Abstract
Molecular analysis of nucleic acid and protein biomarkers is becoming increasingly common in paediatric oncology for diagnosis, risk stratification and molecularly targeted therapeutics. However, many current and emerging biomarkers are based on analysis of tumour tissue, which is obtained through invasive surgical procedures and in some cases may not be accessible. Over the past decade, there has been growing interest in the utility of circulating biomarkers such as cell-free nucleic acids, circulating tumour cells and extracellular vesicles as a so-called liquid biopsy of cancer. Here, we review the potential of emerging circulating biomarkers in the management of neuroblastoma and highlight challenges to their implementation in the clinic.
Keywords: neuroblastoma, circulating biomarkers, cell-free DNA, ALK, MYCN
1. Introduction to neuroblastoma
Neuroblastoma (NB) is the most common and deadly solid extracranial malignancy in children, deriving from precursor sympathoadrenal cells of the neural crest [1,2]. NB is often termed a ‘clinical enigma’, owing to its heterogeneous clinical behaviour ranging from spontaneous regression to treatment resistance, metastasis and death [3,4]. Therefore, in addition to the post-surgical International Neuroblastoma Staging System (INSS; table 1), patients are classified as very low (approx. 28%), low (approx. 27%), intermediate (approx. 9%) or high risk (approx. 36%) based on the likelihood of disease progression and relapse [5–8]. While low-risk disease is usually diagnosed in children younger than 18 months and may spontaneously regress without treatment, high-risk disease generally occurs in children older than 18 months and around half of patients relapse [9]. Despite intensive multi-modal treatment, 5 year survival among high-risk patients remains at 40–50% [10].
Table 1.
International Neuroblastoma Staging System (INSS).
| stage | definition |
|---|---|
| 1 | Localized tumour with complete gross excision, with or without microscopic residual disease; representative ipsilateral lymph nodes negative for tumour microscopically (nodes attached to and removed with the primary tumour may be positive). |
| 2A | Localized tumour with incomplete gross excision; representative ipsilateral non-adherent lymph nodes negative for tumour microscopically. |
| 2B | Localized tumour with or without complete gross excision, with ipsilateral non-adherent lymph nodes positive for tumour. Enlarged contralateral lymph nodes must be negative microscopically. |
| 3 | Unresectable unilateral tumour infiltrating across the midline,a with or without regional lymph node involvement; or localized unilateral tumour with contralateral regional lymph node involvement; or midline tumour with bilateral extension by infiltration (unresectable) or by lymph node involvement. |
| 4 | Any primary tumour with dissemination to distant lymph nodes, bone, bone marrow, liver, skin and/or other organs (except as defined for stage 4S). |
| 4S | Localized primary tumour (as defined for stage 1, 2A or 2B), with dissemination limited to skin, liver and/or bone marrowb (limited to infants < 1 year of age). |
NOTE: Multifocal primary tumours (e.g. bilateral adrenal primary tumours) should be staged according to the greatest extent of disease, as defined above, and followed by a subscript letter M.
aThe midline is defined as the vertebral column. Tumours originating on one side and crossing the midline must infiltrate to or beyond the opposite side of the vertebral column.
bMarrow involvement in stage 4S should be minimal, i.e. less than 10% of total nucleated cells identified as malignant on bone marrow biopsy or on marrow aspirate. More extensive marrow involvement would be considered to be stage 4. The metaiodobenzylguanidine (MIBG) scan (if performed) should be negative in the marrow. Adapted from Brodeur et al. [5].
Genomic amplification of MYCN is reported in around 25% of NB tumours (approx. 40% among high-risk patients) and is generally accepted as the strongest predictor of poor prognosis and rapid tumour progression [11,12]. Other poor prognostic features include chromosome arm-level alterations, namely deletions of 1p (30%) and 11q (45%) and unbalanced gain of 17q (60%), all of which are associated with diploid or near-tetraploid karyotypes [13–16]. In addition, amplification of ALK, encoding the anaplastic lymphoma kinase (ALK) receptor tyrosine kinase, is observed in 1–2% of cases and is often co-amplified with MYCN [17–19]. Recently, massive genomic rearrangement, known as chromothripsis, has been observed in 18% of advanced stage tumours; thus, NB could be considered a predominantly copy number-driven cancer [20,21]. Somatic mutations are less common and include point mutations of ALK (8–10%) as well as point mutations and small, in-frame deletions of alpha thalassaemia/mental retardation syndrome X-linked (ATRX), encoding a SWI/SNF family chromatin remodelling protein, that occur at a frequency correlating with age at diagnosis [20,22–25]. ATRX alterations are associated with poor prognosis [24]. Recent genome-wide sequencing analyses in large NB patient cohorts have identified a relative paucity of recurrent alterations [20,24–26].
Initial investigations for NB involve laboratory testing for full blood count, serum electrolytes, liver function and urine catecholamine metabolites [27]. More general biomarkers such as ferritin, lactate dehydrogenase and neuron-specific enolase (NSE) may also be investigated [28]. For suspected NB in the abdomen, ultrasound is the preferred imaging method [29]. A provisional diagnosis is followed up with cross-sectional imaging such as computed tomography or magnetic resonance imaging and confirmed by histological analysis of tumour tissue obtained from a primary tissue biopsy or bone marrow aspirate [29,30].
The treatment algorithm for NB is dependent on risk stratification, which is defined using parameters such as age, disease stage, tumour histopathology, MYCN status and DNA ploidy [31]. Low-risk patients often require surgery alone or close observation, since spontaneous regression is frequently observed in this risk group [31]. By contrast, intermediate-risk patients require both surgery and chemotherapy of moderate intensity, and high-risk patients are treated with high-intensity chemotherapy, radiotherapy, surgery and autologous haematopoietic stem cell transplant [31,32]. In addition, high-risk patients receive immunotherapy with anti-GD2 antibodies and cytokines, and differentiation therapy with 13-cis-retinoic acid to eliminate minimal residual disease (MRD) [33].
2. Current biomarkers in neuroblastoma
NB is one of few paediatric cancers in which biomarkers are routinely used for diagnosis, prognostication and therapeutic monitoring (table 2).
Table 2.
Current biomarkers in NB.
| biomarker | specimen | inference | studies |
|---|---|---|---|
| catecholamine metabolites (HVA, VMA) | urine | diagnostic; prognostic | [34–39] |
| lactate dehydrogenase | serum | diagnostica; prognostic | [7,37,40,41] |
| neuron-specific enolase | serum | prognostic | [42–46] |
| ferritin | serum | prognostic | [47–50] |
| MYCN amplification | tissue | prognostic | [11,51–53] |
| 1p deletion | tissue | prognostic | [14,54] |
| 11q deletion | tissue | prognostic | [14,55–57] |
| 17q gain | tissue | prognostic | [15,54,58,59] |
| ALK mutation | tissue | prognostic; therapeutic | [22,23,60–62] |
| ALK amplification | tissue | prognostic; therapeutic | [60,63] |
aNot independent.
2.1. Urine catecholamines
The majority of neural crest tumours including NB secrete catecholamines [64]. Elevated urinary levels of the catecholamine metabolites vanillylmandelic acid (VMA) and homovanillic acid (HVA) are observed in 90–95% of NB patients at diagnosis [34,35] and a low VMA-to-HVA ratio is associated with poorly differentiated tumours and poor prognosis [36,37]. These metabolites have been used since the 1970s as non-invasive biomarkers to assist in the diagnosis and therapeutic monitoring of patients with NB [38]. A recent study found the combined diagnostic sensitivity of VMA and HVA in NB to be 84% overall [39], though sensitivity is much lower (33–59%) in stage I tumours [36,39]. To facilitate early detection of NB, a screening programme based on urine catecholamine levels in infants aged six months was trialled and later implemented in Japan [65]. However, the programme was terminated upon publication of evidence from screening trials conducted in other countries, which suggested that NB-specific mortality was not reduced among screened subjects [66–68]. Retrospective analyses have determined that screening for NB results in overdiagnosis; screen-detected patients had a tendency to spontaneously regress [69,70] and many of these tumours showed favourable prognostic features at diagnosis [71].
2.2. Serum proteins
Serum lactate dehydrogenase (LDH) is used as a tumour biomarker in several malignancies [72], although levels can be elevated in non-malignant conditions such as heart failure, kidney disease, hypothyroidism and anaemia [73]. In NB, elevated serum LDH levels have been shown to confer poor prognosis independent of disease stage in patients with MYCN amplification (MNA) [37,40]. Moreover, a recent study identified elevated serum LDH levels as an independent predictor of poorer event-free (EFS) and overall (OS) survival in patients with metastatic disease older than 18 months at diagnosis [74]. LDH levels are typically normal in the early stages of disease, and significantly rise with tumour burden as the rate of cell turnover increases [7,41]. Thus, serum LDH levels may be used as a surrogate for the real-time monitoring of tumour burden.
NSE is a glycolytic enzyme produced in neural tissues [75]. Serum NSE levels are elevated in neuroendocrine tumours such as carcinoids, islet cell tumours, small-cell lung carcinomas and NB [42,43]. While elevated NSE levels are therefore not specific for NB, they can be used in patients with a confirmed diagnosis to provide prognostic information [42,44,45]. Despite extensive tumour burden, serum NSE levels are significantly lower in stage 4S relative to stage 4 disease [44,46].
NB cell lines and tumours produce and secrete glycosylated ferritin, which can be distinguished from the non-glycosylated form secreted by healthy cells [47]. Elevated levels of serum ferritin are observed in patients with stage 3 or 4 NB, serving as a poor prognostic biomarker independent of age at diagnosis and disease stage [48,49]. Moreover, while ferritin levels in tumour tissue of patients with stage 4 and 4S disease are comparable, serum ferritin levels are only significantly elevated in stage 4 patients, thus enabling discrimination between these two clinical stages with markedly different prognoses [50].
2.3. Tumour genetics
Several genetic alterations in NB are routinely detected in diagnostic tissue and serve as important prognostic and therapeutic biomarkers. In addition to MYCN as described above, a plethora of segmental chromosome alterations (SCAs) are associated with poor prognosis in NB, notably deletion of 1p and 11q, and gain of 17q; most high-risk tumours carry at least one such alteration. Both 1p36 deletion and 17q gain are associated with MNA and independently correlate with poor outcome [14]. Gain of 17q is the most frequent genetic alteration in NB and is associated with 1p deletion [54]. Loss of 11q is almost mutually exclusive of MNA but is also a feature of high-risk disease and portends an unfavourable prognosis [14,55]. Interestingly, tumours with 11q loss tend to display numerous SCAs, suggestive of a chromosomal instability phenotype [56,57].
Around 8–10% of sporadic NB tumours present with point mutations in the kinase domain of the full-length ALK receptor [22,23]; this is in contrast to the oncogenic NPM-ALK and EML4-ALK fusions that drive ALK-positive anaplastic large cell lymphoma (ALCL) and non-small cell lung cancer (NSCLC), respectively. ALK mutations in NB are found in equal frequencies across all tumour risk groups but appear to be associated with MNA [60], likely representing a cooperative effect between these oncogenes [61]. Additionally, 1–2% of NB tumours harbour genomic amplification of ALK, which almost exclusively occurs with MNA given their proximal association at chromosome 2p23-24 [60]. Therefore, ALK amplification also tends to afford poor prognosis [63]. ALK alterations serve as important biomarkers in NB because they confer sensitivity to small-molecule kinase inhibitors that are currently undergoing clinical assessment in phase I and II trials [76].
3. Emerging tumour-specific circulating biomarkers in neuroblastoma
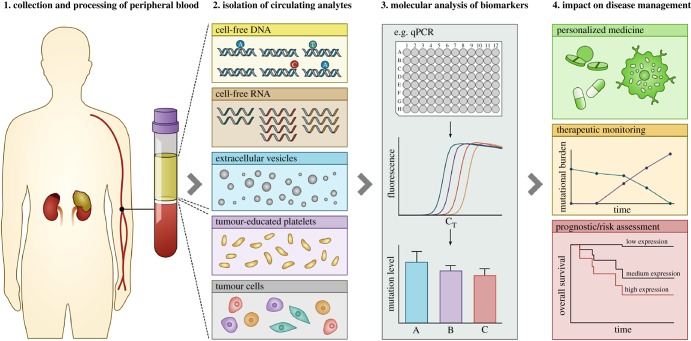
Tissue-based nucleic acid biomarkers are instrumental to risk classification, prognostication and therapeutic assignment in NB and indeed many other cancers [77]. However, surgical acquisition of tumour tissue is invasive, not amenable to sequential application and potentially subject to sampling bias owing to intratumoral genetic heterogeneity, as highlighted in several recent NB studies [78–80]. Over the past two decades, there has been growing interest in the development of other blood-based tissue biomarker analytes such as cell-free DNA and RNA, circulating tumour cells (CTCs) and, more recently, extracellular vesicles and tumour-educated platelets (figure 1). Collectively, these circulating analytes have been termed a ‘liquid biopsy’ of cancer and have been reviewed extensively elsewhere [81–83]. The non-invasive nature of blood sampling overcomes several limitations of conventional tissue-based tumour analysis. Whereas invasive biopsy procedures are not amenable to repeated sampling, a liquid biopsy can be taken at multiple time points such as at diagnosis, during treatment and at relapse, capturing the genetic and proteomic changes that underlie tumour evolution in real time [84]. Some notable clinical applications of circulating biomarkers include the quantitative real-time polymerase chain reaction (qPCR)-based test recently approved by the US Food and Drug Administration (FDA) for EGFR mutations in circulating cell-free DNA from patients with NSCLC, enabling identification of patients likely to respond to EGFR inhibitors [85], and the FDA-approved CELLSEARCH® test for enumeration of CTCs of epithelial origin to aid in the monitoring of patients with metastatic breast, colorectal or prostate cancers [86].
Figure 1.
