Abstract
Several approaches were compared for the entrapment of proteins within hydrazide-activated silica for use in affinity microcolumns and high performance affinity chromatography. Human serum albumin (HSA) and concanavalin A (Con A) were used as model proteins for this work. Items considered in this study included the role played by the solution volume, amount of added protein, and use of slurry vs. on-column entrapment on the levels of solute retention and extent of protein immobilization that could be obtained by means of entrapment. The levels of retention and protein immobilization were evaluated by injecting warfarin or 4-methylumbellipheryl α-D-mannopyranoside as solutes with known binding properties for HSA or Con A. Altering both the solution volume and amount of added protein led to an increase of up to 17-fold in the extent of protein immobilization for HSA in slurry-based entrapment; on-column entrapment provided an additional 3.6-fold increase in protein content vs. the optimized slurry method. Similar general trends were seen for Con A. The protein contents obtained by entrapment for HSA or Con A (i.e., up to ~87 and 46 mg/g silica, respectively) were comparable to or higher than levels reported for the covalent immobilization of these proteins onto silica. The retention of warfarin on the entrapped HSA was at least 1.7-fold higher than has been obtained under comparable support and mobile phase conditions when using covalent immobilization. These results indicated that entrapment can be an attractive alternative to covalent immobilization for proteins such as HSA and Con A, with this approach serving as a potential means for obtaining good solute binding and retention in work with affinity microcolumns or related microscale devices.
Keywords: Entrapment, Protein immobilization, Affinity microcolumn, High performance affinity chromatography, Human serum albumin, Concanavalin A
1. Introduction
High performance affinity chromatography (HPAC) is a method in which an immobilized biological agent, or affinity ligand, is placed onto an HPLC support (e.g., porous silica) and used as a selective binding agent for sample components [1,2]. The combination of such a support with the specificity and strong binding of many affinity ligands makes it possible with HPAC to quickly isolate, measure, or study specific targets even when they are present in complex biological samples. Other advantages of HPAC are its ability to be automated, to study biological interactions, and to reuse the same biological agent for many samples or binding studies [1–10].
The affinity ligand in HPAC is often covalently attached to the support through functional sites such as amines, carboxylates or sulfhydryl groups [2,11,12]. However, covalent coupling methods often have some multisite attachment, improper orientation or immobilization near a binding site, which can decrease the actual or apparent activity of the affinity ligand [2,12]. An alternative approach for immobilization is to use entrapment or encapsulation of the affinity ligand [12–16]. The encapsulation of various binding agents within sol-gels has been used for some time [12–15]. However, the sol-gels that result from traditional entrapment methods are generally not suitable for HPAC because of their band-broadening and inability to be used at the pressures or flow rates that are often encountered in HPLC systems [12,16].
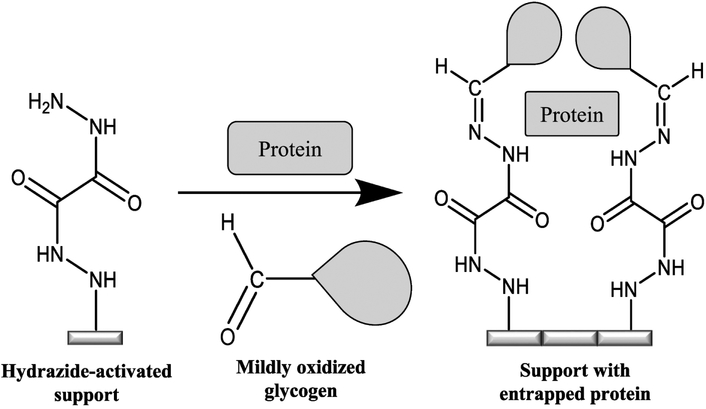
These limitations have been overcome in recent work with HPLC-based silica and the entrapment method shown in Figure 1. In this technique, an affinity ligand such as a protein is mixed with hydrazide-activated silica and a mildly oxidized form of glycogen (molar mass, ~2.5 × 105 kDa), which is used as a large capping agent [16]. The hydrazide groups on the support will not react with the binding agent but will form a stable covalent bond with aldehyde groups on the oxidized glycogen. This process results in some of the affinity ligand being entrapped within the stagnant mobile phase of the support as the glycogen binds to the activated silica and blocks the affinity ligand from leaving the pores or the region near the support’s surface. The result is an immobilization process in which the affinity ligand is kept within the support and in a form that is fully soluble and highly activity [16]. Although previous work has examined the effects of factors such as pore size and glycogen or protein levels on entrapment [16–19], there is still a need to further characterize items that influence the amount of protein that can be immobilized by this approach. This issue is of particular interest in work with affinity microcolumns (i.e., columns containing biological binding agents and with volumes in the low-to-mid μL range) [20], where a high protein content is frequently desirable for strong analyte binding and good retention [20–23].
Figure 1.
Entrapment of a soluble protein using a hydrazide-activated support and mildly oxidized glycogen.
This report will examine the effects of factors such as solution volume, relative protein amounts, and slurry versus on-column entrapment on the amount of protein and levels of solute retention that can be obtained with entrapment. Two model proteins will be used for this work: human serum albumin (HSA) and concanavalin A (Con A) [10,11]. These proteins are of general interest as binding agents in affinity columns for many drugs (i.e., the use of HSA in drug binding studies or chiral separations) or carbohydrate-containing agents (e.g., the use of Con A to bind glycopeptides) [6,7,9–11,21–26]. The retention properties and level of entrapment of these proteins will be evaluated by using zonal elution studies and injected probe compounds with well-characterized binding to these agents. The results that are obtained under various conditions of entrapment will be used to identify conditions that maximize the protein content and solute retention of such supports. The protein content and retention behavior will be compared to prior covalent methods that have been employed with HSA and Con A. The results should provide valuable guidelines for future applications of entrapment with these and other proteins when using affinity microcolumns and related microscale systems [20–23,26].
2. Material and methods
2.1. Reagents
The HSA (essentially fatty acid free, ≥ 96% pure), Con A (type V, lyophilized powder, highly purified), glycogen (bovine liver type IX, total glucose ≥ 85 %, dry basis), racemic warfarin (purity ≥ 98%), 4-methylumbellipheryl α-D-mannopyranoside (MUM, purity ≥ 96%) and 4-methylumbellipheryl α-D-galactopyranoside (MUGA, purity ≥ 98%) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Nucleosil Si-300 silica (7 μm particle diameter, 300 Å pore size) was obtained from Macherey Nagel (Duren, Germany). All other chemicals were of the purest grades available. Vivaspin 6 ultrafiltration tubes (30 kDa cutoff; Sartorius, Gottingen, Germany) were used for purification of the oxidized glycogen. All buffers and aqueous solutions were prepared using water from a Nanopure system (Barnstead, Dubuque, IA, USA). Buffers were filtered using 0.20 μm GNWP nylon membranes from Fisher Scientific (Pittsburgh, PA, USA) and were degassed by sonication under vacuum for at least 30 min prior to use.
