Abstract
Multiple mechanisms of cell death exist (apoptosis, necroptosis, pyroptosis) and the subtle balance of several distinct proteins and inhibitors tightly regulates the cell fate toward one or the other pathway. Here, by combining coimmunoprecipitation, enzyme assays, and molecular simulations, we ascribe a new role, within this entangled regulatory network, to the interleukin-1 receptor antagonist (lL-1Ra). Our study enlightens that IL-1Ra, which usually inhibits the inflammatory effects of IL-1α/β by binding to IL-1 receptor, under advanced pathological states prevents apoptosis and/or necroptosis unprecedentedly reveal that IL-1Ra binds both caspases at their dimeric interface, preventing, in this manner, the formation of their catalytically/signaling active form. The resulting IL-1Ra/caspase-8(9) adducts are stabilized by hydrophobic and by few key hydrogen bonding interactions, formed by residues fully conserved across distinct caspases (−3, −6, −7, −8, and -9), and closely resemble the binding mode of the caspases inhibitors XIAP (X-linked inhibitor of apoptosis) and c-FLIP (cellular FLICE-like inhibitory protein). Tight regulation of the different forms of cell death has a major impact on distinct human illnesses (i.e., cancer, neurodegeneration, ischemic injury, atherosclerosis, viral/bacterial infections, and immune reaction). Hence, our study, pinpointing IL-1Ra as new actor of the intricate cell death regulatory network and gaining an atomic-scale understanding of its mechanism may open new avenues toward innovative therapeutic strategies to tackle major human diseases.
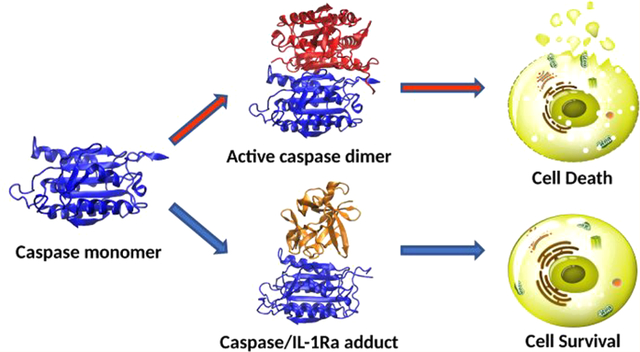
Graphical Abstract
1. INTRODUCTION
Besides passive necrosis, cells may respond to lethal stimuli undergoing a variety of regulated molecular programs, ultimately running cell death.1 Pathways of regulated cell death are (i) apoptosis, a programmed cell death,2 (ii) necroptosis, a form of necrosis executed in a programmed fashion, but occurring in response to extracellular stimuli or to intracellular pathogens,3,4 and (iii) pyroptosis, a programmed cell death of immune cells, caused by intracellular pathogens, and accompanied by the activation of a multimeric enzyme complex called inflammasome. This latter leads to activation and release from the cell of potent inflammatory cytokines, primarily interleukin-1 (IL-1).5,6 Notably, the form of cell death will condition the amplitude of the reaction by the surrounding environment: (i) necroptosis and pyroptosis, likewise necrosis, involve disruption of the cell membrane and release of inflammatory mediators, possibly leading to a systemic reaction by the host; whereas (ii) apoptosis occurs without causing changes in the milieu surrounding the dying cell. Thereafter, the type of cell death bears an actual impact on the feature of important diseases such as is chemic injury, atherosclerosis, cancer, neurodegeneration, viral, and bacterial infections, and of the immune reaction.7,8
Caspases, cysteine aspartate proteases, are the main components of the signaling cascade leading to apoptosis. They are characterized by different activation mechanisms according to their implication in the initial or the terminal part of this signaling cascade (initiator caspases, casp-2, −8, -9, −10 or effector caspases, casp-3, −6, −7). Initiator caspases are produced as monomeric pro-enzymes (pro-caspases) and, after assembling in large multimeric protein complexes, associate into active homodimers, which reach their mature form only after proteolysis of each monomer into a small and a large subunit.9 As a result, initiator caspases transmit their signal to the effector ones, by proteolytically activating them.10 The latter, in turn, trigger release of both cytosol hydrolases and DNA fragmentation enzymes, leading to cellular apoptosis.11 Two canonical apoptotic pathways have been described, that is, the intrinsic and the extrinsic one. The first, triggered by intracellular insults such as DNA damage and hypoxia, involves the release of apoptogenic proteins and cytochrome C from mitochondria, ultimately leading to activation of the initiator casp-9 within a multimeric protein complex called apoptosome. The extrinsic pathway, instead, is induced at the cell membrane level through ligand activation of tumor necrosis factor (TNF)-receptors, by the transduction of this signal via multimeric enzyme complex (i.e., the death inducing signaling complex (DISC)) or via the subsequent activation of enzyme Complex I and II, finally, leading to casp-8 activation. Remarkably, the latter, besides being involved in apoptosis, has recently shown to repress necroptosis and regulate inflammation, suggesting a subtle communication among the distinct death and inflammatory networks.8
Considerable interest has been focused on developing initiator casp-9 and casp-8 inhibitors, capable of withdrawing the cell-scheduled death from apoptosis, thus evolving toward pro-inflammatory forms of cell reaction. X-linked inhibitor of apoptosis (XIAP) potently inhibits the activity of casp-9 and −8.12,13 xiaP prevents the formation of active casp-9 dimer by binding casp-9 monomer at the dimerization surface. Instead in the extrinsic pathway of apoptosis, cellular FLICE-like inhibitory protein (c-FLIP) participates within TNF-receptor multimeric enzyme signal-transduction pathways (DISC or Complex II), inhibiting casp-8 by preventing the formation of active homodimers.7 The mechanism underlying XIAP regulation of casp-8 activity has not yet been clarified, although cells lacking XIAP have been demonstrated to undergo casp-8 mediated apoptotic signal.
Within this complex regulatory network lies also the cytokine interleukin-1 receptor antagonist (IL-1Ra). IL-1Ra exerts an anti-inflammatory activity by competing with IL-1α and IL-1β for binding to IL-1 membrane receptor (IL-1R1).14,15 Besides a secreted protein, three intracellular, unsecreted IL-1Ra isoforms have been described.16 In hypoxic cardiomyocytes, a protective role of intracellular IL-1Ra against apoptosis was recently proposed.17 In particular, IL-1Ra was shown to directly bind casp-9, −3, −6, and −7, inhibiting them with nM (casp-9) and (casp-3, −6, −7) affinity via a noncompetitive mechanism. As a result of this interaction, IL-1Ra was proved to hamper the activation of mitochondria-triggered intrinsic pathway of apoptosis.17,18 Based on clear evidence of IL-1Ra inhibition toward distinct caspases (-9, −3, −6, and −7) and of a direct IL-1Ra involvement in IL-1β activation/regulation, we hypothesized that IL-1Ra may even alter the balance between apoptosis and necroptosis/pyroptosis by directly inhibiting casp-8. Indeed, it seems likely, that the synthesis of intracellular IL-1Ra, by inhibiting casp-8, may play a role in modulating the cellular response to noxious elements and its consequences on the degree of inflammation of the paracellular microenvironment.
