Abstract
DNA damage response Ser/Thr kinases, including ataxia telangiectasia-mutated (ATM) and Rad3-related (ATR), control cell cycle progression, DNA repair, and apoptosis. ATR is activated by ETAA1 activator of ATR kinase (ETAA1) or DNA topoisomerase II binding protein 1 (TOPBP1). Both ETAA1 and TOPBP1 contain experimentally defined ATR activation domains (AADs) that are mostly unstructured and have minimal sequence similarity. A tryptophan residue in both AADs is required for ATR activation, but the other features of these domains and the mechanism by which they activate ATR are unknown. In this study, using bioinformatic analyses, kinase assays, co-immunoprecipitation, and immunofluorescence measures of signaling, we more specifically defined the TOPBP1 and ETAA1 AADs and identified additional features of the AADs needed for ATR activation. We found that both ETAA1 and TOPBP1 contain a predicted coiled-coil motif that is required for ATR activation in vitro and in cells. Mutation of the predicted coiled coils does not alter AAD oligomerization but does impair binding of the AADs to ATR. These results suggest that TOPBP1 and ETAA1 activate ATR using similar motifs and mechanisms.
Keywords: DNA damage response, cell cycle, serine/threonine protein kinase, DNA replication, signaling, cancer, ATR, ATR activation domain (AAD), DNA topoisomerase II binding protein 1 (TOPBP1), ETAA1 activator of ATR kinase (ETAA1), PI3K-related protein kinase (PIKK)
Introduction
Cells are constantly exposed to exogenous and endogenous sources of DNA damage that cause thousands of DNA lesions in each cell every day (1). These lesions can interfere with processes such as DNA replication and transcription and must be accurately repaired to preserve genome stability (2). The recognition and repair of DNA lesions is coordinated by a series of signaling pathways collectively known as the DNA damage response (DDR).3 The DDR also regulates cell cycle progression, cellular senescence, and apoptosis (1). The PI3K-related protein kinases ataxia telangiectasia-mutated (ATM), ATM and Rad3-related (ATR), and DNA-PK activate the DDR (1, 3). ATM and DNA-PK signaling is initiated primarily in response to DNA double-strand breaks, whereas ATR is activated at lesions containing replication protein A (RPA)–coated ssDNA (3, 4).
ATR is recruited to sites of RPA-coated ssDNA through its obligate binding partner ATRIP, which directly binds RPA (5, 6). However, recruitment to RPA-coated ssDNA alone is not sufficient to trigger ATR activation. Several other proteins, including at least one ATR-activating protein, must also be recruited to trigger maximal ATR signaling. One ATR-activating protein is TOPBP1 (7). TOPBP1 is recruited to DNA lesions that possess a ssDNA–dsDNA 5′ junction through its interaction with the MRE11–RAD50–NBS1 complex and the phosphorylated C-terminal tail of RAD9, a component of the RAD9–RAD1–HUS1 (9-1-1) checkpoint clamp (8–10). TOPBP1 also interacts with RAD9–RAD1–HUS1–interacting nuclear orphan (RHINO) at DNA damage sites (11, 12). All of these interactions are required for TOPBP1 to fully stimulate ATR kinase activity, which is specifically mediated by the TOPBP1 ATR activation domain (AAD) (7).
The other known ATR activator is ETAA1. Unlike TOPBP1, ETAA1 is recruited to RPA-coated ssDNA through a direct interaction with RPA, where it then activates ATR through its AAD (13–15). ETAA1 is especially important for ATR activation to regulate cell cycle transitions even in the absence of DNA damage or replication stress (16, 17).
The ATR and ATRIP orthologues in Saccharomyces cerevisiae are Mec1 and Ddc2, respectively. Like ATR–ATRIP, Mec1–Ddc2 localizes to DNA lesions containing RPA-coated ssDNA via a direct interaction between RPA and Ddc2 (18). Mec1 kinase activity can then be stimulated by Dpb11 (TOPBP1 ortholog), Ddc1 (human RAD9 ortholog), or the multifunctional nuclease/helicase Dna2 (19–22). The biochemical mechanism by which these proteins activate Mec1 appears to be the same. Each Mec1 activation domain (MAD) is predicted to be intrinsically disordered and contains two essential aromatic amino acids that, when mutated, abrogate Mec1 activation both in vitro and in vivo (21–23). Additionally, it has been proposed that the Mec1 activators bind Mec1 via a two-step binding mechanism whereby the two aromatic residues bind first, followed by binding of the surrounding residues to stimulate full kinase activation (24).
