Abstract
This chapter describes the identification of the first prokaryotic ubiquitin-like protem modifier, Pup, which covalently attaches to proteins to target them for destruction by a bacterial proteasome in a manner akin to ubiquitin in eukaryotes. Despite using a proteasome as the end point for proteolysis, Pup and ubiquitin differ in sequence, structure and method of activation and conjugation to protein substrates. Pup is so far the only known posttranslational protein modifier in prokaryotes and its discovery opens the door to the possibility that others are present not only for proteolysis, but also to regulate protein function or localization. Here, we discuss the putative mechanism of activation and conjugation of Pup (termed “pupylation”) to target proteins. In addition, because it is unclear whether or not Pup, like ubiquitin, is recycled or degraded during substrate targeting to the proteasome, we propose methods that may identify Pup deconjugation enzymes (“depupylases”). Finally, we outline future directions for Pup research and anti-tuberculosis drug discovery.
INTRODUCTION
Unlike eukaryotes, prokaryotes lack well-defined sub-cellular compartments and therefore have additional requirements for the specificity and regulation of proteolysis. Bacterial ATP-dependent proteases, including ClpP, ClpQ (HslV), Lon and FtsH, provide “mini-compartments” or “barrel-shaped proteases” that tightly regulate the entrance of proteins into chambers enclosing proteolytic active sites (reviewed in refs. 1,2). In some cases bacteria also encode proteasomes that have high structural and chemical similarity to eukaryotic proteasomes.3 As with eukaryotic proteasomes, bacterial proteasomes are likely to form a complex with AAA or AAA+ ATPases (ATPases associated with a variety of cellular activities), which serve as regulatory subunits that recognize, unfold and translocate protein substrates into the proteasome core.
Proteasomes P3">Proteasomesare encoded in all sequenced Archaea but limited to bacteria of the order Actinomycetales, which includes the genera Streptomyces, Rhodococcus, Frankia and Mycobacterium.4 Numerous studies of prokaryotic proteasomes were undertaken with the hope that these proteases would provide a simplified model system for understanding proteolysis by complex eukaryotic proteasomes. Bacterial core particles (CPs) share sequence, structural and functional similarity with eukaryotic CPs. Like eukaryotic 20S CPs, bacterial proteasome CPs are barrel-shaped proteases with 14 alpha (α, PrcA) and 14 beta (β, PrcB) subunits with amino (N)-terminal threonines residing in the β-subunits that provide the catalytic active site nucleophiles (reviewed in ref. 5). Unlike eukaryotic proteasomes, bacterial CPs are usually composed of homo-heptameric rings of α subunits and β subunits. With at least one exception,6 the presence of one type of β-subunit appears to limit prokaryotic proteasomes to having only chymotryptic activity. Proteasome protease activity has been reconstituted in several Actinomycetales in vitro, however, these studies were carried out with model peptide substrates and not with native proteins, suggesting the need for additional factors for full proteasome function.6–9
Putative proteasome-associated genes colocalize with proteasome CP genes in Actinobacteria (Fig. 1A). These genes were initially identified based on comparisons with the genomic region of other proteasome-containing bacteria. Whereas prokaryotic proteasome core subunits were identified based on sequence homology to their eukaryotic counterparts,10 most of the putative proteasome-associated genes in the vicinity of the proteasome genes do not share any similarity with those found in eukaryotes. One exception is arc (AAA ATPase forming ring-shaped complexes), a gene encoding an AAA ATPase with homology to those found in the 19S regulatory particle in eukaryotes (Fig. 1A).11 Several biochemical studies demonstrated that this protein from Rhodococcus erythropolis formed hexameric or dodecameric rings with ATPase activity.11,12 ARC could not form stable or robust interactions with bacterial 20S CPs in vitro, nor could they stimulate protein degradation by 20S CPs. This suggested that the interactions between ARC and CPs were either transient or required additional cofactors. Despite advances in its characterization, the function of proteasomes in Rhodococcus is not well understood, as neither arc nor proteasome mutants have been characterized in this bacterium.
Figure 1.
