How maternal factors in oocytes trigger zygotic genome activation (ZGA) is a long-standing question in developmental biology. Recent studies in 2-cell like embryonic stem cells (2C-like cells) suggest that transcription factors of the DUX family are key regulators of ZGA in placental mammals 1,2. To characterize the role of DUX in ZGA, we generated Dux cluster knockout (KO) mouse lines. Unexpectedly, we found both Dux zygotic KO (Z-KO) and maternal/zygotic KO (MZ-KO) embryos can survive to adulthood despite showing reduced developmental potential. Furthermore, transcriptome profiling of the MZ-KO embryos revealed that loss of DUX has minimal effects on ZGA and most DUX targets in 2C-like cells are normally activated in MZ-KO embryos. Thus, contrary to the key function in inducing 2C-like cells, our data indicate that DUX only has a minor role in ZGA and loss of DUX is compatible with mouse development.
In mammals, early embryonic development is supported first by maternal factors in the egg and later by newly transcribed genes from the zygotic genome. Successful ZGA is essential for embryonic development. In mice, the major wave of ZGA takes place at the 2-cell stage with the activation of thousands of genes and transposable elements (TEs) including the ERVL-family retrotransposons 3–6. Interestingly, ERVL and ERVL-linked genes can also be spontaneously activated in a rare and transient embryonic stem (ES) cell population that termed as 2C-like cells 7,8. Since 2C-like cells mimic 2-cell embryos in terms of the expression of 2-cell transient transcripts and have the capacity to contribute to both embryo and extra-embryonic tissues 7, 2C-like cells have been a useful model for understanding totipotency 9 and early embryonic development 10–13. However, 2C-like cells are not equivalent to 2-cell embryos as the genes induced in 2C-like cells only represent a subset of the mouse ZGA genes that are activated in 2-cell stage embryos 7.
Dux (also known as Duxf3) in mouse and its human homologue DUX4 are double-homeodomain genes that are activated at the onset of ZGA in early embryos 1,2. In mouse, the Dux cluster also includes a truncated variant named Gm4981 (also known as Duxf4), which lacks the first homeodomain and is transcribed as early as during oogenesis 2. In humans, incomplete silencing of DUX4 causes facioscapulohumeral muscular dystrophy (FSHD) 14 characterized by de-repression of genes/repeats, such as ZSCAN4 and ERVL, that are only expressed during ZGA and muscle cells in FSHD patients 15. Additionally, in ES cells, mouse DUX can activate ERVL-family repeats and ERVL-linked genes and is both necessary and sufficient for the ES to 2C-like cell transition 1,2,16. Furthermore, acute depletion of the Dux cluster in zygotes by CRISPR/Cas9 injection leads to down-regulation of a handful of ZGA genes as revealed by RT-qPCR 2. These results suggest that DUX may have an important role in mouse ZGA and embryonic development 2.
To comprehensively define the role of DUX in ZGA, we attempted to acutely deplete the Dux-containing macrosatellite repeats (estimated ~160 kb in C57BL/617) by zygotic CRIPSR-Cas9 injection. The pair of single guide RNA (sgRNA) co-injected with the Cas9 mRNA targets the same flanking sequences of the Dux cluster 2(Supplementary Fig. 1a). Genotyping of the blastocysts revealed that 25% (9/36) and 5.6% (2/36) blastocysts carried the mono-allelic and bi-allelic Dux cluster deletion, respectively (Supplementary Fig. 1b-d), which are comparable to the reported efficiency for CRIPSR-Cas9-mediated large (> 10 kb) genomic fragment deletions 18–22. Since the acute depletion experiment suggests that DUX is not essential for pre-implantation development, we generated mouse lines carrying the Dux KO allele to further characterize the role of DUX in mouse development.
