Abstract
Objective:
To identify commonalities in gene expression data across all ANCA-associated vasculitis (AAV) tissues thus far characterized.
Methods:
Gene expression data were collected from the three AAV tissues thus far characterized (orbit, peripheral leukocytes, and sinus brushings). These data were analyzed to identify commonly expressed genes and disease pathways. The pathways data were adjusted for multiple comparisons using a combined local false discovery rate, which estimates the probability of a false discovery of a given pathway in all three tissues analyzed.
Results:
Only four genes were upregulated in all three tissues—IL1RN, TLR2, SCL11A1, and MMP9. After multiple comparison adjustments, the network pathway analysis revealed 28 pathways associated with all three tissues. The most strongly associated pathway for all three tissues was the neutrophil degranulation pathway (multi-dimensional local false discovery, md-locfdr=1.0×10−12), followed by the osteoclast differentiation (md-locfdr=3.8×10−05), cell surface interactions at the vascular wall (md-locfdr=4.2×10−04), signaling by interleukins (md-locfdr=6.1×10−04), and phagosome (md-locfdr =0.003) pathways. There were no downregulated genes or pathways common to all three tissues.
Conclusion:
This analysis identified individual genes and pathways of disease common to all AAV tissues thus far characterized. The use of a network pathway analysis allowed us to identify pathologic mechanisms that were not readily apparent in the commonly expressed genes alone. Many of these pathways are consistent with current theories about infectious drivers and the crossroads of innate and adaptive immune mechanisms. In addition, this analysis highlights novel pathways, such as vessel wall interactions and platelet activation, which require further investigation.
Keywords: Anti-neutrophil cytoplasmic antibody-associated vasculitis, Vasculitis, Granulomatosis with polyangiitis, Microscopic polyangiitis, Gene expression, Meta-analysis
INTRODUCTION:
Granulomatosis with polyangiitis (GPA) and microscopic polyangiitis (MPA) are life-threatening multi-system vasculitides affecting small to medium sized blood vessels. Both diseases are associated with the anti-neutrophil cytoplasmic antibody (ANCA) and are together referred to as ANCA-associated vasculitis (AAV). In recent years, several studies have characterized the gene expression signature of AAV in affected tissues. Thus far, the gene expression signatures of orbital tissue, peripheral leukocytes, and nasal sinus brushings have been reported. (1-3) The purpose of this study is to compare gene expression data across these three tissues to discover common AAV disease pathways and to validate reported data.
Commonalities in gene expression data can be determined by focusing on individual genes. However, the comparison of pathways can lend additional insight beyond the gene level alone. The comparative pathway analysis approach has been used in meta-analyses of cancers, lupus, rheumatoid arthritis, ankylosing spondylitis, neuropsychiatric disorders, and other diseases. (4-10) Individual genes have multiple functions and can play a role in a large number of different processes. Disease pathways involve many genes that may be utilized at different points in time in different tissues. In the study of systemic lupus, for example, pathway analyses identified the importance of the interferon pathway based on interferon inducible genes, even though the interferons themselves were not upregulated. (9) Through the use of a network analysis system based on knowledge of protein-protein interactions, network pathway analyses allow association of the biologic functions and pathologic mechanisms with a set of expressed genes.
In this analysis, we tested the hypothesis that there are commonly preserved pathways of disease associated with all tissues thus far characterized in AAV. The pathway analysis yielded more robust pathogenic information than the evaluation of commonly expressed individual genes alone. The identified pathways reflect known and plausible mechanisms of AAV disease.
MATERIALS AND METHODS:
Study inclusion criteria: all published gene expression data from studies of patients with AAV compared to controls. Studies were identified through a PubMed search for studies containing at least one AAV term (ANCA, GPA, MPA, or Wegener’s) and at least one gene expression term (gene expression, RNA, or microarray). Ethics and Institutional Review Board approval was not required since this study is a meta-analysis of previously published and de-identified data.
