Abstract
Pseudomonas species thrive in different nutritional environments and can catabolize divergent carbon substrates. These capabilities have important implications for the role of these species in natural and engineered carbon processing. However, the metabolic phenotypes enabling Pseudomonas to utilize mixed substrates remain poorly understood. Here, we employed a multi-omics approach involving stable isotope tracers, metabolomics, fluxomics, and proteomics in Pseudomonas putida KT2440 to investigate the constitutive metabolic network that achieves co-utilization of glucose and benzoate, respectively a monomer of carbohydrate polymers and a derivative of lignin monomers. Despite nearly equal consumption of both substrates, metabolite isotopologues revealed nonuniform assimilation throughout the metabolic network. Gluconeogenic flux of benzoate-derived carbons from the tricarboxylic acid cycle did not reach the upper Embden–Meyerhof–Parnas pathway nor the pentose–phosphate pathway. These latter two pathways were populated exclusively by glucose-derived carbons through a cyclic connection with the Entner–Doudoroff pathway. We integrated the 13C-metabolomics data with physiological parameters for quantitative flux analysis, demonstrating that the metabolic segregation of the substrate carbons optimally sustained biosynthetic flux demands and redox balance. Changes in protein abundance partially predicted the metabolic flux changes in cells grown on the glucose:benzoate mixture versus on glucose alone. Notably, flux magnitude and directionality were also maintained by metabolite levels and regulation of phosphorylation of key metabolic enzymes. These findings provide new insights into the metabolic architecture that affords adaptability of P. putida to divergent carbon substrates and highlight regulatory points at different metabolic nodes that may underlie the high nutritional flexibility of Pseudomonas species.
Keywords: Pseudomonas, bacterial metabolism, metabolomics, proteomics, glucose, benzoate, fluxomics, metabolic flux, metabolic regulation, mixed carbon metabolism
Introduction
A network of metabolic pathways is involved in processing carbon substrates to obtain cellular energy and carbon skeletons toward biomass biosynthesis and product generation. Understanding this cellular physiology for mixed-substrate utilization in Pseudomonas species, including Pseudomonas putida, is of particular interest (1–5). Strains of P. putida are able to catabolize breakdown products of lignocellulose (5–7), which serves as carbon sources in the soil matrix and represents an important feedstock in engineered bioconversion to valuable products. Furthermore, P. putida strains can catabolize oil contaminants such as benzene and toluene (1, 8, 9) and other industrial aromatic contaminants (10–13). Thus, the cellular network of P. putida can accommodate the metabolism of different substrate types. The metabolic pathways in P. putida have been extensively studied for growth on single organic substrates (7, 12, 14–17) including two highly relevant substrates: glucose and benzoate. Glucose is a monomer of various carbohydrate polymers; benzoate is an aromatic derivative of lignin, plant secretions, and petroleum and agricultural contaminants. Little is known, however, about the underlying intracellular metabolism and flux regulation for the co-processing of these substrates.
Nondiauxic biomass growth was observed for P. putida strains grown on different mixtures of glucose with an aromatic substrate: glucose:toluene mixture with strain KT2440 (pWW0) (18), glucose:benzoate mixture with strain KT2440-JD1 (19) and strain KT2440 (14). In accordance with this lack of diauxie, simultaneous consumption of glucose and an aromatic substrate was reported in P. putida KT2440-JD1 (19) and P. putida KT2440 (14). Furthermore, similar biomass growth rates were determined for P. putida KT2440 (pWW0) fed on glucose alone, toluene alone, or a mixture of glucose and toluene (2.3:1 ratio) (18). These previous findings are in line with the proposal that P. putida cells assimilate carbons from both carbohydrate and aromatic substrates in a manner that optimizes biomass growth (18). The cellular physiology, however, that channels these carbons through the different metabolic pathways remains to be fully elucidated.
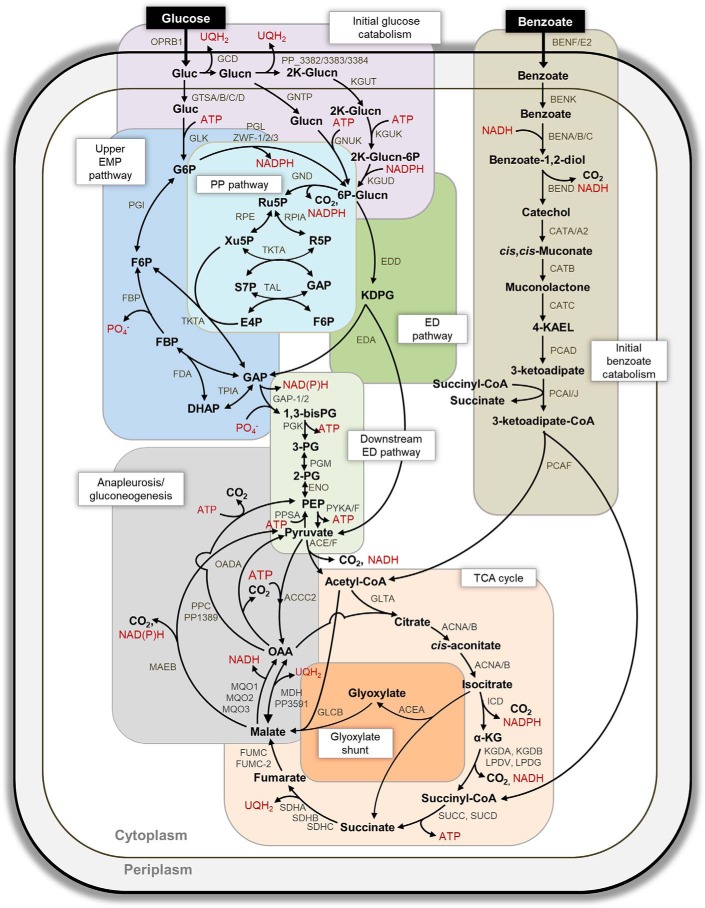
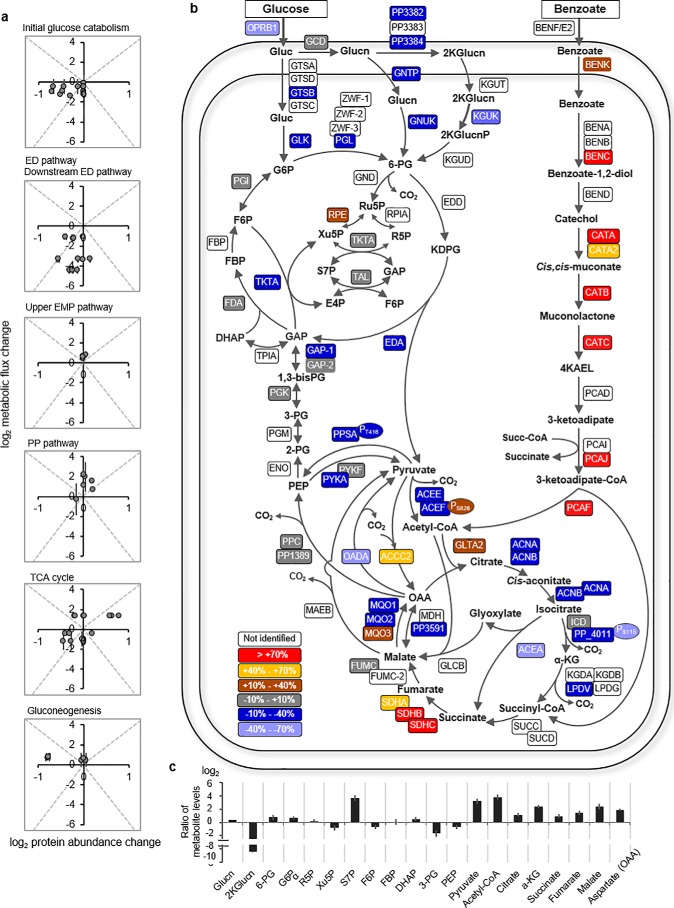
The four principal pathways that channel carbons toward biosynthetic building blocks in P. putida are the Entner–Doudoroff (ED)3 pathway, the pentose–phosphate (PP) pathway, the upper Embden–Meyerof–Parnas (EMP) pathway, and the TCA cycle (Fig. 1). To route glucose-derived carbons toward the ED and PP pathways, glucose catabolism is initiated either by direct glucose phosphorylation to glucose-6-phosphate (G6P) or by glucose oxidation to gluconate (Glucn) and 2-keto-gluconate in the periplasm before subsequent phosphorylation to 6-phosphogluconate (6P-Glucn) (14–16) (Fig. 1). Comparative transcriptional analysis of P. putida KT2440 grown on benzoate alone versus glucose alone showed no change in the transcript levels of all the proteins involved in initial glucose catabolism (14). However, compared to P. putida KT2440 (pWW0) grown on glucose alone, toluene addition led to down-regulation of genes encoding glucose transporters (18). Moreover, the activity of G6P dehydrogenase was reported to be decreased during consumption of an aromatic substrate (naphthalene or benzyl alcohol), relative to glucose alone (20). The consequence of this decrease in either protein activity or associated genes in initial glucose catabolism on the simultaneous catabolism of glucose and an aromatic substrate has not been investigated.
Figure 1.
Metabolic pathways for the catabolism of glucose and benzoate in P. putida KT2440. The reactions involved in initial glucose catabolism, initial benzoate catabolism, PP pathway, ED pathway, reverse EMP pathway, downstream ED pathway, TCA cycle, glyoxylate shunt, and anapleurosis/gluconeogenesis are grouped, respectively, in purple, light brown, green, blue, light green, light orange, dark orange, and gray boxes. The arrows indicate unidirectionality or bidirectionality of the metabolic reactions. The gene annotated to each metabolic protein is placed next to the corresponding reaction arrow; the ORF number is given when gene annotation is not known. The full names of the metabolic proteins are listed in Table S1 (Appendix C). The metabolite abbreviations are as follows: glucose, Gluc; gluconate, 2K-Glucn; glucose-6-phosphate, ribulose 5-phosphate, Ru5P; erythrose 4-phosphate, E4P; 2-keto-3-deoxy-6-phosphogluconate, KDPG; 1,3-biphosphoglycerate, 1,3-bisPG; 3-phosphoglycerate, 3PG; 2-phosphoglycerate, 2PG; acetyl-CoA, Ac-CoA; succinyl CoA, succ-CoA; 4-ketoadipate-enol-lactone, 4KAEL.