To this end, following zygotic CRIPSR-Cas9 injection, 2-cell embryos were transferred to pseudo-pregnant female mice to obtain live pups. Out of the 87 transferred 2-cell embryos, 27 living pups were obtained and six out of the 20 genotyped F0 mice harbored the Dux cluster deletion on one of the alleles (Fig. 1a, Supplementary Fig. 2a-b). When the two F0 mice (i.e., 423 and 426) were backcrossed with wild-type C57BL/6 (WT B6) mice, we obtained 85 (51%) WT and 82 (49%) Dux heterozygous (Dux Het) mice from a total of 22 litters (Supplementary Fig. 2c). The observed WT/Het ratio is consistent with the expected 50:50 Mendelian frequency, indicating that Dux heterozygosity does not impair mouse development.
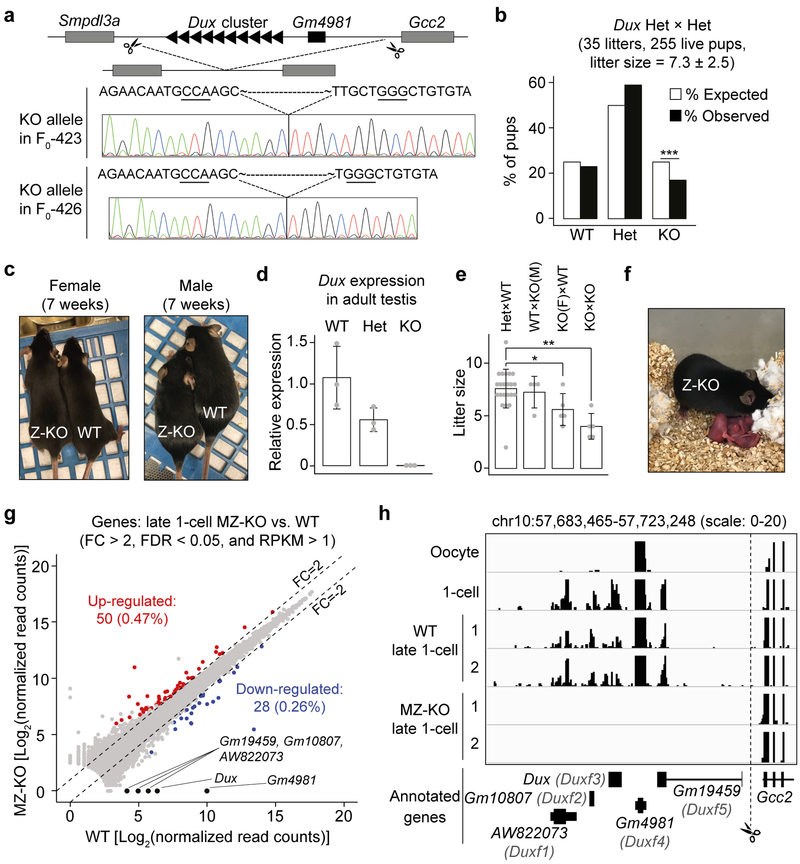
Figure 1. Loss of DUX is compatible with mouse development.
a) Schematics of the Dux cluster in mice (not drawn in scale) and Sanger sequencing results of the KO alleles in the two founder lines. Underlined three nucleotides represent the CRISPR PAM sequences.
b) Bar graph showing the percentage of pups for each genotype from Dux Het × Het crosses. *** P value = 0.005, Chi-squared goodness of fit test.
c) Examples of Dux F2 WT and Z-KO adult mice analyzed in panel B.
d) RT-qPCR results confirming Dux KO in adult testis. The expression level of Dux in WT adult mouse (9–12 weeks) testis was set as 1.0. Three mice were analyzed for each genotype (denoted as grey dots). Measure of center and error bar indicate mean and standard deviation (SD), respectively.
e) Litter sizes of the indicated crosses. Each grey dot represents a single litter analyzed. Number of litters analyzed are 22, 4, 5, 5 for Het × Het, WT × KO(M), KO(F) × WT, and KO × KO mating, respectively. ** P = 0.0003; * P = 0.03, two-tailed Student’s t test. Measure of center and error bar indicate mean and SD, respectively.