The liquid biopsy workflow. In oncology, a liquid biopsy is a minimally invasive alternative to surgical tumour sampling, involving the collection and analysis of a peripheral blood sample. Blood plasma/serum is a source of several circulating analytes with potential clinical utility, namely cell-free DNA, cell-free RNA (mRNA and microRNA), extracellular vesicles (exosomes and microvesicles), tumour-educated platelets (TEPs) and tumour cells. Cell-free nucleic acids are a potential source of tumour-specific genomic alterations such as gene mutations, translocations, copy number alterations (CNAs), DNA methylation and gene expression changes. TEPs are a source of nucleic acids, and extracellular vesicles and tumour cells are a source of nucleic acids and protein. Molecular analysis of circulating biomarkers can impact the clinical management of cancer patients by enabling personalized medicine, monitoring of therapeutic response and assessment of disease prognosis/risk.
3.1. Circulating-free DNA and circulating tumour DNA
DNA was first reported in the circulation 70 years ago [87], but generated little interest until 1977 when levels of circulating free DNA (cfDNA) were shown to be elevated in patients with cancer [88]. Later studies that identified RAS mutations in the blood of patients with pancreatic cancer and acute myelogenous leukaemia provided evidence of a significant tumour-derived component of cfDNA, now termed circulating tumour DNA (ctDNA), and these studies led to further exploration of the clinical potential of cfDNA in patients with cancer [89,90]. Basal levels of cfDNA are thought to arise from turnover of normal haematopoietic cells, whereas elevated levels are sometimes observed in cancer and derive from a combination of apoptosis, necrosis and active secretion from tumour cells [91–93]. Indeed, ctDNA is detectable in most patients with NB [94,95]. Moreover, in many solid cancers, including NB [96,97], cfDNA levels have been shown to reflect tumour burden, rising with disease progression and falling after therapy and surgical resection [80,83,95,96]. Thus, cfDNA levels may be a dynamic biomarker for disease monitoring, though levels can also be elevated in non-malignant conditions [98–100]. In a recent study, cfDNA was shown to be as reliable as NSE and LDH in discriminating NB patients with newly diagnosed (n = 79) versus stable (n = 79) disease (area under the curve (AUC), 0.953, 0.929 and 0.906, respectively). Moreover, in newly diagnosed patients, elevated plasma cfDNA levels were associated with high-risk disease, advanced tumour stage, MNA, abdominal primary site and three or more metastatic sites [96]. This study did not report a significant association between the DNA integrity index (ratio of long to short cfDNA fragments) and these clinical variables. Patients with NB appear to have high levels of cfDNA at diagnosis, of which a high proportion is tumour derived, i.e. ctDNA [94,101], and reports have demonstrated the detection of numerous genomic alterations, including MNA [80,95,97,101–110], ALK mutation [111], 11q deletion [80,95,110,112], 17q gain [80,110,113], and gene methylation [114] in cfDNA of patients with NB at various stages of the clinical course.
3.1.1. MYCN amplification
Owing to its strong association with high-risk NB, MYCN was the first oncogene found to be altered with clinical significance and was consequently the first nucleic acid biomarker in oncology [11,12,51]. Targeted overexpression of N-MYC in neuroectodermal cells is sufficient to induce NB in mice, thus confirming a pathogenic role of MYCN in NB [115]. Indeed, MNA remains the strongest indicator of risk in NB, and its levels correlate with aggressive, metastatic behaviour [116,117].
Interphase fluorescence in situ hybridization (I-FISH) on biopsy and resected tumour tissue remains the gold standard for assessment of MNA in NB [118]. A number of studies have demonstrated the potential to detect MNA in serum and plasma from patients with NB at diagnosis across all tumour stages using PCR methodology (table 3), thus raising the prospect of using MYCN as a circulating biomarker [102–109]. Using conventional PCR, Combaret et al. [102] were the first to show MNA in the sera of 31/32 patients with MNA tumours, with one false positive among 70 patients without MNA and no false positives among 72 healthy patients. Moreover, MYCN levels in eight patients were shown to fall after chemotherapy and rise at relapse, and in one patient elevated levels were observed two months before a clinical diagnosis of relapse [102]. A recent study also reported a decrease in plasma MYCN copy number one week after surgery in five patients with MNA, proportional to the extent of tumour resection [106]. These observations suggest that MYCN in cfDNA may serve as a dynamic biomarker for disease monitoring and early detection of relapse. Indeed, additional studies by Combaret et al. and other groups confirmed the high performance of qPCR and digital PCR (dPCR)-based strategies for detecting MNA in serum [103–105,107,108] and plasma [106,109]. While cohort sizes and INSS disease stages were variable between studies, the reported sensitivities and specificities of MNA detection in cfDNA were 84–100% and 95–100%, respectively (table 3). There was also wide inter-study variation in median blood MYCN copy numbers from patients with tissue-confirmed MNA (range, 2.56–199.32), likely reflecting the aforementioned differences in cohort characteristics. It is also noteworthy that serum generally yields higher ‘apparent’ cfDNA levels than plasma [119] due to release of genomic DNA from haematopoietic cells during the clotting process [120–122]. Gotoh et al. [103] demonstrated this dilution effect in the context of MYCN copy number measurement by spiking white blood cells into serum samples from patients with MNA, resulting in reduced MYCN/reference gene ratios. Therefore, delayed isolation of serum could result in significant genomic DNA contamination, thus reducing apparent MYCN copy number measurements. For this reason, plasma is more suitable than serum for analysis of gene copy number in cfDNA [119].
Table 3.
Summary of NB studies evaluating MYCN amplification status in peripheral blood using PCR methodology.
| serum/plasma | PCR method | ref. gene | MNA status (−/+) by INSS disease stage |
median blood MYCN ratio by tissue status |
overall sens. (%) | overall spec. (%) | study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tissue |
serum/plasma |
||||||||||||||||
| 1 | 2 | 3 | 4 | 4S | 1 | 2 | 3 | 4 | 4S | MNA− | MNA+ | ||||||
| serum | qPCR | RPPH1 | 14/0 | 10/1 | 8/5 | 33/25 | 5/1 | 14/0 | 10/1 | 9/4 | 32/26 | 5/1 | NR | NR | 97 | 99 | [102] |
| serum | qPCR | NAGK | 22/1 | 18/1 | 7/2 | 18/13 | 5/0 | NR | NR | NR | NR | NR | 0.87 | 199.32 | 100 | 100 | [103] |
| serum | qPCR | IL1B | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | 89 | 98 | [104] |
| serum | qPCR | NAGK | 24/10a | 24/10a | 27/16 | 83/41 | 60/6 | 33/1a | 33/1a | 31/12 | 89/35 | 61/5 | NR | NR | 84 | 100 | [105] |
| plasma | qPCR | NAGK | 16/0 | 4/0 | 7/2 | 6/14 | 1/0 | 16/0 | 4/0 | 7/2 | 6/14 | 1/0 | 0.98 | 26.75 | 100 | 100 | [106] |
| serum | qPCR | NAGK | 38/6a | 38/6a | 14/12 | 33/38 | 6/1 | 38/6a | 38/6a | 13/13 | 37/34 | 6/1 | 2.45 | 118.27 | 86 | 95 | [107] |
| serum | qPCR | NAGK | 10/0 | 21/0 | 13/1 | 49/9 | 2/0 | NR | NR | NR | NR | NR | 0.97 | 2.56 | 91 | 98 | [108] |
| plasma | ddPCR | NAGK | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | 2.7 | 49.2 | 100 | 100 | [109] |
aStages 1 and 2 grouped. ddPCR, droplet digital PCR; INSS, International Neuroblastoma Staging System; MNA, MYCN amplification; NR, not reported; qPCR, quantitative real-time PCR; sens., sensitivity; spec., specificity.
A 2005 case report by Combaret et al. [104] highlighted the utility of MYCN evaluation in cfDNA where tumour biopsy is not possible. A 30-day-old infant presented with stage 4S NB, but thrombocytopenia and hypofibrinogenaemia confounded the collection of tumour tissue. Evaluation of serum MYCN by qPCR revealed high-level MNA, and the child was assigned high-intensity chemotherapy. Three weeks after a significant response, blood coagulation parameters had returned to normal and biopsy tissue was obtained, which showed greater than 50 MYCN copies by qPCR. The child achieved remission and was disease free 1 year later [104]. It is estimated that around 29% of patients with NB have unknown MYCN status [123,124], representing a significant proportion of patients who could potentially benefit from MYCN testing in cfDNA.
More recent studies have assessed MYCN status in plasma using whole-genome sequencing, whole-genome copy number profiling (i.e. arrayCGH and OncoScan) [80,95,110] and dPCR [97,109]. dPCR is less prone to technical bias than qPCR for copy number measurement because it relies neither on PCR efficiency nor on the availability of a stable reference gene [125]. Aside from its non-invasive and repeatable nature, MYCN evaluation in cfDNA offers several other advantages over tissue-based methods. First, it enables rapid determination of MYCN status, which is particularly important for patients younger than 18 months at diagnosis where risk grouping is dependent on such information [126,127], and could be implemented in parallel with urine catecholamine tests. Second, the technique is low cost and amenable to a high-throughput format, requiring only DNA extraction from small blood volumes and subsequent PCR amplification of MYCN, alone or along with a reference gene such as NAGK [103,105–109].
3.1.2. Segmental chromosome alterations
Numerous SCAs detected in tumour tissue have been shown to correlate with prognosis in NB by univariate [14,54,128–134] and multivariate [135–137] analyses, and indeed the presence of any SCA is strongly associated with a higher risk of relapse and poorer outcome than in patients with only numerical chromosome alterations [57,138–141]. Several studies have demonstrated the ability to detect recurrent SCAs such as gain of 17q and losses of 11q and 1p in cfDNA isolated from plasma and serum of patients with NB at diagnosis, and have frequently shown high concordances of detection with paired tissue samples [80,110,112,113].
Combaret et al. [113] used a duplex qPCR approach to simultaneously amplify genes at 17q.23.1 and 17q25 and calculate copy numbers relative to TP53 (17p13.1) in cfDNA from patients with NB. With an arbitrary copy number threshold of 1.35, none of 16 cfDNA samples from healthy subjects showed 17q gain, whereas 38% (43/112) of cfDNA samples from patients with NB were positive. Of 58 patients with tissue-confirmed 17q gain, 31 were positive in cfDNA, while 12 of 84 patients with 17q normal tumours showed 17q gain. Consistent with the notion of rising cfDNA levels with disease progression [96,97], diagnostic sensitivity was greater in INSS stage 4 patients (60%) relative to stage 1 and 2 patients (33%), though specificity showed the inverse trend (71.4% versus 88%, respectively) and was, in fact, greater in patients younger than 18 months of age at diagnosis (94.4% versus 71.4%). Indeed, a similar trend was observed upon stratification by MYCN status. It is worth noting that the sensitivity of this assay is lower than that reported for detection of MNA in cfDNA (84–100% and 95–100%, respectively), likely due to the masking effect of healthy cfDNA on moderate 17q gains, particularly in patients with low tumour burden [113]. A recent whole-exome sequencing (WES) analysis in NB demonstrated a high concordance between tumour tissue and plasma cfDNA with respect to 17q status; of 19 patients, 12 patients showed 17q gain in both tissue and cfDNA at diagnosis, and one patient was positive for 17q gain in cfDNA only [110]. Similarly, an earlier study by the same group showed a high concordance of 17q gain between primary tumour tissue and cfDNA by analysis with array CGH and OncoScan microarray, respectively. Of 70 patients at diagnosis, 30 showed 17q gain in both tissue and cfDNA, and two patients showed 17q gain in either tissue or cfDNA [80]. Analysis of 17q gain in cfDNA may be useful as a tool for prognostication and therapeutic decision-making alongside cfDNA-based MYCN testing, particularly in very young patients in whom tissue-based genomic analyses may not be possible or sufficiently reliable.
Reflecting the clinical importance of 11q loss as a negative prognostic indicator in NB patients without MNA [142], several groups have reported the detection of 11q loss in plasma and serum from patients with NB at diagnosis and relapse [80,110,112]. Yagyu et al. [112] used a microsatellite analysis approach to determine allelic status at 11q23 in the serum of 24 patients at diagnosis. Serum allelic intensity scores between tissue-confirmed 11q loss-positive and loss-negative patients did not overlap, and there was full concordance of 11q status between tissue and serum in these patients. Both studies by Chicard et al. [80,110] also demonstrated 11q loss in plasma from patients at diagnosis using OncoScan microarray or WES with high concordance between tissue and plasma. The earlier study used OncoScan microarray and array CGH to analyse plasma and tumour DNA of 70 patients, respectively, and achieved a sensitivity of 91% and specificity of 94% [80]. In the later study of 19 patients, paired WES of tissue and plasma demonstrated 100% sensitivity and specificity [110]. Moreover, these studies were able to detect 1p loss in plasma to high concordance with matched tissue, along with other recurrent SCAs such as gains of 1q, 2p and losses of 3p, 4p and 14q [80,110]. Given that SCAs confer poor prognosis in NB and therefore define patients that require more intensive treatment, larger scale studies employing rapid and targeted methodologies for detecting specific SCAs in cfDNA at diagnosis are warranted.
3.1.3. ALK mutations and amplification
Gain-of-function alterations in ALK, namely point mutations and gene amplification, are observed in around 10–12% of patients with NB at diagnosis and tend to afford a poor prognosis [23,60,62]. Recently, small-molecule inhibitors of ALK currently used to treat ALK fusion-positive NSCLC have entered clinical trials for ALK-positive NB and other ALK-positive paediatric cancers [76]. Therefore, genomic alterations of ALK may serve as important biomarkers in cfDNA for therapeutic stratification and monitoring of treatment response in the near future. Combaret et al. [111] developed dPCR assays for the sensitive detection of point mutations at the two most common mutational hotspots of ALK in NB: F1174L (c.3520T > C and c.3522C > A) and R1275Q (c.3824G > A). Among 111 plasma/serum samples obtained from patients with NB at diagnosis, 20 patients were found to be positive for a single mutation and four patients were positive for both c.3522C > A and c.3824G > A. Mutant-to-wildtype ALK ratios ranged from 0.15% to 43.7%, likely reflecting different tumour burdens and disease stages within the cohort. Detection of the 3520T > C, 3522C > A and 3824G > A point mutations in cfDNA was achieved with a sensitivity of 100%, 85% and 92%, respectively, and a specificity of 100%, 91% and 98%, respectively. However, the specificity could have been underestimated due to potential spatial sampling bias when obtaining biopsies from genetically heterogeneous tumours [111,143]. ALK mutations have also been detected in cfDNA from patients with NB using targeted [97] and whole-exome [110] sequencing approaches, though these studies have used small patient cohorts. Lodrini et al. [109] recently developed a dPCR assay for quantification of ALK copy number and demonstrated its performance in cfDNA isolated from patients and patient-derived mouse xenografts. Of 10 patients in the study, one patient showed ALK gain (copy number greater than or equal to 3) in both cfDNA and tissue by dPCR and two patients showed ALK gain in cfDNA but not in tissue. The latter observation could have been due to ALK gains in subclones of the primary tumours or in metastatic sites undetected at diagnosis [109,143].