2.2. Apparatus
The chromatographic system consisted of a 1200 isocratic pump from Agilent (Santa Clara, CA, USA), a Series 200 UV-Vis detector, and a vacuum degasser from Perkin-Elmer (Waltham, MA, USA). Samples were injected using a Rheodyne LabPro valve (Cotati, CA, USA) equipped with a 5 or 20 μL sample loop. An Isotemp 9100 circulating water bath (Fisher Scientific, Pittsburgh, PA, USA) and a water jacket from Alltech (Deerfield, IL, USA) were used for temperature control of the columns and mobile phases. The chromatographic data were collected and processed using LabView 8 software (National Instruments, Austin, TX, USA), and analyzed using PeakFit 4.12 (Systat Software, San Jose, CA, USA). Two PHD Ultra syringe pumps (Harvard Apparatus, Holliston, MA, USA) were used for the on-column entrapment method. An HPLC slurry packing system from ChromTech (Apple Valley, MN) was used for packing supports into the microcolumns.
2.3. Protein entrapment by the slurry method
The supports containing entrapped proteins were prepared from Nucleosil Si-300 silica that had first been converted into a diol-bonded form, as described previously [27]. This support has been found in prior work to be suitable for the entrapment of HSA and proteins with similar sizes to HSA [16–19]. The diol-bonded silica was later used in preparing hydrazide-activated silica, with oxalic dihydrazide being employed as the activating agent [16,27].
The initial conditions used for slurry-based entrapment were based on prior work with HSA and this method [16] (Note: modifications of this approach that were made and used later in this study are described in Section 3.1). These conditions involved making a slurry that contained 0.08 g of hydrazide-activated silica and 0.80 mL of a 10 mg/mL protein sample that were placed or prepared in a pH 5.0, 0.10 M potassium phosphate solution. This slurry was degassed for 10 min to remove any trapped air within the support. Next, 0.28 mL of a 4.25 mg/ml solution of mildly oxidized glycogen in a pH 5.0, 0.10 M potassium phosphate solution was added to the mixture, giving a slurry that contained 15 mg glycogen/g silica. The glycogen had previously been oxidized in pH 5.0, 20 mM sodium acetate buffer that contained 15 mM sodium chloride, with 17 mg glycogen and 135 mg periodic acid being added to 4 mL of this buffer and allowed to react for 16 h in the dark; these conditions have been shown to oxidize about 0.5% of the glucose units in glycogen [16]. The oxidized glycogen was purified by ultrafiltration [19] and then used for entrapment.
The volume of the entrapment solution containing the hydrazide-activated support, protein and oxidized glycogen was adjusted to 2.0 mL by adding a pH 5.0, 0.10 M potassium phosphate solution. This slurry was mixed slowly for 16 h at 4°C. Finally, 50 μL of oxalic dihydrazide, which was present at a concentration of 1 mg/mL in a pH 5.0, 0.10 M potassium phosphate solution, was added and allowed to mix with the slurry for 1 h. This step was performed to cap any remaining aldehyde groups on the glycogen and to decrease possible non-specific binding that might occur later with such groups in the microcolumn. Control supports were prepared in the same manner, but with a pH 5.0, 0.10 M potassium phosphate solution being used in place of the protein solution during the entrapment step.
Several conditions were varied for the slurry-based entrapment method, with the goal of increasing the amount of entrapped protein and its retention of applied solutes. These conditions, as discussed in Section 3, included the solution volume and the total amount of protein that was used for entrapment. The supports prepared by slurry-based entrapment, or the activated supports used for on-column entrapment, and the corresponding control supports were suspended in pH 7.4, 0.067 M potassium phosphate buffer and downward slurry packed into 1.0 cm × 2.1 mm i.d. stainless steel microcolumns for evaluation and use in chromatographic studies. The packing solution for all supports was pH 7.4, 0.067 M potassium phosphate buffer and the packing pressure was 4000 psi (27.6 MPa). After packing, the microcolumns were placed in an HPLC system, where a pH 7.4, 0.067 M potassium phosphate buffer (for the HSA supports) [16,21–23] or a pH 5.0, 0.50 M sodium acetate that contained 1.0 mM calcium chloride and 1.0 mM manganese chloride (for the Con A supports) [16,28,29] was passed through each microcolumn at 0.5 mL/min for at least 1 h or until a stable baseline was achieved. The microcolumns were stored in the same pH 7.4 or 5.0 buffers and at 4°C when not in use.
2.4. Protein entrapment by an on-column method
An alternative approach to slurry entrapment that was considered for immobilization was on-column entrapment [17]. The hydrazide-activated support was prepared from the same type of silica and using the same general reaction conditions as described in the previous section for slurry-based entrapment [16,27]. This support was packed into 1.0 cm × 2.1 mm i.d. stainless steel microcolumns at 4000 psi using pH 7.4, 0.067 M potassium phosphate buffer, as described in Section 2.3.
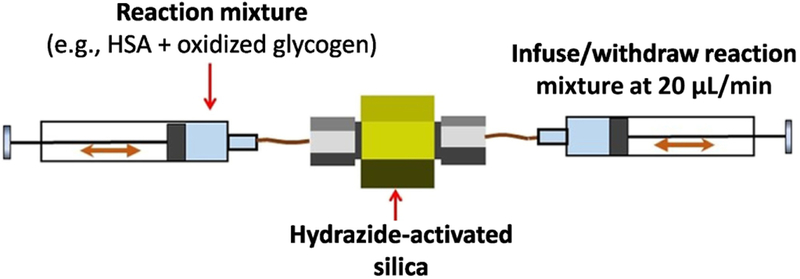
The microcolumns containing hydrazide-activated silica were next used with a dual syringe on-column entrapment procedure, as illustrated in Figure 2. In this method, each microcolumn containing a hydrazide-activated support was connected to two pumps that were each equipped with 250 or 500 μL syringes. These pumps were synchronized to pass a given solution through a microcolumn, with one syringe infusing the solution while the other was used to withdraw the solution at the same flow rate. The total solution volume in the syringes was 350–400 μL, and syringes were used to infuse or withdraw 75 μL of this solution (in the majority of studies) or 200 μL (in later work with 100 mg/mL HSA) during each cycle at a flow rate of 20 μL/min. A stop time of 1 min was included in this program between each change in direction for syringe movement to allow the solution to completely go through the microcolumn and to relieve any pressure buildup that had occurred before a change was made in the direction of solution application.