In contrast to the Mec1 activators, how TOPBP1 and ETAA1 activate ATR is less well understood. The AAD structures have not been solved, and the AAD primary sequences contain no similarities except for a few amino acids that flank a conserved tryptophan (13). Mutation of this tryptophan in both TOPBP1 and ETAA1 prevents ATR activation (7, 13–15). However, whether a second critical aromatic residue or any other motifs in these domains are required for ATR activation is unknown. To better determine how ETAA1 and TOPBP1 activate ATR, we more specifically defined the ETAA1 and TOPBP1 AADs. We found that each of the AADs contains a predicted coiled-coil motif that is required for ATR activation in vitro and in cells. Our data indicate that the predicted coiled coils are required for ATR activation because these motifs bind the ATR–ATRIP complex.
Results
Identification of the ETAA1 ATR activation domain
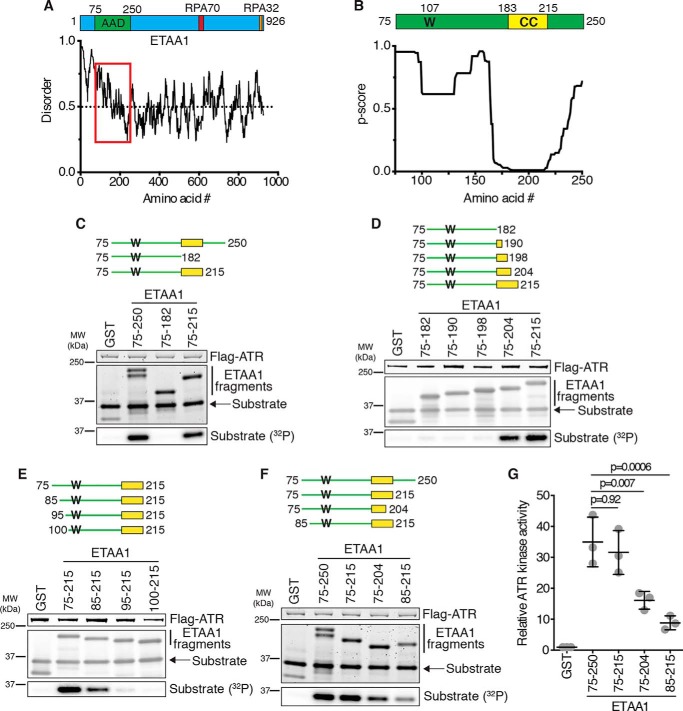
ETAA1 is a 926-amino acid protein that contains two RPA interaction motifs and an AAD (13–15). However, it contains no other defined domains, and several regions within the protein are predicted to be unstructured (Fig. 1A). To better understand how ETAA1 activates ATR, we sought to more specifically define the AAD. We previously identified the ETAA1 AAD to be residues 75–250, as this fragment stimulates robust ATR kinase activity in vitro (13). Like the yeast MADs, the ETAA1 AAD is predicted to be intrinsically disordered without a clear secondary structure except for a predicted coiled-coil motif spanning residues 183–215 (p score = 0.01) (Fig. 1, A and B). To determine whether this predicted coiled coil is important for ATR activation, we incubated purified ATR–ATRIP with recombinant GST-ETAA1 AADs containing or lacking the predicted coiled coil, an ATR substrate (an MCM2 fragment containing Ser-108 (25)), and [γ-32P]ATP and measured substrate phosphorylation. ETAA1 75–250 and 75–215, which contain the predicted coiled coil, stimulate ATR kinase activity whereas ETAA1 75–182, which lacks the predicted coiled coil, does not (Fig. 1C). ETAA1 75–204 has partial activity, but removal of six more residues eliminates virtually all activation (Fig. 1, D and G). We also tested ETAA1 AADs that had amino acids deleted from the N terminus. Compared with ETAA1 75–215, ETAA1 85–215 activates ATR less efficiently, ETAA1 95–215 activates ATR very little, and ETAA1 100–215 does not activate ATR at all (Fig. 1E). A direct comparison of ETAA1 75–250, 75–215, 75–204, and 85–215 revealed that 75–250 and 75–215 activate ATR to a similar extent and more robustly than 75–204 and 85–215 (Fig. 1, F and G). Therefore, we conclude that, in addition to the critical tryptophan (Trp-107), ETAA1 contains a predicted coiled-coil motif that is required for stimulation of maximal ATR kinase activity and that the minimal AAD that retains complete activity contains residues 75–215.
Figure 1.
Identification of the ETAA1 ATR activation domain. A, schematic of ETAA1 with the AAD and RPA interaction motifs indicated. ETAA1 predicted disorder was calculated using IUPred2A (31). The AAD region is indicated by the red box. B, schematic of the ETAA1 AAD, with the critical tryptophan (W) and predicted coiled coil (CC) indicated. Predicted coiled coil per residue scores were calculated using Paircoil2 (29). C–F, Purified ATR–ATRIP complexes were incubated with GST or the indicated GST-ETAA1 AADs, an ATR substrate, and [γ-32P]ATP. Reaction products were separated by SDS-PAGE and detected by Coomassie staining and immunoblotting. Substrate phosphorylation was detected by autoradiography. MW, molecular weight. G, quantification of three experiments as shown in F. Statistical significance was calculated using a one-way analysis of variance and Tukey's multiple comparisons test. Error bars are mean ± S.D.