3.1.4. DNA methylation
Silencing of tumour suppressors by hypermethylation of promoter CpG islands is a key epigenetic event in tumorigenesis [144] and a promising circulating biomarker in diverse cancer types [145]. Several tumour suppressors are frequently silenced by hypermethylation in NB [146]. CpG island hypermethylation of the RAS effector protein RASSF1A is found in the majority of NB tumours at all clinical stages [146–149] and is not associated with prognostic factors such as MYCN status or age at diagnosis [148,150]. By contrast, RASSF1A hypermethylation in the pre-treatment serum of patients with NB has been shown to be significantly associated with age > 12 months at diagnosis and advanced (INSS stage 4) MNA disease [148]. In this study, Misawa et al. [148] used methylation-specific PCR and detected serum RASSF1A hypermethylation in 25% (17/68) of patients, demonstrating its utility as a poor prognostic factor comparable to that of MNA by univariate analysis. The same group also investigated the prognostic potential of serum DCR2 hypermethylation, given its association with poor outcome in primary NB tumours [114,151]. DCR2 hypermethylation was associated with tumour stage, independent of MYCN status, and patients with this alteration had a poorer 5 year EFS, which was particularly significant among patients without MNA [114]. Moreover, hypermethylation levels were found to decrease towards disease remission and become elevated at relapse, thus demonstrating the potential of serum DCR2 hypermethylation as a dynamic biomarker for prognostication and therapeutic monitoring in NB [114].
3.2. Circulating microRNA
MicroRNAs (miRNAs) are a family of endogenous, short (20–25 nt) non-coding RNA molecules that provide post-transcriptional regulation of gene expression [152]. Dysregulation of miRNAs is widely observed in cancer, often caused by mechanisms such as deletion, amplification and changes in gene expression [152]. Over the past decade, miRNAs have been investigated in plasma and serum as non-invasive biomarkers for diagnosis, therapeutic stratification and prognostication across diverse cancer types [153].
Many known oncogenic and tumour-suppressive miRNAs have been shown to be aberrantly expressed in primary NB tumours, with a particular focus on those targeting MYCN [154–156] among other genes implicated in NB pathogenesis [157]. In addition, a limited number of studies have demonstrated the roles of specific miRNAs in mediating chemoresistance in NB cell lines and tumours [157]. However, few studies have investigated the expression and clinical utility of circulating miRNAs in NB. Murray et al. [158] undertook a global (n = 741) reverse transcription (RT)-qPCR-based analysis of miRNA expression in diagnostic serum from 33 paediatric cancer patients and identified a unique expression profile for each tumour type. Sera from NB patients with MNA (n = 2) showed overexpression of miR-124-3p, miR-9-3p, miR-218-5p, miR-490-5p and miR-1538, consistent with a previous study demonstrating MYCN status as a determinant of global miRNA profiles in NB tissue [159]. Moreover, miR-9 is known to be induced by MYCN and its high expression in NB is associated with MNA and metastatic disease [160]. In a recent study, whole-miRNome profiling was conducted in sera from mice bearing favourable, non-metastatic NB xenografts and mice with high-risk, metastatic disease [161]. The authors identified a circulating miRNA signature of high-risk disease, comprising high and low expression of 34 and 46 miRNAs, respectively. Individual miRNAs were functionally validated by analysing expression of their putative target proteins at distant metastatic sites from the high-risk model, and three miRNAs (miR-381, miR-548h and miR-580) were identified as showing greater than 10-fold increased expression in sera from this model, thereby serving as putative biomarkers of high-risk disease for clinical investigation [161]. Another recent study investigated expression of miRNAs in pooled sera from patients with low- and high-risk NB, identifying 743 well-expressed miRNAs [162]. The authors then evaluated expression of these miRNAs in sera from a cohort of 141 patients with NB at diagnosis, identifying tumour stage as the greatest determinant of variance in miRNA levels. Nine miRNAs that showed significantly different expression between patients with low- and high-risk disease were further investigated, and their expression levels were found to increase with tumour stage. Moreover, in mice xenografted with NB tumours, serum expression levels of all nine miRNAs were found to increase with tumour load, an observation subsequently confirmed by longitudinal blood sampling in five patients with high-risk metastatic NB. Interestingly, expression of the nine miRNAs in primary NB tumours was not significantly different between disease stages, leading the authors to conclude that differential expression between disease stages in sera most likely reflects tumour burden and therefore metastatic status [162].
While these studies have provided proof of principle that circulating miRNA profiles can distinguish between favourable and high-risk NB, candidate miRNA biomarkers must be evaluated in large, prospective patient cohorts before their clinical utility can be realized. Moreover, given that miRNA profiles associated with chemoresistance have been identified in NB, further investigation of these miRNAs as circulating biomarkers for therapeutic monitoring is warranted [157].
3.3. CTCs and CTC-derived mRNA
CTCs are malignant cells that disseminate into the bloodstream from primary or metastatic tumours and are responsible for seeding metastatic growth [163]. Together with disseminated tumour cells in bone marrow, CTCs are surrogate markers of sub-clinical metastasis (MRD), in which small numbers of tumour cells persist after therapy in patients in remission, often leading to clinical relapse [164]. CTCs are rare, comprising as few as one cell per billion haematological cells, and this has presented a major challenge to their isolation and molecular characterization. Recent advancements in immunological and size-based enrichment methods have enabled enumeration and molecular analysis of CTCs in peripheral blood isolated from patients with diverse cancer types [165]. CTCs are currently under investigation as biomarkers for diagnosis, therapeutic monitoring, prognostication and assessment of relapse risk [166]. The presence of CTCs in patients with NB was first demonstrated through the establishment of NB cell lines during in vitro culture of peripheral blood samples from patients with disseminated disease [167,168]. These studies highlighted the potential for tumour cell contamination in stem cell harvests from peripheral blood, a concern supported by a later study demonstrating the clonogenic properties of NB CTCs in vitro [169].
3.3.1. CTC detection methods
The first prospective molecular analyses of CTCs employed immunocytochemical methods with monoclonal antibodies against neuronal cell markers such as CD56 (NCAM), CD90 (Thy-1) and GD2 [168,170,171]. CTCs were detected in patients with metastatic disease at diagnosis [168,170,171], during therapy [170,171] and at relapse [168]. In one study, the presence of CTCs in patients with metastatic disease during therapy was found to be an indicator of disease relapse [170]. Subsequent studies have used RT-PCR-based methodologies for the indirect detection of CTCs, targeting mRNA with neuron-specific expression such as UCH-L1 (PGP9.5) [172–175], TH (tyrosine hydroxylase) [173–207], GALGT (GD2 synthase) [200,204,208], DCX (doublecortin) [187,193,199,203–205,207], DDC (DOPA decarboxylase) [192,193,198,202,204,205] and PHOX2B [187,193,202,204,205,207]. Mattano et al. [172] evaluated the performance of an RT-PCR assay targeting UCH-L1, reporting a 100-fold increase in sensitivity relative to immunocytochemical assays, with the ability to detect a single CTC among 107 peripheral blood mononuclear cells (PBMCs) versus 1 or 2 CTCs among 105 PBMCs by immunocytochemistry. Subsequent RT-qPCR analyses targeting other markers such as TH and GALGT have demonstrated variable sensitivities, with detection limits ranging from 1 CTC in 103 to 1 CTC in 107 PBMCs [174,176,178,190,194–202,209].
3.3.2. Association of CTCs with clinical features
As with immunocytochemical analyses, RT-PCR-based studies have consistently identified NB-specific mRNA in peripheral blood at diagnosis in patients with metastatic disease and in a fraction of patients with localized and stage 4S disease (table 4). Diagnostic TH mRNA levels and CTC counts are typically higher in patients with more advanced metastatic disease [185,204,206,212] and in patients with high-risk disease [206,212], as observed in other solid malignancies [166]. However, NB-specific mRNA levels and CTC counts do not correlate with MYCN status [182,184,206,207,212]. In addition to serving as a potential diagnostic biomarker, there is a wealth of evidence supporting the prognostic value of NB-specific mRNA, with numerous studies reporting a correlation between mRNA levels and survival outcomes (table 4). High transcript levels of TH and other NB-specific genes at diagnosis have been associated with poor OS and/or EFS in patients with localized and metastatic disease, independent of risk status [187,193,194,197,203,206,208]. Similarly, high CTC counts (≥ 10 cells per 4 ml blood) have recently been shown to correlate with poor OS [212]. Among patients with high-risk disease, high expression of TH and PHOX2B mRNA at diagnosis has been shown to correlate with remarkably poorer outcome, thus identifying a subset of patients with ultra-high-risk disease who may benefit from novel treatment approaches [187].
Table 4.
Summary of studies reporting direct and indirect detection of CTCs in peripheral blood of patients with NB. CTC, circulating tumour cell; EFS, event-free survival; FISH, fluorescence in situ hybridization; ICC, immunocytochemistry; MRD, minimal residual disease; NB, neuroblastoma; ND, not determined; RT-(q)PCR, reverse transcription (quantitative real-time) PCR; INSS, International Neuroblastoma Staging System; OS, overall survival; PFS, progression-free survival.
