Abstract
Circular RNAs (circRNAs) are a new class of RNA involved in multiple human malignancies. However, limited information exists regarding the involvement of circRNAs in gastric carcinoma (GC). Therefore, we sought to identify novel circRNAs, their functions and mechanisms in gastric carcinogenesis. We analyzed next-generation RNA sequencing data from GC tissues and cell lines, identifying 75,201 candidate circRNAs. Among these, we focused on one novel circRNA, circNF1, that was upregulated in GC tissues and cell lines. Loss- and gain-of-function studies demonstrated that circNF1 significantly promotes cell proliferation. Furthermore, luciferase reporter assays showed that circNF1 binds to miR-16, thereby derepressing its downstream target mRNAs, MAP7 and AKT3. Targeted silencing or overexpression of circNF1 had no effect on levels of its linear RNA counterpart, NF1. Taken together, these results suggest that circNF1 acts as a novel oncogenic circRNA in GC by functioning as a miR-16 sponge.
Keywords: gastric carcinoma, circNF1, proliferation, miR-16
Introduction
Gastric carcinoma (GC) is the fourth most common cancer and third leading cause of cancer death worldwide (Fitzmaurice et al. 2015). Although research on GC has progressed substantially, its 5-year overall survival (OS) rate remains below 30% (Allemani et al. 2015); surgery remains the only potentially curative treatment (Van Cutsem et al. 2011). Thus, improvements in understanding are needed, including biomarkers for early diagnosis.
Recently, circRNAs have been identified as a new type of non-coding RNA. These transcripts comprise covalently closed circular RNA structures without 5’-3’ polarity or a polyadenylated tail, features distinguishing them from linear RNAs. The first description of circRNAs in the cytoplasm of eukaryotic cells occurred in 1979 (Hsu and Coca-Prados 1979). However, due to limited detection technologies, circRNAs were considered errant byproducts of RNA splicing and received little attention. With the advent of high-throughput RNA sequencing and improved bioinformatics, numerous circRNAs have now been identified (Jeck et al. 2013; Rybak-Wolf et al. 2015). Researchers agree that circRNAs represent a novel type of endogenous non-coding RNA, rather than errors. CircRNAs are abundant in eukaryotic cells and exhibit evolutionary sequence conservation. Due to their resistance to exoribonuclease-mediated degradation, circRNAs are more stable than their linear counterparts (Jeck and Sharpless 2014). Based on the origin of their circular configurations, circRNAs can be classified as exonic, intronic, or exonic-intronic. Exonic circRNAs occur principally in the cytoplasm and are the most-studied type to date (Memczak et al. 2013).
Increasing evidence indicates that numerous circRNAs are dysregulated and act as either oncogenic or tumor-suppressive factors in cancer initiation and progression. For example, circCCDC66 expression is elevated in colon cancer and is associated with a poor prognosis (Hsiao et al. 2017). Similarly, overexpression of circ-SMARCA5 promotes cell cycle progression and inhibits apoptosis in prostate adenocarcinoma (Kong et al. 2017). Because they often contain conserved miRNA binding sites, many circRNAs function as endogenous miRNA sponges, reactivating miRNA downstream target genes by binding to and sequestering their corresponding miRNAs. One such cirRNA, CDR1as, contains 73 conserved miRNA-7 binding sites, blocking miR-7-induced tumor suppression by antagonizing miR-7-mediated EGFR/RAF1/MAPK pathway suppression (Weng et al. 2017). In GC, several circRNAs have been implicated in carcinogenesis and progression (Pan et al. 2018; Zhang et al. 2017). However, functions and mechanisms of circRNAs in GC are still poorly understood.
In the current study, we analyzed next-generation sequencing data from GC tissues and cell lines for circRNAs, using the bioinformatic algorithm, circRNA_finder (Westholm et al. 2014). After data analysis and verification, circNF1, derived from exons 2-8 of linear NF1, was chosen for subsequent experiments. NF1, a well-known tumor suppressor gene, is closely associated with neuroendocrine and endocrine tumors including neurofibromas (Gutmann et al. 2017), thyroid cancer (Uhlen et al. 2015) and carcinoid of the gastrointestinal tract (Swinburn et al. 1988). However, the existence of function of circNF1 have not been previously studied or reported. We found that circNF1 was upregulated in GC tissues and cells, and functional assays using interference and overexpression of circNF1 revealed that circNF1 promotes cell proliferation. Finally, circNF1 functioned as a miR-16 sponge, derepressing the miR-16 target genes MAP7 and AKT3. We conclude that circNF1 represents a novel potential biomarker and therapeutic target in GC patients.
Materials and Methods
Bioinformatic identification of circRNAs from next-generation RNA sequencing (RNAseq) data
To identify expression levels of circRNAs in GC, a RNAseq dataset that we had generated (Huang et al. 2016) was studied using the circRNA_finder algorithm (Westholm et al. 2014). CircRNA_finder aligns reads using Spliced Transcripts Alignment to a Reference (STAR) software (Dobin et al. 2013), extracting chimeric reads (reads that do not align continuously in a way that cannot be explained by regular splicing), finding chimeric reads consistent with backsplicing, and outputs these junctions with information regarding how many reads support each of them, as well as whether splice sites are canonical (Westholm et al. 2014). Canonical GT/AG splice sites are usually used as a major filter parameter in current circRNA identification strategies (Jeck and Sharpless 2014). CircRNA_finder does not output which RNAseq reads support each junction, but rather only the number of reads supporting each junction. Next, we compared our results to circBase (www.circbase.org/) (Glazar et al. 2014), finally applying the COSMIC cancer census gene list (https://cancer.sanger.ac.uk/census) to prioritize cancer-related circular RNAs (Forbes et al. 2015). All circRNA junctions were stored in BED files; therefore, we used Bedtools to compare and merge datasets (http://bedtools.readthedocs.io/en/latest/) (Quinlan and Hall 2010).
Cell culture
Human GC cell lines MKN28, NCI-N87, AGS, KATOIII, RF1, RF48 and the normal gastric epithelial cell line HFE145 were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco), incubated in a humidified 5% CO2 atmosphere at 37°C.
Reverse transcription-polymerase chain reaction (RT-PCR) and Sanger sequencing
Total RNA was extracted with TRIzol reagent (Life technologies, Carlsbad, CA) and subjected to reverse transcription with High Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific). Subsequently, divergent and convergent primers for each candidate circRNA were designed and used to amplify synthesized cDNA by TopTaq Master Mix kits (Qiagen). For linear RNA amplification, customary RT-PCR regular primers were applied. Finally, purified circRNA PCR products were subjected to Sanger sequencing to further confirm the correct circRNA backsplice junctions. All primers are contained in Supplemental Methods.
RNase R treatment
Total RNA (20 ug) from MKN28 or NCI-N87 was incubated at 37 °C for 30 minutes with or without 2 U/ug of RNase R (Epicentre Biotechnologies). After purification with QIAprep Spin Miniprep kits (Qiagen), RNA was subjected to RT-PCR.
