Abstract
Peanut is one of the most important oilseed crops grown worldwide. In this study, the mutant ahFAD2 alleles conferring high oleic (HO) content are introgressed into an elite Indian cultivar GPBD4 which is also resistant to the foliar fungal diseases like rust and late leaf spot (LLS). The allele-specific PCR (AS-PCR) and cleaved amplified polymorphic sequences (CAPS) assays were used for the marker-assisted backcross (MABC) approach and 64 HO introgression lines (ILs) were generated. These ILs were tested for the FA compositions under the glasshouse and field conditions. The oleic acid and linoleic acid contents in the ILs were recorded to be between 68.94–82.33% and 1.74–10.87%, respectively, under glasshouse and 67.04–81.71% and 2.00–15.66%, respectively, under field conditions. The increase in the oleic acid content of the ILs over its recurrent parent (RP) was recorded to the tune of 28.78–53.80% and 33.70–62.96% under glasshouse and field conditions, respectively, indicating the stable expression of ahFAD2B gene in two different environments. On the contrary, linoleic acid showed 56.47–93.03% and 40.02–92.34% reduction in the ILs over its RP under glasshouse and field conditions, respectively. These ILs with a healthy FA profile can meet not only the nutritional requirements of a health-conscious society but also the industrial demands for better shelf life of oil and its products.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1774-9) contains supplementary material, which is available to authorized users.
Keywords: Allele-specific PCR, Fatty acid desaturase, Groundnut, High oleate, Introgression lines, MABC
Introduction
Peanut (Arachis hypogaea L.) is one of the major annual legume and oilseed crops, which is grown in nearly 100 countries (Mishra et al. 2015). Worldwide, it grows on 27.66 million ha, yielding 43.98 million metric tons of production, with average productivity of 1.91 metric tons per ha (FAOSTAT 2016). Apart from oil, it is also widely used for the production of snack products, peanut butter, desserts, and soups (Patil et al. 2018). The nutritional quality, shelf life, and flavor of peanut oil and its products are reliant on the presence of different proportions of saturated fatty acids (SFAs), monounsaturated FA (MUFA), and poly UFA (PUFA) (Derbyshire 2014; Nawade et al. 2018). Peanut oil contains nearly 11 different FAs, of which oleic acid (C18:1, Δ9), a MUFA and linoleic acid (C18:2, Δ9, Δ12), a PUFA contribute approximately 80% of the total oil composition (Nawade et al. 2018).
A high proportion of linoleic acid results in off flavors, rancidity, and short shelf life of manufactured food products due to its low oxidative and frying stability (Mondal et al. 2011). On contrary, oleic acid has tenfold higher auto-oxidative stability than linoleic acid due to which high oleic (HO) oils are in great demand by various industries including food (fried products and bakery), cosmetic (emulsions, soaps, and detergents) and oleochemical industries (lubricants, paints, and adhesives) (Abiodun 2017). HO peanut encompasses a relatively longer shelf life along with neutral flavor and odor (O’Keefe et al. 1993), which also makes it an excellent solution for food industries, looking for healthy alternatives to saturated or hydrogenated oils (Cao et al. 2013). A diet with the HO acid can reduce the risk of heart diseases like the reduction of systolic blood pressure and slowing down of atherosclerosis (Vassiliou et al. 2009).
The first milestone in this aspect was achieved by Norden et al. (1987) who identified the first natural high oleate mutant lines, F435 with about 80% oleic acid and 2% linoleic acid, much higher compared to the traditional peanut genotypes having 36–70% oleic and 15–43% linoleic acid (Knauft et al. 1993). In peanut, two homeologous genes, ahFAD2A and ahFAD2B having 99% sequence similarity, are reported to regulate the desaturase activity (Jung et al. 2000b; Lopez et al. 2000). The coding sequences (CDS) of these genes consist of 1140 bp, encoding 379 amino acids with no introns in the coding region. A single base pair (bp) substitution (448G > A) mutation at 448 bp position in ahFAD2A gene, results in a missense amino acid from aspartic acid to asparagine (D150 N). While 1-bp insertion (441_442insA) mutation in ahFAD2B gene, at 442 bp position resulted in frame-shift mutation, which generates a premature stop codon (Jung et al. 2000b; Lopez et al. 2000). Recently, Wang et al. (2015b) identified two natural mutant lines (PI342664 and PI342666) with 80% oleic acid, having substitutions of G448A in FAD2A (same as previously identified) and C301G in FAD2B (new mutation) resulting in a missense amino acid substitution of D150 N, and H101D, respectively. The mutations in ahFAD2 gene were found to affect the histidine motifs of metal ion complex, which is required for oxygen reduction and resulting in reduced enzymatic activity and HO content in the mutant genotypes (Jung et al. 2000a; Yu et al. 2008; Chu et al. 2009).
To enhance the efficiency of HO peanut breeding program, different molecular assays including CAPS for ahFAD2A (Chu et al. 2007) and ahFAD2B alleles (Chu et al. 2009), real-time PCR (Barkley et al. 2010; 2011), AS-PCR (Chen et al. 2010; Yu et al. 2013) and kompetitive allele-specific PCR (KASP) (Zhao et al. 2017) have been developed and successfully utilized for both screening of peanut genotypes (Chu et al. 2007; Wang et al. 2011, 2013; Mukri et al. 2012; Nawade et al. 2016) as well as marker-assisted selection (MAS) studies (Chu et al. 2011; Janila et al. 2016; Bera et al. 2018).
The first HO cultivar, SunOleic95R was released in the USA using the conventional breeding method (Gorbet and Knauft 1997). Since then, over 90 HO peanut cultivars have been developed, of which the majority were bred through traditional methods and some using the chemically induced mutagenesis approach (Nawade et al. 2018). Further, only a few cultivars were developed through the MAS approach namely, Tifguard High O/L (Chu et al. 2011), SA Juweel and ARC Oleic2 (Mienie and Pretorius 2013).
Recently, Nawade et al. (2016) characterized a total of 174 Indian peanut genotypes for ahFAD2 allele polymorphism and its FA compositions, of which 80 were found to have the ahFAD2A mutant (448G > A) allele, while none recorded natural mutation in ahFAD2B (441_442insA) allele. The oleic acid content and O/L (oleic acid to linoleic acid) ratio of these genotypes ranged between 37.81–66.57% and 0.93–3.76%, respectively, which is much lower than the industrially acceptable O/L ratio of > 9 and/or 70% oleic acid content (Davis et al. 2013; Janila et al. 2016). Thus, looking at the increasingly global and domestic market demand for HO peanut, and unavailability of any such variety in India, the present investigation was undertaken to enhance the oleic acid content of an Indian peanut cultivar using the robust molecular breeding approach.
Materials and methods
Plant material
GPBD 4, a leading peanut variety of India with good agronomic features such as early to mid-maturity, and high yield (Sujay et al. 2012), but with a low O/L ratio was used as a recurrent parent (RP). In addition, it is highly resistant to various foliar fungal diseases and thus was used as a disease resistance check in the field trials of All India Coordinated Research Project on Groundnut (AICRP-G) in India (Gowda et al. 2002). Incidentally, in our previous work (Nawade et al. 2016), we found GPBD4 harboring a mutant ahFAD2A (448G > A) allele, while another ahFAD2B allele was normal, which results in the production of low oleic acid (55%) and more linoleic acid (26%) contents (Nawade et al. 2016). The first HO but poor yielder cultivar, SunOleic95R from the USA, having both ahFAD2A (448G > A) and ahFAD2B (441_442insA) mutant alleles in homozygous condition was used as a donor parent (Gorbet and Knauft 1997). The seeds of SunOleic95R were obtained from ICRISAT, Patancheru (India).
Hybridization and backcross breeding
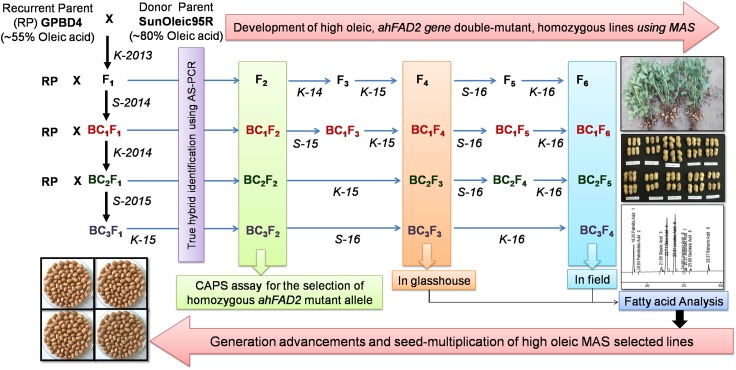
The hybridizations were conducted in a controlled glasshouse, having 60–65% humidity and 30 ± 2 °C temperature at ICAR-Directorate of Groundnut Research, Junagadh, India (21°48′34″N; 70°44′07″E; 67 m amsl). Three seeds were sown per pot (30 × 35 cm dimension), containing a mixture of dry soil, fine sand and well-decomposed farmyard manure (3:1:1 proportion) and only one healthy plant was retained at a later stage for crossing. A total of 15 plants were used as the female parent (GPBD4), while 10 plants were used as the male parent (SunOleic95R) and hybridization was performed in the Kharif season (June–October) in the year 2013. The emasculation was carried out between 16:30 h and 17:30 h, while pollination was conducted on the next day between 06:00 h and 08:00 h (Nigam et al. 1990). For backcrossing, the true hybrids of F1, BC1F1, and BC2F1 generations were used as pollen parents, while RP (GPBD4) was used as the female parent. The hybrids and segregating populations (F2, BC1F2, BC2F2, and BC3F2) generated after each crossing were grown for further molecular studies (Fig. 1).
Fig. 1.
Schematic presentation of the crossing and selection scheme used for the development of ahFAD2 gene double-mutant HO peanut lines in different generations
Mass multiplication of hybrids using the stem cutting technique
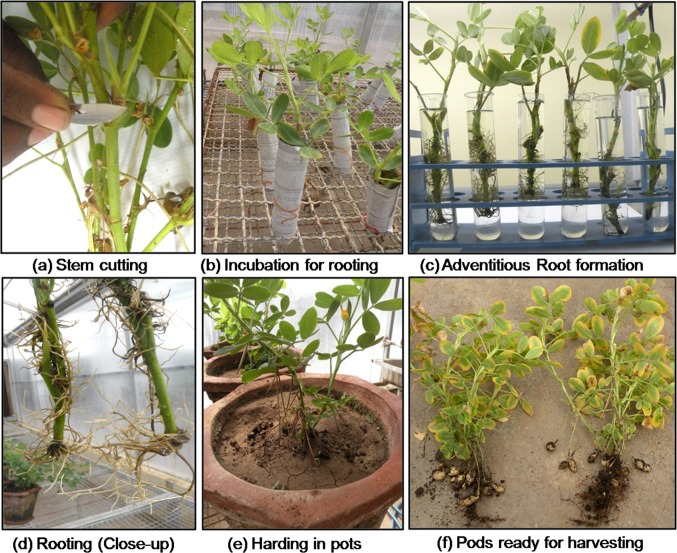
The 10–15-cm-long stem cuttings having 2–3 internodes were prepared from the F1 plants at the time of pod harvesting, as the plants were still green. A slanting cut was made at the base of each cutting, which were then immersed in 0.5% Bavistin (10–15 min), rinsed with distilled water and dipped (4–5 cm) in 1X Hoagland’s solution supplemented with 1% naphthaleneacetic acid (NAA) for rooting, in 50 mL test tubes (Borosil, India) wrapped in aluminum foil to prevent algal contamination. Further, after every 6–7 days, the cuttings were transferred into a new set of tubes filled with fresh growth medium. The cuttings with adventitious roots and 2–3 newly generated shoots were then transplanted into the earthen pots filled with a sterilized mixture of soil and fine sand (1:2, v:v). Hoagland solution was used for watering (6–7 days) the rooted cuttings at the time of hardening, as it simultaneously provides the desired nourishment during establishment (Radhakrishnan et al. 1999). The hardened plants are allowed to complete its life cycle and set the seeds under controlled growth conditions (Fig. 2).
