Abstract
Free-living species vary substantially in the extent of their spatial distributions. However, distributions of parasitic species have not been comprehensively compared in this context. We investigated which factors most influence the geographical extent of mammal parasites. Using the Global Mammal Parasite Database we analysed 17 818 individual geospatial records on 1806 parasite species (encompassing viruses, bacteria, protozoa, arthropods and helminths) that infect 396 carnivore, ungulate and primate host species. As a measure of the geographical extent of each parasite species we quantified the number and area of world ecoregions occupied by each. To evaluate the importance of variables influencing the summed area of ecoregions occupied by a parasite species, we used Bayesian network analysis of a subset (n = 866) of the parasites in our database that had at least two host species and complete information on parasite traits. We found that parasites that covered more geographical area had a greater number of host species, higher average phylogenetic relatedness between host species and more sampling effort. Host and parasite taxonomic groups had weak and indirect effects on parasite ecoregion area; parasite transmission mode had virtually no effect. Mechanistically, a greater number of host species probably increases both the collective abundance and habitat breadth of hosts, providing more opportunities for a parasite to have an expansive range. Furthermore, even though mammals are one of the best-studied animal classes, the ecoregion area occupied by their parasites is strongly sensitive to sampling effort, implying mammal parasites are undersampled. Overall, our results support that parasite geographical extent is largely controlled by host characteristics, many of which are subsumed within host taxonomic identity.
Keywords: biogeography, distributions, emerging diseases, geographical range sizes, host–parasite interactions, macroecology
1. Introduction
The factors governing the distributions of parasites are poorly understood. Attempts to estimate the range of non-human parasites are rare in the literature (but see [1–3]). Even in the case of relatively well-studied parasites such as human pathogens it is unclear why some species remain confined to relatively small regions, whereas others are able to become globally distributed. For example, Zika virus was first reported in Brazil in March 2015, and a little over a year later in July 2016 the first cases of infections from local sources were reported in the US [4,5]. In contrast, Middle East respiratory syndrome was first identified in Saudi Arabia in 2012, and to the present day has failed to establish populations elsewhere [6]. It is also unclear whether parasites tend to inhabit most or only a subset of the geographical ranges of hosts that they infect [2], and thus the extent to which patterns of parasite range variation are driven by variation in host geographical distribution. Detailed information on parasite host-range filling and the chronology of parasite geographical spread that would allow these questions to be addressed directly are still relatively rare. However, there are many pathogens and parasites for which it is possible to derive estimates of their current geographical area, and thus investigate their characteristics that tend to associate with small versus large geographical ranges. These findings would not only enhance our understanding of parasite biogeography, they might also provide insight into some of the factors controlling pathogen outbreaks (e.g. what types of parasites have the greatest potential to become distributed across large geographical regions).
Though there have been a large number of studies modelling the geographical distribution and transmission dynamics of particular parasites and pathogens (e.g. [7–9]), to date there have been no broad studies of the factors that affect variation in geographical range area across multiple parasite taxa. However, a case has been made that a large-scale, or even a macroecological, examination of them would be fruitful [10]. Parasites can only exist in places where they have potential hosts, but precisely what aspects of parasite–host variation most affect parasite geographical range remains unclear. For example, do parasites that infect many hosts, or a broad diversity of hosts, tend to occur over larger areas? It is also unclear what role parasite traits such as taxon or transmission mode have on geographical range. For example, do environmentally transmitted bacteria tend to have larger ranges than vector-borne protozoa?
A number of studies have addressed the variation in geographical ranges in taxonomic groups of multiple free-living species (e.g. [11–14]). There has been relatively little consensus across these studies on universal factors driving variation among species, yet within studies there are often important factors identified for specific groups or geographical regions. For example, various studies have uncovered the influence of species traits such as body size and dispersal ability, interactions between environmental variation and species physiological tolerances, historical biogeographic processes, or even null patterns such as the mid-domain effect (e.g. [15,16]). Findings from these studies now enable us to ask questions about whether species interaction networks change across a species's geographical range, or whether the diversity of habitats that a species occupies across its range can help to buffer that species to future climate change scenarios. Macroecology has formalized the statistical examinations of such processes at large spatial scales [17–19]. Yet it is interesting that few studies have applied large-scale analyses of geographical extent across parasite taxa, perhaps because of the associated challenges in acquiring the relevant data. Many of the factors that affect variation in range area in free-living organisms, such as dispersal ability or population density, might also be important in parasites. However, parasites are unable to reproduce without infecting a host, and so have ranges that are likely to be much more strongly constrained by the ranges of other species than is common in free-living species [20]. Ultimately, the range of a parasite cannot exceed that of its hosts and so represents a unique macroecological phenomenon that has been relatively little studied [10].
To be sure, many efforts have explored spatial patterns of parasite diversity (e.g. [2,21,22]), and such estimates ultimately depend upon making assumptions about the geographical range of each species. Thus these efforts share similarities with those that emphasize geographical distributions, although the former tend to focus more on causes of diversity hotspots, as opposed to factors controlling range distributions and possible expansion. For example, Thieltges et al. [23] addressed whether parasite richness coincided with richness patterns of hosts, and if parasite richness showed latitudinal patterns. In the example that most closely approximates our approach, Krasnov et al. [2] examined geographical ranges in fleas as a function of host specificity and taxonomic distance between host species. Although limited in geographical extent and taxonomic focus, Krasnov et al.'s study is one of the first to analyse the geographical extent of parasites as opposed to just spatial patterns of richness. Studying nine of the world's most widespread helminth parasites, Wells et al. [24] showed that five inhabited a lower phylogenetic or functional diversity of hosts than would be expected based on the host diversity of zoogeographical regions they occur in. This result implies that host breadth is limited by the ability of the parasite to adapt to hosts with differing characteristics. However, the study was limited to nine species and did not consider any species with relatively restricted ranges (e.g. restricted to a single continental region). It remains to be seen what factors influence parasite range area when large numbers of parasite species from taxonomically diverse groups and with a variety of geographical extents are considered.