Patients and tissue specimens
Paired tumor and normal tissues from 23 patients obtained during surgical resections performed for clinical indications were enrolled in this study. This research was approved by the Institutional Review Board at the Johns Hopkins University School of Medicine. An informed consent form was signed by each patient. All age, gender, tumor size, differentiation degree, and TNM stage data were exported from hospital records (see Supplementary Table S4). All tissue samples were pathologically confirmed as GC. Tissues were studied for differential expression of circNF1 and linear NF1 mRNA by qRT-PCR.
Quantitative real-time PCR
After reverse transcription, cDNA was generated and amplified using SYBR PrimeScript™ or iQ™ Supermix kit (Bio-Rad) on an ABI 7900HT Fast Real-Time PCR system. Primers are listed in Supplemental Methods. Messenger RNA expression levels were calculated using the 2−comparative Ct (2−ΔΔCt) method and normalized against the threshold cycle (Ct) of GAPDH or β-Actin. For detecting expression levels of miRNA, cDNA samples were amplified using TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA), and RNU6B was used as a reference gene.
Oligonucleotide transfection
2.0×105 cells/well of MKN28 and NCI-N87 GC cell lines were seeded in 6-well plates before transfection. SiRNA (si-circNF1), targeted to interfere with the backsplice junction of circNF1, was designed and synthesized by Dharmacon (Lafayette, CO). The sequence is in Figure 3A. A negative nonspecific control siRNA (si-NC) was purchased from Dharmacon (Cat # D-001210-05). Subsequently, cells were transfected with a final concentration of 25 nM of either si-circNF1 or si-NC using Lipofectamine RNAiMAX Reagent (Invitrogen) and incubated for 24 hours for functional experiments.
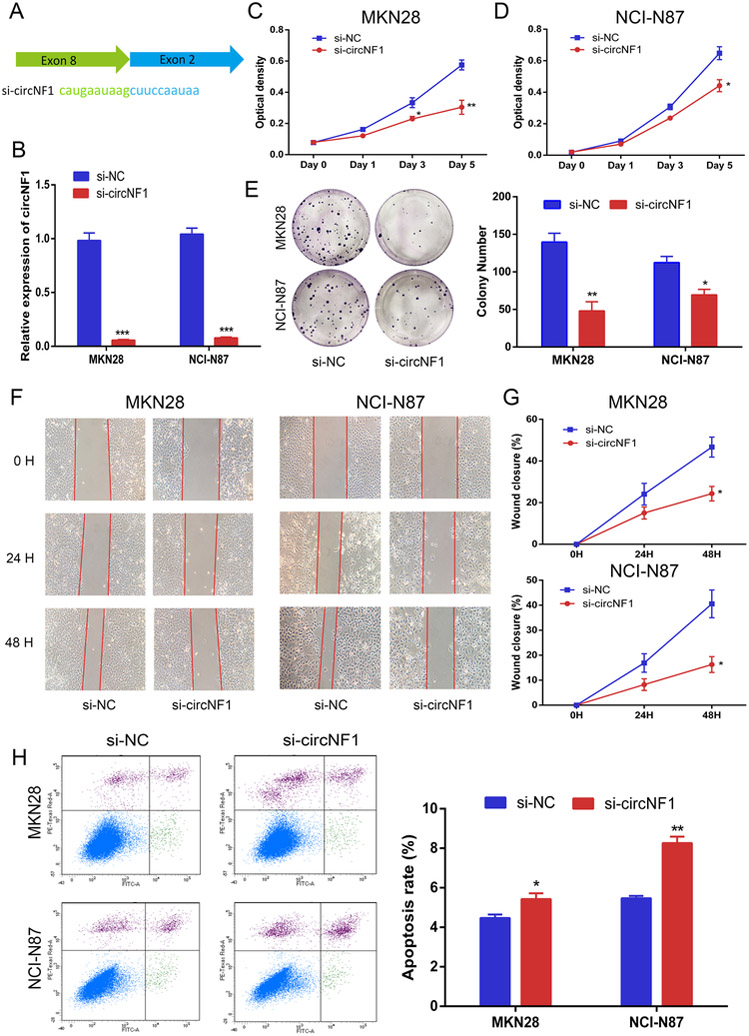
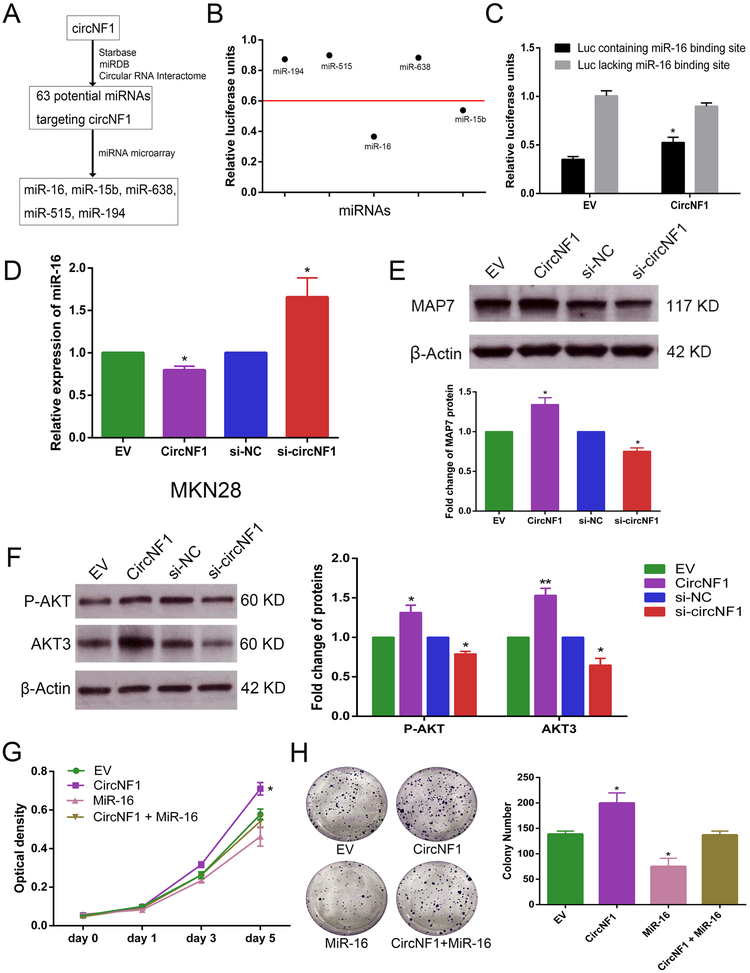
Figure 3. Silencing of circNF1 inhibits GC cell proliferation and migration but promotes apoptosis.