Understanding the molecular mechanism regulating this complex signaling network requires an atomic-level knowledge of the interactions between its principal molecular actors. This information may be gained by solving the three-dimensional structure of the protein adducts. Such a knowledge is, however, challenging to obtain for protein complexes, and is so far lacking for caspases/IL-1Ra adducts. Molecular modeling and simulations, in the last years, have largely contributed to reveal the molecular mechanism of biological molecules.19–25 In this scenario, docking simulations are increasingly used to foretell the binding mode and affinity of protein adducts,26 even though an accurate/reliable prediction of their native (minimum energy) binding pose remains challenging to tackle. This is due to the enormous conformational space that docking algorithms need to sample27 and to the limited accuracy of the scoring functions used to rank the set of predicted binding modes.28 A remarkable improvement in identifying the native binding pose can be achieved by employing extensive force field-based molecular dynamics (MD) simulations, to relax the adducts predicted by docking simulations, followed by molecular mechanics generalized born surface access (MM-GBSA) calculations to estimate their binding free energy (ΔGb), and perfect the relative ranking of the MD-refined poses.29
Here, besides providing evidence of casp-8 and -9 (noncompetitive) inhibition by IL-1Ra at experimental level, we disclose the underlying molecular mechanism by gaining structural information on the casp-8(9)/IL-1Ra adducts. Our simulations unprecedently unveil that IL-1Ra binds to the dimerization site of casp-8/9 monomers, preventing, in this manner, their conversion into the signaling/catalytically active homodimeric states. This finding closely resembles the inhibition/regulatory mechanism exerted by XIAP and c-FLIP toward casp-9 and casp-8, respectively.7,30 In this scenario, IL- 1Ra appears as a key player in the regulatory mechanisms of distinct cellular death types (apoptosis, necroptosis, pyroptosis). Understanding and controlling this mechanism, by exploiting the structural traits enlightened here, may pave the way toward a broad range of novel therapeutic strategies to treat major human diseases.
2. METHODS
2.1. Coimmunoprecipitation of Caspases with IL-1Ra and Western Blots
Immunoprecipitation was conducted on mixtures containing recombinant human IL-1Ra (R&D Systems, Minneapolis, MN) and casp-8 or casp-9 (Alexis Biochemicals, San Diego, CA), using polyclonal Abs to IL-1Ra, or caspases, or control IL-1βeta (Santa Cruz Biotechnology, Santa Cruz, CA) coupled to Sepharose beads plus protein A/G (Santa Cruz Biotechnology, Santa Cruz, CA). Precipitates were washed in PBS and 100 μg fractions were then boiled in SDS buffer and separated on SDS-PAGE. Blots were probed using monoclonal Abs (Santa Cruz Biotechnology, Santa Cruz, CA).
2.2. In Vitro Caspase Activity
Casp-8 and -9 activities were assayed at 22 °C, using a fluorimeter plate reader (BMG Labtech Fluostar, Offenburg, Germany). The fluorimetry assays were conducted in the kinetic mode with excitation and emission wavelengths of 380 and 460 nm, respectively. Activity was measured by the release of 7-amino-4-methylcoumarin (AMC) from the synthetic substrate Ac-LEHD-AMC for casp-9, and Ac-IETD-AMC for casp-8, respectively. Assay mixtures contained 50 Units of rh-caspases (Alexis Biochemicals, San Diego, CA), increasing amounts (1–100 μM) of the specific substrate, and caspase buffer (50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 0.1% CHAPS, 10% sucrose and 5 mM dithiothreitol (DTT)). To determine the effect of rhIL-1Ra on activity of caspases, assays were performed in the absence or presence of 0.02–2.0 ,μM IL-1Ra. Assays were also conducted in the presence or absence of 10 μg/mL polyclonal anti-IL-1Ra blocking antibody (R&D Systems) at a starting dose of IL-1Ra of 100 nmol/L. IL-1β-blocking Abs (R&D Systems) were used as internal controls. Samples were compared to each other based on the activity of control samples. Data were fitted into the reciprocal Michaelis-Menten equation, and the inhibitor concentrations that decreases by 50% the rate of the reaction, i0 5, were then derived from the experimental plot, according to Cornish-Bowden.31 Corrections from blank were performed, and the curves were fitted to a 0.0 intercept assuming the activity to be 0 at time 0. Regression slopes (r > 0.95) were obtained for each experiment at each dose, and coefficients were compared between the three groups (Ll-1Ra, control, IL-1Ra plus blocking antibody). The experiments were performed in triplicates in each condition.
2.3. Statistics
Results are expressed as mean ± SD. Data were compared by analysis of variance using post hoc analysis for paired multiple comparisons with Fisher corrected t test. Twotailed p values < 0.05 were considered statistically significant.
2.4. Protein–protein docking
To build the model of IL- 1Ra/casp-9 and casp-8 adducts we employed their crystal structures (PDB ids 1ILR,32 1JXQ30 and 3KJN33 respectively). Caspases are produced as inactive monomers (zymogens), that becomes active only after a proteolytic cleavage and subsequent dimerization.9 For our protein—protein docking calculations involving casp-8 we have selected a monomer from the dimer crystal structure.33 Instead, in casp-9 the interactions established at the dimeric interface promote the active site formation reorienting the activation loop of one monomer. Thus, for our docking calculations we selected as model the inactive casp-9 monomer taken from its dimeric structure. For the casp-9/XIAP and casp-8/c-FLIP adducts we used the crystal structure deposited with PDB ids 1NW9 34 and 3H11,35 respectively.
In order to rationalize how caspases interact with IL-1Ra we have performed blind protein—protein docking calculations. Due to current limitations of docking algorithms in predicting the native protein—protein binding interface, we have applied a consensus docking strategy. Namely, we employed programs based on different search algorithms and scoring functions (typically divided in three types: (i) force-field (FF) based; (ii) empirical; or (iii) knowledge based) to verify if the predicted binding poses were reproducible.
We initially confronted several docking Web servers such as ZDOCK,36 ClusPro,37 pyDock38 and HDOCK,39 using the suggested parameters and without specifying any constraints to interacting residues. These share search algorithms based on fast-Fourier transform (FFT) methods for grid matching, and similar scoring functions, mainly based on desolvation and electrostatic contributions, with the only exception of HDOCK, which, instead, employs an iterative knowledge-based scoring function.40 As well, we have employed PatchDock,41 whose search algorithm is based on geometric hashing and whose scoring function accounts for geometric fit and atomic desolvation energy.42 As a further control of the programs reliability we tested the ability of ZDOCK and PatchDock to reproduce the crystallographic binding pose of the casp-9 dimer and casp-9/XIAP adduct starting from the isolated proteins. Interestingly, both programs were able to correctly predict the crystallographic structure of the casp-9 dimer (1JXQ) with a backbone RMSD respectively of 1.30 and 0.45 A, whereas only ZDOCK was able to find the correct casp-9/XIAP binding pose with a RMSD of 0.48 A Finally, we employed the RosettaDock server,43 in which a Monte Carlo based algorithm is used to refine docking poses obtained from the rigid docking protocols. In order to identify the most recurrent binding poses, we have selected the largest number of available models produced by each docking software and we have statistically analyzed the results.