Identification of the TOPBP1 ATR activation domain
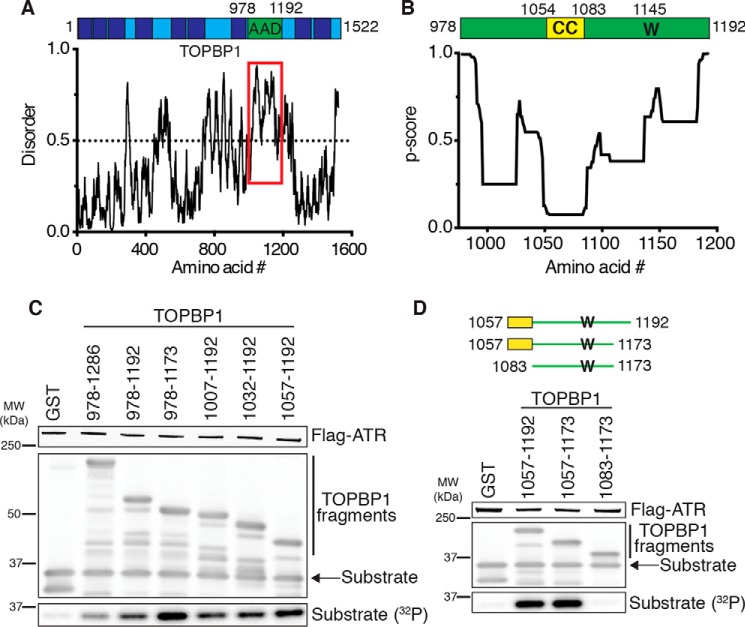
TOPBP1 is a multiple BRCT domain–containing protein with an AAD located between BRCT domains 6 and 7 composed of residues 978–1192 (7). Like the ETAA1 AAD and yeast MADs, the TOPBP1 AAD is predicted to be mostly unstructured (Fig. 2A). Because the ETAA1 AAD contains a predicted coiled coil that is required for ATR activation, we hypothesized that the TOPBP1 AAD may contain a similar motif. Indeed, a secondary structure prediction algorithm identified a possible coiled coil in the TOPBP1 AAD from residues 1054–1083 but with a more modest p score of 0.074 (Fig. 2B). Therefore, we assessed whether this motif was required for TOPBP1-dependent ATR activation. GST-TOPBP1 fragments containing the predicted coiled coil as small as amino acids 1057–1173 stimulate ATR kinase activity (Fig. 2, C and D). However, TOPBP1 1083–1173, which lacks the predicted coiled coil, does not activate ATR (Fig. 2D). Thus, like ETAA1, TOPPB1 contains a critical tryptophan (Trp-1145) and a predicted coiled coil within an otherwise disordered domain that is required for full stimulation of ATR kinase activity.
Figure 2.
Identification of the TOPBP1 ATR activation domain. A, schematic of TOPBP1 with the AAD indicated. BRCT domains are shown as dark blue boxes. TOPBP1 predicted disorder was calculated using IUPred2A (31). The AAD region is indicated by the red box. B, schematic of the TOPBP1 AAD, with the critical tryptophan (W) and predicted coiled coil (CC) indicated. Predicted coiled coil per residue scores were calculated using Paircoil2 (29). C and D, purified ATR–ATRIP complexes were incubated with GST or the indicated GST-TOPBP1 AADs, an ATR substrate, and [γ-32P]ATP. Reaction products were separated by SDS-PAGE and detected by Coomassie staining and immunoblotting. Substrate phosphorylation was detected by autoradiography. MW, molecular weight.