A, Schematic representation of backsplice junction and sequence of siRNA targeted to junction site of circNF1. B, Results of qRT-PCR for circNF1 in MKN28 and NCI-N87 cells treated with either si-NC or si-circNF1. C and D, Results of WST-1 assay for proliferation after transfection with si-NC or si-circNF1. Inhibition of proliferation by knockdown of circNF1 in MKN28 and NCI-N87 cells at day 5 (p=0.0079 and p=0.0212, respectively). E, Representative images and quantification results of colony formation of knock-down circNF1. F, Representative images of scratch assays in MKN28 and NCI-N87 cells transfected with control or circNF1 siRNAs. G, Line chart depicting scratch healing rates of MKN28 and NCI-N87 cells after silencing of circNF1. At hour 48t, wound closure in si-circNF1 group is significantly lower than in si-NC group in both MKN28 and NCI-N87 cells (P=0.0196 and 0.0188, respectively). H, Representative images and quantification results of flow cytometry to evaluate apoptosis induction by circNF1 in MKN28 and NCI-N87 cells transfected with si-NC or si-circNF1. *, p<0.05; **, p<0.01; ***, p<0.001.
For miRNA overexpression, miRNA mimics were synthesized by Dharmacon (Cat # C-300483-03-0002), and transfected into MKN28 cells suing Lipofectamine RNAiMAX Reagent (Invitrogen) at a concentration of 60 nM.
Construction and transfection of circNF1 overexpression vector
To generate a circNF1 overexpression vector, the sequence of circNF1 was first amplified using PCR. For subsequent cloning, a ClaI restriction enzyme site was added to the 5’ end of one primer, while a SacII site was appended to the 5’ end of the other primer. Primer sequences were: forward, 5’-AATCGATGCTTCCAATAAAAACAGGACAG-3’ and reverse, 5’-TCCGCGGCTTATTCATGTTGTTTTCATC-3’. Next, PCR products were cloned into pCR™4-TOPO TA vector (Invitrogen). After verification by Sanger sequencing, verified correct plasmid was digested with ClaI and SacII enzymes (New England Biolabs, Inc.) and electrophoresed on 1% gels. After electrophoresis, bands were excised from gels and purified with NucleoSpin Gel and PCR Clean-up kits (Clontech). Isolated fragments were then ligated with pcDNA3.1 (+) CircRNA Mini Vector, a generous gift of Jeremy Wilusz (Addgene plasmid # 60648) into the ClaI and SacII sites. PcDNA3.1 (+) CircRNA Mini Vector contains several Alu elements, which are considered an important co-factor in forming circRNAs (Liang and Wilusz 2014). Following reconstitution of the circular vector, full-length circNF1 was verified by Sanger sequencing.
After confirming the full-length sequence of circNF1, 2ug of over-expression circNF1 vector (CircNF1) or empty vector (EV) were transfected into 2.0×105 cells/well of MKN28 (gastric cancer) and HFE145 (normal gastric epithelial) cell lines, respectively, using Lipofectamine™ 2000 Reagent (Invitrogen) according the manufacturer’s instructions. Backsplice junctions and efficiency of transfection were confirmed by Sanger sequencing and qRT-PCR, respectively.
Cell proliferation assays
24 hours after transfection, cells were harvested and reseeded onto 96-well plates at a destiny of 1000 cells/well in 100 ul culture medium. 10 ul of WST-1 reagent (Roche, Mannheim, Germany) were added to each well at the indicated time (day 0, 1, 3 and 5), then incubated for 2 h at 37 °C in 5% CO2 before measurement of absorbance. Optical density (OD) values were measured at 660 nm (background) and 440 nm (signal) on a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Colony formation assays
For colony formation assays, 1000 cells/well were seeded onto 6-well plates and cultured for 12 days. Then, the plates were washed twice with PBS and stained with Diff-Quik Fixative, Diff-Quik Solution I and Diff-Quik Solution II (Dade Behring Inc, Newark, DE) for 10 minutes each, in sequence at room temperature. Subsequently, colony numbers were counted and analyzed.
Scratch assays
Cells were transfected on 6-well plates for 24 hours and grown into monolayers. 200 μL filter tips were used to create wound areas. At different timepoints (0, 24 h and 48 h), scratch widths were photographed, and scratch healing rate was analyzed by Image J software.
Cell apoptosis assays
FITC Annexin V/ Dead Cell Apoptosis Kits (Invitrogen) were used to measure apoptosis. Transfected cells were incubated for 24 hours before harvesting. Following washing X 2 with cold PBS, collected cells were resuspended in 1x annexin-binding buffer. Then, 5 uL of FITC annexin V and I ul of 100 ug/mL PI working solution were added, respectively, into each cell suspension. After incubation for 15 minutes, 400 uL of 1x annexin-binding buffer were added before analyzing the stained cells by flow cytometry.
Dual-Glo luciferase assays
First, oligonucleotide pairs containing miR-16, miR-15b, miR-638, miR-515 and miR-194 target regions were ordered and annealed, respectively. Then, five pmirGLO Dual-Luciferase miRNA target expression vectors were constructed by inserting the respective annealed oligonucleotides into the pmirGLO vector (Promega) using PmeI and XbaI restriction enzymes (New England Biolabs, Inc). When fusion rate reached 70%-90%, MKN28 cells were co-transfected with 80 ng of either circNF1 overexpression plasmid (circNF1) or empty vector (EV), along with 80 ng of either pmirGLO vector containing miRNA binding sites or empty pmirGLO vector, into 1.0×104 cells/well using Lipofectamine™ 2000 (Invitrogen). After 24 hours, cells were analyzed for firefly and Renilla luciferase activity using the Dual-Glo® Luciferase Assay Kit (Promega) and VICTOR2 fluorometry (Perkin Elmer, Waltham, MA).
Western blotting
The BCA protein assay procedure (Thermo Fisher Scientific) was conducted to determine protein concentration of the supernatant. Equal amounts of protein lysates were separated on 4% to 15% polyacrylamide gels (Bio-Rad) and transferred to 0.45 μm supported nitrocellulose transfer membranes (Bio-Rad). After blocking with 5% skim milk at room temperature for 1 hour, membranes were incubated with the following specific primary antibodies: AKT3 (1:2000, Cell Signaling, MA), P-AKT (1:2000, Cell Signaling, MA), MAP7 (1:500, Novus Biologicals, CO) and anti-β-actin-peroxidase (1:30,000, Sigma-Aldrich, Bedford, MA) at 4 °C overnight. Subsequently, the membranes were washed three times with TBS containing 0.1% Tween 20 buffer (Sigma-Aldrich) and incubated with sheep anti-rabbit IgG secondary antibodies (1:2000, Cell Signaling, MA) or horseradish peroxidase-labeled anti-mouse IgG (1:20,000, Invitrogen) for 1 hour at 37 °C. Following washing three more times with TBST buffer, protein bands were detected by ECL Western Blotting detection kits (GE Healthcare, Buckinghamshire, UK) according to the manufacturer’s instructions. β-Actin antibody served as a control in these experiments.
Statistical analyses
All experiments were performed at least in triplicate. Discrepancies among group variables were analyzed by Student’s paired or unpaired t-tests. Pearson correlation coefficient was used to compare correlations of circNF1 and linear NF1 with GC. Data were displayed as means ± SEM of three experiments. All statistical analyses were conducted using SPSS 20.0, and statistical significance was defined as p<0.05.