f) An example of a Z-KO × Z-KO litter with live pups analyzed in panel E.
g) Scatter plot comparing the gene expression levels between Dux MZ-KO and WT at late 1-cell stage [~12 hours post in vitro fertilization (hpi)]. Two RNA-seq replicates were generated for differential gene expression analyses.
h) Genome browser view of RNA-seq signal at the Dux cluster in WT and MZ-KO late 1-cell embryos. RNA-seq tracks of oocyte and 1-cell embryos were obtained from 23. Only uniquely aligned reads were used to generate the RNA-seq tracks.
To determine whether DUX deficiency causes developmental arrest, we genotyped 255 pups from 35 litters of Dux Het × Het (F1 × F1 and F2 × F2) mating pairs (Supplementary Fig. 2c). Contrary to the expectation that Dux zygotic KO (Z-KO) embryos arrest during pre-implantation development due to ZGA defects, Dux Z-KO mice can survive to adulthood and were grossly normal although they were born at a reduced frequency (18% vs. expected 25%, P = 0.005) (Fig. 1b and c, Supplementary Fig. 2c). To exclude the possibility that Dux copies outside of the deleted macrosatellite repeats may compensate for DUX deficiency, we determined Dux RNA level in testis, one of the few organs where Dux is expressed in adults 14. As expected, the level of Dux transcript in Het testis is about half of that in WT and is undetectable in the KO testis samples (Fig. 1d). This result is consistent with the previous report that the Dux cluster on chromosome 10 is the only Dux locus in mice 17. Collectively, these data indicate that loss of zygotic DUX is compatible with mouse development.
To determine whether the truncated DUX variant Gm4981 in oocytes might compensate for the deficiency of DUX in Z-KO embryos, we assessed the development of Dux Z-KO × Z-KO offspring, which should lack both zygotic DUX and maternal Gm4981. Dux MZ-KO embryos did not show impaired pre-implantation development (Supplementary Fig. 3) and can also survive to adulthood without obvious abnormalities (Fig. 1e, f, and Supplementary Fig. 2c). Consistent with the reduced frequency of Z-KO pups in Het × Het crosses, the litter sizes of Z-KO × Z-KO mating pairs are also significantly smaller compared to controls (4.0 ± 1.2 vs. 7.6 ± 1.8, P = 0.0003). Interestingly, Dux Z-KO females showed slightly reduced litter size (5.6 ± 1.5, P = 0.03) when their fertility was tested using WT B6 male mice. The reduction in litter size of Dux Z-KO female progeny should occur after implantation as both Z-KO female ovulation and preimplantation development of MZ-KO embryos appear normal when compared to control WT or Het mice (Supplementary Fig. 3). Nevertheless, the fact that both Dux Z-KO and MZ-KO can develop to adulthood indicates that DUX and its truncated variant Gm4981 are not essential for mouse development.
Since lack of DUX is compatible with mouse development, DUX is unlikely to play a major role in ZGA. To investigate this, we generated late 1-cell and late 2-cell Dux MZ-KO embryos by fertilizing F2 Z-KO oocytes with F2 Z-KO sperm, followed by RNA-sequencing (RNA-seq). Embryos generated by fertilizing F2 WT oocytes with F2 WT sperm were used as controls. After confirming data reproducibility (Supplementary Fig. 4), we performed comparative analyses of late 1-cell RNA-seq datasets which revealed that, out of the 10,554 detectable genes (RPKM >1 in either WT or KO), only 50 (0.47%) and 28 (0.26%) were significantly up- and down-regulated, respectively, in Dux MZ-KO embryos [fold change (FC) > 2 and false discovery rate (FDR) < 0.05] (Fig. 1g, Supplementary Table 1), suggesting that DUX and Gm4981 deficiency has little effect on late 1-cell gene expression. Although it is not feasible to assess the expression level of each Dux repeat due to the assembly gap at the Dux cluster, we note that the annotated Dux and the other four known genes (i.e., AW822073/Duxf1, Gm10807/Duxf2, Gm19459/Duxf5, Gm4981/Duxf4) were depleted in the MZ-KO 1-cell embryos (Fig. 1g and h), confirming complete KO of the Dux cluster in our mouse lines.
