Abstract
Na+ K+ ATPase (NKA) is crucial to branchial ion transport as it uses the energy from ATP to move Na+ against its electrochemical gradient. When fish encounter extremely dilute environments the energy available from ATP hydrolysis may not be sufficient to overcome thermodynamic constraints on ion transport. Yet many fish species—including zebrafish—are capable of surviving in dilute environments. Despite much study, the physiological mechanisms by which this occurs remain poorly understood. Here, we demonstrate that zebrafish acclimated to less than 10 µM Na+ water exhibit upregulation of a specific NKA α subunit (zatp1a1a.5) that, unlike most NKA heterotrimers, would result in transfer of only a single Na+ and K+ per ATP hydrolysis reaction. Thermodynamic models demonstrate that this change is sufficient to reduce the activation energy of NKA, allowing it to overcome the adverse electrochemical gradient imposed by dilute freshwater. Importantly, upregulation of zatp1a1a.5 also coincides with the recovery of whole body Na+ post-transfer, which occurs within 24 h. While these structural modifications are crucial for allowing zebrafish to survive in ion-poor environments, phylogenetic and structural analysis of available α subunits from a range of teleosts suggests this adaptation is not widely distributed.
Keywords: ion transport, sodium transport, kidney, FXYD, ionocyte
1. Introduction
Many organisms tolerate wide salinity variation without compromising salt and water balance. In fish, Na+ K+ ATPase (NKA) is crucial for both hypo- and hyperosmotic regulation by catalysing active Na+ absorption across relevant epithelia. In the hyperosmotic marine environment, NKA is abundant in specialized branchial ionocytes where it establishes the thermodynamic gradient required to excrete Na+ and Cl− into the marine environment [1]. Conversely, in hypo-osmotic freshwater environments branchial NKA establishes the conditions necessary for Na+ absorption through branchial ionocytes [1]. The kidneys of freshwater fish are also actively involved in osmoregulation by removing diffusively gained water through the production of copious amounts of dilute urine. NKA throughout the renal tubule system is critical to reabsorbing Na+ and other ions from the glomerular filtrate [2,3], thereby protecting against urinary ion loss [1].
The minimum functional unit of NKA is a heterotrimer of α, β and γ subunits [4]. The α subunits are responsible for the catalytic activity and Na+ affinity of the complete heterotrimer, as these subunits contain the ion binding sites. The β subunits are structurally important chaperones that ensure proper orientation of the catalytic mechanism within the membrane [4]. These subunits also modulate catalytic activity through changes in heterotrimer structure and stability that can impact cation affinity [4,5]. Furthermore, the β subunits can affect transepithelial transport through tight junctions owing to the formation of intercellular bridges (e.g. [6]). The γ subunit—also known as FXYD—contains a single transmembrane domain, and impacts both the cation affinity and maximal transport capacity of the heterotrimer [4,7].
The subunits of NKA have been of intense interest in the study of euryhaline and anadromous fishes—in particular, the phenomenon of NKA α1 isoform switching and the dynamic regulation of FXYD subunits. The general pattern of isoform switching is that α1a (atp1a1a) predominates in freshwater, while α1b (atp1a1b) is present in seawater acclimated fish [3,8–11]. The evidence from Atlantic salmon and rainbow trout suggests the switch in α1 subunits is crucial for survival in freshwater owing to the presence of an Asn/Lys-776 substitution in the freshwater α1a subunit [12]. This structural change prevents Na+ from binding to one of three binding sites in the freshwater heterotrimer, which allows NKA to function under more thermodynamically challenging conditions without the enzyme activation energy exceeding that available from ATP hydrolysis [12,13]. Interestingly, zebrafish have an incredible diversity of NKA subunits, with five α1a subunits originating from a series of gene duplication events [14,15]. This raises questions about the physiological and biochemical significance of the various heterotrimers. Unlike the majority of species that have been studied, zebrafish are not euryhaline as they are unable to transition between seawater and freshwater. However, zebrafish occupy a wide range of osmotic habitats in freshwater, including those that contain less than 10 µM Na+ [16], and recent work has shown that zebrafish exhibit differential regulation of α1a subunits when exposed to these extreme hypo-osmotic environments [17]. While these changes have been localized to the gill and epithelial ionocytes of adult and larval zebrafish, respectively, the physiological significance of the isoform shift is unclear.
On this background, this study was undertaken to explore the physiological significance of NKA isoform switching in zebrafish following hypo-osmotic acclimation. We used a two-stage acclimation protocol and used real-time PCR to assess differential regulation of the α1a, β and γ subunits following acclimation. A structural analysis of the five catalytic α1a subunits was performed to assess theoretical enzyme stoichiometry, which was combined with thermodynamic modelling to provide insight into the physiological implications of any observed isoform switching. Finally, a phylogenetic analysis of NKA α1 subunits explored the prevalence of the reduced NKA reaction stoichiometry in the wider teleost lineage.
2. Results
(a). Influence of environmental salinity on whole body Na+ concentration
An initial transfer from 14 mM to 1.5 mM Na+ was performed and fish were allowed to acclimate for 32 days. No significant changes in whole body Na+ were observed during this acclimation protocol (electronic supplementary material, figure S1). A second protocol transferred 1.5 mM Na+ acclimated fish to ion-poor water containing less than 0.01 mM Na+. A significant 50% reduction in whole body Na+ at 0.5 days post-transfer (dpt) was observed (electronic supplementary material, figure S1). Whole body Na+ returned to control levels by 1 dpt and remained stable for the duration of the experiment.
(b). Differential expression of α, β and γ subunit mRNA after salinity transfer
Expression of five α1a subunits, four β subunits and five γ—or FXYD—subunits was observed in the gills of zebrafish. No expression of β2b or 3b was observed in the gill, nor was any expression detected for FXYD2. In the kidney, no zatp1a1a.3 expression was detected and only FXYD2 and FXYD5, as well as β1a and 2a, were expressed.