In our study, we build upon these previous efforts to statistically model the factors that are correlated with variation in the geographical range of parasites (defined broadly as viruses, bacteria, protozoa, arthropods, helminths and fungi) that infect terrestrial mammals. Such an analysis not only addresses basic questions about species' distributions, but also can inform which parasites may be most sensitive to global changes or most likely to emerge to spread to new locations. Factors influencing parasite geographical distributions may stem from ecological, physiological and epidemiological characteristics of the host, parasite or both [25,26]. Using the Global Mammal Parasite Database (GMPD) v. 2.0 [27] for each parasite species, we extracted its spatial occurrence data to compute the total area of global ecoregions it occupies. We then looked at patterns of variation in multiple parasite traits as well as traits of the hosts that they infect to determine what factors best explain differences in parasites' geographical extent.
We hypothesized that parasites that infect greater numbers of hosts species are likely to have the potential to occupy larger geographical ranges, but it is unclear what aspect of host breadth would be most influential (e.g. host number versus range area). We therefore tested for the influence of simple host richness, host phylogenetic diversity [28] and the summed geographical range area of hosts on parasite range. We also hypothesized that parasite traits such as taxonomic group (e.g. viruses versus helminths) or transmission mode [29] might be influential. Finally, variation in the geographical ranges of free-living species is often correlated with species traits such as body mass, population density or species ecological characteristics [30,31]. Since the potential range of a parasite ultimately depends on the availability of suitable hosts, and presumably hosts also vary in the extent to which susceptibility varies across their range, some patterns of parasite range variation might be related to the characteristics of the hosts that parasites happen to infect. To test for the indirect influence of such factors, which might influence realized parasite ranges, we tested whether parasites that infect hosts in different taxonomic groups (i.e. primates, carnivores and hoofed mammals) show different patterns of range extent. For the first time we consider patterns in the geographical extent of multiple parasite taxa at a global scale in order to increase the understanding of parasite distributions, especially what makes some species relatively widespread and others geographically restricted.
2. Methods
We sought to comprehensively weigh the relative importance of factors that influence parasite geographical extent, including parasite taxonomy, parasite transmission mode, host taxonomy, host geographical range, host species richness and host phylogenetic diversity, while also controlling for sampling effort. We gathered information on parasite (and host) geographical occurrences from sampling locations reported in the GMPD v. 2.0 [27]. The GMPD is a database of the parasites, defined broadly as all disease-causing organisms including viruses, bacteria, protozoa, helminths, fungi and arthropods that infect wild mammals of the orders Carnivora, Primates, Artiodactyla and Perissodactyla. Stephens et al. [27] contains host–parasite associations with presence-only data. The data were published as ‘.csv’ files, and the metadata of Stephens et al. [27] contains detailed explanations on the databases used as taxonomic guidance. In our analyses we included only parasite occurrences that were georeferenced. For every parasite species, we used the raster package [32] in R to overlay its occurrence points on a map of The Nature Conservancy's Terrestrial Ecoregions of the world (2003, http://maps.tnc.org/gis_data.html), which are available as shapefiles. These ecoregions contain a distinct assemblage of communities and species, and were originally defined by Olson et al. [33]. The classification was developed by Olson et al. [33] reviewing all existent biogeographic classifications and consulting with regional experts in biogeography, taxonomy, conservation and ecology. The ecoregions are geographical units with biological importance that were developed to help with macroecological studies and conservation planning at both regional and global scales. We then summed the area of all occupied ecoregions to yield the ecoregion area occupied by each parasite, and used this as the response variable in all subsequent analyses. There are 814 terrestrial ecoregions in The Nature Conservancy's database with mean area of 181 120 km2 (±468 910, s.d.). We chose to focus on ecoregion occupancy as opposed to other potential measures of parasite range area because ecoregions are likely to contain information about the factors that affect the range limits of both parasites and their hosts, without functionally being defined based explicitly on host ranges. For example it allows us to test whether summed host geographical extent or simple host number better predicts the number of ecoregions that parasites occur in. A further advantage of the ecoregion response variable over polygon methods like convex hull is that it can operate even with low sample sizes, and helps to alleviate some bias from under-sampling by assuming that if a parasite is found in an ecoregion it is capable of existing throughout that entire (and presumably largely homogeneous) ecoregion.
We explored response variables in addition to parasite ecoregion area in preliminary analyses. For example, we performed analyses using the number, rather than area, of ecoregions that parasites inhabit as a response variable; however, results using either ecoregion number or ecoregion area are highly similar given the high correlation of both response variables (R2 = 0.84). We settled on ecoregion area as a measure of the maximum geographical range in which a parasite is likely to occur, in part because it is continuous and thus more robust than count data for analyses. To enable uniformity in our comparisons, we excluded parasites of marine mammals and only used terrestrial ecoregions. In total, our data for analysis contained 1806 parasite species with 17 818 total geographical occurrence points.
Our study looks at parasite presence in ecoregions only within their mammal hosts; for a parasite that can infect multiple hosts beyond mammals, this could be smaller than its total range. Specifically, GMPD has no information on non-mammalian hosts. Thus, a parasite with broad host specificity that includes non-mammalian hosts will only show its range within the mammal hosts included in GMPD (ungulates, primates and carnivores).
We examined the following predictor variables to determine their relative influence on parasite ecoregion area.
Number of host species. For each parasite species we counted the total number of host species it was recorded to use. Two other variables related to host breadth—host geographical range area and the total phylogenetic diversity (relatedness) of host species—were considered as potential predictors. However, these two variables were found to have less explanatory power on parasite ecoregion area and were excluded from further analyses (electronic supplementary material, appendix S1 and figure S1).