The set of differentially upregulated and then downregulated genes from each study was analyzed by the Cytoscape software application (11, 12) with the Reactome FI plugin (12). This is a pathway based network analysis system, which uses expert-curated pathways based on available knowledge of protein-protein interactions. The 2016 Reactome FI application incudes 1127 pathways collected from Reactome as well as other pathway databases including the National Cancer Institute Pathway Interactions Database (NCI PID), Kyoto Encyclopedia of Genes and Genomes (KEGG), Panther, CellMap, and Biocarta. (13, 14) Pathways associated with upregulated gene lists from each tissue were collected along with (the conventional Benjamini-Hochberg’s global) false discovery rate (FDR) and p-values. (15) However, since there is no existing method to combine three lists of FDRs directly, we chose to estimate the local false discovery rate (locfdr) for pathways of each tissue and multi-dimensional locfdr (md-locfdr) of common pathways of 3 tissues. (16) The locfdr and md-locfdr of pathway lists were estimated using a semiparametric mixture model. (16) A locfdr is an estimate of the probability of individual pathway being a false discovery in that tissue, while the md-locfdr can be thought of as the probability of individual pathway being a false discovery in all three tissues. The locfdr and the conventional FDR are closely related. A locfdr can be defined as locfdr(z) = Probability{null|Z=z}, where Z is a test statistic, and the conventional FDR as FDR(z) = E{fdr(Z)|Z≤z}. That is, FDR(z) is a (weighted) average of locfdr’s, locfdr(Z) for Z ≤z. The technical details of locfdr can be found in Supplement A as well as in Jeong et al. (16) While there is no widely accepted cutoff value for locfdr, Efron suggested that a locfdr of 0.2 could be used, which corresponds to the conventional FDR of 0.05 to 0.15 under some mild conditions. (17) In this study, a locfdr of <0.2 was used as a cutoff. Twenty-eight pathways with an md-locfdr of <0.2 were identified.
Downregulated genes were those with a fold change (FC) value of ≤−1.5 in ANCA samples compared to healthy controls. A conventional FDR cutoff of <0.25 was used to identify pathways associated with the downregulated gene lists. Because very few pathways were identified and none were associated with all three tissues, locfdr was not used for this analysis.
RESULTS:
Three studies were identified that characterized the gene expression of AAV tissues compared to healthy controls. This included studies of peripheral leukocytes (3), sinus brushings (2), and orbital inflammatory disease (1). All three studies used RNA microarray to characterize these tissues. The characteristics of each study are shown in Table 1. Differences in treatment with glucocorticoid and other immunosuppressant medications is not included in the table, however the nasal sinus brushing and orbital inflammatory data were adjusted for medication use. Medication use in the peripheral leukocyte study is not precisely known. The nasal sinus brushing and orbital inflammatory disease papers included only patients with GPA. The peripheral leukocyte study included patients who had AAV with pauci-immune glomerulonephritis, however the AAV subtype was not reported. (3)
Table 1:
Study characteristics
| Peripheral Leukocytes (3) | Nasal sinus brushings (2) |
Orbital inflammatory disease (1) |
|
|---|---|---|---|
| RNA microarray | Affymetrix HU133 A and B | Affymetrix GeneChip Human Gene 1.0 ST Arrays |
Affymetrix GeneChip Human Genome U133 Plus 2.0 |
| Probe sets (n) | >45,000 | 19,658 | >54,000 |
| AAV samples (n) | 28 | 32 | 6 |
| AAV subtype | Unknown | GPA | GPA |
| Control groups | SLE RA Healthy controls |
Sarcoidosis Allergic rhinitis Healthy controls |
Sarcoidosis Thyroid eye disease Non-specific orbital inflammation Healthy controls |
| Threshold of significance | FC ≥1.8 p-value <0.01 |
FC ≥1.5 FDR <0.05 |
FC ≥1.5 FDR <0.05 |
| Upregulated genes (n) | 256 | 339 | 212 |
FC: fold change, FDR:false discovery rate, GPA: granulomatosis with polyangiitis
Upregulated genes in common:
We identified only four genes that were upregulated across all three studies— interleukin 1 receptor antagonist (IL1RN), matrix metalloproteinase 9 (MMP9), solute carrier family 11 member 1 (SLC11A1), and Toll like receptor 2 (TLR2). These four genes were identified in all three tissues with a FC compared to control tissues of >1.5 and p-values of <0.05.