Results
Filtering of RNAseq data to select circRNAs
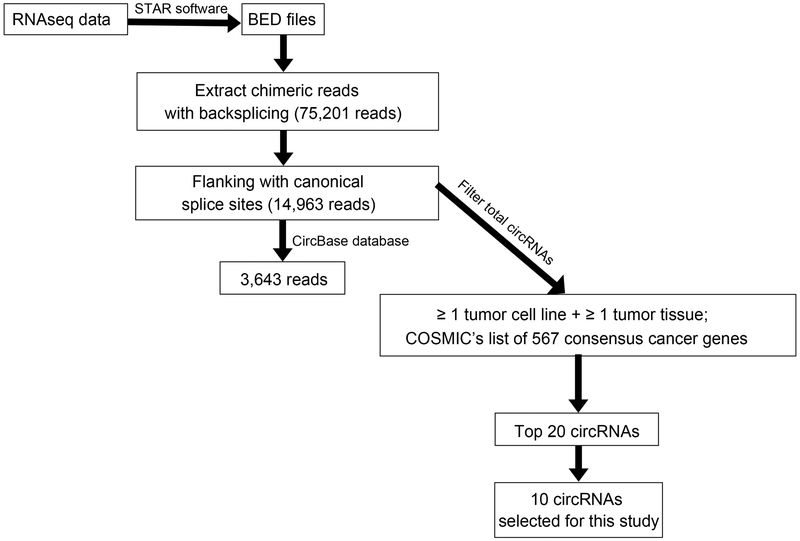
Figure 1 shows steps taken to extract and select circRNAs of interest from RNAseq data. First, we analyzed our RNAseq data (Huang et al. 2016) using the circRNA_finder algorithm (Westholm et al. 2014). These data contained 10 different samples: 2 matched normal (N)-tumor (T) tissues from 2 cancer patients; 1 normal tissue from a non-cancer patient; 4 unmatched GC tissues; 2 GC cell lines, NCI-N87 and MKN28; and 1 immortalized normal gastric epithelial cell line, HFE145 (see Supplementary Table S2). Analysis yielded 75,201 circRNAs (chimeric reads) found in all samples, of which 14,963 circRNAs flanked a canonical splice site, an accepted filter parameter in current circRNA identification (Jeck and Sharpless 2014). Next, we compared our results with online circRNAs found in circBase: this comparison revealed 3,643 circRNAs overlapping our circRNAs. Notably, unique chimeric reads occurred in each paired/unpaired tissue sample. In paired samples 2285 and 3245, overlapping circRNAs in cancer and adjacent tissues numbered 1,205 and 1,471, respectively (Table 1). Subsequently, to identify novel oncogenic circRNAs in GC, we filtered the entire list of circRNAs based on two prerequisites: circRNAs found in at least one tumor cell line and one tumor tissue sample; and circRNAs found in COSMIC’s list of 567 concensus cancer genes. Then, the generated circRNAs were ranked according to smallest BLAST E-value (i.e., the ORF most closely matching a database sequence). Finally, the highest 20 candidates were chosen for subsequent studies (see Supplementary Table S3). For the current study, we randomly selected 10 candidates for further investigation: SETD2, FOXO3, ELK4, AFF4, MYH9, MSH2, CIC, FBXW7, NF1 and ABL2. The remaining 10 circRNAs will be studied in future research.
Figure 1.
Flowchart depicts work steps used to identify circRNA candidates of interest from RNAseq data.
Table 1:
Numbers of circRNA candidates in different samples.
| Sample | Total read counts | Unique counts | With canonical splice sites |
In known DBs | % in DBs |
|---|---|---|---|---|---|
| 2285T | 91,655,404 | 10,436 | 2,942 | 381 | 12.95% |
| 2285N | 115,102,688 | 7,229 | 1,777 | 584 | 32.86% |
| 2285T - 2285N | 9,231 | 2,298 | 227 | 9.88% | |
| 2285N - 2285T | 6,103 | 1,208 | 413 | 34.19% | |
| 2285N ∩ 2285T | 1,205 | 644 | 154 | 23.91% | |
| 3245T | 199,449,428 | 9,036 | 2,065 | 508 | 24.60% |
| 3245N | 164,431,037 | 7,847 | 2,480 | 590 | 23.79% |
| 3245T - 3235N | 7,565 | 1,320 | 328 | 24.85% | |
| 3245N - 3235T | 6,349 | 1,703 | 402 | 23.61% | |
| 3245N ∩ 3245T | 1,471 | 745 | 180 | 24.16% | |
| 2284T | 120,500,864 | 8,503 | 2,082 | 907 | 43.56% |
| 2286T | 95,974,189 | 6,689 | 1,481 | 591 | 39.91% |
| 2289T | 128,941,281 | 9,150 | 2,157 | 886 | 41.08% |
| G008T | 132,821,977 | 7,529 | 1,411 | 561 | 39.76% |
| MKN28 | 103,919,937 | 7,408 | 2,196 | 908 | 41.35% |
| NCI-N87 | 152,408,322 | 7,764 | 2,043 | 831 | 40.68% |
| HFE145 | 117,020,409 | 6,279 | 2,088 | 883 | 42.29% |
| 2759NN | 170,065,701 | 7,884 | 2,335 | 375 | 16.06% |
| Merged | 1,592,291,237 | 75,201 | 14,963 | 3,643 | 24.35% |
10 different samples were used to analyze novel circRNAs. “Total read counts” contains overall chimeric reads (reads of all circRNA candidates ≥1); “Unique counts” represents total number of novel circRNA candidates; “DB” stands for circBase database; “% in DBs” equals the number of circRNAs in known DBs/circRNAs with canonical splice sites. In samples, T: Tumor; N: Normal; T – N: circRNAs present in tumor but not in normal tissues; N –T: circRNAs present in normal tissues but not in tumor tissues; N ∩ T: circRNAs present in both tumor and normal tissues.