In P. putida, benzoate is catabolized through the β-ketoadipate or ortho-cleavage (ortho) pathway to yield acetyl-CoA (acetyl-CoA) and succinate or through the meta-cleavage pathway to yield acetyl-CoA and pyruvate (12, 21, 22); the plasmidless strain KT2440 investigated here only possesses the ortho pathway (Fig. 1). The metabolite products from initial benzoate catabolism are subjected to further metabolism in the TCA cycle, including the glyoxylate shunt (Fig. 1). The glyoxylate shunt bypasses the two decarboxylation reactions, through isocitrate dehydrogenase (ICD) and α-ketoglutarate dehydrogenase (KGD), in the canonical TCA cycle (Fig. 1). Prior investigations of the metabolism of benzoate alone (17) or the co-metabolism of glucose and succinate (a downstream catabolite of benzoate catabolism) (23) in P. putida KT2440 have provided clues on the consequence of benzoate or succinate on flux through the TCA cycle.
The glyoxylate shunt, which was found to be inactive in P. putida KT2440 grown on glucose alone (15, 16), was active during growth on benzoate alone (14, 17). The activation of the glyoxylate shunt in P. putida (14, 17, 24) was shown to be inversely correlated with the activity of or flux through ICD, which catalyzes the decarboxylation of isocitrate to α-KG in the TCA cycle (Fig. 1). Quantitative flux analysis further determined a minimal flux from α-KG to succinate in P. putida grown on benzoate alone (17) or on a glucose:succinate mixture (23), implying a decrease in the activity of both α-ketoglutarate dehydrogenase and succinyl Co-A synthase. Feedback inhibition of ICD by succinyl-CoA, glyoxylate, and oxaloacetate (OAA) was also reported in P. putida (24). How these reprogrammed metabolic nodes in the TCA cycle influence the co-utilization of glucose and benzoate remains to be investigated.
Here we employ a multi-omics approach involving metabolomics, fluxomics, and proteomics to address the aforementioned knowledge gaps regarding the operation of the metabolic network for the co-processing of glucose and benzoate in P. putida KT2440. Using high-resolution LC–MS, we performed stable isotope-assisted metabolomics experiments to track the assimilation of the substrate carbons within the cellular metabolome. The metabolomics data were integrated with genome-scale stoichiometry to quantitate fluxes throughout the metabolic network. Quantitative MS-based proteomics analyses were conducted to profile both protein abundances and protein phosphorylation. We interpret our data to determine relationships between enzyme-level metabolic controls and metabolic flux predictions in the metabolic network.
Results
Growth phenotypes and biosynthetic requirements imply optimal mixed-carbon usage
Growth experiments were conducted with the P. putida KT2440 cells grown on a 1:1 glucose:benzoate mixture at a total of 100 mm carbon; glucose-only growth was also performed at the same total carbon-equivalent concentration for comparative analysis. Glucose consumption rate by cells growing on glucose alone was 48.5 ± 13.8 mmol carbon gCDW−1 h−1 (or 8.09 ± 2.3 mmol glucose gCDW−1 h−1); a similar value was reported in a previous study on glucose-grown P. putida (14) (Fig. 2a and Figs. S1 and S2). During growth on the glucose:benzoate mixture, we obtained carbon-equivalent consumption rates of 19.8 ± 4.4 mmol carbon gCDW−1 h−1 for glucose and 27.2 ± 8.1 mmol carbon gCDW−1 h−1 for benzoate (or 3.3 ± 0.2 mmol glucose gCDW−1 h−1 and 4.5 ± 1.4 mmol benzoate gCDW−1 h−1) (Fig. 2a and Figs. S1 and S2). Thus, the total carbon consumption rate from the glucose:benzoate mixture, 47.0 ± 11.2 mmol carbon gCDW−1 h−1, was similar to the carbon-equivalent consumption rate of glucose alone (48.5 ± 13.8 mmol carbon gCDW−1 h−1) (Fig. 2a). Previous studies (14, 20) have reported a preference for benzoate over glucose in P. putida, but at the same carbon concentration in the glucose:benzoate mixture, we found no difference between the consumption rates of the two substrates (unpaired t test, p = 0.23). Interestingly, we obtained a 37% increase in the growth rate of cells fed on the mixture (0.93 ± 0.10 h−1) versus on glucose alone (0.68 ± 0.09 h−1) (unpaired t test, p = 0.03) (Fig. 2b and Figs. S1 and S2). These data collectively implied that the intracellular processing of both substrate carbons in the P. putida KT2440 cells achieved optimal biomass growth.
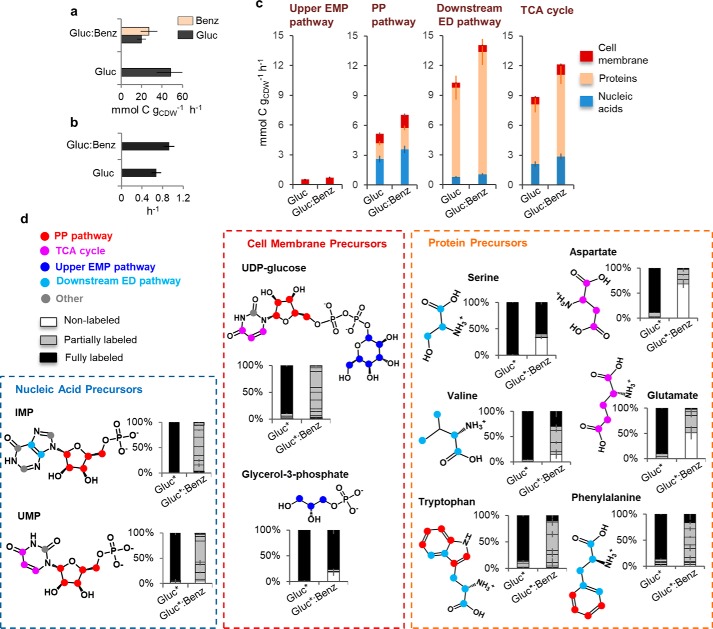
Figure 2.
Predicting and tracing incorporation of substrate carbons into biomass biosynthesis. a, biomass growth rate of cells grown on glucose alone (Gluc) or with benzoate (Benz) at the same total carbon equivalence (100 mm C). b, consumption rate of each substrate during exponential cell growth. c, calculated carbon flux demand required from metabolism to sustain measured biomass growth based on cellular stoichiometries for cell membrane, nucleic acids, and proteins. d, labeling profile of selected biomass precursors during growth on [U-13C6]glucose (Gluc*) alone or with unlabeled benzoate. In d, the structure of the biomass precursor is shown on the left of the bar graphs; the colored circles in the structure indicate the pathway origin of the designated carbon: red circles (PP pathway), pink circles, TCA cycle, dark blue (upper EMP pathway), light blue (downstream ED pathway), and gray circles (involvement of one-carbon units). The error bars represent standard deviation values (n = 3 biological replicates).
The carbon supply required from the different relevant metabolic pathways to support biomass growth would depend on the biomass component (Fig. 2c). Based on the genome-scale stoichiometric composition of the P. putida cell (25) and our measured growth phenotype, we estimated the following carbon effluxes to support biosynthetic demands in cells grown, respectively, on glucose alone versus on the glucose:benzoate mixture: 2.77 ± 0.19 mmol carbon gCDW−1 h−1 and 3.76 ± 0.21 mmol carbon gCDW−1 h−1 for cell membrane biosynthesis; 5.50 ± 0.46 mmol carbon gCDW−1 h−1 and 7.51 ± 0.51 mmol carbon gCDW−1 h−1 for nucleic acid biosynthesis; and, 16.59 ± 1.45 mmol carbon gCDW−1 h−1 and 22.68 ± 1.61 mmol carbon gCDW−1 h−1 for protein biosynthesis (Fig. 2c). For cell membrane, the majority of carbon efflux was derived from the PP pathway (∼35%), and the remainder came from the TCA cycle (28%), upper EMP (20%), and downstream of the ED pathway (18%) (Fig. 2c). For nucleic acids, the majority of carbon efflux was also derived from the PP pathway (∼48%), and the remainder came from the TCA cycle (38%) and downstream of the ED pathway (14%) (Fig. 2c). For proteins, the majority of carbon efflux was derived primarily from downstream of the ED pathway (54%), and the remainder came from the TCA cycle (36%) and the PP pathway (∼10%) (Fig. 2c). Therefore, the highest biosynthetic carbon demand was from downstream of the ED pathway and TCA cycle to make proteins followed by the carbon demand from the PP pathway to make nucleic acids (Fig. 2c). In sum, to support biosynthetic requirements for the observed biomass growth, the equimolar carbon-equivalent supply of glucose and benzoate had to be channeled effectively to different metabolic pathways.
Biosynthetic demands are met by nonuniform metabolic investment of the substrate carbons
We monitored the incorporation of [U-13C6]glucose alone or with unlabeled benzoate into representative metabolite precursors to the different biomass components: inosine monophosphate (IMP) and uridine monophosphate (UMP) for nucleic acid precursors; uridine diphosphate glucose (UDP-glucose) and glycerol-3-phosphate (glycerol-3P) for cell membrane precursors, and a series of amino acids for protein precursors (Fig. 2d). We confirmed that isotopic enrichment had reached pseudo-steady state by examining intracellular metabolite labeling at two different times during the exponential growth phase (Fig. S3).
Ribonucleotide precursors combine metabolite intermediates from three metabolic pathways. The purine IMP is synthesized from the combination of ribose-5-phosphate (R5P) (a PP pathway metabolite) with glycine (an amino acid derived from the 3-phosphoglycerate (3-PG) downstream of the ED pathway), tetrahydrofolate-linked one-carbon units, and dissolved bicarbonate species. The biosynthesis of the pyrimidine UMP combines R5P with aspartate (an amino acid derived from the TCA cycle metabolite OAA) and dissolved bicarbonate species (Fig. 2d). Both IMP and UMP were predominately partially labeled in the glucose:benzoate growth condition, thus indicating incorporation of carbons from both glucose and benzoate in their biosynthesis (Fig. 2d).