Proteasome-associated genes are present in bacteria of the order Actinomycetales. A) Genomic organization of putative proteasome genes in M. tuberculosis (Mt), Streptomyces coelicolor (Sc) and Rhodococcus erythropolis (Re). Proteasome associated genes are in black; homologues are connected by dashed lines. Figure is adapted from Figure 4 in reference 9. B) Alignment of Pup from various Actinomycetales reveals a striking conservation in amino acid sequence at the C-terminus. Identical and similar amino acids are in red and blue, respectively.
mpa (Mycobacterium proteasomal ATPase) is an orthologue of Rhodococcus arc in M. tuberculosis (Mtb). Mutants of this gene were identified in a screen for transposon disruption mutants sensitive to nitric oxide, an anti-microbial molecule made by activated macrophages.13 Mutations in pafA (proteasome accessory factor A), another open reading frame near the prcBA genes, resulted in a similar phenotype to the mpa mutants. PafA was thought to participate in proteasome function because it is usually encoded near proteasome CP genes.14 Two-dimensional protein gel analysis revealed that two proteins, FabD (malonyl coA-acyl carrier protein transacylase) and PanB (ketopantoate hydroxymethyltransferase), displayed altered steady state levels in an mpa mutant compared to wild type. This phenotype was also produced by treating wild type Mtb with eukaryotic proteasome inhibitors.15 Ectopic expression of fabD and panB using a strong, nonnative promoter demonstrated that the over-produced protein accumulated in the mpa and pafA Mtb mutants as well as in proteasome inhibitor treated wild type Mtb, but not in the untreated wild type strain. Although these data strengthened the association of Mpa and PafA with proteasome function, neither had been shown to directly interact with the proteasome CPs or the degradation substrates. Thus it remained to be determined how Mpa and PafA targeted proteins like FabD and PanB for proteasomal degradation.
DISCOVERY OF A BACTERIAL “UBIQUITIN-LIKE” MODIFIER
Although the proteasome, putative proteasome-associated proteins and endogenous substrates had been identified in bacteria, it was unclear how proteins were targeted for degradation by this machinery since ubiquitin-like modifiers had not been identified in prokaryotes. The combination of bacterial 20S CPs and Mpa in vitro did not facilitate the degradation of FabD or PanB (M. Pearce, K.H. Darwin, unpublished). In an effort to understand how proteins were targeted to the proteasome, Pearce and coworkers used an E. coli bacterial two-hybrid system screen to identify Mtb proteins that interact with Mpa. Rv2111c, a 64 amino acid protein of unknown function encoded directly upstream the proteasome CP genes, was identified in this screen.16 The addition of purified Rv2111c to the in vitro system, however, failed to stimulate degradation. Furthermore, expression of recombinant Mtb PrcBA, Mpa and Rv2111c in E. coli failed to degrade FabD (K.H. Darwin, unpublished).
It was possible that additional proteins specific to proteasome-bearing bacteria were required for proteolysis. The development of a mycobacterial two-hybrid system allowed this hypothesis to be tested by looking for interactions between proteasome components and degradation substrates in the proteasome-bearing bacterium M. smegmatis, a nonpathogenic relative of Mtb.17 A positive interaction was detected between the substrate FabD and Rv2111c, a result that was confirmed in a pull down experiment from mycobacterial lysates. Surprisingly, FabD and Rv2111c were isolated as a covalently linked complex, where Rv2111c formed an isopeptide bond between its carboxyl (C) terminus and the є-amino group of a specific lysine (Lys173) in FabD. Mutagenesis of FabD’s modified Lys dramatically stabilized this substrate in wild type mycobacteria.16 In addition, pulse-chase analysis also showed that proteins modified with Rv2111c had longer half-lives in an mpa mutant of M. smegmatis. The modification of FabD with Rv2111c was reminiscent of ubiquitylation of proteasome substrates in eukaryotes, thus Rv2111c was named prokaryotic ubiquitin-likeprotein (Pup).