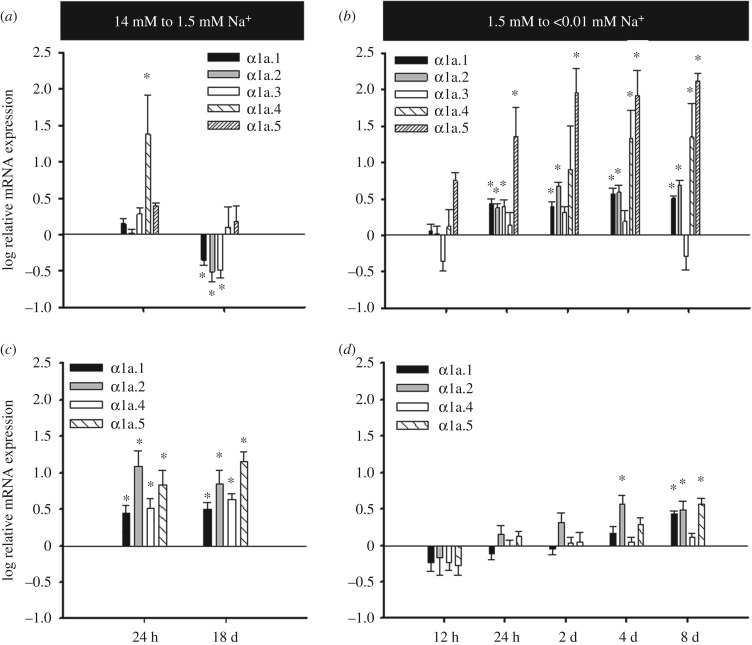
Transfer of zebrafish from 14 to 1.5 mM Na+ resulted in a suite of gene expression changes in the gill. There was a 30-fold upregulation of the zatp1a1a.4 at 1 dpt (figure 1a). Similar upregulation was observed for β1b, FXYD1 and FXYD11 (electronic supplementary material, figure S2). This observed upregulation was transient in nature. At 18 dpt zatp1a1a.4 expression had returned to control levels while zatp1a1a.1, zatp1a1a.2 and zatp1a1a.3 were all significantly downregulated relative to controls. Transfer to 1.5 mM Na+ had no effect on zatp1a1a.5 expression and all β subunit expression was unaffected at 18 dpt. In the kidney only FXYD5 did not show upregulation after transfer to 1.5 mM Na+ (figure 1c; electronic supplementary material, figure S2). These changes were similar at 1 and 18 dpt.
Figure 1.
Gene expression analysis for α subunits of Na+ K+ ATPase in the (a,b) gills and (c,d) kidneys of zebrafish after transfer (a,c) from 14 mM to 1.5 mM Na+ and (b,d) from 1.5 mM to 0.01 mM Na+. All values represent the mean ± s.e.m. of log-transformed relative expression. An asterisk denotes a significant difference from pre-transfer expression (n = 5–10 for gills and n = 8–10 for kidneys).
Transfer from 1.5 mM to less than 0.01 mM Na+ resulted in upregulation of all five α1a subunits (figure 1b) in the gill; however, the response of α1a.3 was transient and returned to control levels by 2 dpt. Conversely, the α1a.4 response was delayed when compared with the other subunits as significant upregulation was not observed until 4 dpt. The most dramatic responses were observed for α1a.4 and α1a.5, whose expression increased by approximately 25- and 100-fold, respectively. All four β subunits showed upregulation by 1 dpt, which was maintained throughout the experiment (electronic supplementary material, figure S2). A general pattern of upregulation was also observed for the FXYD subunits with only FXYD5 not showing a response (electronic supplementary material, figure S2). In contrast to the gill, the kidney showed a more muted response when transferred to less than 0.01 mM Na+. While only α1a.4 and FXYD5 expression was unchanged, most of the responses in the kidney did not occur until 8 dpt (figure 1d; electronic supplementary material, figure S2).
Overall the most dramatic expression changes in NKA subunit distribution were observed for the gill α1a subunits. At 14 mM Na+ the α1a subunit distribution was dominated by zatp1a1a.2 and zatp1a1a.3 (electronic supplementary material, figure S3). However, at less than 0.01 mM Na+ the expression of zatp1a1a.3 had dropped to 10% that of zatp1a1a.2 and zatp1a1a.5 had become the second most abundant transcript. By contrast, upregulation of α1a subunits in the kidney did not result in a dramatic shift in relative transcript abundance, as zatp1a1a.4 was at least 15-fold more abundant than any other transcript under all conditions (electronic supplementary material, figure S4).
(c). Structural analysis of NKA α1 paralogues in zebrafish
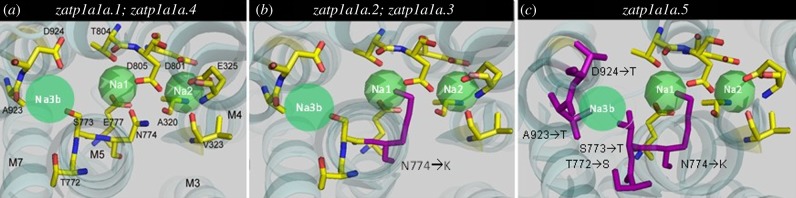
Structural models generated in PyMol revealed three different NKA reaction stoichiometries as a consequence of the specific α1 subunit. Both zatp1a1a.1 and zatp1a1a.4 have models representative of the standard 3 Na+ and 2 K+ per ATP reaction cycle (figure 2a); however, the Asn/Lys-774 substitution found in zatp1a1a.2 and zatp1a1a.3 causes the Lys-774 side chain to insert into the center of Na+ binding site 1 (figure 2b). This blocks the Na+ binding site, resulting in 2 Na+ and 1 K+ per ATP cycle. A further series of alterations are found in zatp1a1a.5 (figure 2c), which effectively blocks the third Na+ binding site, resulting in only a single Na+ and K+ to be transported per ATP reaction cycle. Two of these substitutions (Ser/Thr-773 and Thr/Ser-772) occur in TM5 while two additional changes (Ala/Thr-923 and Asp/Thr-924) are found in the TM8 region (electronic supplementary material, table S1).