2.5. Molecular Dynamics (MD) Simulations
MD simulations were performed by PMEMD (Particle Mesh Ewald MD) module of AMBER 12,44 using periodic boundary conditions and the Amber ff99SB-ILDN force field.45Physiological protonation states were calculated using the H+ + server.46 The different IL-lRa/caspase adducts (three models for casp-9 and two for casp-8), generated with distinct docking programs, were solvated in a truncated octahedron box with a distance between solute atoms and walls set to 12 A and filled with TIP3P water molecules.47 Na+ ions were added to neutralize the charge of the systems. The same procedure was done for the casp-9 dimer and casp-9/XIAP adduct. The systems counted up to 58 559, 44 629, and 51 687 atoms, respectively. Bonds involving hydrogen atoms were constrained using SHAKE algorithm.48 A time step of 2 fs and a cutoff of 10 A for the nonbonded interaction were used. The system was gradually heated in 1 ns to a final temperature of 300 K. The temperature control was performed by a Langevin thermostat.49The box size was equilibrated using the NPT ensemble for 1 ns. The pressure control (l atm) was accomplished using a Berendsen barostat.50 Finally, production NPT simulations were performed for 1 μs for each system, for a cumulative simulation time of 10 μs.
2.6. Binding Free Energy Calculations
The binding free energies (ΔGb) of the adducts were calculated with the MM-GBSA method51 using MMPBSA.py program52 and for protein–protein complexes can be calculated using the following equations29:
| (1) |
| (2) |
where ΔGbind represents the free energy of binding, which is the sum of the molecular mechanics energy terms from bonded, electrostatic and van der Waals contributions. ΔGpolar is here obtained by using the generalized Born (GB) model, whereas ΔGnonpolar is usually obtained from a linear relation to the solvent accessible surface area (SASA). Finally, –TΔS represents the conformational entropy contribution upon binding and is usually calculated by normal-mode analysis.
The calculations were done for all considered adducts by taking 100 frames of the equilibrated part of the MD trajectory. For solvation an improved generalized born solvation model was used (igb = 8)53 together with the mbondi3 radii set and a salt concentration of 0.1 M. The entropic contribution of the free energy was not taken into account, as it was suggested that this term does not improve the quality of the results using the MM-G(P)BSA.54,55
2.7. Analysis
Hydrogen (H–)bond analysis and root mean square (RMSD) calculations were performed with the cpptraj module of ambertools 16. In order to generalize also to other caspases the binding mode observed for casp-8(9)/IL-1Ra adducts we performed a sequence alignment ofcaspase-3, −6, −7, −8, and -9 with the Schrodinger Multiple Sequence Viewer and ClustalW.56 Electrostatic potential analysis was performed using APBS software.57 Images of protein structures were rendered using VMD.58
3. RESULTS AND DISCUSSION
3.1. Interaction of IL-1RA with Caspase-8 and -9
Co-immunoprecipitation experiments using anti-IL-1Ra or anticaspase Abs coupled to sepharose beads demonstrated a direct interaction of IL-1Ra with casp-8 (Figure 1a) and with casp-9 (Figure 1b). No interaction with caspases was evidenced for IL- 1β, used as internal controls in our assays (data not shown). These results indicate that IL-1Ra specifically binds casp-8 and confirm previous evidence of IL-1Ra binding to casp-9.17
Figure 1.
Physical binding of IL-1Ra with (a) casp-8 and (b) casp-9 are shown by means of coimmunoprecipitation. IL-1Ra was incubated with recombinant human casp-8 or -9. Mixtures was then precipitated by addition to three different sets of Sepharose beads coupled with anti-IL- 1Ra, anticasp-8, or anticasp-9 antibody. The coprecipitated proteins in each setting were then analyzed by Western blot. Anti-IL-1Ra and anticasp-8 or -9 antibodies were used to detect immunoreactivity of IL- 1Ra and of casp-8 or -9 in the coprecipitate. IL-1Ra and casp-8 or -9 were run in the Western blot in additional lines and used as positive controls.
Notably, the present study is focused on intracellular IL-1Ra. Of the two isoforms of IL-1Ra, that is, the secreted (sIL-1Ra) and the intracellular (icIL-1Ra) isoforms, sIL-1Ra was described as a 18 kDa peptide bearing a variable degree (up a MW of 22–24) of glycosylation,59 as foreseen in pioneering studies on inhibition of the endogenous pyrogen by Dinarello in the 1980s.60,61 In contrast, no glycosylation was evidenced for 17 kDa icIL-1Ra.62,63 Actually, in our precipitation assays, icIL-Ra constantly ran at the predicted mW of 17 kDa,17 and caspase inhibition was obtained in our in vitro enzyme assays by a recombinant IL-1Ra peptide. Therefore, binding studies of icIL- 1Ra to casp-8 described below are limited to the peptide, not considering any potential glycosylation of it.
3.2. IL-1Ra Inhibition of Activated Caspase-8 and -9
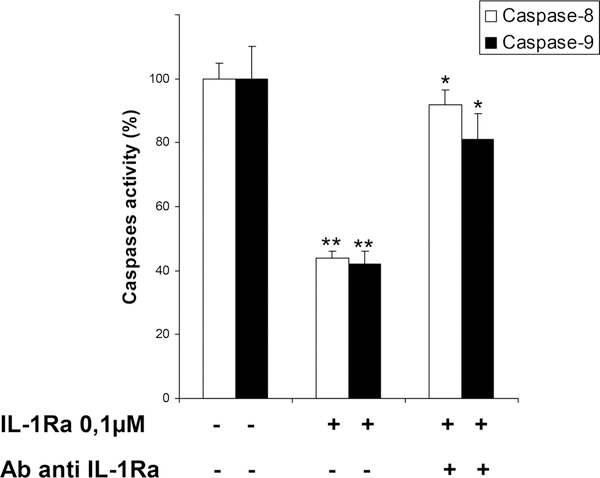
To analyze the inhibitory effect by IL-1Ra on activated caspases, activity of rh-caspases was measured by spectrofluorimetry in the presence or absence of rhIL-1Ra (Figure 2). In our conditions, we obtained Km values of 85.0 μM for casp-9, and of 42.9 μM, for casp-8, (which were in accordance with previously published data.64 At substrate Km-concentrations, IL-1Ra inhibited casp-9 with i05 values of 0.31 μM, (Ki competitive, Kic = 0.35 μM, K uncompetitive, Kiu = 0.28 μM), and casp-8 with i0 5 values of 0.57 μM (Kic 0.85 μM, Kiu 0.37 μM). Notably, IL-1Ra inhibition of casp-8 and -9 activities was abolished in the presence of anti-IL-1Ra blocking Abs, used as internal controls in our assays. These results indicate a noncompetitive inhibition by IL-1Ra of both casp-8 and -9 activity with i0 5 values in the nanomolar range.
Figure 2.
Casp-8 and -9 inhibition by IL-1Ra. IL-1Ra 100 nmol/L significantly inhibits casp-8 and -9 activities, respectively, by #50% (data shown represent mean values in triplicate samples ± SD;**P < 0.001 for IL-1Ra 100 nmol/L vs control for both experiments). Addition of IL-1Ra-blocking antibodies reverses the casp-8 and -9 inhibition by IL-1Ra (*P < 0.05 for IL-1Ra plus antibody vs IL-1Ra for casp-8 or casp-9).
3.3. Modeling the Caspases/IL-1Ra Adducts
3.3.1. Cas-pase-9/IL-1Ra Binding Model
In order to rationalize at atomic- scale the noncompetitive inhibition mechanism exerted by IL- 1Ra on casp-9 and −8 we have performed blind docking calculations with different programs to obtain structural information on the corresponding adducts.