In a separate study, Burns and colleagues independently noticed thatpup encoded a small protein with a di-glycine motif at the penultimate position of the C-terminus, followed by either glutamine (Gln) or glutamate (Glu) (depending on the organism) (Fig. 1B).18 They speculated that Pup could covalently attach to bacterial proteasome substrates, despite the lack of overall sequence homology to ubiquitin. Using epitope-tagged Pup from M. smegmatis, two covalently linked proteins, super oxide dismutase (SodA) and myo-inositol-1-phosphate synthase (Ino1), were identified. Burns and coworkers also showed that several pupylated proteins were more stable in a proteasome CP mutant when compared to wild type M. smegmatis,18 consistent with the Pearce et al study.16
By analogy with ubiquitin processing and activation,19 it was hypothesized that the C-terminal residue of Pup is removed to expose the di-glycine (Gly-Gly) motif for activation by an E1-like enzyme. Mass spectrometry analysis revealed that this is not the case. Not only had Pup retained its C-terminal amino acid upon conjugation to its substrates, it was shown that the C-terminal Gln was deamidated, converting it to a Glu.16,18 Deletion of the C-terminal Glu or penultimate Gly abrogated pupylation.18 When unconjugated Pup was purified from mycobacteria and then analyzed by mass spectrometry, nearly all molecules were deamidated (“PupGGE”); in sharp contrast, the majority of Pup purified from E. coli ended in Gln (“PupGGQ”).16 This result suggested that a specific activity is present in mycobacteria that deamidates Pup prior to covalent attachment to substrate proteins. Alternatively, this result may indicate that Pup-target complexes are hydrolyzed, releasing PupGGE for recycling. These studies showed that proteasome substrates are posttranslationally modified with Pup, which is first processed at the C-terminus in a manner different than the proteolytic processing of ubiquitin and therefore likely requires a different activation pathway for conjugation onto substrates.
PUP CONJUGATION (“PUPYLATION”)
Pup appears to be deamidated in Mycobacteria prior to conjugation to target proteins. Striebel and coworkers confirmed this observation and showed that the reaction was catalyzed by Dop (deamidase of Pup), which is encoded upstream of pup in several bacterial genomes and is highly similar to PafA (Figs. 1A and 2).20 Dop shares no homology to ubiquitin-activating enzymes (E1) or ligases (E2, E3). Bioinformatics analysis suggests structural homology to the glutamine synthetase fold family, with Dop and PafA most likely belonging to the carboxylate-amine/ammonia ligase super family, similar to γ-glutamyl-cysteine synthetases.21 This family of enzymes catalyzes ligation reactions involving phosphorylation of a carboxylate group followed by ligation of an amino group, resulting in an amide linkage. The deamidation reaction generates ammonia and ATP is not hydrolyzed during the reaction but serves as a cofactor.20 The deamidation step may serve as a regulatory mechanism in organisms where Pup terminates in Gln. It is furthermore of note that bacteria encoding PupGGE have retained the dop gene, possibly suggesting roles in addition to deamidation for Dop.
Figure 2.
Pup conjugation. Following a deamidation reaction catalyzed by Dop, Pup is conjugated to a substrate (e.g., FabD) via an isopeptide linkage between a Lys of the substrate and either the C-terminal side chain carboxylate of Glu (i) or the C-terminal backbone carboxylate (ii). The ligation reaction is catalyzed by PafA and is ATP-dependent. Ligation is shown through the γ-carboxylate of Glu for simplicity.
The Mtb pafA mutant, which had previously been shown to have a defect in proteasome function,13 was unable to pupylate target proteins and was thereby implicated in the activation and/or conjugation of Pup to substrates.16 PafA was shown to catalyze the conjugation of Pup to a known proteasome substrate, FabD, in the presence of ATP and Dop (Fig. 2).20 It is unclear which C-terminal carboxylate (the backbone carboxylate or the y-carboxylate on the Glu) is conjugated to substrates (Fig. 2). PupGGE is a substrate for PafA-catalyzed conjugation in the absence of Dop, suggesting deamidation precedes conjugation and that Dop and PafA are not necessarily coupled. ATP is hydrolyzed during the course of the reaction and one molecule of ADP is generated per molecule of conjugated Pup.20 These data suggest that the PafA-catalyzed ligation reaction proceeds through a phosphorylated intermediate (Fig. 2) as hypothesized by bioinformatics analysis,21 although this intermediate has yet to be detected.
Both in vitro as well as in vivo experiments indicate that a single Pup moiety is conjugated onto a particular Lys residue on a target; chains of Pup have not been observed.16,18,20 Because Pup has three Lys, it is quite possible that “polypupylation” occurs. In addition, a single substrate may have multiple pupylated Lys.
More than 600 distinct mammalian proteins are thought to be involved in the ligation of ubiquitin to substrates.22 It is the multitude, diversity and combination of these ubiquitin ligases that allows a variety of substrates to be ubiquitylated in a specific and regulated manner. In Mtb, a pafA mutation abrogates pupylation,16 raising the obvious question as to how pupylation is regulated. Preliminary data suggest that there are potentially up to 155 pupylation targets in M. smegmatis (J. Watrous, P. Dorrestein, unpublished) and over 600 in Mtb (F. McAllister, J. Mintseris, S. Gygi, unpublished). The number and diversity of putative pupylation targets suggest the requirement for additional factors to accommodate selective protein targeting through pupylation. Furthermore, the ligation of Pup catalyzed by PafA on FabD in vitro was slow (17 h),20 perhaps suggesting the requirement of additional cofactors required for optimal pupylation.
