Abstract
Mediator is an evolutionarily conserved multi-subunit complex, bridging transcriptional activators and repressors to the general RNA polymerase II (Pol II) initiation machinery. Though the Mediator complex is crucial for transcription of almost all Pol II promoters in eukaryote organisms, the phenotypes of individual Mediator subunit mutants are each distinct. Here we report for the first time, the essential role of subunit Med20 in early mammalian embryo development. Although Med20 mutant mouse embryos exhibit normal morphology at E3.5 blastocyst stage, they cannot be recovered at early post-gastrulation stages. Outgrowth assays show that mutant blastocysts cannot hatch from the zona pellucida, indicating impaired blastocyst function. Assessments of cell death and cell lineage specification reveal that apoptosis, inner cell mass, trophectoderm, and primitive endoderm markers are normal in mutant blastocysts. However, the epiblast marker Nanog is ectopically expressed in the trophectoderm of Med20 mutants, indicative of defects in trophoblast specification. These results suggest that Med20 specifically, and the Mediator complex in general, are essential for the earliest steps of mammalian development and cell lineage specification.
Keywords: Mediator Med20, Mouse embryo, Nanog, Trophectoderm, Cell lineage specification
Introduction
Mammalian preimplantation development refers to the period from fertilization to implantation, during which the fertilized oocyte progresses through a number of cleavage divisions and three major transcriptional and morphogenetic events that lead to a blastocyst capable of implantation (Cockburn and Rossant 2010). The first well-defined event is the Maternal-to-Zygotic Transition (MZT), which includes degradation of maternal mRNAs and replacement with zygotic transcripts. This dramatic reprogramming of gene expression is indispensable for establishment of totipotency and embryo development (Latham et al. 1991). The second major event is embryo compaction and polarization, and inheritance of cell polarity in daughter cells during subsequent divisions has been demonstrated as critical for solidification of cell-fate acquisition (Leung et al. 2016). The third critical event is blastomere outer/inner configuration and the cell fate allocation when the outer polar cells differentiate exclusively into the trophectoderm (TE), whereas the apolar cells located inside of the morula give rise to the inner cell mass (ICM) (Arnold and Robertson 2009). Well-characterized gene expression patterns occur within these two distinct lineages. For example, the transcription factor (TF) Oct4 (also known as Pou5f1) is enriched in ICM, while the TF Cdx2 becomes highly expressed in TE (Nichols et al. 1998, Niwa et al. 2005). After the first cell fate determination, when blastocysts reach more than 32 cells, the second cell fate determination occurs, to segregate the ICM into epiblast (EPI) and primitive endoderm (PE). Well-defined profiles of gene expression demarcate these two populations. For example, Nanog only localizes to EPI cells, while TF Sox17 is expressed exclusively in PE lineage (Frum and Ralston 2015, Molotkov and Soriano 2018, Morgani et al. 2018). Finally, these three lineages EPI, PE, and TE will contribute to the embryo, parietal yolk sac, and placenta, respectively.
Although distinct localizations of transcription factors within ICM/TE and EPI/PE/TE lineages have been well illustrated, their upstream regulatory networks are not fully delineated (Cui and Mager 2018, Lokken and Ralston 2016). Among multiple signaling pathways involved in early cell fate decisions, Hippo signaling was demonstrated to play a critical role through the analysis of mutant mouse embryos lacking transcription factor TEA domain family member 4 (Tead4) (Yagi et al. 2007). Other experiments also showed the indispensable role of Hippo signaling in regulation of TE-specific transcription factor Cdx2 (Strumpf et al. 2005) and Gata3 (Ralston et al. 2010). Interestingly, recent studies indicate that Hippo signaling promotes ICM fate acquisition as well, though the regulatory mechanisms are still unknown (Wicklow et al. 2014). In addition to the Hippo pathway, the function of Notch signaling in TE lineage specification was also recently uncovered (Rayon et al. 2014). By using double knockouts for Tead4 and the Notch effector Rbpj, Rayon et al. demonstrated Hippo and Notch signals converge on Cdx2 to cooperatively promote TE lineage specification. Additional transcription factors continue to emerge as crucial regulators of early cell fate decisions. For example, Tfap2c can directly regulate Cdx2 expression through an enhancer in intron 1 during early cleavage stages to promote TE lineage specification (Cao et al. 2015). Although many other mechanisms, such as epigenetic regulation (Marcho et al. 2015, Paul and Knott 2014) and newly discovered genes (Cui et al. 2016a), contribute to and dictate these unique cellular identities, the full cadre of cellular mechanisms that controls these events remains unresolved.