Figure 2.
The three-dimensional reconstructions for the five zebrafish α1a paralogues as generated from the backbone of the high resolution (2.4 Å) structure of the E1-form of Ca-ATPase (see Methods). (a) The structure of paralogues exhibiting typical three Na+ per ATP hydrolysis reaction stoichiometry. (b) The structural modifications (purple amino acids) that block one Na+ binding site. (c) The modifications that block two Na+ binding sites.
(d). Thermodynamic modelling
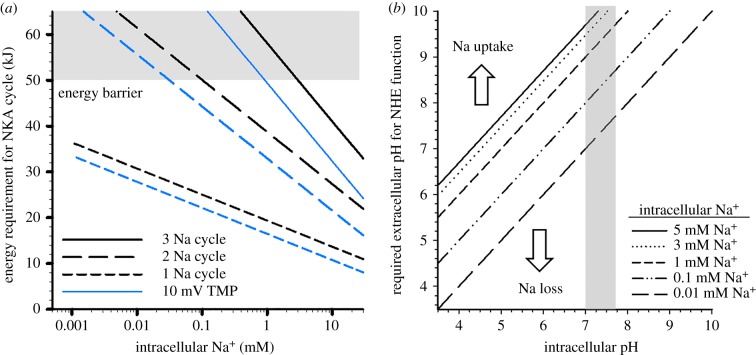
To explore the physiological relevance of the apparent hypo-osmotic α1a subunit ‘switch' in zebrafish, a series of thermodynamic calculations were performed. The first series assessed the effects of reduced Na+ transport per unit ATP on NKA activation energy as a consequence of intracellular Na+. Under typical stoichiometry, the lower intracellular Na+ threshold for NKA function using the energy available from ATP hydrolysis, assuming 75% conversion efficiency and a TMP ranging from 40 to 70 mV, is between 1 and 3 mM (figure 3a). The 2 Na+ stoichiometry provides sufficient energy from ATP to power NKA down to 0.04 to 0.1 mM; however, the single Na+ stoichiometry allows sufficient activation energy for an NKA cycle even at nM Na+ concentrations (figure 3a).
Figure 3.
(a) The calculated activation energy of a single Na+ K+ ATPase reaction cycle at varying intracellular Na+ concentrations as a consequence of the number of Na+ ions transferred per cycle. The blue and black lines denote a basolateral transmembrane potential of 40 mV and 70 mV, respectively. A temperature of 24°C was used for all calculations. The shaded area represents the upper limit of available energy assuming a 75% (lower) to 100% (upper) energy transfer efficiency. (b) A plot representing the external micro-domain pH required to drive Na+ uptake through NHE at various intracellular pH and Na+ concentrations. The shaded area represents the physiologically relevant intracellular pH values.
A second series of calculations assessed the effects of reduced intracellular Na+ on apical NHE function in 10 µM Na+ environments at varying pH values. At physiologically relevant intracellular pH, the extracellular micro-environment pH would have to exceed 9.6 to allow Na+ uptake, assuming an intracellular Na+ concentration of 5 mM. The ability to reduce intracellular Na+ would allow Na+ uptake to occur in 10 µM Na+ environments at neutral extracellular pH (figure 3b).
(e). Phylogenetic analysis of NKA α1a isoforms
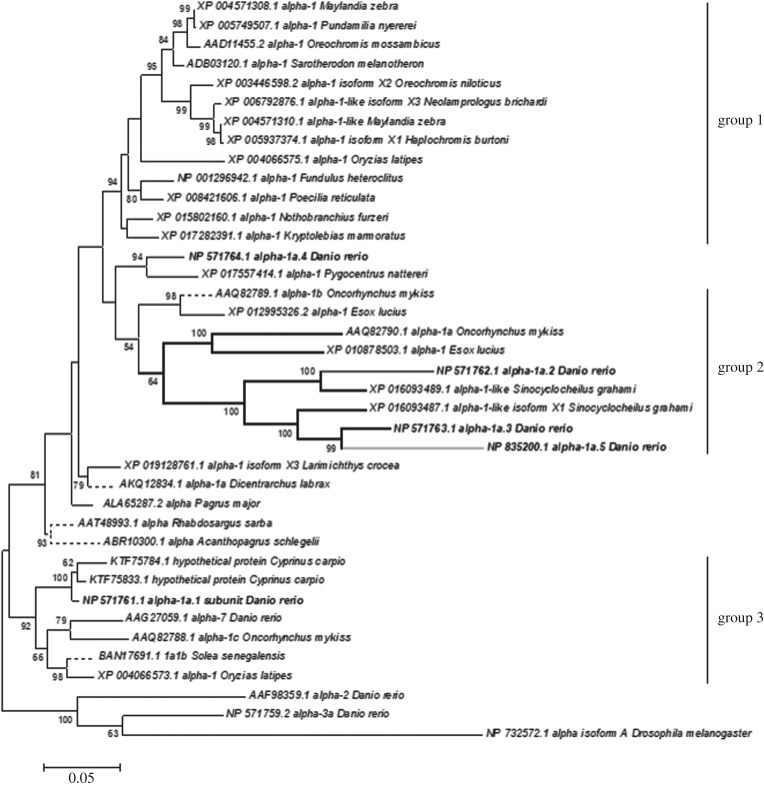
A maximum-likelihood phylogenetic analysis of available NKA α1-subunit isoforms was performed to explore the prevalence of hypo-osmotic adapted isoforms in a diverse array of teleosts (figure 4). Three relatively well-supported groups were identified, which were taxon specific. Group 1 consisted of species from the Acanthopterygii superorder, while group 2 consisted of Otocephala and Protacanthopterygii superorders. Group 3 is a basal isoform clade and contained isoforms from a wide array of teleost species. All isoforms were assessed for the structural modifications found in TM5 and TM8 (as in electronic supplementary material, table S1); however, these were confined to a subset of group 2. The vast majority of sequences included in this analysis were from freshwater lineages.