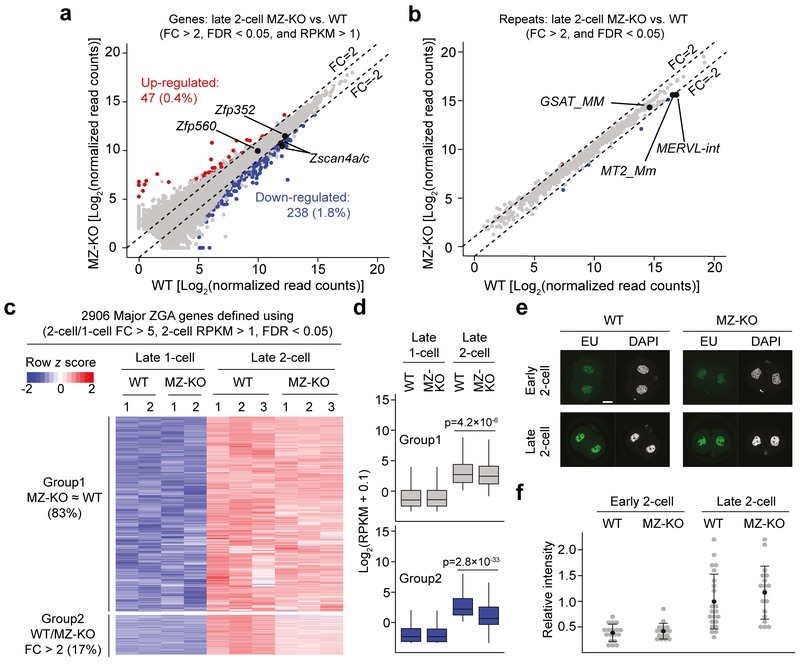
We next analyzed late 2-cell WT and MZ-KO RNA-seq datasets and identified that, out of the 12,960 detectable genes (RPKM > 1 in either WT or MZ-KO), 47 (0.4%) and 238 (1.8%) were significantly up- and down-regulated, respectively, in Dux MZ-KO embryos (FC > 2 and FDR < 0.05) (Fig. 2a, Supplementary Table 2). Consistent with the few gene expression changes, most repeats also showed comparable expression levels between the two groups (Fig. 2b). To define to what extent DUX contributes to major ZGA, we identified 2,906 major ZGA genes by comparing the transcriptome of WT late 2-cell to late 1-cell embryos (2-cell/1-cell FC > 5, RPKM in WT 2-cell > 1, FDR < 0.05) (Fig. 2c, Supplementary Table 3). Even using a relaxed cutoff to define differential gene expression (FC > 2 and FDR < 1), only 493 (16.9%) ZGA genes showed decreased expression in Dux MZ-KO 2-cell embryos (Fig. 2c, Supplementary Table 3). Importantly, these affected genes/repeats are still activated in Dux KO MZ-KO 2-cells when compared with that in WT 1-cell embryos (Fig. 2d, Supplementary Table 3), indicating that ZGA of these genes still takes place in Dux KO embryos although at a lesser extent. Consistently, global transcription levels are comparable between WT and MZ-KO early and late 2-cell embryos as revealed by EU incorporation assay (Fig. 2e and f). Nonetheless, activation of a small group of ZGA genes were affected at certain level, which may account for the reduced frequency of Dux Z-KO mice from F1 Het × F1 Het and smaller litter sizes for Z-KO × Z-KO mating pairs (Fig. 1b and e). Taken together, these data support that DUX only plays a minor role in mouse ZGA.