Lineage specification and cell differentiation are complicated and highly regulated processes relying on the differential expression of various genes within distinct cell populations. For all eukaryote organisms and nearly all RNA polymerase II (Pol II) promoters, a crucial pathway to finely tune these regulatory signals and appropriate transcriptome activation is through the following: enhancer - activator - Mediator - Pol II - promoter (Kornberg 2005). As a core molecular signaling mechanism, the Mediator complex was originally identified in budding yeast (Kelleher et al. 1990), with subsequent identification of many protein subunits (25 in yeast and 30 in human) (Tsai et al. 2014). Functioning as the bridge, Mediator can convey regulatory signaling information to the basal RNA Pol II transcription machinery, eliciting both positive and negative regulation of gene transcription (Beyer et al. 2007). While the core Mediator complex seems to be universally required at all genes, subunit phenotypes can be distinct from one another, suggesting functional redundancy and specificity (Risley et al. 2010). Furthermore, although Mediator is evolutionarily conserved at the protein level, mutants in the same Mediator subunit can display dissimilar phenotypes in different organisms (Hentges 2011). Additionally, studies have confirmed that Mediator can interact with diverse transcription factors and co-factors to ensure specific genes are expressed with appropriate temporal and cell type specificity (Yin and Wang 2014).
In the present study, we explored the role of Med20, one of the most conserved proteins in the Mediator complex, during mouse embryo development, using both knockout (KO) and knockdown (KD) strategies. Our data show that Med20 is essential for hatching of the blastocyst from the zona pellucida. Moreover, outgrowth, apoptosis and lineage specification assays revealed that mutant blastocysts exhibit severe ectopic expression of Nanog, an epiblast marker, in the outer putative trophectoderm cells, demonstrating a failure to appropriately implant and establish the trophectoderm lineage.
2. Material and methods
2.1. Generation of Med20 mutants
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts, Amherst (2015–0011, 2018–0003). Med20 KO allele (C57BL/6NJ-Med20<em1J>/J, Stock #: 027272) was generated on C57BL/6NJ background in The Jackson Laboratory (JAX) using CRISPR-Cas9 system, with two guide RNAs targeting AGGAACTCTTGGGGACTGAT and GCTTAGAGTATTTACGTTAA. The founder with 348bp deletion beginning in intron 1 at GGGGACTGATGGGTGGGGAT at Chromosome 17 positive strand position 47,612,827 bp (GRCm38) and ending after GCTTAGAGTATTTACGTTAAT at position 47,613,174 bp in intron 2 (Fig. 1A), which causes a short truncated protein with only 11 amino acids, was selected to establish the colony. To expand the colony, heterozygous (Het) mice from JAX were backcrossed again with C57BL/6NJ wildtype (WT) for the following heterozygous intercrosses to generate Med20 mutants (Mut). Genotyping primers are used as follows (Fig. 1A): common Forward primer for both WT allele and Mut allele: TGATGCCTTTGATTCCAACA; WT Reverse: CACTCAATTCCCCAACAGGT; Mut Reverse: CCCTTGACAGAAAAGCAAGC.
Fig. 1.
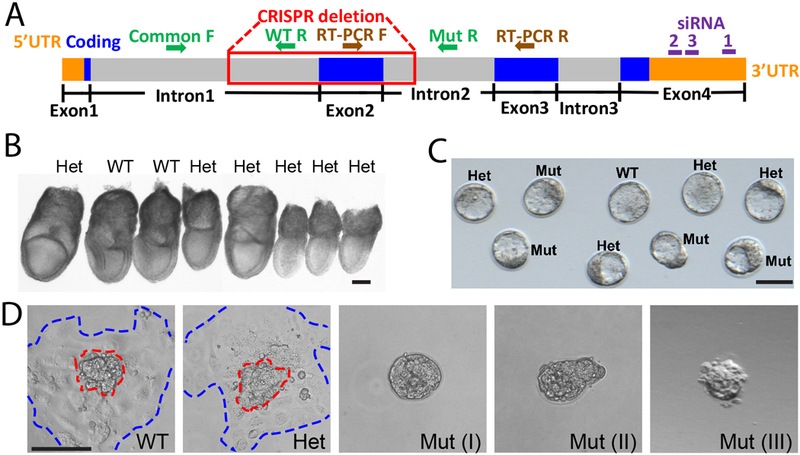
(A) Schematic of Med20 gene, CRISPR-Cas9 mediated deletion, genotyping primers for WT allele and Mut allele, RT-PCR primers (flanking intron2, which is 5,691 bp), and 3 different siRNAs. F, forward; R, reverse. (B) Representative genotyped embryos at E7.5. (C) Representative genotyped embryos at E3.5. (D) The outgrowths produced by different genotypes. Outgrowths from WT and Het displayed a distinct ICM colony (red dashed line) surrounded by robustly proliferating trophoblast cells (blue dashed line). Scale bars, 100μm.
2.2. Embryo recovery, culture and genotyping
Med20 heterozygous females 8- to 14-weeks old were caged with Med20 heterozygous males for natural matings and the presence of a vaginal plug was defined as embryonic day 0.5 (E0.5). Embryos were then collected from uteri of heterozygous females by dissection or flushing to collect E7.5 or E3.5 embryos, respectively. Embryos were imaged as a group and carefully collected into individual tubes in the order presented, then lysed for PCR genotyping using the primers mentioned above.