Biosynthesis of the cell membrane precursors UDP-glucose and glycerol-3P also makes use of metabolites from three different pathways: UDP-glucose is synthesized by combining the pyrimidine UMP with G6P (an upper-EMP pathway metabolite); glycerol-3P is synthesized by reducing dihydroxy-acetone-phosphate (DHAP) (a metabolite downstream of ED pathway) (Fig. 2d). During growth on the mixture, glycerol-3P was made exclusively either from the labeled glucose (∼25%) or from the nonlabeled benzoate (∼75%), whereas the carbon skeleton of UDP-glucose contained carbons derived from both substrates (Fig. 2d).
The labeling patterns of amino acids in cells grown on the glucose:benzoate mixture depended on the source of the metabolite precursors (Fig. 2d). For serine (derived from 3-PG downstream of the ED pathway), a large portion (69%) was made exclusively from glucose-derived 13C-labeled carbons, and the remainder was made from nonlabeled benzoate carbons (Fig. 2d). A previous study (18) of P. putida KT2440 (pWW0) fed on nonlabeled toluene and 13C-labeled glucose also reported toluene-derived nonlabeled carbons in serine. Derived from 2 mol of pyruvate further downstream of the ED pathway, valine was ∼30% 13C-labeled and ∼60% partially labeled, indicating the incorporation of both substrates into the majority of the valine pool (Fig. 2d). For the aromatic amino acids tryptophan and phenylalanine, which are derived from metabolites of the PP and downstream ED pathways, a high fraction (80 to 90%) of partial 13C labeling indicated that the contribution of both substrates (Fig. 2d). However, the two amino acids derived from the TCA cycle (aspartate and glutamate) were 50–75% nonlabeled, reflecting that the TCA cycle was primarily populated by benzoate-derived carbons (Fig. 2d). Taken collectively, the 13C-labeling patterns of the biosynthetic precursors revealed that the substrate carbons from the mixture were not distributed uniformly toward the biosynthetic pathways.
Quantitative tracing of the metabolic network reveals segregated routing of the substrate carbons through central carbon metabolism
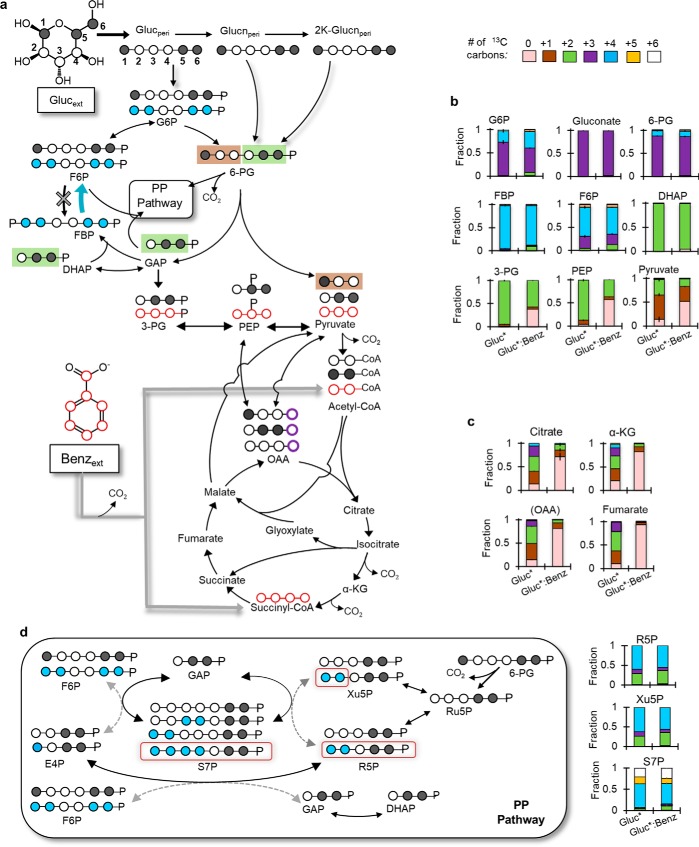
To elucidate the specific metabolic routing of the substrate carbons, we performed additional long-term isotopic enrichment with [1,5,6-13C3]glucose alone or with unlabeled benzoate (Fig. 3). For both growth conditions, we obtained similar labeling patterns for the metabolites in initial glucose catabolism, the upper EMP pathway, and the PP pathway (Fig. 3). In accordance with the periplasmic oxidation of [1,5,6-13C3]glucose, Glucn was nearly completely triply 13C-labeled (Fig. 3, a and b). The phosphorylated metabolites in initial glucose catabolism, G6P and 6P-Glucn, were also primarily triply 13C-labeled, respectively 53–71% and 84–88% (Fig. 3, a and b).
Figure 3.
Elucidating the segregated metabolic network structure through the assimilation of [1,5,6-13C3]glucose (Gluc*) and unlabeled benzoate (Benz). a–c, carbon mapping (a) and metabolite labeling in b the periplasm, the EMP pathway, the ED pathway, and in c the TCA cycle. d, carbon mapping (left) and metabolite labeling (right) in the PP pathway. In a and d, the white- and black-filled circles represent, respectively, nonlabeled and 13C-labeled carbons. In d, the dashed arrows indicate the minor formation routes of the metabolites; the red squares highlight how sextuply 13C-labeled S7P is produced from the two pentose–phosphate metabolites. Color coding for the 13C-labeling patterns is as follows: nonlabeled (light pink), singly labeled (brown), doubly labeled (green), triply labeled (purple), quadruply labeled (light blue), quintuply labeled (yellow), and sextuply labeled (white). The error bars represent standard deviation values (n = 3 biological replicates). Metabolite abbreviations are as detailed in the Fig. 1 legend.
Quadruply 13C-labeled fractions (12–36%) of G6P and 6P-Glucn were due to carbon cycling from the ED pathway to the upper EMP pathway (Fig. 3b). Through the ED pathway, triply 13C-labeled 6P-Glucn generated singly 13C-labeled pyruvate (up to 50%) and doubly 13C-labeled (>90%) glyceraldehyde-3-phosphate (GAP) and DHAP (an isomer of GAP) (Fig. 3, a and b). Through backward flux, doubly 13C-labeled GAP and DHAP combined to produce over 85% quadruply 13C-labeled fructose-1,6-bisphosphate (FBP) (Fig. 3, a and b). The dephosphorylation of FBP led to up to 65% quadruply 13C-labeled fructose-6-phosate (F6P) and the isomerization of G6P to F6P generated triply 13C-labeled F6P (up to 25%) (Fig. 3, a and b). Because of the lack of 6-phosphofructokinase in P. putida to convert F6P to FBP (26), there was no triply 13C-labeled FBP as would be expected from the phosphorylation of triply 13C-labeled F6P to FBP (Fig. 3, a and b). Thus, for both growth conditions, the 13C mapping thus showed that glucose-derived carbons were catabolized through a cyclic flux connection from the ED pathway toward the upper EMP pathway and the nonoxidative PP pathway (Fig. 3, a and b). This cyclic connection was also shown previously for glucose-grown P. putida KT2440 (15, 16, 27), Pseudomonas protegens Pf-5 (28), and Pseudomonas aeruginosa PAO1 (27) (Fig. 3, a and b).
Through the nonoxidative PP pathway, F6P and GAP combined through a series of ketolase and aldolase reactions to generate up to 60% quadruply 13C-labeled xylulose-5-phosphate (Xu5P) and R5P; the oxidative PP pathway generated up to 33% doubly 13C-labeled PP metabolites (Fig. 3d). Thus, there was approximately a two-to-one involvement of the nonoxidative route relative to the oxidative route of the PP pathway (Fig. 3d). A similar finding was reported by a previous analysis (16) of glucose-grown P. putida. The labeling of sedoheptulose-7-phosphate (S7P), primarily quadruply 13C-labeled (∼50%) and sextuply 13C-labeled S7P (∼25%), corroborated carbon flux primarily through the nonoxidative PP pathway for both growth conditions (Fig. 3d).
For the downstream metabolic pathways (i.e. downstream ED pathway and the TCA cycle), there were different labeling patterns in the absence versus the presence of benzoate (Fig. 3, a–c). In addition to singly 13C-labeled pyruvate from the ED pathway, doubly 13C-labeled pyruvate (up to 33%) was due to carbon flux through metabolites (3-PG and phosphoenolpyruvate (PEP)) downstream of doubly 13C-labeled GAP, and nonlabeled pyruvate (∼50%) indicated the incorporation of benzoate-derived carbons (Fig. 3, a and b). During growth on the glucose:benzoate mixture, gluconeogenic flux of benzoate-derived carbons further led to the appearance of nonlabeled fraction in 3-PG and PEP (38–56%; Fig. 3c). In agreement with the canonical routing of carbons toward and through the TCA cycle in the glucose-grown cells, the metabolites in the TCA cycle were primarily singly, doubly, and triply 13C-labeled (Fig. 3, a and c). However, in cells grown on the glucose:benzoate mixture, the influx of nonlabeled benzoate-derived carbons via the ortho pathway led to enrichment in nonlabeled fractions (71–93%) in the TCA-cycle metabolites (Figs. 1 and 3, a and c). The similarity between the labeling pattern of citrate and the labeling pattern of OAA (deduced from aspartate labeling) further indicated that the acetyl moiety in acetyl-CoA was primarily nonlabeled from benzoate catabolism (Figs. 1 and 3b).
In sum, the carbon mapping demonstrated that the metabolism of glucose and benzoate was segregated into different pathways in the metabolic network. Notably, the upper EMP pathway, the ED pathway, and the PP pathway were populated exclusively by glucose-derived carbons (Fig. 3, a and b). Benzoate-derived carbons primarily populated the TCA cycle and directly upstream of the TCA cycle (Fig. 3, a and c).
Multi-omics analysis highlights regulation of metabolic fluxes by metabolite levels and post-translational modifications
We profiled changes in metabolic fluxes in conjunction with changes in both protein levels and metabolite pools in cells grown on the glucose:benzoate mixture versus those grown on glucose alone. Where appropriate, we also examined relative extent of protein phosphorylation, which serves as a switch on–off mechanism to control protein activity (Figs. 4 and 5 and Tables S2–S4). In the presence of benzoate, we obtained only a 4% increase in the global regulator CRC (18, 29), which is known to repress carbohydrate uptake and catabolism (Fig. S4). However, of the total 83 profiled proteins in the central carbon metabolism, the abundances of 15 were increased (by up to 200%), and 25 were decreased (by up to −60%) in the presence of benzoate; the remaining protein levels were either unchanged or not quantifiable (Fig. 5b). Correlation matrices of flux changes versus protein abundance changes illustrated that changes in protein levels were not always predictive of changes in metabolic fluxes between the two growth conditions (Fig. 5a and Table S4).