Fig. 2.
Clonal multiplication of F1 plants using stem cuttings for increasing the number of F2 progenies
DNA extraction, polymerase chain reaction and genotyping
DNA was extracted from fresh leaves of the parental genotypes, F1s and segregation populations using the cetyltrimethylammonium bromide (CTAB) extraction method (Cuc et al. 2008). The quality of DNA was checked on the agarose gel (0.8%, w/v), quantification was performed using NanoDropND-1000 (NanoDrop products, DE, USA) and working concentration was adjusted to 20 ng µL−1. For the identification of mutant ahFAD2A and ahFAD2Balleles, all the hybrid plants were first screened using AS-PCR markers (Chen et al. 2010). Further, CAPS (Chu et al. 2007, 2009) and AS-PCR (Yu et al. 2013) assays (Table 1) were performed to identify the zygosity of ILs for the mutant ahFAD2 alleles.
Table 1.
Details of true hybrids, and homozygous ahFAD2B mutant lines obtained in each generation
| Target | Generation | Seeds obtained | Plants germinated and screened | Plants found positive/homozygous | Season |
|---|---|---|---|---|---|
| Hybrid identification | F1 | 8 | 6 (75.0%) | 04 (66.7%) | Summer-2014 |
| BC1F1 | 6 | 5 (83.3%) | 03 (60.0%) | Kharif-2014 | |
| BC2F1 | 6 | 5 (83.3%) | 03 (60.0%) | Summer-2015 | |
| BC3F1 | 10 | 8 (80.0%) | 05 (62.5%) | Kharif-2015 | |
| Total | – | 30 | 24 (80.0%) | 15 (62.5%) | – |
| Selection of ahFAD2B homozygous mutant | F2 | 56 | 52 (92.9%) | 09 (17.3%) | Kharif-2014 |
| F2 (cutting) | 148 | 112 (75.7%) | 18 (16.1%) | Summer-2015 | |
| BC1F2 | 62 | 58 (93.5%) | 08 (13.8%) | Summer-2015 | |
| BC1F2 (cutting) | 70 | 69 (98.6%) | 10 (14.5%) | Kharif-2015 | |
| BC2F2 | 40 | 38 (95.0%) | 04 (10.5%) | Kharif-2015 | |
| BC3F2 | 86 | 83 (96.5%) | 15 (18.1%) | Summer-2016 | |
| Total | – | 462 | 412 (89.2%) | 64 (15.5%) | – |
For AS-PCR analysis, PCR mixture (10.0 µL) containing template DNA (1.0 µL, 20.0 ng), 5 × PCR buffer (2.0 μL, Promega, USA), 25.0 mM MgCl2 (0.8 μL, Promega, USA), 2.0 mM dNTP (0.7 µL, Thermo Fisher Scientific, USA), primers (0.5 μL, 25.0p moles), 5U Taq polymerase (0.2 µL, Promega, USA) and sterile ddH2O (4.3 µL) were used. Amplification was performed in a thermal cycler (Eppendorf, USA) using thin-walled 96-well PCR plates (Sorenson™ Bioscience, USA). The touchdown PCR was done with an initial denaturation at 94.0 °C/3.0 min and then 5 cycles of 94.0 ºC/30 s (− 1.0 ºC reduction per cycle), 65–60 ºC/30 s and 72.0 °C/1 min. This was followed by another 35 cycles of 94.0 °C/30 s, 60 °C/30 s and 72.0 ºC/1.0 min of denaturation, annealing, and extension, respectively. The final extension was done at 72.0 ºC/10 min. Amplification was performed twice and amplified products were analyzed using 2% agarose gel in 1 × TBE buffer at 225 volts for 2.5–3.0 h and stained with ethidium bromide. The gels were documented in an automated gel documentation system (Fujifilm FLA-5000) and scored.
For CAPS analysis of ahFAD2A and ahFAD2B mutations, the PCR was performed with 4.0 μl 5 × PCR buffer (Promega, USA), 1.6 μL MgCl2 (25 mM, Promega-USA), 1.4 μL dNTPs (2 mM, Thermo Fisher Scientific-USA), 1.0 μL each of forward and reverse primers (10 pmol), 0.2 μL Taq DNA polymerase (5U per μL, Promega-USA) polymerase and 6.6 μL sterile ddH2O in a total PCR mixture volume of 20.0 μL.
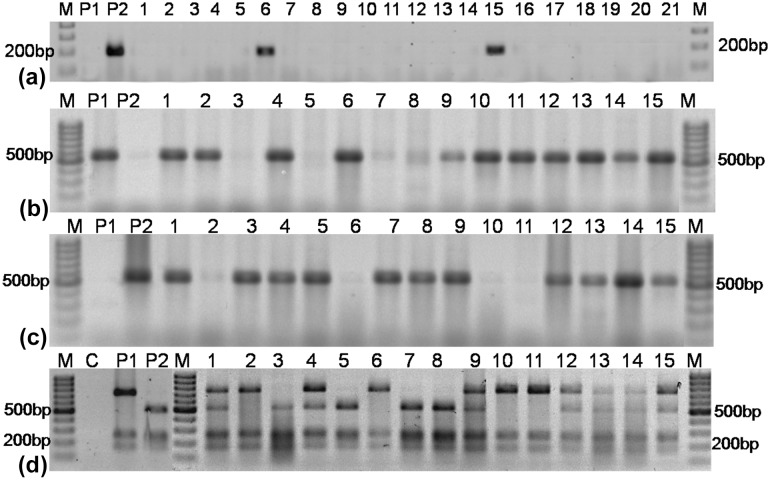
Amplification conditions were initial denaturation at 94 °C/5.0 min followed by 35 cycles of 94 °C/30 s, 55 °C/30 s, and 72 °C/1.0 min denaturation, annealing and extension, respectively. The final extension step was done at 72 °C/10 min. After confirmation of 1200 bp PCR amplification on a 2.0% agarose gel, 10.0 μL PCR products were digested with 0.4 μL of Hpy188I (4 U) restriction enzyme (New England, Biolabs, Ipswich, MA), in a solution containing digestion buffer (2.0 μL), 0.1% BSA (2.0 μL; Takara, Japan) and distilled water (5.6 μL); restriction digestion was done at 37 °C overnight. The digested products were separated on a 2% (w/v) agarose gel and wild-type allele resulted in five fragments of 736, 263, 171, 32, and 12 bp, while in the mutant allele the 736-bp fragment got further digested into 505 and 213 bp, producing six fragments of 505, 263, 213, 171, 32, and 12 bp. However, the larger three fragments could be resolved on a 2% (w/v) agarose gel (Fig. 3).
Fig. 3.
Identification of homozygous ahFAD2B mutant allele a AS-PCR (Yu et al. 2013) for non-mutant ahFAD2B allele (557 bp) identification; b AS-PCR (Yu et al. 2013) for mutant ahFAD2B (441_442insA; 539 bp) allele identification; c CAPS assays for selection of heterozygous and homozygous (441_442insA) alleles; Lanes: M (100 bp DNA ladder); C (Control with water); P1 (GPBD4); P2 (SunOleic95R); 3, 5, 7, 8 (lines homozygous for theahFAD2B mutant allele); 1, 4, 9, 12–15 (lines heterozygous for the ahFAD2B mutant allele); 2, 6, 10,11 (lines homozygous for the ahFAD2B wild allele)
Fatty acid (FA) profiling
The FAs were analyzed using the gas chromatography system (Thermo Fisher, Trace GC 1100) equipped with flame ionization detector (FID), by passing the FA methyl esters through a capillary column (TR wax) (Misra and Mathur 1998). The temperature of the inlet and FID detector was set at 240 ºC, with oven at 190 ºC, whereas, the flow of carrier gas (nitrogen) and fuel gas (hydrogen) was maintained at 30 mL per min. Total run time for each sample was 12 min, and the peaks were identified by comparing to a FAME standard mix RM-3 (Sigma-Aldrich, St. Louis, MO). FA profiles of ILs and parents were estimated from the seeds of the plants which are grown under, (1) controlled conditions, in F4, BC1F4, BC2F3, and BC3F3 generations; and (2) field conditions, in F6, BC1F6, BC2F5, and BC3F4 generations.
Statistical analysis
The data were analyzed using the one-way analysis of variance (ANOVA) by XLSTAT software (Addinsoft XLSTAT 2017) to test the statistical differences (p < 0.05), and Tukey’s HSD multiple comparison test was done to evaluate the significant differences in the means.
Results
Identification of true hybrids and implementation of MABC
Crosses were attempted between SunOleic95R and GPBD4 during Kharif-2013, F1s were raised during the summer of 2014, and were tested for ahFAD2B insertion mutation using AS-PCR assay (Fig. 3). Further, various hybrids from different generations (BC1F1, BC2F1, and BC3F1), and the segregating populations (F2, BC1F2, BC2F2 and BC3F2) were screened using AS-PCR and CAPS assays at 8–10 days after germination (DAG) for the presence of ahFAD2B mutant alleles and its zygosity was confirmed (Fig. 3). True hybrids and the plants homozygous for ahFAD2 alleles from the segregating generations were transferred to the earthen pots (30 × 35 cm) for the completion of its life cycle, while other plants were discarded.
The true hybrid plants having a mutant ahFAD2B allele were then used as a pollen parent and GPBD4 used as RP at each backcross generation, thus the successive backcrossing-generated BC1F1, BC2F1, and BC3F1 generations were also shelfed to get segregation populations. Details about the number of seeds generated, plants analyzed for identification of true hybrids using AS-PCR marker in various F1 generations (F1, BC1F1, BC2F1, and BC3F1) and homozygosity test in different F2 (F2, BC1F2, BC2F2 and BC3F2) are given in Table 1. Only the confirmed homozygous ahFAD2B mutant allele lines identified from each segregation population through molecular maker assays were selected and multiplied as an individual line. Finally, a total of 64 lines were found homozygous for the mutant ahFAD2A and ahFAD2B alleles from all segregating generations Table 1. Pod and kernel features of some of the introgression lines are shown in supplementary figure S1.