A single point mutation in the predicted coiled coils disrupts ATR activation
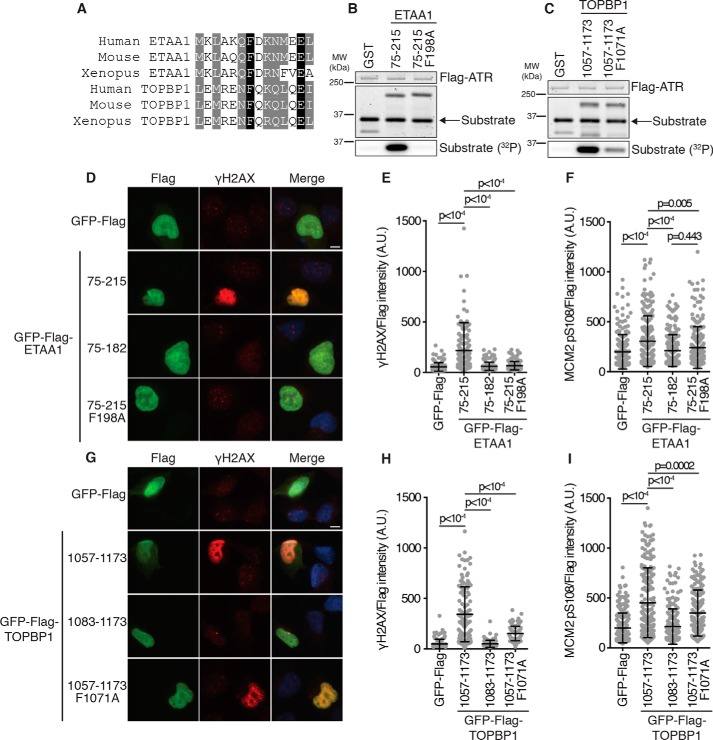
A sequence alignment of the ETAA1 and TOPBP1 predicted coiled-coil motifs reveals some sequence similarity (Fig. 3A). Both contain a conserved phenylalanine (Phe-198 in ETAA1 and Phe-1071 in TOPBP1), which we hypothesized could be a second critical aromatic residue in each AAD required for ATR activation, similar to the requirement in the Mec1 activators. Mutation of ETAA1 Phe-198 to alanine completely abolishes ATR activation (Fig. 3B). Mutation of TOPBP1 Phe-1071 to alanine also reduces ATR activation, although not to the extent observed for ETAA1 (Fig. 3C).
Figure 3.
Mutation of a conserved phenylalanine disrupts ETAA1- and TOPBP1-dependent ATR activation. A, sequence alignment of ETAA1 and TOPBP1 predicted coiled coil residues flanking ETAA1 Phe-198 and TOPBP1 Phe-1071. B and C, purified ATR–ATRIP complexes were incubated with GST or the indicated GST-ETAA1 or GST-TOPBP1 AADs, an ATR substrate, and [γ-32P]ATP. Reaction products were separated by SDS-PAGE and detected by Coomassie staining. Substrate phosphorylation was detected by autoradiography. D–I, empty vector or the indicated ETAA1 or TOPBP1 AAD proteins were expressed in U2OS cells. γH2AX (D, E, G, and H) or MCM2 pSer-108 (F and I) was visualized in FLAG-positive nuclei. Scale bars = 10 μm. A.U., arbitrary units. γH2AX and MCM2 pSer-108 intensity is normalized to the FLAG expression level. Statistical significance was calculated using a one-way analysis of variance and Tukey's multiple comparisons test. Error bars are mean ± S.D.
Overexpression of either the TOPBP1 or ETAA1 AAD in cells results in ATR activation, which can be assessed by measuring histone variant H2AX phosphorylation at Ser-139 (γH2AX) (13, 14, 18). We expressed the full-length AADs, AADs lacking the predicted coiled coils, or AADs with the phenylalanine mutations in U2OS cells and measured γH2AX induction by immunofluorescence. Expression of ETAA1 75–215 induced a large increase in γH2AX whereas ETAA1 75–182 and ETAA1 75–215 F198A expression did not (Fig. 3, D and E). We also assessed MCM2 Ser-108 phosphorylation as a second measure of ATR activation and observed that expression of ETAA1 75–215, but not ETAA1 75–182 or ETAA1 75–215 F198A, causes a modest increase in MCM2 Ser-108 phosphorylation (Fig. 3F).
Next we measured γH2AX induction and MCM2 Ser-108 phosphorylation in cells expressing our newly defined TOPBP1 AADs. Expression of TOPBP1 1057–1173 causes a large increase in γH2AX, whereas expression of TOPBP1 1083–1173, which lacks the predicted coiled coil, results in no increase in γH2AX (Fig. 3, G and H). Expression of TOPBP1 1057–1173 F1071A causes moderate γH2AX induction (Fig. 3, G and H), consistent with the result of the in vitro kinase assay, where the F1071A mutation reduces, but does not abolish, ATR activation by TOPBP1 (Fig. 3C). Expression of TOPBP1 1057–1173 also causes a modest increase in MCM2 Ser-108 phosphorylation whereas expression of TOPBP1 1083–1173 does not. Expression of TOPBP1 1057–1173 F1071A, as for γH2AX, causes an intermediate level of MCM2 Ser-108 phosphorylation (Fig. 3I). Taken together, these results indicate that the ETAA1 and TOPBP1 predicted coiled coils are required for efficient ATR activation in cells.
