Abstract
Background
Intra-tumor heterogeneity implies that sub-populations of cancer cells that differ in genetic, phenotypic, or behavioral characteristics coexist in a single tumor 1,2. Tumor heterogeneity drives progression, metastasis and treatment resistance, but its relationship with tumor infiltrating immune cells is a matter of debate where some argue that tumors with high heterogeneity may generate neo-antigens that attract immune cells, and the others claim that immune cells provide selection pressure that shapes tumor heterogeneity 3,4. Here we sought to study the association between tumor heterogeneity and immune cells in a real-world cohort utilizing The Cancer Genome Atlas (TCGA).
Methods
Mutant Allele Tumor Heterogeneity (MATH) was calculated to estimate intra-tumoral heterogeneity, and immune cell compositions were estimated by CIBERSORT. Survival analyses were demonstrated by Kaplan Meir curves.
Results
Tumors with high heterogeneity (High MATH) associated with worse overall survival (p=0.049) as well as ER+ (p=0.011) and non-triple negative tumors (p=0.01). High MATH tumors associated with less infiltration of anti-tumor CD8 (p<0.013) and CD4 T cells (p<0.00024), more tumor promoting regulatory T cells (p<4e-04), lower expression of T cell exhaustion markers; PDL-1 (p=0.0031), IDO2 (p=0.34), ADORA2A (p=0.018), VISTA (p=0.00013), and CCR4 (p<0.00001), lower expression of cytolytic enzymes granzyme-A (p=0.0056) and perforin 1 (p=0.053) as well as low cytolytic activity score (p=0.0028).
Conclusions
High heterogeneity tumors are associated with less immune cell infiltration, less activation of the immune response, and worse survival in breast cancer. Our results support the notion that tumor heterogeneity is shaped by selection pressure of tumor infiltrating immune cells.
INTRODUCTION
Breast cancer is a heterogeneous disease with genetic and phenotypic variability. Two breast cancers that appear to be similar based on clinical, pathologic, and biomarker signatures can behave differently because of differences in underlying biology. Recently it has been suggested that tumor biology, at least in part, may be determined by intra-tumor heterogeneity 3. Intra-tumor heterogeneity implies that subpopulations of cancer cells that differ in their genetic, phenotypic, or behavioral characteristics coexist within the same tumor 1,2 Tumor heterogeneity has been linked to cancer progression and therapeutic resistance 1,2,4–6. Evaluation of intra-tumor heterogeneity of individual tumors, which impacts disease progression and efficacy of therapies, is essential to overcome treatment challenges of the primary tumors and subsequent metastasis in breast cancer.
Mutant-allele tumor heterogeneity (MATH) is a bioinformatic algorithm that provides a measurable and quantitative assessment of intra-tumor heterogeneity, which was generated from whole-exome sequencing of tumors and their matched normal DNA 1. The prognostic impact of MATH has been studied in a variety of cancers, such as head and neck, colorectal, and breast cancer cohorts 1,7,8. Mroz et. al analyzed next generation sequencing results for 74 head and neck squamous cell cancers and found that MATH was higher in tumors with a mutated TP53 gene 7. In a retrospective analysis of head and neck squamous cancers in The Cancer Genome Atlas (TCGA), a relationship was found between high MATH and worse overall survival 9. Rajput et al. used the next-generation sequencing approach to analyze mutations in stage II and III colon cancer patient samples and found a strong correlation between higher MATH and risk of metastases 8. In addition, Ma et al. found that higher stage, triple negative breast cancer, and p53 mutations were associated with higher MATH in breast cancer patients 1. Taken together, these studies suggest that intra-tumor heterogeneity is associated with worse cancer outcomes and has prognostic relevance.
Immune surveillance of cancer is important to suppress tumor growth, progression, and metastasis. Tumors evolve as a result of genomic instability leading to dominant mutations, which can drive cancer progression 3,10. Genetic heterogeneity is suggested to generate neo-antigens that attract immune cells. It has been shown that clonal neo-antigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade 3. PDL-1, the programmed death ligand, is an immune checkpoint protein for T cells. Specifically, CD8+ tumor-infiltrating lymphocytes (TILs) reactive to clonal neo-antigens were identified in early-stage non–small cell lung cancer, expressed high levels of PD-1, and tumors that expressed more clonal neo-antigens were more sensitive to PD-1 blockade 3. Alternatively, there is evidence that TILs themselves function as a selective pressure resulting in tumor clonality and intra-tumor heterogeneity 3. In the first model, high heterogeneity tumors should attract high number of TILs with high cytolytic activity, as opposed to the latter model which should have less TILs with low cytolytic activity because high heterogeneity should be the consequence.