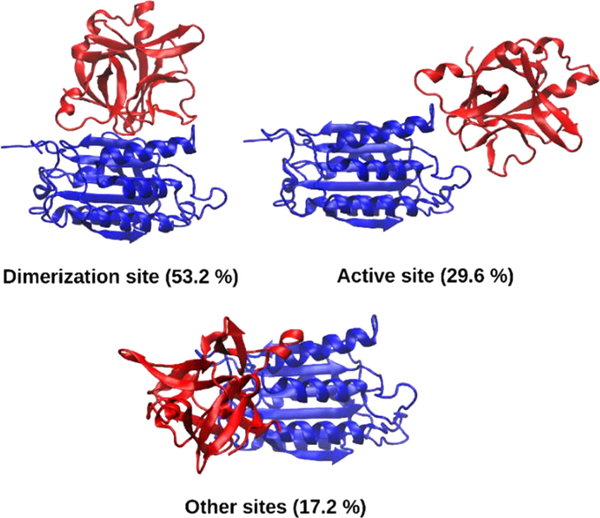
Strikingly, our consensus docking strategy led to a coherent view on the contact surfaces involved in the formation of the casp-9/IL-1Ra adduct (Table 1). As a result, the most energetically favored and populated binding pose (among the best top-ranked models), always predicts IL-1Ra binding to the casp-9 dimerization site. In a second type of interaction, less frequently observed and corresponding to a lower energy score, IL-1Ra binds to the casp-9 active site (Figure 3).
Table 1.
Relative Percentages (Calculated on the Best- Ranked Models) Of the Caspases Sites Involved in IL-1Ra Binding As Predicted by the Different Docking Programs Listed
| software | dimerization site | active site | other sites | |
|---|---|---|---|---|
| Casp-9/IL-1Ra | FFT based | |||
| ZDOCK | 53.2 | 29.6 | 17.2 | |
| ClusPro | 73.3 | 20.0 | 6.7 | |
| PyDock | 88.7 | 2.8 | 8.5 | |
| HDOCK | 61.0 | 15.5 | 23.5 | |
| geometric hashing | ||||
| PatchDock | 57.0 | 16.0 | 27.0 | |
| Casp-8/IL-1Ra | FFT based | |||
| ZDOCK | 58.6 | 1.8 | 39.6 | |
| ClusPro | 82.8 | 0.0 | 17.2 | |
| PyDock | 66.7 | 0.0 | 32.3 | |
| HDOCK | 44.5 | 19.7 | 35.8 | |
| geometric hashing | ||||
| PatchDock | 70.0 | 10.0 | 20.0 |
Figure 3.
Representative casp-9/IL-1Ra adducts as predicted from ZDOCK. Casp-9 and IL-1Ra are shown as blue and red ribbons, respectively.
The relative percentages of the different binding poses obtained by distinct docking algorithms are very similar for ZDOCK and PatchDock, predicting IL-1Ra binding to the caspases dimerization site as the most recurrent pose in 53.2 and 57.0% of cases, respectively. This binding mode has even a larger preference when using PyDock (i.e., 88.7% of models). Additionally, the best-ranked and consensually predicted casp- 9/IL-1Ra adducts, in which IL-1Ra inhibits casp-9 activation by preventing its conversion into an active dimeric form, is fully consistent with a noncompetitive inhibition mechanism predicted by coimmunoprecipitation and spectrofluorimetric experiment. As such this binding pose was selected for further studies.
Although a striking preference for the casp-9 dimerization site is envisioned by docking calculations, a more detailed comparison of the best poses obtained by the different algorithms does not furnish a consensus view on the IL-1Ra residues involved in casp-9 binding. All the best-ranked models are principally stabilized by hydrophobic interactions, and by few specific H-bonds. Remarkably, IL-1Ra residues, such as Trp16, Gln20, Tyr34, Gln36, and Tyr147, predicted here to interact with casp-9, are crucial also for IL-1Ra binding to the IL- 1R1 receptor.65 Conversely, PatchDock predicts a slightly different relative orientation of the two proteins in which a polar region of IL-1Ra, comprising Glu139, Lys96, Glu112, Asp138, interacts with casp-9.
To further assess the reliability of these predictions we have selected the top-ranked models as obtained from ZDOCK and PatchDock and we have relaxed them by performing 1 μs of MD simulations for each system. The model predicted by ZDOCK gained a stable binding pose only after 750 ns, while the PatchDock one reached stability earlier (Supporting Information (SI) Figure S1). Although the interaction between the two proteins is mainly driven by hydrophobic interactions, we observed the formation of few H-bonds along the MD runs, further contributing to stabilize the adducts (SI Figure S2). The most relevant residues involved in H-bonds formation and their contribution to the ΔGb are reported in Table 2.66,67
Table 2.
Residues Involved into Persistent H-bonds, as Observed During the MD Simulations of the casp-9(8)/IL- 1Ra Adductsa
| casp-9 residues | casp-9 residues MM-GBSA | IL-1Ra residues | IL-1Ra residues MM-GBSA | conserved across caspases |
| Lys398 | −4.6 ± 2.6 | Glu90 | −0.1 ± 0.1 | −3, −6, −7, −8, −9 |
| Gln399 | −3.1 ± 0.9 | Val18 | −1.7 ± 0.4 | −3, −6, −7, −8, −9 |
| Ser392 | −1.3 ± 0.6 | Gln36 | −5.2 ± 0.8 | 8, 9 |
| Asn405 | −0.4 ± 0.2 | Gln36 | −5.2 ± 0.8 | no |
| Ser242 | −3.8 ± 1.0 | Gln149 | −0.2 ± 0.4 | no |
| casp-8 residues | casp-8 residues MM-GBSA | IL-1Ra residues | IL-1Ra residues MM-GBSA | conserved across caspases |
| Gln462 | −0.1 ± 0.1 | Gln20 | −1.2 ± 0.4 | −3, −6, −7, −8, −9 |
| Lys472 | −2.4 ± 1.1 | Glu139 | −1.2 ± 1.2 | −3, −6, −7, −8, −9 |
| Asn447 | −1.5 ± 0.5 | Asn19 | −1.1 ± 0.6 | −3, −6, −7, −8 |
| Arg435 | −4.3 ± 0.6 | Glu139 | −0.9 ± 0.8 | no |
| Thr467 | −4.3 ± 0.9 | Glu126 | −0.9 ± 1.6 | no |
Contribution of these residues to the ΔGb (kcal/mol) is also reported. In the last column we show whether of casp-9/8 residues interacting with IL-1Ra are conserved across other caspases (−3, −6, −7, −8, and −9), which are also non-competitively inhibited by IL-1Ra.
In a final attempt to improve the reliability of our prediction, we have even refined the best-ranked pose obtained by ZDOCK using the RosettaDock server. The resulting model has been relaxed by 1 μs-long MD simulation (RMSD is shown in SI Figure S1c). In this model IL-1Ra interacts with casp-9 similarly to the ZDOCK model, however, during the MD simulation, the two proteins establish a larger number of hydrophobic and H- bonding interactions (Table S1), displaying, as a result, the largest ΔGb (Table 3 and S1). Hence, this model results to be the best prediction of the casp-9/IL-1Ra adduct, among those obtained in this work.
Table 3.
