig. 4C). In contrast, enzalutamide did not induce the same effects on C4-2R–shNOTCH2 cells when compared with shCtrl (Fig. S1). This suggests that knockdown of NOTCH1 is sufficient to reduce cell proliferation and increase apoptosis, indicating a resensitization of C4-2R to enzalutamide.
Figure 4.
Knockdown of NOTCH1 induces an increase in cell death and a decrease in cell proliferation. A, C4-2R cells were infected with shCtrl or shNOCH1 virus particles and selected with puromycin. Cells were collected and subjected to Western blotting analysis with indicated antibodies. B, stable C4-2R shCtrl and shNOTCH1 cells were treated with DMSO or 40 μm enzalutamide for 72 h and subjected to Western blotting analysis. C, stable C4-2R shCtrl and shNOTCH1 cells were treated with DMSO or increasing concentrations of enzalutamide and subjected to MTT assay. Optical densities were measured and normalized to DMSO-treated samples. *, p < 0.05; **, p < 0.01.
Inhibition of Notch signaling pathway resensitizes enzalutamide-resistant cells to enzalutamide and increases apoptosis and decreases colony formation ability
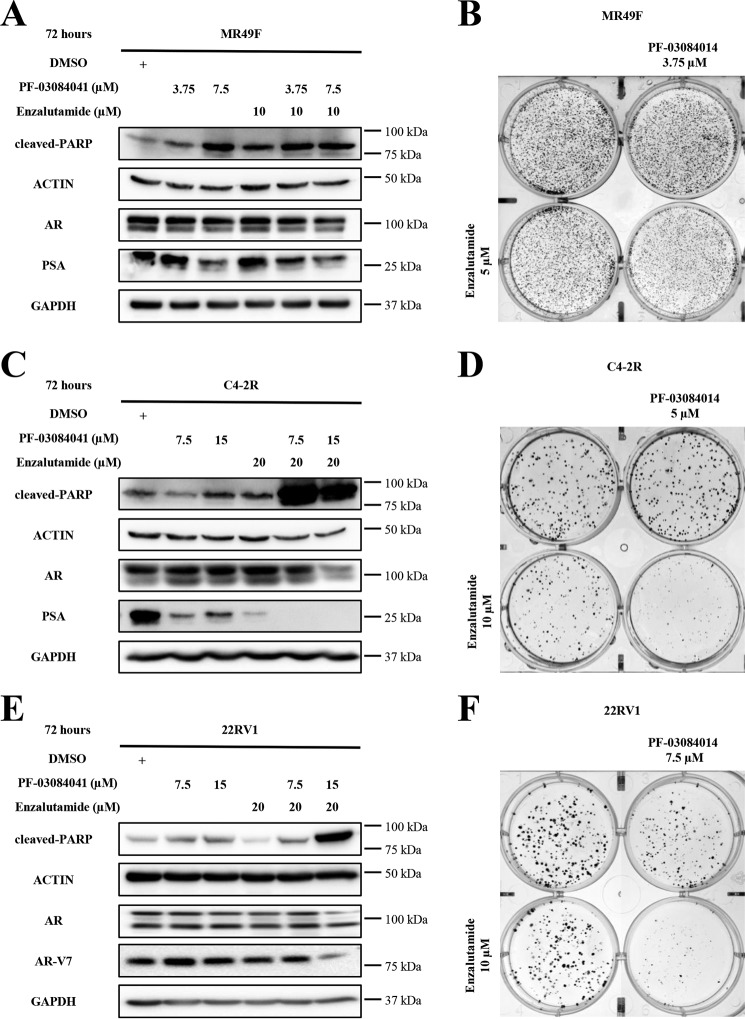
To test whether inhibiting Notch signaling in prostate cancer enzalutamide-resistant cells resensitizes these cells to enzalutamide, we treated MR49F, C4-2R, and 22RV1 cells with enzalutamide, PF-03084014 (γ-secretase inhibitor), or both. Treatment with PF-03084014 induced a decrease in HES1 and c-MYC expression, indicating an inhibition of Notch signaling in all cell lines (Fig. S2, A–C). Exposure to both PF-03084014 and enzalutamide induced an increase in cleaved PARP expression levels, indicating an increased apoptosis after 72 h (Figs. 5, A, C, and E). In addition, in the same groups we observed a decrease in AR and PSA (prostate specific antigen) levels in MR49F and C4-2R compared with enzalutamide exposure alone (Fig. 5, A and C). In 22RV1, the combination of enzalutamide and PF-03084014 caused a reduction in AR and AR-V7 protein levels (Fig. 5E). Furthermore, Fig. 5, B, D, and F show a decrease in colony numbers and colony size in cells treated with both PF-03084014 and enzalutamide compared with cells treated with DMSO, enzalutamide, or PF-03084014 alone. These results suggest that inhibition of Notch signaling resensitizes resistant cells to enzalutamide by blocking the AR signaling and reducing AR expression.