To prepare zygotes for siRNA microinjection or in vitro culture, B6D2F1 female mice 8- to 10-weeks old were induced to superovulate with 7.5 IU pregnant mare serum gonadotropin (PMSG, Sigma-Aldrich), followed 48 hr later by 7.5 IU human chorionic gonadotropin (hCG, Sigma-Aldrich). Females were mated with B6D2F1 males and euthanized at 20 hr post-hCG injection. Oviductal ampullae were dissected to release zygotes, and cumulus cells were removed by pipetting in M2 medium containing hyaluronidase (EMD Millipore). Zygotes were then washed in M2 medium (EMD Millipore) and cultured in KSOM medium (EMD Millipore) at 37 °C in a humidified atmosphere of 5% CO2/5% O2 balanced in N2.
2.3. Outgrowth assay
Blastocysts were collected and transferred gently into culture plate (Nunclon Delta, Thermo Fisher) and cultured in DMEM (Lonza, Allendale, NJ) containing 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) and 1X GlutaMAX (Thermo Fisher). Outgrowth assay was conducted at 37 °C in a humidified atmosphere of 5% CO2 for 3 days. Outgrowths were then imaged and genotyped.
2.4. Immunofluorescence and confocal microscopy
Immunofluorescence was performed as previously described (Cui et al. 2016a, Cui et al. 2016b). In vivo derived blastocysts were flushed at E3.5, and then cultured overnight before fixation and immunofluorescence (to ensure embryos had undergone EPI/PE/TE specification). In vitro blastocysts were harvested at 4 days post microinjection. Primary antibodies used in this study include: mouse anti-Cdx2 (BioGenex, MU392A-UC); rabbit anti-Nanog (abcam, ab80892); rabbit anti-Trp53 (Cell Signaling Technology, #9284); goat anti-Sox17 (R&D Systems, AF1924); goat anti-Oct4 (abcam, ab27985). After secondary antibodies (Alexa Fluor, Life Technologies) and DAPI (Sigma) staining, embryos were transferred to chambered slides (BD Falcon) with 1 embryo per well for imaging. Embryos were imaged using Nikon A1 Spectral Detector Confocal with FLIM Module. Z-stacks (20X objective, 8 μm sections) were collected and maximum projection was applied. Blastocysts collected from heterozygous intercrosses were imaged prior to knowledge of their genotypes. After imaging, embryos were individually recovered and lysed for genotyping.
2.5. Microinjection
Microinjection was performed as previously described (Cui et al. 2016a, Cui et al. 2016b). A volume of 5–10 pl of 50 μM Scrambled Control (5′‐CAGGGTATCGACGATTACAAA, Qiagen) or Med20 siRNA (siRNA1 target: 5′‐CGCAGACGTTAATTTAATTAA, siRNA2 target: 5′‐TACAGAGACATTTAACACAAA, siRNA3 target: 5′‐CTCGGGAAAGCTGTTAATCTA, Qiagen, Fig. 1A) was microinjected into the cytoplasm of zygotes.
2.6. RNA extraction and Reverse Transcription PCR (RT-PCR)
Total RNA extraction was performed with a Roche High Pure RNA Isolation Kit (#11828665001). cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad Laboratories, 170–8891). Specific intron-spanning primers were used for RT-PCR (Actb: 5′-GGCCCAGAGCAAGAGAGGTATCC and 5′-ACGCACGATTTCCCTCTCAGC; Med20: 5′-AGTGGAGCTCCTCACCAAGA and 5′-CCTTGGCACTCTGGAAGAAG, Fig. 1A).
2.7. Simultaneous extraction of RNA and DNA from single blastocyst
Blastocysts collected from heterozygous intercrosses were lysed individually (10 μl lysis buffer per embryo) following the manual of Roche Kit (#11828665001), with DNase treatment step skipped. A volume of 13 μl Elution Buffer was applied and the eluted mixture of RNA and DNA was used as follows: 6 μl mixture for genotyping PCR with Platinum SuperFi Green PCR Master Mix (Thermo Fisher) and genotyping primers as listed above; the other 6 μl mixture for cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad, 170–8891). Regarding the resultant 8 μl cDNA, 2 μl was used for Actb RT-PCR and 6 μl was used for Med20 RT-PCR, with Platinum SuperFi PCR Mix (Thermo Fisher) and RT-PCR primers listed above.
2.8. Statistical Analysis
All experiments were repeated at least three times. Percentage data were analyzed by ANOVA, and a value of P < 0.05 was considered statistically significant. Data are expressed as mean ± standard error of the mean.
3. Results
3.1. Med20 mutants cannot be recovered in vivo after E3.5
A CRISPR-mediated Med20 knock-out allele was generated for the Knockout Mouse Phenotyping Program (KOMP2) at The Jackson Laboratory. During the initial phenotyping pipeline of The International Mouse Phenotyping Consortium (IMPC, http://www.mousephenotype.org), no homozygous Med20 mutants were born, nor found at E15.5 or E9.5. Considering this, we first dissected embryos at E7.5 early post-gastrulation stage. Twenty-nine embryos were recovered from 5 heterozygous intercrosses and genotyped. Genotyping results showed 20 Het and 9 WT embryos all with normal gastrulation morphology (Fig. 1B). No Med20 mutant embryos were found at E7.5, nor were excessive empty decidua (n=6), suggesting Med20 mutants failed to implant. Therefore, we switched to collection and genotyping at the E3.5 blastocyst stage, where we recovered mutant Med20 embryos at the expected Mendelian ratios. A total of 24 WT, 69 Het, and 18 Mut embryos were found in 15 litters. Compared with WT and Het littermates, Mut blastocysts could not be identified by morphology alone (Fig. 1C). Combined, the normal blastocyst morphology plus the complete absence of mutants during gastrulation suggested implantation failure.