Validation of selected circRNAs
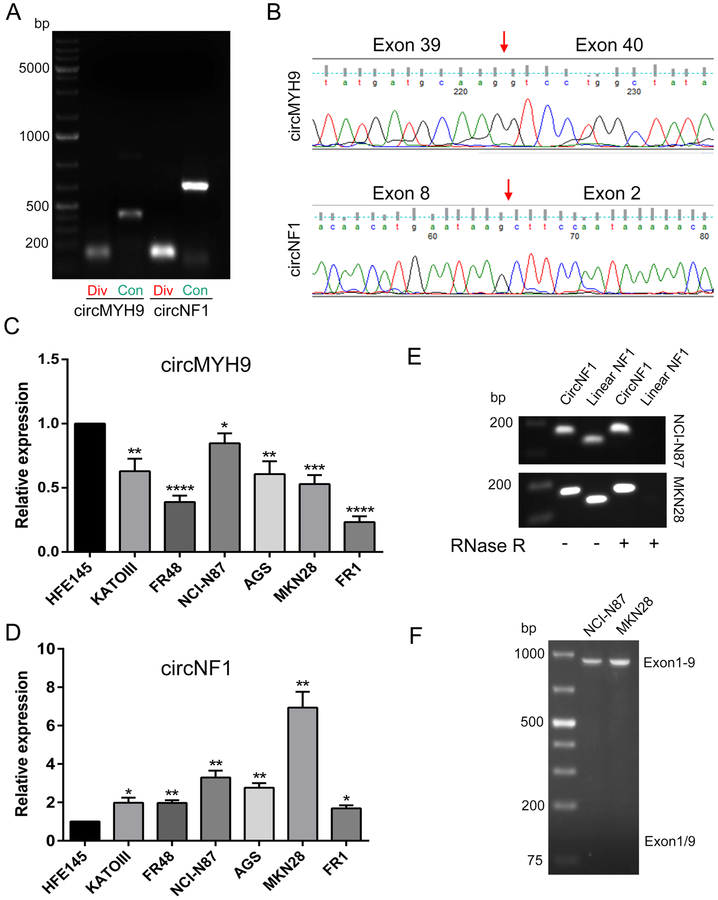
To confirm backsplice junctions and spliced sequence lengths of the 10 candidate circRNAs, both divergent and convergent primers were designed for RT-PCR. Results in NCI-N87 GC cells showed that with divergent primers, circRNAs derived from the genes MSH2 and ABL2 generated no PCR products of the expected size; circRNAs originating from the genes ELK4 and CIC generated multiple amplicons; and the remaining candidates yielded single amplification products (see Supplementary Figure 1A). Three of the 10 candidates (MSH2, MYH9, and NF1) generated clear PCR products with convergent primers (Supplementary Figure 1B). Thus, MYH9 and NF1 were the only two genes generating unique, correctly-sized amplicons from both divergent and convergent primers; these were named circMYH9 and circNF1 (Figure 2A). Next, Sanger sequencing of circMYH9 and circNF1 with divergent RT-PCR products was conducted to further validate and elucidate the backsplice junctions. After blasting sequence results in GenBank, we found that circMYH9 contained exons 39-40 of the antisense strand of MYH9, while circNF1 consisted of the sense strand of exons 2-8 of NF1 (Figure 2B). Finally, we compared expression levels of circMYH9 and circNF1 in 6 GC cell lines vs. an immortalized normal gastric epithelial cell line, HFE145. circMYH9 was downregulated in GC cells compared with HFE145 cells (Figure 2C), while circNF1 was upregulated in all GC cell lines (Figure 2D). Since NF1 is a well-known tumor suppressor gene (Xu et al. 1990), we hypothesized that circNF1 was likely to participate in gastric carcinogenesis. Therefore, circNF1 was chosen for further investigation.
Figure 2. Verification strategy of circRNA selection.
A, RT-PCR amplification products of circMYH9 and circNF1 using divergent (‘Div’) or convergent (‘Con’) primers. PCR products with divergent primers contain backsplice junction sites. Convergent primers’ amplicons reflecting full-length circMYH9 and circNF1 are approximately 500 and 800 bp, respectively. B, Head-to-tail splicing of circMYH9 and circNF1 was confirmed by Sanger sequencing of divergent PT-PCR products. The red arrows refer to backsplice junction sites. C, Expression levels of circMYH9 in gastric cancer cells vs. the immortalized normal gastric epithelial cell line, HFE145. D, Expression levels of circNF1 in gastric cancer cells vs. HFE145. E, RNAs extracted from NCI-N87 and MKN28 gastric cancer cell lines were treated without or with (− / +) RNase R prior to reverse transcription, then amplified with circNF1 or linear NF1 primers. Only circNF1 resists RNase R treatment. F, RT-PCR with forward primer on exon 1 and reverse primer on exon 9 (to amplify exons 1 to 9 of linear NF1) and exon-skipped transcripts (exons 1 / 9, not detected) in NCI-N87 and MKN28 cells. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.00001.
In the UCSC genome browser, circNF1 (chr17:31155982-31182665), derived from the NF1 locus on chromosome 17(q11.2), contains exons 2-8. After Sanger sequencing with convergent primers, we concluded that the full length of circNF1 was 828 bp, consistent with findings in the circBase database (hsa_circ_0042881). Because of its unique structure, circNF1 was treated with RNase R prior to reverse transcription to verify the lack of a 5’ cap and 3’ tail, and corresponding linear NF1 transcript was used to demonstrate the efficacy of RNase R digestion (Figure 2E). Then, we designed a traditional primer to amplify exons 1 to 9 of linear NF1 to detect whether the biological mechanism of circNF1 is exon-skipping; however, no exon-skipped product was found (Figure 2F), indicating that circNF1 was not a product of exon-skipping.
Silencing of circNF1 inhibits GC cell proliferation and migration but promotes apoptosis in vitro
To investigate the biological functions of circNF1, a specific siRNA targeting the backsplice junction of circNF1 was designed (Figure 3A) and transfected into MKN28 and NCI-N87 cells, the 2 GC lines that most highly expressed circNF1 vs. HFE145. Results of qRT-PCR showed that relative mRNA levels of circNF1 were successfully and specifically knocked down, from a baseline of 1.00 to 0.06 and 0.08 in MKN28 and NCI-N87 cells, respectively (p<0.001, Figure 3B). Subsequently, WST-1 assays showed that knockdown of circNF1 significantly decreased proliferation of MKN28 and NCI-N87 cells at day 5 (p=0.0079, p=0.0212, respectively; Figures 3C and D). Similarly, colony formation ability was inhibited by transfection with si-circNF1 vs. transfection with si-NC in MKN28 and NCI-N87 cells (p<0.01 and p<0.05, respectively; Figure 3E). Moreover, cell migration was also inhibited by transient transfection with specific anti-circNF1 siRNA. Specifically, scratch assays revealed that silencing of circNF1 caused a substantial reduction in cell migration at 48h in both cell lines (p=0.0196, p=0.0188; Figures 3F and G). Next, flow cytometry assays were conducted to examine whether knockdown of circNF1 induced programmed cell death or cell cycle arrest. Compared with si-NC, si-circNF1 increased apoptosis to varying degrees of both cell lines (p<0.05 and p<0.01, Figures 3H). However, no significant changes in cell cycle were observed (data not shown).