Figure 5.
Inhibition of Notch signaling pathway resensitizes enzalutamide-resistant cells to enzalutamide and causes cell death and decreased colony formation ability. A, C, and E, MR49F, C4-2R, and 22RV1 cells were treated with DMSO, PF-03084014, enzalutamide, or both at the indicated concentrations for 72 h. The cells were collected and subjected to Western blotting analysis. B, D, and F, low seeding density MR49F, C4-2R, and 22RV1 cells were treated with DMSO, PF-03084014, enzalutamide, or both at the indicated concentrations until colonies were formed. The cells were fixed with 4% formaldehyde and stained with crystal violet.
PF-03084014 and enzalutamide treatment induces a decrease in 22RV1 tumor xenografts
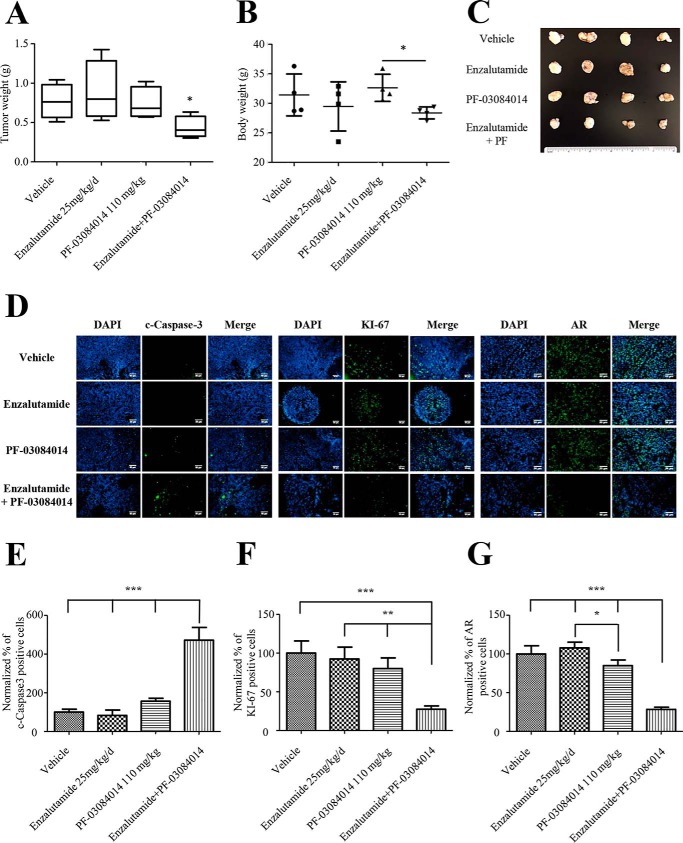
To further validate our in vitro data, we generated an enzalutamide-resistant mouse xenograft model using 22RV1 cells. Nude mice were castrated and inoculated subcutaneously with 22RV1 cells. Upon tumor formation mice were treated for 4 weeks with a vehicle, enzalutamide, PF-03084014, or the combination of enzalutamide and PF-03084014. Mice in the combination group showed a significant decrease in tumor weight compared with vehicle or single-treatment groups (Fig. 6, A and C). There were no significant signs of toxicity in mice. Mice from the combination group showed a significant decrease in body weight only when compared with mice treated with PF-03084014 alone (Fig. 6B).