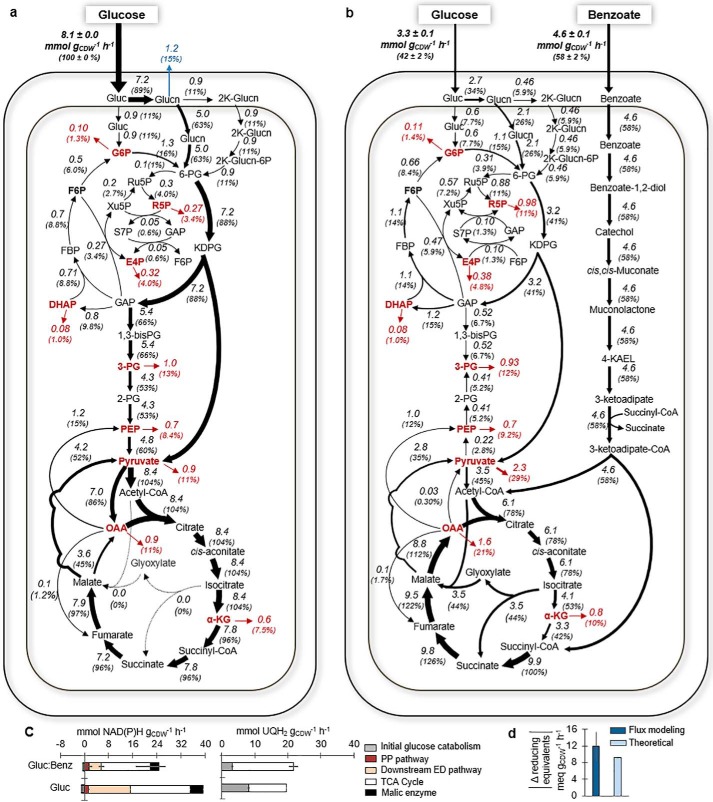
Figure 4.
Cellular metabolic flux distributions. a and b, net metabolic fluxes both in absolute values (mmol gCDW−1 h−1) and normalized to the total uptake flux (%) during feeding on glucose alone (a) or glucose and benzoate (b). c, relative contribution of different pathways to generate NAD(P)H and UQH2 calculated from the metabolic fluxes. d, absolute difference in the yield of reducing equivalents determined from the quantitative flux modeling (dark blue) and theoretically from carbon oxidation state of the substrates (light blue). In a and b, arrow widths are proportional to absolute magnitudes of fluxes. The cellular fluxes were determined using data from biological replicates (n = 3). The full statistics of the data are presented in Tables S2 and S3. Metabolite abbreviations are as detailed in the Fig. 1 legend.
Figure 5.
Relationship of proteomics to fluxomics and metabolomics. Shown are the correlation matrices of relative changes in metabolic fluxes versus relative changes in abundance levels of metabolic proteins (a), percentage of change of metabolic proteins (b), and fold change (expressed in log2) in the abundance of selected intracellular metabolites (c). In a–c, changes or ratios are determined by comparing data obtained with cells fed on the glucose:benzoate mixture to data obtained with cells fed on glucose alone. In b, relative extent of phosphorylation site is shown with a colored oval for the following proteins: ACEF, PPSA, and PP_4401. The data in a and c (average ± standard deviation) were obtained from biological replicates (n = 3); the statistics of the data presented in a and b are listed in Table S4.
Initial glucose catabolism, upper EMP pathway, and pentose-phosphate pathway
In both growth conditions, periplasmic oxidation of glucose to Glucn represented over 80% of the glucose uptake flux; the direct uptake of glucose to G6P represented a small flux (less than 15%) (Fig. 4, a and b). A 13-fold increase in the S7P pool was consistent with a 2-fold increase in the flux from PP metabolites to produce S7P during growth on the glucose:benzoate mixture (Fig. 5c). Compared with growth on glucose alone, the levels of 9 of the 10 quantified proteins involved in initial glucose catabolism were decreased (−20% to −60%) and the 2-keto-gluconate level was depleted (by 300-fold) during growth on the glucose:benzoate mixture (Fig. 5, b and c). Relative to glucose-grown cells, a significant increase both in the backward fluxes through the upper EMP pathway (up to 80%) and the fluxes in the PP pathway (nearly up to 3-fold) did not match the relatively minor changes (within ±16%) in the associated protein levels in the cells grown on the mixture (Figs. 4, a and b, and 5b). In addition to the aforementioned cyclic connection between the ED pathway and upper EMP pathway, decreased abundance (by over 30%) of GAP dehydrogenase may have contributed to the retention of glucose-derived carbons in these pathways to feed carbon fluxes therein during growth on the glucose:benzoate mixture (Figs. 4, a and b, and 5b).
ED pathway and gluconeogenic flux
In the glucose-grown cells, 89% of the glucose uptake was channeled through the ED pathway to sustain biosynthetic fluxes in downstream pathways (Fig. 4a). However, despite a near 2-fold decrease in the ED pathway flux and a 10- to 20-fold decrease in the flux from 3-PG to pyruvate in cells grown on the glucose:benzoate mixture, the biosynthetic flux demands (from 3-PG, PEP, and pyruvate) were still satisfied due to gluconeogenic flux of benzoate-derived carbons upstream of the TCA cycle (Fig. 4, a and b). This flux directionality in the presence of benzoate could not be deduced from the protein abundances of the corresponding reactions, which remained unchanged or reduced by up to 20% (Fig. 5, a and b). However, the observed decrease (by 36%) in a tyrosine phosphorylation of PEP synthase (PPSA), which would turn on the activation of PPSA, would thus facilitate backward flux from pyruvate to PEP (Fig. 5b). Moreover, the measured increase (by 25%) in a serine phosphorylation of acetyl-CoA carboxylase (ACEF), which would repress the activation of ACEF, would impair flux from pyruvate to acetyl-CoA (Fig. 5b). Moreover, we obtained a 10-fold accumulation in pyruvate pool, whereas there were up to 63% reduction in the PEP pool and a 3-fold decrease in the 3-PG pool (Fig. 5c), illustrating both the decrease in ACEF activity and that flux directionality was in the direction that favored metabolic flow from high concentration to low concentration of metabolites.
TCA cycle
Growth on the glucose:benzoate mixture resulted in a nearly 3-fold increase in the protein levels in the benzoate catabolic ortho pathway to generate acetyl-CoA and succinate (Fig. 5b). In accordance with the benzoate-derived carbon influx toward the TCA cycle, we obtained a 14-fold increase in the acetyl-CoA pool, a 2-fold increase in the succinate pool, and a 2- to 5-fold accumulation in the pools of the other TCA cycle metabolites (citrate, α-KG, fumarate, malate, aspartate) (Fig. 5c). The 2-fold increase in the flux from succinate to OAA in the presence of benzoate was accompanied by an increase (albeit to a lesser extent) in the abundance of most of the associated proteins: subunits of succinate dehydrogenase (SDH) (SDHA, +50%; SDHB, +73%; SDHC, +158%), one subunit of malate:quinone oxidoreductase (MQO) (MQO1, +55%), and fumarate hydratase (+1.8%) (Figs. 4, a and b, and 5b); the levels of the other subunits of MQO were decreased (MQO2, −11.9%; MQO3, −13%) (Fig. 5b). Along with the high accumulation of acetyl-CoA in cells grown on the glucose:benzoate mixture, there was an increased abundance (by nearly 15%) of citrate synthase (GLTA2), which combines acetyl-CoA with OAA to form citrate (Fig. 5, b and c). Because of the activation of the glyoxylate shunt, which has been known to be induced by an elevated acetyl-CoA (30), the flux from citrate to α-KG in the TCA cycle was decreased by up to 60% during growth on the glucose:benzoate mixture, and the abundances of the corresponding proteins were also decreased (by up to 36%) (Figs. 4, a and b, and 5b).
Both the active glyoxylate shunt and higher fluxes from succinate to OAA during growth on the glucose:benzoate mixture observed here were reported by a previous flux analysis (13) of P. putida KT2440 grown on benzoate alone compared with growth on fructose, but no change was observed in the levels of both gene expression and transcripts of the involved proteins in the previous study. Here, we further observed that the active glyoxylate shunt was not consistent with decreased abundances of isocitrate lyase (ACEA, −44% %) (Figs. 4b and 5b). However, consistent with reported feedback inhibition of ICD (24, 31, 32) and the co-regulation of glyoxylate shunt and ICD activity (24, 33, 34), we obtained both decreased flux from isocitrate to α-KG (−54%) and decreased abundance in one of the ICD-associated proteins (PP_4011, −33.1%) (Figs. 4, a and b, and 5b). Interestingly, we also obtained a decrease (by over 50%) in the serine phosphorylation of PP_4011, which would promote ICD activity and thus would counteract to some extent the decreased flux because of decreased protein abundance in the presence of benzoate (Fig. 5b).
In sum, metabolic flux changes in cells grown on the glucose:benzoate mixture versus those grown on only glucose were partially in agreement with the corresponding changes in protein abundances (Fig. 5a). However, changes in metabolite levels and protein phosphorylation could explain flux magnitude and directionality, which were not predictable from the changes in protein levels (Figs. 4 and 5).