The coiled-coil point mutations do not alter AAD oligomerization
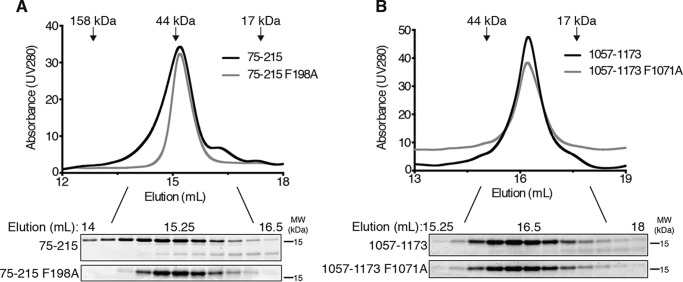
Coiled coils often function as oligomerization domains (26). To determine whether mutation of the predicted coiled coils altered AAD oligomerization, we compared the elution profiles of ETAA1 75–215 and ETAA1 75–215 F198A on a size exclusion column. Both ETAA1 75–215 and ETAA1 75–215 F198A elute at a retention volume of 15.2 ml, indicating that the F198A mutation does not disrupt function by preventing homo-oligomerization (Fig. 4A). Both the WT and mutant proteins elute at a position that does not match their predicted molecular mass (∼38.6 kDa compared with the predicted 16.3 kDa). However, because the ETAA1 AAD is predicted to be intrinsically disordered (Fig. 1A), it is unlikely to be globular, likely explaining this difference.
Figure 4.
Mutation of the AADs does not alter AAD oligomerization. A and B, the indicated ETAA1 (A) or TOPBP1 (B) AADs were eluted from a SuperdexTM 200 Increase 10/300 GL column while measuring UV absorbance at 280 nm. Equal amounts of fractions corresponding to peaks were separated by SDS-PAGE and visualized by Coomassie staining. The elution volume of molecular mass standards is indicated. MW, molecular weight.
We also assessed TOPBP1 AAD oligomerization using gel filtration chromatography. TOPBP1 1057–1173 elutes at a retention volume of 16.23 ml, whereas TOPBP1 1057–1173 F1071A elutes at a retention volume of 16.2 ml (Fig. 4B). These retention volumes correspond to molecular masses of 21.8 kDa and 22.2 kDa, respectively, compared with the predicted molecular masses of 13.4 kDa and 13.3 kDa. These results indicate that the F1071A mutation does not alter TOPBP1 AAD oligomerization despite impairing ATR activation.
The predicted coiled coils promote the AAD–ATR interaction
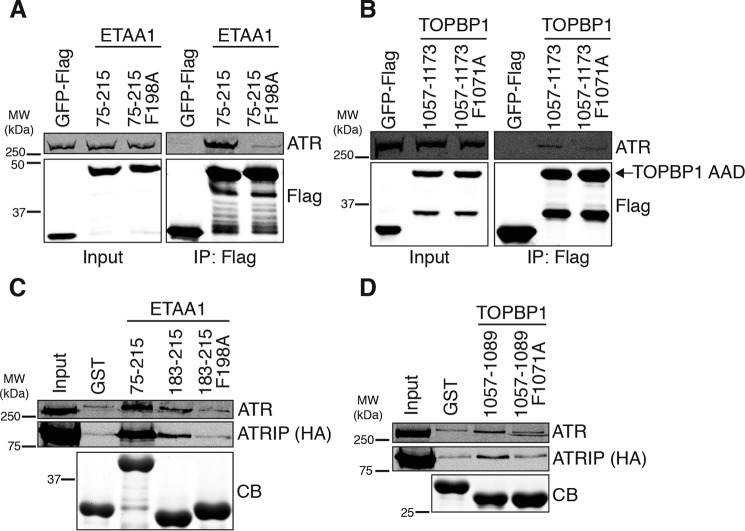
Because mutation of the predicted coiled coils had no apparent effect on AAD oligomerization, we next tested how these mutations affect the AAD–ATR interactions. The F198A mutation almost completely abolishes the ETAA1 AAD–ATR interaction, as measured by co-immunoprecipitation (Fig. 5A). Similarly, the F1071A mutation diminished the TOPBP1 AAD–ATR interaction (Fig. 5B).
Figure 5.
The predicted coiled coils promote the ETAA1 and TOPBP1 AAD–ATR interaction. A and B, empty vector or the indicated ETAA1 (A) or TOPBP1 (B) AADs were immunoprecipitated (IP) from HEK293T cell nuclear extracts. Immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting. MW, molecular weight. C and D, purified GST or recombinant GST-ETAA1 (C) or GST-TOPBP1 (D) proteins bound to GSH beads were incubated with HEK293T cell nuclear extracts expressing FLAG-ATR and HA-ATRIP. Bound proteins were eluted, separated by SDS-PAGE, and detected by Coomassie staining (CB) or immunoblotting.