Figure 6.
PF-03084014 combined with enzalutamide treatment induces a decrease in enzalutamide-resistant tumor xenograft. A and B, 22RV1 xenograft mice were treated with a vehicle control, enzalutamide, PF-03084014, or a combination of enzalutamide and PF-03084014 (PF). Upon completion, tumor weight and body weight were measured (n = 4 mice). C, representative image of tumors from different treatment groups. D, immunofluorescent images from tumor sections stained with cleaved Caspase-3, KI-67, AR, and 4′,6′-diamino-2-phenylindole (DAPI). E–G, quantification of cleaved Caspase-3, KI-67 and AR positively stained cells (n = 3). All groups were normalized to the vehicle control group. *, p [lt ]0.05; **, p < 0.01; ***, p < 0.001.
Immunofluorescent staining of tumors showed effective inhibition of the Notch signaling pathway by PF-03084014 represented by a significant decrease in the percentage of HES1-expressing cells in the PF-03084014 and combination-treatment groups (Fig. S2D). In addition, a significant increase in cleaved caspase 3–expressing cells in the combination-treatment group compared with the other groups indicated an increase in apoptosis (Fig. 6, D and E). KI-67 staining was significantly decreased in tumors from the combination-treatment group compared with the other groups, suggesting a reduced proliferation (Fig. 6, D and F). Combination of enzalutamide and PF-03084014 induced a significant decrease in AR-expressing cells within the tumor (Fig. 6, D and G). These results suggest that blocking Notch signaling in vivo resensitizes 22RV1 cells to enzalutamide by increasing apoptosis, reducing cell proliferation and AR expression.
Discussion
The already Food and Drug Administration–approved enzalutamide is currently under investigation in clinical trials with early-stage prostate cancer patients. If the outcome of these trials turns out to be a success, enzalutamide will become widely used for treatment of prostate cancer patients at all stages of the disease. However, one of the major obstacles is development of enzalutamide resistance in patients. Most of the therapy-resistance studies in prostate cancer focus on the role of the AR signaling pathway in overcoming the resistance to ADT and enzalutamide (13). In this study, our aim was to identify pathways that are independent of the AR signaling pathway and may play a role enzalutamide resistance. We compared enzalutamide-resistant cells MR49F and C4-2R to their enzalutamide-sensitive counterparts LNCaP and C4-2, respectively. Our analysis revealed hundreds of genes that are up- or down-regulated in the enzalutamide-resistant cells (Fig. 1). The combination of our RNA-Seq with patient sample bioinformatics analysis helped identify Notch signaling pathway as a potential deregulated pathway in enzalutamide-resistant cells.
In prostate cancer, studies have shown contradicting roles of NOTCH signaling where it either plays a tumor-suppressive or a tumor-promoting role (14, 15). Our results indicate that the Notch signaling pathway plays a tumor-promoting role in prostate cancer by contributing to enzalutamide resistance and promoting cell survival. Furthermore, NOTCH proteins have been shown to contribute to therapy resistance in prostate cancer (16). In addition, studies have shown synergistic positive results in prostate cancer cells when Notch signaling inhibitors were combined with ADTs (17, 18). Our data support the observations above where we show that Notch signaling is activated in enzalutamide-resistant cells, and inhibition of this pathway restores sensitivity to enzalutamide.
In addition, Notch signaling has been shown to be a driver of stemness and contributor to survival and maintenance of cancer stem cells, which are believed to a driver of cancer therapy resistance (15, 19). There is evidence that ADT and enzalutamide treatment promote the acquisition of stem cell-like features in prostate cancer cells (20, 21). Deregulation of Notch signaling in enzalutamide-resistant cells may play a role in maintenance of stem-like features, leading to therapy resistance.
To investigate whether Notch signaling is active in resistant cells, expression of cleaved Notch isoforms was tested. Our data suggest that Notch1 signaling is active because of a high expression of cleaved NOTCH1 in the resistant cells (Fig. 3B). Furthermore, downstream targets such as HES1 and c-MYC were overexpressed in resistant cells, indicating an active pathway (Fig. 3C). However, because of the lack of a specific cleaved NOTCH3 antibody, we were unable to verify the expression of this protein. Thus, Notch3 may also be playing a role in the activation of downstream targets.