We next performed in vitro outgrowth assays, a technique used as a model for implantation (Armant 2005, Cui et al. 2016a, Qin et al. 2005). Another 42 blastocysts collected from 6 females were subjected to a 3-day outgrowth assay where each outgrowth was individually cultured, imaged and subsequently genotyped. As expected, successful hatching and outgrowth rates were high in both WT blastocysts (8/10, 80%) and Het blastocysts (21/23, 91.3%), displaying a distinct ICM colony surrounded by robustly proliferating trophoblast cells after 72 hours in culture (Fig. 1D). However, all of the mutant blastocysts failed to hatch or grow normally (0/9). Three of 9 mutant blastocysts failed to hatch out of the zona pellucida (Fig 1D, type I), 2 arrested during the process of hatching (type II), and 4 hatched free of the zona pellucida but did not form a monolayer of attached trophoblast cells (type III). Each of these subtle phenotypes suggested impaired function of TE cells in mutant blastocysts, that lead to failures of hatching or implantation. These results are also consistent with a complete absence of mutant embryos at E7.5.
3.2. Med20 mutants have normal expression of Oct4 and Cdx2
To explore the cause of the outgrowth and implantation failure, we first examined markers for apoptosis (Trp53) and first cell lineage choices including Oct4 for ICM and Cdx2 for TE using immunofluorescence (IF). Blastocysts were collected, imaged, and then genotyped. Of 25 blastocysts examined (5 WT, 15 Het, 5 Mut), all genotypes showed no apoptosis and normal robust expression of Oct4 in ICM and mutually exclusive expression of Cdx2 in TE (Fig. 2). These results show that Med20 null blastocysts are not dying via apoptosis, and their ICM and TE has been appropriately specified.
Fig. 2.
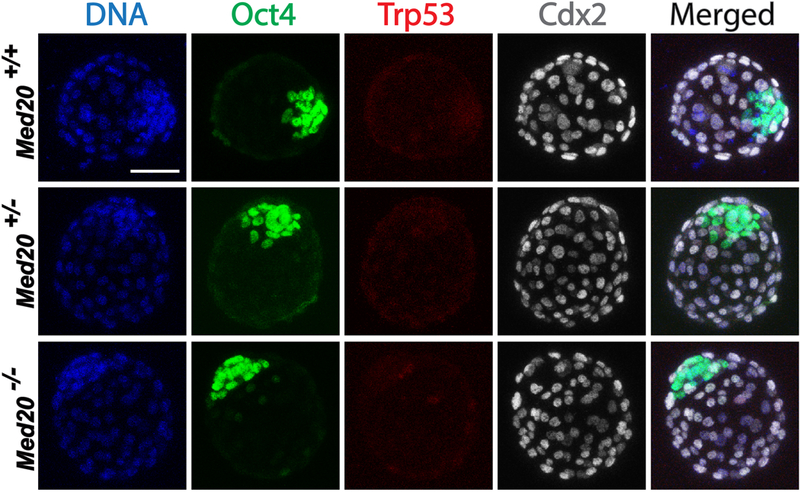
Knock-out of Med20 did not affect apoptosis index or ICM/TE lineage specification. Blastocysts in this experiment were flushed at E3.5, and then cultured overnight before fixation and immunofluorescence. Blastocysts of all genotypes showed low apoptosis index (Trp53 as the marker) and that ICM cells were tightly arranged with robust expression of Oct4, while TE cells were uniformly arranged with specific expression of Cdx2. Scale bar, 50μm.
3.3. Ectopic Nanog in Med20 mutant TE
We next investigated the second cell fate specification, the segregation of ICM into epiblast (EPI) and primitive endoderm (PE). Blastocysts were collected, assessed for Nanog and Sox17 localization (EPI and PE markers, respectively) via IF, and then genotyped. From 31 blastocysts (8 WT, 16 Het, 7 Mut), all genotypes showed regular expression and localization of both Sox17 and Cdx2. However, the majority of Med20 mutant blastocysts (5/7, 71.4%) exhibited widespread ectopic expression of Nanog in TE cells (Fig. 3) compared with WT (1 from 8, 12.5%) and Het (2 from 16, 12.5%), suggesting that Med20 regulates early embryo development in part through repression of Nanog in Cdx2-positive TE cells. Combined with the outgrowth failure of mutant embryos, we can conclude that deletion of Med20 results in defective function of TE cells.
Fig. 3.