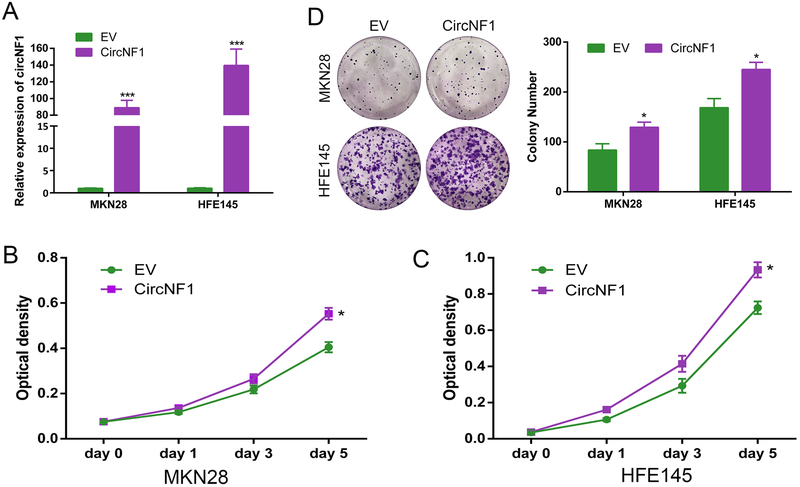
Overexpression of circNF1 promotes cell proliferation in vitro
To further characterize the participation of circNF1 in gastric carcinogenesis, we constructed a synthetic circRNA containing exons 2 to 8 of NF1 using pcDNA3.1 (+) CircRNA Mini Vector. HFE145, exhibiting the lowest native expression level of circNF1, along with MKN28, were chosen to transfect with either overexpression vector (CircNF1) or empty vector (EV), after which circNF1 expression levels were quantified by qRT-PCR. Results showed that this circNF1 vector had been successfully constructed; relative to EV control, circNF1 RNA levels were upregulated by 88.95- and 139.67-fold in MKN28 and HFE145 cells, respectively (p<0.001, Figure 4A). Next, we showed that overexpression of circNF1 increased proliferation of both MKN28 and HFE145 cells at day 5 (p=0.0128 and p=0.0181, Figure 4B and C). In addition, colony formation assays showed that overexpression of circNF1 caused a significant increase in colony number, suggesting that circNF1 promotes cell survival and proliferation (p<0.05, Figure 4D).
Figure 4. Over-expression of circNF1 promotes cell proliferation.
A, Results of qRT-PCR for circNF1 in MKN28 and HFE145 cells without or with exogenous overexpression of circNF1. B and C, WST-1 proliferation assay results in MKN28 and HFE145 cells transfected with either circNF1 (CircNF1) or empty vector (EV) at day 0, 1, 3 and 5. Proliferation increase at day 5 in cells treated by exogenous circNF1 overexpression. (p=0.0128 and p=0.0181, respectively). D, Representative images for colony formation after transfecting circNF1 or EV. *, p<0.05; ***, p<0.001.
CircNF1 acts as a miR-16 sponge
To explore molecular mechanisms exerted by circNF1, we performed online circRNA bioinformatics databases engines, including starBase v2.0 (http://starbase.sysu.edu.cn/starbase2/mirCircRNA.php), miRDB (http://www.mirdb.org/) and Circular RNA Interactome (https://circinteractome.nia.nih.gov/), which revealed that circNF1 was potentially targeted by 63 different miRNAs, of which miR-194 and miR-515 appeared simultaneously in two of these databases. Next, our own previous miRNA microarray results generated from gastric cell line RNAs (Jin et al. 2011) were used to help identify the most likely carcinogenic miRNAs. In these data, 22 miRNAs were not detected in GC cells and baseline expression levels of 32 miRNAs were too low (cutoff level less than 0.5) to exert effects on cells, even if they became bound to circNF1. After this screening process, 5 putative miRNAs (miR-16, miR-15b, miR-638, miR-515 and miR-194) were determined to be likely candidates for further investigation. A flowchart describing this screening process of miRNAs targeted to circNF1 is shown in Figure 5A. Next, five luciferase plasmids, each containing a respective miRNA binding site, were constructed and subjected to luciferase assays (firefly luciferase activity / Renilla luciferase activity) to evaluate luciferase activity of each miRNA. Results showed that relative to the control luciferase plasmid, luciferase plasmids containing either miR-16 or miR-15b binding sites significantly inhibited firefly luciferase activity (by at least 40%, Figure 5B), indicating that endogenous miR-16 and miR-15b can be efficiently conjugated to luciferase reporter plasmids containing either miR-16 or miR-15b binding sites. Subsequently, specific circNF1 overexpression vector or empty vector was co-transfected with luciferase reporters containing miR-16 or miR-15b binding sites into MKN28 gastric cancer cells. CircNF1 overexpression increased luciferase reporter activity of the luciferase plasmid containing the miR-16 binding site, suggesting that circNF1 competitively inhibited endogenous miR-16 binding to luciferase plasmid by “sponging” miR-16 (p<0.05, Figure 5C). However, co-transfection of circNF1 and the luciferase plasmid containing the miR-15b binding site had no significant effect on luciferase activity (data not shown). Next, we explored expression levels of miR-16 after either knocking down or over-expressing circNF1. QRT-PCR confirmed that circNF1 over-expression decreased miR-16 levels, while silencing of circNF1 had the opposite effect in MKN28 gastric cancer cells (P<0.05, Figure 5D). We next investigated whether circNF1 affected protein translation of downstream miR-16 target genes. In MKN28, over-expressed circNF1 increased MAP7 protein levels, whereas translation levels of MAP7 was decreased by silencing of circNF1 (Figure 5E). Similarly, we found expression levels of AKT3 protein were significantly increased by introduction of circNF1, but reduced after transfecting with si-circNF1. Moreover, AKT phosphorylation levels were also varied with either over-expressed or under-expressed circNF1 (Figure 5F). These data supported the hypothesis that circNF1 functions as a miR-16 sponge and protects translational modification of MAP7 and AKT3 from attack by miR-16 (Spinetti et al. 2013; Yan et al. 2013). In addition, WST-1 assays demonstrated that a miR-16 mimic reduced the pro-proliferative effect of circNF1 at day 5 (p=0.022, Figure 5G). Similar results occurred with colony formation assays (p<0.05, Figure 5H). Taken together, these findings indicated that circNF1 bound to miR-16 and inhibited its activity.
Figure 5. circNF1 functions as a sponge of miR-16 in GC cells.
A, Schematic flowchart shows pipelines for screening miRNAs binding to circNF1. B. Luciferase activity of luciferase vector containing each candidate miRNA binding site to detect endogenous miRNAs that were able to competitively bind to corresponding miRNA binding sequences in MKN28 cells. C, Activity of luciferase plasmid (“Luc”) containing or lacking a miR-16 binding site in MKN28 cells with vs. without circNF1 overexpression. D, Results of qRT-PCR for miR-16 in MKN28 gastric cancer cells with either exogenous overexpression of circNF1 or si-circNF1. E, Representative images and quantitative analysis from Western blot of MAP7 protein with either over-expressed or under-expressed circNF1 in MKN28 cells. F, Representative images and quantitative analysis from Western blot of P-AKT and AKT3 in MKN28 cells treated with overexpression or silencing of circNF1, respectively. β-Actin antibody served as a control in these experiments. G, WST-1 proliferation assay in cells transfected with circNF1 or miR-16 as indicated. Proliferation decrease at day 5 in cells treated by combination of circNF1 vector and miR-16 mimic relative to circNF1 overexpression (p=0.022). H, Representative images for colony formation after either transfecting circNF1 and miR-16 independently or co-transfecting circNF1 and miR-16. *, p<0.05, **, p<0.01.