| detection method(s) | marker(s) | sensitivity (tumour cell n/non-tumour cell n) | main observation(s) in peripheral blood | study |
|---|---|---|---|---|
| ICC | NCAM, unknown NB-specific antigen | ND | CTCs detected in patients with metastatic disease at diagnosis and relapse. | Lanino et al. [168] |
| ICC | GD2, Thy-1, unknown NB-specific antigen | 2/105 | CTCs detected in patients with metastatic disease at diagnosis and during therapy. The presence of CTCs in patients during therapy associated with disease relapse. |
Moss & Sanders [170] |
| ICC | GD2, Thy-1, unknown NB-specific antigen | ND | CTCs detected in patients with metastatic disease at diagnosis. CTCs detected during therapy in one patient. |
Sanders et al. [171] |
| ICC; RT-PCR | ICC: NCAM, GD2, Thy-1; RT-PCR: UCH-L1 | ICC: ND; RT-PCR: 1/107 | CTCs/NB-mRNA detected in patients with localized and metastatic disease, and in patients with no evidence of disease. RT-PCR more sensitive than ICC, though analytical sensitivity of ICC not determined. |
Mattano et al. [172] |
| ICC | NCAM, GD2, Thy-1 | ND | CTCs detected in patients with metastatic disease at diagnosis and during therapy. Around half of ICC-positive blood samples and several negative samples showed clonogenic growth of NB cells in vitro. |
Moss et al. [169] |
| RT-PCR | TH | 1–10/106 |
TH mRNA detected in patients with metastatic disease at diagnosis, with some patients undetectable 6–8 weeks into therapy. TH mRNA detected in patients with relapsed disease and patients in remission, one of whom later relapsed. |
Burchill et al. [176] |
| RT-PCR | TH | ND |
TH mRNA detected in patients with metastatic disease at diagnosis, with some patients undetectable 6–8 weeks into therapy. TH mRNA detected in patients with relapsed disease and patients in remission, some of whom later relapsed. Some patients in remission and with no detectable TH mRNA eventually relapsed. |
Burchill et al. [177] |
| RT-PCR | TH | 1/105–106 | TH mRNA detected in patients with localized and metastatic disease, including INSS stage 4S patients. | Miyajima et al. [178] |
| RT-PCR | TH | ND |
TH mRNA detected in patients with localized and metastatic disease, and in patients in remission. Significantly higher TH mRNA levels in samples taken before complete remission. |
Miyajima et al. [179] |
| RT-PCR | TH | ND | TH mRNA detected in a patient with metastatic disease and undetectable by conventional histology. | Kuroda et al. [180] |
| RT-PCR | TH, UCH-L1 | ND | NB-mRNA detected in patients of unknown tumour stage, some of whom tested negative following treatment. | Gao et al. [175] |
| RT-PCR | GAGE, TH | GAGE: 1/106; TH: ND | GAGE mRNA, but not TH mRNA, detected in some patients with metastatic disease at diagnosis. | Cheung & Cheung [190] |
| RT-PCR | TH, UCH-L1 |
TH: 10−4 µg RNA UCH-L1: 10−6 µg RNA |
NB-mRNA detected at diagnosis in patients aged 1 year or older with metastatic disease. RT-PCR detection of UCH-L1 more sensitive than that for TH. | Yanagisawa et al. [173] |
| ICC | GD2 | 1/106 | CTCs detected in patients with metastatic disease and INSS stage 4S disease at diagnosis. | Faulkner et al. [209] |
| RT-PCR | TH | 1 cell/2 ml blood | TH mRNA detected in patients with metastatic disease at diagnosis. | Burchill et al. [191] |
| RT-PCR | TH | ND | TH mRNA detected in patients with metastatic disease at diagnosis. | Kuroda et al. [181] |
| RT-PCR | TH | 1/106 | Detection of TH mRNA in patients older than 1 year with metastatic disease associated with poorer OS and EFS. Detection of TH mRNA in patients with no evidence of disease associated with increased risk of relapse and death. |
Burchill et al. [194] |
| ICC; RT-PCR | ICC: GD2; RT-PCR: TH, CHGA | GD2: 1/106; TH: 1/103; CHGA: 1/106 | TH RT-PCR less sensitive than CHGA RT-PCR and GD2 ICC. | Pagani et al. [195] |
| RT-qPCR | TH | 70 transcripts/1 ml blood | At diagnosis, TH mRNA levels significantly higher in patients with metastatic/INSS stage 4S disease than limited disease. Some patients positive for TH mRNA before, during and 24 h after surgery. |
Träger et al. [185] |
| RT-qPCR | TH | 1/106 | TH mRNA detected in patients with localized and metastatic diagnosis, and in a patient after seven months of chemotherapy. | Lambooy et al. [196] |
| RT-qPCR | TH | 1/106 | TH mRNA detected at diagnosis, during chemotherapy and at relapse in patients aged 1 year or older with metastatic disease. High frequency of tumour cell contamination in autologous PBSC harvests; high TH mRNA levels associated with poorer survival. | Tchirkov et al. [197] |
| ICC; RT-PCR | ICC: GD2; RT-PCR: TH, UCH-L1 | GD2: 1/106 TH: 1/105 UCH-L1: 1/106 |
Detection of CTCs by GD2 ICC at diagnosis in patients with localized disease correlated with unfavourable outcome. No correlation of TH mRNA status with outcome in patients with metastatic disease. GD2 ICC showed greatest accuracy and UCH-L1 RT-PCR showed poor accuracy. | Corrias et al. [174] |
| RT-PCR | GALGT | 1/106 | GALGT mRNA detected in patients with metastatic disease at diagnosis and associated with higher rates of relapse and death. | Cheung et al. [208] |
| RT-PCR | GAGE | ND | GAGE mRNA detected in high proportion of healthy subjects; thus, GAGE mRNA not specific for NB. | Oltra et al. [210] |
| RT-PCR | DDC, TH | 1/106 | DDC and TH mRNA detected at similar frequencies in patients with metastatic disease at diagnosis. | Bozzi et al. [198] |
| RT-PCR | DCX, TH | 1/105 |
DCX and TH mRNA detected at similar frequencies in patients with high-risk disease at diagnosis. High correlation between DCX and TH mRNA levels in positive samples. |
Oltra et al. [199] |
| RT-qPCR | TH | ND |
TH mRNA detected in patients with localized and metastatic disease at diagnosis. Detection of TH mRNA after therapy associated with poorer prognosis. |
Parareda et al. [183] |
| RT-PCR | TH | ND | TH mRNA detected in patients with metastatic disease at diagnosis, and all deaths in mRNA-positive patients related to systemic metastases. No association of TH mRNA positivity with MYCN status. | Kuroda et al. [184] |
| RT-PCR | TH, GALGT, ELAVL4 | TH: 1/106; GALGT: 1/104; ELAVL4: 1/106 | Persistence of high ELAVL4 mRNA levels during therapy has prognostic value in patients with metastatic disease. GALGT RT-PCR less sensitive than that for TH or ELAVL4. |
Swerts et al. [200] |
| RT-qPCR | TH | 1/106 | For CTC detection, blood should be collected into PAXgene or EDTA tubes and stored for 48 h or less. Implementation of standardized protocols increased sensitivity of TH RT-PCR from 58% to 90%. |
Viprey et al. [201] |
| RT-PCR | TH | ND |
TH mRNA detected in patients with metastatic disease during treatment, but not in patients with stage 3 disease. No association of TH mRNA with MYCN status. All mRNA-positive patients died of disseminated disease. |
Kuroda et al. [211] |
| RT-qPCR | TH, DDC, GD2S | ND | High NB-mRNA levels associated with poor prognosis in patients with various disease stages. RT-qPCR for GD2S less specific than that for TH and DDC. |
Träger et al. [192] |
| RT-qPCR | TH, DDC, DBH, PHOX2B |
TH: 1/106; DDC: 1/106–107; DBH: 1/107 PHOX2B: 1/106–107 |
Panel of RT-qPCR markers more sensitive than PHOX2B alone in detecting MRD. | Stutterheim et al. [202] |
| RT-qPCR | TH, DCX | ND | TH and/or DCX mRNA detected in some patients with localized disease and associated with poorer EFS but not OS. | Yáñez et al. [203] |
| RT-qPCR | TH, GALGT, DDC, DCX, ELAVL4, STX, PHOX2B | ND | All markers except PHOX2B expressed in healthy donors. TH mRNA levels significantly lower in patients with localized versus metastatic disease. Weak correlation between detection of different markers in patients with localized disease, but perfect agreement between TH mRNA and GALGT mRNA in patients with metastatic disease. | Corrias et al. [204] |
| RT-qPCR | CHRNA3, CRMP1, DBH, DCX, DDC, GABRB3, GAP43, ISL1, KIF1A, PHOX2B, TH | ND | Sensitivity of MRD detection is greater with multiple RT-qPCR markers. | Hartomo et al. [205] |
| RT-qPCR | TH, PHOX2B, DCX | ND | High levels of TH, PHOX2B or DCX mRNA at diagnosis in patients with metastatic disease strongly associated with poorer EFS and OS. High levels of TH and PHOX2B at diagnosis identify patients with ultra-high-risk disease who may benefit from novel treatment approaches. | Viprey et al. [187] |
| RT-qPCR | TH | ND | Higher TH mRNA levels at diagnosis in high-risk patients and patients with metastatic disease, but no association with MYCN status. Expression of TH mRNA at diagnosis correlated with poorer 5 year PFS, both independent of risk stage and also when assessed in high-risk patients. No significant correlations with EFS were made after induction therapy. | Lee et al. [206] |
| RT-qPCR | CHGA, DCX, DDC, PHOX2B, TH | ND | NB-mRNA detectable in patients with relapsed/refractory disease. Lower RT-qPCR ΔCT values correlated with poorer PFS, independent of disease stage or MYCN status. |
Marachelian et al. [193] |
| RT-qPCR | TH, PHOX2B, DCX | ND | Levels of PHOX2B and DCX mRNAs, but not TH mRNA, in the highest tertile were associated with shorter EFS in a cohort of high-risk patients with metastatic disease. Correlation of mRNA and EFS was independent of MYCN status. Prognostic power of PHOX2B mRNA lost when combined with TH mRNA. | Corrias et al. [207] |
| FISH | CD45, CEP8, DAPI | ND | CTCs identified by depletion of non-tumour cells with magnetic beads coated with anti-CD45 antibodies, followed by analysis of non-bound cells with CEP8 probe and DAPI. CTC = CD45-negative, DAPI-positive, CEP8 ≥ 3 spots. Significantly more CTCs identified in high-risk patients and patients with metastatic disease. Patients with greater than or equal to 3 CTCs per 4 ml blood had greater likelihood of having metastases, and patients with greater than or equal to 10 per 4 ml blood had poorer OS. No correlation of CTC count with MYCN status. Higher CTC counts in patients with partial response to therapy or progressive disease relative to patients with complete response. | Liu et al. [212] |
Detection of CTCs or NB-specific mRNA in peripheral blood of patients in remission following completion of therapy may indicate MRD and is associated with poorer prognosis in NB [183]. Consistent with the eradication of systemic disease, several studies have shown that mRNA may decrease and/or become undetectable during and after chemotherapy [175–177,212]. CTCs or detectable levels of mRNA that remain upon completion of chemotherapy or after surgical resection are associated with an increased risk of relapse [170,176,177,183,185,197,200]. Thus, mRNA in peripheral blood has potential as a dynamic biomarker for monitoring response to therapy and evaluation of relapse risk in patients with NB.
Several groups have demonstrated an increase in sensitivity of MRD detection in peripheral blood and bone marrow by evaluating multiple mRNAs simultaneously, which is perhaps expected given the genetic heterogeneity of NB [193,202,204,205]. However, some mRNAs such as GALGT have been shown to lack sensitivity [200] and contribute false positivity [189,192], and hence should be validated individually before incorporation into multi-marker panels. Corrias et al. [204] evaluated a panel of seven mRNAs and found no correlation of any individual mRNA with specific clinical features in patients with localized disease. Moreover, a high rate of false positivity was observed upon combined analysis of all mRNAs, leading the authors to conclude that multi-marker analysis may not offer benefit over analysis of a single mRNA in patients with low tumour burden [204].
Towards clinical implementation of mRNA-based MRD analysis in patients with NB, Viprey et al. [201] coordinated an initiative to standardize the methodology for detection of NB cells by RT-qPCR. Original methodologies on blood collection, RNA isolation and PCR protocols were evaluated from several reference laboratories across Europe and a standardized, quality-controlled protocol was devised. This protocol led to an increase in sensitivity of TH mRNA detection by RT-qPCR from 58% to 90% [201] and has since been implemented by the same group for MRD detection in patients with metastatic disease enrolled on the international phase 3 HR-NBL-1/SIOPEN trial [187,207]. In the first of two studies, Viprey et al. [187] reported that levels of TH, PHOX2B and DCX mRNAs in peripheral blood (and bone marrow) of patients at diagnosis and at the end of induction therapy were predictive of EFS. A recently published follow-up study evaluated levels of TH, PHOX2B and DCX mRNAs in patients < 18 months of age at diagnosis, since this age group was not sufficiently represented in the first study. In these patients, levels of PHOX2B and DCX mRNA, but not TH mRNA, in the highest tertile at diagnosis were associated with shorter EFS, and PHOX2B mRNA alone showed prognostic power in patients during the first year of follow-up [207]. Studies into MRD detection in patients with NB (table 4) have clearly demonstrated the potential for NB-mRNAs in peripheral blood as non-invasive predictive and prognostic biomarkers within defined patient subsets. The clinical significance of NB-mRNAs in peripheral blood is worthy of further exploration in comparison with established biomarkers in large multi-centre trials using standardized protocols before integration into the clinic [201,213].
4. Clinical implementation of circulating biomarkers in neuroblastoma
A major obstacle to the discovery and validation of clinically applicable biomarkers in NB, and indeed paediatric cancers in general, is the scarcity of patient material available for correlative research. There is a tendency to take core needle biopsies rather than tissue sections, which limits the number of tests that can be performed. In addition, protocols used for blood collection, plasma/serum isolation and specimen storage may not permit optimal recovery of biomarker analytes such as nucleic acids [119]; these methodological factors may be a significant cause of inter-study variability, as exemplified by the choice of plasma or serum for analysis of MNA in blood [102–109]. Furthermore, retrospective biomarker analysis requires access to patient clinicopathological data, which is not always possible or easy unless in the context of a clinical trial. Indeed, inferences made from prospective biomarker analyses in patients enrolled on clinical trials are often limited due to small cohort sizes that result from stratification of patients into treatment arms or into groups based on clinicopathological features such as age, risk status and disease stage [201,207]. Despite these limitations, it has been possible to develop prognostic assays.
There is sufficient retrospective evidence to indicate that analysis of MNA in blood by qPCR can determine MYCN status in the majority of NB patients with advanced disease, thus serving as a biomarker for prognostication and potentially response to therapy [102–109]. The most obvious benefit of blood-based MYCN analysis is the rapid determination of risk status in patients at diagnosis, enabling immediate application of appropriate treatment. Blood-based MYCN assessment should now be incorporated into large-scale prospective trials in patients at diagnosis, during/after induction therapy and at relapse. A limited number of retrospective studies have demonstrated successful detection of various SCAs in plasma and serum of patients with advanced disease [80,110,112,113]. Given that SCAs are currently used as tissue-based indicators of poor prognosis, further analysis of their detectability in blood in larger patient cohorts is warranted, perhaps in tandem with MYCN analysis [113]. The relatively ‘quiet’ nature of NB genomes has not provided a plethora of potential aberrations for the development of further blood-based assays, which has further confounded the implementation of this approach to patient management.
To date, ALK is the only druggable gene product that is frequently mutated in NB, and small-molecule inhibitors of ALK currently approved for the treatment of ALK-positive NSCLC are undergoing clinical assessment in patients with NB and other ALK-positive paediatric malignancies. However, a wealth of preclinical studies and a few clinical case reports in patients with NB are highlighting the issue of therapeutic resistance, thus compromising the long-term efficacy of these compounds [76]. The ability to detect ALK mutations in the blood at diagnosis and relapse, and to monitor mutations during treatment, would enable the rapid assignment of ALK inhibitors to eligible patients and to monitor treatment efficacy in real time. In particular, given that many children are too unwell for re-biopsy on relapse, an assay that can determine the identity of the ALK mutation on relapse would be highly beneficial in the clinic. For example, emergence of secondary ALK mutations in the blood would indicate resistance and may provide the rationale for switching to other structurally related compounds with distinct resistance profiles. Combaret et al. [111] have demonstrated the detection of mutations at the F1174 and R1275 hotspots of ALK in cfDNA of patients at diagnosis using a droplet digital PCR (ddPCR) methodology [111], and several other groups have detected point mutations in ALK by next-generation sequencing in small patient cohorts [97,110]. The utility of ALK mutations in cfDNA as therapeutic biomarkers will only be realized upon integration of cfDNA analysis into large, well-designed prospective trials that include an ALK inhibitor treatment arm, several of which are currently underway [214,215].
Another clinical application of circulating biomarkers is the detection of MRD, a strong predictor of relapse in cancer [164]. To this end, a variety of MRD detection methods have been developed based on direct and indirect detection of tumour cells in peripheral blood [216]. Early studies in patients with NB employed immunocytochemical methods to detect CTCs, and later studies revealed the increased sensitivity offered by qualitative and quantitative RT-PCR-based strategies targeting NB-specific mRNAs (table 4). Numerous retrospective analyses have amassed a substantial evidence base around the utility of qualitative and quantitative RT-PCR for detection of rare CTCs in NB, with a sensitivity of a single tumour cell in up to 10 million normal haematopoietic cells [172,202]. Of particular clinical interest is the ability of NB-specific mRNA levels in blood to predict OS and/or EFS outcomes among patients with metastatic disease, as determined in a series of small-cohort studies [183,192,193,197,200,203,206] and more recently in larger studies of patients enrolled on the HR-NBL-1/SIOPEN trial [187,207]. RT-qPCR-based MRD analysis must now be incorporated into well-powered prospective clinical trials to assess the predictive and prognostic value of NB-specific mRNA levels in blood, both independently and in combination with established biomarkers.