To our knowledge, the relationship between intra-tumor heterogeneity and the immunogenic landscape in breast cancer has not been fully studied in a large cohort of patients. The current study sought to identify the association of tumor heterogeneity and infiltrating immune cell compositions as well as its cytolytic activity, immune response genes, and survival in breast cancer. We hypothesized that high intra-tumor heterogeneity calculated by the MATH algorithm associated with low infiltrating immune cells, low cytolytic activity, and worse survival.
METHODS
Patient Cohort
The Cancer Genome Atlas (TCGA), which is a project supervised by the National Cancer Institute (NCI) and the National Human Genome Research Institute, is a publicly available database that includes clinical and genomic data on breast cancer patient samples collected worldwide 11. Many data parameters are included in the TCGA dataset. However, grade is not included. Therefore, we discovered that a majority of the grade data of the patient tumors used in TCGA are available in TIES database. We reviewed all pathology reports in TIES and associated the grade data to the tumor in TCGA. The primary endpoint was overall survival (OS), defined as the time from date of diagnosis to death by any cause. There are 1093 patients with mRNA expression from RNA sequence in the breast cancer cohort (BRCA) of TCGA-Provisional cohort. Out of 1093, overall survival data were available in 959 patients. The gene expression level quantification data (mRNA expression z-score from RNA sequence) for TCGA cohort was downloaded through cBioPortal 12,13 and used as previously described 14–22.
MATH
The MATH level was calculated from the median absolute deviation (MAD) and the median of its mutant-allele fractions at tumor-specific mutated loci as described previously 7,23.
Survival Analysis
The overall survival between MATH high and low tumors were analyzed using Kaplan-Meier curves with log-rank test. The patients were divided based on degree of heterogeneity as calculated by the MATH algorithm. The threshold of dichotomization of MATH high and low groups was determined by comparing differences in the overall survival between the two groups at multiple candidate cutoff points within the range of MATH, which is similar to the method previously published 24–27.
CIBERSORT
CIBERSORT, a bioinformatic algorithm to calculate immune cell composition from their gene expression profiles, was used to estimate tumor infiltrating cell composition in tumors 28. Immune cell fraction data was downloaded through TCIA (https://tcia.at/home) 29. Each immune cell fraction was compared between MATH high and low tumors in TCGA cohort using same cutoff of overall survival analysis.
Statistical Analysis
All statistical analyses were performed using R software (http:///www.r-project.org/) and Bioconductor (http://bioconductor.org/). The Kaplan-Meier method with log-rank tests and Cox proportional hazards models were used to compare MRS-BM high and low groups. Pearson correlations were calculated based on expression levels of each interested genes and plotted. Gene expression comparison was analyzed using a Student t-test, immune cell fraction comparison was analyzed using Wilcoxon signed-rank test. In all analysis, a two-sided p<0.05 was considered as statistically significant.
RESULTS
Patient and tumor characteristics
Tumor characteristics of low and high MATH groups were overall similar. The majority of patients in both groups was less than 60 years of age, and had Stage II, ER positive, PR positive, HER-2 negative breast cancers. Factors found to be statistically significant included ER, PR, and triple negative breast cancer (TNBC) status (p<=0.0002, p<0.0001, p<0.0003 respectively), stage (p=0.0354), tumor size (p=0.0001), and grade (p=0.0002). Specifically, ER negative, PR negative, and TNBC were more associated with high tumor heterogeneity (Table 1). High MATH tumors were associated with T2 status and N2 nodal positivity. High MATH tumors were also found to have higher grades. The grade of breast carcinoma is a prognostic factor, and different grades have different profiles by proteomic, and genomic analysis 30.
Table 1.