Figure 4.
A maximum-likelihood analysis of Na+ K+ ATPase α subunits from teleost fish. Group 1 is a well-supported clade that includes only species of the Acanthopterygii superorder, while Group 2 includes only members of the closely related Otocephala and Protacanthopterygii superorders. Group 3 is a well-supported basal clade that is widely distributed among the teleost lineages. The analysed zebrafish α1 sequences are highlighted in bold. The heavier weighted dark lines show those sequences with the structural modifications for a 2 Na+ per ATP stoichiometry, while the grey line represents a single Na+ per ATP stoichiometry. The tree is rooted with the outgroup containing a Drosophila α subunit.
3. Discussion
Acclimation to extreme hypo-salinity requires that fish counteract ion loss with increased uptake at the gills and efficient re-absorption in the kidneys [1]. Zebrafish are incredibly adept in their ability to tolerate extreme hypo-osmotic environments, which stems in part from an ability to increase the affinity and capacity for ion transport following acclimation [16]. The current study builds on this finding and suggests that NKA isoform switching—particularly that related to the α1 subunit—is a crucial adaptation for acclimation of zebrafish to dilute freshwater. This adaptation probably contributes to the ability of zebrafish to overcome the thermodynamic constraints imposed by the large concentration gradient between the environment and extracellular fluid.
The function of NKA in osmoregulatory tissues is to move Na+ against its electrochemical gradient using the energy released from ATP hydrolysis. Low environmental Na+ concentrations require more energy to perform an enzyme cycle [13], which represents a significant thermodynamic constraint for osmoregulation in freshwater. NKA typically transports 3 Na+ ions and 2 K+ per cycle, and this is the estimated reaction stoichiometry for complexes containing zatp1a1a.1 and zatp1a1a.4. At this stoichiometry the energy barrier is reached at approximately 1 mM environmental Na+ (figure 3a), assuming a 70 mV basolateral TMP and 75% energy transfer from ATP hydrolysis [13]. However, the predominant α subunit in zebrafish gills was zatp1a1a.2, which was nearly 10-fold more abundant than other subunits. This is consistent with our findings, as the 14 mM to 1.5 mM Na+ transfer did not result in wide-scale changes in relative NKA α1a transcript abundance, probably because the 2 Na+ reaction cycle of zatp1a1a.2 alleviates any thermodynamic constraint.
By contrast, a shift in the relative α subunit expression was observed when fish were transferred to less than 0.01 mM Na+. Most noteworthy was the 100-fold increase in the expression of zatp1a1a.5. This conformed to previous work [17] and resulted in zatp1a1a.5 becoming the second most abundant α1a transcript. The protein structure of zatp1a1a.5 suggests that only one Na+ is transported per enzyme cycle. The most abundant pre-transfer α1a isoforms reach an energy limitation around 0.1 mM Na+, and therefore the upregulation of zatp1a1a.5 is crucial for maintaining osmotic balance in extreme hypo-osmotic environments. This is supported by the trend in whole body Na+, which was significantly reduced at 0.5 days after transfer but returned to control values by 1 dpt, coincident with the onset of zatp1a1a.5 upregulation. While the current data only provide transcriptional support for isoform switching, we would stress that the relative protein abundance is not as important as the mere presence of zatp1a1a.5 in sufficient quantity. This is simply because the lower activation energy of zatp1a1a.5 will allow it to function in an ion-poor environment, while zatp1a1a.3 will not function.
Our interpretation of the significance of isoform switching is dependent on the zatp1a1a.5 being located in the proper ionocyte cell type, as well as the validity of structural and thermodynamic modelling. Previous work has demonstrated that the zatp1a1a.5 isoform is found in HR-type ionocytes, which are defined by the presence of NHE and V-type ATPase [17]. The abundant zatp1a1a.2 isoform is not expressed in this cell type. While the zatp1a1a.3 localization has not yet been defined, the current data lead to the hypothesis that the HR cell type may undergo a switch from zatp1a1a.3 to zatp1a1a.5 expression following hypo-osmotic acclimation. Our structural modelling approach is also common practice in the study of structure–function relationships in P-type ATPases [12,18–20], and its utility has been consistently validated using site-directed mutagenesis. While we have not performed such experiments for this study, they have been performed when studying mammalian ATPases. Most notable are a series of studies that demonstrated a Thr-774/Ser mutation or a Ser-775/Thr mutation significantly reduced Na+ binding, as indicated by reduced activity and changes in the Gibbs free energy of Na+ binding [19]. The latter also impaired K+ binding [19]. Importantly, these are the identical substitutions observed in zatp1a1a.5, and are analogous to the previously described structural changes that impair Na+ binding site 2 (figure 2b) [12]. Similar findings were also observed for Asp-925/Asn [20], which is similar to the Thr substitution observed in zebrafish. Taken together, these studies provide strong justification for our suggestion that the third Na+ binding site is disrupted in zatp1a1a.5. The final question is whether the significance of isoform switching is subject to change under different thermodynamic assumptions. As demonstrated in figure 3a, a reduction in TMP reduces the activation energy for NKA. But it would take a TMP of only 10 mV to allow NKA to function using 50 kJ of energy at a 2 Na+ per ATP stoichiometry. Similarly, a greater ATP energy conversion rate would also reduce the overall activation energy for an enzyme cycle at any stoichiometry, but as shown in figure 3a, it would take 90% efficiency for NKA to work at 0.01 mM Na+ and a 40 mV TMP using a 2 Na+ stoichiometry (assuming 63 kJ mol−1 ATP) [15]. As pointed out previously, such high conversion efficiency from ATP hydrolysis is unlikely [13].