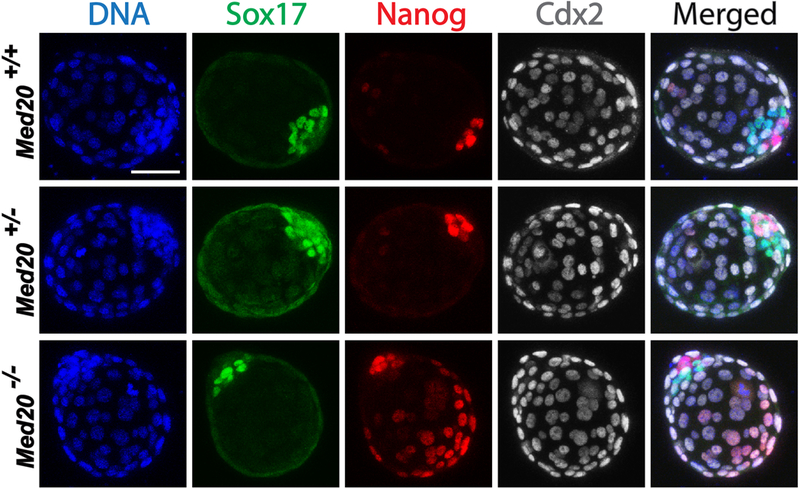
Knock-out of Med20 did not affect expression or localization of neither Sox17 (marker of primitive endoderm) nor Cdx2 (marker of trophectoderm); however, KO of Med20 led to severe ectopic expression of Nanog (marker of epiblast) in outside Cdx2-positive TE cells. Blastocysts in this experiment were flushed at E3.5, and then cultured overnight before fixation and immunofluorescence. Scale bar, 50μm.
3.4. Med20 knockdown embryos phenocopy genetic knockout mutants
Due to the lack of available antibodies suitable for immunofluorescence, we examined the expression of Med20 by RT-PCR to verify the success of both knock-out and knock-down approaches (location of primers is shown in Fig. 1A). RT-PCR using cDNA from different stages of wildtype embryos shows that Med20 is expressed at all preimplantation stages, from oocyte to blastocyst (Fig. 4A). We then extracted both RNA and DNA from single blastocysts to both genotype embryos and assess Med20 expression (details in methods section). As expected, genotyped mutant blastocysts contain no Med20 mRNA (Fig. 4B), confirming functional knockout of the deletion allele. To further verify the deletion phenotype was due to lack of Med20 and to establish a more efficient system to study Med20 function in embryos, 3 distinct commercial Med20 siRNAs (Fig. 1A) were individually microinjected into zygotes. Satisfactory KD efficiency was confirmed at early morula stage (2 days after microinjection) with each siRNA (Fig. 4C). Similar to KO phenotype, KD of Med20 did not affect blastocyst formation or morphology, but resulted in overall outgrowth failure (Fig. 4D).
Fig. 4.
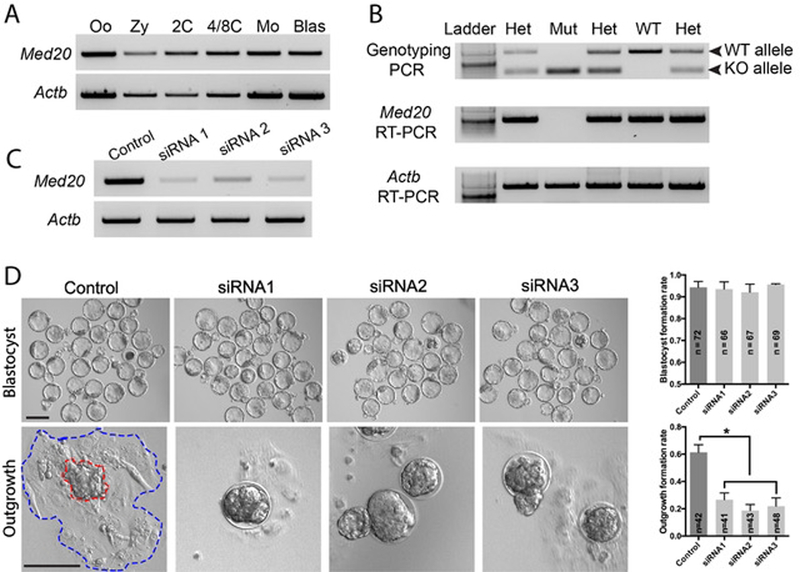
(A) Expression pattern of Med20 in wild-type preimplantation embryos. Actb was used as loading control. Oo, metaphase II oocyte; Zy, zygote; 2C, 2-cell embryo; 4/8C, mix of 4- and 8-cell stage embryos; Mo, morula; Blas, Blastocyst. (B) Simultaneous extraction of both RNA and DNA from single blastocyst to perform both genotyping PCR and Med20 RT-PCR, confirming KO was successful. Actb was used as loading control. (C) Endogenous Med20 mRNA was significantly depleted by 3 distinct siRNAs after microinjection. (D) KD of Med20 using distinct siRNAs did not affect blastocyst formation or morphology, but significantly altered embryo outgrowth potential. Red and blue dashed lines indicate ICM colony and trophoblast cells, respectively. Control: scrambled siRNA. n, number of embryos; *, P < 0.05. Scale bars, 100μm.