Figure 2. Loss of DUX causes minor defects in ZGA.
a, b) Scatter plots comparing the genes (A) and repeats (B) expression levels of late 2-cell embryos (~30 hpi) of Dux MZ-KO and WT. Three RNA-seq replicates were generated for differential gene expression analyses.
c) Heatmap illustrating the expression levels of major ZGA genes at late 1-cell and late 2-cell stages of Dux WT and MZ-KO embryos. Group 1 represents genes that showed similar expression (FC < 2) between WT and MZ-KO 2-cell embryos, while Group 2 represents genes that showed decreased expression (FC > 2 and FDR < 1) in MZ-KO 2-cell embryos.
d) Boxplots illustrating the expression levels of Group 1 (n = 2,413) and 2 (n = 493) genes in panel C. Mann-Whitney-Wilcoxon test (two-sided) was used to calculate the P values between WT and MZ-KO. The middle lines in the boxes represent medians. Box hinges indicate the 25th/75th percentiles and the whiskers indicate the hinge ± 1.5 × inter-quartile range.
e) The EU-staining assay showing the global transcriptional activity in early (~22 hpi) and late (~30 hpi) 2-cell embryos. Scale bar = 20 μm, hpi = hours post in vitro fertilization.
f) Quantification of the EU signal intensity shown in panel E. The average signal intensity of late 2-cell was set as 1.0. Each grey dot represents a single embryo analyzed. Measure of center and error bar indicate mean and SD, respectively. Two-tailed Student’s t test was used to compare the signal intensity between WT and MZ-KO (1-cell: P = 0.58; 2-cell: P = 0.29). The total number of embryos analyzed were 18, 23, 16, and 18 for WT (~22 and ~30 hpi) and MZ-KO (~22 and ~30 hpi), respectively.
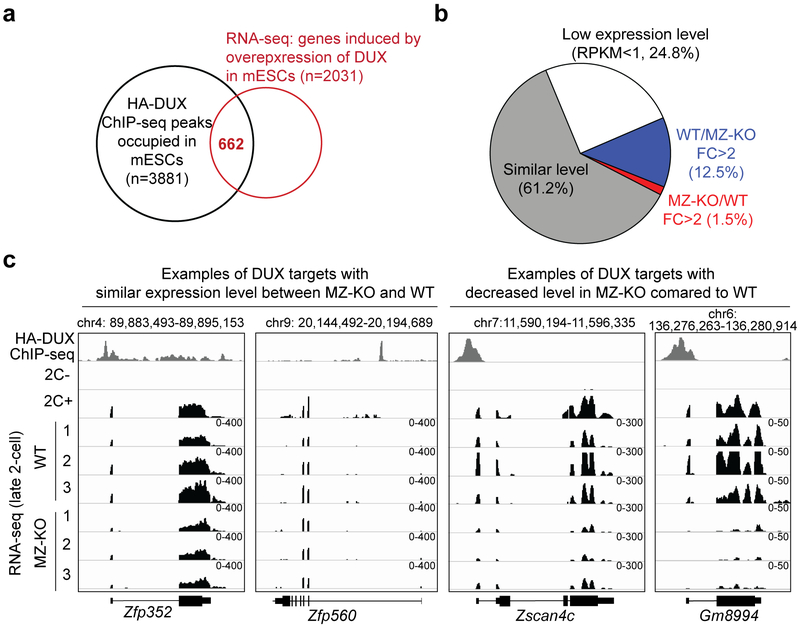
Because of the subtle effect of loss of DUX on 2-cell transcriptome, we hypothesized that most DUX targets identified in 2C-like cells should be normally activated in Dux MZ-KO embryos. Indeed, out of the 662 genes that are activated by exogeneous DUX and are also associated with HA-DUX ChIP-seq peaks 1, only 12.5% showed more than 2-fold of decrease in Dux MZ-KO embryos, while the majority (61.2%) exhibited comparable expression levels between WT and MZ-KO (Fig. 3, Supplementary Fig. 5 and Supplementary Table 4). This suggests that either DUX targets identified in 2C-like cells are not targeted by DUX in 2-cell embryos or that factors other than DUX can activate them in 2-cell embryos.