Average host phylogenetic diversity. Even though total phylogenetic diversity of hosts (i.e. the summed total branch lengths of a phylogeny of hosts in units of millions of years divergence) was excluded due to its high collinearity with the number of host species (electronic supplementary material, appendix S1), we reasoned that host phylogenetic breadth might contain some information on parasite ecoregion area that was not captured by number of host species alone. We therefore sought a measure of phylogenetic diversity that was independent of the number of host species to use as a potential additional predictor in further analyses. Two measures of phylogenetic diversity were considered: (i) average phylogenetic diversity between hosts (i.e. total phylogenetic diversity of hosts divided by number of host species), and (ii) deviations of observed total phylogenetic diversity of hosts from expected total phylogenetic diversity of hosts using null models, where the same number of hosts that are infected by a given parasite species are selected at random from the mammalian supertree [34,35]. The mammalian supertree was pruned down to only those host species that were infected by a given parasite species. The latter measure proved empirically to be strongly negatively correlated with host richness (which is already included as a variable), whereas the former—average phylogenetic diversity—was weakly correlated with the number of host species (R2 = 0.17). Therefore, to capture variation in host phylogenetic diversity that was not simply driven by the number of host species, we used average phylogenetic diversity as an additional predictor. Because average phylogenetic diversity can only be calculated for parasites with more than one host species, the inclusion of phylogenetic diversity as a variable results in a smaller sample size of parasites for analyses that include it.
Citation count. Variation in sampling effort among parasite species can be a strong predictor of the number of hosts and localities in which the parasites are reported to occur (e.g. [36,37]). To control for the degree of sampling effort on the size of a parasite's ecoregion area, we used the overall citation count for each parasite species within the GMPD. We chose this variable as our measure of sampling effort over other measures that we considered, primarily because it could be applied unambiguously to each parasite. Other measures such as number of samples or number of hosts sampled vary considerably in their sensitivity and accuracy depending upon the method used (e.g. visual examination versus antibodies versus PCR). Further, the latter methods would have in some cases greatly overestimated the effective sampling effort that has been applied to understanding the geographical extent of a parasite. For example, O'Brien et al. [38] sampled more than 62 000 individual hosts for Mycobacterium bovis, all from the state of Michigan.
Host taxonomy. For each parasite in our database, we determined whether it infected any hosts in each of three taxonomic groups: Carnivora, Ungulates (Artiodactyla + Perissodactyla) and Primates. These three binary variables (one for each group) were included as indicator variables. Although ‘ungulate’ is a paraphyletic grouping, ungulates have important ecological similarities and have been considered as a group in numerous past comparative and disease ecology studies (e.g. [39–41]).
Other host traits. We considered incorporating into the model other predictive variables related to host traits such as sociality, home range, body mass, maximum longevity, social group size and trophic level that could influence the rates of parasite infection and transmission, and thus parasite ecoregion area. We ultimately decided against this for several reasons. First, because each parasite often has multiple hosts, we would have had to average the information on traits for all hosts, which was often nonsensical, especially for categorical variables. Second, the number of hosts for which we had trait data strongly varied across traits, making the data fairly noisy. Third, traits correlate strongly with host taxonomy which is a variable already included in the model.
Parasite taxonomy and transmission modes. Information on parasite taxonomy and transmission mode was extracted from the GMPD v. 2.0 [27]. Parasites were grouped taxonomically into one of six groups: arthropods, helminths, protozoa, fungi, viruses and bacteria. Transmission modes for parasites (when known, as was the case for 68%) were grouped into the following non-mutually exclusive categories:
—Close: highly contagious and communicable by close proximity or direct contact such as biting, scratching, mating contact or other touching.
—Non-close: transmission via soil, water, faeces, fomites or other environmental contamination.
—Vector: when transmission is via a biting arthropod (e.g. mosquito) or other vector.
—Intermediate: parasites with intermediate hosts, including the presence of complex life cycle and/or trophic transmission.
(a). Analysis
Initial analyses were performed using boosted regression trees (BRT) to predict parasite ecoregion area [42,43]. Because BRT are robust to incomplete data, this method allowed us to perform analyses using our entire dataset (e.g. including parasites for which we had no transmission mode data and single host parasites for which we could not estimate phylogenetic diversity). We built 20 000 trees and applied 10-fold cross-validation with a learning rate of 0.001 and an interaction depth of 5 during model building to prevent overfitting, and used permutation procedures to generate relative importance scores for each predictor variable. Analyses were performed using the gbm package in R (electronic supplementary material, appendix S2). To better explore the causal relationships among predictor variables in our study we next applied Bayesian network analysis, which is a type of directed acyclic graph similar to family trees that structures the data as a network of conditional probabilities with linear relationships between variables [44,45]. To learn the network structure, we used number of host species, citation count, average host phylogenetic diversity, host taxonomy, parasite taxonomy and parasite ecoregion area to perform Tabu Search [46], a modified hill-climbing algorithm. The models are fitted using maximum-likelihood estimation and the best-fitting model is selected by the lowest Bayesian information criterion (BIC). Because the network is learned directly from the data with minimal prior assumption, Bayesian networks are useful for inferring causal relationships from variables and to summarize the structure of the data (electronic supplementary material, appendix S2). Finally, to explore quantitatively how the two best variables from the Bayesian network analysis individually affected parasite ecoregion area (perhaps non-linearly), we used Gaussian general additive models (GAMs) [47] of citation count and the number of host species against parasite ecoregion area (all variables log10-transformed). For additional details about these analyses, further justification for choice of methods, packages used for analyses and sample R code for Bayesian network analyses, see electronic supplementary material, appendix S2.
3. Results
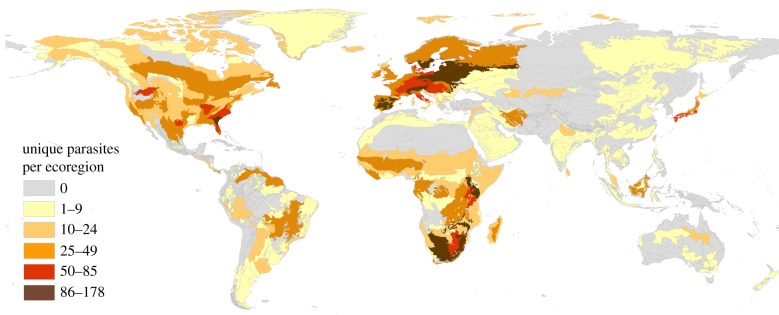
On average, a parasite in our full database (n = 1806) had 5.2 (±11.61, s.d.) geographical occurrence points, was spread across 3.49 ecoregions (an area equivalent to 1 023 293 km2), and had 3.18 hosts and 4 citation counts; however, there was considerable variation in these values, with coefficients of variation between 150% and 200% (electronic supplementary material, figure S4). The richness of mammalian parasite species was heterogeneously spread around the globe, with hotspots including Europe, southern Africa, Japan and the southeastern United States (figure 1). The richest ecoregions had up to 178 parasites, while many ecoregions had no recorded parasite occurrences (figure 1).