3.5. KD of Med20 also induces severe ectopic Nanog expression in TE cells
Considering the overlapping expression of Nanog + Cdx2 observed in Med20 KO blastocysts, we also performed lineage specification assessment in KD blastocysts. Compared with control embryos (Fig. 5), KD of Med20 does not alter Sox17 (PE) or Cdx2 (TE) localization. However, as with KO embryos, ectopic Nanog in outer TE cells was detected in all KD groups (ectopic Nanog rate: Control, 2 from 19, 10.5%; siRNA1, 16 from 20, 80%; siRNA2, 16 from 18, 88.9%; siRNA3, 15 from 16, 93.8%), indicating that phenotypes of CRISPR-Cas9 mediated Med20 KO can be fully recapitulated by microinjection of single Med20 siRNAs. Importantly, these results confirm that loss of Med20 results in TE defects and outgrowth failure in vivo and in vitro.
Fig. 5.
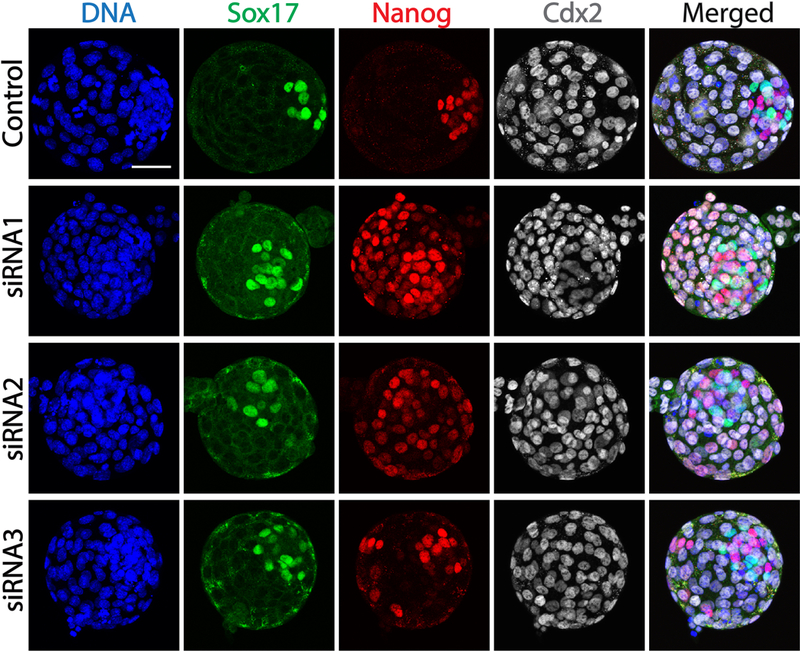
Knock-down of Med20 by 3 distinct siRNAs did not affect the expression or localization of either Sox17 (primitive endoderm marker) or Cdx2 (trophectoderm marker), but resulted in ectopic expression of Nanog in Cdx2-positive TE cells. Blastocysts in this experiment were harvested at 4 days post microinjection, and then fixed for immunofluorescence. Scale bar, 50μm.
4. Discussion
Cell differentiation and lineage specification is a complex and highly regulated process during development of all multicellular eukaryotic organisms. To obtain unique profiles of gene expression and distinct cellular identity, complexity in transcriptional control between regulatory elements and RNA polymerase must exist. Among these complicated networks, the Mediator complex, is a key component of the RNA polymerase II (Pol II) transcriptional machinery. It can convey distal regulatory information to basal Pol II transcription machinery, playing a crucial role in not only activation, but also repression of eukaryotic mRNA synthesis (Beyer et al. 2007). The Mediator complex was originally detected in yeast, then it was identified in mammalian species comprising up to 30 subunits (Tsai et al. 2014). Though the whole Mediator complex is required for all tissues and cell lineages, different Mediator subunits have distinct target genes and phenotypes of individual subunit mutants can be distinct from each other (Westerling et al. 2007), indicating a multifaceted role of Mediator complex during organismal development.
Though it is generally believed that the head module of Mediator is involved in interactions with the core Pol II machinery, studies have demonstrated that some regulators can directly target subunits in the head module, for example, Med17 (Park et al. 2001). Med20, another core component in head module, has not been well studied yet. The function of Med20 has been restricted to the regulation of tRNA and some non-coding RNAs transcription in fission yeast (Carlsten et al. 2016). In plants, Med20 controls the balance of salicylic acid and jasmonate associated defense pathways (Fallath et al. 2017). The role of Med20 in mammalian development has not been studied. In this study, we took advantage of both KO and KD strategies, to demonstrate that Med20 is essential for early mouse development. Consistent with our findings, other Mediator subunits also exhibit specific embryonic lethal phenotypes; however, only CDK8 and Med21 have similar early lethality with all other documented mutants showing post gastrulation phenotypes (reviewed in (Yin and Wang 2014)).