ΔGb (kcal/mol) of the casp-9/IL-1Ra and casp-8/IL- 1Ra Adducts, casp-9 Dimer and the Known Inhibitor XIAP Predicted by MM-GBSA Calculations. Standard Deviations Are Reported
| system | MM-GBSA |
|---|---|
| casp-9/IL-1Ra (ZDOCK) | −35.3 ± 3.1 |
| casp-9/IL-1Ra (PatchDock) | −48.6 ± 5.3 |
| casp-9/IL-1Ra (refinement) | −59.0 ± 5.8 |
| casp-8/IL-1Ra (ZDOCK) | −31.4 ± 5.5 |
| casp-8/IL-1Ra (PatchDock) | −36.1 ± 5.1 |
| casp-8/IL-1Ra (refinement) | −47.1 ± 5.0 |
| casp-9/XIAP | −85.9 ± 6.4 |
| casp-9 dimer | −131.0 ± 6.7 |
3.3.2. Caspase-8/IL-1Ra binding model
The same computational protocol was applied in order to dissect the inhibition mechanism of IL-1Ra toward casp-8. Consistently with the findings detailed above, all the FFT-based algorithms predict the binding of IL-1Ra to casp-8 dimerization site (Table 1) as the most favorable and recurrent binding pose. Remarkably, in those poses IL-1Ra interacts with casp-8 exploiting the same surface implicated in binding to the casp-9 (SI Figure S2), validating and generalizing our previous findings.
As well, we have relaxed the casp-8/IL-1Ra adduct by performing 1 μs ofMD simulation on the best models obtained with ZDOCK, RosettaDock and PatchDock programs (RMDS are shown in SI Figure S3). As a result, IL-1Ra anchors tightly to the dimerization surface of casp-8 via H-bonds (see Table 2 and SI Table S1). Strikingly, the ΔGb of IL-1Ra to casp-8 is lower than that to casp-9 (Table 3) in agreement with their relative i05 values of 0.31 and 0.57 μM, respectively (see Sections 3.1 and 3.2). In line with the results obtained for casp-9, the local refinement performed by RosettaDock determines an increased number of interactions in the adduct, suggesting that a local sampling of the conformational landscape using a Monte Carlo- based algorithm, followed by μs-long MD simulations, is needed to improve the quality of the blind protein–protein docking prediction.
3.4. Bioinformatics Analysis
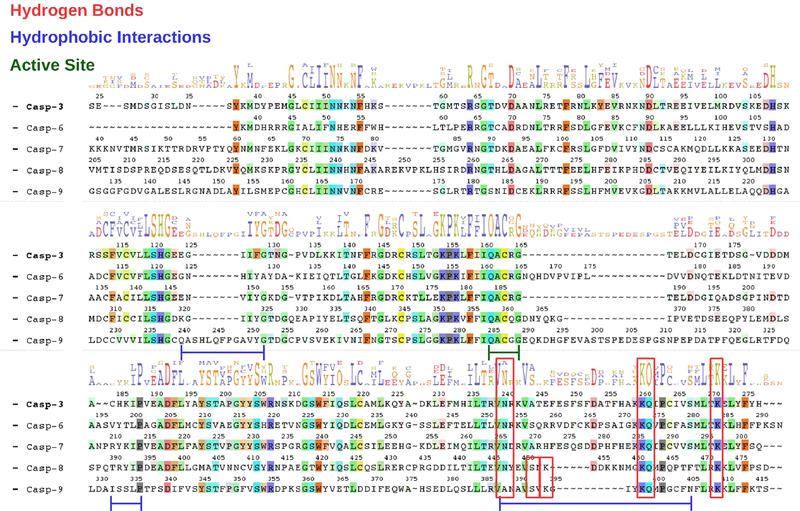
To further validate our models and explain why IL-1Ra displays a largest inhibitory power on casp-9 and −8 with respect to the effector casp-3, −6, and −7, we have verified if the residues stabilizing the casp-9(8)/ IL-1Ra adduct are conserved across all inhibited caspases (Figure 4). An alignment of their primary sequences interestingly reveals that Lys398 and Gln399 of casp-9, forming H-bonds that strongly stabilize the casp-9/IL-1Ra adduct during MD simulations (Table 2), are conserved in casp-8 and even in the effector caspases-3, −6, and −7. This extends also to Gln462, Lys472 and Asn447 in casp-8, which contributes to stabilize casp-8/IL-1Ra (see Table 2 and SI Table S1), As such our sequence alignment reveals that a similar H-bond network, although involving a lower number of residues, may be formed even between the effector caspases and IL-1Ra, justifying the experimentally observed binding of IL-1Ra to casp-3, −6, and −7 and suggesting a possible general mechanism of caspases inhibition as exerted by IL-1Ra. Conversely, the interaction established exclusively between IL-1Ra and a loop of casp-9, composed by the residues ranging from Gln240 to Gly252, accounts for the largest binding affinity and, possibly, the higher inhibitory power of IL-1Ra toward casp-9.
Figure 4.
Alignment of casp-3, −6, −7, −8, and -9 sequences (starting at Ser24 Tyr37, Lys39, Val205, and Gly132 residues, respectively). Residues not conserved in the other caspases are represented with a dashed line. Consensus residues are highlighted with their letter code, whose height is proportional to their conservation extent.68 Conserved residues involved in H-bonds, and hydrophobic interactions, stabilizing the adducts during the MD simulations, and composing the caspases active site, are highlighted in red square, and blue and green lines, respectively.
3.5. Generalizing the Noncompetitive Inhibition Mechanism of Caspases
As a further last check, we have also performed MD simulations on the adduct between casp-9 and XIAP, which hampers cell death by blocking the enzymatic activity of casp-9 at 10−9 M concentrations,69 being therefore a more potent inhibitor than IL-1Ra. Consistently with our results, in the casp-9/XIAP crystal structure, the third BIR domain (BIR3) of XIAP sequesters the casp-9 monomer, preventing its conversion into an active dimeric state.70 In order to compare the binding mode and affinity of casp-9/IL-1Ra with that of casp-9/XIAP adducts and casp-9 dimer we have even performed a 500 ns long MD simulation on the latter two systems (Table 3, RMSD is shown in SI Figure S4). The calculated AGb confirmed that casp-9 has a larger affinity toward XIAP than IL-1Ra, consistently with the measured i05. Moreover, the relative ΔGb between IL-1Ra and the two investigated caspases is in line with the largest inhibitory power exerted by IL-1Ra on casp-9. These calculations reveal that casp- 9 dimer has the largest ΔGb among the adducts considered. In order to validate our simulations we monitored the agreement between the experimentally determined X-ray temperature factors (B-factors) with the ones obtained from our MD simulations of the casp-9 dimer and the casp-9/XIAP adduct (SI Figure S5), as previously reported obtaining a good qualitative agreement.71,72
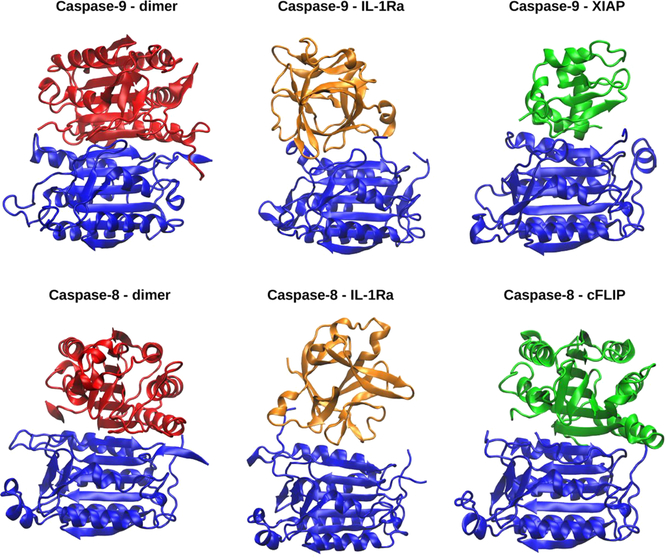
Interestingly, the crystal structure of c-FLIP, a key regulator of casp-8 in the extrinsic pathway of apoptosis,35 reveals that this binds at casp-8 dimerization site, resembling the mechanism observed experimentally for XIAP and predicted in this study for IL-1Ra (Figure 5). Thus, our findings provide clear evidence of a similar caspases regulatory/inhibitory mechanism lying at the crosstalk of different cell death pathways.