Patient demographics characterized by MATH level in TCGA breast cancer cohort.
| Factor | MATH |
p-value | ||
|---|---|---|---|---|
| Low (n) | High (n) | |||
| Age | ≤60 | 222 | 288 | |
| >60 | 184 | 253 | 0.658 | |
| ER | negative | 66 | 140 | |
| positive | 325 | 376 | 0.0002 | |
| PR | negative | 99 | 195 | |
| positive | 291 | 319 | <0.0001 | |
| HER2 | negative | 340 | 432 | |
| positive | 58 | 92 | 0.2239 | |
| TNBC | no | 361 | 435 | |
| yes | 46 | 108 | 0.0003 | |
| Stage | I | 86 | 76 | |
| II | 218 | 330 | ||
| III | 94 | 122 | ||
| IV | 6 | 8 | 0.0354 | |
| T-Stage | 1 | 134 | 118 | |
| 2 | 215 | 338 | ||
| 3 | 54 | 65 | ||
| 4 | 8 | 27 | 0.0001 | |
| N-stage | negative | 187 | 268 | |
| positive | 218 | 272 | 0.3238 | |
| Grade | 1 | 41 | 27 | |
| 2 | 116 | 113 | 0.0002 (chi-square test) | |
| 3 | 69 | 128 | <0.0001 (Cochran-Armitage trend test) | |
High tumor heterogeneity is associated with worse survival in hormone positive breast cancer
Previous studies demonstrated that patients with highly heterogeneous cancers have worse survival 31. We sought to explore this association further in TCGA breast cancer cohort. We found that high MATH tumors had a higher mutation burden, higher KI-67 gene expression, and more intra-tumor heterogeneity (Fig. 1A). This correlated to high MATH tumors having worse overall survival compared to low MATH tumors (MATH high, n=548; low, n=411; p=0.0499) (Fig.IB). Subgroup analysis revealed that this survival disadvantage in the high MATH group was statistically significant for ER-positive (MATH high, n=376; low, n=325; p=0.011), PR-positive (MATH high, n=319; low, n=291; p=0.024), and non-triple negative breast cancers (MATH high, n=435; low, n=361; p=0.01) (Fig. IC). This result is in agreement with the previous reports and validates that TCGA cohort is similar to the previous cohorts.
Figure I:
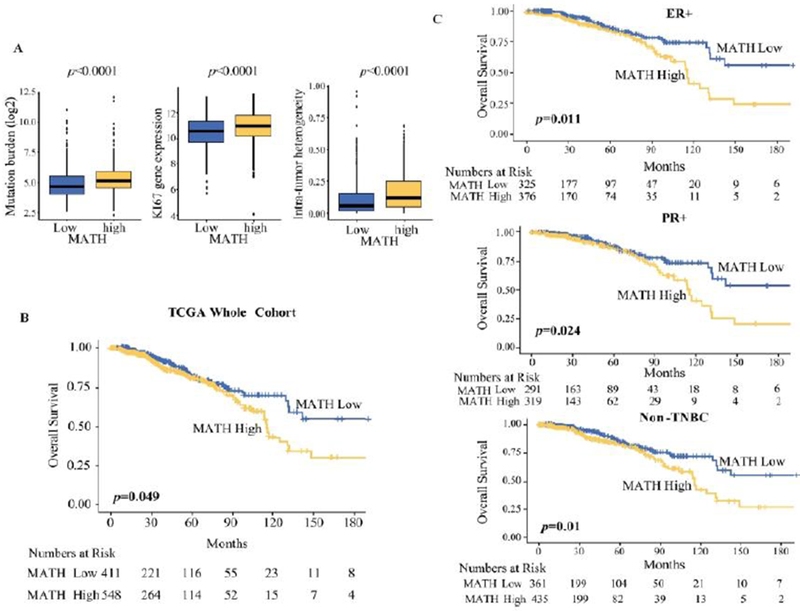
MATH level and patient survival in TCGA breast cancer cohort. (A) Box plots correlating MATH to mutation burden, KI-67 gene expression, and intra-tumor heterogeneity. (B) MATH level and overall survival in the TCGA whole cohort. (C) MATH level and patient survival for estrogen receptor (ER)-positive, progesterone receptor (PR)-positive, and non-triple negative breast cancers (non-TNBC).
Tumors with high heterogeneity are associated with less immune cell infiltration
We assessed immune cell composition in TCGA cohort and studied its association with tumor heterogeneity. Each immune cell fraction was compared between MATH high and low tumors using CIBERSORT. High MATH tumor associated with significantly lower fractions of activated CD8+ T cells (p=0.013), B cells (p=0.0016), dendritic cells (p=0.0019), activated memory CD4+ T cells (p=0.0002), and mast cells (p=0.0007), which all play anti-tumor roles in breast cancer (Fig.II). Interestingly, high MATH tumor also associated with significantly higher fractions of immunosuppressive regulatory T cells (Tregs) (p=<0.0001) compared to tumors with low MATH (Fig.II). Our finding that tumors with high heterogeneity have not only less anti-tumor immune cells but also more immunosuppressive Tregs is in agreement with the notion that tumor heterogeneity is multifactorial with immune cells being an important component.