The segregated metabolic network maintains redox balance
Relative to the metabolic fluxes for the glucose:benzoate mixture, the flux analysis determined that the metabolism of glucose alone led to an 11% increase in the yield of UQH2 and a 37% increase in the yield of NAD(P)H (Fig. 4c). For UQH2, the higher contribution (by 61%) from the periplasmic oxidation reactions in the glucose-grown cells was compensated by the higher contribution (by 63%) from the increased TCA cycle flux in the presence of benzoate (Fig. 4c). For NAD(P)H, there was a lower contribution from the malic enzyme (by 35%), the TCA cycle (by 18%), and downstream of the ED pathway (by 71%) during growth on the mixture compared with growth on only glucose (Fig. 4c). In sum, the total yield of reducing equivalents was decreased by 12.0 ± 3.4 meq gCDW−1 h−1 in the presence of benzoate (Fig. 4d). Accounting for the carbon oxidation states (−2 per mol of benzoate versus 0 per mol of glucose) and the measured carbon consumption rates, we calculated a theoretical surplus of 9.2 ± 0.2 meq gCDW−1 h−1 from the influx of electrons from benzoate (Fig. 4d). This theoretical difference was remarkably consistent with the flux-determined surplus of reducing equivalents by glucose metabolism (Fig. 4d). Therefore, the segregated metabolic flux network in P. putida KT2440 cells grown on the glucose:benzoate mixture maintained approximately the same redox balance as the cells grown on only glucose.
Discussion
Here, using P. putida as a prototypical species of the Pseudomonas genus, we analyzed the metabolome and proteome to unravel the mechanisms by which the unique metabolic attributes of Pseudomonas species channel simultaneously sugar and aromatic carbons. Our 13C-metabolomics mapping demonstrated the compartmentalization of each substrate into two cyclic pathway networks in P. putida KT2440. Glucose-derived carbons were compartmentalized within the cyclic connection of the ED pathway to the upper EMP pathway and the nonoxidative PP pathway (15, 16); benzoate-derived carbons were compartmentalized within the TCA cycle and the glyoxylate shunt (Fig. 6). This metabolic segregation evidently represented an efficient strategy to meet biosynthetic flux demands both in terms of redox balance and carbon skeletons (Fig. 4).
Figure 6.
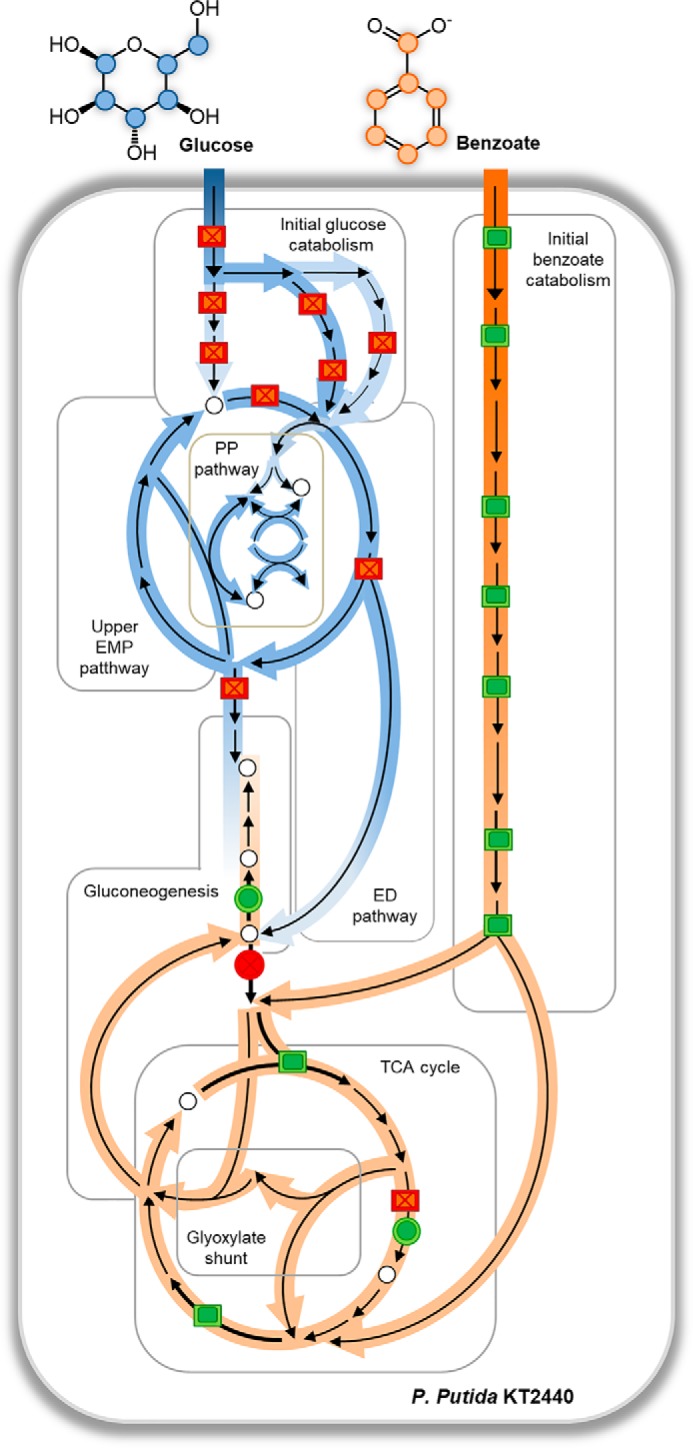
Schematic of the segregated metabolic network tuned for the co-processing of glucose and benzoate in P. putida KT2440. Blue- and orange-shaded arrows illustrate, respectively, the major metabolic flows of glucose-derived carbons and benzoate-derived carbons. Colored rectangles and circles show regulatory points of metabolic flux reactions by, respectively, protein abundance and protein phosphorylation: green for up-regulation and red for down-regulation. White-filled circles show points of biosynthetic flux demands from the central carbon metabolism.
A recent report (35) of metabolite profiling of different organisms demonstrated that over two-thirds of intracellular metabolite concentrations were above enzyme-binding affinity, which thus maintained favorable reaction thermodynamics. In the absence of absolute metabolite quantitation, we were not able to conduct a similar evaluation in our study. However, our profiling of relative metabolite levels in P. putida KT2440 across the two growth conditions implied fluctuations in reaction quotients to meet energetically favorable metabolic fluxes (Fig. 5c). Notably, compared with P. putida KT2440 feeding on glucose alone, the fluxomics of the metabolic network during feeding on the glucose:benzoate mixture was not fully predictable by relative protein abundances (Figs. 4 and 5); a similar finding was reported by a previous study of P. aeruginosa grown on single substrates (36).
Consistent with reported down-regulation of genes in glucose catabolism in P. putida strains grown on aromatic substrates (18, 37, 38), we obtained a decrease in both protein abundances and metabolic fluxes in initial glucose catabolism in cells grown on the glucose:benzoate mixture compared with those grown on only glucose (Figs. 4, a and b, and 5, a and b). However, biosynthetic flux demands in the upper EMP and PP pathways were still satisfied, which we attributed to the exclusive retention of glucose-derived carbons in these pathways (Figs. 4b and 6). Remarkably, there were minimal changes in the levels of metabolites in the upper EMP and PP pathways, with the exception of one metabolite (S7P) in the PP pathway and one metabolite (2-ketogluconate) in the periplasm (Fig. 5c).
In the TCA cycle, discrepancies between changes in protein levels and changes in metabolic fluxes stressed the importance of metabolite-level regulation (24, 31–34, 39) (Figs. 4, a and b, and 5, a and b). Flux analysis in a previous study (14) of P. putida KT2440 grown on benzoate alone determined an active glyoxylate shunt, despite a lack of change in both gene expression and transcripts of proteins in the glyoxylate shunt when compared with cells grown on a hexose sugar. Here, consistent with the well-known induced activation of the glyoxylate shunt by elevated acetyl-CoA levels (30), our profiling of metabolite levels in cells grown on the glucose:benzoate mixture captured a greater than 10-fold increase in acetyl-CoA (Figs. 4b and 5c). We also recorded a decrease in the phosphorylation of an ICD-associated protein (PP_4011), which may serve to counter the expected overwhelming metabolite-level inhibition of ICD activity from metabolites of the glyoxylate shunt and pyruvate accumulation (30) (Figs. 4b and 5, b and c).
The gluconeogenic flux of benzoate-derived carbons immediately upstream of the TCA cycle was in the direction of favorable cascade of metabolic flow, from high concentrations to low concentrations of metabolites (Fig. 6). Additional regulation in support of this flux directionality was provided by the phosphorylation levels of PPSA and ACEF (Fig. 6). In sum, our results highlighted that both metabolite levels and post-translational switch of protein activity via phosphorylation modifications could be implicated in establishing thermodynamic favorability (40, 41) of flux magnitude and directionality in the metabolic network (Fig. 6). Further investigation is needed to gain more insights on the manifestation of these regulatory controls under different nutrient conditions or in engineered strains.
Because of its diverse metabolic capabilities, P. putida has been of particular interest as an attractive bacterial cell factory for various functionalities in industrial applications (42, 43). Subsequent research with mutant or engineered strains is needed to explore whether leveraging in part or in whole the segregated metabolism detailed here may enhance mixed-substrate processing in biotechnological applications of Pseudomonas species. Toward informing these potential engineering efforts, our multi-omics investigation provides new insights into the constitutive tuning of the metabolic architecture that achieves efficient co-utilization of carbohydrate and aromatic substrates in P. putida KT2440. This metabolic architecture may be instrumental in conferring the nutritional flexibility of Pseudomonas species and the ubiquitous survival of these bacterial species in different environmental niches.
Experimental procedures
Culturing conditions
P. putida KT2440 was obtained from American Type Culture Collection (Manassas, VA) as freeze-dried cultures. The cells were resuspended and grown in nutrient-rich liquid Luria–Bertani medium. For carbon source-specific growth, liquid cultures of P. putida KT2440 were grown in 125-ml baffled flasks (cell suspension did not exceed one-fifth of the total flask volume) shaken vigorously at 220 rpm at 30 °C in a G24 environmental incubator shaker (New Brunswick Scientific, Edison, NJ). The pH-adjusted (pH 7.0) and filter-sterilized (0.22-μm nylon; Waters) growth medium contained major and minor salts as previously reported (44). The total carbon-equivalent substrate concentration of 100 mm C was added as glucose alone (equivalent to 16.7 mm or 3 g liter−1 glucose) or as a 1:1 glucose:benzoate mixture (equivalent to 1.5 g liter−1 glucose and 2.0 g liter−1 benzoate). The cells were transferred twice into fresh growth medium before conducting the experiments to ensure that cells were conditioned in their respective growth medium. Cell growth (three biological replicates) was monitored by measuring the A600 using an Agilent Cary UV-visible spectrophotometer (Santa Clara, CA). The initial A600 of the cells at each transfer was between 0.05 and 0.07. The cells were transferred during mid-exponential phase, between A600 0.5 and 1.0. An A600 measurement was obtained every 60 min until stationary phase; we performed dilutions of cell suspensions to get accurate readings at A600 above 0.5. Cell dry weight in grams (gCDW) was also determined following drying of 1-ml aliquots of cell suspensions using a Labconco freeze-dryer (Kansas City, MO).