Our studies revealed ectopic Nanog (epiblast marker) expression in Cdx2-positive TE cells of Med20 KO and KD blastocysts. It is well established that Cdx2 is essential for segregation of ICM and TE lineages at the blastocyst stage by repressing Oct4 and Nanog in the TE, although the detailed mechanism underlying this repression is still largely unknown (Bassalert et al. 2018, Carey et al. 2015, Niwa et al. 2005, Piliszek et al. 2017, Strumpf et al. 2005, Wang et al. 2010). Interestingly, in our study, both KO and KD of Med20 only causes ectopic Nanog in the outer TE cells, without effects on Oct4 expression or localization, suggesting that Cdx2 represses Oct4, at least partially, through different pathways. Indeed, previous studies have illustrated that Oct4 can bind to the Nanog promoter and this cis-regulatory machinery is essential for Nanog pluripotent transcription, suggesting Oct4 is at the top of this regulatory hierarchy (Kuroda et al. 2005, Rodda et al. 2005).
Embryonic stem (ES) cells have been used extensively to explore the functions of Mediator complex in cell lineage commitment. For example, a functional role of Med12 in regulation of Nanog expression and maintenance of ES cell pluripotency has been debated (Rocha et al. 2010, Tutter et al. 2009), and mounting evidence indicates that Med12 and Med1, together with cohesin complex and loading factor, can contribute to ES cell state through DNA loops that directly link enhancers and promoters (Apostolou et al. 2013, Kagey et al. 2010, Phillips-Cremins et al. 2013). Additionally, many Mediator subunits have been identified as regulators of ES cell maintenance: Med6, Med7, Med10, Med12, Med14, Med15, Med17, Med21, Med24, Med27, Med28 and Med30 (Kagey et al. 2010).
Our results suggest that unlike these other subunits, Med20 is essential for repression of Nanog in TE cells, to maintain TE identity and function. However, the exact mechanism by which Med20 and/or Mediator coordinates multiple transcription factors and co-factors to regulate Nanog expression is unknown. Complicating any mechanistic conclusions are studies that have shown certain Mediator subunits have selective affinity among different activators and transcription factors, such that the absence of Med20 may allow for Mediator interaction with activators of Nanog and other loci (Miao et al. 2018, Niwa 2014, Yin and Wang 2014). Other novel functions of Mediator have recently been documented. For example, it can interact directly with non-coding RNAs to influence transcription (Carlsten et al. 2013), regulate alternative mRNA processing (Huang et al. 2012) and alter epigenetic silencing of selected genes (Ding et al. 2008). In order to fully understand the mechanism and specificity of Med20 function, we will likely need to assess transcriptome wide effects specifically in TE cells of KO or KD embryos. Alternatively, trophoblast stem cells may serve as a good model to explore Med20 function, however, in vivo results may differ from in vitro cell line studies.
In summary, by using both KO and KD strategies, our study suggests that Med20 plays a significant role in proper trophectoderm development that is essential for hatching and implantation. In addition, Med20 is indispensable for repression of Nanog in TE cells during early murine development to maintain TE identity and function.
Acknowledgment
We thank the Knockout Mouse Phenotyping Program (KOMP2) and The Jackson Laboratory for providing Med20 knock-out allele. We thank Yuran Tsuchida for assistance in genotyping. The confocal microscopy data was gathered in the Light Microscopy Facility and Nikon Center of Excellence at the Institute for Applied Life Sciences, UMass Amherst with support from the Massachusetts Life Sciences Center.
Funding
This work was supported by R01HD083311 to JM and JAR-P and faculty start-up fund to WC.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML et al. 2013. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 12 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armant DR 2005. Blastocysts don’t go it alone. Extrinsic signals fine-tune the intrinsic developmental program of trophoblast cells. Dev Biol 280 260–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ & Robertson EJ 2009. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10 91–103. [DOI] [PubMed] [Google Scholar]
- Bassalert C, Valverde-Estrella L & Chazaud C 2018. Primitive Endoderm Differentiation: From Specification to Epithelialization. Curr Top Dev Biol 128 81–104. [DOI] [PubMed] [Google Scholar]
- Beyer KS, Beauchamp RL, Lee MF, Gusella JF, Naar AM & Ramesh V 2007. Mediator subunit MED28 (Magicin) is a repressor of smooth muscle cell differentiation. J Biol Chem 282 32152–32157. [DOI] [PubMed] [Google Scholar]
- Cao Z, Carey TS, Ganguly A, Wilson CA, Paul S & Knott JG 2015. Transcription factor AP-2gamma induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 142 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey TS, Cao Z, Choi I, Ganguly A, Wilson CA, Paul S & Knott JG 2015. BRG1 Governs Nanog Transcription in Early Mouse Embryos and Embryonic Stem Cells via Antagonism of Histone H3 Lysine 9/14 Acetylation. Mol Cell Biol 35 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsten JO, Zhu X & Gustafsson CM 2013. The multitalented Mediator complex. Trends Biochem Sci 38 531–537. [DOI] [PubMed] [Google Scholar]
- Carlsten JO, Zhu X, Lopez MD, Samuelsson T & Gustafsson CM 2016. Loss of the Mediator subunit Med20 affects transcription of tRNA and other non-coding RNA genes in fission yeast. Biochim Biophys Acta 1859 339–347. [DOI] [PubMed] [Google Scholar]