Figure II:
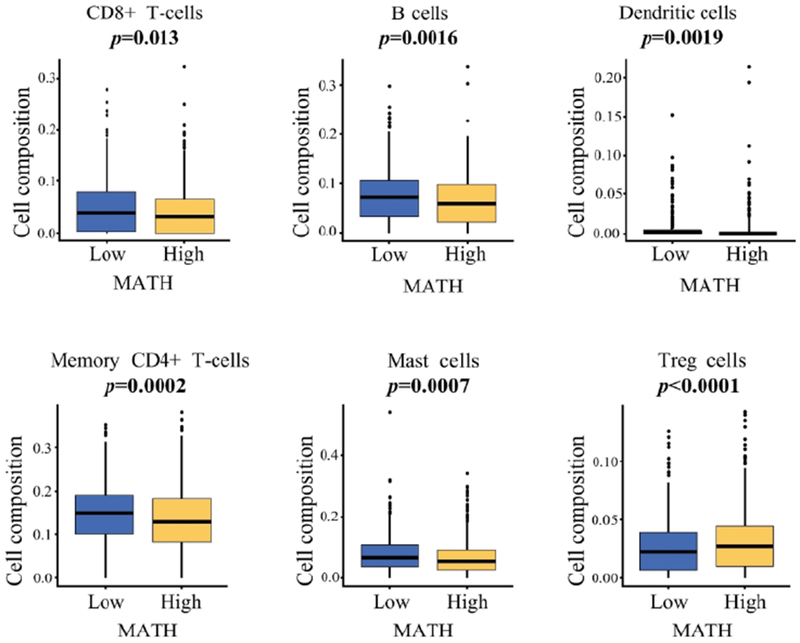
MATH level and immune cell composition by CIBERSORT. Cell composition fraction comparison of activated memory CD8 T cells, B cells, dendritic cells, memory CD4 cells, mast cells, and TREG cells between MATH high and low expression in TCGA breast cancer cohort.
Tumors with high heterogeneity are associated with decreased immune response genes
Immune checkpoint molecules are also known as T cell exhaustion markers and reflect immunogenicity, i.e. their expression levels reflect the existence of humoral or cell mediated immune responses. We found that high MATH tumor associated with significantly lower mRNA expression of T cell exhaustion markers, specifically PDL-1 (p=0.0031), IDO2 (Indoleamine 2,3-dioxygenase 2) (p=0.34), ADORA2A (Adenosine A2a receptor) (p=0.018), VISTA (V-domain immunoglobulin (Ig)-containing suppressor of T-cell activation) (p=0.00013), and CCR4 (C-C chemokine receptor type 4 ) (p<0.00001); (Fig.III). This result is consistent with the previous finding that tumors with high heterogeneity are associated with lower numbers of active immune cell infiltrations.
Figure III:
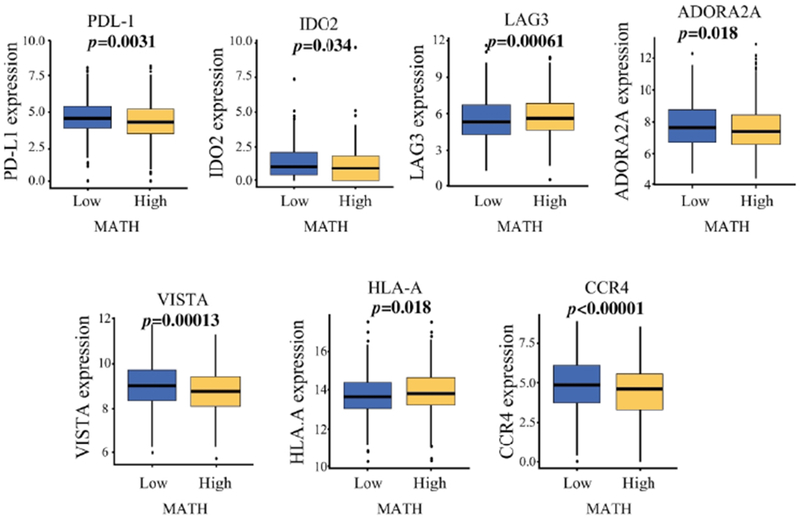
MATH level and expression of immune response genes. mRNA expression of PDL-1 PD-L1 (programmed death ligand), IDO2 (Indoleamine 2, 3-dioxygenase 2), LAG3 (lymphocyte activation gene 3), ADORA2A (Adenosine A2a receptor), VISTA (V-domain immunoglobulin (Ig)-containing suppressor of T-cell activation), HLA-A (human leukocyte antigen-A), and CCR4 (C-C chemokine receptor type 4) proteins in TCGA breast cancer cohort.