Figure 2.
Scheme for dual syringe method for on-column protein entrapment.
The on-column entrapment method was used with several microcolumns and with HSA concentrations that ranged from 12.5 to 100 mg/mL and Con A concentrations that ranged from 1.0 to 10 mg/mL. These solutions were each entrapped at room temperature. The on-line entrapment procedure first involved applying for 4 h to the microcolumn a 350–400 μL solution that contained only the desired protein (e.g., HSA) in a pH 5.0, 0.10 M potassium phosphate solution. This step was followed by applying 400 μL of a mixture of the same protein plus 4.25 mg/mL of oxidized glycogen for 16 h. Finally, 350–400 μL of a 1 mg/mL oxalic dihydrazide solution that was prepared in a pH 5.0, 0.10 M potassium phosphate solution was passed through the microcolumn for 2 h. A control microcolumn was prepared under the same conditions, with the exception of using only a pH 5.0, 0.10 M potassium phosphate solution in place of the protein solution. Immediately after the entrapment procedure was completed, the microcolumn was connected to an HPLC pump and washed at 0.50 mL/min for 1 h with either pH 7.4, 0.067 M potassium phosphate buffer (for the HSA microcolumns) or pH 5.0, 0.50 M sodium acetate that contained 1.0 mM calcium chloride and 1.0 mM manganese chloride (for the Con A microcolumns) to wash away any excess reactants (e.g., unreacted oxidized glycogen or oxalic dihydrazide, and any remaining, non-entrapped protein). The microcolumns were stored in the same pH 7.4 or 5.0 buffers at 4°C when not in use.
2.5. Chromatographic studies
The mobile phase that was used in the chromatographic studies with the entrapped HSA microcolumns was pH 7.4, 0.067 M potassium phosphate buffer [16,21–23]. The mobile phase for the entrapped Con A microcolumns was pH 5.0, 0.50 M sodium acetate that contained 1.0 mM calcium chloride and 1.0 mM manganese chloride [28,29]. All microcolumns were equilibrated with their corresponding buffer at 0.50 mL/min for at least 1 h before any sample injections were made onto the column. During these experiments, the temperature was maintained at 37 °C for the HSA microcolumns and at 20 °C for the Con A microcolumns, as well as for their respective control microcolumns.
The probes that were injected onto these microcolumns to measure their retention and to estimate their protein content were racemic warfarin for HSA and MUM for Con A. The wavelengths used for detection were 308 nm for warfarin, 316 nm for MUM or MUGA, and 205 or 220 nm for sodium nitrate, with sodium nitrate and MUGA being used as non-retained solutes and markers for the void time of the system in work with HSA and Con A, respectively [16–19,28,29]. Sample solutions containing these probes were prepared in the same mobile phase that was used for each corresponding microcolumn. The injection volume was 5 μL or 20 μL.The samples were injected at 0.50 mL/min and contained 10–20 μM warfarin, 5–10 μM MUM or MUGA, and 2.5–5.0 μM sodium nitrate. No significant variations in the retention factors for the injected probes or non-retained solutes were seen over this range of sample concentrations, indicating that linear elution conditions were present. The retention factors also showed no variation with small changes in the flow rate, in agreement with previous observations made for warfarin and related solutes in prior studies with columns containing covalently immobilized HSA [9,21,30].
The retention time for each injected solute was estimated by analyzing the chromatograms with PeakFit 4.12. These peaks were processed by using the exponentially-modified Gaussian and half-Gaussian modified Gaussian function of PeakFit, followed by determination of each peak’s first statistical moment. All of the retention factors were determined for data acquired over at least three injections of the probe compounds or non-retained solutes on the protein microcolumns and control microcolumns. The extra-column void time of the system was found by measuring the elution time of sodium nitrate or MUGA when a zero-volume union was placed instead of a microcolumn into the chromatographic system [21].
3. Results and discussion
3.1. Optimization of slurry-based entrapment for HSA
The slurry-based method of entrapment is the original form of this immobilization approach [16] and is the format that has been used in most prior work with this technique [18,19]. This procedure uses a slurry that contains a mixture of hydrazide-activated silica, oxidized glycogen and the protein of interest for the entrapment process [16]. Previous studies have examined the effects of using different types of supports and pore sizes on the amount of protein that can be entrapped by this method [16,19]. The effects of varying the amount of oxidized glycogen vs. silica and the means for preparing the hydrazide-activated silica or oxidized glycogen have also been considered [16,19]. In addition, some preliminary work has examined the effect of varying the total amount of protein that is added to the entrapment mixture [16]. One factor that has not been examined in these prior studies is the way in which the final protein content of the support is altered when varying the total volume of solution that is used for slurry-based entrapment.
HSA was used as the test protein for this work because it has been employed as a model binding agent in numerous previous studies aimed at the creation or optimization of covalent immobilization methods [5,31–34]. In addition, HSA has been of ongoing interest in binding studies based on HPAC because this protein is a serum transport agent for many drugs and can act as a chiral stationary phase for a number of these solutes [5–10]. This protein, which consists of a single polypeptide chain with a molar mass of 66.5 kDa [24], also has well-characterized binding to the drug warfarin [5,16]. This last fact was used in this report by employing warfarin as a probe to estimate and compare, through retention factor measurements, the amounts of active HSA that were present in supports and microcolumns prepared by entrapment (Note: this approach, and related methods using warfarin as a probe, have been found in previous studies to give good agreement with direct measurements of entrapped HSA content based on protein assays) [16,18].
Table 1 shows the results that were obtained in this study when the total volume of the HSA/glycogen solution and/or the total added amount of HSA were varied in this entrapment method. Some adjustments in the amounts of silica and oxidized glycogen that were added to this mixture were also made during this process to provide consistent relative amounts for these agents. In going from Procedure 1 to 2 in Table 1, the solution volume was decreased by 70% from 1.0 to 0.30 mL. The masses of glycogen and silica were decreased to keep their ratio in the range of 15–18 mg glycogen/g silica. The added amount of HSA was also decreased slightly from that used in Procedure 1 (i.e., from 8.0 to 6.0 mg); this gave a mixture that contained a 20% increase in the relative amount of HSA vs. silica (i.e., 120 vs. 100 mg HSA/g silica). Under these conditions, the final protein content, as calculated from the measured retention of warfarin [16,18,31], gave a 1.4-fold apparent increase from 1.9 to 2.6 mg HSA/g silica; however, this change was not significant at the 95% confidence level. A further reduction in the solution volume to 0.20 mL, as made in Procedure 3 and with no change in the amount of added HSA from Procedure 1 (8.0 mg), provided an increase in the entrapped protein content of 3.2-fold from the initial conditions. This difference was significant at the 95% confidence level and indicated that a decrease in solution volume could alter and be used to increase the final protein content of a support that was made by slurry-based entrapment.