Production of a large number of F2 seeds in different generations, using the stem cutting method
Unlike Arabidopsis or rice, where a single F1 plant can produce hundreds of seeds, in peanut, only 20–25 seeds can be obtained per plant under normal growth conditions. Therefore, to get more number of seeds of a segregating generation, we have resorted to our own stem cutting method for mass multiplication of any generation. We used plants of F1 and BC1F1 generations, immediately after harvesting its seeds for the stem cutting experiment. A total of 52 (F1) and 38 (BC1F1) cuttings were attempted which generated 40 and 25 cuttings, respectively, with well-elongated roots within 15–20 days after incubation in the rooting media (Table 2). The cuttings having adventitious roots and 2–3 new leaflets were selected for transplanting in the earthen pots (Fig. 2). On average, 12–15 stem cuttings were made from each hybrid plant. During hardening, 70% F1 and 63% BC1F1 plants could survive and perform like a normal peanut plant for flowering, pegging, and pod development, but overall plant growth habit was quite poor. It was interesting to note that the cuttings from the primary and secondary branches get elongated, while main stem cuttings produced both primary and secondary branches. After 90–100 days, the plants get matured and yielded 148 F2 and 70 BC1F2 seeds from 28 F1 and 15 BC1F1 plants (Table 2). The use of stem cutting in peanut is reported only for the perennial Arachis species, soon after flowering, where seed setting is a major problem (Nigam 2014). However, we have optimized and successfully used the method of stem cutting using mature cultivated peanut plants for increasing the number of F2 seeds. Although this approach needs an extra season, it is a good alternative to increase the number of F2 seeds, especially when the number of plants in any F1 generation is less.
Table 2.
Details of the stem cutting used for the multiplication of F1 and BC1F1 plants
| Generation | Number of SC made | Days to rooting | Number of cutting with AR | Plants survived during hardening | Total number of seeds |
|---|---|---|---|---|---|
| F1 | 52 | 12–15 | 40 (76.9%) | 28 (70%) | 148 |
| BC1F1 | 38 | 18–20 | 25 (65.8%) | 15 (63) | 70 |
where SC Stem cuttings; AR adventitious roots. F1 and BC1F1 were raised during Rainy-2014 and Summer-2015, respectively
Fatty acid analysis
The GC analysis of parents and 64 ILs detected various FAs, among which oleic, linoleic, and palmitic acids constituted more than 80% of total FAs (Table 3). The oleic acid content in ILs ranged from 68.94 to 82.33% with the mean of 78.54% under controlled conditions (Table 3), while under field conditions, it ranged from 67.04 to 81.71% with a mean of 77.63% (Table 4). The linoleic acid content ranged from 1.74 to 10.87% and 2.00 to 15.66% under controlled and field conditions, respectively. With respect to the RP, an increment of 28.78–53.80% has been recorded among the ILs for oleic acid content with an average increase of 46.74% under controlled condition. On the contrary, in field conditions, this increment was 33.70–62.96% with a mean of 54.82% over RP. While the linoleic acid showed 56.47–93.03% and 40.02–92.34% reduction over RP under controlled and field conditions, respectively, with the mean reduction of 86.66 and 82.81% in both the situations. The palmitic acid content varied from 5.85 to 9.92% with an average of 7.06%, which is a reduction of 31.49% compared to the RP in glasshouse conditions (Table 3), while it ranged from 5.99 to 9.75% with a mean of 6.94% under field conditions (Table 4).
Table 3.
Fatty acid composition (%) and O/L ratio of 64 introgression lines (ILs) and their parents grown under glasshouse conditions
| ILS | Palmitic acid (C16:0) | Palmitoleic acid (C16:1) | Stearic acid (C18:0) | Oleic acid (C18:1) | Linoleic acid (C18:2) | Linolenic acid (C18:3) | Arachidic acid (C20:0) | Gadoleic acid (C20:1) | Behenic acid (C22:0) | Erucic acid (C22:1) | Lignoceric acid (C24:0) | O/L ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GPBD4 | 10.30 | 0.07 | 3.14 | 53.53 | 24.97 | 0.04 | 1.68 | 1.31 | 3.45 | 0.07 | 1.45 | 2.14 |
| SunOleic95R | 7.30 | 0.06 | 1.74 | 80.21 | 3.52 | 0.05 | 1.05 | 1.95 | 2.59 | 0.12 | 1.44 | 22.81 |
| MAS_HOL_1 | 6.69 | 0.04 | 2.48 | 80.91 | 2.59 | 0.03 | 1.37 | 1.68 | 2.93 | 0.09 | 1.19 | 31.29 |
| MAS_HOL_2 | 6.75 | 0.07 | 3.46 | 76.82 | 3.99 | 0.05 | 1.55 | 1.98 | 3.50 | 0.18 | 1.66 | 19.23 |
| MAS_HOL_3 | 7.02 | 0.10 | 3.39 | 79.91 | 1.94 | 0.03 | 1.51 | 1.67 | 2.83 | 0.12 | 1.49 | 41.28 |
| MAS_HOL_4 | 5.96 | 0.09 | 3.47 | 80.15 | 1.93 | 0.03 | 1.53 | 1.86 | 3.11 | 0.13 | 1.76 | 41.62 |
| MAS_HOL_5 | 6.61 | 0.02 | 3.28 | 78.47 | 3.89 | 0.05 | 1.50 | 1.58 | 3.14 | 0.09 | 1.37 | 20.19 |
| MAS_HOL_6 | 6.50 | 0.02 | 4.10 | 77.63 | 2.75 | 0.04 | 1.90 | 1.59 | 3.65 | 0.09 | 1.75 | 28.26 |
| MAS_HOL_7 | 7.20 | 0.14 | 2.70 | 77.11 | 3.69 | 0.07 | 1.36 | 2.02 | 3.75 | 0.16 | 1.80 | 20.88 |
| MAS_HOL_8 | 7.10 | 0.06 | 2.10 | 81.66 | 2.39 | 0.06 | 1.09 | 1.82 | 2.34 | 0.13 | 1.27 | 34.14 |
| MAS_HOL_9 | 7.08 | 0.06 | 2.68 | 81.42 | 3.00 | 0.04 | 1.05 | 1.74 | 2.21 | 0.12 | 1.22 | 27.13 |
| MAS_HOL_10 | 7.33 | 0.07 | 2.87 | 79.53 | 3.09 | 0.07 | 1.50 | 1.39 | 2.87 | 0.08 | 1.20 | 25.73 |
| MAS_HOL_11 | 6.42 | 0.08 | 3.76 | 75.34 | 4.14 | 0.03 | 1.74 | 2.08 | 4.25 | 0.19 | 1.97 | 18.20 |
| MAS_HOL_12 | 6.10 | 0.05 | 3.36 | 79.77 | 2.21 | 0.04 | 1.48 | 1.87 | 3.26 | 0.17 | 1.69 | 36.03 |
| MAS_HOL_13 | 6.15 | 0.10 | 3.58 | 78.00 | 2.86 | 0.03 | 1.76 | 1.72 | 3.71 | 0.12 | 1.99 | 27.30 |
| MAS_HOL_14 | 6.15 | 0.07 | 4.06 | 79.47 | 2.17 | 0.04 | 1.62 | 1.57 | 3.25 | 0.11 | 1.48 | 36.64 |
| MAS_HOL_15 | 6.81 | 0.06 | 2.72 | 76.25 | 4.59 | 0.07 | 1.41 | 2.17 | 3.73 | 0.19 | 2.00 | 16.63 |
| MAS_HOL_16 | 6.97 | 0.09 | 3.38 | 76.50 | 3.54 | 0.05 | 1.69 | 1.82 | 3.97 | 0.16 | 1.83 | 21.59 |
| MAS_HOL_17 | 7.36 | 0.07 | 3.64 | 75.23 | 4.10 | 0.06 | 1.77 | 1.77 | 4.15 | 0.15 | 1.70 | 18.34 |
| MAS_HOL_18 | 6.42 | 0.08 | 3.72 | 76.63 | 3.02 | 0.04 | 1.74 | 2.06 | 3.99 | 0.16 | 2.12 | 25.37 |
| MAS_HOL_19 | 6.29 | 0.08 | 3.49 | 78.29 | 2.85 | 0.04 | 1.69 | 1.79 | 3.60 | 0.12 | 1.77 | 27.51 |
| MAS_HOL_20 | 7.36 | 0.09 | 3.03 | 79.95 | 2.48 | 0.05 | 1.37 | 1.64 | 2.61 | 0.12 | 1.29 | 32.21 |
| MAS_HOL_21 | 6.58 | 0.03 | 3.60 | 77.56 | 3.29 | 0.05 | 1.78 | 1.75 | 3.60 | 0.11 | 1.67 | 23.55 |
| MAS_HOL_22 | 6.60 | 0.07 | 3.68 | 76.01 | 3.32 | 0.07 | 1.94 | 1.79 | 4.46 | 0.14 | 1.94 | 22.87 |
| MAS_HOL_23 | 6.21 | 0.08 | 3.90 | 75.63 | 3.94 | 0.05 | 1.84 | 2.19 | 3.99 | 0.21 | 1.97 | 19.22 |
| MAS_HOL_24 | 6.55 | 0.07 | 4.15 | 73.83 | 4.94 | 0.09 | 1.87 | 2.11 | 4.15 | 0.23 | 2.02 | 14.96 |
| MAS_HOL_25 | 6.49 | 0.06 | 4.02 | 77.50 | 3.50 | 0.04 | 1.67 | 1.77 | 3.36 | 0.13 | 1.48 | 22.15 |
| MAS_HOL_26 | 6.74 | 0.07 | 3.66 | 79.46 | 2.27 | 0.02 | 1.68 | 1.52 | 3.01 | 0.09 | 1.48 | 34.97 |
| MAS_HOL_27 | 5.85 | 0.09 | 4.14 | 74.13 | 4.37 | 0.07 | 1.95 | 2.58 | 4.00 | 0.27 | 2.55 | 16.97 |