Tumors with high heterogeneity are associated with decreased immune cytolytic activity
The interplay of immune system activation and heterogeneity was further investigated by analyzing the expression of immune cytolysis genes as well as measuring the cytolytic activity score (CYT). High MATH tumors associated with significantly lower expression of GZMA (granzyme-A) (p=0.0056) and PRF-1 (perforin 1) (p=0.053) in TCGA breast cancer cohort, both of which are key genes of immune cytolysis. Indeed, CYT was also significantly lower in high MATH tumors consistent with less cytolitic immune activity (p=0.0028) (Fig. IV). This finding further supports the notion that when there are fewer active immune cell infiltration and less cytolytic activity, the tumor is allowed to clonally evolve and thus develops heterogeneity.
Figure IV:
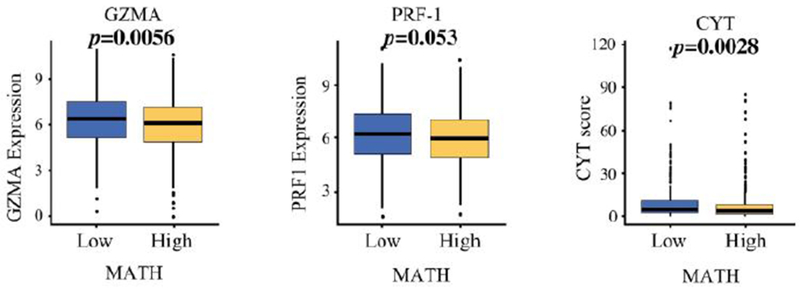
MATH level and expression of cytolysis proteins. Box plot of mRNA expression of GZMA (granzyme-A), PRF-1 (perforin 1), and CYT (cytolysis) protein in high and low MATH tumors in TCGA breast cancer cohort.
DISCUSSION
Tumor heterogeneity accounts for differences in tumor behavior among individual patients and even within an individual tumor that may not be explained by clinical and pathologic features alone. Currently the relationship between tumor heterogeneity and immune cells is a matter of debate where some argue that tumor heterogeneity generates neo-antigens that attract immune cells 10, and others claim that immune cells function as a selective pressure that controls tumor heterogeneity 3,4. To this end, the current study was aimed at studying the association between tumor heterogeneity and immune cells and its impact on survival. Evaluation of breast tumor heterogeneity has advanced from whole tumor analysis to single cell analysis, allowing examination of more than 90% of the genome in a single cell 32,33. MATH served to stratify patients into low and high risk groups based on a quantitative assessment of their tumor heterogeneity. Based on this study, high MATH tumors are synonymous with high heterogeneity. We show that high heterogeneity correlates with worse prognosis and we show data suggesting that immune system interaction with the tumors plays an important role in shaping the cellular makeup of the tumors.
This study demonstrates that high MATH tumors are more mutation laden, have the ability to be more proliferative, and are more heterogeneous overall (Fig. 1A). The survival analysis in this study is similar to that of Ma et. al where hormone receptor-positive patients with high MATH showed a tendency toward a worse overall survival 1. In general, patients with ER-positive breast cancer are considered to have a better prognosis confounded by the risk of recurrence occurring long after initial treatment. Lordstrom et. al explored heterogeneity among hormone positive tumors by examining the ER intensity level among 1,780 postmenopausal lymph node negative breast cancer patients with and without endocrine therapy 34. They found that patients with low tumor heterogeneity of ER in the tamoxifen treated arm had an excellent 25 year breast cancer specific survival of 88.3%, while patients with high heterogeneity of ER had a 79.6% survival independent of other known tumor markers 34. We have shown that high heterogeneity in ER+ cancers correlates with worse prognosis.