5. Summary
The poor prognosis of patients with high-risk NB necessitates the development of biomarkers to facilitate therapeutic stratification, prognostication and assessment of relapse risk. There is substantial evidence from retrospective studies to warrant investigation of MNA and CTC-derived mRNAs in peripheral blood of patients enrolled on large-scale prospective trials to confirm the clinical utility of these biomarkers in risk stratification and prognostic assessment, respectively. The detection of ALK mutations in cfDNA of patients enrolled on current ALK inhibitor trials will establish the feasibility of real-time non-invasive monitoring of treatment response and detection of resistance.
Supplementary Material
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O'Leary M, Smith FO, Reaman GH. 2010. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J. Clin. Oncol. 28, 2625–2634. ( 10.1200/JCO.2009.27.0421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spix C, Pastore G, Sankila R, Stiller CA, Steliarova-Foucher E. 2006. Neuroblastoma incidence and survival in European children (1978–1997): report from the Automated Childhood Cancer Information System project. Eur. J. Cancer 42, 2081–2091. ( 10.1016/j.ejca.2006.05.008) [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM. 2003. Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer 3, 203–216. ( 10.1038/nrc1014) [DOI] [PubMed] [Google Scholar]
- 4.Maris JM, Hogarty MD, Bagatell R, Cohn SL. 2007. Neuroblastoma. Lancet 369, 2106–2120. ( 10.1016/S0140-6736(07)60983-0) [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM, et al. 1993. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 11, 1466–1477. ( 10.1200/JCO.1993.11.8.1466) [DOI] [PubMed] [Google Scholar]
- 6.Brodeur GM, et al. 1988. International criteria for diagnosis, staging, and response to treatment in patients with neuroblastoma. J. Clin. Oncol. 6, 1874–1881. ( 10.1200/JCO.1988.6.12.1874) [DOI] [PubMed] [Google Scholar]
- 7.Cohn SL, et al. 2009. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J. Clin. Oncol. 27, 289–297. ( 10.1200/JCO.2008.16.6785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto NR, et al. 2015. Advances in risk classification and treatment strategies for neuroblastoma. J. Clin. Oncol. 33, 3008–3017. ( 10.1200/JCO.2014.59.4648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodeur GM, Bagatell R. 2014. Mechanisms of neuroblastoma regression. Nat. Rev. Clin. Oncol. 11, 704–713. ( 10.1038/nrclinonc.2014.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK, Hogarty M, COG Neuroblastoma Committee. 2013. Children's Oncology Group's 2013 blueprint for research: neuroblastoma. Pediatr. Blood Cancer 60, 985–993. ( 10.1002/pbc.24433) [DOI] [PubMed] [Google Scholar]
- 11.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. 1984. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124. ( 10.1126/science.6719137) [DOI] [PubMed] [Google Scholar]
- 12.Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D. 1985. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med. 313, 1111–1116. ( 10.1056/NEJM198510313131802) [DOI] [PubMed] [Google Scholar]
- 13.Maris JM, et al. 2001. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med. Pediatr. Oncol. 36, 32–36. () [DOI] [PubMed] [Google Scholar]
- 14.Attiyeh EF. et al. 2005. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 353, 2243–2253. ( 10.1056/NEJMoa052399) [DOI] [PubMed] [Google Scholar]
- 15.Bown N, et al. 2001. 17q gain in neuroblastoma predicts adverse clinical outcome. U.K. Cancer Cytogenetics Group and the U.K. Children's Cancer Study Group. Med. Pediatr. Oncol. 36, 14–19. () [DOI] [PubMed] [Google Scholar]
- 16.Janoueix-Lerosey I, et al. 2000. Molecular analysis of chromosome arm 17q gain in neuroblastoma. Genes Chromosomes Cancer 28, 276–284. () [DOI] [PubMed] [Google Scholar]
- 17.Weiser D, Laudenslager M, Rappaport E, Carpenter E, Attiyeh EF, Diskin S, London WB, Maris JM, Mosse YP. 2011. Stratification of patients with neuroblastoma for targeted ALK inhibitor therapy. J. Clin. Oncol. 29 ( 10.1200/jco.2011.29.15_suppl.9514) [DOI] [Google Scholar]
- 18.Miyake I, Hakomori Y, Shinohara A, Gamou T, Saito M, Iwamatsu A, Sakai R. 2002. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene 21, 5823–5834. ( 10.1038/sj.onc.1205735) [DOI] [PubMed] [Google Scholar]
- 19.Osajima-Hakomori Y, Miyake I, Ohira M, Nakagawara A, Nakagawa A, Sakai R. 2005. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am. J. Pathol. 167, 213–222. ( 10.1016/S0002-9440(10)62966-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molenaar JJ, et al. 2012. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593. ( 10.1038/nature10910) [DOI] [PubMed] [Google Scholar]
- 21.Boeva V, et al. 2013. Breakpoint features of genomic rearrangements in neuroblastoma with unbalanced translocations and chromothripsis. PLoS ONE 8, e72182 ( 10.1371/journal.pone.0072182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosse YP, et al. 2008. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455, 930–935. ( 10.1038/nature07261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janoueix-Lerosey I, et al. 2008. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455, 967–970. ( 10.1038/nature07398) [DOI] [PubMed] [Google Scholar]
- 24.Cheung NK, et al. 2012. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307, 1062–1071. ( 10.1001/jama.2012.228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugh TJ, et al. 2013. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 45, 279–284. ( 10.1038/ng.2529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sausen M, et al. 2013. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 45, 12–17. ( 10.1038/ng.2493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colon NC, Chung DH. 2011. Neuroblastoma. Adv. Pediatr. 58, 297–311. ( 10.1016/j.yapd.2011.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Chung DH. 2006. Pediatric solid malignancies: neuroblastoma and Wilms' tumor. Surg. Clin. North Am. 86, 469–487. ( 10.1016/j.suc.2005.12.008) [DOI] [PubMed] [Google Scholar]
- 29.Papaioannou G, McHugh K. 2005. Neuroblastoma in childhood: review and radiological findings. Cancer Imaging 5, 116–127. ( 10.1102/1470-7330.2005.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qualman SJ, et al. 2005. Protocol for the examination of specimens from patients with neuroblastoma and related neuroblastic tumors. Arch. Pathol. Lab. Med. 129, 874–883. ( 10.1043/1543-2165(2005)129[874:PFTEOS]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 31.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA. 2016. Neuroblastoma. Nat. Rev. Dis. Primers. 2, 16078 ( 10.1038/nrdp.2016.78) [DOI] [PubMed] [Google Scholar]
- 32.Sung KW. 2012. Treatment of high-risk neuroblastoma. Korean J. Pediatr. 55, 115–120. ( 10.3345/kjp.2012.55.4.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds CP. 2004. Detection and treatment of minimal residual disease in high-risk neuroblastoma. Pediatr. Transplant. 8(Suppl. 5), 56–66. ( 10.1111/j.1398-2265.2004.00216.x) [DOI] [PubMed] [Google Scholar]
- 34.Laug WE, Siegel SE, Shaw KN, Landing B, Baptista J, Gutenstein M. 1978. Initial urinary catecholamine metabolite concentrations and prognosis in neuroblastoma. Pediatrics 62, 77–83. ( 10.1203/00006450-197610000-00158) [DOI] [PubMed] [Google Scholar]
- 35.LaBrosse EH, Com-Nougue C, Zucker JM, Comoy E, Bohuon C, Lemerle J, Schweisguth O. 1980. Urinary excretion of 3-methoxy-4-hydroxymandelic acid and 3-methoxy-4-hydroxyphenylacetic acid by 288 patients with neuroblastoma and related neural crest tumors. Cancer Res. 40, 1995–2001. [PubMed] [Google Scholar]
- 36.Strenger V, Kerbl R, Dornbusch HJ, Ladenstein R, Ambros PF, Ambros IM, Urban C. 2007. Diagnostic and prognostic impact of urinary catecholamines in neuroblastoma patients. Pediatr. Blood Cancer 48, 504–509. ( 10.1002/pbc.20888) [DOI] [PubMed] [Google Scholar]
- 37.Cangemi G, et al. 2012. Prognostic value of ferritin, neuron-specific enolase, lactate dehydrogenase, and urinary and plasmatic catecholamine metabolites in children with neuroblastoma. Onco Targets Ther. 5, 417–423. ( 10.2147/OTT.S36366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond JV. 1975. Clinical significance of catecholamine excretion levels in diagnosis and treatment of neuroblastoma. Arch. Dis. Child. 50, 691–695. ( 10.1136/adc.50.9.691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verly IR, et al. 2017. Catecholamines profiles at diagnosis: increased diagnostic sensitivity and correlation with biological and clinical features in neuroblastoma patients. Eur. J. Cancer 72, 235–243. ( 10.1016/j.ejca.2016.12.002) [DOI] [PubMed] [Google Scholar]
- 40.Thompson D, Vo KT, London WB, Fischer M, Ambros PF, Nakagawara A, Brodeur GM, Matthay KK, DuBois SG. 2016. Identification of patient subgroups with markedly disparate rates of MYCN amplification in neuroblastoma: a report from the International Neuroblastoma Risk Group project. Cancer 122, 935–945. ( 10.1002/cncr.29848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgenstern DA, et al. 2016. Prognostic significance of pattern and burden of metastatic disease in patients with stage 4 neuroblastoma: a study from the International Neuroblastoma Risk Group database. Eur. J. Cancer 65, 1–10. ( 10.1016/j.ejca.2016.06.005) [DOI] [PubMed] [Google Scholar]
- 42.Zeltzer PM, Marangos PJ, Parma AM, Sather H, Dalton A, Hammond D, Siegel SE, Seeger RC. 1983. Raised neuron-specific enolase in serum of children with metastatic neuroblastoma. A report from the Children's Cancer Study Group. Lancet 2, 361–363. ( 10.1016/S0140-6736(83)90342-2) [DOI] [PubMed] [Google Scholar]
- 43.Nobels FR, Kwekkeboom DJ, Coopmans W, Schoenmakers CH, Lindemans J, De Herder WW, Krenning EP, Bouillon R, Lamberts SW. 1997. Chromogranin A as serum marker for neuroendocrine neoplasia: comparison with neuron-specific enolase and the alpha-subunit of glycoprotein hormones. J. Clin. Endocrinol. Metab. 82, 2622–2628. ( 10.1210/jcem.82.8.4145) [DOI] [PubMed] [Google Scholar]
- 44.Zeltzer PM, Marangos PJ, Evans AE, Schneider SL. 1986. Serum neuron-specific enolase in children with neuroblastoma. Relationship to stage and disease course. Cancer 57, 1230–1234. () [DOI] [PubMed] [Google Scholar]
- 45.Tsuchida Y, et al. 1987. Serial determination of serum neuron-specific enolase in patients with neuroblastoma and other pediatric tumors. J. Pediatr. Surg. 22, 419–424. ( 10.1016/S0022-3468(87)80261-0) [DOI] [PubMed] [Google Scholar]
- 46.Massaron S, et al. 1998. Neuron-specific enolase evaluation in patients with neuroblastoma. Tumour Biol. 19, 261–268. ( 10.1159/000030016) [DOI] [PubMed] [Google Scholar]
- 47.Hann HW, Stahlhut MW, Evans AE. 1986. Source of increased ferritin in neuroblastoma: studies with concanavalin A-sepharose binding. J. Natl Cancer Inst. 76, 1031–1033. [PubMed] [Google Scholar]
- 48.Hann HW, Evans AE, Siegel SE, Wong KY, Sather H, Dalton A, Hammond D, Seeger RC. 1985. Prognostic importance of serum ferritin in patients with stages III and IV neuroblastoma: the Childrens Cancer Study Group experience. Cancer Res. 45, 2843–2848. [PubMed] [Google Scholar]
- 49.Meany HJ, et al. 2014. Significance of clinical and biologic features in stage 3 neuroblastoma: a report from the International Neuroblastoma Risk Group project. Pediatr. Blood Cancer 61, 1932–1939. ( 10.1002/pbc.25134) [DOI] [PubMed] [Google Scholar]
- 50.Hann HW, Evans AE, Cohen IJ, Leitmeyer JE. 1981. Biologic differences between neuroblastoma stages IV-S and IV. Measurement of serum ferritin and E-rosette inhibition in 30 children. N. Engl. J. Med. 305, 425–429. ( 10.1056/NEJM198108203050803) [DOI] [PubMed] [Google Scholar]
- 51.Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. 1983. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305, 245–248. ( 10.1038/305245a0) [DOI] [PubMed] [Google Scholar]
- 52.Shimada H, Stram DO, Chatten J, Joshi VV, Hachitanda Y, Brodeur GM, Lukens JN, Matthay KK, Seeger RC. 1995. Identification of subsets of neuroblastomas by combined histopathologic and N-myc analysis. J. Natl Cancer Inst. 87, 1470–1476. ( 10.1093/jnci/87.19.1470) [DOI] [PubMed] [Google Scholar]
- 53.Tonini GP, et al. 1997. MYCN oncogene amplification in neuroblastoma is associated with worse prognosis, except in stage 4S: the Italian experience with 295 children. J. Clin. Oncol. 15, 85–93. ( 10.1200/JCO.1997.15.1.85) [DOI] [PubMed] [Google Scholar]
- 54.Bown N, et al. 1999. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N. Engl. J. Med. 340, 1954–1961. ( 10.1056/NEJM199906243402504) [DOI] [PubMed] [Google Scholar]