Figure 1.
Map of 814 world ecoregions coded by the number of parasite species found in ungulate, carnivore and primate hosts (n = 1806 parasite species).
For parasites with more than one host and for which we had complete information including average phylogenetic diversity (n = 866), the average parasite had 9.1 geographical occurrence points, was spread across 5.85 ecoregions (which corresponded to an average area of 1 698 244 km2), and had 5.56 host species and 6.9 citation counts. The documented average host phylogenetic diversity was 28.09 million years (electronic supplementary material, figure S5). Among the mammal host groups, many metrics, including geographical occurrence points per parasite species, number of ecoregions, citation count and average phylogenetic diversity of hosts, trended higher in the carnivores (electronic supplementary material, figure S5).
The BRT on the full dataset achieved a good fit to the observed data, explaining 59% of variance in parasite geographical range area. Important predictors were positive effects of citation count and host species per parasite, and a negative effect of average host phylogenetic diversity, with relative influence scores of 62.3, 10.2 and 12.6 respectively. BRT analyses showed that parasite traits such as taxonomy and transmission mode influenced variation in ecoregion area among parasite species much less than factors related to the hosts that parasites infect, such as average host phylogenetic diversity and host taxonomic group (electronic supplementary material, table S1).
In the Bayesian network analysis the strongest direct links to parasite ecoregion area were the number of host species and citation count, which both had positive effects, and average host phylogenetic diversity, which had a negative effect (figure 2; electronic supplementary material, table S2). These three variables explained the majority of the variation in parasite ecoregion area (electronic supplementary material, table S2). The boosted regression trees also confirmed the strongest influence from these same variables (electronic supplementary material, table S1). Two host taxonomic categories (carnivore and ungulate) and one parasite taxonomic category (arthropod) slightly affected parasite ecoregion area (electronic supplementary material, table S1), but were too weak to show up in the Bayesian network analysis (figure 2). Citation count, in addition to positively predicting parasite ecoregion area, also strongly predicted the number of host species per parasite. The GAM models of citation count and number of host species on parasite ecoregion area isolate the influence of these particularly strong variables, which indicated no signs of asymptoting (electronic supplementary material, figure S2).
Figure 2.
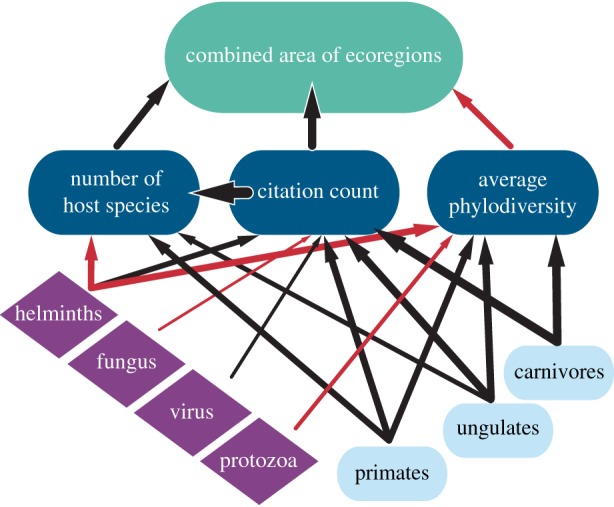
Bayesian network analysis on variables affecting the amount of ecoregion area occupied by 866 mammal parasite species. All continuous variables were log10-transformed for this analysis. For predictor variables, dark blue boxes are the three non-taxonomic variables (average host phylogenetic diversity, number of host species, and citation count); purple boxes are parasite groups; and light blue boxes are host groups. Thickness of arrows represent the strength of log-normalized conditional probabilities of significant dependencies between variables (2.5 × (ln (|x|) + 2)) and the direction of effect is indicated by colour (black = positive; red = negative effects). Electronic supplementary material, table S2 reports the overall structure of these relationships.
Host and parasite taxonomic categories only affected parasite ecoregion area indirectly through host phylogenetic diversity, number of host species, and citation count. The significant effects of host and parasite groups indicate differences across these categories in both sampling effort and biology. For example, citation count and host phylogenetic diversity were most positively influenced by parasites infecting carnivores. Citation count was significantly lower for fungi, while host phylogenetic diversity was significantly lower (i.e. hosts are more related) among helminths and protozoan parasites (figure 2; electronic supplementary material, table S2). One of the predictor variables (bacteria) did not significantly affect citation count, host phylogenetic diversity, number of host species or parasite ecoregion, and hence is not included in figure 2.
4. Discussion
The most widespread mammal parasites are well studied (high citation count), have many host species and have close genetic relatedness among their host species (low phylogenetic diversity given the number of hosts they infect). The variable with the strongest direct effect on ecoregion area was citation count (figure 2). Across the 866 species of parasites that our Bayesian network analysis considers, those that have been more thoroughly studied are known to occur in more ecoregions. For example, the three parasite species with the largest identified ecoregion areas are Toxoplasma gondii, Morbillivirus Canine morbillivirus (canine distemper) and Lyssavirus Rabies lyssavirus, which have the first, second and sixth most studies, respectively. Plots of citation count versus ecoregion area (electronic supplementary material, figure S2a) also showed no evidence of an asymptote, which implies that the known ranges of most parasite species would be even larger if they were better sampled. Taken together, these results indicate that the current picture of the range limits of most parasites is highly incomplete; parasites generally occur in more regions than those in which they have been documented. It is sobering that mammals are collectively one of the best-studied host groups, yet still remain highly under-sampled for parasites. However, our results also provide insights into the biological drivers of variation in range area, and the correlations we observed with these predictors do not appear to be driven solely by sampling effort.