Table 1.
Effect on protein content when varying reaction conditions for slurry-based entrapment of HSAa
| Procedure | |||||
|---|---|---|---|---|---|
| Entrapment conditions | 1 | 2 | 3 | 4 | 5 |
| Solution and support in entrapment mixture | |||||
| Total added solution volume (mL) | 1.00 | 0.30 | 0.20 | 0.72 | 0.20 |
| Total added silica (mg) | 80 | 50 | 50 | 63 | 50 |
| HSA in entrapment mixture | |||||
| Conc. HSAC (mg/mL) | 8.0 | 20 | 40 | 35 | 100 |
| Total Added HSA (mg) | 8.0 | 6.0 | 8.0 | 25.2 | 20.0 |
| Added HSA vs. silica (mg/g) | 100 | 120 | 160 | 400 | 400 |
| Oxidized glycogen in entrapment mixture | |||||
| Cone, oxidized glycogen (mg/mL) | 1.2 | 3.0 | 4.2 | 1.3 | 3.8 |
| Total added glycogen (mg) | 1.2 | 0.90 | 0.84 | 0.94 | 0.75 |
| Added glycogen vs. silica (mg/g) | 15 | 18 | 17 | 15 | 15 |
| Properties of entrapped HSA supportb | |||||
| Specific retention factor, warfarin | 3.0 (±0.6) | 4.0 (±0.9) | 9.5 (±2.6) | 15.8 (±1.4) | 49.9 (±7.9) |
| Protein content (mg/g silica) | 1.9(± 0.5) | 2.6(± 0.8) | 6.1(± 2.1) | 10.2(± 2.2) | 32.2(± 8.1) |
Average retention factor for triplicate injections of racemic warfarin, where the numbers in parentheses represent a range of ± 1 S.D. For Procedure 5, the average retention factor was calculated from the results for two different batches of entrapped HSA supports.
The specific retention factors were measured at pH 7.4 and 0.5 mL/min at 37 °C and have been corrected for any binding by warfarin to the support in the absence of entrapped HSA. The protein content of each support was calculated from the specific retention factor for warfarin by using a molar mass for HSA of 66.5 kDa, the measured void volume of the column, the known packing density for the support (0.45 g/cm3) and an association equilibrium constant of 2.3 (± 0.4) × 105 M−1 for racemic warfarin with HSA at pH 7.4 and 37 °C [31].
The effect of altering the total amount of protein, with a small change in the solution volume, in the entrapment procedure was next considered. This is demonstrated in going from the conditions in Procedure 1 to those Procedure 4, in which the amount of added HSA was increased from 8.0 to 25.2 mg and the solution volume was decreased slightly (28%) from 1.00 to 0.72 mL. To compensate for the decrease in solution volume, the masses of silica and glycogen were decreased to provide the same mass ratio between these agents as was present in Procedure 1. However, the relative amount of added HSA vs. silica was now increased by 4-fold. The combined effect of this increase in the relative amount of HSA and the modest decrease in solution volume was a 5.4-fold increase in the amount of entrapped HSA. This increase was significant at the 95% confidence level and indicated that an increase in the amount of added protein along with a decrease in solution volume could be used to provide a large increase in the level of protein immobilization during entrapment.
The results obtained with Procedure 2–4 were next used to identify a new set of conditions (i.e., Procedure 5) for examining the effect of significantly altering both the amount of added protein and the solution volume in the slurry entrapment method. This set of conditions used the same mass ratios of HSA vs. silica and glycogen vs. silica as were used in Procedure 4 but now used a solution volume that was decreased 3.6-fold from 0.72 to 0.20 mL. This modification in the entrapment method also resulted in a large increase in the amount of entrapped protein, now giving a final protein content of over 32 mg HSA/g silica. This amount was 3.2-fold higher than was seen in Procedure 4 and 17-fold higher than the protein content that was obtained with the initial conditions used in Procedure 1. Based on the increase that was noted in this last group of studies, the conditions employed in Procedure 5 (i.e., in which the solution volume was minimized and the amount of added protein in this solution was maximized) were selected for further use in the slurry-based entrapment, with a modified version of this method being employed with HSA in Section 3.2 and with Con A in Section 3.3.
3.2. Optimization of on-column entrapment for HSA
An on-column format is another possible approach for protein entrapment [17]. In this method, proteins and other reagents are circulated through hydrazide-activated silica that has been packed into a column. The advantage of this approach is that the reaction mixture contains the support within a region that can now make use of the smallest possible volume for the entrapment process. This reduced volume promotes better contact of the applied protein with the pores or region near the surface of the support, which in turn should result in a larger amount of entrapped protein than the slurry-based method [17].
In this report the on-column entrapment system was initially compared to the slurry-based method for the entrapment of HSA. This work was carried out by using a slurry-based method that employed 100 mg/mL HSA as a reference, as based on conditions obtained from Table 1. The concentration of HSA was varied in the on-column method from 12.5 to 100 mg/mL. The same concentration of oxidized glycogen that was present in the slurry-based method was used in the on-column method (i.e., 4.25 mg/mL). In addition, the total volume for the HSA/glycogen solution was kept at 0.35–0.40 mL in both the slurry and on-column methods. Although these solution volumes were slightly higher than the final value of 0.20 mL that was used for the slurry method in Procedure 5 of Table 1, this volume did make it possible to now make a direct comparison between the slurry-based and on-column entrapment methods under equivalent solution reaction conditions.
Table 2 summarizes the final protein contents of the supports that were prepared by the slurry and on-column methods. For the on-column method, a 1.8-fold increase in protein content was seen as the concentration of the applied HSA increased from 12.5 to 50 mg/mL, and a 3.2-fold increase was seen when going from 12.5 to 100 mg/mL HSA, with both of these increases being significant at the 95% confidence level. The results obtained when going from 25 to 100 mg/mL HSA in the on-column entrapment method gave protein contents that were 1.8-to 3.6-fold higher than was obtained when using slurry-based entrapment with 100 mg/mL HSA, with all of these differences being significant at the 95% confidence level. In addition, the protein content obtained when using only 12.5 mg/mL HSA for on-column entrapment was comparable and statistically equivalent to that obtained when using 100 mg/mL HSA in the slurry-based method. Finally, the highest protein content that was obtained here with on-column entrapment was over 46-fold larger than the protein content that was obtained with the initial conditions used in Procedure 1 of Table 1 for the slurry-based entrapment of HSA.
Table 2.