Figure 3. The majority of DUX targets identified in 2C-like cells are activated normally in Dux MZ-KO 2-cell embryos.
a) Identification of known DUX targets by overlapping the HA-DUX ChIP-seq peak-associated genes and DUX-overexpression-induced genes in mESCs. Both HA-DUX ChIP-seq and 2C-like cells RNA-seq datasets were obtained from 1.
b) Expression level changes of known DUX targets in MZ-KO 2-cell embryos. Known DUX targets were defined as genes that are associated with HA-DUX ChIP-seq peaks and are upregulated (FC > 2 and FDR < 0.05) in 2C-like cells. The average of three RNA-seq replicates of WT and MZ-KO late 2-cell embryos were used for the analyses.
c) Genome browser views illustrating the RNA levels of known DUX targets in 2C-like cells and 2-cell embryos. HA-DUX ChIP-seq track and RNA-seq tracks of non-2C (2C-) and 2C-like (2C+) cells were obtained from 1. Only uniquely aligned reads were used to generate the ChIP-seq and RNA-seq tracks.
Overall, our results demonstrate that mouse ZGA genes, including many exogenous DUX targets identified in mouse ESCs, can be activated in Dux MZ-KO embryos and thereby loss of DUX is compatible with mouse development. It is possible that other transcription factors and/or chromatin remodelers play a redundant role in 2-cell embryos for successful ZGA. Our study in mice seems to be in direct contrast to the observations in ES cells, in which DUX is essential for the entry of ESCs into the 2C-like state 1,2. Therefore, despite the simplicity of the 2C-like state, caution should be taken in using the ESC system to study the totipotent state as there are fundamental differences between the in vitro 2C-like cell state and 2-cell embryos.
Methods
Generation of Dux KO mice
All animal experiments were performed in accordance with the protocols of the Institutional Animal Care and Use Committee at Harvard Medical School. For superovulation, B6D2F1 (BDF1) female mice (6–8 weeks) (Jackson Laboratory, 100006) were injected interperitoneally with 7.5 IU of pregnant mare serum gonadotropin (PMSG, Millipore) on day 1 and human chorionic gonadotropin (hCG, Millipore) on day 3 (44–48 hours after PMSG injection). For in vitro fertilization (IVF), the oocytes collected 12–16 hours after hCG injection were inseminated with the activated spermatozoa collected from the caudal epididymis of BDF1 males (9–10 weeks) in HTF medium supplemented with 10 mg/ml bovine serum albumin (BSA; Sigma). The spermatozoa were capacitated by pre-incubation in HTF medium for one hour. At 2 h post-IVF (hpi), Cas9 mRNA (100 ng/μl) and sgRNA (50 ng/μl each) were injected into cytoplasm of fertilized eggs using a Piezo impact-driven micromanipulator (Primer Tech, Ibaraki, Japan). Following injection, zygotes were cultured in HTF medium for another four hours and then cultured in KSOM (Millipore) at 37°C under 5% CO2 with air. At ~24 h hpi, 2-cell embryos were transferred into oviducts of surrogate ICR strain mothers. The synthesis of Cas9 mRNA and sgRNA were as previously described 24. The sgRNA sequences were the same as previously reported 2.