Figure 5.
A comparison between the crystallographic structures of casp-9 (pdb ID 1JXQ) and casp-8 dimers (3KJN), the most representative cluster obtained from our MD simulation of casp-9(8)/IL-1Ra and the adducts with the known caspases inhibitors XIAP (1NW9), and c-FLIP (3H11).
4. CONCLUSIONS
Building on the recent identification of the inhibitory effect of IL-1Ra on casp-9, underlying the regulation of ischemia induced apoptosis,17 here we disclose that IL-1Ra can even inhibit the initiator casp-8. This finding is in line with a recent study suggesting a novel nonapoptotic involvement of casp-8 at the crosstalk between cell death and innate immune cell inflammatory signaling.8
Consensus docking followed by multijμs MD simulations and free energy calculations predict that IL-1Ra binds to casp-8/9 at their dimerization surface, preventing, in this manner, their conversion into a catalytically/signaling active homodimeric form. This is fully consistent with the noncompetitive inhibition mechanism of IL-1Ra toward both caspases observed experimentally. The resulting casp-8(9)/IL-1Ra adducts are mainly stabilized by cumulative hydrophobic interactions and by key H- bonds with IL-1Ra, whose residues are at tract conserved across all initiator (casp-8, -9) and the effector caspases (−3, −6, −7) (Table 2 and SI Table S1) inhibited by IL-1Ra. The observed binding mode closely resembles that of XIAP and c-FLIP to casp-9 and −8, respectively, suggesting a general regulatory mechanism of casp-8/9 apoptosis, necroptosis and inflammation signaling exerted by preventing their conversion into their active homodimeric form. This mechanism is also in line with recent evidence pinpointing an allosteric site at the caspases dimeric interface, which may be exploited by small-molecule inhibitors.73
Strikingly, our outcomes provide clear evidence that beyond XIAP/c-FLIP other similar modulators of caspases exist, among which we identify IL-1Ra as a novel actor of this entangled regulatory network. Since IL-1Ra production has been monitored to increase by 160-fold in ischemic cardiomyocytes, where it plays a protective antiapoptotic role, it is tempting to suggest that IL-1Ra overexpression may be a signature of critical pathological states (ischemic injuries, cancer, neurodegeneration or other inflammatory disorders) in which caspases modulate different cell death types. As such our findings, contribute to move a step forward toward an in-depth understanding of cell death regulation, and provide unprecedented atomic-level structural information on the underlying mechanism. This knowledge may be exploited to devise novel therapeutic strategies in the fight against major human diseases.
Supplementary Material
ACKNOWLEDGMENTS
A.M. thanks the Italian Cancer Research Association (AIRC: My first AIRC grant no 17134) for financial support. AM. thanks CINECA, the Italian supercomputing center, for computational resources.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.8b00565.
Supplementary Figures S1–S5 showing the root-mean- square deviation (RMSD) for all the simulated models, the electrostatic properties of the adducts and a comparison of calculated and crystallographic protein B- factors. Table S1 contains an accurate description of the residues involved in the formation of all the persistent H- bonds during MD simulations (PDF)
REFERENCES
- (1).Hotchkiss RS; Strasser A; McDunn JE; Swanson PE Cell Death. N. Engl. J. Med 2009, 361, 1570–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Elmore S Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol 2007, 35, 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Linkermann A; Green DR Necroptosis. N. Engl. J. Med 2014, 370, 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhou W; Yuan J Necroptosis in Health and Diseases. Semin. Cell Dev. Biol 2014, 35, 14–23. [DOI] [PubMed] [Google Scholar]
- (5).Bergsbaken T; Fink SL; Cookson BT Pyroptosis: Host Cell Death and Inflammation. Nat. Rev. Microbiol 2009, 7, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ting JP; Willingham SB; Bergstralh DT Nlrs at the Intersection of Cell Death and Immunity. Nat. Rev. Immunol 2008, 8, 372–379. [DOI] [PubMed] [Google Scholar]
- (7).Tummers B; Green D R Caspase-8: Regulating Life and Death. Immunol. Rev 2017, 277, 76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Feltham R; Vince JE; Lawlor KE Caspase-8: Not So Silently Deadly. Clin. Transl. Immunol 2017, 6, No. e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pop C; Salvesen GS Human Caspases: Activation, Specificity, and Regulation. J. Biol. Chem 2009, 284, 21777–21781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Parrish AB; Freel CD; Kornbluth S Cellular Mechanisms Controlling Caspase Activation and Function. Cold Spring Harbor Perspect. Biol 2013, 5, a008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shalini S; Dorstyn L; Dawar S; Kumar S Old, New and Emerging Functions of Caspases. Cell Death Differ 2015, 22, 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Srinivasula SM; Hegde R; Saleh A; Datta P; Shiozaki E; Chai JJ; Lee RA; Robbins PD; Fernandes-Alnemri T; Shi YG; Alnemri ES A Conserved Xiap-Interaction Motif in Caspase-9 and Smac/Diablo Regulates Caspase Activity and Apoptosis. Nature 2001, 410, 112–116. [DOI] [PubMed] [Google Scholar]
- (13).Lawlor KE; Feltham R; Yabal M; Conos SA; Chen KW; Ziehe S; Grass C; Zhan Y; Nguyen TA; Hall C; Vince AJ; Chatfield SM; D’Silva DB; Pang KC; Schroder K; Silke J; Vaux L; Jost PJ; Vince JE Xiap Loss Triggers Ripk3- and Caspase-8- Driven Il-1beta Activation and Cell Death as a Consequence of Tlr- Myd88-Induced Ciap1-Traf2 Degradation. Cell Rep. 2017, 20, 668–682. [DOI] [PubMed] [Google Scholar]
- (14).Dinarello CA Anti-Inflammatory Agents: Present and Future. Cell 2010, 140, 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dinarello CA Overview of the Il-1 Family in Innate Inflammation and Acquired Immunity. Immunol. Rev 2018, 281, 8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gabay C; Porter B; Fantuzzi G; Arend WP Mouse Il-1 Receptor Antagonist Isoforms: Complementary DNA Cloning and Protein Expression of Intracellular Isoform and Tissue Distribution of Secreted and Intracellular Il-1 Receptor Antagonist in Vivo. J. Immunol 1997, 159, 5905–5913. [PubMed] [Google Scholar]