Effects on protein concentration and use of slurry vs on-column entrapment for HSAa
| Entrapment method and HAS concentration |
Specific retention factor, warfarin |
Protein content (mg/g silica) |
Protein content (nmol/g silica) |
|---|---|---|---|
| Slurry methodb 100 mg/mL HSA | 37.6 (± 5.2) | 24.2 (±5.9) | 0.36 (±0.09) |
| On-column method 12.5 mg/mL HSA | 42.4 (±± 1.0) | 27.3 (±5.4) | 0.41 (±0.08) |
| On-column method 25 mg/mL HSA | 68.2 (±3.8) | 44.0 (±9.0) | 0.66 (±0.14) |
| On-column method 50 mg/mL HSA | 75.6 (±3.5) | 48.7 (±9.9) | 0.73 (±0.15) |
| On-column method 100 mg/mL HSA | 134.5 (±5.3) | 86.7 (±17.5) | 1.30 (±0.26) |
The numbers in parentheses represent a range of ± 1 S.D. for three sample injections. The specific retention factors were measured at pH 7.4 and 0.5 mL/min at 37 °C and have been corrected for any binding by warfarin to the support in the absence of entrapped HSA. The protein content of each support was calculated in the same manner as described in Table 1.
The total volume of the HSA/glycogen solution (or HSA/buffer mixture) that was used in the listed slurry method was 0.40 mL to make it equivalent to the HSA/glycogen solution volume that was used for on-column entrapment.
The results in Table 2 clearly indicated that the on-column method allowed for more effective entrapment than the slurry method. This feature made it possible with the on-column method to provide much higher levels of entrapment than the slurry method when using the same total amount of protein, as seen in Table 2 for these two approaches when both used 100 mg/mL HSA. This feature also made it possible to obtain comparable levels of entrapment at much lower levels of applied protein in on-column entrapment, as indicated in Table 2 by the on-column result at 12.5 mg/mL HSA vs. the slurry method result at 100 mg/mL HSA. In the later situation, the slurry method used 40 mg HSA (i.e., a 0.40 mL portion of 100 mg/mL HSA) to prepare the HSA microcolumn, while the on-column method only needed 5 mg HSA (0.4 mL of 12.5 mg/mL HSA). This made the on-column method a more efficient and cost-effective approach for entrapment than the slurry method for the immobilization of HSA in affinity microcolumns.
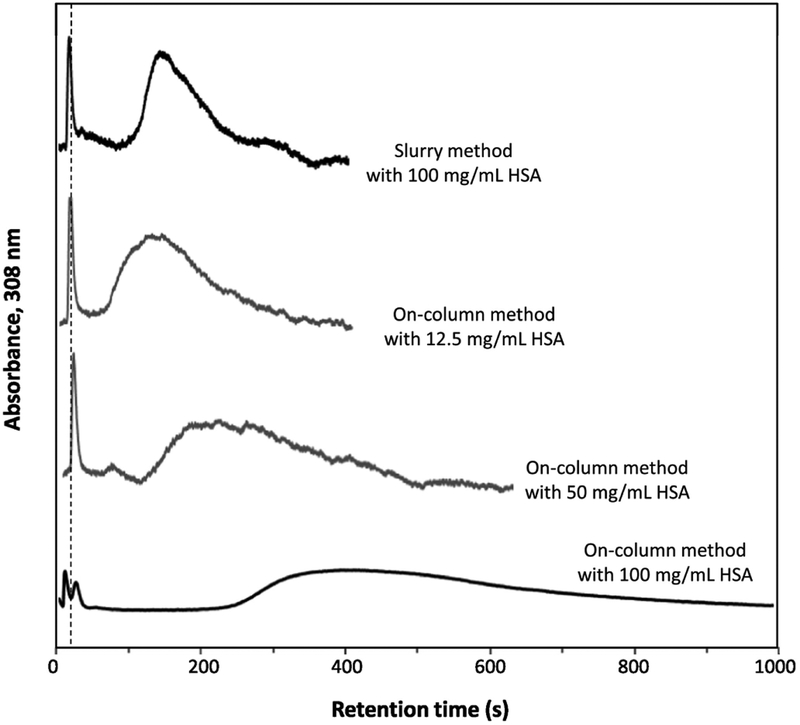
Figure 3 shows some typical chromatograms that were obtained for HSA microcolumns that were prepared by the slurry-based and on-column procedures. This figure indicates that warfarin could easily access and be retained by the entrapped protein in these microcolumns. This observation agrees with prior work that has examined the retention of warfarin and other drugs with columns containing entrapped HSA or alpha1-acid glycoprotein (i.e., another serum transport protein) [16–19]. The chromatograms in Figure 3 also illustrate the ability of on-column entrapment to give higher retention than slurry-based entrapment when both methods use the same total amount of protein (i.e., see top and bottom chromatograms, in which both are based on the use of 100 mg/mL HSA for entrapment). In addition, Figure 3 demonstrates how comparable or even higher amounts of retention can be obtained by on-column entrapment when using less added amounts of protein than slurry-based entrapment. This last situation can be seen by comparing by the top chromatogram, which is for the slurry method at 100 mg/mL HSA, and the middle two chromatograms, which are for the on-column method when using 12.5 or 50 mg/mL HSA and which give equivalent or higher retention for warfarin than the slurry method.
Figure 3.
Chromatograms obtained for racemic warfarin on columns that contained entrapped HSA, as prepared by slurry-based or on-column entrapment using various concentrations of HSA. Conditions: sample, 40 μM racemic warfarin, 5 μL; column size, 1.0 cm × 2.1 mm i.d.; flow rate, 0.50 mL/min; temperature, 37 °C. The total solution volume was 400 μL in the entrapment methods represented by the top three chromatograms and 350 μL in the procedure represented by the bottom chromatogram. Other conditions are provided in the Materials and methods. The dashed vertical line shows the mean elution time of sodium nitrate, which was used as a non-retained solute.
Many previous studies have used HSA as a model protein to optimize and develop new immobilization methods for silica supports in HPAC. Examples of these other immobilization techniques have included the Schiff base method, the N-hydroxysuccinimide method, and various sulfhydryl-reactive techniques [31–34]. Use of these methods with the same type of silica as employed in this study have resulted in final protein contents that have ranged from 8–54 mg HSA/g silica [31–34]. These amounts are comparable to those obtained in this study when using on-column entrapment, with entrapment even reaching a level of almost 87 mg HSA/g silica (or 1.30 μmol/g silica) with the largest amount of HSA that was employed. The potential of entrapment to immobilize more protein than these covalent methods is linked to the use of a three-dimensional region (e.g., the pore volume) to contain the protein instead of using only the surface area of the support. This difference would be expected to provide entrapment with a larger level of immobilization particularly in cases where a protein is quite soluble and can be added to the entrapment solution at high concentrations, as is the case with HSA [24].