While the results presented here clearly have implications for basolateral Na+ transport, they also have implications for transepithelial transport. The two proposed pathways for apical Na+ uptake in dilute water are a Na+ channel—such as the acid-sensing ion channels [21]—paired with a V-type H+ ATPase (VHA), or via NHE (see review [22]). The former would not be impacted by NKA isoform switching, as the thermodynamic constraints are overcome by VHA; however, there is an additional energetic cost. For example, it costs 0.33 ATP per Na+ under normal 3 Na+ NKA stoichiometry, and this would increase to 1.33 ATP per Na+ after recruiting VHA for apical Na+ uptake. Conversely, the cost per Na+ is only 0.5 and 1 ATP for the 2 Na+ and 1 Na+ reaction cycles, respectively, through apical NHE. It has also been argued that the thermodynamic constraints on Na+ uptake in very low Na+ environments preclude the function of NHE [23]. Our data suggest that the activation energy for NKA in zebrafish expressing zatp1a1a.5 could allow intracellular Na+ concentrations to reach below 0.01 mM without compromising NKA function. The importance of intracellular Na+ to NHE function was well demonstrated by Parks et al. [23]. More recently, apical micro-domain alkalization caused by ammonia excretion has been proposed, and validated, as a means to allow Na+ uptake through NHE. Yet as demonstrated here (figure 3b), alkalization would have to exceed a pH of 10 to effectively drive uptake in a 10 µM Na+ environment, assuming 5 mM intracellular Na+. This would require ammonia excretion to greatly exceed the combined acidifying effects of NHE and CO2 on micro-domain pH.
The β and γ subunits of NKA are also known to impact Na+ transport function [4]. In mammals, the expression of FXYD4 in NKA complexes increases Na+ affinity and this role is presumably co-opted by the structurally similar FXYD11 in fish gills [4,7,24]. Previous work has demonstrated that the predominant subunit present in teleost gills is FXYD11 [24], and this was confirmed here for zebrafish. However, five FXYD isoforms were expressed in the gills of zebrafish, with FXYD5 and FXYD1 being the second and third most abundant. Similar to the α subunits, the initial 1.5 mM transfer step resulted in selective transient upregulation of FXYD1 and FXYD11 after 24 h followed by a wide spread downregulation by 18 dpt. By contrast, transfer to less than 0.01 mM Na+ resulted in upregulation in four of five FXYD isoforms beginning at 0.5–1 dpt. The largest responses were found for FXYD11 and FXYD1; however, no dramatic shifts in relative isoform distributions were observed. This trend was also observed for the gill β subunits with all isoforms upregulated by 1 dpt; however, with no change in relative subunit distribution. As such, it appears that subunit abundance, rather than changing isoform identity, is the important factor for the role of γ and β subunits in osmoregulation in severely hypo-osmotic environments.
Zebrafish kidneys contained a less diverse array of NKA subunit isoforms than the gills. Only four α subunits were found, with no detectable expression of zatp1a1a.3. Furthermore, only two β and two γ subunits were detected. Previous work on Atlantic salmon showed similar expression with respect to FXYD2 and FXYD5; however, salmon also expressed FXYD9 and FXYD12 [24]. Similarly, two species of medaka showed significant expression of FXYD9 and FXYD12, as well as detectable levels of FXYD6 and FXYD8 [25]. Wide-scale upregulation of NKA subunits was observed in the kidney after transfer to 1.5 mM, with only FXYD5 being unaffected. This suggests a general increase in the demand for osmoregulatory machinery in the kidney following transfer. This is not surprising as we would hypothesize that the kidney would become more active in hypo-osmotic environments to prevent ion loss during increased urine production [1]. A similar trend was observed following transfer to less than 0.01 mM Na+. By 8 dpt six subunits showed significant upregulation, with only the FXYD5 and zatp1a1a.4 showing no change in expression. Notably, there was no evidence of α1a isoform switching in the kidney. This is not surprising as the ion composition of the glomerular filtrate is dependent on the plasma ion composition.
While much of this discussion has focused on the impact of dynamic isoform regulation on Na+ transport, we must acknowledge that all ions were diluted during the hypo-osmotic exposures, which is representative of ion-poor waters in the natural environment. The fundamental role of NKA in establishing electrochemical gradients for all active ion transport processes means that some of the observed changes may relate to other ions, or to the hypo-osmotic environment as a whole. For example, the wide-scale upregulation in the kidney should be attributed to the hypo-osmotic environment as a whole, and not simply as a consequence of reduced Na+. Similarly, the interpretation of changes in β subunit expression in the gill should relate to the hypo-osmotic environment as a whole because these isoforms play an important role in forming tight junctions [6]. A tighter branchial epithelium in hypo-osmotic environments would presumably benefit overall ion loss. Yet it seems unlikely that α1a isoform switching in the gill would be driven by other ions, and this is supported by work that demonstrated low Cl− and low Ca2+ did not alter gill zatp1a1a.5 expression in zebrafish [17]. So while single isoform responses (e.g. zatp1a1a.1) in the gill may respond to other ions, such as Cl− and Ca2+ [17], the broader conclusion relates to Na+.