- (17).Vecile E; Dobrina A; Salloum FN; Van Tassell BW; Falcione A; Gustini E; Secchiero S; Crovella S; Sinagra G; Finato N; Nicklin MJ; Abbate A Intracellular Function of Interleukin-1 Receptor Antagonist in Ischemic Cardiomyocytes. PLoS One 2013, 8, No. e53265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Abbate A; Salloum FN; Vecile E; Das A; Hoke NN; Straino S; Biondi-Zoccai GG; Houser JE; Qureshi IZ; Ownby DD; Gustini E; Biasucci LM; Severino A; Capogrossi MC; Vetrovec GW; Crea F; Baldi A; Kukreja RC; Dobrina A Anakinra, a Recombinant Human Interleukin-1 Receptor Antagonist, Inhibits Apoptosis in Experimental Acute Myocardial Infarction. Circulation 2008, 117, 2670–2683. [DOI] [PubMed] [Google Scholar]
- (19).Pavlin M; Spinello A; Pennati M; Zaffaroni N; Gobbi S; Bisi A; Colombo G; Magistrato A A Computational Assay of Estrogen Receptor Alpha Antagonists Reveals the Key Common Structural Traits of Drugs Effectively Fighting Refractory Breast Cancers. Sci. Rep 2018, 8, 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Keskin O; Tuncbag N; Gursoy A Predicting Protein-Protein Interactions from the Molecular to the Proteome Level. Chem. Rev. 2016, 116,4884–4909. [DOI] [PubMed] [Google Scholar]
- (21).Spinello A; Pavlin M; Casalino L; Magistrato A A Dehydrogenase Dual Hydrogen Abstraction Mechanism Promotes Estrogen Biosynthesis. Can We Expand the Functional Annotation of the Aromatase Enzyme? Chem. - Eur. J 2018, 24, 10840–10849. [DOI] [PubMed] [Google Scholar]
- (22).Bucur O; Gaidos G; Yatawara A; Pennarun B; Rupasinghe C; Roux J; Andrei S; Guo B; Panaitiu A; Pellegrini M; Mierke DF; Khosravi-Far R A Novel Caspase 8 Selective Small Molecule Potentiates Trail-Induced Cell Death. Sci. Rep 2015, 5, 9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Casalino L; Palermo G; Spinello A; Rothlisberger U; Magistrato A All-Atom Simulations Disentangle the Functional Dynamics Underlying Gene Maturation in the Intron Lariat Spliceosome. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 6584–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Spinello A; Barone G; Grunenberg J Molecular Recognition of Naphthalene Diimide Ligands by Telomeric Quadruplex-DNA: The Importance of the Protonation State and Mediated Hydrogen Bonds. Phys. Chem. Chem. Phys 2016, 18, 2871–2877. [DOI] [PubMed] [Google Scholar]
- (25).Vargiu AV; Ruggerone P; Magistrato A; Carloni P Sliding of Alkylating Anticancer Drugs Along the Minor Groove of DNA: New Insights on Sequence Selectivity. Biophys. J 2008, 94, 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Huang SY Search Strategies and Evaluation in Protein-Protein Docking: Principles, Advances and Challenges. Drug Discovery Today 2014, 19, 1081–1096. [DOI] [PubMed] [Google Scholar]
- (27).Kastritis PL; Bonvin AM On the Binding Affinity of Macromolecular Interactions: Daring to Ask Why Proteins Interact. J. R. Soc. Interface 2013, 10, 20120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kastritis PL; Bonvin AM Are Scoring Functions in Protein-Protein Docking Ready to Predict Interactomes? Clues from a Novel Binding Affinity Benchmark. J. Proteome Res. 2010, 9, 2216–2225. [DOI] [PubMed] [Google Scholar]
- (29).Chen F; Liu H; Sun H; Pan P; Li Y; Li D; Hou T Assessing the Performance of the Mm/Pbsa and Mm/Gbsa Methods. 6. Capability to Predict Protein-Protein Binding Free Energies and Re-Rank Binding Poses Generated by Protein-Protein Docking. Phys. Chem. Chem. Phys 2016, 18, 22129–22139. [DOI] [PubMed] [Google Scholar]
- (30).Renatus M; Stennicke HR; Scott FL; Liddington RC; Salvesen GS Dimer Formation Drives the Activation of the Cell Death Protease Caspase 9. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Cortes A; Cascante M; Cardenas ML; Cornish-Bowden A Relationships between Inhibition Constants, Inhibitor Concentrations for 50% Inhibition and Types of Inhibition: New Ways of Analysing Data. Biochem. J 2001, 357, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Schreuder HA; Rondeau JM; Tardif C; Soffientini A; Sarubbi E; Akeson A; Bowlin TL; Yanofsky S; Barrett RW Refined Crystal Structure of the Interleukin-1 Receptor Antagonist. Presence of a Disulfide Link and a Cis-Proline. Eur. J. Biochem 1995, 227, 838–847. [DOI] [PubMed] [Google Scholar]
- (33).Wang Z; Watt W; Brooks NA; Harris MS; Urban J; Boatman D; McMillan M; Kahn M; Heinrikson RL; Finzel BC; Wittwer AJ; Blinn J; Kamtekar S; Tomasselli AG Kinetic and Structural Characterization of Caspase-3 and Caspase-8 Inhibition by a Novel Class of Irreversible Inhibitors. Biochim. Biophys. Ada, Proteins Proteomics 2010, 1804, 1817–1831. [DOI] [PubMed] [Google Scholar]
- (34).Shiozaki EN; Chai J; Rigotti DJ; Riedl SJ; Li P; Srinivasula SM; Alnemri ES; Fairman R; Shi Y Mechanism of Xiap-Mediated Inhibition of Caspase-9. Mol. Cell 2003, 11, 519–527. [DOI] [PubMed] [Google Scholar]
- (35).Yu JW; Jeffrey PD; Shi Y Mechanism of Procaspase-8 Activation by C-Flipl. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pierce BG; Wiehe K; Hwang H; Kim BH; Vreven T; Weng Z Zdock Server: Interactive Docking Prediction of Protein-Protein Complexes and Symmetric Multimers. Bioinformatics 2014, 30, 1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kozakov D; Hall DR; Xia B; Porter KA; Padhorny D; Yueh C; Beglov D; Vajda S The Cluspro Web Server for Protein-Protein Docking. Nat. Protoc 2017, 12, 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Jimenez-Garcia B; Pons C; Fernandez-Recio J Pydockweb: A Web Server for Rigid-Body Protein-Protein Docking Using Electrostatics and Desolvation Scoring. Bioinformatics 2013, 29, 1698–1699. [DOI] [PubMed] [Google Scholar]
- (39).Yan Y; Zhang D; Zhou P; Li B; Huang SY Hdock: A Web Server for Protein-Protein and Protein-DNA/Rna Docking Based on a Hybrid Strategy. Nucleic Acids Res. 2017, 45, W365–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Huang SY; Zou X An Iterative Knowledge-Based Scoring Function for Protein-Protein Recognition. Proteins: Struct., Fund., Genet 2008, 72, 557–579. [DOI] [PubMed] [Google Scholar]
- (41).Schneidman-Duhovny D; Inbar Y; Nussinov R; Wolfson HJ Patchdock and Symmdock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res. 2005, 33, W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhang C; Vasmatzis G; Cornette JL; DeLisi C Determination of Atomic Desolvation Energies from the Structures of Crystallized Proteins. J. Mol. Biol 1997, 267, 707–726. [DOI] [PubMed] [Google Scholar]