IL1RN is upregulated in peripheral leukocytes (FC=1.7, p=1.9×10−4), sinus tissue (FC=2.5, p=1.9×10−4), and orbital tissue (FC=3.69 and 4.06, p=4.68×10−3 and 8.97 × 10−3)—the orbital study shows two values each for FC and p-values because in this study two sets of samples were processed at different time points. SLC11A1 encodes an iron transport membrane protein and is upregulated in peripheral leukocytes (FC=2.3, p=8.4×10−7), sinus tissue (FC=2.5, p=1.9×10−4), and orbital tissue (FC=3.5 and 3.3, p=3.6×10−4 and 1.9×10−2). TLR2 encodes a cell surface protein of the TLR family; it is upregulated in peripheral leukocytes (FC=2.2, p=2.1×10−6), sinus tissue (FC=1.7, p=2.3×10−3), and orbital tissue (FC=3.1 and 2.5, p=3.3×10−2 and 2.6×10−2). (18) MMP9 is upregulated in peripheral leukocytes (FC=4.0, p=1.5×10−6), nasal tissue (FC=2.2,p=2.0×10−4), and the orbit (FC=10.4 and 4.75, p=2.2×10−5 and 1.5×10−2). A further 45 genes were identified that were present in at least two of the three tissues; these are listed in Table 2.
Table 2:
Individual genes upregulated in two or more AAV studies
| Studies: | Orbit and nasal sinus brushings |
Orbit and leukocytes |
Nasal sinus brushings and leukocytes |
||
|---|---|---|---|---|---|
| Genes in common: | IL1RN | FN1 | IL1RN | IL1RN | MMP25 |
| MMP9 | ITGAX | MMP9 | MMP9 | NFE2 | |
| SLC11A1 | ITGB2 | SLC11A1 | SLC11A1 | NFIL3 | |
| TLR2 | LILRB3 | TLR2 | TLR2 | OSM | |
| ADAM8 | NLRP3 | SLC8A1 | ABCA1 | PFKFB3 | |
| ALOX5 | SERPINA1 | ACSL1 | PROK2 | ||
| CHI3L1 | SPP1 | ADM | QPCT | ||
| CLEC4D | TLR8 | APOBEC3A | RASGRP4 | ||
| CLEC7A | TREML2 | BCL2A1 | S100A12 | ||
| CXCR4 | CDA | SAMSN1 | |||
| DEFA3 | SLC2A3 | ||||
| DYSF | SOCS3 | ||||
| EMR2 | SSH2 | ||||
| IL18RAP | TNFAIP6 | ||||
| IL1R2 | |||||
Associated upregulated pathways in common:
Out of 1127 identifiable pathways, 28 pathways had an md-locfdr of <0.2 across all three tissues. (16, 17) These 28 disease pathways are represented in Table 3. The individual genes associated with each pathway in each tissue are shown in the supplemental material Table S1.