PUP DECONJUGATION (“DEPUPYLATION”)
Unlike ubiquitin and other ubiquitin-related modifiers, the processing of de novo synthesized Pup prior to substrate ligation may not require a Pup protease (“depupylase”) activity. In addition, although there is no evidence for poly-Pup chains that would necessitate a depupylating activity, the recycling of Pup from substrates would provide an energy efficient means of protein degradation and regulation. There are no homologues of eukaryotic DUBs (deubiquitinating protease) or ULPs (ubiquitin-like specific protease) present in the vicinity of proteasome genes in bacteria and it is unknown if Pup is deconjugated prior to target degradation, or if it is simply degraded in the process.
Pup attaches to a target via an isopeptide bond, most likely at the γ-carboxylate position of the C-terminal glutamate. One would anticipate putative depupylases to be proteases that can recognize and hydrolyze the amide bond between Pup and Lys residues of target proteins. Hydrolysis by a depupylase then results in a PupGGE sequence, primed for additional substrate conjugation reactions (Fig. 3A). Alternatively, a transamidation reaction with ammonia would regenerate PupGGQ (Fig. 3A), which would require deamidation by Dop prior to conjugation to substrates. Both the peptidase and transamidase reactions could in theory proceed through a protein-substrate intermediate complex that can be exploited for developing activity-based probes to trap depupylases, if present. The probes would be designed to crosslink the depupylases with Pup, similar to those used for the general identification of DUBs and ULPs that belong to the cysteine family of proteases.23,24 An example of such a probe would be Pup vinyl methylesters (Fig. 3B), analogous to the ubiquitin vinyl methylester probe generated to trap DUBs, which has been instrumental to the identification of numerous novel DUBs, including novel DUB families.24 Alternatively, Pup could be modified at the C-terminus with acivicin or DON (6-diazo-5-oxo-L-norleucine) (Fig. 3B), known traps for deamidases.25,26
Figure 3.

A depupylase? A) Proposed depupylase reaction by either hydrolysis (i) or transamidation (ii). B) Possible probes to test and trap putative depupylase activity.
Due to the similarity in chemistry proposed for depupylation and deamidation (Fig. 2 and Fig. 3A), it is possible that these probes will react with Dop. As mentioned previously, bioinformatics analysis suggests that organisms that naturally encode PupGGE instead of PupGGQ also have dop, suggesting a possible depupylase role for Dop in addition to deamidation. Whether or not Dop can serve as a depupylase, its observed deamidation activity should enable it to react with one or more of the proposed probes.
Over 100 DUBs have been identified in mammalian cells,27 with functions ranging from recycling ubiquitin prior to target degradation by the proteasome to rescuing proteins from degradation.28 Some DUBs simply bind ubiquitin with high affinity. Certain DUBs, however, do not function in concert with the proteasome, as extensively discussed in this book. Although it is currently unknown whether depupylases exist in bacteria, it is imperative to investigate whether such proteins, if identified, play a more sophisticated role in protein homeostasis, similar to their eukaryotic counterparts. Thus, in addition to trapping proteasome-associated depupylases, the probes highlighted in Fig. 3B may trap proteasome-independent depupylases.
CONCLUSION AND FUTURE PERSPECTIVES
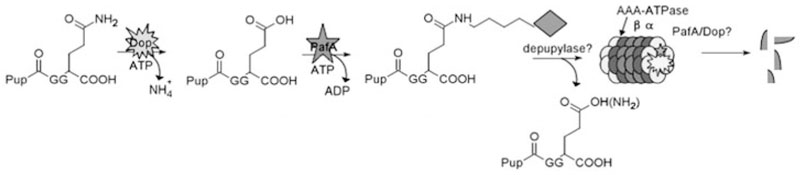
Pup posttranslationally tags proteins for degradation by the proteasome and it is the only currently known prokaryotic protein that is functionally similar to ubiquitin. Despite the functional homology, Pup differs from ubiquitin in many other aspects. With little sequence and no structural homology to ubiquitin, Pup is first deamidated by Dop and subsequently conjugated to a variety of substrates by PafA, two proteins highly similar to each other but bearing no resemblance to eukaryotic ubiquitin/proteasome-associated proteins (Fig. 4). Similar to ubiquitin, pupylation is through a C-terminal carboxylate to substrate lysines. Pupylation dooms proteins to the proteasome for destruction, however, additional roles for pupylation cannot be ruled out.