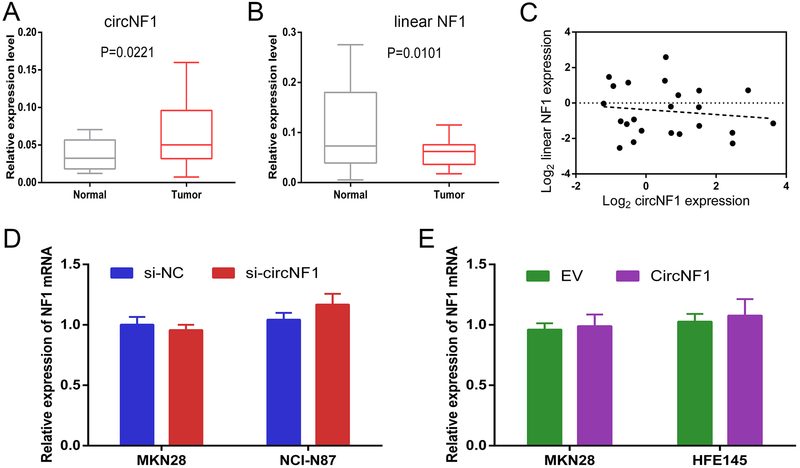
Expression of circNF1 vs. linear NF1 mRNA in GC
Finally, we studied circNF1 in 23 paired GC and normal tissues. Overall, expression circNF1 was significantly elevated in GC tissues compared with adjacent normal tissues (p=0.0221, Figure 6A). Next, to understand whether the high levels of circNF1 originated from its parental transcripts, we quantified levels of linear NF1 mRNA in GC tissues. Surprisingly, unlike circNF1, expression levels of linear NF1 were actually lower in GC tissues (p=0.0101, Figure 6B). We hypothesized that low expression of linear NF1 could have resulted from its depletion to generate circNF1. Therefore, expression correlation of linear vs. circular NF1 in GC patients was analyzed by Pearson correlation coefficient. However, there was no significant negative correlation between linear NF1 and circNF1 (r=−0.1348, p=0.5397, Figure 6C). Similarly, consistent with the results in GC tissues, there was no correlation between linear and circular NF1 transcript levels in GC cell lines (see Supplementary Figure 2). Moreover, neither targeted silencing nor overexpression of circNF1 had any effects on linear NF1 levels (Figures 6D and E). Taken together, these results suggested that linear NF1 and circNF1 act independently roles in the development of GC.
Figure 6. Expression of circNF1 and linear NF1 in GC tissues and cell lines.
A and B, Results of qRT-PCR of circNF1 and linear NF1 in 23 GC tissues. β-Actin served as an internal control. C, Pearson correlation coefficient analysis of correlation between expression levels of circNF1 (x) and linear NF1 (y) (r=−0.1348, p=0.5397). D, Results of qRT-PCR for linear NF1 transcript levels in MKN28 and NCI-N87 cells transfected with either si-NC or si-circNF1. E, Results of qRT-PCR for linear NF1 transcript levels in MKN28 and HFE145 cells treated with either empty vector (EV) or circular NF1 vector (CircNF1).
Discussion
Recently, the involvement of circRNAs in human cancer has received intense attention. In the current study, we detected circRNAs enriched in GC by analyzing RNASeq data, identifying circNF1 and circMYH9 as bona fide exonic circRNAs. Moreover, we found that expression levels of circNF1 were elevated in GC tissues as well as GC cells, and that circNF1 functioned oncogenically in cellular functions including proliferation, migration, and apoptosis. To our knowledge, this is the first report to explore the expression, function, or regulation of circNF1 in any human cancer.
Previous studies have demonstrated that circRNAs are abundant, diverse molecules expressed in a complex tissue- and cell-type-specific manner (Guo et al. 2014). To explore the expression and function of circRNAs in GC, we analyzed RNASeq data using bioinformatic algorithms, finding 75,201 distinct circRNA candidates. Notably, levels of circRNAs varied both inter- and intra-individually, suggesting that only a minority of circRNAs are shared between tumor samples in this particular tumor type, and that differentially expressed circRNAs may exert potential carcinogenic functions in GC. Lin et al. found almost no overlapping circRNA candidates in 16 liver cancer samples. Similarly, only 0.5% of brain circRNA candidates were shared among 30 brain samples (Lin et al. 2017). One possibility underlyting this apparent diversity may be that circRNA isoforms are commonly spliced into very specific pairs of exons (Salzman et al. 2013).
Both circRNAs and linear RNAs are generated from precursor mRNA. Unlike linear RNA, circRNAs are derived by canonical back-splicing. In addition, most reported circRNAs are generated exclusively via exonic circularization. Currently, proposed mechanisms of biogenesis of circRNAs include intron pairing, RNA binding protein-driven circularization, and exon-skipping (Ivanov et al. 2015; Kelly et al. 2015). In our study, we found that circNF1 was derived from exons 2-8 of the NF1 gene. Furthermore, with traditional primers for exons 1-9 of NF1, no exon-skipping product was found; thus, circNF1 was apparently not derived from exon-skipping. It is worth noting that by using the GenBank and UCSC genome browsers we found abundant Alu repeats in the flanking regions of exons 2 and 8, which have been viewed as a necessary co-factor for the process of circularization (Ashwal-Fluss et al. 2014). In the formation of circHIPK3, long flanking introns with highly complementary Alu elements are indispensable to support circularization (Zheng et al. 2016). Therefore, we hypothesized that the formation of circNF1 may be due in part to back-splicing with the help of intronic RNA pairing. However, the specific formation mechanism underlying circNF1 deserves further exploration.
Accumulating research has indicated that circRNAs play important roles in carcinogenesis and cancer progression. In several studies, circRNAs function as tumor suppressors. For example, in bladder carcinoma, circHIPK3 was downregulated and correlated negatively with cancer grade, invasion, and lymph node metastasis (Li et al. 2017). Similarly, circLARP4 inhibited biological behaviors, including proliferation and invasion of GC cells, by “sponging” miR-424, a known independent prognostic factor for tumor recurrence in GC (Zhang et al. 2017). In contrast, some studies showed that circRNAs may function as oncogenes. For example, circ-ABCB10 was significantly upregulated in breast cancer tissues, promoted proliferation and suppressed apoptosis. This same study also confirmed miR-1271 as a circABCB10-associated miRNA, which rescued the function of circABCB10 (Liang et al. 2017). Currently, the ability to function as a molecular sponge of native miRNA is the most well-documented function of circRNAs, protecting genes from attack by miRNAs. Our results demonstrated that circNF1 was highly expressed in GC tissues as well as cell lines. We also found that exogenous overexpression of circNF1 affected multiple tumor-associated cellular processes, including proliferation, migration and apoptosis, suggesting that circNF1 functions as a gastric oncogene. In addition, online circRNA bioinformatics databases predicted that circNF1 was potentially targeted by 63 different miRNAs, any of which could be involved in circNF1 mechanisms. However, we found that expression levels of most of these 63 were too low to exert substantial effects mediated by circNF1. Therefore, we narrowed our exploration to 5 highly expressed miRNAs, focusing first on miR-16. The remaining 58 miRNAs and their involvement in other types of tumors expressing circNF1 should prompt future studies to explore mechanisms of action of circNF1, for example by constructing luciferase vectors.