We further explored tumor heterogeneity among HER2 positive and negative breast cancers. This is because a subset of HER2 positive cancers may have also HER2 negative regions, and the subpopulations of cancer cells in these tumors may differ depending on many factors. For instance, tumors tend to lose biomarker expression as they become more poorly differentiated, but it may retain a mix of clones with positive or negative expression depending on distinct genetic alterations. We were unable to find a correlation between heterogeneity and prognosis neither in tumors that were HER2 positive or negative by pathology nor in the PAM50 subgroup (Supplemental Fig.S1).
However, cancer mutation calls may vary depending on the algorithm used for the determination. PyClone is one such algorithm that determines clonal populations in tumors 35. Another statistical model, known as PhyloSub, has the ability to infer the clonal evolution of tumors from single nucleotide somatic mutations 36. THetA (Tumor Heterogeneity Analysis), infers not only the number of clones and clonal alterations but also the most likely collection of genes in those clonal populations using high-throughput DNA sequencing data 37. ABSOLUTE is a method that quantifies tumor heterogeneity directly from analysis of somatic DNA alterations 38. Heterogeneity, as measured by MATH, can be affected by the accuracy of the mutation calls. Together, all of these individual algorithms can determine cellular clonality using genetic information. The application of more than one algorithm in this study may have yielded additional data supporting the association between MATH and survival. Ascertaining detailed information on sub-populations of clones can provide insight into a tumors metastatic potential and possible therapeutic resistance.
We have also shown that tumors with high heterogeneity associate with fewer numbers of immune cell infiltrations. Immune cells may serve as a selective pressure and shape tumor heterogeneity. TILs in ER+ HER2 negative cancers are rare. Gu-Trantien et al. found that 75% of the leukocyte infiltration in breast cancer were T cells, 20% were B cells, <10% were monocytes, and <5% were natural killer cells 39. Given its amount, the association between TILs and tumor heterogeneity was of interest because it could be driving the observed survival differences. Our finding is consistent with those of Morris et. al where breast cancers with high heterogeneity tended to have lower levels of immune cell infiltration 31. We found that high MATH tumor associated with increased tumor promoting Tregs and with less anti-tumor CD4+ and CD8+ T cells as well as mast cells, B cells, and dendritic cells. This result is in agreement with the previous report that mast cells are associated with favorable prognosis in invasive breast cancer 40. Since neo-antigens are detected by immune cells such as dendritic cells and B cells that form the antibodies against those antigens 3, our result does not support the notion that tumors with high heterogeneity provide high levels of neo-antigens that attract immune cells. In fact, TILs have not been shown to be prognostic in other studies. Our data is different because tumors with high heterogeneity are associated with less infiltration of immune cells, less activation of the immune response, and worse overall survival in breast cancer. Thus, supporting the notion that tumor heterogeneity may be shaped by the selection pressure of tumor infiltrating immune cells and this ultimately can influence survival.
Others authors have explored the correlation between heterogeneity and immune cells in TNBC. Some TNBC have T cell related immune signature that is associated with less likelihood to have distant metastasis, and more likely to achieve a pathologic complete response to neoadjuvant chemotherapy 39. We were unable to identify any other report that demonstrated a direct correlation between TILs and heterogeneity in TNBC. Several potentially actionable mutations have been found in TNBC, such as P13k/mTOR, RAS/REF/MEK, however, none have been shown to definitively result in the TNBC phenotype 39.
We have also found that tumors with high heterogeneity have less immune response genes. PDL-1, the programmed death ligand, is an immune checkpoint protein for T cells. IDO2, indoleamine 2, 3-dioxygenase 2, is an enzyme that breaks down tryptophan which results in immunosuppression 42. ADORA2A, adenosine A2a receptor, suppresses CD8+ T cells in an in vitro model of melanoma 43. VISTA (V-domain immunoglobulin (Ig)-containing suppressor of T-cell activation), and CCR4 (C-C chemokine receptor type 4) also play a role in immunosuppression in cancer 44. Furthermore, the notion that tumor heterogeneity is shaped by selection pressure of immune cells was further supported by the demonstration of less cytolytic activity. Perforin/granzyme-induced apoptosis is the main pathway used by cytotoxic lymphocytes to eliminate cancer cells 45. High MATH tumors associated with decreased expression of GZMA and PRF-1; both cytolysis related proteins, as well as less CYT and more cell death.