Sampling effort had a strong influence on ecoregion area but an even stronger influence on the number of host species (figure 2). The latter result is concerning because host number was also the strongest biological predictor of ecoregion area. We therefore took sampling bias into account by two methods: (i) using citation count as a covariate (i.e. as an additional predictor variable, figure 2), and (ii) using residuals of a regression model of host number on citation count as a predictor and using residuals of ecoregion area on citation count as the response variable. The latter approach is similar to how studies of variation in parasite richness among hosts have generally dealt with sampling bias (e.g. [40,48]). We obtained qualitatively similar results using both methods to account for sampling bias (figure 2; electronic supplementary material, appendix S3 and figure S3). Regardless of the fact that the range areas of most parasites included in our analyses appear underestimated, our picture of the biological factors that are important in driving variation in range area among species is statistically robust.
The strongest biological predictor of ecoregion area occupied by parasites was number of host species. Surprisingly, this simple metric in our preliminary analyses outperformed two alternative measures of host breadth: total phylogenetic diversity and the summed area of hosts (electronic supplementary material, appendix S1), both of which presumably contain more information than host richness alone. Parasites that infect more hosts occur in a greater area of ecoregions. Whether they infect distantly related or closely related hosts, or happen to infect hosts with large geographical ranges, is less important. The latter result probably indicates a mismatch between host range area and parasite range in some cases. Parasites that happen to infect an extremely wide-ranging species such as red fox do not necessarily occur throughout every ecoregion that their host occurs in. Even more surprisingly, our measure of phylogenetic diversity that was independent of host number—average phylogenetic diversity—was actually slightly negatively correlated with ecoregion area. This indicates that parasites that infect distantly related hosts (i.e. high average phylogenetic diversity) often do so in hosts that occur within fewer and/or smaller ecoregions. This is opposite to the pattern demonstrated by Krasnov et al. [2] in fleas. Furthermore, our pattern initially seems counterintuitive since one might guess that parasites with a phylogenetically diverse host spectrum might be capable of occupying diverse ecoregions, and thus increasing parasite ecoregion area. However, taken together, our results indicate that environmental limits on parasite ranges might be more important than previously recognized [49,50]. Parasites able to overcome these environmental challenges are also generally able to infect many host species. Perhaps the ability to make an evolutionary jump to distantly related hosts does not provide as much ecological opportunity as simply being able to bypass climatological or geographical barriers (especially important for ectoparasites or parasites with intermediate hosts). The fact that ecoregion area and average host phylogenetic diversity were negatively correlated might even indicate a trade-off between the ability to cross phylogenetic and geographical distance (i.e. spread to hosts within or between ecoregions).
Among the other biological predictors that we considered, none of the host or parasite taxonomic groups directly affected ecoregion area (figure 2), and they only influenced ecoregion area indirectly through other factors that had direct effects on the parasite ecoregion area. For example, all three of the mammal host groups affected citation count, phylogenetic diversity and number of hosts, which all had strong effects on parasite ecosystem area. In addition, fungal pathogens tended to be relatively poorly studied and viral pathogens tended to be well studied, but there was no evidence of a direct effect of these taxonomic groups on ecoregion area or other predictor variables.
Of all the factors that we considered in this study, parasite transmission mode had the weakest influence on parasite range area. Perhaps the influence of transmission mode is weakened by different directions of effects between parasite taxa. Pederson et al. [51] found that the transmission mode of primate parasites correlated with host specificity within parasite groups, but not across parasite groups, due to contrasting and variable relationships among parasite groups. In our preliminary boosted regression tree analyses, transmission modes always had relative influence scores of 2% or much less. Boosted regression trees allowed us to incorporate missing data, and so allowed us to include transmission mode in an analysis of our full dataset. During early analyses where we included transmission modes in Bayesian network analyses, we confirmed that transmission modes have no direct effect on ecoregion area, and only affected other variables to the extent that parasites in different taxonomic groups tend to exhibit similar patterns of transmission (i.e. for any given transmission mode the great majority of parasites in any given group had the same score of 0 or 1). However, this method uses complete case analysis, which forced us to throw out a large percentage of our data, adding considerable noise to our dataset, reducing model fits and obscuring some of the statistical relationships among predictors. For this reason we excluded transmission mode from our final Bayesian network analyses (figure 2). Regardless, we found little evidence that parasite traits apart from taxonomy have much influence on parasite range area (electronic supplementary material, table S1). Even parasites restricted to transmission by close contact can have extremely large geographical ranges, and even environmentally transmitted parasites can have quite small ranges.
Compared with data pooled across all hosts, parasites that infect members of any particular host group show higher average host phylogenetic diversity (e.g. the overall average host phylogenetic diversity was 28.09 million years, but that of carnivore-hosted parasites was 37.11 million years). This is partially a statistical artefact related to binning parasites into lists of those that infect hosts of a particular group. The parasites that infect members of any one host group (i.e. primates, carnivores or ungulates) will include roughly two-thirds generalist species that infect hosts in multiple groups (i.e. generalist parasites with a high host phylogenetic diversity) and one-third parasites are endemic to a single group (i.e. specialists with low phylogenetic diversity). In contrast, when the global parasite pool is considered, the list will include roughly the same number of generalist parasites, but also all of the specialist parasites, driving down average host breadth. In addition, carnivore parasites seem to infect the highest average phylogenetic diversity of hosts, whereas primate parasite species infect hosts with the lowest phylogenetic diversity. The former result is possibly related to the ecology of carnivores. Almost all carnivores are mammalian predators to some extent [52]. These direct ecological interactions between carnivores and members of other taxa may provide opportunities for parasites to spread between distantly related hosts. Thus it might be expected that parasites that infect carnivores also tend to infect an unusually wide phylogenetic range of hosts.