Table 3:
Pathways commonly associated with all ANCA-associated vasculitis gene expression studies
| Pathway | Peripheral leukocytes |
Nasal sinus brushing |
Orbital tissue | md-locfdr * |
|---|---|---|---|---|
| Innate Immunity | ||||
| Neutrophil degranulation(R) | p=3.14×10−13 | p= 1.11×10−16 | p=3.28×10−09 | 1.05×10−12 |
| Antimicrobial peptides(R) | p=7.69×10−06 | p=1.13×10−03 | p=1.14×10−03 | 8.23×10−03 |
| Toll-Like Receptors Cascades(R) | p=1.85×10−05 | p=2.78×10−04 | p=3.39×10−03 | 0.026 |
| Phagosome(K) | p=0.477 | p=2.88×10−08 | p=0.021 | 3.165×10−03 |
| Interferon gamma signaling(R) | p=0.533 | p=0.733 | p=6.9×10−03 | 0.134 |
| Adaptive Immunity | ||||
| Signaling by the B Cell Receptor (BCR)(R) | p=0.774 | p=0.867 | p=6.68×10−03 | 0.022 |
| Vascular wall interactions | ||||
| Cell surface interactions at the vascular wall(R) | p=0.887 | p=4.49×10−04 | p=5.86×10−05 | 4.19×10−04 |
| amb2 Integrin signaling(N) | p=0.276 | p=1.90×10−06 | p=0.041 | 0.086 |
| Leukocyte transendothelial migration(K) | p=0.34 | p=2.94×10−04 | p=6.98×10−03 | 0.178 |
| Cellular signaling | ||||
| Signaling by Interleukins(R) | p=8.91×10−05 | p=8.24×10−08 | p=1.88×10−05 | 6.13×10−04 |
| Cytokine-cytokine receptor interaction(K) | p=6.66×10−03 | p=7.78×10−06 | p=4.08×10−04 | 0.036 |
| Chemokine signaling pathway(K) | p=0.308 | p=1.97×10−06 | p=0.013 | 0.041 |
| IL4-mediated signaling events(N) | p=5.47×10−04 | p=0.107 | p=5.04×10−04 | 0.021 |
| Complement Activation | ||||
| Complement and coagulation cascades(K) | p=0.199 | p=2.70×10−07 | p=0.048 | 0.044 |
| Tissue Damage/Tissue Repair | ||||
| Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling(N) | p=0.354 | p=9.81×10−08 | p=9.39×10−03 | 7.77×10−03 |
| Extracellular matrix organization(R) | p=0.497 | p=2.51×10−04 | p=1.27×10−03 | 0.024 |
| Platelet Activation | ||||
| Response to elevated platelet cytosolic Ca2+(R) | p=0.670 | p=4.50×10−06 | p=0.095 | 0.038 |
| GPVI-mediated activation cascade(R) | p=0.400 | p=2.1×10−03 | p=1.69×10−03 | 0.079 |
| Platelet homeostasis(R) | p=0.024 | p=0.656 | p=3.28×10−03 | 0.091 |
| Infectious Disease Pathways | ||||
| Leishmaniasis(K) | p=0.041 | p=1.22×10−09 | p=0.172 | 3.54×10−03 |
| Malaria(K) | p=0.340 | p=3.77×10−06 | p=1.69×10−03 | 0.012 |
| Tuberculosis(K) | p=0.040 | p=3.79×10−08 | p=2.52×10−03 | 0.019 |
| Measles(K) | p=0.759 | p=0.012 | p=2.84×10−03 | 0.035 |
| Amoebiasis(K) | p=5.97×10−04 | p=1.42×10−05 | p=0.080 | 0.142 |
| Other | ||||
| Osteoclast differentiation(K) | p=0.013 | p=6.19×10−12 | p=0.391 | 3.75×10−05 |
| Inflammatory bowel disease (IBD)(K) | p=4.91×10−03 | p=0.031 | p=5.8 ×10−04 | 0.093 |
| Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds(R) | p=1.12×10−03 | p=0.540 | p=0.01 | 0.124 |
| Syndecan-4-mediated signaling events(N) | p=0.283 | p=3.18×10−04 | p=4.47×10−03 | 0.155 |
md-localFDR = multi-dimensional local false discovery rate which can be thought of as the probability of individual pathway being a false discovery in all three tissues.
C = CellMap, R = Reactome, K = Kyoto Encyclopedia of Genes and Genomes (KEGG), N = National Cancer Institute Pathway Interaction Database (NCI PID)
The 28 pathways discovered in this analysis can be grouped into seven functional categories including innate immunity, adaptive immunity, vascular wall interactions, cellular signaling, complement activation, tissue damage/repair, infectious disease pathways, and platelet activation (Table 3). The most strongly associated pathway in all three tissues is the neutrophil degranulation pathway (locfdr=1.0×10−12). After this, the most strongly associated pathways are osteoclast differentiation (locfdr=3.8×10−05), cell surface interactions at the vascular wall (locfdr=4.2×10−04), signaling by interleukins (locfdr=6.1×10−04), and the phagosome (locfdr=0.003) pathways.