It is also possible that the reduced NKA stoichiometry can carry negative consequences for other ion transport processes. For example, the proposed stoichiometry of zatp1a1a.5 is electroneutral, and therefore the ability of the cell to maintain TMP may be compromised. This would have implications for other ion transport processes, such as Cl−. This ion is taken up from ion-poor water in part through slc26a6 [26], which is an electrogenic transporter that benefits from a negative internal charge of which NKA is considered the primary generator. Reduced function of slc26a6 would compromise organismal osmoregulation regardless of the ability of zebrafish to absorb Na+. It seems possible that zebrafish are able to maintain ionocyte TMP in part by modifying basolateral K+ conductance. Furthermore, the presence of VHA provides an alternative avenue for TMP generation. In fact, VHA is the primary electromotive force driving ion and fluid excretion in the Malpighian tubules in the mosquito Aedes aegypti [27], and has been implicated in Cl− uptake in zebrafish [16].
It is curious that the adaptations to NKA α1a isoforms that allow zebrafish to perform well in ion-poor water are not found in a broader spectrum of freshwater fish. Of the 85 sequences examined (figure 4) only zatp1a1a.5 contained the structural modifications that would allow for a 1 Na+ per ATP reaction cycle. In fact, this analysis suggests that the α1 adaptations that allow for any reduced stoichiometry are absent from Acanthopterygii species, many of which can effectively survive at Na+ concentrations below 1 mM. While this analysis is limited to the available genomic information, the lack of lysine substitutions in other freshwater teleosts remains compelling, and would argue that adaptation to low ion environments has evolved via several distinct pathways. This can also be extended to euryhalinity in general, which is exemplified by research in salmonids [3,8,9,28]. Many of these species exhibit an isoform switch from NKA α1b (3 Na+ per cycle) to α1a (2 Na+ per cycle) when they move from seawater to freshwater, or vice versa. It seems likely that the NKA α1 isoform switching associated with euryhalinity in salmonids, as well as other members of the Protacanthopterygii [11], may not extend to many euryhaline fish. This is supported by work on Japanese medaka, a euryhaline fish of the Acanthopterygii superorder, which demonstrated that this species does not exhibit isoform switching upon salinity transfer [29]. Overall, this analysis would suggest that NKA isoform switching is only one possible route to extreme hypo-osmotic acclimation and euryhalinity in teleost fishes.
4. Methods
(a). Animals
Zebrafish were obtained from local suppliers (unknown pet store strain) and housed at the University of Miami Rosenstiel School of Marine and Atmospheric Science. Fish were split into two groups and maintained for one month under flow-through conditions in dechlorinated freshwater with either 14 or 1.5 mM Na+ (see electronic supplementary material, table S2 for water quality). Each group was held in a 120 l aquarium at 23–25°C with a 16 : 8 h light/dark cycle and fed Tetramin flake food daily. After the one-month acclimation period, fish were acutely transferred to a lower salinity regime (14 mM to 1.5 mM and 1.5 mM to 0.01 mM Na+) and maintained under static-renewal conditions for the remainder of the experiment. In each group, fish were sampled (n = 10) at 0, 0.5, 1, 2, 4, 8, 16 and 32 d after the acute transfer to obtain gill and kidney samples for RNA. Sampled fish were euthanized by an overdose of MS-222 (pH balanced with NaHCO3), and then both gill baskets and the kidney were excised and flash frozen in liquid N2 and stored at −80°C for later RNA extraction. The remainder of the fish were weighed and frozen at −20°C for subsequent analysis of whole body Na+ content. Water samples were collected periodically (n = 6–8) throughout each exposure for determination of Na+, Ca2+, Cl− and pH.
(b). Analytical procedures
To determine whole body Na+ content, fish were digested in 5 volumes (fish mass: volume ratio) of 1 N HNO3 at 70°C for 24 h. Digests were then centrifuged at 2000g for 10 min and an aliquot of the supernatant was removed for Na+ analysis by flame atomic absorption spectrophotometry. Water Na+ and Ca2+ as well as fish digest Na+ was determined by flame atomic absorption spectrophotometry (Varian 220) using an air/acetylene flame. Water Cl− was determined by anion chromatography (Dionex 120) and pH by a Radiometer pH meter (PHM210) and combination electrode (PHC2401-8).
(c). Molecular procedures
See supplemental information for detailed information. In brief, manufacturer protocols were followed for RNA extraction (RNA-STAT 60; Tel-Test), cDNA synthesis (Revertaid Reverse Transcriptase; Thermo Scientific), DNase treatment (Turbo DNA-free Kit; Applied Biosystems) and real-time PCR (Maxima SYBR Green Master Mix Kit; Thermo Scientific). cDNA synthesis was performed using random hexamers, and cDNA dilution curves were used to define the PCR reaction efficiency for each primer pair (93–109%). A no-reverse transcriptase control was included to assess potential genomic DNA contamination, as well as a no-template control to assess other potential sources of contamination. All primer pairs were generated using Primer 3 or Primer Express 2.0 software (Applied Biosystems) with a target annealing temperature of 60°C (electronic supplementary material, table S3). Primer specificity was assessed using NCBI Blast: zebrafish nucleotide sequences as well as dissociation curves after completion of 40 PCR cycles. In all cases only a single dissociation peak was observed. Gene expression was analysed using the ΔΔCt method [30] and normalized to elongation factor 1α. Within-gene analyses are expressed relative to pre-transfer treatment groups. Across-gene analyses for the purposes of relative abundance within a tissue were made relative to the most abundant isoform.