To further understand how Notch signaling is activated in enzalutamide-resistant cells, we investigated the expression of upstream proteases involved in Notch receptor activation and downstream targets of the pathway. Our results show that ADAM10 is activated, and TACE is up-regulated in the resistant cells (Fig. 3C). ADAM10 and TACE overexpression contributes to therapy resistance, and their inhibition leads to resensitizing tumor cells to existing therapies in different cancers (22–24). Here, we believe that the overexpression of the metalloproteases might be involved in the activation of the Notch1 signaling pathway. According to our data, inhibition of ADAM10 with GI254023X reduced the expression of cleaved NOTCH1 exclusively (Fig. 3D), which suggests a higher sensitivity of Notch1 cleavage to changes in ADAM10 expression or activation. Other explanations may be attributed to selectivity in ligands toward Notch1 over other isoforms (25) and the elevated number of reported mutations in the heterodimerization domain of Notch1 that can affect its ligand independent cleavage compared with the other isoforms (26).
Notch1 signaling is shown to become activated and to play a role in cell death and drug sensitivity after cells acquire resistance in other cancers (27, 28). To investigate whether Notch1 signaling plays a significant role in enzalutamide resistance, we generated shNotch1 C4-2R cells. These cells have a better response to enzalutamide treatment compared with shCtrl cells (Fig. 4). In contrast, knockdown of NOTCH2 in C4-2R did not yield the same results.
Inhibitors and blockers of Notch signaling are being investigated in clinical trials in multiple cancers. To investigate whether blockade of Notch signaling pathway reverses enzalutamide resistance in MR49F, C4-2R, and 22RV1, we used the γ-secretase inhibitor PF-03084014. Combination of enzalutamide and PF-03084014 showed overexpression of cleaved PARP expression in all cell lines (Fig. 5, A, C, and E). One of the established mechanisms of resistance to ADT is increased AR and AR-variant expression in prostate cancer cells (5, 6). Our data indicate that exposure to enzalutamide and PF-03084014 decreased full-length AR, PSA (prostate specific antigen), and AR-V7 expression, potentially explaining the restoration of enzalutamide function in the resistant cells (Fig. 5, A, C, and E). Additionally, increased levels of c-MYC have been shown to promote ligand-independent prostate cancer survival (29). Our data show that combination of enzalutamide and the γ-secretase inhibitor decreases c-MYC expression in the resistant cells, offering another mechanism by which resistance is reversed in those cells (Fig. S2, A–C).
Furthermore, the inhibition of AR and Notch signaling together induced a significant decrease in colony formation ability of MR49F, C4-2R, and 22RV1 (Fig. 5, B, D, and F). In the xenograft model, the combination therapy showed growth inhibition in tumors compared with vehicle and single treatment groups, with no signs of significant toxicity. This observation was accompanied by an increase in cleaved Caspase-3 expression, a decrease in KI-67 expression and a decrease in AR expressing cells, which replicated our in vitro cell line data (Fig. 6).
Finally, our study adds up to existing data on the oncogenic role of Notch signaling in prostate cancer. Notch1 signaling is activated in enzalutamide-resistant cells, and inhibition of this pathway may restore enzalutamide function in vitro and in vivo, leading to a better outcome. These data offer a precedent for the combination of Notch signaling inhibitors with enzalutamide and potentially other ADTs.
Materials and methods
Mammalian cell lines
LNCaP, C4-2, and 22RV1 cells were purchased from ATCC in 2016. MR49F and C4-2R cells were kindly provided by Dr. Amina Zoubeidi (5) and Dr. Allen Gao (6). LNCaP and C4-2 were used as enzalutamide-sensitive cell lines, whereas MR49F, C4-2R, and 22RV1 were considered enzalutamide-resistant cell lines. For this study LNCaP and C4-2 were paired with their enzalutamide-resistant counterparts MR49F and C4-2R, respectively. 22RV1 was used as an independent cell line with no enzalutamide-sensitive pair. All cell lines were maintained in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin at 37 °C in a humidified incubator with 5% carbon dioxide. MR49F and C4-2R were constantly maintained in medium supplemented with 10 and 20 μm enzalutamide, respectively.