Intracellular metabolite labeling and substrate consumption
For long-term intracellular labeling, culturing conditions were as described with two transfers of cultures (three biological replicates) in fresh growth medium with labeled substrates purchased from Cambridge Isotopes (Tewskbury, MA) or Omicron Biochemicals (South Bend, IN). The labeled substrates were the following: [U-13C6]glucose alone, [U-13C6]glucose with unlabeled glucose, [1,5,6-13C3]glucose alone, or [1,5,6-13C3]glucose with unlabeled benzoate. At two time points during mid-exponential phase (A600 of 0.5 and 1.0), cell suspensions (3 ml) were collected and filtered following immediate quenching of the cell-containing filters in a cold (4 °C) 2-ml solution of methanol:acetonitrile:water (40:40:20) solution. Solutions with the lysed cells were subsequently filter-centrifuged (Sigma–Aldrich Spin-X, 0.22-μm filters). Aliquots of the supernatants were dried under nitrogen gas and resuspended in ultrapure LC-MS water (Fisher Scientific) before analysis via LC-MS. Metabolite levels were normalized to biomass quantity at the time of sampling.
To quantify depletion of extracellular glucose and benzoate from the medium, samples (three biological replicates) were harvested throughout the growth of the cells and stored at 4 °C until analysis time. The samples were analyzed by 1H NMR using a Varian Unity INOVA 600-MHz NMR spectrometer at 25 °C (relaxation delay of 5 s, recording of 16 scans/sample, receiver gain of 32 dB) (45). The extracellular samples were prepared for NMR analysis by mixing 250 μl of filtered sample (0.22-μm pore size nylon filters; Fisher Scientific) with 60 μl of 100% D2O, 50 μl 2,2-dimethyl-2-silapentane-5-sulfonate as a chemical shift reference compound, and 50 μl of NaN3 as an antimicrobial agent. The 13C-tracer experiments confirmed that extracellular depletion of the substrates correlated with substrate consumption. Substrate consumption rates (in mmol gCDW−1 h−1) during exponential growth were then determined by regression analysis.
Sampling of extracellular metabolite levels
To monitor metabolite excretion rates, metabolites in the extracellular medium were quantified during exponential phase. Aliquots (250 μl) of the cell suspensions were collected for each of the three biological replicates followed by centrifugation (21,130 × g for 5 min) and filtration (0.22-μm pore size nylon filters) to remove cell particulates. The samples were diluted (by 1:100 or 1:1000) before LC-MS analysis as described below. Metabolite excretion rates (μmol gCDW−1 h−1) were determined by regression analysis.
Metabolomics analysis via LC-MS
Metabolite extracts were analyzed by reversed-phase ion-pairing LC-MS using ultra-HPLC (UHPLC; Thermo Scientific DionexUltiMate 3000) coupled with high-resolution/accurate-mass mass spectrometer (Thermo Scientific Q Exactive quadrupole-Orbitrap hybrid MS) with electrospray ionization operated in negative mode. Details on solvent composition and gradient for the LC protocol through a Waters Acquity UPLC BEH C18 column (1.7 μm with a column size of 2.1 × 100 mm) (Waters) were reported previously (44). The following metabolites were monitored: G6P, Glucn, F6P, R5P, Xu5P, S7P, FBP, DHAP, 3-PG, PEP, pyruvate, citrate, α-KG, succinate, fumarate, malate, and aspartate. All metabolite identification and isotopic enrichment were determined using the Metabolomics Analysis and Visualization Engine (MAVEN) software package (46). The 13C-labeled fractions were corrected for natural 13C abundance.
Metabolic flux quantitation
Using the software 13CFLUX2 (47), a metabolic flux analysis of the central carbon metabolism was performed using experimental data (average ± standard deviation) as constraints. The labeling patterns of the intracellular metabolites following growth on [1,5,6-13C3]glucose alone or with unlabeled benzoate were integrated with the growth phenotype data, measured metabolite excretions, and genome-scale determination of biosynthetic flux demands to quantitate explicitly the fluxes through 38 reactions in the metabolic network and 57 reactions collectively by considering that a flux remains unchanged in the absence of carbon loss from one metabolic reaction to the next. Using the biomass growth rates determined experimentally and published biomass composition of P. putida KT2440 (25), conversion rates were calculated for metabolite precursors to the biosynthesis of proteins, nucleic acids, and cell membrane. The 13C-labeling data used in the metabolic flux analysis were for the following metabolites: gluconate, G6P, F6P, R5P, Xu5P, S7P FBP, DHAP, 3-PG, PEP, pyruvate, citrate, α-KG, succinate, fumarate, and aspartate for OAA. Initial flux values were set based on published values (15, 16) and subsequently optimized based on the aforementioned experimentally obtained data. Optimization of the metabolic flux analysis was achieved by satisfying carbon balance and evaluated agreement between experimentally determined and model-estimated labeling patterns (Figs. S5 and S6). The quality of the fit to experimental data were measured by calculating sum of squared residuals based on comparisons of model-estimated metabolite labeling patterns to the measured values (16, 48).
Protein extraction method
All steps for cell harvest and protein extraction were conducted at 4 °C. At mid-exponential phase (A600 = 0.5–0.7), 25-ml culture aliquots were harvested from biological replicates (n = 3). Each sample was centrifuged (at 4 °C), and the supernatant was carefully removed before washing the cell pellet twice with high-purity deionized (Milli-Q grade) water. Pellets were then stored at −20 °C until further use. For extraction, the thawed pellet was resuspended in 4-ml of 2.5 mm phosphate buffer (pH 7) and then homogenized by vortexing. The cells were lysed in a French Pressure cell press (Thermo Spectronics, Waltham, MA) with two 3-min cycles at 900 p.s.i. The lysates were centrifuged (at 16,000 × g for 10 min at 4 °C), and an aliquot of the supernatant (2 ml) was concentrated to 200 μl using a Spin-X® UF 500 Concentrator (Corning, Tewksbury, MA). The proteins were precipitated overnight at −20 °C in 9 volumes of cold 10% TCA in acetone. The solution was centrifuged at 16,000 × g (4 °C) for 15 min to form a protein pellet; the supernatant was discarded. The pellet was washed two times in cold acetone. Urea (7 m) was used to initiate protein dissolution. Each dissolved pellet was then diluted in 50 mm triethylammonium bicarbonate (TEAB) buffer (pH 8). The final sample had less than 1 m urea contamination. The protein concentration for each sample was determined by Bradford assay and further quantified by running on a precast NOVEX 12% Tris/glycine mini-gel (Invitrogen) along with a series of amounts of E. coli lysates (2, 5, 10, and 20 μg/lane). The SDS gel was visualized with colloidal Coomassie Blue stain (Invitrogen) and imaged by Typhoon 9400 scanner followed by ImageQuant TL 8.1 (GE Healthcare). Further processing of the proteins was then performed according to Thermo Scientific's TMT mass tagging kits and reagents protocol with a slight modification (49, 50).
A total of 200 μg protein of each sample in 70 μl of 7 m urea and 50 mm TEAB was reduced with 11 mm tris (2-carboxyethyl)phosphine for 1 h at room temperature, alkylated with 37 mm iodoacetamide for 1 h in the dark, and then quenched by addition of 40 mm DTT. The alkylated proteins were diluted with 240 μl of 100 mm TEAB. Each sample was digested with 18 μg of trypsin for 18 h at 35 °C, and an additional 2 μg of trypsin was added and incubated for 4 h. The TMT 10-plex labels were reconstituted with 45 μl of anhydrous acetonitrile (ACN) prior to labeling and added with 1:2 ratio to each of the tryptic digest samples. The samples were incubated for 1 h at room temperature to TMT label. The peptides from 10 samples were mixed with each tag of TMT10plex separately. After checking the label incorporation using Orbitrap Fusion (Thermo Fisher Scientific) by mixing 1-μl aliquots from each sample and desalting with SCX ziptip (Millipore, Billerica, MA), equal amounts of peptide from the 10 digested samples were pooled together. The pooled peptides were evaporated to 200 μl and subsequently purified by solid-phase extraction on Sep-Pak Cartridges (Waters), according to the manufacturer recommendations and dried at reduced pressure in a SpeedVac concentrator (Savant).
Proteomics analysis using nano-scale reverse phase LC and tandem MS
The tryptic peptides prepared as described above were subjected to a first-dimensional fractionation via high-pH reverse-phase chromatography, carried out using a Dionex UltiMate 3000 HPLC system (Thermo Scientific, Sunnyvale, CA) as reported previously (49, 50). Specifically, the TMT 10-plex tagged tryptic peptides were reconstituted in 20 mm ammonium formate at pH 9.5 (eluent A) and loaded onto an XTerra MS C18 column (3.5 μm, 2.1 × 150 mm) from Waters equilibrated with eluant A. Eluant B consisted of 80% ACN, 20% 20 mm ammonium formate (pH 9.5). The chromatography was carried out using a gradient from 10–45% eluent B over 30 min at a flow rate 200 μl/min. Forty-eight fractions were collected at 1-min intervals and pooled into a total of 10 fractions based on the absorbance at 214 nm and with a multiple fraction concatenation strategy (49). 10% of each fraction was pooled, dried, and reconstituted in 120 μl of 2% CAN, 0.5% formic acid (FA) for low pH nanoLC-MS/MS analysis. The remaining 90% of each sample was pooled into five fractions, dried, and subjected to TiO2 enrichment.
TiO2 enrichment was conducted using a TiO2 Mag-Sepharose kit (GE Healthcare). The TMT 6-plex tagged tryptic peptides were reconstituted in 400 μl of binding buffer (1 m glycolic acid in 80% acetonitrile, 5% TFA). The TiO2 slurry (75 μl) was used and incubated with the sample for 30 min at 1800 rpm vortex. After washing the beads with washing buffer (80% acetonitrile, 1% TFA), the phosphopeptides were eluted with 100 μl of elution buffer (5% ammonium hydroxide) twice. The eluted fraction was dried and reconstituted in 25 μl of 0.5% FA for subsequent nano scale LC-MS/MS analysis.