| MAS_HOL_28 | 6.28 | 0.08 | 3.39 | 76.63 | 4.18 | 0.09 | 1.57 | 2.02 | 3.66 | 0.20 | 1.89 | 18.33 |
| MAS_HOL_29 | 6.48 | 0.06 | 3.54 | 78.47 | 2.96 | 0.06 | 1.56 | 1.75 | 3.49 | 0.12 | 1.51 | 26.51 |
| MAS_HOL_30 | 7.04 | 0.03 | 2.59 | 76.47 | 5.13 | 0.06 | 1.41 | 2.12 | 3.53 | 0.16 | 1.46 | 14.90 |
| MAS_HOL_31 | 7.42 | 0.06 | 2.06 | 80.08 | 2.94 | 0.04 | 1.18 | 1.86 | 2.77 | 0.13 | 1.47 | 27.23 |
| MAS_HOL_32 | 7.20 | 0.07 | 1.95 | 79.74 | 3.62 | 0.06 | 1.10 | 1.94 | 2.53 | 0.16 | 1.64 | 22.00 |
| MAS_HOL_33 | 6.89 | 0.07 | 1.97 | 80.12 | 2.83 | 0.05 | 1.14 | 2.15 | 2.81 | 0.19 | 1.80 | 28.36 |
| MAS_HOL_34 | 6.41 | 0.04 | 2.35 | 80.72 | 2.22 | 0.03 | 1.27 | 2.16 | 2.93 | 0.17 | 1.70 | 36.33 |
| MAS_HOL_35 | 7.66 | 0.12 | 2.13 | 81.04 | 2.47 | 0.06 | 1.06 | 1.71 | 2.26 | 0.11 | 1.38 | 32.76 |
| MAS_HOL_36 | 7.52 | 0.09 | 2.32 | 79.68 | 2.67 | 0.04 | 1.25 | 1.88 | 2.96 | 0.14 | 1.46 | 29.90 |
| MAS_HOL_37 | 6.23 | 0.03 | 1.90 | 81.96 | 2.72 | 0.05 | 1.04 | 1.98 | 2.55 | 0.15 | 1.40 | 30.13 |
| MAS_HOL_38 | 7.73 | 0.12 | 2.02 | 80.63 | 2.27 | 0.08 | 1.17 | 1.94 | 2.38 | 0.13 | 1.56 | 35.49 |
| MAS_HOL_39 | 6.75 | 0.05 | 1.94 | 81.20 | 2.68 | 0.04 | 1.17 | 1.92 | 2.71 | 0.13 | 1.40 | 30.36 |
| MAS_HOL_40 | 6.82 | 0.04 | 2.23 | 81.70 | 1.80 | 0.03 | 1.33 | 1.78 | 2.90 | 0.11 | 1.26 | 45.29 |
| MAS_HOL_41 | 7.79 | 0.13 | 2.81 | 80.41 | 1.77 | 0.05 | 1.51 | 1.51 | 2.69 | 0.11 | 1.22 | 45.41 |
| MAS_HOL_42 | 6.93 | 0.13 | 2.75 | 79.30 | 3.33 | 0.04 | 1.62 | 1.64 | 3.31 | 0.08 | 1.31 | 23.82 |
| MAS_HOL_43 | 7.95 | 0.07 | 2.78 | 71.29 | 10.62 | 0.04 | 1.45 | 1.52 | 2.96 | 0.06 | 1.28 | 6.71 |
| MAS_HOL_44 | 7.01 | 0.07 | 2.69 | 80.26 | 2.82 | 0.05 | 1.44 | 1.53 | 2.66 | 0.08 | 1.39 | 28.48 |
| MAS_HOL_45 | 6.34 | 0.07 | 3.28 | 75.11 | 4.49 | 0.07 | 1.64 | 2.60 | 3.88 | 0.27 | 2.26 | 16.73 |
| MAS_HOL_46 | 6.67 | 0.06 | 2.87 | 78.40 | 3.83 | 0.05 | 1.38 | 1.94 | 3.21 | 0.15 | 1.60 | 20.45 |
| MAS_HOL_47 | 7.14 | 0.08 | 2.73 | 80.33 | 2.04 | 0.05 | 1.34 | 1.79 | 2.74 | 0.11 | 1.66 | 39.35 |
| MAS_HOL_48 | 8.13 | 0.09 | 2.66 | 79.62 | 1.76 | 0.05 | 1.47 | 1.48 | 3.14 | 0.08 | 1.47 | 45.13 |
| MAS_HOL_49 | 6.84 | 0.07 | 2.69 | 80.93 | 2.26 | 0.04 | 1.43 | 1.54 | 2.68 | 0.08 | 1.44 | 35.83 |
| MAS_HOL_50 | 9.92 | 0.10 | 3.06 | 68.94 | 10.87 | 0.05 | 1.47 | 1.30 | 2.85 | 0.12 | 1.33 | 6.34 |
| MAS_HOL_51 | 7.91 | 0.12 | 2.77 | 71.32 | 9.96 | 0.06 | 1.45 | 1.61 | 3.10 | 0.10 | 1.60 | 7.16 |
| MAS_HOL_52 | 8.04 | 0.08 | 2.46 | 79.48 | 2.07 | 0.02 | 1.36 | 1.73 | 3.03 | 0.13 | 1.59 | 38.40 |
| MAS_HOL_53 | 7.19 | 0.07 | 2.47 | 79.85 | 2.72 | 0.05 | 1.28 | 1.81 | 2.77 | 0.16 | 1.50 | 29.36 |
| MAS_HOL_54 | 6.58 | 0.07 | 2.72 | 81.55 | 2.11 | 0.04 | 1.40 | 1.58 | 2.62 | 0.08 | 1.25 | 38.67 |
| MAS_HOL_55 | 8.30 | 0.14 | 2.35 | 80.11 | 2.01 | 0.03 | 1.29 | 1.54 | 2.72 | 0.08 | 1.43 | 39.80 |
| MAS_HOL_56 | 7.99 | 0.11 | 2.34 | 75.39 | 5.85 | 0.05 | 1.40 | 1.80 | 3.13 | 0.12 | 1.84 | 12.90 |
| MAS_HOL_57 | 8.00 | 0.11 | 2.47 | 80.36 | 1.93 | 0.06 | 1.18 | 1.73 | 2.70 | 0.10 | 1.38 | 41.62 |
| MAS_HOL_58 | 9.33 | 0.25 | 2.37 | 78.18 | 2.85 | 0.07 | 1.29 | 1.37 | 2.80 | 0.07 | 1.44 | 27.41 |
| MAS_HOL_59 | 7.27 | 0.06 | 2.61 | 79.57 | 2.69 | 0.04 | 1.33 | 1.83 | 2.88 | 0.13 | 1.58 | 29.56 |
| MAS_HOL_60 | 6.26 | 0.03 | 1.92 | 82.33 | 1.74 | 0.04 | 1.28 | 1.93 | 2.98 | 0.11 | 1.41 | 47.43 |
| MAS_HOL_61 | 7.85 | 0.09 | 2.29 | 80.18 | 2.51 | 0.07 | 1.28 | 1.66 | 2.67 | 0.10 | 1.33 | 31.96 |
| MAS_HOL_62 | 6.56 | 0.04 | 1.91 | 80.92 | 3.02 | 0.04 | 1.11 | 2.09 | 2.63 | 0.16 | 1.50 | 26.78 |
| MAS_HOL_63 | 7.00 | 0.10 | 1.63 | 81.87 | 2.29 | 0.03 | 1.13 | 1.85 | 2.50 | 0.12 | 1.47 | 35.72 |
| MAS_HOL_64 | 8.85 | 0.20 | 2.04 | 79.52 | 2.38 | 0.04 | 1.15 | 1.74 | 2.50 | 0.12 | 1.47 | 33.45 |
| Meana | 7.06 | 0.08 | 2.87 | 78.54 | 3.33 | 0.05 | 1.44 | 1.81 | 3.13 | 0.13 | 1.58 | 27.97 |
| Minimum | 5.85 | 0.02 | 1.63 | 68.94 | 1.74 | 0.02 | 1.04 | 1.30 | 2.21 | 0.06 | 1.19 | 6.34 |
| Maximum | 9.92 | 0.25 | 4.15 | 82.33 | 10.87 | 0.09 | 1.95 | 2.60 | 4.46 | 0.27 | 2.55 | 47.43 |
| % increase/decreaseb | − 31.49 | 10.56 | − 8.57 | 46.74 | − 86.66 | 13.90 | − 14.21 | 38.04 | − 9.31 | 97.59 | 9.38 | 1205.19 |
aValues of both the parents are not considered
bPercent increase or decrease in mean fatty acid content of ILs with respect to recurrent parent
Table 4.
Fatty acid composition (%) and O/L ratio of 64 introgression lines (ILs) and their parents grown under open-field conditions
| ILS | Palmitic acid (C16:0) | Palmitoleic acid (C16:1) | Stearic acid (C18:0) | Oleic acid (C18:1) | Linoleic acid (C18:2) | Linolenic acid (C18:3) | Arachidic acid (C20:0) | Gadoleic acid (C20:1) | Behenic acid (C22:0) | Erucic acid (C22:1) | Lignoceric acid (C24:0) | O/L ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GPBD4 | 11.56 | 0.06 | 3.37 | 50.14 | 26.11 | 0.04 | 1.63 | 1.16 | 4.28 | 0.08 | 1.16 | 1.92 |
| SunOleic95R | 6.49 | 0.03 | 2.16 | 78.68 | 5.20 | 0.05 | 1.11 | 2.06 | 2.74 | 0.16 | 1.32 | 15.12 |
| MAS_HOL_1 | 6.01 | 0.04 | 2.61 | 79.44 | 3.93 | 0.04 | 1.34 | 1.95 | 3.06 | 0.14 | 1.44 | 20.21 |
| MAS_HOL_2 | 6.54 | 0.03 | 2.59 | 77.10 | 5.33 | 0.05 | 1.35 | 2.01 | 3.45 | 0.13 | 1.42 | 14.46 |
| MAS_HOL_3 | 6.66 | 0.04 | 2.79 | 78.78 | 3.36 | 0.04 | 1.45 | 1.95 | 3.41 | 0.11 | 1.42 | 23.48 |
| MAS_HOL_4 | 6.45 | 0.03 | 2.24 | 77.13 | 4.98 | 0.07 | 1.25 | 2.45 | 3.58 | 0.21 | 1.63 | 15.49 |
| MAS_HOL_5 | 6.54 | 0.04 | 3.26 | 76.40 | 4.42 | 0.06 | 1.62 | 1.86 | 3.90 | 0.13 | 1.77 | 17.29 |
| MAS_HOL_6 | 6.69 | 0.05 | 3.05 | 77.48 | 3.90 | 0.06 | 1.64 | 1.91 | 3.52 | 0.13 | 1.58 | 19.87 |
| MAS_HOL_7 | 6.86 | 0.04 | 2.90 | 76.82 | 4.54 | 0.06 | 1.50 | 1.90 | 3.63 | 0.15 | 1.62 | 16.92 |
| MAS_HOL_8 | 6.03 | 0.06 | 3.10 | 79.41 | 3.06 | 0.04 | 1.61 | 1.87 | 3.33 | 0.11 | 1.38 | 25.97 |
| MAS_HOL_9 | 7.02 | 0.05 | 2.24 | 78.56 | 4.79 | 0.05 | 1.06 | 2.11 | 2.82 | 0.18 | 1.44 | 16.41 |
| MAS_HOL_10 | 6.56 | 0.05 | 4.05 | 77.54 | 3.88 | 0.04 | 1.66 | 1.52 | 3.52 | 0.07 | 1.12 | 19.99 |
| MAS_HOL_11 | 7.34 | 0.04 | 2.94 | 75.37 | 5.64 | 0.07 | 1.42 | 2.17 | 3.37 | 0.16 | 1.49 | 13.36 |
| MAS_HOL_12 | 6.87 | 0.04 | 2.27 | 78.48 | 4.65 | 0.04 | 1.34 | 1.81 | 3.07 | 0.12 | 1.33 | 16.89 |
| MAS_HOL_13 | 6.47 | 0.06 | 3.37 | 78.53 | 2.92 | 0.06 | 1.61 | 1.87 | 3.47 | 0.11 | 1.53 | 26.90 |
| MAS_HOL_14 | 6.86 | 0.05 | 2.97 | 77.41 | 3.83 | 0.07 | 1.50 | 1.94 | 3.71 | 0.13 | 1.54 | 20.21 |
| MAS_HOL_15 | 7.35 | 0.04 | 2.84 | 75.89 | 5.02 | 0.07 | 1.47 | 1.96 | 3.78 | 0.13 | 1.45 | 15.10 |
| MAS_HOL_16 | 7.29 | 0.04 | 2.71 | 78.00 | 4.27 | 0.05 | 1.31 | 1.77 | 3.13 | 0.12 | 1.31 | 18.28 |
| MAS_HOL_17 | 7.21 | 0.05 | 2.81 | 76.46 | 4.35 | 0.08 | 1.61 | 1.91 | 3.88 | 0.13 | 1.52 | 17.57 |
| MAS_HOL_18 | 6.70 | 0.03 | 2.39 | 77.26 | 4.91 | 0.06 | 1.39 | 1.98 | 3.63 | 0.11 | 1.57 | 15.74 |
| MAS_HOL_19 | 6.78 | 0.04 | 3.16 | 76.92 | 4.45 | 0.06 | 1.55 | 1.91 | 3.56 | 0.10 | 1.48 | 17.27 |
| MAS_HOL_20 | 6.45 | 0.04 | 2.98 | 78.53 | 3.55 | 0.05 | 1.50 | 1.92 | 3.31 | 0.12 | 1.56 | 22.10 |
| MAS_HOL_21 | 7.21 | 0.06 | 3.57 | 76.50 | 4.02 | 0.06 | 1.75 | 1.67 | 3.63 | 0.12 | 1.42 | 19.04 |