Conversely, the unusually low host phylogenetic diversity of primate parasites may be related to the biogeography and phylogenetic history of primates. Most primates in the GMPD belong to either the New World monkeys (Haplorrhini) or Old World monkeys and apes (a clade consisting of Cercopithecoidea + Hominoidea). These sister clades diverged less than 40 million years ago, and both have crown ages of less than 30 million years [53]. In contrast, most regional assemblages of carnivores and ungulates contain representatives of lineages that diverged in the much more distant past. For example, any regional assemblage of carnivores that contains at least one species of fox and bear includes species that diverged roughly 60 million years ago [54], and any ungulate assemblage that contains at least one artiodactyl and one perissodactyl includes hosts that diverged 85 million years ago [35]. Thus parasites that infect similar numbers of hosts would be expected to infect a lower phylogenetic range of primate hosts than carnivore or ungulate hosts, particularly if endemic to a single hemisphere.
The factors that seem to drive variation in geographical range in parasites are quite different from those that have been reported in free-living organisms (reviewed in [31]). One of the major factors that contributes to variation in geographical range size among free-living species is body size, with larger species tending to have larger geographical ranges [30]. Although some parasite groups such as helminths exhibit substantial body size variation, it is doubtful that this is an important source of variation in the range size of single cell and viral parasites. Preliminary analyses also showed that the geographical area of hosts was a weak predictor of parasite ecoregion area (electronic supplementary material, appendix S1), making it seem unlikely that parasites that happen to infect larger hosts (i.e. which presumably tend to have larger ranges) have wider geographical ranges. In free-living species, taxonomic group is also a strong predictor of variation in geographical range (e.g. [55]), whereas in our analyses parasite taxonomy had no direct effects on ecoregion area (figure 2). There is evidence in some groups that free-living species with greater dispersal ability tend to have larger geographical ranges (e.g. [31,56,57]). In parasites, dispersal ability is presumably related to transmission mode, and transmission mode had almost no influence on ecoregion area or host number in our boosted regression tree analyses (electronic supplementary material, table S1). However, the ability to infect a wider range of hosts could itself be considered greater ‘dispersal’ ability. Finally, a common pattern in free-living organisms is that species that occur at higher latitudes also tend to have larger geographical ranges (i.e. Rapoport's rule [21,58]). There is some evidence that this pattern also occurs in parasites [59], and this pattern probably does occur in our data. However it would be driven at least partially by the fact that smaller ecoregions tend to be concentrated at low to mid-latitudes [33].
In sum, although mammals are one of the best-studied animal classes, the ecoregion area occupied by their parasites is strongly sensitive to sampling effort, implying mammal parasites are undersampled. Nonetheless, influences of important variables still emerge, the most important of which is the strong positive effect of the number of hosts a parasite has on the total area of ecoregions occupied by the parasite. Mechanistically, a greater number of host species likely increases both the collective abundance and habitat breadth of hosts, providing more opportunities for a parasite to have an expansive range. Thus, parasites that infect many host species and certain types of host species have big areas; parasite geographical extent is largely controlled by host characteristics, many of which are subsumed within host taxonomic identity. Parasite traits such as transmission mode have little direct explanatory power after accounting for these dominant influences of hosts.
Supplementary Material
Supplementary Material
Acknowledgements
For useful discussion and feedback, we thank Max Farrell, John Drake and the participants in the Macroecology of Infectious Disease Research Coordination Network.
Data accessibility
The data used in this study are accessible in the GMPD v. 2.0 [27] and are part of the electronic archives of the Ecology Society of America: https://esajournals.onlinelibrary.wiley.com/doi/10.1002/ecy.1799/suppinfo.Manuscript. The dataset and model code supporting this article are uploaded as part of the electronic supplementary material and available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bd3v5gk [60].
Competing interests
We declare we have no competing interests.
Funding
The study was funded by NSF, DEB no. 1316223, Macroecology of Infectious Disease Research Coordination Network (P. Stephens, PI).
References
- 1.Robinson AF, Inserra RN, Caswell-Chen EP, Vovlas N, Troccoli A. 1997. Rotylenchulus species: identification, distribution, host ranges, and crop plant resistance. Nematropica 27, 127–180. [Google Scholar]
- 2.Krasnov BR, Poulin R, Shenbrot GI, Mouillot D, Khokhlova IS. 2005. Host specificity and geographic range in haematophagous ectoparasites. Oikos 108, 449–456. ( 10.1111/j.0030-1299.2005.13551.x) [DOI] [Google Scholar]
- 3.Aiken HM, Bott NJ, Mladineo I, Montero FE, Nowak BF, Hayward CJ. 2007. Molecular evidence for cosmopolitan distribution of platyhelminth parasites of tunas (Thunnus spp.). Fish Fish. 8, 167–180. ( 10.1111/j.1467-2679.2007.00248.x) [DOI] [Google Scholar]