- (43).Lyskov S; Gray JJ The Rosettadock Server for Local Protein-Protein Docking. Nucleic Acids Res. 2008, 36, W233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Case DA; T. A. D; Cheatham TE III; Simmerling CL; Wang J; Duke RE; Luo R; Walker RC; Zhang W; Merz KM; Roberts B; Hayik S; Roitberg A; Seabra G; Swails J; Gotz AW; Kolossvary I; Wong KF; Paesani F; Vanicek J; Wolf RM; Liu J; Wu X; Brozell SR; Steinbrecher T; Gohlke H; Cai Q; Ye X; Wang J; Hsieh M-J; Cui G; Roe DR; Mathews DH; Seetin MG; Salomon-Ferrer R; Sagui C; Babin V; Luchko T; Gusarov S; Kovalenko A; Kollman PA AMBER 12; University of California: San Francisco, 2012. [Google Scholar]
- (45).Lindorff-Larsen K; Piana S; Palmo K; Maragakis P; Klepeis JL; Dror RO; Shaw DE Improved Side-Chain Torsion Potentials for the Amber Ff99sb Protein Force Field. Proteins: Struct., Funct. Genet 2010, 78, 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Anandakrishnan R; Aguilar B; Onufriev AV H++ 3.0: Automating Pk Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012, 40, W537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- (48).Ryckaert JP; Ciccotti G; Berendsen HJC Numerical- Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics ofN-Alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- (49).Davidchack RL; Ouldridge TE; Tretyakov MV New Langevin and Gradient Thermostats for Rigid Body Dynamics. J. Chem. Phys 2015, 142, 144114–144127. [DOI] [PubMed] [Google Scholar]
- (50).Berendsen HJC; Postma JPM; Vangunsteren WF; Dinola A; Haak JR Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys 1984, 81, 3684–3690. [Google Scholar]
- (51).Kollman PA; Massova I; Reyes C; Kuhn B; Huo SH; Chong L; Lee M; Lee T; Duan Y; Wang W; Donini O; Cieplak P; Srinivasan J; Case DA; Cheatham TE Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res 2000, 33, 889897. [DOI] [PubMed] [Google Scholar]
- (52).Miller BR; McGee TD; Swails JM; Homeyer N; Gohlke H; Roitberg AE Py Mmpbsa.: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput 2012, 8, 3314–3321. [DOI] [PubMed] [Google Scholar]
- (53).Nguyen H; Roe DR; Simmerling C Improved Generalized Born Solvent Model Parameters for Protein Simulations. J. Chem. Theory Comput 2013, 9, 2020–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Genheden S; Ryde U The Mm/Pbsa and Mm/Gbsa Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discovery 2015, 10,449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Yang T; Wu JC; Yan C; Wang Y; Luo R; Gonzales MB; Dalby KN; Ren P Virtual Screening Using Molecular Simulations. Proteins: Struct., Funct., Genet 2011, 79, 1940–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Thompson JD; Higgins DG; Gibson TJ Clustal W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Jurrus E; Engel D; Star K; Monson K; Brandi J; Felberg LE; Brookes DH; Wilson L; Chen J; Liles K; Chun M; Li P; Gohara DW; Dolinsky T; Konecny R; Koes DR; Nielsen JE; Head-Gordon T; Geng W; Krasny R; Wei GW; Holst MJ; McCammon JA; Baker NA Improvements to the Apbs Biomolecular Solvation Software Suite. Protein Sci 2018, 27, 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (59).Hannum CH; Wilcox CJ; Arend WP; Joslin FG; Dripps DJ; Heimdal PL; Armes LG; Sommer A; Eisenberg SP; Thompson RC Interleukin-1 Receptor Antagonist Activity of a Human Interleukin-1 Inhibitor. Nature 1990, 343, 336–340. [DOI] [PubMed] [Google Scholar]
- (60).Dinarello CA; Rosenwasser LJ; Wolff SM Demonstration of a Circulating Suppressor Factor of Thymocyte Proliferation During Endotoxin Fever in Humans. J. Immunol 1981, 127, 2517–2519. [PubMed] [Google Scholar]
- (61).Granowitz EV; Santos AA; Poutsiaka DD; Cannon JG; Wilmore DW; Wolff SM; Dinarello CA Production of Interleukin-1-Receptor Antagonist During Experimental Endotoxae- mia. Lancet 1991, 338, 1423–1424. [DOI] [PubMed] [Google Scholar]
- (62).Haskill S; Martin G; Van Le L; Morris J; Peace A; Bigler CF; Jaffe GJ; Hammerberg C; Sporn SA; Fong S Cdna Cloning of an Intracellular Form of the Human Interleukin 1 Receptor Antagonist Associated with Epithelium. Proc. Natl. Acad. Sci. U. S. A 1991, 88, 3681–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Bigler CF; Norris DA; Weston WL; Arend WP Interleukin-1 Receptor Antagonist Production by Human Keratino- cytes. J. Invest. Dermatol 1992, 98, 38–44. [DOI] [PubMed] [Google Scholar]
- (64).Garcia-Calvo M; Peterson EP; Rasper DM; Vaillancourt JP; Zamboni R; Nicholson DW; Thornberry NA Purification and Catalytic Properties of Human Caspase Family Members. Cell Death Differ. 1999, 6, 362–369. [DOI] [PubMed] [Google Scholar]
- (65).Evans RJ; Bray J; Childs JD; Vigers GP; Brandhuber BJ; Skalicky JJ; Thompson RC; Eisenberg SP Mapping Receptor Binding Sites in Interleukin (Il)-1 ReceptorAntagonist and Il-1 Beta by Site-Directed Mutagenesis. Identification of a Single Site in Il-1ra and Two Sites in Il-1 Beta. J. Biol. Chem 1995, 270, 11477–11483. [DOI] [PubMed] [Google Scholar]
- (66).Bren M; Florian J; Mavri J; Bren U Do All Pieces Make a Whole? Thiele Cumulants and the Free Energy Decomposition. Theor. Chem. Acc 2007, 117, 535–540. [Google Scholar]
- (67).Bren U; Martinek V; Florian J Decomposition of the Solvation Free Energies of Deoxyribonucleoside Triphosphates Using the Free Energy Perturbation Method. J. Phys. Chem. B 2006, 110, 12782–12788. [DOI] [PubMed] [Google Scholar]
- (68).Schneider TD; Stephens RM Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Salvesen GS; Duckett CS Iap Proteins: Blocking the Road to Death’s Door. Nat. Rev. Mol. Cell Biol 2002, 3, 401–410. [DOI] [PubMed] [Google Scholar]
- (70).Fesik SW; Shi Y Structural Biology. Controlling the Caspases.Science 2001, 294, 1477–1478. [DOI] [PubMed] [Google Scholar]
- (71).Graf MM; Bren U; Haltrich D; Oostenbrink C Molecular Dynamics Simulations Give Insight into D-Glucose Dioxidation at C2 and C3 by Agaricus Meleagris Pyranose Dehydrogenase. J. Comput.- Aided Mol. Des 2013, 27, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Bren U; Oostenbrink C Cytochrome P450 3a4 Inhibition by Ketoconazole: Tackling the Problem of Ligand Cooperativity Using Molecular Dynamics Simulations and Free-Energy Calculations. J. Chem. Inf. Model 2012, 52, 1573–1582. [DOI] [PubMed] [Google Scholar]
- (73).Murray J; Renslo AR Modulating Caspase Activity: Beyond the Active Site. Curr. Opin. Struct. Biol 2013, 23, 812–819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.