| MAS_HOL_22 | 6.84 | 0.05 | 3.40 | 77.45 | 3.49 | 0.06 | 1.70 | 1.78 | 3.57 | 0.11 | 1.55 | 22.18 |
| MAS_HOL_23 | 7.07 | 0.02 | 2.30 | 77.16 | 5.03 | 0.07 | 1.23 | 2.19 | 3.51 | 0.16 | 1.27 | 15.34 |
| MAS_HOL_24 | 7.64 | 0.04 | 2.86 | 75.84 | 5.26 | 0.05 | 1.35 | 2.07 | 3.40 | 0.14 | 1.36 | 14.41 |
| MAS_HOL_25 | 7.23 | 0.03 | 3.10 | 77.91 | 4.00 | 0.05 | 1.58 | 1.72 | 3.16 | 0.07 | 1.16 | 19.50 |
| MAS_HOL_26 | 6.81 | 0.03 | 2.40 | 76.53 | 5.68 | 0.07 | 1.26 | 2.30 | 3.25 | 0.20 | 1.50 | 13.48 |
| MAS_HOL_27 | 6.87 | 0.05 | 2.41 | 76.23 | 4.61 | 0.07 | 1.33 | 2.57 | 3.71 | 0.25 | 1.92 | 16.55 |
| MAS_HOL_28 | 6.34 | 0.02 | 2.29 | 77.64 | 5.35 | 0.06 | 1.09 | 2.18 | 3.49 | 0.18 | 1.36 | 14.50 |
| MAS_HOL_29 | 6.86 | 0.04 | 3.07 | 77.09 | 4.42 | 0.06 | 1.52 | 1.89 | 3.46 | 0.11 | 1.49 | 17.45 |
| MAS_HOL_30 | 5.99 | 0.06 | 4.87 | 75.78 | 3.94 | 0.07 | 1.87 | 1.95 | 3.44 | 0.18 | 1.87 | 19.22 |
| MAS_HOL_31 | 7.02 | 0.05 | 2.24 | 78.56 | 4.79 | 0.05 | 1.06 | 2.11 | 2.82 | 0.18 | 1.44 | 16.41 |
| MAS_HOL_32 | 6.35 | 0.04 | 2.54 | 77.91 | 4.72 | 0.06 | 1.27 | 2.19 | 3.10 | 0.19 | 1.64 | 16.52 |
| MAS_HOL_33 | 6.04 | 0.02 | 2.78 | 79.60 | 3.78 | 0.05 | 1.33 | 2.01 | 2.87 | 0.15 | 1.37 | 21.04 |
| MAS_HOL_34 | 6.47 | 0.05 | 2.77 | 78.00 | 3.83 | 0.05 | 1.65 | 1.92 | 3.54 | 0.13 | 1.59 | 20.36 |
| MAS_HOL_35 | 6.36 | 0.03 | 3.37 | 78.58 | 3.65 | 0.05 | 1.59 | 1.76 | 3.12 | 0.12 | 1.36 | 21.55 |
| MAS_HOL_36 | 6.62 | 0.05 | 3.26 | 77.63 | 3.91 | 0.06 | 1.56 | 1.92 | 3.52 | 0.13 | 1.35 | 19.85 |
| MAS_HOL_37 | 6.55 | 0.04 | 2.83 | 78.79 | 3.75 | 0.05 | 1.41 | 1.99 | 3.03 | 0.14 | 1.43 | 21.03 |
| MAS_HOL_38 | 6.63 | 0.04 | 2.58 | 77.13 | 5.04 | 0.07 | 1.32 | 2.20 | 3.15 | 0.16 | 1.67 | 15.31 |
| MAS_HOL_39 | 6.69 | 0.05 | 2.97 | 78.64 | 3.74 | 0.05 | 1.48 | 1.89 | 2.99 | 0.13 | 1.38 | 21.05 |
| MAS_HOL_40 | 6.60 | 0.05 | 2.75 | 79.93 | 3.47 | 0.04 | 1.41 | 1.58 | 2.96 | 0.08 | 1.14 | 23.05 |
| MAS_HOL_41 | 6.79 | 0.06 | 2.91 | 81.27 | 2.10 | 0.05 | 1.48 | 1.53 | 2.62 | 0.07 | 1.13 | 38.79 |
| MAS_HOL_42 | 6.38 | 0.10 | 2.64 | 79.96 | 2.80 | 0.05 | 1.63 | 1.70 | 3.34 | 0.09 | 1.32 | 28.53 |
| MAS_HOL_43 | 8.29 | 0.07 | 2.76 | 68.91 | 13.02 | 0.04 | 1.45 | 1.39 | 2.88 | 0.03 | 1.18 | 5.29 |
| MAS_HOL_44 | 6.99 | 0.08 | 2.62 | 81.29 | 2.25 | 0.05 | 1.45 | 1.41 | 2.46 | 0.07 | 1.34 | 36.18 |
| MAS_HOL_45 | 9.75 | 0.18 | 2.93 | 74.27 | 5.62 | 0.06 | 1.54 | 1.29 | 3.05 | 0.12 | 1.28 | 13.22 |
| MAS_HOL_46 | 6.76 | 0.06 | 2.23 | 78.86 | 4.16 | 0.06 | 1.17 | 2.07 | 2.81 | 0.17 | 1.64 | 18.94 |
| MAS_HOL_47 | 6.70 | 0.05 | 2.67 | 78.61 | 4.33 | 0.06 | 1.36 | 2.13 | 3.28 | 0.18 | 1.63 | 18.17 |
| MAS_HOL_48 | 6.75 | 0.10 | 3.79 | 79.57 | 2.00 | 0.04 | 1.72 | 1.52 | 2.97 | 0.14 | 1.46 | 39.75 |
| MAS_HOL_49 | 6.94 | 0.06 | 3.48 | 78.56 | 3.46 | 0.05 | 1.58 | 1.44 | 3.20 | 0.07 | 1.15 | 22.71 |
| MAS_HOL_50 | 8.03 | 0.07 | 2.83 | 67.04 | 15.66 | 0.03 | 1.40 | 1.17 | 2.54 | 0.05 | 1.19 | 4.28 |
| MAS_HOL_51 | 7.67 | 0.02 | 2.31 | 71.22 | 10.42 | 0.07 | 1.18 | 2.16 | 3.30 | 0.16 | 1.48 | 6.83 |
| MAS_HOL_52 | 6.56 | 0.06 | 2.53 | 81.71 | 2.01 | 0.04 | 1.28 | 1.55 | 2.73 | 0.10 | 1.42 | 40.71 |
| MAS_HOL_53 | 7.00 | 0.05 | 2.28 | 78.55 | 4.10 | 0.06 | 1.24 | 2.03 | 2.98 | 0.13 | 1.55 | 19.14 |
| MAS_HOL_54 | 6.71 | 0.07 | 2.75 | 80.45 | 2.72 | 0.04 | 1.41 | 1.65 | 2.78 | 0.10 | 1.32 | 29.60 |
| MAS_HOL_55 | 6.91 | 0.08 | 2.61 | 80.68 | 2.51 | 0.05 | 1.40 | 1.64 | 2.73 | 0.10 | 1.29 | 32.09 |
| MAS_HOL_56 | 7.50 | 0.07 | 2.56 | 72.34 | 9.84 | 0.05 | 1.40 | 1.64 | 2.91 | 0.11 | 1.58 | 7.35 |
| MAS_HOL_57 | 7.52 | 0.09 | 2.58 | 80.36 | 2.29 | 0.06 | 1.25 | 1.67 | 2.27 | 0.11 | 1.39 | 35.09 |
| MAS_HOL_58 | 7.02 | 0.08 | 2.70 | 80.21 | 2.76 | 0.07 | 1.33 | 1.62 | 2.68 | 0.13 | 1.40 | 29.08 |
| MAS_HOL_59 | 7.05 | 0.04 | 2.46 | 77.24 | 5.58 | 0.03 | 1.39 | 1.77 | 2.91 | 0.10 | 1.43 | 13.84 |
| MAS_HOL_60 | 8.30 | 0.14 | 2.35 | 80.11 | 2.01 | 0.03 | 1.29 | 1.54 | 2.72 | 0.08 | 1.43 | 39.80 |
| MAS_HOL_61 | 8.64 | 0.11 | 2.30 | 78.15 | 3.00 | 0.04 | 1.25 | 1.83 | 2.85 | 0.15 | 1.66 | 26.01 |
| MAS_HOL_62 | 6.61 | 0.06 | 2.55 | 79.92 | 3.27 | 0.06 | 1.17 | 1.97 | 2.57 | 0.15 | 1.69 | 24.42 |
| MAS_HOL_63 | 7.02 | 0.09 | 1.97 | 80.68 | 2.85 | 0.04 | 1.22 | 1.85 | 2.69 | 0.12 | 1.48 | 28.30 |
| MAS_HOL_64 | 6.97 | 0.08 | 2.85 | 78.17 | 3.82 | 0.04 | 1.44 | 1.75 | 3.03 | 0.11 | 1.48 | 20.46 |
| Minimum | 5.99 | 0.02 | 1.97 | 67.04 | 2.00 | 0.03 | 1.06 | 1.17 | 2.46 | 0.03 | 1.12 | 4.28 |
| Maximum | 9.75 | 0.18 | 4.87 | 81.71 | 15.66 | 0.08 | 1.87 | 2.57 | 3.90 | 0.25 | 1.92 | 40.71 |
| Meana | 6.94 | 0.06 | 2.81 | 77.63 | 4.49 | 0.05 | 1.42 | 1.86 | 3.18 | 0.13 | 1.45 | 20.31 |
| % increase/decreaseb | − 39.98 | − 3.49 | − 16.73 | 54.82 | − 82.81 | 33.73 | − 12.46 | 61.28 | − 25.73 | 70.34 | 25.52 | 957.27 |
aValues of both the parents are not considered
bPercent increase or decrease in mean fatty acid content of ILs with respect to recurrent parent
Correlation among various FAs under different growth conditions
Pearson’s correlation coefficient among all the ILs was carried out to find the effect of growth conditions, viz., controlled and field conditions on the variations recorded for different FAs. A highly significant negative correlation was observed between oleic and linoleic acid contents under both field (r = − 0.975) and glasshouse (r = − 0.940) conditions. Further, significant negative correlations have been recorded for oleic acid content with arachidic and behenic acids under both controlled and field conditions (Table S2). Palmitic acid content under controlled conditions showed a significantly negative correlation with SFAs such as stearic (r = − 0.390), arachidic (r = − 0.310) and lignoceric acids (r = − 0.486). Further, behenic acid showed a significant positive correlation with stearic acid (r = 0.725) and arachidic acid (r = 0.856) under controlled conditions, while no such strong correlation was observed under field conditions. Moreover, under controlled and field conditions, erucic acid showed a significant positive correlation with gadoleic acid (r = 0.779 and 0.877, respectively) and lignoceric acid (r = 0.725 and 0.703, respectively). The linolenic acid also recorded a significantly positive correlation with gadoleic (r = 0.279), behenic (r = 0.294), erucic (r = 0.465), and lignoceric acid (r = 0.366), under controlled conditions. Similarly, stearic acid showed a significantly positive correlation with behenic (r = 0.725), erucic (r = 0.312), and lignoceric acid (r = 0.537), under controlled conditions, but under field conditions it reflected a significant negative correlation with gadoleic (r = − 0.468), erucic (r = − 0.423), and lignoceric acids (r = − 0.266) (Table S2). Wang et al. (2015a) also reported a significant negative correlation of stearic acid with gadoleic and also with lignoceric acid under filed conditions, indicating more use of stearic and linolenic acid for long-chain FA production. But it still needs a further in-depth analysis to pinpoint the exact factors responsible for such expression.