Another difference between the covalent and entrapment methods is that the entrapment process results in immobilized HSA that is present in an essentially fully active form [16,18], while covalent immobilization methods have typically given activities of 7–87% for this same protein [31,32,34]. This difference means entrapment can not only be used to potentially produce a larger amount of immobilized protein but to also provide higher retention than a covalent method by providing a higher level of protein activity. For example, the specific retention factors reported for warfarin on HSA columns that have been prepared by various covalent methods have ranged from 7.6–80 [31,32,34]. In this study, specific retention factors for warfarin as high as almost 135 were obtained when using the same type of support and mobile phase/temperature conditions as used in the prior cited work with covalent immobilization schemes.
3.3. Optimization of entrapment for Con A
Con A was another protein that was used as a model to test and optimize the entrapment scheme that was employed in this report. This work was done to see if the trends observed for entrapment with HSA also applied to other binding agents. Con A was entrapped and used at pH 5.0, a condition at which this protein mainly exists as a dimer (molar mass of 53 kDa, with each monomer having a mass of 26.5 kDa) [10,28,29]. Although the dimer of Con A has a slightly lower mass than HSA, the hydrodynamic radius of the Con A dimer is comparable to and slightly larger than that of HSA (i.e., 4.1 nm vs. 3.5 nm) [35,36]. Furthermore, Con A is similar to HSA in that it has well-described binding for some solutes, one of which is the fluorescent sugar MUM [28,29]. This last feature made it again possible to use retention measurements with MUM as a probe compound to estimate the content of active Con A in affinity microcolumns and within supports that were prepared under various entrapment conditions.
The results that were obtained for the entrapment of Con A are summarized in Table 3. The lower solubility of Con A compared to HSA [24,37,38] did limit the range of concentrations for Con A that could be used for the entrapment process (i.e., values up to 10 mg/mL). However, the general trends seen in Table 2 for HSA also appeared in the data for Con A in Table 3. For instance, an increase in the total amount of Con A that was used for on-column entrapment again resulted in an increase in the final level of entrapped protein; the amount of entrapped protein increased by 2.2-fold (i.e., a result significant at the 95% confidence level) when going from an applied concentration of 1.0 to 10 mg/mL Con A. This increase, which occurred over a 10-fold range in the applied concentration of Con A, was smaller than the 3.2-fold increase seen in Table 2 over an 8-fold change in the concentration of HSA that was used for entrapment. This difference may be due to the loss of some Con A before immobilization, as some cloudiness was noted to occur during the preparation of a 10 mg/mL Con A solution, which was then filtered prior to its use in entrapment.
Table 3.
Effects of protein concentration and use of slurry vs. on-column entrapment for Con Aa
| Entrapment method and Con A concentration |
Specific retention factor, MUM |
Protein content (mg/g silica) |
Protein content (μmol dimer/g silica) |
|---|---|---|---|
| Slurry method lOmg/mLCon A | 13.4 (±1.2) | 17.6 (±2.3) | 0.33 (±0.04) |
| On-column method 1.0 mg/mLCon A | 16.0 (±1.2) | 21.0 (±2.6) | 0.40 (±0.05) |
| On-column method 10 mg/mLCon A | 34.7 (±2.9) | 45.6 (±5.7) | 0.86 (±0.11) |
The numbers in parentheses represent a range of ± 1 S.D., as obtained for a set of three injections. The specific retention factors were measured at 0.5 mL/min and 20 °C and have been corrected for any binding by MUM with the support in the absence of MUM. The protein content of each support was calculated from the specific retention factor for MUM by using a molar mass for the monomer of Con A of 26.5 kDa, the measured void volume of the column, the known packing density for the support (0.45 g/cm3), and an association equilibrium constant of 4.5 × 105 M−1 for MUM with Con A at pH 5.0 and 25 °C [28].
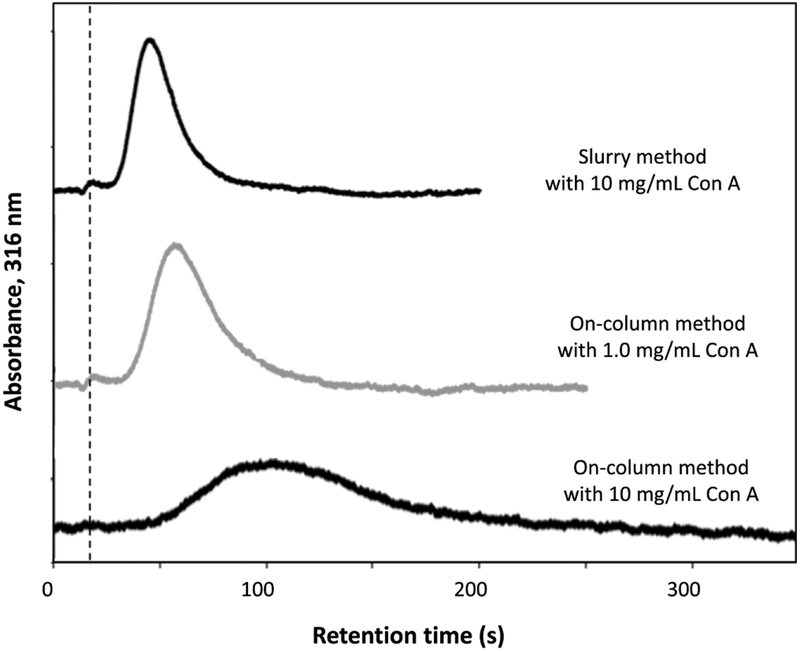
Another similarity in the results for Con A and HSA was in the ability of the on-column approach to give more effective entrapment than the slurry method. For Con A, the use of on-column entrapment and a 1.0 mg/mL Con A solution gave a statistically-equivalent level of entrapment (at the 95% confidence level) to the use of slurry-based entrapment with 10 mg/mL Con A. In addition, use of 10 mg/mL Con A for both slurry entrapment and on-column entrapment resulted in a 2.6-fold higher amount of immobilized Con A for the on-column method. The effect of these different levels of entrapment on the retention of MUM is illustrated by the chromatograms in Figure 4. These chromatograms show how the use of only 1.0 mg/mL Con A in the on-column entrapment gave a support that was similar in retention behavior for MUM to that obtained with the slurry method when using 10 mg/mL Con A. This figure also shows how the use of 10 mg/mL Con A for on-column entrapment created a microcolumn that had higher retention for MUM than when using the same concentration of Con A in the slurry method.
Figure 4.