The LC-MS/MS analysis was carried out using an Orbitrap Fusion (Thermo Fisher Scientific) mass spectrometer equipped with a nanospray Flex Ion Source similar to previous reports (51, 52). The mass spectrometer was coupled to an UltiMate3000 RSLCnano HPLC (Thermo Scientific). A portion of each reconstituted fraction (2–3 μl for global proteomics fractions and 5–8 μl for enriched phosphor fractions) was injected onto a PepMap C-18 RP nano-trap column (100-μm × 20-mm dimensions), 3-μm particle size; Dionex) at 20 μl/min flow rate for concentration and on-line desalting. The bound peptides were then separated on a PepMap C-18 RP column (75 μm × 25 cm, 3.5-μm particle size) by elution at a flow rate of 300 nl/min. using a gradient of 5% to 35% ACN in 0.1% formic acid. The column was re-equilibrated with 5% CAN, 0.1% FA for 25 min prior to the next run.
The Orbitrap Fusion was operated in positive ion mode with the nano spray voltage set at 1.6 kV and the source temperature at 275 °C. The Fourier transform, ion-trap (IT), and quadrupole mass analyzers were externally calibrated. The mass spectrometer was operated in the data-dependent acquisition mode. The Fourier transform mass analyzer was used for survey scans to select precursor ions. These were followed by 3-s top speed data-dependent higher-energy collision dissociation MS/MS scans using a normalized collision energy of 37.5% for all precursor ions with at least two but no more than seven charges per ion. The threshold ion count was >10,000. MS survey scans were conducted at a resolving power of 120,000 full-width-at-half-maximum (measured at m/z 200) for all the masses in the range of m/z 400–1600 using the settings AGC = 3e5 and Max IT = 50 ms. The MS/MS scans were conducted using higher-energy collision dissociation at 60,000 full-width-at-half-maximum resolution for the mass range m/z 105–2000. The AGC setting, Max IT, and Q isolation window were set to 1e5, 120 ms, and ±1.6 Da, respectively. The dynamic exclusion parameters were set at 1 with a 45-s exclusion duration with a ±10 ppm exclusion mass width. All data were acquired using the Xcalibur 3.0 operation software and Orbitrap Fusion Tune 2.0 (Thermo Fisher Scientific).
Proteomics data processing and protein identification
All MS and MS/MS raw spectra were processed and searched using Proteome Discoverer 2.2 (Thermo) with Sequest HT. The Pseudomonas_putidaKT2440.fasta database containing 5528 entries (downloaded June 12, 2017) was used to query the MS data. The default search settings used for 10-plex TMT quantitative processing and protein identification were as follows: tryptic proteolysis, allowing two missed cleavages, with fixed carbamidomethyl modification of cysteine, fixed 10-plex TMT modifications of lysine and N-terminal amines and variable modifications of methionine oxidation and deamidation of asparagine/glutamine residues. The peptide and fragment mass tolerance values were 10 ppm and 50 mDa, respectively. Identified peptides were filtered for a maximum 1% false discovery rate using the Benjamini–Hochberg procedure (53) incorporated in the Percolator algorithm in PD 2.2, and the peptide confidence was set to high. The TMT10-plex quantification method within the PD 2.2 software was used to calculate the reporter ratios with mass tolerance ±10 ppm without applying the isotopic correction factors. Only peptide spectra containing all reporter ions were designated as quantifiable and used for peptide/protein quantitation. Summed intensity normalization was used to estimate expression ratios. To minimize the ratio distortion expected because of the co-isolation of near isobaric precursor ions, a co-isolation filter of 50% was applied.
Statistical analysis
For comparisons of data from growth and metabolomics experiments at the two growth conditions (glucose:benzoate mixture versus glucose alone), statistical analysis was done using unpaired two-tailed t test. Statistically significant difference was determined at p ≤ 0.05.
To estimate the thresholds of significant change from the proteomics data, the expression ratios from each of the data sets were log2 transformed and fit to 60 different statistical distributions using the program EasyFit (MathWave Technologies). Goodness of fit was determined by the Kalmogrov/Smirnov, Anderson Darling, and χ2 tests (54–56). In each case the best fit was determined to be to the four-parameter Dagum distribution (57). The 95% confidence intervals were estimated by generating distributions randomly using the exact parameters of the experimentally derived distributions. A two-tailed heteroscedastic t test was conducted to evaluate the difference between the means of the samples being compared. An expression ratio is considered to have changed significantly if the magnitude of the change is greater than what would be expected to be observed by chance with a probability of 0.05 and if its associated p value was < 0.01.
Author contributions
M. A. K. and L. A. conceptualization; M. A. K., C. M. M., M. S., and T. W. T. data curation; M. A. K. investigation; M. A. K., C. M. M., M. S., and R. A. W. methodology; M. A. K., C. M. M., and L. A. writing-original draft; M. A. K., C. M. M., M. S., R.A.W., T. W. T., and L. A. writing-review and editing; C. M. M., M. S., and T. W. T. formal analysis; L. A. resources; L. A. supervision; L. A. funding acquisition; L. A. project administration.
Supplementary Material
Acknowledgments
We thank Sheng Zhang of the Cornell Proteomics facility for preliminary curation of the proteomics data. We thank James Shapleigh and Tara Fish for helpful discussions in the preparation of the protein extractions and in the consideration of redox balance in carbon metabolism, respectively.
This work was supported by an Integrative Graduate Education and Research Traineeship Fellowship from the National Science Foundation (to M. A. K.), graduate fellowships from Cornell University (to M. S. and R. A. W.), and a start-up package from Cornell University. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4 and Figs. S1–S6.
- ED
- Entner–Doudoroff
- PP
- pentose–phosphate
- EMP
- Embden–Meyerof–Parnas
- G6P
- glucose-6-phosphate
- Glucn
- gluconate
- 6P-Glucn
- 6-phosphogluconate
- ICD
- isocitrate dehydrogenase
- OAA
- oxaloacetate
- IMP
- inosine monophosphate
- UMP
- uridine monophosphate
- UDP-glucose
- uridine diphosphate glucose
- glycerol-3P
- glycerol-3-phosphate
- PEP
- phosphoenolpyruvate
- R5P
- ribose-5-phosphate
- 3-PG
- 3-phosphoglycerate
- DHAP
- dihydroxy-acetone-phosphate
- GAP
- glyceraldehyde-3-phosphate
- FBP
- fructose-1,6-bisphosphate
- F6P
- fructose-6-phosate
- Xu5P
- xylulose-5-phosphate
- S7P
- sedoheptulose-7-phosphate
- PPSA
- PEP synthase
- SDH
- succinate dehydrogenase
- MQO
- malate:quinone oxidoreductase
- TEAB
- triethylammonium bicarbonate
- ACN
- acetonitrile
- FA
- formic acid
- IT
- ion-trap
- α-KG
- α-ketogluconate.
References
- 1. Poblete-Castro I., Becker J., Dohnt K., dos Santos V. M., and Wittmann C. (2012) Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 93, 2279–2290 10.1007/s00253-012-3928-0 [DOI] [PubMed] [Google Scholar]
- 2. Vardon D. R., Franden M. A., Johnson C. W., Karp E. M., Guarnieri M. T., Linger J. G., Salm M. J., Strathmann T. J., and Beckham G. T. (2015) Adipic acid production from lignin. Energy Environ. Sci. 8, 617–628 10.1039/C4EE03230F [DOI] [Google Scholar]
- 3. Belda E., van Heck R. G., José Lopez-Sanchez M., Cruveiller S., Barbe V., Fraser C., Klenk H.-P., Petersen J., Morgat A., Nikel P. I., Vallenet D., Rouy Z., Sekowska A., Martins dos Santos V. A., de Lorenzo V., et al. (2016) The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 18, 3403–3424 10.1111/1462-2920.13230 [DOI] [PubMed] [Google Scholar]
- 4. Johnson C. W., Salvachúa D., Khanna P., Smith H., Peterson D. J., and Beckham G. T. (2016) Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 3, 111–119 10.1016/j.meteno.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palazzolo M. A., and Kurina-Sanz M. (2016) Microbial utilization of lignin: available biotechnologies for its degradation and valorization. World J. Microbiol. Biotechnol. 32, 173 10.1007/s11274-016-2128-y [DOI] [PubMed] [Google Scholar]
- 6. Bugg T. D., Ahmad M., Hardiman E. M., and Singh R. (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400 10.1016/j.copbio.2010.10.009 [DOI] [PubMed] [Google Scholar]
- 7. Fang W., Fang Z., Zhou P., Chang F., Hong Y., Zhang X., Peng H., and Xiao Y. (2012) Evidence for lignin oxidation by the giant panda fecal microbiome. PLoS One 7, e50312 10.1371/journal.pone.0050312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., and Timmis K. N. (1987) Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science 238, 1395–1398 10.1126/science.3479842 [DOI] [PubMed] [Google Scholar]
- 9. George K. W., and Hay A. (2012) Less is more: reduced catechol production permits Pseudomonas putida F1 to grow on styrene. Microbiology 158, 2781–2788 10.1099/mic.0.058230-0 [DOI] [PubMed] [Google Scholar]
- 10. Johnson G. R., and Spain J. C. (2003) Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62, 110–123 10.1007/s00253-003-1341-4 [DOI] [PubMed] [Google Scholar]
- 11. Akkaya Ö., Pérez-Pantoja D. R., Calles B., Nikel P. I., and de Lorenzo V. (2018) The metabolic redox regime of Pseudomonas putida tunes its evolvability toward novel xenobiotic substrates. MBio, 9, e01512–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J., Pérez-Pantoja D., Silva-Rocha R., Oliveros J. C., and de Lorenzo V. (2016) High-resolution analysis of the m-xylene/toluene biodegradation subtranscriptome of Pseudomonas putida mt-2. Environ. Microbiol. 18, 3327–3341 10.1111/1462-2920.13054 [DOI] [PubMed] [Google Scholar]