The ahFAD2 gene double mutant plant uses its resources for the production of more linoleic acid by mobilizing the palmitic acid towards the oleic acid formation (Wang et al. 2015a). Thus, increase in oleic acid also acts as a signal to trigger a negative feedback loop to deal with an excess of SFAs, which might lead to the negative correlation between oleic acid and SFAs (Lim et al. 2013; Harvey et al. 2010).
Discussion
The alteration of FA profiles aimed for higher oxidative stability and better dietary properties is an important and evolving theme to meet the nutritional needs and industrial criteria of the modern market. Henceforth, a concentrated effort has been made in our peanut breeding program, and an array of HO lines has been generated by introgressing mutant ahFAD2B gene through MAS.
Peanut being an allotetraploid crop contains two sets of alleles of any gene in its A and B genomes, and mutation in both ahFAD2A and ahFAD2B allele is required for the expression of HO trait in peanut line (Nawade et al. 2018). The understanding of the genetics and molecular basis of ahFAD2 gene has led to the development of several molecular marker systems such as CAPS and allele-specific PCR (AS-PCR) assays for the accurate genotyping of ahFAD2 genes. Based on our previous result (Nawade et al. 2016), we have selected GPBD4, multiple foliar fungal disease-resistant and high-yielding variety, having a natural mutation in the ahFAD2A gene. Furthermore, GPBD4 is extremely popular among the south Indian farmers because of its durable resistance to various foliar fungal diseases including rust and late leaf spot (LLS). The rust and LLS are prevalent in groundnut growing regions across the world causing yield loss up to 70% (Sujay et al. 2012). Further, the extent of economic losses to the tune of 467 m USD by rust and 599 m USD by LLS was estimated (Khera et al. 2016). Besides, adversely affecting productivity, they also affect the quality of the seeds and fodder, making it unsuitable for consumption. Henceforth, several popular groundnut varieties have been phased out of farmer’s fields in the recent past due to heavy yield losses caused by foliar fungal diseases. We have focused on the selection and transfer of only one mutant allele, ahFAD2B from SunOleic95R into GPBD4 using the MABC approach (Fig. 1). Moreover, we have confirmed the presence and homozygosity of ahFAD2A allele using AS-PCR and CAPS markers in introgression lines.
Hybridization is the most critical step for the success of any peanut breeding program, as the peanut is a self-pollinated crop with cleistogamous pollination system (Othman 1979; Lim et al. 1980). Consequently, significant efforts are required during hybridization programs to produce sufficient true hybrid seeds for the development of mapping populations or to improve the desired trait of selection (Chu et al. 2016; Norden 1980). We got a hybridization success rate in the range of 3–6% (data not shown) which is in tune with the previous reports (Norden and Rodriguez 1971; Banks 1976). However, a high pollination success rate (25–70%) was also reported (Norden 1980; Kale and Mouli 1984; Nigam et al. 1990). This large variation could be due to the fact that the pollination in peanut is greatly influenced by a number of factors like humidity, temperature, crossing schedule, the integrity of emasculated flowers, the skill of the operator and the parental combinations (Chu et al. 2016).
The AS-PCR assay proficiently identified the true hybrids from putative F1s, while, CAPS helped in timely identification of homozygous plants for both ahFAD2 alleles, in the segregating generations within a week of peanut germination and enabled the availability of pollen parent for subsequent backcrossing. The AS-PCR assay (Yu et al. 2013) targeting the mutant and non-mutant ahFAD2B alleles in separate reactions was also validated in these populations (Fig. 3) and was found more convenient to perform over CAPS assay. The CAPS assay was first utilized by Chu et al. (2011) to generate ‘Tifguard High O/L’ genotype through three rounds of accelerated backcrossing. While Janila et al. (2016) introgressed the ahFAD2 mutant alleles from SunOleic95R in the background of elite genotypes (ICGV06110, ICGV06142, and ICGV06420) using AS-PCR (Chen et al. 2010) and CAPS assay (Chu et al. 2007, 2009) and generated a total of 469 ILs. Further, real-time PCR (Barkley et al. 2010; 2011) was also exploited to transfer the HO trait in South African peanut cultivars, ‘SA Juweel’ and ‘ARC Oleic2’ (Mienie and Pretorius 2013), whereas, Koilkonda et al. (2013) identified 9 ahFAD2 homozygous lines from 205 BC2F2 plants. In this experiment, we have successfully introgressed high oleic trait through MABC and developed a total of 64 HO ILs.
Further, among the 64 ILs generated, only 3 lines; MAS_HOIL_43, 50 and 51 (from glasshouse) and 4 lines; MAS_HOIL_43, 50, 51 and 56 (from field conditions) recorded O/L ratio of the below industrially acceptable value of 9.0 (Tables 3, 4). Similarly, Janila et al. (2016) also reported considerable variations in the oleic and linoleic acid contents in the ahFAD2 gene double mutant ILs which ranged from 62 to 82% and 2 to 20%, respectively. A 0.5–1.1-fold increase in the oleic acid content with concomitant reduction of linoleic acid by 0.4–1.0-fold among 82 MABC and 387 MAS-derived ILs compared to recurrent parents was also recorded by Janila et al. (2016). Moreover, many reports on ahFAD2 gene double mutant recombinant inbred lines (RILs) also revealed substantial phenotypic variations for oleic and linoleic acid contents (Wang et al. 2015a; Pandey et al. 2014; Sarvamangala et al. 2011). Pandey et al. (2014) reported phenotypic variance for ahFAD2B gene (26.54%, 25.59% and 41.02%) and ahFAD2A gene (8.08%, 6.86% and 3.78%) for oleic acid (C18:1), linoleic acid (C18:2), and O/L ratio, respectively.
Furthermore, the double mutant line SunOleic95R when grown under US conditions was reported to have 79–81% oleic acid and 2.5–4.7% linoleic acid (Gorbet and Knauft, 1997; Andersen et al. 1998; Barkley et al. 2010, 2011). However, when same genotype is grown under Junagadh, India (21º47′73″N, 70º44′80″E; Nawade et al. 2016) and ICRISAT-Patancheru, India (17º50′28″N, 78º27′79″E; Janila et al. 2016) conditions, recorded 78.68%–80.21% and 78.30% oleic acid and 3.20%–7.34% and 5.00–6.00% linoleic acid, respectively. Furthermore, a few seeds having normal O/L were also identified in the seed lots of HO peanut cultivar ‘Brantley’ (Chamberlin et al. 2011). All these convincingly prove that multiple factors are involved in the regulation of oleic to linoleic acid flux, but a major role was played by the ahFAD2 gene.
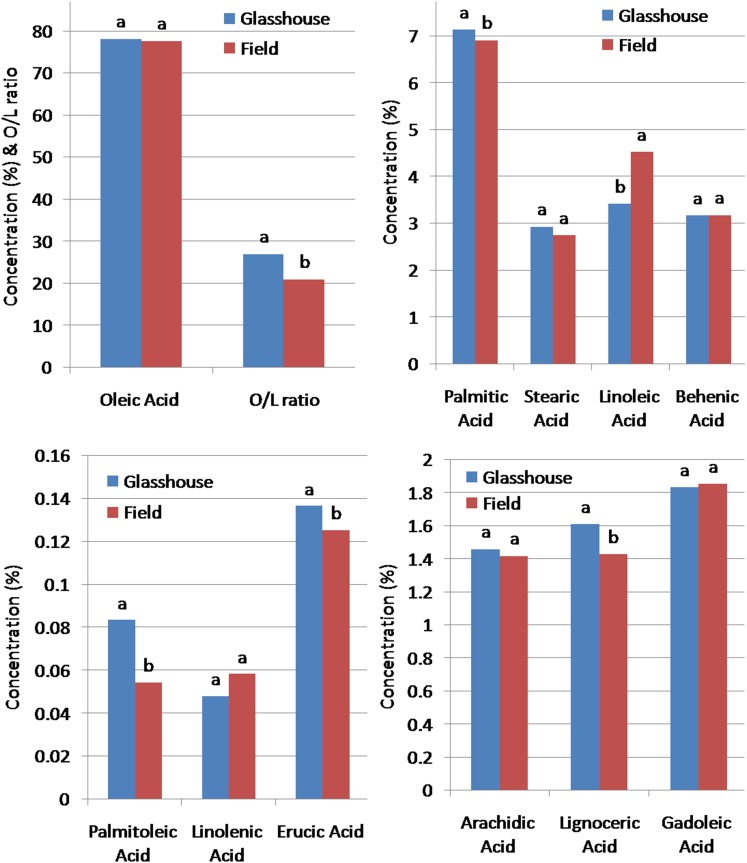
The ILs recorded higher mean SFA and MUFA contents under controlled conditions than under field conditions, which recorded a higher PUFA content (Fig. 4). The mean linoleic acid content of ILs showed an increment of 1.24% under field conditions, whereas palmitoleic, oleic, and lignoceric acid contents which along with O/L ratio showed a significant reduction under field conditions. Furthermore, other FAs did not show any significant change under both conditions. Thus, these HO lines under two different growth conditions showed the HO content indicating successful transfer and expression of the targeted trait in the RP. Moreover, the variations recorded are within the range of HO classification, which could be attributed to the growth conditions and other minor factors (Hinds 1995; Golombek et al. 1995; Singkham et al. 2010; Sun et al. 2014). The genetic factors and its interaction with the environmental factors are known to play a significant role in the formation of different FAs including oleic acid and linoleic acid (Singkham et al. 2010; Isleib et al. 2008; Andersen and Gorbet 2002).
Fig. 4.