For genotyping of blastocysts, each embryo collected at 120 hpi was lysed in 8 μl lysis buffer (50 mM Tirs-HCl [pH 8.0], 0.5% Triton, 400 μg/ml Proteinase K) (Sigma) at 60°C for one hour. Following heat inactivation at 90°C for 5 min, 2 μl of lysis buffer containing genomic DNA were used as template for nested PCR. The primers used for genotyping are included in Supplementary Table 6 (WT allele 268 bp and KO allele is ~320 bp). For both rounds of PCR, the following program was used: initial denaturation: 5 min at 95°C; 30 cycles of 30s at 95°C, 30s at 60°C, and 30s at 72°C; final extension: 5 min at 72°C.
For genotyping of colonies, a mouse tail tip was lysed in the same lysis buffer (70 μl) at 60°C overnight and the supernatants were used as template for PCR (only inner primers were used, WT allele 268bp, KO allele is 322 bp and 318 bp for line 423 and 426, respectively).
RNA-seq
For embryos collected for RNA-seq (i.e., Dux F2 × F2), IVF was performed as described above except that the micro-injection steps were omitted. Late 1-cell and late 2-cell were collected at ~12 and ~30 hpi, respectively. For each biological replicate, 11–13 embryos were pooled for RNA-seq analyses. Specifically, the embryos were briefly incubated in Acid Tyrode (Millipore) to remove zona pellucida and then washed three times in 0.2% BSA/PBS prior for library construction.
RNA-seq libraries were prepared as previously described 25. Briefly, SMARTer Ultra Low Input RNA cDNA preparation kit (Clontech, 643890) was used for reverse transcription and cDNA amplification (11 cycles). cDNA were then fragmented, adaptor-ligated and amplified using Nextera XT DNA Library Preparation Kit (Illumina) according to the manufacturer’s instructions. Single-end 100-bp sequencing was performed on a HiSeq 2500 sequencer (Illumina). The summary of the generated datasets can be found in Supplementary Table 5.
RNA-seq analyses
RNA-seq reads were first trimmed to remove adaptor sequences and low-quality bases using Trimgalore (version 0.4.5). Reads (>35bp) were aligned to mm9 reference genome using HISAT2 (version 2.1.0) 26 with default parameters and RPKM values for each gene were computed using Cufflinks (version 2.2.1) 27. For differential gene/repeats expression analyses, TEtranscripts (version 1.5.1) 28 was used to generate read counts for genes (uniquely aligned reads only) and repeats (including both unique- and multi-aligned reads) and DESeq package 29 was used to compute false discovery rate using the ‘nbinomTest’ function.
For the comparative analyses between WT and MZ-KO late 1-cell and 2-cell embryos, only genes with both FC > 2 and FDR < 0.05 were considered as differentially expressed (Supplementary Tables 1 and 2). For the determination of whether known DUX targets or major ZGA genes were affected in KO embryos, a more relaxed criterion that only considers fold change (FC > 2 and FDR < 1) was used (Supplementary Tables 3 and 4).
Detection of RNA synthesis by EU incorporation
Early (~21 hpi) and late (~29 hpi) 2-cell embryos were incubated in KSOM supplemented with 500 μM EU (Invitrogen) for 1 h prior to fixation in 3.7% paraformaldehyde (Sigma). Following permeabilization in PBS containing 0.5% Triton X-100 (Sigma), embryos were stained using Click-iT RNA Alexa Fluor 488 Imaging Kit (Invitrogen). Fluorescence was detected using a laser scanning confocal microscope (Zeiss LSM800) and the images were acquired using Axiovision software (Carl Zeiss). Signal intensity of nuclei and cytoplasm of two blastomeres were acquired and the cytoplasmic signal was subtracted from the nuclei signal as background. The averaged signal intensity of the WT late 2-cell (~30 hpi) was set as 1.0.