Downregulated genes and pathways in common:
There were no downregulated genes or pathways found in all tissues. Only one gene, the G0S2 gene, was downregulated in two of the three tissues; peripheral leukocytes (FC=−1.9, p=0.26), and the orbit (FC=−13.7 and −24.9, p= 4.7×10−3 and 1.4×10−4). Downregulated pathways common to two out of three tissues are show in the supplemental material Table S2.
Discussion:
This is the first analysis of gene expression pathways in common across multiple tissues in AAV. The use of a network analysis system has allowed us to identify biologic functions and pathologic mechanisms that were not readily apparent by analysis of upregulated genes alone. Many of the common genes and pathways identified in this analysis support current knowledge and theories about AAV pathophysiology, while other pathways have not previously been implicated in AAV and thus, they may offer novel insights into disease pathogenesis. The pathophysiology of AAV based on gene expression pathways is represented in Figure 1.
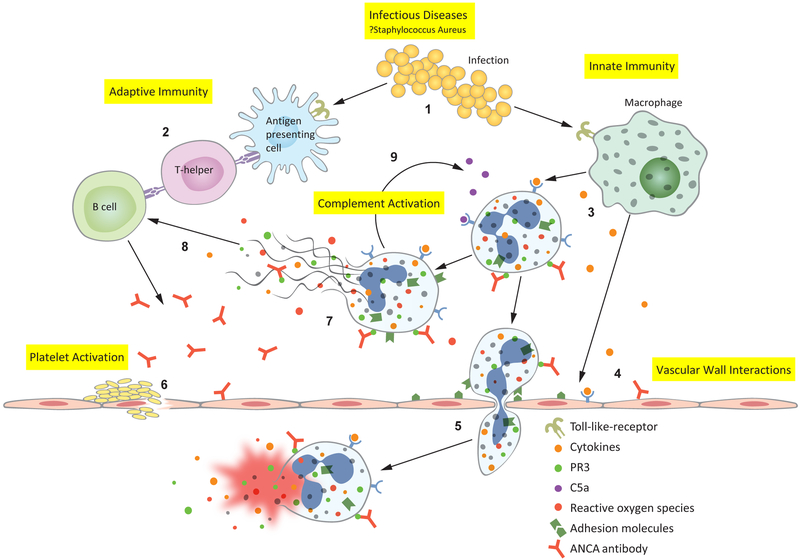
Figure 1:
Anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis pathophysiology revealed through gene expression pathways: 1. Infectious diseases, such as Staphylococcus aureus, activate Toll-like receptors (TLR) on antigen presenting cells and macrophages.(19) 2. Antimicrobial peptide homology with human antigens leads to the formation of pathogenic ANCAs.(20) 3. Macrophages secrete inflammatory cytokines which prime neutrophils— primed neutrophils express proteinase 3 (PR3) and myeloperoxidase (MPO) on their cell surface, allowing ANCA to bind and activate the neutrophil. 4. Cytokines and ANCAs activate endothelial cells, which express leukocyte adhesion molecules.(21-24) 5. ANCA-activated neutrophils migrate through the vascular wall where they degranulate, releasing reactive oxygen species and damage the vessel wall—thus causing the characteristic necrotizing vasculitis. 6. Platelets are activated at the sites of damaged endothelium, however their precise contribution to AAV pathophysiology is not well understood. 9. Activated neutrophils release neutrophil extracellular traps (NETs); NETs contain all of the intracellular contents of a neutrophil including MPO and PR3.(25) 8. Exposure of MPO and PR3 to the extracellular environment during NETosis drives further antibody formation. 9. Activated neutrophils also activate the alternative complement cascade, resulting in cleavage of C5 into C5a and C5b—C5a goes on to prime and attract more neutrophils leading to an amplification loop during active disease.(26)
The results of this analysis support the central roles of an infectious trigger, molecular mimicry, and the dysregulation of both innate and adaptive immune mechanisms in AAV (Figure 1). Infections are thought to drive AAV through molecular mimicry. This is supported by the antimicrobial peptide pathway (locfdr=8.2×10−03), infectious disease pathways, and the clinical finding that nasal carriage of Staphylococcus aureus, which shares peptide homology with human PR3 antigen, increases the risk of relapse in GPA. (20, 27) Three of the four genes expressed in common, IL1RN, SLC11A1, and TLR2, are also important innate immune genes. TLR2 is particularly interesting since this is the predominant TLR recognizing Staphylococcus aureus ligands (Figure 1). (19)
In addition to innate and infectious disease mechanisms, AAV is also an adaptive immune disease associated with pathogenic antibodies. Myeloperoxidase (MPO) antibodies have been found to be pathogenic in murine transfer models and in one transplacental human transfer case report. (28, 29) However perhaps the strongest evidence for ANCA pathogenicity is the success of rituximab treatment, which is increasingly becoming the standard of care in severe pathology. (30)