Inhibitors
PF-03084014 and enzalutamide were purchased from Selleckchem. GI254023X was purchased from Sigma–Aldrich. For in vitro experiments, PF-03084014, enzalutamide, and GI254023X were suspended in DMSO. For in vivo experiments, PF-03084014 was suspended in 0.5% methylcellulose, and enzalutamide was formulated in 5% DMSO and 0.1% Tween 80 suspended in 1% carboxymethyl cellulose.
Western blotting
The cells were harvested and washed with PBS. Pellets were resuspended in TBSN buffer (20 mmol/liter Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 5 mm EGTA, 1.5 mmol/liter EDTA, 0.5 mm sodium vanadate, and 150 mm NaCl) supplemented with protease and phosphatase inhibitor mixture, and subjected to sonication. Sample protein concentrations were measured using Pierce BCA protein assay kit. Equal amounts of proteins were mixed with 4× SDS loading buffer and loaded onto SDS-PAGE gels. Upon protein separation, proteins were transferred to polyvinylidene difluoride membranes and subjected to detection using appropriate primary and secondary antibodies. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and β-actin were used as housekeeping genes.
RNA-Seq
RNA was extracted from LNCaP, MR49F, C4-2, and C4-2R after 4-h treatment with enzalutamide using the RNeasy kit (Qiagen). Libraries were prepared using TruSeq Stranded kit (Illumina) at the Purdue Genomics Facility. Two 100-bp reads were sequenced in two lanes using the HiSeq2500 on high-output mode. Before library preparation, the dscDNA quality was checked using an Agilent Bioanalyzer with a high-sensitivity DNA chip. All plots were checked to ensure that read quality for the reads that would be used in the remainder of the analysis were of high quality and had no problems. Tophat2 (30, 31) was used to align reads to the Ensembl Homo sapiens genome database version GRCh38.p5. The mitochondrial chromosome and the nonchromosomal sequences were excluded from the analysis. The htseq-count script in HTSeq v.0.6.1 (32) was run to count the number of reads mapping to each gene. DESeq2 (33), edgeR (34), and Cufflinks2 (35, 36) were used for differential expression analysis.
Gene set enrichment analysis
Clinical data were collected from level 3 (for segmented or interpreted data; IllumninaHiSeq_RNASeqV2) of the TCGA database. The samples were divided into low grade (Gleason score of <8) and high grade (Gleason score of ≥8). Of the high-grade samples, we looked at 72 of 497 cases that had been treated with anti-hormone therapy. We then separate the 72 patients into AR high and AR low based on the expression of the genes from the AR signaling pathway. AR high cases were considered to mimic enzalutamide-resistant cells, and AR low cases were considered to mimic enzalutamide-sensitive cells. Using GSEA we detected pathway enrichment in AR high (26 samples) versus AR low (46 samples).
Colony formation assay
The cells were seeded at equal densities in 6-well plates, followed by treatment with DMSO as a control, PF-03084014, enzalutamide, or the combination of both. After colonies became of size, the cells were subjected to fixation with 4% paraformaldehyde followed by staining with 0.5% crystal violet solution.
MTT cell proliferation assay
To evaluate cell proliferation, the cells were seeded in 96-well plates and treated with increasing concentrations of enzalutamide. At the end of incubation time, the cells were then treated with 0.5 mg/ml MTT for 1 h. The supernatant was removed, and DMSO was added to dissolve crystals. Optical density was measured at a wavelength of 560 nm. The values were normalized as 100% to the control group treated with DMSO.