- 4.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. 2016. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res. 130, 69–80. ( 10.1016/j.antiviral.2016.03.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grubaugh ND, et al. 2017. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 546, 401–405. ( 10.1038/nature22400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zumla A, Hui DS, Perlman S. 2015. Middle East respiratory syndrome. Lancet 386, 995–1007. ( 10.1016/S0140-6736(15)60454-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensch S, Akesson S. 2003. Temporal and spatial variation of hematozoans in Scandinavian willow warblers. J. Parasitol. 89, 388–391. ( 10.1645/0022-3395(2003)089[0388:TASVOH]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 8.Colizza V, Barrat A, Barthelemy M, Valleron AJ, Vespignani A. 2007. Modeling the worldwide spread of pandemic influenza: baseline case and containment interventions. PLoS Med. 4, 95–110. ( 10.1371/journal.pmed.0040013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, Reference ES. 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7, e1000215 ( 10.1371/journal.pmed.1000215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PR, et al. 2016. The macroecology of infectious diseases: a new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 19, 1159–1171. ( 10.1111/ele.12644) [DOI] [PubMed] [Google Scholar]
- 11.Blackburn TM, Gaston KJ, Quinn RM, Arnold H, Gregory RD. 1997. Of mice and wrens: the relation between abundance and geographic range size in British mammals and birds. Phil. Trans. R. Soc. Lond. B 352, 419–427. ( 10.1098/rstb.1997.0030) [DOI] [Google Scholar]
- 12.Jetz W, Rahbek C. 2002. Geographic range size and determinants of avian species richness. Science 297, 1548–1551. ( 10.1126/science.1072779) [DOI] [PubMed] [Google Scholar]
- 13.Byers JE, et al. 2015. Invasion expansion: time since introduction best predicts global ranges of marine invaders. Sci. Rep. 5, 12436 ( 10.1038/srep12436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pappalardo P, Pringle JM, Wares JP, Byers JE. 2015. The location, strength, and mechanisms behind marine biogeographic boundaries of the east coast of North America. Ecography 38, 722–731. ( 10.1111/ecog.01135) [DOI] [Google Scholar]
- 15.Colwell RK, Lees DC. 2000. The mid-domain effect: geometric constraints on the geography of species richness. Trends Ecol. Evol. 15, 70–76. ( 10.1016/S0169-5347(99)01767-X) [DOI] [PubMed] [Google Scholar]
- 16.Hawkins BA, Diniz JAF, Weis AE. 2005. The mid-domain effect and diversity gradients: is there anything to learn? Am. Nat. 166, E140–E143. ( 10.1086/491686) [DOI] [PubMed] [Google Scholar]
- 17.Brown JH, Maurer BA. 1989. Macroecology—the division of food and space among species on continents. Science 243, 1145–1150. ( 10.1126/science.243.4895.1145) [DOI] [PubMed] [Google Scholar]
- 18.Brown JH, Stevens GC, Kaufman DM. 1996. The geographic range: size, shape, boundaries, and internal structure. Annu. Rev. Ecol. Syst. 27, 597–623. ( 10.1146/annurev.ecolsys.27.1.597) [DOI] [Google Scholar]
- 19.Gaston KJ. 1996. Species-range-size distributions: patterns, mechanisms and implications. Trends Ecol. Evol. 11, 197–201. ( 10.1016/0169-5347(96)10027-6) [DOI] [PubMed] [Google Scholar]
- 20.Poulin R. 2014. Parasite biodiversity revisited: frontiers and constraints. Int. J. Parasitol. 44, 581–589. ( 10.1016/j.ijpara.2014.02.003) [DOI] [PubMed] [Google Scholar]
- 21.Rohde K. 1999. Latitudinal gradients in species diversity and Rapoport's rule revisited: a review of recent work and what can parasites teach us about the causes of the gradients? Ecography 22, 593–613. ( 10.1111/j.1600-0587.1999.tb00509.x) [DOI] [Google Scholar]
- 22.Guernier V, Hochberg ME, Guegan JF. 2004. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2, 740–746. ( 10.1371/journal.pbio.0020141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thieltges DW, Ferguson MAD, Jones CS, Noble LR, Poulin R. 2009. Biogeographical patterns of marine larval trematode parasites in two intermediate snail hosts in Europe. J. Biogeogr. 36, 1493–1501. ( 10.1111/j.1365-2699.2008.02066.x) [DOI] [Google Scholar]
- 24.Wells K, Gibson DI, Clark NJ. 2019. Global patterns in helminth host specificity: phylogenetic and functional diversity of regional host species pools matter. Ecography 42, 416–427. ( 10.1111/ecog.03886) [DOI] [Google Scholar]
- 25.Dallas T, Park AW, Drake JM. 2017. Predictability of helminth parasite host range using information on geography, host traits and parasite community structure. Parasitology 144, 200–205. ( 10.1017/S0031182016001608) [DOI] [PubMed] [Google Scholar]
- 26.Wells K, Gibson DI, Clark NJ, Ribas A, Morand S, McCallum HI. 2018. Global spread of helminth parasites at the human–domestic animal–wildlife interface. Glob. Change Biol. 24, 3254–3265. ( 10.1111/gcb.14064) [DOI] [PubMed] [Google Scholar]
- 27.Stephens PR, et al. 2017. Global Mammal Parasite Database version 2.0 Ecology 98, 1476 ( 10.1002/ecy.1799) [DOI] [PubMed] [Google Scholar]
- 28.Faith DP, Baker AM. 2006. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol. Bioinform. Online 2, 121–128. [PMC free article] [PubMed] [Google Scholar]
- 29.Poulin R. 2011. The many roads to parasitism: a tale of convergence. Adv. Parasit. 74, 1–40. ( 10.1016/B978-0-12-385897-9.00001-X) [DOI] [PubMed] [Google Scholar]
- 30.Gaston KJ, Blackburn TM. 1996. Conservation implications of geographic range size—body size relationships. Conserv. Biol. 10, 638–646. ( 10.1046/j.1523-1739.1996.10020638.x) [DOI] [Google Scholar]
- 31.Gaston KJ. 2003. The structure and dynamics of geographic ranges. Oxford, UK: Oxford University Press. [Google Scholar]
- 32.Hijmans RJ. 2016. raster: Geographic Data Analysis and Modeling. R package version 2.5-8 See https://CRAN.R-project.org/package=raster.