- 55.George RE, et al. 2007. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS ONE 2, e255 ( 10.1371/journal.pone.0000255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T. 2010. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl Acad. Sci. USA 107, 4323–4328. ( 10.1073/pnas.0910684107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schleiermacher G, et al. 2007. Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br. J. Cancer 97, 238–246. ( 10.1038/sj.bjc.6603820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abel F, Ejeskar K, Kogner P, Martinsson T. 1999. Gain of chromosome arm 17q is associated with unfavourable prognosis in neuroblastoma, but does not involve mutations in the somatostatin receptor 2(SSTR2) gene at 17q24. Br. J. Cancer 81, 1402–1409. ( 10.1038/sj.bjc.6692231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lastowska M, Cotterill S, Pearson AD, Roberts P, McGuckin A, Lewis I, Bown N. 1997. Gain of chromosome arm 17q predicts unfavourable outcome in neuroblastoma patients. U.K. Children's Cancer Study Group and the U.K. Cancer Cytogenetics Group. Eur. J. Cancer 33, 1627–1633. ( 10.1016/S0959-8049(97)00282-7) [DOI] [PubMed] [Google Scholar]
- 60.De Brouwer S, et al. 2010. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin. Cancer Res. 16, 4353–4362. ( 10.1158/1078-0432.CCR-09-2660) [DOI] [PubMed] [Google Scholar]
- 61.Zhu S, et al. 2012. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 21, 362–373. ( 10.1016/j.ccr.2012.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bresler SC, et al. 2014. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 26, 682–694. ( 10.1016/j.ccell.2014.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M, Zhou C, Sun Q, Cai R, Li Y, Wang D, Gong L. 2013. ALK amplification and protein expression predict inferior prognosis in neuroblastomas. Exp. Mol. Pathol. 95, 124–130. ( 10.1016/j.yexmp.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 64.Greer M, Anton AH, Williams CM, Echevarria RA. 1965. Tumors of neural crest origin; biochemical and pathological correlation. Arch. Neurol. 13, 139–148. ( 10.1001/archneur.1965.00470020029004) [DOI] [PubMed] [Google Scholar]
- 65.Sawada T. 1992. Past and future of neuroblastoma screening in Japan. Am. J. Pediatr. Hematol. Oncol. 14, 320–326. ( 10.1097/00043426-199211000-00007) [DOI] [PubMed] [Google Scholar]
- 66.Tsubono Y, Hisamichi S. 2004. A halt to neuroblastoma screening in Japan. N. Engl. J. Med. 350, 2010–2011. ( 10.1056/NEJM200405063501922) [DOI] [PubMed] [Google Scholar]
- 67.Schilling FH, et al. 2002. Neuroblastoma screening at one year of age. N. Engl. J. Med. 346, 1047–1053. ( 10.1056/NEJMoa012277) [DOI] [PubMed] [Google Scholar]
- 68.Woods WG, et al. 2002. Screening of infants and mortality due to neuroblastoma. N. Engl. J. Med. 346, 1041–1046. ( 10.1056/NEJMoa012387) [DOI] [PubMed] [Google Scholar]
- 69.Hero B, et al. 2008. Localized infant neuroblastomas often show spontaneous regression: results of the prospective trials NB95-S and NB97. J. Clin. Oncol. 26, 1504–1510. ( 10.1200/JCO.2007.12.3349) [DOI] [PubMed] [Google Scholar]
- 70.Fritsch P, Kerbl R, Lackner H, Urban C. 2004. ‘Wait and see’ strategy in localized neuroblastoma in infants: an option not only for cases detected by mass screening. Pediatr. Blood Cancer 43, 679–682. ( 10.1002/pbc.20126) [DOI] [PubMed] [Google Scholar]
- 71.Goodman SN. 1991. Neuroblastoma screening data. An epidemiologic analysis. Am. J. Dis. Child. 145, 1415–1422. ( 10.1001/archpedi.1991.02160120083024) [DOI] [PubMed] [Google Scholar]
- 72.Wulaningsih W, et al. 2015. Serum lactate dehydrogenase and survival following cancer diagnosis. Br. J. Cancer 113, 1389–1396. ( 10.1038/bjc.2015.361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrelli F, Cabiddu M, Coinu A, Borgonovo K, Ghilardi M, Lonati V, Barni S. 2015. Prognostic role of lactate dehydrogenase in solid tumors: a systematic review and meta-analysis of 76 studies. Acta. Oncol. 54, 961–970. ( 10.3109/0284186X.2015.1043026) [DOI] [PubMed] [Google Scholar]
- 74.Morgenstern DA, et al. 2018. Risk stratification of high-risk metastatic neuroblastoma: a report from the HR-NBL-1/SIOPEN study. Pediatr. Blood Cancer 65, e27363 ( 10.1002/pbc.27363) [DOI] [PubMed] [Google Scholar]
- 75.Isgro MA, Bottoni P, Scatena R. 2015. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv. Exp. Med. Biol. 867, 125–143. ( 10.1007/978-94-017-7215-0_9) [DOI] [PubMed] [Google Scholar]
- 76.Trigg RM, Turner SD. 2018. ALK in neuroblastoma: biological and therapeutic implications. Cancers (Basel) 10, 113 ( 10.3390/cancers10040113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henry NL, Hayes DF. 2012. Cancer biomarkers. Mol. Oncol. 6, 140–146. ( 10.1016/j.molonc.2012.01.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schramm A, et al. 2015. Mutational dynamics between primary and relapse neuroblastomas. Nat. Genet. 47, 872–877. ( 10.1038/ng.3349) [DOI] [PubMed] [Google Scholar]
- 79.Bellini A, et al. 2015. Deep sequencing reveals occurrence of subclonal ALK mutations in neuroblastoma at diagnosis. Clin. Cancer Res. 21, 4913–4921. ( 10.1158/1078-0432.CCR-15-0423) [DOI] [PubMed] [Google Scholar]
- 80.Chicard M, et al. 2016. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin. Cancer Res. 22, 5564–5573. ( 10.1158/1078-0432.CCR-16-0500) [DOI] [PubMed] [Google Scholar]
- 81.Rapisuwon S, Vietsch EE, Wellstein A. 2016. Circulating biomarkers to monitor cancer progression and treatment. Comput. Struct. Biotechnol. J. 14, 211–222. ( 10.1016/j.csbj.2016.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. 2017. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238. ( 10.1038/nrc.2017.7) [DOI] [PubMed] [Google Scholar]
- 83.Perakis S, Speicher MR. 2017. Emerging concepts in liquid biopsies. BMC Med. 15, 75 ( 10.1186/s12916-017-0840-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siravegna G, Marsoni S, Siena S, Bardelli A. 2017. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548. ( 10.1038/nrclinonc.2017.14) [DOI] [PubMed] [Google Scholar]
- 85.Malapelle U, Sirera R, Jantus-Lewintre E, Reclusa P, Calabuig-Farinas S, Blasco A, Pisapia P, Rolfo C, Camps C. 2017. Profile of the Roche cobas(R) EGFR mutation test v2 for non-small cell lung cancer. Expert Rev. Mol. Diagn. 17, 209–215. ( 10.1080/14737159.2017.1288568) [DOI] [PubMed] [Google Scholar]
- 86.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. 2004. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904. ( 10.1158/1078-0432.CCR-04-0378) [DOI] [PubMed] [Google Scholar]
- 87.Mandel P, Metais P. 1948. Les acides nucléiques du plasma sanguin chez l'homme. C. R. Seances Soc. Biol. Fil. 142, 241–243. [PubMed] [Google Scholar]
- 88.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. 1977. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650. [PubMed] [Google Scholar]
- 89.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. 1994. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomarkers Prev. 3, 67–71. ( 10.1016/0169-5002(94)93830-x) [DOI] [PubMed] [Google Scholar]
- 90.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. 1994. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 86, 774–779. ( 10.1111/j.1365-2141.1994.tb04828.x) [DOI] [PubMed] [Google Scholar]
- 91.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. 1998. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 17, 89–97. ( 10.1097/00006676-199807000-00012) [DOI] [PubMed] [Google Scholar]
- 92.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. 2001. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665. [PubMed] [Google Scholar]
- 93.Jiang P, et al. 2015. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl Acad. Sci. USA 112, E1317–E1325. ( 10.1073/pnas.1500076112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bettegowda C, et al. 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 ( 10.1126/scitranslmed.3007094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Roy N, et al. 2017. Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clin. Cancer Res. 23, 6305–6314. ( 10.1158/1078-0432.CCR-17-0675) [DOI] [PubMed] [Google Scholar]
- 96.Wang X, et al. 2018. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 7, 3022–3030. ( 10.1002/cam4.1586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kurihara S, Ueda Y, Onitake Y, Sueda T, Ohta E, Morihara N, Hirano S, Irisuna F, Hiyama E. 2015. Circulating free DNA as non-invasive diagnostic biomarker for childhood solid tumors. J Pediatr. Surg. 50, 2094–2097. ( 10.1016/j.jpedsurg.2015.08.033) [DOI] [PubMed] [Google Scholar]
- 98.Rumore PM, Steinman CR. 1990. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J. Clin. Invest. 86, 69–74. ( 10.1172/JCI114716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. 2006. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit. Care. 10, R60 ( 10.1186/cc4894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rainer TH, Wong LK, Lam W, Yuen E, Lam NY, Metreweli C, Lo YM. 2003. Prognostic use of circulating plasma nucleic acid concentrations in patients with acute stroke. Clin. Chem. 49, 562–569. ( 10.1373/49.4.562) [DOI] [PubMed] [Google Scholar]
- 101.Klega K, et al. 2018. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis. Oncol. 2, 1–13. ( 10.1200/PO.17.00285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Combaret V, Audoynaud C, Iacono I, Favrot MC, Schell M, Bergeron C, Puisieux A. 2002. Circulating MYCN DNA as a tumor-specific marker in neuroblastoma patients. Cancer Res. 62, 3646–3648. [PubMed] [Google Scholar]
- 103.Gotoh T, et al. 2005. Prediction of MYCN amplification in neuroblastoma using serum DNA and real-time quantitative polymerase chain reaction. J. Clin. Oncol. 23, 5205–5210. ( 10.1200/JCO.2005.02.014) [DOI] [PubMed] [Google Scholar]
- 104.Combaret V, Bergeron C, Noguera R, Iacono I, Puisieux A. 2005. Circulating MYCN DNA predicts MYCN-amplification in neuroblastoma. J. Clin. Oncol. 23, 8919–8920; author reply 8920 ( 10.1200/JCO.2005.04.0170) [DOI] [PubMed] [Google Scholar]
- 105.Combaret V, et al. 2009. Influence of neuroblastoma stage on serum-based detection of MYCN amplification. Pediatr. Blood Cancer 53, 329–331. ( 10.1002/pbc.22009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kojima M, Hiyama E, Fukuba I, Yamaoka E, Ueda Y, Onitake Y, Kurihara S, Sueda T. 2013. Detection of MYCN amplification using blood plasma: noninvasive therapy evaluation and prediction of prognosis in neuroblastoma. Pediatr. Surg. Int. 29, 1139–1145. ( 10.1007/s00383-013-3374-9) [DOI] [PubMed] [Google Scholar]
- 107.Yagyu S, et al. 2016. Serum-based quantification of MYCN gene amplification in young patients with neuroblastoma: potential utility as a surrogate biomarker for neuroblastoma. PLoS ONE 11, e0161039 ( 10.1371/journal.pone.0161039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma Y, Lee JW, Park SJ, Yi ES, Choi YB, Yoo KH, Sung KW, Koo HH. 2016. Detection of MYCN amplification in serum DNA using conventional polymerase chain reaction. J. Korean Med. Sci. 31, 1392–1396. ( 10.3346/jkms.2016.31.9.1392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lodrini M, et al. 2017. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget 8, 85 234–85 251. ( 10.18632/oncotarget.19076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chicard M, et al. 2018. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin. Cancer Res. 24, 939–949. ( 10.1158/1078-0432.CCR-17-1586) [DOI] [PubMed] [Google Scholar]
- 111.Combaret V, et al. 2015. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med. 4, 540–550. ( 10.1002/cam4.414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yagyu S, et al. 2011. Preoperative analysis of 11q loss using circulating tumor-released DNA in serum: a novel diagnostic tool for therapy stratification of neuroblastoma. Cancer Lett. 309, 185–189. ( 10.1016/j.canlet.2011.05.032) [DOI] [PubMed] [Google Scholar]
- 113.Combaret V, Brejon S, Iacono I, Schleiermacher G, Pierron G, Ribeiro A, Bergeron C, Marabelle A, Puisieux A. 2011. Determination of 17q gain in patients with neuroblastoma by analysis of circulating DNA. Pediatr. Blood Cancer 56, 757–761. ( 10.1002/pbc.22816) [DOI] [PubMed] [Google Scholar]
- 114.Yagyu S, et al. 2008. Circulating methylated-DCR2 gene in serum as an indicator of prognosis and therapeutic efficacy in patients with MYCN nonamplified neuroblastoma. Clin. Cancer Res. 14, 7011–7019. ( 10.1158/1078-0432.CCR-08-1249) [DOI] [PubMed] [Google Scholar]
- 115.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. 1997. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 16, 2985–2995. ( 10.1093/emboj/16.11.2985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zaizen Y, Taniguchi S, Noguchi S, Suita S. 1993. The effect of N-myc amplification and expression on invasiveness of neuroblastoma cells. J. Pediatr. Surg. 28, 766–769. ( 10.1016/0022-3468(93)90321-B) [DOI] [PubMed] [Google Scholar]