Given that mutation of the predicted coiled coils disrupts the ETAA1 and TOPBP1 AAD–ATR interactions, we considered the possibility that the predicted coiled coils might directly contact ATR and/or ATRIP. To test this hypothesis, we purified the full-length ETAA1 AAD, the ETAA1 predicted coiled coil, and the ETAA1 predicted coiled coil containing the F198A mutation as recombinant GST fusion proteins, bound the proteins to GSH beads, and incubated the beads with nuclear extracts expressing FLAG-ATR and HA-ATRIP. As expected, the full-length ETAA1 AAD bound the ATR–ATRIP complex. The ETAA1 predicted coiled coil also bound a small amount of ATR–ATRIP, whereas no appreciable binding above background was observed with the F198A mutant (Fig. 5C). We performed the same experiment with the TOPBP1 predicted coiled coil and, similarly, observed that this protein bound a small amount of ATR–ATRIP. In contrast, the TOPBP1 predicted coiled coil with the F1071A mutation exhibited no ATR–ATRIP binding above background (Fig. 5D). Taken together, these results suggest that the ETAA1 and TOPBP1 predicted coiled coils are important for ATR activation because these motifs contribute to the AAD interaction with ATR–ATRIP.
Discussion
Combined with data published previously, our results suggest that the defining features of an ATR activation domain include the presence of an evolutionary conserved tryptophan within a region predicted to be mostly disordered and a predicted coiled-coil motif that mediates binding to ATR–ATRIP. Both ATR and ATRIP are thought to contain AAD interaction surfaces needed for ATR activation (27), perhaps explaining the requirement of more than one ATR–ATRIP–binding motif in the AADs. With this more specific definition of an AAD, it may be possible to combine phenotypic data, secondary structure predictions, and primary sequence alignments to identify or evaluate additional AADs.
NBS1 has been proposed previously to be a direct ATR activator (28). The ATR interaction domain on NBS1 was mapped to BRCT domain 2, which does not have any similarity to the ETAA1 or TOPBP1 AAD motifs. The authors concluded that NBS1 activates ATR differently than TOPBP1. Therefore, we cannot rule out the possibility that additional ATR activators, such as NBS1, may activate ATR using mechanisms other than through AADs.
Like the MADs, which contain two aromatic residues required for Mec1 activation (21–23), we identified a second critical aromatic residue, a phenylalanine, in the ETAA1 and TOPBP1 AADs within the predicted coiled coils. The critical aromatic residues in the Mec1 activators are always either tryptophan or tyrosine, and interestingly, although one of the critical aromatics in Dna2, Trp-128, can be substituted with Tyr, mutation of this residue to Phe causes a defect in Mec1 activation (24). The MADs also contain no obvious predicted coiled coils (29). Thus, there may be some differences between the yeast MAD and vertebrate AAD domains. Atomic-resolution structural information will be required to fully determine how the MADs and AADs actually bind to and activate Mec1 and ATR, respectively.
ATR inhibitors are currently being tested as single agents and in conjunction with genotoxic chemotherapies to treat various malignancies (30). Identification of ATR-activating proteins and the mechanism by which they activate ATR should reveal more detailed information about how, when, and where ATR is activated and about downstream activator-specific signaling cascades. This information, in turn, may be useful in identifying which patients would most benefit from ATR inhibitor treatment regimens and contribute to improved efficacy of these drugs.
Experimental procedures
Cell culture
HEK293T and U2OS cells were maintained in DMEM and 7.5% FBS. Transfections were performed with polyethyleneimine.
Bioinformatic analysis
ETAA1 and TOPBP1 predicted disorder was determined using IUPred2A (31), and coiled-coil predictions were determined using Paircoil2 (29). ETAA1 and TOPBP1 predicted coiled coil sequence alignments were assembled using ClustalOmega.
Protein purification
GST-tagged AADs were purified from ArcticExpress Escherichia coli (Agilent Technologies). Bacteria were resuspended in NET buffer (25 mm Tris (pH 8), 50 mm NaCl, 0.1 mm EDTA, 5% glycerol, 1 mm DTT, 5 μg/ml aprotinin, and 5 μg/ml leupeptin) and sonicated. Triton X-100 was then added to a final concentration of 1%, and lysates were incubated on ice for 30 min. Following centrifugation, cleared lysates were incubated with GSH-Sepharose beads (GE Healthcare) for 2.5 h at 4 °C. Beads were then washed three times with NET buffer and 1% Triton X-100. Bound proteins were eluted using elution buffer (75 mm Tris (pH 8), 15 mm GSH, and 5 μg/ml leupeptin). For AADs used in gel filtration chromatography experiments, GST was removed by incubating beads with cleavage buffer (50 mm Tris (pH 7), 150 mm NaCl, 1 mm EDTA, and 1 mm DTT) containing Prescission Protease (GE Healthcare) overnight at 4 °C. Purified proteins were dialyzed (20 mm HEPES-KOH (pH 7.5), 50 mm NaCl, and 1 mm DTT) twice, once for 2 h, and then again overnight.