- 13. Worsey M. J., and Williams P. A. (1975) Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sudarsan S., Dethlefsen S., Blank L. M., Siemann-Herzberg M., and Schmid A. (2014) The functional structure of central carbon metabolism in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 80, 5292–5303 10.1128/AEM.01643-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikel P. I., Chavarría M., Fuhrer T., Sauer U., and de Lorenzo V. (2015) Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, embden-meyerhof-parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932 10.1074/jbc.M115.687749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sasnow S. S., Wei H., and Aristilde L. (2016) Bypasses in intracellular glucose metabolism in iron-limited Pseudomonas putida. Microbiologyopen 5, 3–20 10.1002/mbo3.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudarsan S., Blank L. M., Dietrich A., Vielhauer O., Takors R., Schmid A., and Reuss M. (2016) Dynamics of benzoate metabolism in Pseudomonas putida KT2440. Metab. Eng. Commun. 3, 97–110 10.1016/j.meteno.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. del Castillo T., and Ramos J. L. (2007) Simultaneous catabolite repression between glucose and toluene metabolism in Pseudomonas putida is channeled through different signaling pathways. J. Bacteriol. 189, 6602–6610 10.1128/JB.00679-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Duuren J. B., Wijte D., Karge B., dos Santos V. A., Yang Y., Mars A. E., and Eggink G. (2012) pH-stat fed-batch process to enhance the production of cis, cis-muconate from benzoate by Pseudomonas putida KT2440-JD1. Biotechnol. Prog. 28, 85–92 10.1002/btpr.709 [DOI] [PubMed] [Google Scholar]
- 20. Basu A., Apte S. K., and Phale P. S. (2006) Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl. Environ. Microbiol. 72, 2226–2230 10.1128/AEM.72.3.2226-2230.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burlage R. S., Hooper S. W., and Sayler G. S. (1989) The TOL (pWWO) catabolic plasmid. Appl. Environ. Microbiol. 55, 1323–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silva-Rocha R., Pérez-Pantoja D., and de Lorenzo V. (2013) Decoding the genetic networks of environmental bacteria: regulatory moonlighting of the TOL system of Pseudomonas putida mt-2. ISME J. 7, 229–232 10.1038/ismej.2012.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. La Rosa R., Nogales J., and Rojo F. (2015) The Crc/CrcZ-CrcY global regulatory system helps the integration of gluconeogenic and glycolytic metabolism in Pseudomonas putida. Environ. Microbiol. 17, 3362–3378 10.1111/1462-2920.12812 [DOI] [PubMed] [Google Scholar]
- 24. Nikel P. I., Kim J., and de Lorenzo V. (2014) Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ. Microbiol. 16, 239–254 10.1111/1462-2920.12224 [DOI] [PubMed] [Google Scholar]
- 25. van Duuren J. B., Puchałka J., Mars A. E., Bücker R., Eggink G., Wittmann C., Dos Santos V. A. (2013) Reconciling in vivo and in silico key biological parameters of Pseudomonas putida KT2440 during growth on glucose under carbon-limited condition. BMC Biotechnol. 13, 93 10.1186/1472-6750-13-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson K. E., Weinel C., Paulsen I. T., Dodson R. J., Hilbert H., Martins dos Santos V. A., Fouts D. E., Gill S. R., Pop M., Holmes M., Brinkac L., Beanan M., DeBoy R. T., Daugherty S., Kolonay J., et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808 10.1046/j.1462-2920.2002.00366.x [DOI] [PubMed] [Google Scholar]
- 27. Kohlstedt M., and Wittmann C. (2019) GC-MS-based 13C metabolic flux analysis resolves the parallel and cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. Metab. Eng. 54, 35–53 10.1016/j.ymben.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 28. Wilkes R. A., Mendonca C. M., and Aristilde L. (2018) A cyclic metabolic network in Pseudomonas protegens Pf-5 prioritizes the Entner–Doudoroff pathway and exhibits substrate hierarchy during carbohydrate co-utilization. Appl. Environ. Microbiol. 85, e02084–18 10.1128/AEM.02084-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruiz-Manzano A., Yuste L., and Rojo F. (2005) Levels and activity of the Pseudomonas putida global regulatory protein Crc vary according to growth conditions. J. Bacteriol. 187, 3678–3686 10.1128/JB.187.11.3678-3686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LaPorte D. C. (1993) The isocitrate dehydrogenase phosphorylation cycle: regulation and enzymology. J. Cell. Biochem. 51, 14–18 10.1002/jcb.240510104 [DOI] [PubMed] [Google Scholar]
- 31. Smith C. M., Bryla J., and Williamson J. R. (1974) Regulation of mitochondrial α-ketoglutarate metabolism by product inhibition at α-ketoglutarate dehydrogenase. J. Biol. Chem. 249, 1497–1505 [PubMed] [Google Scholar]
- 32. Tretter L., and Adam-Vizi V. (2005) α-Ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 360, 2335–2345 10.1098/rstb.2005.1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao B., and Loh K.-C. (2008) Catabolic pathways and cellular responses of Pseudomonas putida P8 during growth on benzoate with a proteomics approach. Biotechnol. Bioeng. 101, 1297–1312 10.1002/bit.21997 [DOI] [PubMed] [Google Scholar]
- 34. Walsh K., and Koshland D. E. (1985) Branch point control by the phosphorylation state of isocitrate dehydrogenase: a quantitative examination of fluxes during a regulatory transition. J. Biol. Chem. 260, 8430–8437 [PubMed] [Google Scholar]
- 35. Park J. O., Rubin S. A., Xu Y.-F., Amador-Noguez D., Fan J., Shlomi T., and Rabinowitz J. D. (2016) Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 12, 482–489 10.1038/nchembio.2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lassek C., Berger A., Zühlke D., Wittmann C., and Riedel K. (2016) Proteome and carbon flux analysis of Pseudomonas aeruginosa clinical isolates from different infection sites. Proteomics 16, 1381–1385 10.1002/pmic.201500228 [DOI] [PubMed] [Google Scholar]
- 37. Hester K. L., Lehman J., Najar F., Song L., Roe B. A., MacGregor C. H., Hager P. W., Phibbs P. V. Jr., Sokatch J. R., and Sokatch J. R. (2000) Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182, 1144–1149 10.1128/JB.182.4.1144-1149.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moreno R., Marzi S., Romby P., and Rojo F. (2009) The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 37, 7678–7690 10.1093/nar/gkp825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hackett S. R., Zanotelli V. R., Xu W., Goya J., Park J. O., Perlman D. H., Gibney P. A., Botstein D., Storey J. D., and Rabinowitz J. D. (2016) Systems-level analysis of mechanisms regulating yeast metabolic flux. Science 354, aaf2786 10.1126/science.aaf2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., and Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 10.1038/nchembio.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tepper N., Noor E., Amador-Noguez D., Haraldsdóttir H. S., Milo R., Rabinowitz J., Liebermeister W., and Shlomi T. (2013) Steady-state metabolite concentrations reflect a balance between maximizing enzyme efficiency and minimizing total metabolite load. PLoS One 8, e75370 10.1371/journal.pone.0075370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Görke B., and Stülke J. (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 43. Dvorák P., and de Lorenzo V. (2018) Refactoring the upper sugar metabolism of Pseudomonas putida for co-utilization of cellobiose, xylose, and glucose. Metab. Eng. 48, 94–108 10.1016/j.ymben.2018.05.019 [DOI] [PubMed] [Google Scholar]
- 44. Aristilde L., Reed M. L., Wilkes R. A., Youngster T., Kukurugya M. A., Katz V., and Sasaki C. R. S. (2017) Glyphosate-induced specific and widespread perturbations in the metabolome of soil Pseudomonas species. Front. Environ. Sci. 5, 1–13 [Google Scholar]
- 45. Aristilde L., Lewis I. A., Park J. O., and Rabinowitz J. D. (2015) Hierarchy in pentose sugar metabolism in Clostridium acetobutylicum. Appl. Environ. Microbiol. 81, 1452–1462 10.1128/AEM.03199-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Melamud E., Vastag L., and Rabinowitz J. D. (2010) Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 10.1021/ac1021166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weitzel M., Nöh K., Dalman T., Niedenführ S., Stute B., and Wiechert W. (2013) 13CFLUX2: high-performance software suite for 13C-metabolic flux analysis. Bioinformatics 29, 143–145 10.1093/bioinformatics/bts646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zamboni N., Fendt S.-M., Rühl M., and Sauer U. (2009) 13C-Based metabolic flux analysis. Nat. Protoc. 4, 878–892 10.1038/nprot.2009.58 [DOI] [PubMed] [Google Scholar]
- 49. Yang Y., Qiang X., Owsiany K., Zhang S., Thannhauser T. W., and Li L. (2011) Evaluation of different multidimensional LC–MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J. Proteome Res. 10, 4647–4660 10.1021/pr200455s [DOI] [PubMed] [Google Scholar]
- 50. Chen J.-W., Scaria J., Mao C., Sobral B., Zhang S., Lawley T., and Chang Y.-F. (2013) Proteomic comparison of historic and recently emerged hypervirulent Clostridium difficile strains. J. Proteome Res. 12, 1151–1161 10.1021/pr3007528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thomas C. J., Cleland T. P., Zhang S., Gundberg C. M., and Vashishth D. (2017) Identification and characterization of glycation adducts on osteocalcin. Anal. Biochem. 525, 46–53 10.1016/j.ab.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang S., Li X., Liu X., Ding X., Xin X., Jin C., Zhang S., Li G., and Guo H. (2018) Parallel comparative proteomics and phosphoproteomics reveal that cattle myostatin regulates phosphorylation of key enzymes in glycogen metabolism and glycolysis pathway. Oncotarget 9, 11352–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 [Google Scholar]
- 54. Stephens M. A. (1974) EDF statistics for goodness of fit and some comparisons. J. Am. Stat. Assoc. 69, 730 10.1080/01621459.1974.10480196 [DOI] [Google Scholar]
- 55. Snedecor G. W., and Cochran W. G. (1989) Statistical Methods, 8th Ed., Iowa State University Press, Ames, IA [Google Scholar]
- 56. Chakravarti I. M., Laha R. G., and Roy J. (1967) Handbook of Methods of Applied Statistics, 69th Ed., John Wiley and Sons [Google Scholar]
- 57. Kleiber C. (2008) A Guide to the Dagum Distributions. In Modeling Income Distributions and Lorenz Curves, pp. 97–117, Springer; New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.