Figure 4.
Model of the Pup-proteasome system. Following deamidation by Dop, Pup is ligated to substrate Lys via an isopeptide linkage to the C-terminal carboxylate of Glu. Ligation is shown on the γ-carboxylate for simplicity. Pup interacts with the AAA-ATPase Mpa, which presumably unfolds substrates for delivery into the catalytic chamber of the proteasome core for degradation. It is currently unknown whether additional factors are required for optimal conjugation and delivery to the proteasome, if PafA and Dop interact with the proteasome, or if Pup is recycled or degraded
Many aspects of the Pup-proteasome system remain to be resolved. How is Pup recognized by the proteasome system in bacteria? Pup is an intrinsically disordered protein,29,30 very different from the highly structured ubiquitin.31 Pup binds Mpa,16,20,29,30 most likely to target proteins for proteasomal degradation, but detecting interactions between Mpa and the 20S proteasome have been elusive in prokaryotes. It is unclear if this interaction is transient or whether additional factors are involved in directing substrate specificity at the proteasome. Pup also stably binds to Dop and PafA20 (F. Cerda, K.H. Darwin, unpublished), however, similar to Mpa, it is unclear what these associations mean in the context of the proteasome. It is intriguing to hypothesize the presence a Pup interaction motif, analogous to the ubiquitin interacting motifs, whereby Pup becomes ordered upon binding distinct motifs on proteasomal components. Due to the lack of structural information for Dop and PafA, we are unsure whether this motif exists. The presence of a Pup interaction motif may help guide the identification of additional proteasome components, including potential depupylases, regulators and specificity factors.
It should be mentioned that various organisms have only parts of the Pup-proteasome system. For example in Archaea, proteasome-dependent proteolysis has been demonstrated in vitro and proteolysis was stimulated by the presence of the AAA+ ATPase PAN (proteasome activating nucleotidase).32 Archaea, however, do not have homologues of Pup or bacterial proteasome-associated proteins such as PafA. The PAN-proteasome complex may serve as a general protease in Archaea, similar to Clp proteases in bacteria. In addition, Corynebacteria encode homologues of proteasome-associated proteins, including PafA, Dop, AAA ATPase and Pup, however, proteasomes are absent from these organisms. It is unknown whether Corynebacteria Pup is able to conjugate proteins and if so, what purpose it serves. It is intriguing to hypothesize a signaling or regulatory role for pupylation in the absence of a proteasome.
Although the chemistries ofthe ubiquitylation and pupylation systems differ, many of the principles and techniques used to study the ubiquitin-proteasome system can be applied to unravel the Pup-proteasome system. Similar to the ubiquitin system, we will not only begin to understand key players involved in target recognition, but also the significance of the Pup signal in bacterial physiology and disease, opening novel options for therapeutic intervention of Mtb. The Pup-proteasome system is essential for the pathogenesis of Mtb, one of the most deadly bacterial pathogens in the world (WHO; http://www.who.int/en). Thus the identification of players in this pathway may also provide ideal drug targets for the development of novel tuberculosis chemotherapies.
ACKNOWLEDGEMENTS
We are grateful to F. Cerda-Maira, A. Darwin, T. Huang and H. Ovaa for critical review of this chapter. K.H. Darwin received support from National Institutes of Health grants AI065437 and HL092774 and is a Burroughs Wellcome Fund Investigator in the Pathogenesis ofInfectious Disease.