Lentivirus production and Notch1 knockdown in C4-2R cells
A lentivirus system encoding shRNA targeting a scrambled sequence, NOTCH1 and NOTCH2 mRNA was used for knockdown. Sequences targeting Ctrl (5′-TTCTCCGAACGTGTCACGT-3′), NOTCH1 (5′-GCATGTGTAACATCAACAT-3′), and NOTCH2 (5′-CAGGTAGCTCAGACCATTC-3′) were cloned in pLVshRNA-puro. Lentiviral particles were generated by transfecting shCtrl, shNOTCH1, and shNOTCH2 with psPAX2 and PMD.2G plasmids at a ratio of 4:3:1 in 293T cells. The medium was collected and used to infect C4-2R cells with corresponding shRNA to generated C4-2R–shCtrl, C4-2R–shNOTCH1, and C4-2R–shNOTCH2.
22RV1 mouse xenografts
Castrated nu/nu mice were subjected to a right flank subcutaneous inoculation of 2 × 105 22RV1 cells suspended in PBS and mixed with Matrigel (1:1). Tumor-bearing mice were randomly assigned into four groups (n = 4) in which mice from different groups received through oral gavage a vehicle control, 20 mg/kg/day of enzalutamide, 110 mg/kg of PF-03084014 daily on alternate weeks, or the combination of both. Upon necropsy, tumors were collected and weighed. In vivo experiments were approved by the Purdue Animal Care and Use Committee.
Immunofluorescent chemistry
Xenograft tumors were fixed in 4% neutral buffered paraformaldehyde, sectioned, and stained using specific antibodies. Staining was accomplished with the M.O.M.TM kit from Vector Laboratories. Positively stained cells were quantified using NIS-Elements AR 3.2. All groups were normalized to vehicle control.
Statistical analysis
Standard two-tailed Student's t tests were performed to analyze statistical significance of the results. Two-way analysis of variance tests were performed to analyze statistical significance of data sets with grouped observations. A one-way analysis of variance test was performed to analyze statistical significance of immunofluorescence quantification. A p value of less than 0.05 indicates statistical significance.
Author contributions
E. F., Y. K., and X. L. conceptualization; E. F. and C. L. data curation; E. F., C. L., L. C., and N. A. L. formal analysis; E. F. and C. L. validation; E. F., Y. K., and X. L. investigation; E. F., C. L., L. C., Y. K., N. A. L., P. P., and L. L. visualization; E. F., L. C., Y. K., N. A. L., G.R.L., L. L., and X. L. methodology; E. F. writing-original draft; L. C., N. A. L., and P. P. software; Y.Z. resources; N.A. and X. L. funding acquisition; T.R. writing-review and editing; X. L. supervision; X. L. project administration.
Supplementary Material
Acknowledgments
We acknowledge Purdue Bioinformatics, DNA sequencing core, and Walther Cancer Foundation.
This work was supported by National Institutes of Health Grants R01 CA157429, R01 CA192894, R01 CA196835, and R01 CA196634 (to X. L.). The work was also supported by Purdue University Center for Cancer Research Grant P30 CA023168. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE123379.
This article contains Table S1 and Figs. S1 and S2.
- ADT
- androgen deprivation therapy
- AR
- androgen receptor
- PCA
- principle component analysis
- GSEA
- gene set enrichment analysis
- MTT
- 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide
- FDR
- false discovery rate
- TACE
- TNFα converting enzyme.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2017) Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2. Bahl A., Masson S., Birtle A., Chowdhury S., and de Bono J. (2014) Second-line treatment options in metastatic castration-resistant prostate cancer: a comparison of key trials with recently approved agents. Cancer Treat. Rev. 40, 170–177 10.1016/j.ctrv.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 3. Alumkal J. J., Chowdhury S., Loriot Y., Sternberg C. N., de Bono J. S., Tombal B., Carles J., Flaig T. W., Dorff T. B., Phung Forer D., Noonberg S. B., Mansbach H., Beer T. M., and Higano C. S. (2015) Effect of visceral disease site on outcomes in patients with metastatic castration-resistant prostate cancer treated with enzalutamide in the PREVAIL trial. Clin. Genitourin. Cancer 15, 610–617.e3 10.1016/j.clgc.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 4. Bianchini D., Lorente D., Rodriguez-Vida A., Omlin A., Pezaro C., Ferraldeschi R., Zivi A., Attard G., Chowdhury S., and de Bono J. S. (2014) Antitumour activity of enzalutamide (MDV3100) in patients with metastatic castration-resistant prostate cancer (CRPC) pre-treated with docetaxel and abiraterone. Eur. J. Cancer 50, 78–84 10.1016/j.ejca.2013.08.020 [DOI] [PubMed] [Google Scholar]