We found that circNF1 bound to miR-16 and affected translational levels of this miR’s downstream target genes, MAP7 and AKT3, by “sponging” miR-16. MiR-16 has been reported as a tumor suppressor in multiple tumor types; its downregulation may enhance tumor cell proliferation, apoptosis and cell cycle progression by inhibiting miR-16’s target activity on the 3’-UTR of oncogenes such as Bcl-2, Smad3, and Yap-1 (Cimmino et al. 2005; Kang et al. 2015; Zhang et al. 2018). Moreover, MAP7 (Yan et al. 2013) and AKT3 (Spinetti et al. 2013) has been previously validated as a direct target of miR-16, and decrease in the phosphorylated form of AKT pointed to dysregulation of the AKT-associated signaling pathway induced by miR-16 (Spinetti et al. 2013). Consistent with this previous observation, we observed increased translational levels of MAP7 and AKT3 as well as enhancive phospho-AKT after sponging of miR-16 by circNF1.
In previous studies, linear NF1, a classic tumor suppressor gene, has been extensively studied; its product, neurofibromin, is an important negative regulator of Ras signaling (Xu et al. 1990). However, unlike linear NF1, our results suggest that circNF1 functions as an oncogene. Moreover, there was no negative correlation between circNF1 and linear NF1 expression in GC tissues or cell lines, suggesting that regulation of NF1 transcription and processing into circular RNA act independently during cancer development. CircNF1 was apparently not derived from exon-skipping, which may explain our finding that abnormal expression of circNF1 did not affect linear NF1 levels. Consistent with this result, circCCDC66 and circPVT1 are dysregulated in colon and gastric cancer, respectively, but expression levels of circular and linear RNAs of both CCDC66 and PVT1 are poorly correlated (Chen et al. 2017; Hsiao et al. 2017). However, a positive correlation was observed between circular and linear transcripts of circITCH and circFOXO3 (Huang et al. 2015; Yang et al. 2016). In cell lines, the current study showed that expression of circNF1 has no effect on levels of linear NF1 in either knockdown or exogenous overexpression experiments. This finding differs from previous studies showing that knockdown of exon-intron circRNAs, circEIF3J and circPAIP2, promoted EIF3J and PAIP2 transcription levels, respectively (Li et al. 2015). Taken together, our results suggest that circNF1 and linear NF1 represent independent transcripts in GC, and that circNF1 is not generated from linear NF1. The relationship between circRNAs and their counterpart linear RNAs is worthy of further investigation. This relationship may provide additional insights into mechanisms of circRNA production.
In conclusion, based on RNASeq data bioinformatics analysis, we identified novel circRNAs expressed in GC. Among them, circNF1 was highly expressed in GC. The biological functions of circNF1 included stimulation of cell proliferation. Moreover, one molecular function of circNF1 was shown to be regulation of the expression of MAP7 and AKT3 by functioning as a miR-16 molecular sponge. In summary, these findings support the belief that circRNAs are an important class of tumor biomarkers and potential therapeutic targets. Thus, studying molecular mechanisms of circRNAs should be a high priority for future researchers.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH grants CA190040, CA211457, DK118250, and the Emerson Research Foundation.
Footnotes
Disclosures
The authors disclose no conflicts of interest.
References
- Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang X-S, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, et al. 2015. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2). The Lancet 385 977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N & Kadener S 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56 55–66. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. 2017. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 388 208–219. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 102 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, et al. 2015. The Global Burden of Cancer 2013. JAMA Oncol 1 505–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43 D805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P & Rajewsky N 2014. circBase: a database for circular RNAs. RNA 20 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H & Bartel DP 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biology 15 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann DH, Ferner RE, Listernick RH, Korf BR, Wolters PL & Johnson KJ 2017. Neurofibromatosis type 1. Nat Rev Dis Primers 3 17004. [DOI] [PubMed] [Google Scholar]
- Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS & Tsai SJ 2017. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res 77 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT & Coca-Prados M 1979. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280 339–340. [DOI] [PubMed] [Google Scholar]
- Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z, Ke X & Meltzer SJ 2016. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene 35 4927–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhu H, Shi Y, Wu W, Cai H & Chen X 2015. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One 10 e0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. 2015. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10 170–177. [DOI] [PubMed] [Google Scholar]
- Jeck WR & Sharpless NE 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF & Sharpless NE 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, Olaru AV, Yang J, David S, Hamilton JP, et al. 2011. MicroRNA-192 and −215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene 30 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al. 2015. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer 14 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Greenman C, Cook PR & Papantonis A 2015. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol 427 2414–2417. [DOI] [PubMed] [Google Scholar]
- Kong Z, Wan X, Zhang Y, Zhang P, Zhang Y, Zhang X, Qi X, Wu H, Huang J & Li Y 2017. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun 493 1217–1223. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, et al. 2017. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 18 1646–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22 256–264. [DOI] [PubMed] [Google Scholar]
- Liang D & Wilusz JE 2014. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HF, Zhang XZ, Liu BG, Jia GT & Li WL 2017. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 7 1566–1576. [PMC free article] [PubMed] [Google Scholar]
- Lin L, Zheng YC, Kayani MUR, Xu W, Wang GQ, Sun P, Ao N, Zhang LN, Gu ZQ, Wu LC, et al. 2017. Comprehensive analysis of circRNA expression profiles in humans by RAISE. Int J Oncol 51 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. [DOI] [PubMed] [Google Scholar]
- Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H & Kong D 2018. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem 119 440–446. [DOI] [PubMed] [Google Scholar]
- Quinlan AR & Hall IM 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. 2015. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 58 870–885. [DOI] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL & Brown PO 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9 e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, Descamps B, Floris I, Sangalli E, Vono R, et al. 2013. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res 112 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburn BA, Yeong ML, Lane MR, Nicholson GI & Holdaway IM 1988. Neurofibromatosis associated with somatostatinoma a report of two patients. Clin Endocrinol (Oxf). 28 353–359. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347 1260419. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Dicato M, Geva R, Arber N, Bang Y, Benson A, Cervantes A, Diaz-Rubio E, Ducreux M, Glynne-Jones R, et al. 2011. The diagnosis and management of gastric cancer: expert discussion and recommendations from the 12th ESMO/World Congress on Gastrointestinal Cancer, Barcelona, 2010. Ann Oncol 22 Suppl 5 v1–9. [DOI] [PubMed] [Google Scholar]
- Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y & Goel A 2017. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res 23 3918–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR & Lai EC 2014. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9 1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, O’Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. 1990. The Neurofibromatosis Type 1 Gene Encodes a Protein Related to GAP. Cell 62 599–608. [DOI] [PubMed] [Google Scholar]
- Yan X, Liang H, Deng T, Zhu K, Zhang S, Wang N, Jiang X, Wang X, Liu R, Zen K, et al. 2013. The identification of novel targets of miR-16 andcharacterization of their biological functions incancer cells. Mol Cancer 12 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Du WW, Li X, Yee AJ & Yang BB 2016. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 35 3919–3931. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang K, Ren T, Huang Y, Tang X & Guo W 2018. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis 9 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X & Zhu J 2017. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 16 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.