Kinase assays
FLAG–ATR–HA–ATRIP complexes were purified from HEK293T cell nuclear extracts using monoclonal anti-HA–agarose beads (Sigma-Aldrich). Beads were washed three times with TGN buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 10% glycerol, 1% Tween 20, 0.5 mm DTT, 1 mm NaF, 1 mm sodium orthovanadate, 10 mm β-glycerol phosphate, 5 μg/ml aprotinin, and 5 μg/ml leupeptin). Beads were then washed once in TGN buffer containing 500 mm LiCl and twice with 1× kinase buffer (10 mm HEPES-KOH (pH 7.5), 50 mm NaCl, 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, and 50 mm β-glycerol phosphate). Kinase reactions containing ∼16 nm ATR–ATRIP, 0.33 μm GST or GST-AAD, 0.95 μm substrate (GST-MCM2 79–138), 10 μm ATP, and 27.8 nm [γ-32P]ATP were incubated for 20 min at 30 °C and stopped upon addition of 2× SDS sample buffer. Reaction products were separated by SDS-PAGE and detected by Coomassie staining or immunoblotting. Substrate phosphorylation was detected by autoradiography.
Gel filtration chromatography
Proteins were loaded onto a SuperdexTM 200 Increase 10/300 GL column (GE Healthcare) equilibrated previously with buffer (20 mm HEPES-KOH (pH 7.5), 50 mm NaCl, and 10 mm DTT) and eluted at 0.25 ml min−1. 0.25-ml fractions were collected, and equal volumes of fractions corresponding to peaks were separated by SDS-PAGE and visualized by Coomassie staining.
Co-immunoprecipitation
Equal amounts of HEK293T cell nuclear extracts expressing GFP-FLAG AADs were incubated with EZviewTM Red Anti-FLAG M2 affinity gel for 1–2 h at 4 °C. Beads were washed three times with dialysis buffer (20 mm HEPES-KOH (pH 7.9), 100 mm KCl, 20% glycerol, and 0.5 mm DTT) and once with FLAG elution buffer (10 mm HEPES-KOH (pH 7.9), 300 mm KCl, 1.5 mm MgCl2, 0.05% NP-40, and 0.5 mm DTT). Bound proteins were eluted with FLAG elution buffer containing 0.3 μg/ml 3× FLAG peptide (Sigma-Aldrich), separated by SDS-PAGE, and detected by immunoblotting.
Binding assays
GST-tagged proteins were purified as described above. After washes with NET buffer and 1% Triton X-100, GSH beads were incubated with equal amounts of HEK293T cell nuclear extracts expressing FLAG–ATR–HA–ATRIP overnight at 4 °C. Beads were washed three times with low-salt buffer (20 mm HEPES-KOH (pH 7.9), 150 mm NaCl, 20% glycerol, 0.05% Tween20, and 0.5 mm DTT). Bound proteins were eluted, separated by SDS-PAGE, and detected by Coomassie staining or immunoblotting.
Immunofluorescence
U2OS cells were fixed in 3% paraformaldehyde/2% sucrose in PBS and blocked with 5% BSA in PBS prior to incubation with antibodies. Images were obtained using a Nikon microscope, and FLAG, γH2AX, and MCM2 Ser-108 nuclear intensities were quantitated using microscope software.
Author contributions
V. T. and D. C. conceptualization; V. T. investigation; V. T. writing-original draft; D. C. supervision; D. C. funding acquisition; D. C. project administration; D. C. writing-review and editing.
Funding was provided by National Institutes of Health grants CA102729 and CA239161 (to D.C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DDR
- DNA damage response
- ATM
- ataxia telangiectasia-mutated
- ATR
- ATM and Rad3-related
- RPA
- replication protein A
- ssDNA
- single-stranded DNA
- ATRIP
- ATR-interacting protein
- AAD
- ATR activation domain
- MAD
- Mec1 activation domain
- BRCT
- BRCA1 C-terminal.