Chromatograms obtained for MUM on columns that contained entrapped Con A, as prepared by slurry-based or on-column entrapment. Conditions: sample, 10 μM MUM, 5 μL; column size, 1.0 cm × 2.1 mm I.D.; flow rate, 0.50 mL/min; temperature, 20 °C. Other conditions are provided in the Materials and methods. The dashed vertical line shows the mean elution time of MUGA, which was used as a non-retained solute.
These results were compared to those obtained in prior studies that have used a covalent immobilization method to couple Con A to silica. The highest Con A content that was obtained in this study (i.e., ~46 mg/g silica, or 0.86 μmol Con A dimer/g silica) was comparable to values of 40–50 mg Con A/g silica that have been reported for the Schiff base method of covalent immobilization [25,26]. This was the case even though the silica used in these earlier reports either had a lower [26] or larger surface area [25] than the support that was used for entrapment in this current work. These results indicated that entrapment could be used as a viable alternative to covalent immobilization methods for the preparation of Con A supports and columns. It was further noted that the levels of entrapment seen in Table 3 for the slurry-based and on-column entrapment of Con A (i.e., 0.33 and 0.40–0.86 μmol dimer/g silica, respectively) were in the same general range as obtained in Table 2 when these entrapment methods were used with HSA (i.e., 0.36 and 0.41–1.30 μmol/g silica).
4. Conclusion
This report examined several factors that may influence the extent of entrapment for proteins such as HSA and Con A within HPLC-grade porous silica. Of particular interest was the use of entrapment to create affinity microcolumns [20]. The retention properties and levels of entrapment of the resulting microcolumns were evaluated by using injections of warfarin or MUM as probes for HSA and Con A, respectively [5,16,28,29]. It was found with HSA that altering both the solution volume and amount of added protein could lead to a significant increase in the extent of protein immobilization that was obtained in slurry-based entrapment. The amount of added protein was also found to be important when using on-column entrapment with HSA, where a large increase in the final protein content of the supports was noted when comparing on-column and slurry-based entrapment. Similar results were obtained with Con A when comparing slurry-based vs. on-column entrapment.
The final, optimized conditions identified for the slurry entrapment of HSA gave a 17-fold higher protein content than conditions based on a previous approach for the entrapment of this protein (i.e., Procedure 1 in Table 1) [16]. The conditions that were optimized for the on-column entrapment of HSA gave over a 45-fold higher protein content than was obtained with the original procedure for slurry-based entrapment [16] and a 3.6-fold higher protein content than the optimized slurry method, when the latter approach used a comparable protein concentration and solution volume for the immobilization process. The largest protein content that was obtained for either the slurry-based or on-column entrapment of HSA (i.e., ~32 and 86 mg/g silica, respectively) was comparable to or larger than values of 8–54 mg/g silica that have been reported for a number of covalent methods with the same type of silica as employed in this report [31–34]. In addition, the highest specific retention factor observed here for warfarin on an entrapped HSA microcolumn was 18-to 1.7-fold higher than values that have been obtained when using covalent immobilization [31,32,34]. For Con A, the highest protein content that was measured in this study (i.e., ~46 mg/g silica) was in the same range as values that have been reported for covalent immobilization using other types of silica [25,26].
These results indicate that entrapment can be a valuable alternative to covalent immobilization for proteins such as HSA and Con A. These data also demonstrate how entrapment can provide higher levels of protein immobilization than covalent coupling techniques and result in supports and columns with increased levels of solute retention. These differences are believed to be due to two factors: the use of a three-dimensional region in entrapment instead of a surface for immobilization, and the loss of actual or apparent activity of the binding agent during covalent immobilization [16,18,31,32,34]. The trends noted in this study should provide valuable guidelines for future applications of HSA, Con A and other binding agents in HPAC. As illustrated in this report, the ability to prepare supports by entrapment should be especially attractive as a means to provide strong binding and good retention in future work with affinity microcolumns or related microscale devices that employ affinity ligands as immobilized agents [20–23,26,39].
Highlights.
Several procedures were evaluated for protein entrapment in hydrazide-silica.
The protein content of each support was evaluated for use in affinity microcolumns.
Human serum albumin and concanavalin A were used as model proteins.
On-column entrapment was found to be more effective than a slurry-based method.
The protein contents seen with entrapment met or exceeded those of covalent methods.
Acknowledgements
This work was funded by the National Institutes of Health under grant R01 DK069629.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hage DS (Ed.), Handbook of Affinity Chromatography, CRC Press, Boca Raton, FL, 2005, pp. 3–14. [Google Scholar]
- [2].Walters RR, Affinity chromatography, Anal. Chem 57 (1985) 1099A–1114A. [DOI] [PubMed] [Google Scholar]
- [3].Larsson PO, High-performance liquid affinity chromatography, Methods Enzymol. 104 (1984) 212–223. [DOI] [PubMed] [Google Scholar]
- [4].Singh P, Madhaiyan K, Duong-Thi M-D, Dymock BW, Ohlson S, Analysis of protein target interactions of synthetic mixtures by affinity-LC/MS, SLAS Discovery 22 (2017) 440–446. [DOI] [PubMed] [Google Scholar]
- [5].Zhang C, Rodriguez E, Bi C, Zheng X, Suresh D, Suh K, Li Z, Elsebaei F, Hage DS, High performance affinity chromatography and related methods for the analysis of biological and pharmaceutical agents: a review, Analyst 143 (2018) 374–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hage DS, Anguizola JA, Bi C, Li R, Matsuda R, Papastavros E, Pfaunmiller E, Vargas J, Zheng X, Pharmaceutical and biomedical applications of affinity chromatography: recent trends and developments, J. Pharm. Biomed. Anal 69 (2012) 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kratochwil NA, Huber W, Muller F, Kansy M, Gerber PR, Predicting plasma protein binding to drugs: a new approach, Biochem. Pharmacol 64 (2002) 1355–1374. [DOI] [PubMed] [Google Scholar]
- [8].Li Z, Beeram SR, Bi C, Suresh D, Zheng X, Hage DS, High-performance affinity chromatography: applications in drug-protein binding studies and personalized medicine, in: Donev R (Ed.), Advances in Protein Chemistry and Structural Biology, Vol. 102, Elsevier, Amsterdam, 2016, pp. 1–39. [DOI] [PubMed] [Google Scholar]