REFERENCES
- 1.Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 2003; 19:565–87. [DOI] [PubMed] [Google Scholar]
- 2.Butler SM, Festa RF, Pearce MJ et al. Self-compartmentalized Bacteria Proteases and Pathogenesis. Mol Microbiol 2006; 60:553–62. [DOI] [PubMed] [Google Scholar]
- 3.Lupas A, Zuhl F, Tamura T et al. Eubacterial proteasomes. Mol Biol Reports 1997;24:125–31. [DOI] [PubMed] [Google Scholar]
- 4.Darwin KH. Prokaryotic Ubiquitin-Like Protein, Proteasomes and Pathogenesis. Nat Rev Microbiol 2009; 7:485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumeister W, Walz J, Zuhl F et al. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 1998; 92:367–80. [DOI] [PubMed] [Google Scholar]
- 6.Lin G, Hu G, Tsu C et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol 2006; 59:1405–16. [DOI] [PubMed] [Google Scholar]
- 7.Pouch M-N, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol Microbiol 2000; 35:368–77. [DOI] [PubMed] [Google Scholar]
- 8.Tamura T, Nagy I, Lupas A et al. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. CurrBiol 1995; 5:766–74. [DOI] [PubMed] [Google Scholar]
- 9.Nagy I, Tamura T, Vanderleyden J et al. The 20S proteasome of Streptomyces coelicolor. J Bacteriol 1998; 180:5448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupas A, Zwickl P, Baumeister W. Proteasome sequences in eubacteria. Trends Biochem Sci 1994; 19:533–34. [DOI] [PubMed] [Google Scholar]
- 11.Wolf S, Nagy I, Lupas A et al. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol 1998; 277:13–25. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Stoffels K, Wurzbacher S et al. The N-terminal coiled coil of*the Rhodococcus erythropolis ARC AAA ATPase is neither necessary for oligomerization nor nucleotide hydrolysis. J Struct Biol 2004; 146:155–65. [DOI] [PubMed] [Google Scholar]
- 13.Darwin KH, Ehrt S, Weich N et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003; 302:1963–6. [DOI] [PubMed] [Google Scholar]
- 14.Nagy I, Geert S, Vanderleyden J et al. Further sequence analysis of the DNA regions with the Rhodococcus 20S proteasome structural genes reveals extensive homolgy with Mycobacterium leprae. DNA Seq 1997; 7:225–8. [DOI] [PubMed] [Google Scholar]
- 15.Pearce MJ, Arora P, Festa RA et al. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J 2006; 25:5423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce MJ, Mintseris J, Ferreyra J et al. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 2008; 322:1104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Mai D, Kumar A et al. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc Natl Acad Sci USA 2006; 103:11346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns KE, Liu WT, Boshoff HI et al. Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem 2009; 284:3069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickart CM. Mechanisms Underlying Ubiquitination. Annual Review of Biochemistry 2001; 70:503–33. [DOI] [PubMed] [Google Scholar]
- 20.Striebel F, Imkamp F, Sutter M et al. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol 2009. [DOI] [PubMed]
- 21.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct 2008; 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshaies RJ, Joazeiro CAP. RING Domain E3 Ubiquitin Ligases. Annual Review of Biochemistry 2009; 78:399–434. [DOI] [PubMed] [Google Scholar]
- 23.Borodovsky A, Ovaa H, Meester WJ et al. Small-molecule inhibitors and probes for ubiquitin- and ubiquitin-like-specific proteases. Chembiochem 2005; 6:287–91. [DOI] [PubMed] [Google Scholar]
- 24.Borodovsky A, Ovaa H, Kolli N et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol 2002; 9:1149–59. [DOI] [PubMed] [Google Scholar]
- 25.Hartman SC. Glutaminase from Escherichia coli. Journal ofBiologica Chemistry 1968;243:853. [PubMed] [Google Scholar]
- 26.Prajda N. Enzyme targets of antiglutamine agents in cancer chemotherapy. Adv Enzyme Regul 1985; 24:207–23. [DOI] [PubMed] [Google Scholar]
- 27.Nijman SM, Luna-Vargas MP, Velds A et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005; 123:773–86. [DOI] [PubMed] [Google Scholar]
- 28.Sowa ME, Bennett EJ, Gygi SP et al. Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell 2009; 138:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao S, Shang Q, Zhang X et al. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J 2009; in press. [DOI] [PubMed]
- 30.Chen X, Solomon WC, Kang Y et al. Prokaryotic ubiquitin-like protein Pup is intrinsically disordered. J Mol Biol 2009; in press. [DOI] [PMC free article] [PubMed]
- 31.Vijay-Kumar S, Bugg CE, Wilkinson KD et al. Three-dimensional struture of ubiquitin at 2.8 A resolution. PNAS 1985; 82:3582–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benaroudj N, Goldberg AL. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol 2000; 2:833–9. [DOI] [PubMed] [Google Scholar]