RNA isolation, reverse transcription, and quantitative PCR
Total RNA was isolated from adult testis (9–12 weeks) using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. Following RQ1 DNase (Promega) treatment, RNA was used as template to synthesize cDNA with the use of SuperScriptIII First-Strand Synthesis System (Invitrogen). To ensure no genomic DNA contamination, a minus reverse-transcriptase control were also included. SYBR green gene expression assay (Invitrogen) were used to determine Dux transcript abundance in a ViiA 7 Real-Time PCR System (ThermoFisher Scientific). The threshold cycles were normalized to the housekeeping gene Gapdh and the relative abundance in each sample was calculated using the comparative CT method. The primers used are included in Supplementary Table 6.
Statistical analyses and data visualization
All statistical analyses were performed with R (http://www.r-project.org/). Pearson’s r co-efficient was computed using ‘cor’ function. Figure 2C was generated using the R function ‘heatmap.2’. Smoothed scatter plots (Supplementary Fig. 4) were generated with the R function ‘smoothScatter’ and all other plots were generated using ggplot2 package. The RNA-seq and ChIP-seq bigwig tracks were generated with uniquely aligned reads using deeptools (version 3.0.2) 30 with the following parameters “--skipNonCoveredRegions --binSize 10 –scaleFactor 1/DESeq’s sizeFactor”. The bigwig tracks were visualized in the Integrative Genomic Viewer genome browser 31.
Data availability
All RNA-seq data sets generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE121746. Oocyte and 1-cell RNA-seq data were obtained from a previous publication 23. HA-DUX ChIP-seq data and Dux overexpression RNA-seq data in mESCs were downloaded from a previous report 1.
Life Sciences Reporting Summary
Further detailed summary of the methods and reagents can be found in the Reporting Summary document.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Azusa Inoue for training Z.C. to manipulate mouse embryos and his advice on generating the Dux KO mouse lines. We acknowledge Drs. Xudong Fu, Chunxia Zhang, Xiaoji Wu, and Wenhao Zhang for helpful discussion. We thank Dr. Nadhir Djekidel for his advice on bioinformatic analyses. This project was supported by NIH (R01HD092465) and HHMI. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR INFORMATION
The authors declare no competing financial interests.
References:
- 1.Hendrickson PG et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49, 925–934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Iaco A et al. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49, 941–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamatani T, Carter MG, Sharov AA & Ko MS Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6, 117–31 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Wang QT et al. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 6, 133–44 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Zeng F, Baldwin DA & Schultz RM Transcript profiling during preimplantation mouse development. Dev Biol 272, 483–96 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Svoboda P et al. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269, 276–85 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Macfarlan TS et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zalzman M et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu F & Zhang Y Cell totipotency: molecular features, induction, and maintenance. Natl Sci Rev 2, 217–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Percharde M et al. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 174, 391–405 e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiuchi T et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22, 662–71 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Terrones D et al. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat Genet 50, 106–119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckersley-Maslin MA et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep 17, 179–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snider L et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 6, e1001181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng LN et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22, 38–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiddon JL, Langford AT, Wong CJ, Zhong JW & Tapscott SJ Conservation and innovation in the DUX4-family gene network. Nat Genet 49, 935–940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clapp J et al. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet 81, 264–79 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii W, Kawasaki K, Sugiura K & Naito K Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res 41, e187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J et al. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol 11, 829–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue K et al. The Rodent-Specific MicroRNA Cluster within the Sfmbt2 Gene Is Imprinted and Essential for Placental Development. Cell Rep 19, 949–956 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Wang L et al. Large genomic fragment deletion and functional gene cassette knock-in via Cas9 protein mediated genome editing in one-cell rodent embryos. Sci Rep 5, 17517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L et al. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One 10, e0120396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–7 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Wang H et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue A, Chen Z, Yin Q & Zhang Y Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Y, Tam OH, Paniagua E & Hammell M TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S & Huber W Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data sets generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE121746. Oocyte and 1-cell RNA-seq data were obtained from a previous publication 23. HA-DUX ChIP-seq data and Dux overexpression RNA-seq data in mESCs were downloaded from a previous report 1.