- 117.Goodman LA, Liu BC, Thiele CJ, Schmidt ML, Cohn SL, Yamashiro JM, Pai DS, Ikegaki N, Wada RK. 1997. Modulation of N-myc expression alters the invasiveness of neuroblastoma. Clin. Exp. Metastasis 15, 130–139. ( 10.1023/A:1018448710006) [DOI] [PubMed] [Google Scholar]
- 118.Ambros PF, et al. 2009. International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br. J. Cancer 100, 1471–1482. ( 10.1038/sj.bjc.6605014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trigg RM, Martinson LJ, Parpart-Li S, Shaw JA. 2018. Factors that influence quality and yield of circulating-free DNA: a systematic review of the methodology literature. Heliyon 4, e00699 ( 10.1016/j.heliyon.2018.e00699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. 2002. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 48, 421–427. [PubMed] [Google Scholar]
- 121.Lee TH, Montalvo L, Chrebtow V, Busch MP. 2001. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 41, 276–282. ( 10.1046/j.1537-2995.2001.41020276.x) [DOI] [PubMed] [Google Scholar]
- 122.Jung M, Klotzek S, Lewandowski M, Fleischhacker M, Jung K. 2003. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin. Chem. 49, 1028–1029. ( 10.1373/49.6.1028) [DOI] [PubMed] [Google Scholar]
- 123.Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, Gerbing RB, London WB, Villablanca JG. 2009. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a Children's Oncology Group study. J. Clin. Oncol. 27, 1007–1013. ( 10.1200/JCO.2007.13.8925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schneiderman J, London WB, Brodeur GM, Castleberry RP, Look AT, Cohn SL. 2008. Clinical significance of MYCN amplification and ploidy in favorable-stage neuroblastoma: a report from the Children's Oncology Group. J. Clin. Oncol. 26, 913–918. ( 10.1200/JCO.2007.13.9493) [DOI] [PubMed] [Google Scholar]
- 125.Olmedillas-Lopez S, Garcia-Arranz M, Garcia-Olmo D. 2017. Current and emerging applications of droplet digital PCR in oncology. Mol. Diagn. Ther. 21, 493–510. ( 10.1007/s40291-017-0278-8) [DOI] [PubMed] [Google Scholar]
- 126.Bagatell R, et al. 2009. Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J. Clin. Oncol. 27, 365–370. ( 10.1200/JCO.2008.17.9184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iehara T, et al. 2006. MYCN gene amplification is a powerful prognostic factor even in infantile neuroblastoma detected by mass screening. Br. J. Cancer 94, 1510–1515. ( 10.1038/sj.bjc.6603149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maris JM, White PS, Beltinger CP, Sulman EP, Castleberry RP, Shuster JJ, Look AT, Brodeur GM. 1995. Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res. 55, 4664–4669. [PubMed] [Google Scholar]
- 129.Caron H, et al. 1996. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N. Engl. J. Med. 334, 225–230. ( 10.1056/NEJM199601253340404) [DOI] [PubMed] [Google Scholar]
- 130.Schleiermacher G, et al. 1996. Clinical relevance of loss heterozygosity of the short arm of chromosome 1 in neuroblastoma: a single-institution study. Int. J. Cancer 69, 73–78. () [DOI] [PubMed] [Google Scholar]
- 131.Spitz R, Hero B, Ernestus K, Berthold F. 2003. Deletions in chromosome arms 3p and 11q are new prognostic markers in localized and 4S neuroblastoma. Clin. Cancer Res. 9, 52–58. [PubMed] [Google Scholar]
- 132.Pezzolo A, et al. 2009. Presence of 1q gain and absence of 7p gain are new predictors of local or metastatic relapse in localized resectable neuroblastoma. Neuro. Oncol. 11, 192–200. ( 10.1215/15228517-2008-086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jeison M, et al. 2010. 2p24 Gain region harboring MYCN gene compared with MYCN amplified and nonamplified neuroblastoma: biological and clinical characteristics. Am. J. Pathol. 176, 2616–2625. ( 10.2353/ajpath.2010.090624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pinto N, et al. 2016. Segmental chromosomal aberrations in localized neuroblastoma can be detected in formalin-fixed paraffin-embedded tissue samples and are associated with recurrence. Pediatr. Blood Cancer 63, 1019–1023. ( 10.1002/pbc.25934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lastowska M, et al. 2001. Comprehensive genetic and histopathologic study reveals three types of neuroblastoma tumors. J. Clin. Oncol. 19, 3080–3090. ( 10.1200/JCO.2001.19.12.3080) [DOI] [PubMed] [Google Scholar]
- 136.Vandesompele J, et al. 2005. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J. Clin. Oncol. 23, 2280–2299. ( 10.1200/JCO.2005.06.104) [DOI] [PubMed] [Google Scholar]
- 137.Uryu K, et al. 2017. Identification of the genetic and clinical characteristics of neuroblastomas using genome-wide analysis. Oncotarget 8, 107 513–107 529. ( 10.18632/oncotarget.22495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Defferrari R, et al. 2015. Influence of segmental chromosome abnormalities on survival in children over the age of 12 months with unresectable localised peripheral neuroblastic tumours without MYCN amplification. Br. J. Cancer 112, 290–295. ( 10.1038/bjc.2014.557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schleiermacher G, et al. 2010. Accumulation of segmental alterations determines progression in neuroblastoma. J. Clin. Oncol. 28, 3122–3130. ( 10.1200/JCO.2009.26.7955) [DOI] [PubMed] [Google Scholar]
- 140.Schleiermacher G, et al. 2011. Segmental chromosomal alterations lead to a higher risk of relapse in infants with MYCN-non-amplified localised unresectable/disseminated neuroblastoma (a SIOPEN collaborative study). Br. J. Cancer 105, 1940–1948. ( 10.1038/bjc.2011.472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schleiermacher G, et al. 2012. Segmental chromosomal alterations have prognostic impact in neuroblastoma: a report from the INRG project. Br. J. Cancer 107, 1418–1422. ( 10.1038/bjc.2012.375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Luttikhuis ME, Powell JE, Rees SA, Genus T, Chughtai S, Ramani P, Mann JR, McConville CM. 2001. Neuroblastomas with chromosome 11q loss and single copy MYCN comprise a biologically distinct group of tumours with adverse prognosis. Br. J. Cancer 85, 531–537. ( 10.1054/bjoc.2001.1960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heitzer E, Ulz P, Geigl JB. 2015. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 61, 112–123. ( 10.1373/clinchem.2014.222679) [DOI] [PubMed] [Google Scholar]
- 144.Nebbioso A, Tambaro FP, Dell'Aversana C, Altucci L. 2018. Cancer epigenetics: moving forward. PLoS Genet. 14, e1007362 ( 10.1371/journal.pgen.1007362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zeng H, He B, Yi C, Peng J. 2018. Liquid biopsies: DNA methylation analyses in circulating cell-free DNA. J. Genet. Genomics 45, 185–192. ( 10.1016/j.jgg.2018.02.007) [DOI] [PubMed] [Google Scholar]
- 146.Michalowski MB, de Fraipont F, Plantaz D, Michelland S, Combaret V, Favrot MC. 2008. Methylation of tumor-suppressor genes in neuroblastoma: the RASSF1A gene is almost always methylated in primary tumors. Pediatr. Blood Cancer 50, 29–32. ( 10.1002/pbc.21279) [DOI] [PubMed] [Google Scholar]
- 147.Wong IH, Chan J, Wong J, Tam PK. 2004. Ubiquitous aberrant RASSF1A promoter methylation in childhood neoplasia. Clin. Cancer Res. 10, 994–1002. ( 10.1158/1078-0432.CCR-0378-3) [DOI] [PubMed] [Google Scholar]
- 148.Misawa A, Tanaka S, Yagyu S, Tsuchiya K, Iehara T, Sugimoto T, Hosoi H. 2009. RASSF1A hypermethylation in pretreatment serum DNA of neuroblastoma patients: a prognostic marker. Br. J. Cancer 100, 399–404. ( 10.1038/sj.bjc.6604887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Harada K, et al. 2002. Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene 21, 4345–4349. ( 10.1038/sj.onc.1205446) [DOI] [PubMed] [Google Scholar]
- 150.Astuti D, et al. 2001. RASSF1A promoter region CpG island hypermethylation in phaeochromocytomas and neuroblastoma tumours. Oncogene 20, 7573–7577. ( 10.1038/sj.onc.1204968) [DOI] [PubMed] [Google Scholar]
- 151.Yang Q, Kiernan CM, Tian Y, Salwen HR, Chlenski A, Brumback BA, London WB, Cohn SL. 2007. Methylation of CASP8, DCR2, and HIN-1 in neuroblastoma is associated with poor outcome. Clin. Cancer Res. 13, 3191–3197. ( 10.1158/1078-0432.CCR-06-2846) [DOI] [PubMed] [Google Scholar]
- 152.Peng Y, Croce CM. 2016. The role of microRNAs in human cancer. Signal Transduct. Target Ther. 1, 15004 ( 10.1038/sigtrans.2015.4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.O'Brien KP, Ramphul E, Howard L, Gallagher WM, Malone C, Kerin MJ, Dwyer RM. 2017. Circulating microRNAs in cancer. Methods Mol. Biol. 1509, 123–139. ( 10.1007/978-1-4939-6524-3_12) [DOI] [PubMed] [Google Scholar]
- 154.Powers JT, et al. 2016. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 535, 246–251. ( 10.1038/nature18632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flaegstad T, Einvik C. 2011. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer 105, 296–303. ( 10.1038/bjc.2011.220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao Z, et al. 2014. A high-content morphological screen identifies novel microRNAs that regulate neuroblastoma cell differentiation. Oncotarget 5, 2499–2512. ( 10.18632/oncotarget.1703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Galardi A, Colletti M, Businaro P, Quintarelli C, Locatelli F, Di Giannatale A. 2018. MicroRNAs in neuroblastoma: biomarkers with therapeutic potential. Curr. Med. Chem. 25, 584–600. ( 10.2174/0929867324666171003120335) [DOI] [PubMed] [Google Scholar]
- 158.Murray MJ, Raby KL, Saini HK, Bailey S, Wool SV, Tunnacliffe JM, Enright AJ, Nicholson JC, Coleman N. 2015. Solid tumors of childhood display specific serum microRNA profiles. Cancer Epidemiol. Biomarkers Prev. 24, 350–360. ( 10.1158/1055-9965.EPI-14-0669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen Y, Stallings RL. 2007. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 67, 976–983. ( 10.1158/0008-5472.CAN-06-3667) [DOI] [PubMed] [Google Scholar]
- 160.Ma L, et al. 2010. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256. ( 10.1038/ncb2024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ramraj SK, Aravindan S, Somasundaram DB, Herman TS, Natarajan M, Aravindan N. 2016. Serum-circulating miRNAs predict neuroblastoma progression in mouse model of high-risk metastatic disease. Oncotarget 7, 18 605–18 619. ( 10.18632/oncotarget.7615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zeka F, et al. 2018. Circulating microRNA biomarkers for metastatic disease in neuroblastoma patients. JCI Insight 3, 97021 ( 10.1172/jci.insight.97021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gupta GP, Massague J. 2006. Cancer metastasis: building a framework. Cell 127, 679–695. ( 10.1016/j.cell.2006.11.001) [DOI] [PubMed] [Google Scholar]
- 164.Alix-Panabieres C, Riethdorf S, Pantel K. 2008. Circulating tumor cells and bone marrow micrometastasis. Clin. Cancer Res. 14, 5013–5021. ( 10.1158/1078-0432.CCR-07-5125) [DOI] [PubMed] [Google Scholar]
- 165.Agarwal A, Balic M, El-Ashry D, Cote RJ. 2018. Circulating tumor cells: strategies for capture, analyses, and propagation. Cancer J. 24, 70–77. ( 10.1097/PPO.0000000000000310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chinen LTD, Abdallah EA, Braun AC, Flores B, Corassa M, Sanches SM, Fanelli MF. 2017. Circulating tumor cells as cancer biomarkers in the clinic. Adv. Exp. Med. Biol. 994, 1–41. ( 10.1007/978-3-319-55947-6_1) [DOI] [PubMed] [Google Scholar]
- 167.Gerson JM, Schlesinger HR, Sereni P, Moorhead PS, Hummeler K. 1977. Isolation and characterization of a neuroblastoma cell line from peripheral blood in a patient with disseminated disease. Cancer 39, 2508–2512. () [DOI] [PubMed] [Google Scholar]
- 168.Lanino E, Melodia A, Casalaro A, Cornaglia-Ferraris P. 1989. Neuroblastoma cells circulate in peripheral blood. Pediatr. Hematol. Oncol. 6, 193–195. ( 10.3109/08880018909034286) [DOI] [PubMed] [Google Scholar]
- 169.Moss TJ, Cairo M, Santana VM, Weinthal J, Hurvitz C, Bostrom B. 1994. Clonogenicity of circulating neuroblastoma cells: implications regarding peripheral blood stem cell transplantation. Blood 83, 3085–3089. [PubMed] [Google Scholar]
- 170.Moss TJ, Sanders DG. 1990. Detection of neuroblastoma cells in blood. J. Clin. Oncol. 8, 736–740. ( 10.1200/JCO.1990.8.4.736) [DOI] [PubMed] [Google Scholar]
- 171.Sanders DG, Wiley FM, Moss TJ. 1991. Serial immunocytologic analysis of blood for tumor cells in two patients with neuroblastoma. Cancer 67, 1423–1427. () [DOI] [PubMed] [Google Scholar]
- 172.Mattano LA Jr, Moss TJ, Emerson SG. 1992. Sensitive detection of rare circulating neuroblastoma cells by the reverse transcriptase-polymerase chain reaction. Cancer Res. 52, 4701–4705. [PubMed] [Google Scholar]