References
- 1. Ciccia A., and Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeman M. K., and Cimprich K. A. (2014) Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blackford A. N., and Jackson S. P. (2017) ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 10.1016/j.molcel.2017.05.015 [DOI] [PubMed] [Google Scholar]
- 4. Saldivar J. C., Cortez D., and Cimprich K. A. (2017) The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 10.1038/nrm.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cortez D., Guntuku S., Qin J., and Elledge S. J. (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716 10.1126/science.1065521 [DOI] [PubMed] [Google Scholar]
- 6. Zou L., and Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
- 7. Kumagai A., Lee J., Yoo H. Y., and Dunphy W. G. (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 10.1016/j.cell.2005.12.041 [DOI] [PubMed] [Google Scholar]
- 8. Duursma A. M., Driscoll R., Elias J. E., and Cimprich K. A. (2013) A Role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol. Cell 50, 116–122 10.1016/j.molcel.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., and Karnitz L. M. (2007) The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21, 1472–1477 10.1101/gad.1547007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rappas M., Oliver A. W., and Pearl L. H. (2011) Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 39, 313–324 10.1093/nar/gkq743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cotta-Ramusino C., McDonald E. R. 3rd, Hurov K., Sowa M. E., Harper J. W., and Elledge S. J. (2011) A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 332, 1313–1317 10.1126/science.1203430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindsey-Boltz L. A., Kemp M. G., Capp C., and Sancar A. (2015) RHINO forms a stoichiometric complex with the 9-1-1 checkpoint clamp and mediates ATR-Chk1 signaling. Cell Cycle 14, 99–108 10.4161/15384101.2014.967076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bass T. E., Luzwick J. W., Kavanaugh G., Carroll C., Dungrawala H., Glick G. G., Feldkamp M. D., Putney R., Chazin W. J., and Cortez D. (2016) ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 18, 1185–1195 10.1038/ncb3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haahr P., Hoffmann S., Tollenaere M. A., Ho T., Toledo L. I., Mann M., Bekker-Jensen S., Räschle M., and Mailand N. (2016) Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 18, 1196–1207 10.1038/ncb3422 [DOI] [PubMed] [Google Scholar]
- 15. Lee Y. C., Zhou Q., Chen J., and Yuan J. (2016) RPA-binding protein ETAA1 is an ATR activator involved in DNA replication stress response. Curr. Biol. 26, 3257–3268 10.1016/j.cub.2016.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saldivar J. C., Hamperl S., Bocek M. J., Chung M., Bass T. E., Cisneros-Soberanis F., Samejima K., Xie L., Paulson J. R., Earnshaw W. C., Cortez D., Meyer T., and Cimprich K. A. (2018) An intrinsic S/G2 checkpoint enforced by ATR. Science 361, 806–810 10.1126/science.aap9346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bass T. E., and Cortez D. (2019) Quantitative phosphoproteomics reveals mitotic function of the ATR activator ETAA1. J. Cell Biol. 218, 1235–1249 10.1083/jcb.201810058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ball H. L., Ehrhardt M. R., Mordes D. A., Glick G. G., Chazin W. J., and Cortez D. (2007) Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 27, 3367–3377 10.1128/MCB.02238-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mordes D. A., Nam E. A., and Cortez D. (2008) Dpb11 activates the Mec1-Ddc2 complex. Proc. Natl. Acad. Sci. U.S.A. 105, 18730–18734 10.1073/pnas.0806621105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Navadgi-Patil V. M., and Burgers P. M. (2008) Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 283, 35853–35859 10.1074/jbc.M807435200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Navadgi-Patil V. M., and Burgers P. M. (2009) The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol. Cell 36, 743–753 10.1016/j.molcel.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar S., and Burgers P. M. (2013) Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 27, 313–321 10.1101/gad.204750.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Navadgi-Patil V. M., Kumar S., and Burgers P. M. (2011) The unstructured C-terminal tail of yeast Dpb11 (human TopBP1) protein is dispensable for DNA replication and the S phase checkpoint but required for the G2/M checkpoint. J. Biol. Chem. 286, 40999–41007 10.1074/jbc.M111.283994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wanrooij P. H., Tannous E., Kumar S., Navadgi-Patil V. M., and Burgers P. M. (2016) Probing the Mec1(ATR) checkpoint activation mechanism with small peptides. J. Biol. Chem. 291, 393–401 10.1074/jbc.M115.687145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cortez D., Glick G., and Elledge S. J. (2004) Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 101, 10078–10083 10.1073/pnas.0403410101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burkhard P., Stetefeld J., and Strelkov S. V. (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 10.1016/S0962-8924(00)01898-5 [DOI] [PubMed] [Google Scholar]
- 27. Mordes D. A., Glick G. G., Zhao R., and Cortez D. (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22, 1478–1489 10.1101/gad.1666208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kobayashi M., Hayashi N., Takata M., and Yamamoto K. (2013) NBS1 directly activates ATR independently of MRE11 and TOPBP1. Genes Cells 18, 238–246 10.1111/gtc.12031 [DOI] [PubMed] [Google Scholar]
- 29. McDonnell A. V., Jiang T., Keating A. E., and Berger B. (2006) Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics 22, 356–358 10.1093/bioinformatics/bti797 [DOI] [PubMed] [Google Scholar]
- 30. Forment J. V., and O'Connor M. J. (2018) Targeting the replication stress response in cancer. Pharmacol. Ther. 188, 155–167 10.1016/j.pharmthera.2018.03.005 [DOI] [PubMed] [Google Scholar]
- 31. Mészáros B., Erdos G., and Dosztányi Z. (2018) IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 10.1093/nar/gky384 [DOI] [PMC free article] [PubMed] [Google Scholar]