ANCA antibodies have diverse functions in this disease, however their main function is activating primed neutrophils (Figure 1). (23, 31, 32) All three tissues gene lists were most strongly associated with the neutrophil degranulation pathway (locfdr=1.05 ×10−12). ANCA-activated neutrophils may degranulate or extrude their intracellular contents in the form of a neutrophil extracellular trap (NET). (25, 33) AAV neutrophils also activate the alternative complement cascade—consistent with the complement and coagulation cascade pathway (locfdr=0.044) in this analysis (Figure 1). (26) A C5a inhibitor is currently in clinical trials as a steroid sparing agent in AAV. (26)
Platelet activation and vascular wall interaction pathways are not well understood with regard to their role in AAV and require further exploration. Patients with AAV are at increased risk for venous thrombosis, ischemic heart disease, and cardiovascular (CV) events. (34, 35) Platelets are acute phase reactants, but can also secrete cytokines and mediate inflammatory responses. (36, 37) Despite the increased CV risk associated with AAV, at this time there are no specific recommendations for CV risk reduction in these patients.
The interaction of the peripheral immune mechanisms with the vascular wall represents an important gap in our understanding of this disease. Endothelial cells (ECs) lining the vessel wall are heterogeneous and immunologically active cells, yet our understanding of how these cells function in AAV remains limited. (38) Enhancing our understanding of the vascular wall in AAV may allow us to understand patterns of organ involvement and potentially identify novel therapeutic targets.
Our analysis has limitations. Network-based pathway analyses are based on current knowledge of protein interactions, and some pathways may be associated due to diverse functions of genes rather than specific AAV physiology. For example, while infections are likely important, pathogens such as malaria, amoebiasis, and measles (see Table 3) are unlikely to be the inciting pathogens. This analysis is also limited by variability in methodologies. Each study has a different number of samples and used a microarray with different numbers of probes, which may prevent the detection of more subtle pathways of interest. The control groups studied for these tissues were also different, which may result in variability in which genes are considered differentially regulated when compared to controls. Finally, comparing peripheral leukocytes to end organ tissue is problematic, since end organ tissue includes a large number of other cell types that are not represented in the leukocyte study. Vascular wall interaction and tissue damage/repair pathways are more strongly associated with the orbital and nasal studies, which is most likely due to the inclusion of end organ tissue in these studies.
Despite these limitations, this pathway analysis has successfully identified pathways of disease common to all AAV tissues thus far characterized. Many of these pathways are consistent with current theories about infectious drivers and the crossroads of innate and adaptive immune mechanisms. In addition, this analysis highlights novel pathways that require further investigation, such as vessel wall interactions and platelet activation. Further study of these pathways may lead to novel biomarkers and therapeutic strategies that improve the care of patients with this disease.