- 5. Kuruma H., Matsumoto H., Shiota M., Bishop J., Lamoureux F., Thomas C., Briere D., Los G., Gleave M., Fanjul A., and Zoubeidi A. (2013) A novel antiandrogen, compound 30, suppresses castration-resistant and MDV3100-resistant prostate cancer growth in vitro and in vivo. Mol. Cancer Ther. 12, 567–576 10.1158/1535-7163.MCT-12-0798 [DOI] [PubMed] [Google Scholar]
- 6. Liu C., Lou W., Zhu Y., Nadiminty N., Schwartz C. T., Evans C. P., and Gao A. C. (2014) Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin. Cancer Res. 20, 3198–3210 10.1158/1078-0432.CCR-13-3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. D'Souza B., Meloty-Kapella L., and Weinmaster G. (2010) Canonical and non-canonical notch ligands. Curr. Top. Dev. Biol. 92, 73–129 10.1016/S0070-2153(10)92003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sprinzak D., Lakhanpal A., Lebon L., Santat L. A., Fontes M. E., Anderson G. A., Garcia-Ojalvo J., and Elowitz M. B. (2010) cis-Interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90 10.1038/nature08959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cordle J., Johnson S., Tay J. Z., Roversi P., Wilkin M. B., de Madrid B. H., Shimizu H., Jensen S., Whiteman P., Jin B., Redfield C., Baron M., Lea S. M., and Handford P. A. (2008) A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 15, 849–857 10.1038/nsmb.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kopan R., and Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolfe M. S., and Kopan R. (2004) Intramembrane proteolysis: theme and variations. Science 305, 1119–1123 10.1126/science.1096187 [DOI] [PubMed] [Google Scholar]
- 12. Lubman O. Y., Korolev S. V., and Kopan R. (2004) Anchoring notch genetics and biochemistry: structural analysis of the ankyrin domain sheds light on existing data. Mol. Cell 13, 619–626 10.1016/S1097-2765(04)00120-0 [DOI] [PubMed] [Google Scholar]
- 13. Kita Y., Goto T., Akamatsu S., Yamasaki T., Inoue T., Ogawa O., and Kobayashi T. (2018) Castration-resistant prostate cancer refractory to second-generation androgen receptor axis-targeted agents: opportunities and challenges. Cancers (Basel) 10, E345 10.3390/cancers10100345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo H., Lu Y., Wang J., Liu X., Keller E. T., Liu Q., Zhou Q., and Zhang J. (2014) Targeting the Notch signaling pathway in cancer therapeutics. Thorac. Cancer 5, 473–486 10.1111/1759-7714.12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng G., Ma L., Meng Q., Ju X., Jiang K., Jiang P., and Yu Z. (2016) Notch signaling in the prostate: critical roles during development and in the hallmarks of prostate cancer biology. J. Cancer Res. Clin. Oncol. 142, 531–547 10.1007/s00432-015-1946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Domingo-Domenech J., Vidal S. J., Rodriguez-Bravo V., Castillo-Martin M., Quinn S. A., Rodriguez-Barrueco R., Bonal D. M., Charytonowicz E., Gladoun N., de la Iglesia-Vicente J., Petrylak D. P., Benson M. C., Silva J. M., and Cordon-Cardo C. (2012) Suppression of acquired docetaxel resistance in prostate cancer through depletion of Notch- and Hedgehog-dependent tumor-initiating cells. Cancer Cell 22, 373–388 10.1016/j.ccr.2012.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohamed A. A., Tan S. H., Xavier C. P., Katta S., Huang W., Ravindranath L., Jamal M., Li H., Srivastava M., Srivatsan E. S., Sreenath T. L., McLeod D. G., Srinivasan A., Petrovics G., Dobi A., et al. (2017) Synergistic activity with NOTCH inhibition and androgen ablation in ERG-positive prostate cancer cells. Mol. Cancer Res. 15, 1308–1317 10.1158/1541-7786.MCR-17-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui J., Wang Y., Dong B., Qin L., Wang C., Zhou P., Wang X., Xu H., Xue W., Fang Y. X., and Gao W. Q. (2018) Pharmacological inhibition of the Notch pathway enhances the efficacy of androgen deprivation therapy for prostate cancer. Int. J. Cancer 143, 645–656 10.1002/ijc.31346 [DOI] [PubMed] [Google Scholar]