In conclusion, high MATH tumors have lower levels of effector T cells, lower expression of exhaustion markers, and lower levels of expression of genes associated with T cell cytolitic activity. This data is suggestive that high MATH tumors are associated with a lower, active anti-tumor response and worse overall survival in breast cancer. Our results are consistent with the notion that tumor heterogeneity is at least partially shaped by selection pressure of immune response to the tumor. Further understanding of the molecular, genetic, and cellular changes that influence tumor heterogeneity will allow us to better understand changes leading to high heterogeneity and develop methods to prevent it with resulting improvement of prognosis of patients with breast cancer.
Supplementary Material
Supplemental Figure S1: MATH level and patient survival for HER2-positive and negative breast cancers in the (A) TCGA and (B) PAM50 cohort.
SYNOPSIS.
Intra-tumor heterogeneity is a known mechanism of breast cancer progression and metastasis. We found that it correlates with less immune cell infiltration, less cytolytic activity, and worse survival in breast cancer utilizing computational analyses including Mutant Allele Tumor Heterogeneity (MATH) algorithm.
Acknowledgments
Conflicts of Interest and Source of Funding
Kazuaki Takabe, M.D. is funded by the United States National Institute of Health – National Cancer Institute (R01CA160688) and Susan G. Komen Foundation (Investigator Initiated Research Grant (IIR12222224). This work was also supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Cancer Institute’s Bioinformatics and Biostatistics Shared Resources. Biospecimens or research pathology services for this study were provided by the Pathology Resource Network. Clinical Data Delivery and Honest Broker services for this study were provided by the Clinical Data Network. For the remaining authors none were declared.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Ma D, Jiang YZ, Liu XY, Liu YR, Shao ZM. Clinical and molecular relevance of mutant-allele tumor heterogeneity in breast cancer. Breast Cancer Res Treat. 2017;162(1):39–48. doi: 10.1007/s10549-017-4113-z [doi]. [DOI] [PubMed] [Google Scholar]
- 2.Martelotto LG, Ng CK, Piscuoglio S, Weigelt B, Reis-Filho JS. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014;16(3):210. doi: 10.1186/bcr3658 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell. 2017;168(4):613–628. doi: S0092-8674(17)30066-1 [pii]. [DOI] [PubMed] [Google Scholar]
- 5.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi: 10.1038/nrc3261 [doi]. [DOI] [PubMed] [Google Scholar]
- 6.Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22(1):105–113. doi: 10.1038/nm.3984 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49(3):211–215. doi: 10.1016/j.oraloncology.2012.09.007 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajput A, Bocklage T, Greenbaum A, Lee JH, Ness SA. Mutant-allele tumor heterogeneity scores correlate with risk of metastases in colon cancer. Clin Colorectal Cancer. 2017;16(3):e165–e170. doi: S1533-0028(16)30258-4 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PLOS Medicine Staff. Correction: Intra-tumor genetic heterogeneity and mortality in head and neck cancer: Analysis of data from the cancer genome atlas. PLoS Med. 2015;12(6):e1001844. doi: 10.1371/journal.pmed.1001844 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625 [doi]. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramanathan R, Raza A, Sturgill J, et al. Paradoxical association of postoperative plasma sphingosine-1-phosphate with breast cancer aggressiveness and chemotherapy. Mediators Inflamm. 2017;2017:5984819. doi: 10.1155/2017/5984819 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat. 2017;162(1):191–198. doi: 10.1007/s10549-017-4102-2 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SY, Kawaguchi T, Yan L, Young J, Qi Q, Takabe K. Clinical relevance of microRNA expressions in breast cancer validated using the cancer genome atlas (TCGA). Ann Surg Oncol. 2017;24( 10):2943–2949. doi: 10.1245/s10434-017-5984-2 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018. doi: 10.1245/s10434-018-6506-6 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moro K, Kawaguchi T, Tsuchida J, et al. Ceramide species are elevated in human breast cancer and are associated with less aggressiveness. Oncotarget. 2018;9(28):19874–19890. doi: 10.18632/oncotarget.24903 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi T, Yan L, Qi Q, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Sci Rep. 2017;7(1):15945-017-16112-y. doi: 10.1038/s41598-017-16112-y [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young J, Kawaguchi T, Yan L, Qi Q, Liu S, Takabe K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget. 2017;8(59):99978–99989. doi: 10.18632/oncotarget.21577 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi T, Yan L, Qi Q, et al. Novel MicroRNA-based risk score identified by integrated analyses to predict metastasis and poor prognosis in breast cancer. Ann Surg Oncol. 