- 173.Yanagisawa TY, et al. 1998. Detection of the PGP9.5 and tyrosine hydroxylase mRNAs for minimal residual neuroblastoma cells in bone marrow and peripheral blood. Tohoku J. Exp. Med. 184, 229–240. ( 10.1620/tjem.184.229) [DOI] [PubMed] [Google Scholar]
- 174.Corrias MV, et al. 2004. Detection of neuroblastoma cells in bone marrow and peripheral blood by different techniques: accuracy and relationship with clinical features of patients. Clin. Cancer Res. 10, 7978–7985. ( 10.1158/1078-0432.CCR-04-0815) [DOI] [PubMed] [Google Scholar]
- 175.Gao Y, Li G, Zhang X, Xu Q, Zheng B. 1997. Detection of neuroblastoma cells in blood by reverse transcriptase-polymerase chain reaction. Chin. Med. J. (Engl) 110, 341–345. [PubMed] [Google Scholar]
- 176.Burchill SA, Bradbury FM, Smith B, Lewis IJ, Selby P. 1994. Neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Int. J. Cancer 57, 671–675. ( 10.1002/ijc.2910570510) [DOI] [PubMed] [Google Scholar]
- 177.Burchill SA, Bradbury FM, Selby P, Lewis IJ. 1995. Early clinical evaluation of neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Eur. J. Cancer 31A, 553–556. ( 10.1016/0959-8049(95)00053-L) [DOI] [PubMed] [Google Scholar]
- 178.Miyajima Y, Kato K, Numata S, Kudo K, Horibe K. 1995. Detection of neuroblastoma cells in bone marrow and peripheral blood at diagnosis by the reverse transcriptase-polymerase chain reaction for tyrosine hydroxylase mRNA. Cancer 75, 2757–2761. () [DOI] [PubMed] [Google Scholar]
- 179.Miyajima Y, Horibe K, Fukuda M, Matsumoto K, Numata S, Mori H, Kato K. 1996. Sequential detection of tumor cells in the peripheral blood and bone marrow of patients with stage IV neuroblastoma by the reverse transcription-polymerase chain reaction for tyrosine hydroxylase mRNA. Cancer 77, 1214–1219. () [DOI] [PubMed] [Google Scholar]
- 180.Kuroda T, Saeki M, Nakano M, Mizutani S. 1997. Clinical application of minimal residual neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction. J. Pediatr. Surg. 32, 69–72. ( 10.1016/S0022-3468(97)90097-X) [DOI] [PubMed] [Google Scholar]
- 181.Kuroda T, Saeki M, Nakano M, Mizutani S, Endo M, Akiyama H. 2000. Surgical treatment of neuroblastoma with micrometastasis. J. Pediatr. Surg. 35, 1638–1642. ( 10.1053/jpsu.2000.18341) [DOI] [PubMed] [Google Scholar]
- 182.Burchill SA, Lewis IJ, Abrams KR, Riley R, Imeson J, Pearson ADJ, Pinkerton R, Selby P. 2001. Circulating neuroblastoma cells detected by reverse transcriptase polymerase chain reaction for tyrosine hydroxylase mRNA are an independent poor prognostic indicator in stage 4 neuroblastoma in children over 1 year. J. Clin. Oncol. 19, 1795–1801. ( 10.1200/Jco.2001.19.6.1795) [DOI] [PubMed] [Google Scholar]
- 183.Parareda A, Gallego S, Roma J, Llort A, Sabado C, Gros L, De Toledo JS. 2005. Prognostic impact of the detection of microcirculating tumor cells by a real-time RT-PCR assay of tyrosine hydroxylase in patients with advanced neuroblastoma. Oncol. Rep. 14, 1021–1027. ( 10.3892/or.14.4.1021) [DOI] [PubMed] [Google Scholar]
- 184.Kuroda T, et al. 2005. Tumor cell dynamics and metastasis in advanced neuroblastoma. Pediatr. Surg. Int. 21, 859–863. ( 10.1007/s00383-005-1503-9) [DOI] [PubMed] [Google Scholar]
- 185.Träger C, Kogner P, Lindskog M, Ponthan F, Kullman A, Kagedal B. 2003. Quantitative analysis of tyrosine hydroxylase mRNA for sensitive detection of neuroblastoma cells in blood and bone marrow. Clin. Chem. 49, 104–112. ( 10.1373/49.1.104) [DOI] [PubMed] [Google Scholar]
- 186.Ifversen MRS, Kagedal B, Christensen LD, Rechnitzer C, Petersen BL, Heilmann C. 2005. Comparison of immunocytochemistry, real-time quantitative RT-PCR and flow cytometry for detection of minimal residual disease in neuroblastoma. Int. J. Oncol. 27, 121–129. ( 10.3892/ijo.27.1.121) [DOI] [PubMed] [Google Scholar]
- 187.Viprey VF, et al. 2014. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: a European HR-NBL1/SIOPEN Study. J. Clin. Oncol. 32, 1074–1083. ( 10.1200/Jco.2013.53.3604) [DOI] [PubMed] [Google Scholar]
- 188.Yanez Y, et al. 2016. TH and DCX mRNAs in peripheral blood and bone marrow predict outcome in metastatic neuroblastoma patients. J. Cancer Res. Clin. Oncol. 142, 573–580. ( 10.1007/s00432-015-2054-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Stutterheim J, et al. 2008. PHOX2B is a novel and specific marker for minimal residual disease testing in neuroblastoma. J. Clin. Oncol. 26, 5443–5449. ( 10.1200/Jco.2007.13.6531) [DOI] [PubMed] [Google Scholar]
- 190.Cheung IY, Cheung NK. 1997. Molecular detection of GAGE expression in peripheral blood and bone marrow: utility as a tumor marker for neuroblastoma. Clin. Cancer Res. 3, 821–826. [PubMed] [Google Scholar]
- 191.Burchill SA, Lewis IJ, Selby P. 1999. Improved methods using the reverse transcriptase polymerase chain reaction to detect tumour cells. Br. J. Cancer 79, 971–977. ( 10.1038/sj.bjc.6690155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Träger C, Vernby A, Kullman A, Ora I, Kogner P, Kagedal B. 2008. mRNAs of tyrosine hydroxylase and dopa decarboxylase but not of GD2 synthase are specific for neuroblastoma minimal disease and predicts outcome for children with high-risk disease when measured at diagnosis. In. J. Cancer 123, 2849–2855. ( 10.1002/ijc.23846) [DOI] [PubMed] [Google Scholar]
- 193.Marachelian A, et al. 2017. Expression of five neuroblastoma genes in bone marrow or blood of patients with relapsed/refractory neuroblastoma provides a new biomarker for disease and prognosis. Clin. Cancer Res. 23, 5374–5383. ( 10.1158/1078-0432.Ccr-16-2647) [DOI] [PubMed] [Google Scholar]
- 194.Burchill SA, Lewis IJ, Abrams KR, Riley R, Imeson J, Pearson AD, Pinkerton R, Selby P. 2001. Circulating neuroblastoma cells detected by reverse transcriptase polymerase chain reaction for tyrosine hydroxylase mRNA are an independent poor prognostic indicator in stage 4 neuroblastoma in children over 1 year. J. Clin. Oncol. 19, 1795–1801. ( 10.1200/JCO.2001.19.6.1795) [DOI] [PubMed] [Google Scholar]
- 195.Pagani A, Macri L, Faulkner LB, Tintori V, Paoli A, Garaventa A, Bussolati G. 2002. Detection procedures for neuroblastoma cells metastatic to blood and bone marrow: blinded comparison of chromogranin A heminested reverse transcription polymerase chain reaction to tyrosine hydroxylase nested reverse transcription polymerase chain reaction and to anti-GD2 immunocytology. Diagn. Mol. Pathol. 11, 98–106. ( 10.1097/00019606-200206000-00006) [DOI] [PubMed] [Google Scholar]
- 196.Lambooy LH, Gidding CE, van den Heuvel LP, Hulsbergen-van de Kaa CA, Ligtenberg M, Bokkerink JP, De Abreu RA. 2003. Real-time analysis of tyrosine hydroxylase gene expression: a sensitive and semiquantitative marker for minimal residual disease detection of neuroblastoma. Clin. Cancer Res. 9, 812–819. [PubMed] [Google Scholar]
- 197.Tchirkov A, Paillard C, Halle P, Bernard F, Bordigoni P, Vago P, Demeocq F, Kanold J. 2003. Significance of molecular quantification of minimal residual disease in metastatic neuroblastoma. J. Hematother Stem Cell Res. 12, 435–442. ( 10.1089/152581603322286060) [DOI] [PubMed] [Google Scholar]
- 198.Bozzi F, Luksch R, Collini P, Gambirasio F, Barzano E, Polastri D, Podda M, Brando B, Fossati-Bellani F. 2004. Molecular detection of dopamine decarboxylase expression by means of reverse transcriptase and polymerase chain reaction in bone marrow and peripheral blood: utility as a tumor marker for neuroblastoma. Diagn. Mol. Pathol. 13, 135–143. ( 10.1097/01.pdm.0000128699.14504.06) [DOI] [PubMed] [Google Scholar]
- 199.Oltra S, Martinez F, Orellana C, Grau E, Fernandez JM, Canete A, Castel V. 2005. The doublecortin gene, a new molecular marker to detect minimal residual disease in neuroblastoma. Diagn. Mol. Pathol. 14, 53–57. ( 10.1097/01.pas.0000149876.32376.c0) [DOI] [PubMed] [Google Scholar]
- 200.Swerts K, De Moerloose B, Dhooge C, Vandesompele J, Hoyoux C, Beiske K, Benoit Y, Laureys G, Philippe J. 2006. Potential application of ELAVL4 real-time quantitative reverse transcription-PCR for detection of disseminated neuroblastoma cells. Clin. Chem. 52, 438–445. ( 10.1373/clinchem.2005.059485) [DOI] [PubMed] [Google Scholar]
- 201.Viprey VF, Corrias MV, Kagedal B, Oltra S, Swerts K, Vicha A, Ladenstein R, Burchill SA. 2007. Standardisation of operating procedures for the detection of minimal disease by QRT-PCR in children with neuroblastoma: quality assurance on behalf of SIOPEN-R-NET. Eur. J. Cancer 43, 341–350. ( 10.1016/j.ejca.2006.08.007) [DOI] [PubMed] [Google Scholar]
- 202.Stutterheim J, et al. 2009. Detecting minimal residual disease in neuroblastoma: the superiority of a panel of real-time quantitative PCR markers. Clin. Chem. 55, 1316–1326. ( 10.1373/clinchem.2008.117945) [DOI] [PubMed] [Google Scholar]
- 203.Yáñez Y, Grau E, Oltra S, Canete A, Martinez F, Orellana C, Noguera R, Palanca S, Castel V. 2011. Minimal disease detection in peripheral blood and bone marrow from patients with non-metastatic neuroblastoma. J. Cancer Res. Clin. Oncol. 137, 1263–1272. ( 10.1007/s00432-011-0997-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Corrias MV, et al. 2012. Multiple target molecular monitoring of bone marrow and peripheral blood samples from patients with localized neuroblastoma and healthy donors. Pediatr. Blood Cancer 58, 43–49. ( 10.1002/pbc.22960) [DOI] [PubMed] [Google Scholar]
- 205.Hartomo TB, et al. 2013. Minimal residual disease monitoring in neuroblastoma patients based on the expression of a set of real-time RT-PCR markers in tumor-initiating cells. Oncol. Rep. 29, 1629–1636. ( 10.3892/or.2013.2286) [DOI] [PubMed] [Google Scholar]
- 206.Lee NH, Son MH, Choi YB, Yi E, Lee JW, Yoo KH, Sung KW, Koo HH. 2016. Clinical significance of tyrosine hydroxylase mRNA transcripts in peripheral blood at diagnosis in patients with neuroblastoma. Cancer Res. Treat. 48, 1399–1407. ( 10.4143/crt.2015.481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Corrias MV, et al. 2018. Event-free survival of infants and toddlers enrolled in the HR-NBL-1/SIOPEN trial is associated with the level of neuroblastoma mRNAs at diagnosis. Pediatr. Blood Cancer 65, e27052 ( 10.1002/pbc.27052) [DOI] [PubMed] [Google Scholar]
- 208.Cheung IY, Sahota A, Cheung NKV. 2004. Measuring circulating neuroblastoma cells by quantitative reverse transcriptase-polymerase chain reaction analysis—correlation with its paired bone marrow and standard disease markers. Cancer 101, 2303–2308. ( 10.1002/cncr.20660) [DOI] [PubMed] [Google Scholar]
- 209.Faulkner LB, et al. 1998. High-sensitivity immunocytologic analysis of neuroblastoma cells in paired blood and marrow samples. J. Hematother 7, 361–366. ( 10.1089/scd.1.1998.7.361) [DOI] [PubMed] [Google Scholar]
- 210.Oltra S, Martinez F, Orellana C, Grau E, Fernandez JM, Canete A, Castel V. 2004. Minimal residual disease in neuroblastoma: to GAGE or not to GAGE. Oncol. Res. 14, 291–295. ( 10.3727/096504003773994824) [DOI] [PubMed] [Google Scholar]
- 211.Kuroda T, Morikawa N, Matsuoka K, Fujino A, Honna T, Nakagawa A, Kumagai M, Masaki H, Saeki M. 2008. Prognostic significance of circulating tumor cells and bone marrow micrometastasis in advanced neuroblastoma. J. Pediatr. Surg. 43, 2182–2185. ( 10.1016/j.jpedsurg.2008.08.046) [DOI] [PubMed] [Google Scholar]
- 212.Liu X, Zhang Z, Zhang B, Zheng Y, Zheng C, Liu B, Zheng S, Dong K, Dong R. 2018. Circulating tumor cells detection in neuroblastoma patients by EpCAM-independent enrichment and immunostaining-fluorescence in situ hybridization. EBioMed. 35, 244–250. ( 10.1016/j.ebiom.2018.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Beiske K, et al. 2009. Consensus criteria for sensitive detection of minimal neuroblastoma cells in bone marrow, blood and stem cell preparations by immunocytology and QRT-PCR: recommendations by the International Neuroblastoma Risk Group Task Force. Br. J. Cancer 100, 1627–1637. ( 10.1038/sj.bjc.6605029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mosse Y.2018. Next Generation Personalized Neuroblastoma Therapy (NEPENTHE). https://clinicaltrials.gov/ct2/show/NCT02780128. (accessed 22 November 2018).
- 215.Curie I.2018. Use of Specific Genetic Alterations of Tumoral Cells Identified by the Next Generation Sequencing Techniques (NGS) to Follow Peripheral Samples of Children With Metastatic and/or High Risk Solid Tumor - NGSKids (NGSKids). https://clinicaltrials.gov/ct2/show/NCT02546453. (accessed 22 November 2018).
- 216.Tanaka F, Yoneda K, Hasegawa S. 2010. Circulating tumor cells (CTCs) in lung cancer: current status and future perspectives. Lung Cancer (Auckl) 1, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.