(d). Structural, energetic and phylogenetic analysis
Structural models of NKA based on α subunit protein sequences were generated in PyMol using the backbone of the high resolution (2.4 Å) structure of the E1-form of Ca-ATPase [31], as previously described [12]. The theoretical reaction stoichiometry of the various α1 structures was used to calculate the effect of intracellular Na+ on NKA activation energy according to the methods of Kirschner [13]. Thermodynamic modelling of NHE activity was based on the methods of Parks et al. [23]. Maximum-likelihood analysis was performed using MEGA v. 6.06 following the methods outlined in Esbaugh et al. [32]. Preliminary analyses contained 85 sequences identified through a combination of manual and BLAST searches; however, for illustrative purposes, the final tree was pruned to contain 45. No differences in clade orientation or branch support were noted between the preliminary and final trees. All GenBank accession numbers are presented on the tree, and sequence designations are as reported by GenBank.
Supplementary Material
Acknowledgements
The authors wish to dedicate this paper to Prof. Peter Leth Jørgensen, who sadly passed away during the final stages of its preparation. Peter proposed the original ideas tested and contributed the reported structural analyses of NKA α subunits.
Data accessibility
All data collected as part of this work are directly presented in the text of the manuscript and electronic supplementary material. All real-time PCR primers are made available in the electronic supplementary material alongside the relevant NCBI GenBank accession numbers. All accession numbers for the phylogenetic analysis are included directly in figure 4.
Authors' contributions
K.V.B. performed all animal experimentation and whole body Na+ determinations. A.J.E. and K.V.B. performed all sample processing. A.J.E. performed molecular, phylogenetic and thermodynamic analyses. K.V.B. and M.G. conceived of the study design, and all authors contributed to the critical review of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
Funding for this work was provided in part by a National Science Foundation grant to A.J.E. (EF-1315290). M.G. is a Maytag Professor of Ichthyology.
References
- 1.Marshall WS, Grosell M. 2006. Ion transport, osmoregulation, and acid–base balance. In The physiology of fishes (eds Evans DH, Claiborne JB), 3rd edn, pp. 177–230. New York, NY: Taylor and Francis Group. [Google Scholar]
- 2.Tang CH, Wu WY, Tsai SC, Yoshinaga T, Lee TH. 2010. Elevated Na+/K+-ATPase responses and its potential role in triggering ion reabsorption in kidneys for homeostasis of marine euryhaline milkfish (Chanos chanos) when acclimated to hypotonic fresh water. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 180, 813–824. ( 10.1007/s00360-010-0458-x) [DOI] [PubMed] [Google Scholar]
- 3.Gilmour KM, Perry SF, Esbaugh AJ, Genz J, Taylor JR, Grosell M. 2012. Compensatory regulation of acid-base balance during salinity transfer in rainbow trout (Oncorhynchus mykiss). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 182, 259–274. ( 10.1007/s00360-011-0617-8) [DOI] [PubMed] [Google Scholar]
- 4.Geering K. 2008. Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 17, 526–532. ( 10.1097/MNH.0b013e3283036cbf) [DOI] [PubMed] [Google Scholar]
- 5.Hasler U, Wang X, Crambert G, Béguin P, Jaisser F, Horisberger J-D, Geering K. 1998. Role of beta-subunit domains in the assembly, stable expression, intracellular routing, and functional properties of Na,K-ATPase. J. Biol. Chem. 273, 30 826–30 835. ( 10.1074/jbc.273.46.30826) [DOI] [PubMed] [Google Scholar]
- 6.Vagin O, Dada LA, Tokhtaeva E, Sachs G. 2012. The Na-K-ATPase α1β1 heterodimer as a cell adhesion molecule in epithelia. Am. J. Physiol. Cell Physiol. 302, C1271–C1281. ( 10.1152/ajpcell.00456.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geering K. 2006. FXYD proteins: new regulators of Na-K-ATPase. Am. J. Physiol. Renal. Physiol. 290, F241–F250. ( 10.1152/ajprenal.00126.2005) [DOI] [PubMed] [Google Scholar]
- 8.Richards JG, Semple JW, Bystriansky JS, Schulte PM. 2003. Na+/K+-ATPase α-isoform switching in gills of rainbow trout (Oncorhynchus mykiss) during salinity transfer. J. Exp. Biol. 206, 4475–4486. ( 10.1242/jeb.00701) [DOI] [PubMed] [Google Scholar]
- 9.Bystriansky JS, Richards JG, Schulte PM, Ballantyne JS. 2006. Reciprocal expression of gill Na+/K+-ATPase α subunit isoforms α1a and α1b during seawater acclimation of three salmonid fishes that vary in their salinity tolerance. J. Exp. Biol. 209, 1848–1858. ( 10.1242/jeb.02188) [DOI] [PubMed] [Google Scholar]
- 10.Bystriansky JS, Schulte PM. 2011. Changes in gill H+-ATPase and Na+/K+-ATPase expression and activity during freshwater acclimation of Atlantic salmon (Salmo salar). J. Exp. Biol. 214, 2435–2442. ( 10.1242/jeb.050633) [DOI] [PubMed] [Google Scholar]
- 11.Urbina MA, Schulte PM, Bystriansky JS, Glover CN. 2013. Differential expression of Na+, K+-ATPase α-1 isoforms during seawater acclimation in the amphidromous galaxiid fish Galaxias maculatus. J. Comp. Physiol. B 183, 345–357. ( 10.1007/s00360-012-0719-y) [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen PL. 2008. Importance for absorption of Na+ stop from freshwater of lysine, valine and serine substitutions in the α1a-isoform of Na,K-ATPase in the gills of rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar). J. Membr. Biol. 223, 37–47. ( 10.1007/s00232-008-9111-y) [DOI] [PubMed] [Google Scholar]