Supplementary Material
ACKNOWLEDGEMENT:
none
Sources of support:
NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases: 5K23AR068440
NIH National Heart, Lung, and Blood Institute: 3T32HL094294-08S1
NIH National Eye Institute: EY010572, EY020249, and EY265722
Research to Prevent Blindness
Stan and Madelle Rosenfeld Family Trust
William and Mary Bauman Foundation
Footnotes
Conflict of interest: We have no conflicts of interest to disclose.
REFERENCES:
- 1.Rosenbaum JT, Choi D, Wilson DJ, Grossniklaus HE, Harrington CA, Sibley CH, et al. Orbital pseudotumor can be a localized form of granulomatosis with polyangiitis as revealed by gene expression profiling. Exp Mol Pathol 2015;99:271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayson PC, Steiling K, Platt M, Berman JS, Zhang X, Xiao J, et al. Defining the nasal transcriptome in granulomatosis with polyangiitis (wegener's). Arthritis Rheumatol 2015;67:2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcorta DA, Barnes DA, Dooley MA, Sullivan P, Jonas B, Liu Y, et al. Leukocyte gene expression signatures in antineutrophil cytoplasmic autoantibody and lupus glomerulonephritis. Kidney Int 2007;72:853–64. [DOI] [PubMed] [Google Scholar]
- 4.Xu W, Huang H, Yu L, Cao L. Meta-analysis of gene expression profiles indicates genes in spliceosome pathway are up-regulated in hepatocellular carcinoma (hcc). Med Oncol 2015;32:96. [DOI] [PubMed] [Google Scholar]
- 5.Fang F, Pan J, Xu L, Li G, Wang J. Identification of potential transcriptomic markers in developing ankylosing spondylitis: A meta-analysis of gene expression profiles. Biomed Res Int 2015;2015:826316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YH, Bae SC, Song GG. Meta-analysis of gene expression profiles to predict response to biologic agents in rheumatoid arthritis. Clin Rheumatol 2014;33:775–82. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y, Chen Y, et al. Meta-analysis of differentially expressed genes in osteosarcoma based on gene expression data. BMC Med Genet 2014;15:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microrna expression in lung cancer. Int J Cancer 2013;132:2884–93. [DOI] [PubMed] [Google Scholar]
- 9.Arasappan D, Tong W, Mummaneni P, Fang H, Amur S. Meta-analysis of microarray data using a pathway-based approach identifies a 37-gene expression signature for systemic lupus erythematosus in human peripheral blood mononuclear cells. BMC Med 2011;9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018;359:693–7.29439242 [Google Scholar]
- 11.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol 2010;11:R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2018;46:D649–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reactomefiviz. [cited 2018. 4.2.2018]; Available from: https://reactome.org/tools/reactome-fiviz.
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. 1995. [Google Scholar]
- 16.Jeong S-O, Choi D, Jang W A semiparametric mixture method for local false discovery rate estimation. Arxivorg 2016;Available from: http://arxiv.org/abs/1604.04264. [Google Scholar]
- 17.Efron B Microarrays, empirical bayes and the two-groups model. Statist Sci 2008;23:1–22. [Google Scholar]
- 18.Liu Y, Yin H, Zhao M, Lu Q. Tlr2 and tlr4 in autoimmune diseases: A comprehensive review. Clin Rev Allergy Immunol 2014;47:136–47. [DOI] [PubMed] [Google Scholar]
- 19.Uematsu S, Akira S. Toll-like receptors (tlrs) and their ligands. Handb Exp Pharmacol 2008:1–20. [DOI] [PubMed] [Google Scholar]
- 20.Tadema H, Heeringa P, Kallenberg CG. Bacterial infections in wegener's granulomatosis: Mechanisms potentially involved in autoimmune pathogenesis. Curr Opin Rheumatol 2011;23:366–71. [DOI] [PubMed] [Google Scholar]
- 21.Al-Hussain T, Hussein MH, Conca W, Al Mana H, Akhtar M. Pathophysiology of anca-associated vasculitis. Adv Anat Pathol 2017;24:226–34. [DOI] [PubMed] [Google Scholar]