Effect of growth conditions on FA composition and O/L ratio of peanut IL
The significant increment in the linoleic acid content under open conditions could be the effect of lower mean temperature during pod-filling stage during August, September and October months (27.4, 27.9 and 27.1 °C, respectively) of the year 2016 (Table S3 and Fig. 4). The lower temperature range (22–29 °C) is known to boost the oleate desaturase activity, thus promoting linoleic acid synthesis in peanut (Sogut et al. 2016; Andersen and Gorbet 2002). Moreover, the temperature in the glasshouse was set between 30 and 32 °C during the experiment period, which seems responsible for the lower linoleic acid content, due to the poor desaturase activity (Chaiyadee et al. 2013; Dwivedi et al. 1996; Golombek et al. 1995). This appears one of the very relevant reasons for peanuts generally showing higher O/L ratio when grown in warmer climatic conditions. Accordingly, Sun et al. (2014) have also reported a significant decrease in O/L ratio of HO cultivars under lower temperature, but in normal oleate cultivars, the corresponding decrease was not significant. It means there are certainly some other factors regulating the O/L ratio, along with the temperature. Thus, considering the influence of various abiotic factors on O/L flux underlines the importance of the selection of well-characterized parental lines and cultural practices according to the area so that it will minimize the environment-dependent negative modifications in the oil composition including O/L ratio (Nawade et al. 2018).
The variations in the oleic (67.04–82.33%) and linoleic acid (1.74–15.66%) contents in different ILs even in the presence of homozygous ahFAD2 mutant alleles could also be due to the presence of some modifying genes and/or ahFAD2 gene families (Nawade et al. 2016; Janila et al. 2016; Wang et al. 2015c). Recently, Wang et al. (2015c) reported six novel members of the ahFAD2 gene family in peanut with varying expression in different plant parts and ahFAD2-1 showed the highest expression. Besides, they also predicted the presence of more candidate genes controlling the oleate levels in developing seeds and/or presence of complex gene networks controlling the fluxes between the endoplasmic reticulum and the chloroplast within the peanut cells. The peanut whole genome sequence data also revealed the presence FAD2 gene family consisting of two genes from A. duranensis and four from A. ipaensis (https://peanutbase.org/). The availability of peanut genome sequence and identification of different ahFAD2 gene families is expediting the research for detailed understanding of the O/L flux of HO peanut genotypes (Nawade et al. 2018).
Conclusions
In this age of capitalism and globalization, the role of edible oils and fats in health and related issues continues to evolve as further knowledge is gained about the significant interplay between health and dietary fats, FAs and chronic diseases (Huth et al. 2015). HO oils are increasingly demanded as a value-added product with wide applicability across industries. We have successfully developed high oleic ILs in the background of foliar fungal disease-resistant cultivar GPBD-4. The ILs lines identified through MAS performed excellently well in two different growth conditions for HO content. These improved lines could be the potential breeding material for further HO breeding programs and can also be released as a variety. These ILs having customized FA profile with lower SFA and higher MUFA contents are health friendly and simultaneously it will expand the applicability of peanut oil in different industries.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1 Pod, kernel and plant features of the parents and a few introgression lines along with the fatty acid analysis peaks. (TIFF 13526 kb)
Acknowledgements
Financial support received from Indian Council of Agricultural Research (ICAR), New Delhi, India, and Science and Engineering Board (SERB) of Department of Science and Technology, New Delhi, India for the project SERB/LS-50/2013 is gratefully acknowledged.
References
- Abiodun Olufunmilola A. Oilseed Crops. Chichester, UK: John Wiley & Sons, Ltd; 2017. The role of oilseed crops in human diet and industrial use; pp. 249–263. [Google Scholar]
- Addinsoft XLSTAT (2017) Data analysis and statistics with Microsoft Excel Paris France MacOS ed. 2017
- Andersen PC, Gorbet DW. Influence of year and planting date on fatty acid chemistry of high oleic acid and normal peanut genotypes. J Agric Food Chem. 2002;50:1298–1305. doi: 10.1021/jf0113171. [DOI] [PubMed] [Google Scholar]
- Andersen PC, Hill K, Gorbet DW, Brodbeck BV. Fatty acid and amino acid profiles of selected peanut cultivars and breeding lines. J Food Compost Anal. 1998;11:100–111. doi: 10.1006/jfca.1998.0565. [DOI] [Google Scholar]
- Banks DJ. Hybridization of peanuts in growth chambers. Peanut Sci. 1976;3:66–69. doi: 10.3146/i0095-3679-3-2-3. [DOI] [Google Scholar]
- Barkley NA, Chamberlin KD, Wang ML, Pittman RN. Development of a real-time PCR genotyping assay to identify high oleic acid peanuts (Arachis hypogaea L.) Mol Breed. 2010;25:541–548. doi: 10.1007/s11032-009-9338-z. [DOI] [Google Scholar]
- Barkley NA, Wang ML, Pittman R. A real-time PCR genotyping assay to detect FAD2A SNPs in peanuts (Arachis hypogaea L.) Electron J Biotechnol. 2011;14:1–12. doi: 10.2225/vol14-issue1-fulltext-12. [DOI] [Google Scholar]
- Bera SK, Kamdar JH, Kasundra SV, Dash P, Maurya AK, Jasani MD, et al. Improving oil quality by altering levels of fatty acids through marker-assisted selection of ahfad2 alleles in peanut (Arachis hypogaea L.) Euphytica. 2018;214:162. doi: 10.1007/s10681-018-2241-0. [DOI] [Google Scholar]
- Cao S, Zhu QH, Shen W, Jiao X, Zhao X, Liu L, et al. Comparative profiling of miRNA expression in developing seeds of high linoleic and high oleic safflower (Carthamus tinctorius L.) plants. Front Plant Sci. 2013;4:489. doi: 10.3389/fpls.2013.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyadee S, Jogloy S, Songsri P, Singkham N, Vorasoot N, Sawatsitang P, et al. Soil moisture affects fatty acids and oil quality parameters in peanut. Int J Plant Prod. 2013;7:81–96. [Google Scholar]
- Chamberlin KD, Melouk HA, Madden R, Dillwith JW, Bannore Y, Rassi ZE, et al. Determining the oleic/linoleic acid ratio in a single peanut seed: a comparison of two methods. Peanut Sci. 2011;38:78–84. doi: 10.3146/PS11-3.1. [DOI] [Google Scholar]
- Chen ZB, Wang ML, Barkley NA, Pittman RN. A simple allele-specific PCR assay for detecting FAD2 alleles in both A and B genomes of the cultivated peanut for high-oleate trait selection. Plant Mol Biol Rep. 2010;28:542–548. doi: 10.1007/s11105-010-0181-5. [DOI] [Google Scholar]
- Chu Y, Ramos L, Holbrook CC, Ozias-Akins P. Frequency of a loss-of-function mutation in oleoyl-PC desaturase (ahFAD2A) in the mini-core of the US peanut germplasm collection. Crop Sci. 2007;47:2372–2378. doi: 10.2135/cropsci2007.02.0117. [DOI] [Google Scholar]
- Chu Y, Holbrook CC, Ozias-Akins P. Two alleles of ahFAD2B control the high oleic acid trait in cultivated peanut. Crop Sci. 2009;49:2029–2036. doi: 10.2135/cropsci2009.01.0021. [DOI] [Google Scholar]
- Chu Y, Wu CL, Holbrook CC, Tillman BL, Person G, Ozias-Akins P. Marker assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome. 2011;4:110–117. doi: 10.3835/plantgenome2011.01.0001. [DOI] [Google Scholar]
- Chu Y, Wu CL, Holbrook CC, Ozias-Akins P. Conditions that impact artificial hybridization of Arachis hypogaea L. Peanut Sci. 2016;43:106–115. doi: 10.3146/PS16-11.1. [DOI] [Google Scholar]
- Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea L.) BMC Plant Biol. 2008;8:55–65. doi: 10.1186/1471-2229-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JP, Sweigart DS, Price KM, Dean LL, Sanders TH. Refractive index and density measurements of peanut oil for determining oleic and linoleic acid contents. J Am Oil Chem Soc. 2013;90:199–206. doi: 10.1007/s11746-012-2153-4. [DOI] [Google Scholar]
- Derbyshire EJ. A review of the nutritional composition organoleptic characteristics and biological effects of the high oleic peanut. Int J Food Sci Nutr. 2014;65:781–790. doi: 10.3109/09637486.2014.937799. [DOI] [PubMed] [Google Scholar]
- Dwivedi SL, Nigam SN, Nageswara Rao RC, Singh U, Rao KVS. Effect of drought on oil fatty acids and protein contents of groundnut (Arachis hypogaea L.) seeds. Field Crops Res. 1996;48:125–133. doi: 10.1016/s0378-4290(96)01027-1. [DOI] [Google Scholar]
- FAOSTAT (2016). http://faostat.fao.org/. Accessed 23 Nov 2018
- Golombek SD, Sridhar R, Singh U. Effect of soil temperature on seed composition of three Spanish cultivars of groundnut (Arachis hypogaea L.) J Agric Food Chem. 1995;43:2067–2070. doi: 10.1021/jf00056a021. [DOI] [Google Scholar]
- Gorbet DW, Knauft DA. Registration of ‘SunOleic95R’ peanut. Crop Sci. 1997;37:1392. doi: 10.2135/cropsci2000.0032rcv. [DOI] [Google Scholar]
- Gowda MVC, Motagi BN, Naidu GK, Diddimani SB, Sheshagiri R. GPBD 4: a Spanish bunch groundnut genotype resistant to rust and late leaf spot. Int Arachis Newsl. 2002;22:29–32. [Google Scholar]
- Harvey KA, Walker CL, Xu Z, Whitley P, Pavlina TM, Hise M, et al. Oleic acid inhibits stearic acid-induced inhibition of cell growth and pro-inflammatory responses in human aortic endothelial cells. J Lipid Res. 2010;51:3470–3480. doi: 10.1194/jlr.M010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds MJ. Fatty acid composition of Caribbean-grown peanut (Arachis hypogaea L.) at three maturity stages. Food Chem. 1995;53:7–14. doi: 10.1016/0308-8146(95)95779-6. [DOI] [Google Scholar]
- Huth PJ, Fulgoni VL, III, Larson BT. A systematic review of high-oleic vegetable oil substitutions for other fats and oils on cardiovascular disease risk factors: implications for novel high-oleic soybean oils. Adv Nutr. 2015;6:674–693. doi: 10.3945/an.115.008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isleib TG, Tillman BL, Pattee HE, Sanders TH, Hendrix KW, Dean LO. Genotype-by-environment interactions for seed composition traits of breeding lines in the uniform peanut performance test. Peanut Sci. 2008;35:130–138. doi: 10.3146/PS08-001.1. [DOI] [Google Scholar]