- 13.Kirschner LB. 2004. The mechanism of sodium chloride uptake in hyperregulating aquatic animals. J. Exp. Biol. 207, 1439–1452. ( 10.1242/jeb.00907) [DOI] [PubMed] [Google Scholar]
- 14.Saez AG, Lozano E, Zaldivar-Riveron A. 2009. Evolutionary history of Na,K-ATPases and their osmoregulatory role. Genetica 136, 479–490. ( 10.1007/s10709-009-9356-0) [DOI] [PubMed] [Google Scholar]
- 15.Serluca FC, Sidow A, Mably JD, Fishman MC. 2001. Partitioning of tissue expression accompanies multiple duplications of the Na+/K+ ATPase α subunit gene. Genome Res. 11, 1625–1631. ( 10.1101/gr.192001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boisen AM, Amstrup J, Novak I, Grosell M. 2003. Sodium and chloride transport in soft water and hard water acclimated zebrafish (Danio rerio). Biochim. Biophys. Acta 1618, 207–218. ( 10.1016/j.bbamem.2003.08.016) [DOI] [PubMed] [Google Scholar]
- 17.Liao BK, Chen RD, Hwang PP. 2009. Expression regulation of Na+K+-ATPase α1-subunit subtypes in zebrafish gill ionocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 296, R1897–R1906. ( 10.1152/ajpregu.00029.2009) [DOI] [PubMed] [Google Scholar]
- 18.Håkansson KO, Jorgensen PL. 2003. Homology Modeling of Na,K-ATPase. Ann. N. Y. Acad. Sci. 986, 163–167. ( 10.1111/j.1749-6632.2003.tb07155.x) [DOI] [PubMed] [Google Scholar]
- 19.Pedersen PA, Nielsen JM, Rasmussen JH, Jorgensen PL. 1998. Contribution to Tl+, K+, and Na+ binding of Asn776, Ser775, Thr774, Thr772, and Tyr771 in cytoplasmic part of fifth transmembrane segment in α subunit of renal Na,K-ATPase. Biochemistry 37, 17 818–17 827. ( 10.1021/bi981898w) [DOI] [PubMed] [Google Scholar]
- 20.Jewell-Motz EA, Lingrel JB. 1993. Site-directed mutagenesis of the Na,K-ATPase: consequences of substitutions of negatively-charged amino acids localized in the transmembrane domains. Biochemistry 32, 13 523–13 530. ( 10.1021/bi00212a018) [DOI] [PubMed] [Google Scholar]
- 21.Dymowska AK, Boyle D, Schultz AG, Goss GG. 2015. The role of acid-sensing ion channels in epithelial Na+ uptake in adult zebrafish (Danio rerio). J. Exp. Biol. 218, 1244–1251. ( 10.1242/jeb.113118) [DOI] [PubMed] [Google Scholar]
- 22.Kwong RW, Kumai Y, Perry SF. 2014. The physiology of fish at low pH: the zebrafish as a model system. J. Exp. Biol. 217, 651–662. ( 10.1242/jeb.091603) [DOI] [PubMed] [Google Scholar]
- 23.Parks SK, Tresguerres M, Goss GG. 2008. Theoretical considerations underlying Na+ uptake mechanisms in freshwater fishes. Comp. Biochem. Physiol. C. 148, 411–418. ( 10.1016/j.cbpc.2008.03.002) [DOI] [PubMed] [Google Scholar]
- 24.Tipsmark CK. 2008. Identification of FXYD protein genes in a teleost: tissue-specific expression and response to salinity change. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1367–R1378. ( 10.1152/ajpregu.00454.2007) [DOI] [PubMed] [Google Scholar]
- 25.Yang WK, Kang C-K, Chang C-H, Hsu A-D, Lee T-H, Hwang P-P. 2013. Expression profiles of branchial FXYD proteins in the brackish medaka Oryzias dancena: a potential saltwater fish model for studies of osmoregulation. PLoS ONE 8, e55470 ( 10.1371/journal.pone.0055470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayaa M, Vulesevic B, Esbaugh A, Braun M, Ekker ME, Grosell M, Perry SF. 2009. The involvement of SLC26 anion transporters in chloride uptake in zebrafish (Danio rerio) larvae. J. Exp. Biol. 212, 3283–3295. ( 10.1242/jeb.033910) [DOI] [PubMed] [Google Scholar]
- 27.Beyenbach KW. 2001. Energizing epithelial transport with the vacuolar H+-ATPase. Physiology 16, 145–151. ( 10.1152/physiologyonline.2001.16.4.145) [DOI] [PubMed] [Google Scholar]
- 28.Esbaugh AJ, Kristensen T, Takle H, Grosell M. 2014. The effects of sustained aerobic swimming on osmoregulatory pathways in Atlantic salmon Salar salar smolts. J. Fish Biol. 85, 1355–1368. ( 10.1111/jfb.12475) [DOI] [PubMed] [Google Scholar]
- 29.Bollinger RJ, Madsen SS, Bossus MC, Tipsmark CK. 2016. Does Japanese medaka (Oryzias latipes) exhibit a gill Na+/K+-ATPase isoform switch during salinity change? J. Comp. Physiol. B 186, 485–501. ( 10.1007/s00360-016-0972-6) [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 45 ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyoshima C, Nakasako M, Nomura H, Ogawa H. 2000. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655. ( 10.1038/35015017) [DOI] [PubMed] [Google Scholar]
- 32.Esbaugh AJ, Tufts BL. 2006. Evidence of a high activity carbonic anhydrase isozyme in the red blood cells of an ancient vertebrate, the sea lamprey (Petromyzon marinus). J. Exp. Biol. 209, 1169–1178. ( 10.1242/jeb.02111) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data collected as part of this work are directly presented in the text of the manuscript and electronic supplementary material. All real-time PCR primers are made available in the electronic supplementary material alongside the relevant NCBI GenBank accession numbers. All accession numbers for the phylogenetic analysis are included directly in figure 4.