- 22.Taekema-Roelvink ME, Kooten C, Kooij SV, Heemskerk E, Daha MR. Proteinase 3 enhances endothelial monocyte chemoattractant protein-1 production and induces increased adhesion of neutrophils to endothelial cells by upregulating intercellular cell adhesion molecule-1. J Am Soc Nephrol 2001;12:932–40. [DOI] [PubMed] [Google Scholar]
- 23.Mayet WJ, Schwarting A, Orth T, Duchmann R, Meyer zum Buschenfelde KH. Antibodies to proteinase 3 mediate expression of vascular cell adhesion molecule-1 (vcam-1). Clin Exp Immunol 1996;103:259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arrizabalaga P, Sole M, Abellana R, Ascaso C. Renal expression of adhesion molecules in anca-associated disease. J Clin Immunol 2008;28:411–9. [DOI] [PubMed] [Google Scholar]
- 25.Soderberg D, Segelmark M. Neutrophil extracellular traps in anca-associated vasculitis. Front Immunol 2016;7:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallenberg CG, Heeringa P. Complement system activation in anca vasculitis: A translational success story? Mol Immunol 2015;68:53–6. [DOI] [PubMed] [Google Scholar]
- 27.Salmela A, Rasmussen N, Tervaert JWC, Jayne DRW, Ekstrand A, European Vasculitis Study G. Chronic nasal staphylococcus aureus carriage identifies a subset of newly diagnosed granulomatosis with polyangiitis patients with high relapse rate. Rheumatology (Oxford) 2017;56:965–72. [DOI] [PubMed] [Google Scholar]
- 28.Jennette JC, Xiao H, Falk R, Gasim AM. Experimental models of vasculitis and glomerulonephritis induced by antineutrophil cytoplasmic autoantibodies. Contrib Nephrol 2011;169:211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bansal PJ, Tobin MC. Neonatal microscopic polyangiitis secondary to transfer of maternal myeloperoxidase-antineutrophil cytoplasmic antibody resulting in neonatal pulmonary hemorrhage and renal involvement. Ann Allergy Asthma Immunol 2004;93:398–401. [DOI] [PubMed] [Google Scholar]
- 30.Stone JH, Merkel PA, Spiera R, Seo P, Langford CA, Hoffman GS, et al. Rituximab versus cyclophosphamide for anca-associated vasculitis. N Engl J Med 2010;363:221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kettritz R How anti-neutrophil cytoplasmic autoantibodies activate neutrophils. Clin Exp Immunol 2012;169:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H, Hu P, Falk RJ, Jennette JC. Overview of the pathogenesis of anca-associated vasculitis. Kidney Dis (Basel) 2016;1:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grayson PC, Carmona-Rivera C, Xu L, Lim N, Gao Z, Asare AL, et al. Neutrophil-related gene expression and low-density granulocytes associated with disease activity and response to treatment in antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol 2015;67:1922–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Springer J, Villa-Forte A. Thrombosis in vasculitis. Curr Opin Rheumatol 2013;25:19–25. [DOI] [PubMed] [Google Scholar]
- 35.Huang YM, Wang H, Wang C, Chen M, Zhao MH. Promotion of hypercoagulability in antineutrophil cytoplasmic antibody-associated vasculitis by c5a-induced tissue factor-expressing microparticles and neutrophil extracellular traps. Arthritis Rheumatol 2015;67:2780–90. [DOI] [PubMed] [Google Scholar]
- 36.Willeke P, Kumpers P, Schluter B, Limani A, Becker H, Schotte H. Platelet counts as a biomarker in anca-associated vasculitis. Scand J Rheumatol 2015;44:302–8. [DOI] [PubMed] [Google Scholar]
- 37.Miao D, Li DY, Chen M, Zhao MH. Platelets are activated in anca-associated vasculitis via thrombin-pars pathway and can activate the alternative complement pathway. Arthritis Res Ther 2017;19:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zielinska KA, Van Moortel L, Opdenakker G, De Bosscher K, Van den Steen PE. Endothelial response to glucocorticoids in inflammatory diseases. Front Immunol 2016;7:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.