- Cockburn K & Rossant J 2010. Making the blastocyst: lessons from the mouse. J Clin Invest 120 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Dai X, Marcho C, Han Z, Zhang K, Tremblay KD & Mager J 2016a. Towards Functional Annotation of the Preimplantation Transcriptome: An RNAi Screen in Mammalian Embryos. Sci Rep 6 37396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W & Mager J 2018. Transcriptional Regulation and Genes Involved in First Lineage Specification During Preimplantation Development. Adv Anat Embryol Cell Biol 229 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Pizzollo J, Han Z, Marcho C, Zhang K & Mager J 2016b. Nop2 is required for mammalian preimplantation development. Mol Reprod Dev 83 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S et al. 2008. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell 31 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallath T, Kidd BN, Stiller J, Davoine C, Bjorklund S, Manners JM, Kazan K & Schenk PM 2017. MEDIATOR18 and MEDIATOR20 confer susceptibility to Fusarium oxysporum in Arabidopsis thaliana. PLoS One 12 e0176022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T & Ralston A 2015. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet 31 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges KE 2011. Mediator complex proteins are required for diverse developmental processes. Semin Cell Dev Biol 22 769–775. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J & Wang G 2012. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell 45 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ 3rd, Flanagan PM& Kornberg RD 1990. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61 1209–1215. [DOI] [PubMed] [Google Scholar]
- Kornberg RD 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30 235–239. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N & Tada T 2005. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol 25 2475–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Solter D & Schultz RM 1991. Activation of a two-cell stage-specific gene following transfer of heterologous nuclei into enucleated mouse embryos. Mol Reprod Dev 30 182–186. [DOI] [PubMed] [Google Scholar]
- Leung CY, Zhu M & Zernicka-Goetz M 2016. Polarity in Cell-Fate Acquisition in the Early Mouse Embryo. Curr Top Dev Biol 120 203–234. [DOI] [PubMed] [Google Scholar]
- Lokken AA & Ralston A 2016. The Genetic Regulation of Cell Fate During Preimplantation Mouse Development. Curr Top Dev Biol 120 173–202. [DOI] [PubMed] [Google Scholar]
- Marcho C, Cui W & Mager J 2015. Epigenetic dynamics during preimplantation development. Reproduction 150 R109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YL, Gambini A, Zhang Y, Padilla-Banks E, Jefferson WN, Bernhardt ML, Huang W, Li L & Williams CJ 2018. Mediator complex component MED13 regulates zygotic genome activation and is required for postimplantation development in the mouse. Biol Reprod 98 449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A & Soriano P 2018. Distinct mechanisms for PDGF and FGF signaling in primitive endoderm development. Dev Biol 442 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani SM, Metzger JJ, Nichols J, Siggia ED & Hadjantonakis AK 2018. Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H & Smith A 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95 379–391. [DOI] [PubMed] [Google Scholar]
- Niwa H 2014. The pluripotency transcription factor network at work in reprogramming. Curr Opin Genet Dev 28 25–31. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R & Rossant J 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123 917–929. [DOI] [PubMed] [Google Scholar]
- Park JM, Werner J, Kim JM, Lis JT & Kim YJ 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell 8 9–19. [DOI] [PubMed] [Google Scholar]
- Paul S & Knott JG 2014. Epigenetic control of cell fate in mouse blastocysts: the role of covalent histone modifications and chromatin remodeling. Mol Reprod Dev 81 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piliszek A, Madeja ZE & Plusa B 2017. Suppression of ERK signalling abolishes primitive endoderm formation but does not promote pluripotency in rabbit embryo. Development 144 3719–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Takahashi Y, Isuzugawa K, Imai M, Yamamoto S, Hirai Y & Imakawa K 2005. Regulation of embryo outgrowth by a morphogenic factor, epimorphin, in the mouse. Mol Reprod Dev 70 455–463. [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS & Rossant J 2010. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137 395–403. [DOI] [PubMed] [Google Scholar]
- Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Canon S, Sasaki H, Hadjantonakis AK, de la Pompa JL et al. 2014. Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 30 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risley MD, Clowes C, Yu M, Mitchell K & Hentges KE 2010. The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Dev Biol 342 146–156. [DOI] [PubMed] [Google Scholar]
- Rocha PP, Scholze M, Bleiss W & Schrewe H 2010. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development 137 2723–2731. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH & Robson P 2005. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem 280 24731–24737. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F & Rossant J 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132 2093–2102. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW & Asturias FJ 2014. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutter AV, Kowalski MP, Baltus GA, Iourgenko V, Labow M, Li E & Kadam S 2009. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem 284 3709–3718. [DOI] [PubMed] [Google Scholar]
- Wang K, Sengupta S, Magnani L, Wilson CA, Henry RW & Knott JG 2010. Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One 5 e10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerling T, Kuuluvainen E & Makela TP 2007. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 27 6177–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H & Ralston A 2014. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 10 e1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML & Buonanno A 2007. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134 3827–3836. [DOI] [PubMed] [Google Scholar]
- Yin JW & Wang G 2014. The Mediator complex: a master coordinator of transcription and cell lineage development. Development 141 977–987. [DOI] [PubMed] [Google Scholar]