2018;25(13):4037–4046. doi: 10.1245/s10434-018-6859-x [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi T, Narayanan S, Takabe K. ASO author reflections: “From computer to bedside”: A new translational approach to immunogenomics. Ann Surg Oncol. 2018;25(Suppl 3):846–847. doi: 10.1245/s10434-018-6957-9 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocco JW. Mutant allele tumor heterogeneity (MATH) and head and neck squamous cell carcinoma. Head Neck Pathol. 2015;9(1):1–5. doi: 10.1007/s12105-015-0617-1 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budczies J, Klauschen F, Sinn BV, et al. Cutoff finder: A comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. doi: 10.1371/journal.pone.0051862 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C, Hsieh MK, Chang WY, Chiang AJ, Chen J. Determining the optimal number and location of cutoff points with application to data of cervical cancer. PLoS One. 2017;12(4):e0176231. doi: 10.1371/journal.pone.0176231 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazumdar M, Glassman JR. Categorizing a prognostic variable: Review of methods, code for easy implementation and applications to decision-making about cancer treatments. Stat Med. 2000;19(1):113–132. doi: 10.1002/(SICI)1097-0258(20000115)19:13.0.CO;2-O [pii]. [DOI] [PubMed] [Google Scholar]
- 27.Brondum L, Eriksen JG, Singers Sorensen B, et al. Plasma proteins as prognostic biomarkers in radiotherapy treated head and neck cancer patients. Clin Transl Radiat Oncol. 2017;2:46–52. doi: 10.1016/j.ctro.2017.01.001 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: S2211-1247(16)31709-0 [pii]. [DOI] [PubMed] [Google Scholar]
- 30.Turashvili G, Brogi E. Tumor heterogeneity in breast cancer. Front Med (Lausanne). 2017;4:227. doi: 10.3389/fmed.2017.00227 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris LG, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7(9):10051–10063. doi: 10.18632/oncotarget.7067 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellsworth RE, Blackburn HL, Shriver CD, Soon-Shiong P, Ellsworth DL. Molecular heterogeneity in breast cancer: State of the science and implications for patient care. Semin Cell Dev Biol. 2017;64:65–72. doi: S1084-9521(16)30266-X [pii]. [DOI] [PubMed] [Google Scholar]
- 33.Szulwach KE, Chen P, Wang X, et al. Single-cell genetic analysis using automated microfluidics to resolve somatic mosaicism. PLoS One. 2015;10(8):e0135007. doi: 10.1371/journal.pone.0135007 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindstrom LS, Yau C, Czene K, et al. Intratumor heterogeneity of the estrogen receptor and the long-term risk of fatal breast cancer. J Natl Cancer Inst. 2018. doi: 10.1093/jnci/djx270 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth A, Khattra J, Yap D, et al. PyClone: Statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao W, Vembu S, Deshwar AG, Stein L, Morris Q. Inferring clonal evolution of tumors from single nucleotide somatic mutations. BMC Bioinformatics. 2014;15:35-2105-15-35. doi: 10.1186/1471-2105-15-35 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oesper L, Mahmoody A, Raphael BJ. THetA: Inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol. 2013;14(7):R80-2013-14-7-r80. doi: 10.1186/gb-2013-14-7-r80 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu-Trantien C, Loi S, Garaud S, et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123(7):2873–2892. doi: 10.1172/JCI67428 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajput AB, Turbin DA, Cheang MC, et al. Stromal mast cells in invasive breast cancer are a marker of favourable prognosis: A study of 4,444 cases. Breast Cancer Res Treat. 2008;107(2):249–257. doi: 10.1007/s10549-007-9546-3 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34(3):137–143. doi: 10.1016/j.it.2012.10.001 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cekic C, Linden J. Adenosine A2A receptors intrinsically regulate CD8+ T cells in the tumor microenvironment. Cancer Res. 2014;74(24):7239–7249. doi: 10.1158/0008-5472.CAN-13-3581 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lines JL, Sempere LF, Broughton T, Wang L, Noelle R. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer immunotherapy. Cancer Immunol Res. 2014;2(6):510–517. doi: 10.1158/2326-6066.CIR-14-0072 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapani JA, Smyth MJ. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol. 2002;2(10):735–747. doi: 10.1038/nri911 [doi]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: MATH level and patient survival for HER2-positive and negative breast cancers in the (A) TCGA and (B) PAM50 cohort.