- 33.Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GV, Underwood EC, Loucks CJ. 2001. Terrestrial ecoregions of the world: a new map of life on earth: a new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. Bioscience 51, 933–938. ( 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2) [DOI] [Google Scholar]
- 34.Bininda-Emonds ORP, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512. ( 10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 35.Fritz SA, Bininda-Emonds ORP, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549. ( 10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 36.Walther BA, Cotgreave P, Price RD, Gregory RD, Clayton DH. 1995. Sampling effort and parasite species richness. Parasitol. Today 11, 306–310. ( 10.1016/0169-4758(95)80047-6) [DOI] [PubMed] [Google Scholar]
- 37.Blakeslee AMH, Byers JE. 2008. Using parasites to inform ecological history: comparisons among three congeneric marine snails. Ecology 89, 1068–1078. ( 10.1890/07-0832.1) [DOI] [PubMed] [Google Scholar]
- 38.O'Brien DJ, et al. 2002. Epidemiology of Mycobacterium bovis in free-ranging white-tailed deer, Michigan, USA, 1995–2000. Prev. Vet. Med. 54, 47–63. ( 10.1016/S0167-5877(02)00010-7) [DOI] [PubMed] [Google Scholar]
- 39.Geist V. 1974. On the relationship of social evolution and ecology in ungulates. Am. Zool. 14, 205–220. ( 10.1093/icb/14.1.205) [DOI] [Google Scholar]
- 40.Ezenwa VO, Price SA, Altizer S, Vitone ND, Cook KC. 2006. Host traits and parasite species richness in even- and odd-toed hoofed mammals, Artiodactyla and Perissodactyla. Oikos 115, 526–536. ( 10.1111/j.2006.0030-1299.15186.x) [DOI] [Google Scholar]
- 41.Turner WC, Getz WM. 2010. Seasonal and demographic factors influencing gastrointestinal parasitism in ungulates of Etosha National Park. J. Wildl. Dis. 46, 1108–1119. ( 10.7589/0090-3558-46.4.1108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elith J, Leathwick JR, Hastie T. 2008. A working guide to boosted regression trees. J. Anim. Ecol. 77, 802–813. ( 10.1111/j.1365-2656.2008.01390.x) [DOI] [PubMed] [Google Scholar]
- 43.Ridgeway G, with contributions from others . 2013. gbm: Generalized Boosted Regression Models. R package, version 21.
- 44.Korb K, Nicholson AE. 2010. Bayesian artificial intelligence, 2nd edn Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- 45.Scutari M. 2010. Learning Bayesian Networks with the bnlearn R Package. J. Stat. Softw. 35, 1–22. ( 10.18637/jss.v035.i03)21603108 [DOI] [Google Scholar]
- 46.Glover F. 1986. Future paths for integer programming and links to artificial intelligence. Comput. Oper. Res. 13, 533–549. ( 10.1016/0305-0548(86)90048-1) [DOI] [Google Scholar]
- 47.Wood SN. 2006. Generalized additive models: an introduction with R. Boca Raton, FL: Chapman and Hall. [Google Scholar]
- 48.Huang S, Drake JM, Gittleman JL, Altizer S. 2015. Parasite diversity declines with host evolutionary distinctiveness: a global analysis of carnivores. Evolution 69, 621–630. ( 10.1111/evo.12611) [DOI] [PubMed] [Google Scholar]
- 49.Clark NJ, Clegg SM, Sam K, Goulding W, Koane B, Wells K. 2018. Climate, host phylogeny and the connectivity of host communities govern regional parasite assembly. Divers. Distrib. 24, 13–23. ( 10.1111/ddi.12661) [DOI] [Google Scholar]
- 50.Gehman AM, Hall RJ, Byers JE. 2018. Host and parasite thermal ecology jointly determine the effect of climate warming on epidemic dynamics. Proc. Natl Acad. Sci. USA 115, 744–749. ( 10.1073/pnas.1705067115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen AB, Altizer S, Poss M, Cunningham AA, Nunn CL. 2005. Patterns of host specificity and transmission among parasites of wild primates. Int. J. Parasitol. 35, 647–657. ( 10.1016/j.ijpara.2005.01.005) [DOI] [PubMed] [Google Scholar]
- 52.Hunter L, Barrett P. 2011. Carnivores of the world. Princeton, NJ: Princeton University Press. [Google Scholar]
- 53.Chatterjee HJ, Ho SY, Barnes I, Groves C. 2009. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol. Biol. 9, 259 ( 10.1186/1471-2148-9-259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nyakatura K, Bininda-Emonds OR. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol. 10, 12 ( 10.1186/1741-7007-10-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson S, Marcus LF. 1992. Aerography of Australian tetrapods. Aust. J. Zool. 40, 627–651. ( 10.1071/ZO9920627) [DOI] [Google Scholar]
- 56.Malmqvist B. 2000. How does wing length relate to distribution patterns of stoneflies (Plecoptera) and mayflies (Ephemeroptera)? Biol. Conserv. 93, 271–276. ( 10.1016/S0006-3207(99)00139-1) [DOI] [Google Scholar]
- 57.Dennis RL, Donato B, Sparks TH, Pollard E. 2000. Ecological correlates of island incidence and geographical range among British butterflies. Biodivers. Conserv. 9, 343–359. ( 10.1023/A:1008924329854) [DOI] [Google Scholar]
- 58.Ruggiero A, Werenkraut V. 2007. One-dimensional analyses of Rapoport's rule reviewed through meta-analysis. Glob. Ecol. Biogeogr. 16, 401–414. ( 10.1111/j.1466-8238.2006.00303.x) [DOI] [Google Scholar]
- 59.Krasnov BR, Vinarski MV, Korallo-Vinarskaya NP, Khokhlova IS. 2013. Ecological correlates of body size in gamasid mites parasitic on small mammals: abundance and niche breadth. Ecography 36, 1042–1050. ( 10.1111/j.1600-0587.2012.00140.x) [DOI] [Google Scholar]
- 60.Byers JE, Schmidt JP, Pappalardo P, Haas SE, Stephens PR. 2019. Data from: What factors explain the geographical range of mammalian parasites? Dryad Digital Repository. ( 10.5061/dryad.bd3v5gk) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Byers JE, Schmidt JP, Pappalardo P, Haas SE, Stephens PR. 2019. Data from: What factors explain the geographical range of mammalian parasites? Dryad Digital Repository. ( 10.5061/dryad.bd3v5gk) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data used in this study are accessible in the GMPD v. 2.0 [27] and are part of the electronic archives of the Ecology Society of America: https://esajournals.onlinelibrary.wiley.com/doi/10.1002/ecy.1799/suppinfo.Manuscript. The dataset and model code supporting this article are uploaded as part of the electronic supplementary material and available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bd3v5gk [60].