- 19. Venkatesh V., Nataraj R., Thangaraj G. S., Karthikeyan M., Gnanasekaran A., Kaginelli S. B., Kuppanna G., Kallappa C. G., and Basalingappa K. M. (2018) Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 5, 5 10.21037/sci.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ojo D., Lin X., Wong N., Gu Y., and Tang D. (2015) Prostate cancer stem-like cells contribute to the development of castration-resistant prostate cancer. Cancers (Basel) 7, 2290–2308 10.3390/cancers7040890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z., Cheng L., Li J., Farah E., Atallah N. M., Pascuzzi P. E., Gupta S., and Liu X. (2018) Inhibition of the Wnt/β-catenin pathway overcomes resistance to enzalutamide in castration-resistant prostate cancer. Cancer Res. 78, 3147–3162 10.1158/1538-7445.AM2018-3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Atapattu L., Saha N., Chheang C., Eissman M. F., Xu K., Vail M. E., Hii L., Llerena C., Liu Z., Horvay K., Abud H. E., Kusebauch U., Moritz R. L., Ding B. S., Cao Z., et al. (2016) An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J. Exp. Med. 213, 1741–1757 10.1084/jem.20151095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li D. D., Zhao C. H., Ding H. W., Wu Q., Ren T. S., Wang J., Chen C. Q., and Zhao Q. C. (2018) A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-fluorouracil by reversing Notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif. 51, e12480 10.1111/cpr.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller M. A., Sullivan R. J., and Lauffenburger D. A. (2017) Molecular pathways: receptor ectodomain shedding in treatment, resistance, and monitoring of cancer. Clin. Cancer Res. 23, 623–629 10.1158/1078-0432.CCR-16-0869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tveriakhina L., Schuster-Gossler K., Jarrett S. M., Andrawes M. B., Rohrbach M., Blacklow S. C., and Gossler A. (2018) The ectodomains determine ligand function in vivo and selectivity of DLL1 and DLL4 toward NOTCH1 and NOTCH2 in vitro. Elife 7, e40045 10.7554/eLife.40045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Habets R. A., Groot A. J., Yahyanejad S., Tiyanont K., Blacklow S. C., and Vooijs M. (2015) Human NOTCH2 is resistant to ligand-independent activation by metalloprotease Adam17. J. Biol. Chem. 290, 14705–14716 10.1074/jbc.M115.643676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou W., Tan W., Huang X., and Yu H. G. (2018) Doxorubicin combined with Notch1-targeting siRNA for the treatment of gastric cancer. Oncol. Lett. 16, 2805–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prabakaran D. S., Muthusami S., Sivaraman T., Yu J. R., and Park W. Y. (2019) Silencing of FTS increases radiosensitivity by blocking radiation-induced Notch1 activation and spheroid formation in cervical cancer cells. Int. J. Biol. Macromol. 126, 1318–1325 10.1016/j.ijbiomac.2018.09.114 [DOI] [PubMed] [Google Scholar]
- 29. Gao L., Schwartzman J., Gibbs A., Lisac R., Kleinschmidt R., Wilmot B., Bottomly D., Coleman I., Nelson P., McWeeney S., and Alumkal J. (2013) Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One 8, e63563 10.1371/journal.pone.0063563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trapnell C., Pachter L., and Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25, 1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anders S., Pyl P. T., and Huber W. (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberts A., Pimentel H., Trapnell C., and Pachter L. (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 27, 2325–2329 10.1093/bioinformatics/btr355 [DOI] [PubMed] [Google Scholar]
- 36. Roberts A., Trapnell C., Donaghey J., Rinn J. L., and Pachter L. (2011) Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.