- Janila P, Pandey MK, Shasidhar Y, Variath MT, Sriswathi M, Khera P, et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016;242:203–213. doi: 10.1016/j.plantsci.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Jung S, Powell G, Moore K, Abbott A. The high oleate trait in the cultivated peanut (Arachis hypogaea L.) II. Molecular basis and genetics of the trait. Mol Gen Genet. 2000;263:806–811. doi: 10.1007/s004380000243. [DOI] [PubMed] [Google Scholar]
- Jung S, Swift D, Sengoku E, Patel M, Teule F, Powell G, et al. The high oleate trait in the cultivated peanut [Arachis hypogaea L.]. I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases. Mol Gen Genet. 2000;263:796–805. doi: 10.1007/s004380000244. [DOI] [PubMed] [Google Scholar]
- Kale DM, Mouli C. Hybridization technique in groundnut. Indian J Genet Plant Breed. 1984;44:379–384. [Google Scholar]
- Khera P, Pandey MK, Wang H, Feng S, Qiao L, Culbreath AK, et al. Mapping quantitative trait loci of resistance to tomato spotted wilt virus and leaf spots in a recombinant inbred line population of peanut (Arachis hypogaea L.) from SunOleic97R and NC94022. PLoS One. 2016;11:e0158452. doi: 10.1371/journal.pone.0158452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauft DA, Moore KM, Gorbet DW. Further studies on the inheritance of fatty acid composition in peanut. Peanut Sci. 1993;20:74–76. doi: 10.3146/i0095-3679-20-2-2. [DOI] [Google Scholar]
- Koilkonda Padmalatha, Kuwata Chikara, Fukami Masanobu, Shirasawa Kenta, Aoki Koh, Tabata Satoshi, Hasegawa Makoto, Kiyoshima Hiroyuki, Suzuki Shigeru, Sasamoto Shigemi, Kurabayashi Atsushi, Tsuruoka Hisano, Wada Tsuyuko, Isobe Sachiko. Translational Genomics for Crop Breeding. Chichester, UK: John Wiley & Sons Ltd; 2013. Marker-Assisted Backcrossing Selection for High O/L Ratio in Cultivated Peanut; pp. 177–191. [Google Scholar]
- Lim ES, Surjit S, Amartalingam (1980) A reproductive efficiency of groundnuts. In: Proc. legumes in the tropics. pp 87–96
- Lim HJ, Hines ZG, Dominy JE, Lee Y, Kim S, Tabata M, et al. Oleic acid stimulates complete oxidation of fatty acids through PKA-dependent activation of SIRT1/PGC1 complex. J Biol Chem. 2013;288:7117–7126. doi: 10.1074/jbc.M112.415729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Y, Nadaf HL, Smith OD, Connell JP. Isolations and characterization of the ∆12 fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphism for the high oleate trait in Spanish market-type lines. Theor Appl Genet. 2000;101:1131–1138. doi: 10.1007/s001220051589. [DOI] [Google Scholar]
- Mienie CMS, Pretorius AE. Application of marker-assisted selection for ahFAD2A and ahFAD2B genes governing the high-oleic acid trait in South African groundnut cultivars (Arachis hypogaea L.) Afr J Biotechnol. 2013;12:4283–4289. doi: 10.5897/ajb2012.2976. [DOI] [Google Scholar]
- Mishra GP, Radhakrishnan T, Kumar A, Thirumalaisamy PP, Kumar N, Bosamia TC, et al. Advancements in molecular marker development and their applications in the management of biotic stresses in peanuts. Crop Prot. 2015;77:74–86. doi: 10.1016/j.cropro.2015.07.019. [DOI] [Google Scholar]
- Misra JB, Mathur RS. A simple and economic procedure for transmethylation of fatty acids of groundnut oil for analysis by GLC. Int Arachis Newsl. 1998;18:40–42. [Google Scholar]
- Mondal S, Badigannavar AM, Dsouza SF. Induced variability for fatty acid profile and molecular characterization of high oleate mutant in cultivated groundnut (Arachis hypogaea L.) Mol Breed. 2011;130:242–247. doi: 10.1111/j.1439-0523.2010.01787.x. [DOI] [Google Scholar]
- Mukri G, Nadaf HL, Bhat RS, Gowda MVC, Upadhyaya HD, Sujay V. Phenotypic and molecular dissection of ICRISAT mini core collection of peanut (Arachis hypogaea L.) for high oleic acid. Plant Breed. 2012;131:418–422. doi: 10.1111/j.1439-0523.2012.01970.x. [DOI] [Google Scholar]
- Nawade B, Bosamia TC, Thankappan T, Rathnakumar AL, Kumar A, Dobaria JR, et al. Insights into the Indian peanut genotypes for ahFAD2 gene polymorphism regulating its oleic and linoleic acid fluxes. Front Plant Sci. 2016;7:1271. doi: 10.3389/fpls.2016.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawade B, Mishra GP, Radhakrishnan T, Dodia SM, Ahmad S, Kumar A, et al. High oleic peanut breeding: achievements perspectives and prospects. Trends Food Sci Technol. 2018;78:107–119. doi: 10.1016/j.tifs.2018.05.022. [DOI] [Google Scholar]
- Nigam SN (2014) Groundnut at a glance, pp 121. ICRISAT, Patancheru. http://oar.icrisat.org/8455/
- Nigam SN, Vasudeva Rao MJ, Gibbons RW (1990) Artiicial hybridization in groundnut. Information bulletin no. 29. International Crops Research Institute for the Semi-Arid Tropics, Patancheru, AP-502 324, India. http://oar.icrisat.org/959/1/RA_00166.pdf
- Norden AJ. Peanut. In: Fehr WR, Hadley HH, editors. Hybridization of crop plants. Madison: American Society of Agronomy and Crop Science Society of America; 1980. pp. 443–456. [Google Scholar]
- Norden AJ, Rodriguez VA. Artifical hybridization of peanuts. Oleagineux. 1971;26:159–162. [Google Scholar]
- Norden AJ, Gorbet DW, Knauft DA, Young CT. Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci. 1987;14:7–11. doi: 10.3146/i0095-3679-14-1-3. [DOI] [Google Scholar]
- O’Keefe SF, Wiley VA, Knauft DA. Comparison of oxidative stability of high and normal oleic peanut oils. J Am Oil Chem Soc. 1993;70:489–492. doi: 10.1007/BF02542581. [DOI] [Google Scholar]
- Othman H (1979) Reproductive efficiency of groundnuts (Arachis hypogaea L.). Project Pap Uniy Pertanian Malaysi, vol l, 1978/79
- Pandey MK, Wang ML, Qiao L, Feng S, Khera P, Wang H, et al. Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.) BMC Genet. 2014;15:133. doi: 10.1186/s12863-014-0133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil AS, Hedvat I, Levy Y, Galili S, Hovav R. Genotype-by-environment effects on the performance of recombinant inbred lines of virginia-type peanut. Euphytica. 2018;214:83. doi: 10.1007/s10681-018-2159-6. [DOI] [Google Scholar]
- Radhakrishnan T, Murthy TGK, Bandopadhyay A. Multiple shoot induction in groundnut (Arachis hypogaea L.) In: Srivastava GC, Singh K, Pal M, editors. Plant physiology for agriculture. Jaipur: Pointer Publishers; 1999. pp. 433–439. [Google Scholar]
- Sarvamangala C, Gowda MVC, Varshney RK. Identification of quantitative trait loci for protein content oil content and oil quality for groundnut (Arachis hypogaea L.) Field Crop Res. 2011;122:49–59. doi: 10.1016/j.fcr.2011.02.010. [DOI] [Google Scholar]
- Singkham N, Jogloy S, Kesmala T, Swatsitang P, Jaisil P, Puppala N. Genotypic variability and genotype by environment interactions in oil and fatty acids in high intermediate and low oleic acid peanut genotypes. J Agric Food Chem. 2010;58:6257–6263. doi: 10.1021/jf903728e. [DOI] [PubMed] [Google Scholar]
- Sogut T, Ozturk F, Kizil S. Effect of sowing time on peanut (Arachis hypogaea L.) cultivars: II. Fatty acid composition. Agric Agric Sci Procedia. 2016;10:76–82. doi: 10.1016/j.aaspro.2016.09.018. [DOI] [Google Scholar]
- Sujay V, Gowda MVC, Pandey MK, Bhat RS, Khedikar YP, Nadaf HL, et al. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.) Mol Breed. 2012;30:773. doi: 10.1007/s11032-011-9661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Spears JF, Isleib TG, Jordan DL, Penny B, Johnson D, et al. Effect of production environment on seed quality of normal and high-oleate large seeded Virginia-type peanut (Arachis hypogaea L.) Peanut Sci. 2014;41:90–99. doi: 10.3146/PS12-16.1. [DOI] [Google Scholar]
- Vassiliou EK, Gonzalez A, Garcia C, Tadros JH, Chakraborty G, Toney JH. Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-α both in vitro and in vivo system. Lipids Health Dis. 2009;8:25. doi: 10.1186/1476-511X-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Sukumaran S, Barkley NA, Chen Z, Chen CY, Pittman RN, et al. Population structure and marker–trait association analysis of the US peanut (Arachis hypogaea L.) mini-core collection. Theor Appl Genet. 2011;123:1307–1317. doi: 10.1007/s00122-011-1668-7. [DOI] [PubMed] [Google Scholar]
- Wang ML, Chen CY, Tonnis B, Barkley NA, Pinnow DL, Pittman RN, et al. Oil fatty acid flavonoid and resveratrol content variability and FAD2A functional SNP genotypes in the U.S. peanut mini-core collection. J Agric Food Chem. 2013;61:2875–2882. doi: 10.1021/jf305208e. [DOI] [PubMed] [Google Scholar]
- Wang ML, Khera P, Pandey MK, Wang H, Qiao L, Feng S, et al. Genetic mapping of QTLs controlling fatty acids provided insights into the genetic control of fatty acid synthesis pathway in peanut (Arachis hypogaea L.) PLoS One. 2015;10(4):e0119454. doi: 10.1371/journal.pone.0119454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Pinnow D, Tonnis B, Tishchenko V, An YQC, Pederson GA. Newly identified natural high-oleate mutant from Arachis hypogaea L. subsp. hypogaea. Mol Breed. 2015;35:186. doi: 10.1007/s11032-015-0377-3. [DOI] [Google Scholar]
- Wang Y, Zhang X, Zhao Y, Prakash CS, He G, Yin D. Insights into the novel members of the FAD2 gene family involved in high-oleate fluxes in peanut. Genome. 2015;58:375–383. doi: 10.1139/gen-2015-0008. [DOI] [PubMed] [Google Scholar]
- Yu S, Pan L, Yang Q, Min P, Ren Z, Zhang H. Comparison of the delta (12) fatty acid desaturase gene between high-oleic and normal-oleic peanut genotypes. J Genet Genom. 2008;35:679–685. doi: 10.1016/s1673-8527(08)60090-9. [DOI] [PubMed] [Google Scholar]
- Yu HT, Yang WQ, Tang YY, Wang XZ, Wu Q, Hu DQ, et al. An AS-PCR assay for accurate genotyping of FAD2A/FAD2B genes in peanuts (Arachis hypogaea L.) Grasas Aceites. 2013;64:395–399. doi: 10.3989/gya.118712. [DOI] [Google Scholar]
- Zhao S, Li A, Li C, Xia H, Zhao C, Zhang Y, et al. Development and application of KASP marker for high throughput detection of AhFAD2 mutation in peanut. Electron J Biotechnol. 2017;25:9–12. doi: 10.1016/j.ejbt.2016.10.010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Pod, kernel and plant features of the parents and a few introgression lines along with the fatty acid analysis peaks. (TIFF 13526 kb)