- [9].Kim HS, Wainer I, Rapid analysis of the interactions between drugs and human serum albumin (HSA) using high performance affinity chromatography (HPAC), J. Chromatogr. B 870 (2008) 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hage DS, Bian M, Burks R, Karle E, Ohnmacht C, Wa C, Bioaffinity chromatography, in: Hage DS (Ed.), Handbook of Affinity Chromatography, CRC Press, Boca Raton, FL, 2005, pp. 101–126. [Google Scholar]
- [11].Hermanson GT, Mallia AK, Smith PK, Immobilized Affinity Ligand Techniques, Academic Press, New York, 1992. [Google Scholar]
- [12].Kim HS, Hage DS, Immobilization methods for affinity chromatography, in: Hage DS (Ed.), Handbook of Affinity Chromatography, CRC Press, Boca Raton, FL, 2006, pp. 35–78. [Google Scholar]
- [13].Avnir D, Coradin T, Lev O, Livage J, Recent bio-applications of sol-gel materials, J. Mater. Chem 16 (2006) 1013–1030. [Google Scholar]
- [14].Livage J, Coradin T, Roux C, Encapsulation of biomolecules in silica gels, J. Phys.: Condens. Matter 13 (2001) R673–R691. [Google Scholar]
- [15].Betancor L, Luckarift HR, Bioinspired enzyme encapsulation for biocatalysis, Trends Biotechnol. 25 (2008) 566–572. [DOI] [PubMed] [Google Scholar]
- [16].Jackson AJ, Xuan H, Hage DS, Entrapment of proteins in glycogen-capped and hydrazide-activated supports, Anal. Biochem 404 (2010) 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Anguizola J, Bi C, Koke M, Jackson A, Hage DS, On-column entrapment of alpha1-acid glycoprotein for studies of drug-protein binding by high-performance affinity chromatography, Anal. Bioanal. Chem 408 (2016) 5745–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jackson AJ, Anguizola J, Pfaunmiller EL, Hage DS, Use of entrapment and high-performance affinity chromatography to compare the binding of drugs and site-specific probes with normal and glycated human serum albumin, Anal. Bioanal. Chem 405 (2013) 5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bi C, Jackson A, Vargas-Badilla J, Li R, Rada G, Anguizola J, Pfaunmiller E, Hage DS, Entrapment of alpha1-acid glycoprotein in high-performance affinity columns for drug-protein binding studies, J. Chromatogr. B 1021 (2016) 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zheng X, Li Z, Beeram S, Matsuda R, Pfaunmiller EL, Podariu M, White CJ II, Carter N, Hage DS, Analysis of biomolecular interactions using affinity microcolumns: a review, J. Chromatogr. B 968 (2014) 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yoo M, Schiel JE, Hage DS, Evaluation of affinity microcolumns containing human serum albumin for rapid analysis of drug-protein binding, J. Chromatogr. B 878 (2010) 1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anguizola J, Joseph KS, Barnaby OS, Matsuda R, Alvarado G, Clarke W, Cerny RL, Hage DS, Development of affinity microcolumns for drug-protein binding studies in personalized medicine: interactions of sulfonylurea drugs with in vivo glycated human serum albumin, Anal. Chem 85 (2013) 4453–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pfaunmiller EL, Hartmann M, Dupper CM, Soman S, Hage DS, Optimization of human serum albumin monoliths for chiral separations and high-performance affinity chromatography, J. Chromatogr. A 1269 (2012) 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peters T Jr., All About Albumin: Biochemistry, Genetics and Medical Applications, Academic Press, San Diego, 1996. [Google Scholar]
- [25].Mann BF, Mann AKP, Skrabalak SE, V Novotny M, Sub 2-μm macroporous silica particles derivatized for enhanced lectin affinity enrichment of glycoproteins, Anal. Chem 85 (2013) 1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Madera M, Mechref Y, Novotny MV, Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides, Anal. Chem 77 (2005) 4081–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ruhn PF, Garver S, Hage DS, Development of dihydrazide-activated silica supports for high-performance affinity chromatography, J. Chromatogr. A 669 (1994) 9–19. [DOI] [PubMed] [Google Scholar]
- [28].Anderson DJ, Walters RR, Equilibrium and rate constants of immobilized concanavalin A determined by high-performance affinity chromatography, J. Chromatogr 376 (1986) 69–85. [Google Scholar]
- [29].Anderson DJ, Walters RR, Affinity chromatographic examination of a retention model for macromolecules, J. Chromatogr 331 (1985), 1–10. [DOI] [PubMed] [Google Scholar]
- [30].Loun B, Hage DS, Characterization of thyroxine-albumin binding using high-performance affinity chromatography, II. Comparison of the binding of thyroxine, triiodothyronines and related compounds at the warfarin and indole sites of human serum albumin, J. Chromatog. B 665 (1995) 303–314. [DOI] [PubMed] [Google Scholar]
- [31].Loun B, Hage DS, Chiral separation mechanism in protein-based HPLC columns. I. Thermodynamic studies of (R)- and (S)-warfarin binding to immobilized human serum albumin, Anal. Chem 66 (1994) 3814–3822. [DOI] [PubMed] [Google Scholar]
- [32].Kim HS, Kye YS, Hage DS, Development and evaluation of N-hydroxysuccinimide- activated silica for immobilizing human serum albumin in liquid chromatography columns, J. Chromatogr. A 1049 (2004) 51–61. [PubMed] [Google Scholar]
- [33].Kim HS, Mallik R, Hage DS, Chromatographic analysis of carbamazepine binding to human serum albumin. II. Comparison of the Schiff base and N-hydroxysuccinimide immobilization methods, J. Chromatogr. B 837 (2006) 138–146. [DOI] [PubMed] [Google Scholar]
- [34].Mallik R, Wa C, Hage DS, Development of sulfhydryl-reactive silica for protein immobilization in high-performance affinity chromatography, Anal. Chem 79 (2007) 1411–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Khan MV, Ishtikhar M, Siddiqui MK, Zaman M, Chandel TI, Majid N, Ajmal MR, Abdelhameed AS, Shahein YE, Khan RH, Biophysical insight reveals tannic acid as amyloid inducer and conformation transformer from amorphous to amyoid aggregates in Concanavalin A (ConA), J. Biomol. Struct. Dynam, 36 (2018) 1261–1273. [DOI] [PubMed] [Google Scholar]
- [36].Armstrong JK, Wenby RB, Meiselman HJ, Fisher TC, The hydrodynamic radii of macromolecules and their effect on red blood cell aggregation, Biophys. J 87 (2004) 4259–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Olson MO, Liener IE, Some physical and chemical properties of concanavalin A, the phytohemagglutinin of the jack bean, Biochemistry 6 (1967) 105–111. [DOI] [PubMed] [Google Scholar]
- [38].Mikol V, Giege R, Phase diagram of a crystalline protein: determination of the solubility of concanavalin A by a microquantitation assay, J. Crystal Growth 97 (1989) 324–332. [Google Scholar]
- [39].Phillips TM, Microanalytical methods based on affinity chromatography, in: Hage DS (Ed.), Handbook of Affinity Chromatography, CRC Press, Boca Raton, FL, 2006, pp. 763–788. [Google Scholar]