Abstract
A person’s lipid profile is influenced by genetic variants and alcohol consumption, but the contribution of interactions between these exposures has not been studied. We therefore incorporated gene-alcohol interactions into a multiancestry genome-wide association study of levels of high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and triglycerides. We included 45 studies in stage 1 (genome-wide discovery) and 66 studies in stage 2 (focused follow-up), for a total of 394,584 individuals from 5 ancestry groups. Analyses covered the period July 2014–November 2017. Genetic main effects and interaction effects were jointly assessed by means of a 2–degrees-of-freedom (df) test, and a 1-df test was used to assess the interaction effects alone. Variants at 495 loci were at least suggestively associated (P < 1 × 10−6) with lipid levels in stage 1 and were evaluated in stage 2, followed by combined analyses of stage 1 and stage 2. In the combined analysis of stages 1 and 2, a total of 147 independent loci were associated with lipid levels at P < 5 × 10−8 using 2-df tests, of which 18 were novel. No genome-wide-significant associations were found testing the interaction effect alone. The novel loci included several genes (proprotein convertase subtilisin/kexin type 5 (PCSK5), vascular endothelial growth factor B (VEGFB), and apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1) complementation factor (A1CF)) that have a putative role in lipid metabolism on the basis of existing evidence from cellular and experimental models.
Keywords: alcohol consumption, cholesterol, gene-environment interactions, gene-lifestyle interactions, genome-wide association studies, lipids, triglycerides
Serum concentrations of high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG) are modifiable risk factors for cardiovascular disease, the leading cause of death globally (1). Lipid levels are influenced by multiple exposures, including genetic and lifestyle factors. The genetic factors influencing lipid levels have been widely studied (2–8), and large-scale genome-wide association studies (GWAS) have identified 236 loci associated with HDL-C, LDL-C, and TG, which account for up to approximately 12% of the total trait variance in the studied populations (5, 7).
Lifestyle factors, such as alcohol consumption, are also considerably associated with lipid levels: In epidemiologic studies, greater alcohol consumption is associated with an improved lipid profile, including associations with HDL-C levels, high-density lipoprotein particle concentrations, and HDL-C subfractions (9, 10). The relationship between alcohol use and LDL-C or TG is less clear, with some studies reporting positive associations while others have reported negative associations (11–20). A causal role of low-to-moderate alcohol consumption in improving overall lipid profile is supported by intervention studies (19), and more recently by Mendelian randomization studies (21, 22).
Potential modification of genetic effects on lipid levels by lifestyle exposures, including alcohol consumption, is relatively unexplored (23). Genetic association studies accounting for potential gene-alcohol interactions may lead to the identification of novel lipid loci and may reveal new biological insights that can potentially be explored for treatment or prevention of dyslipidemia. In order to investigate the potential modulating role of alcohol consumption in the genetic architecture of lipid levels and to identify novel HDL-C, LDL-C, and TG loci, we performed genome-wide gene-alcohol interaction meta-analyses of LDL-C, HDL-C, and TG.
METHODS
Overall design
Table 1 shows the overall design of this study, which was conducted within the setting of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Gene-Lifestyle Interactions Working Group (24, 25). In order to decrease the computational burden, we carried out genome-wide analyses in stage 1 and followed up suggestively associated variants in stage 2, with the combined results of analyses carried out in stage 1 and stage 2 serving as the primary analysis (26). We used 2 complementary approaches to model interactions: 1) a 2–degrees-of-freedom (df) test was used to jointly assess both the genetic main effect and the interaction effect on lipid levels, and 2) a 1-df test was used to assess the effect of interactions alone. The 2-df test is more powerful when there is both a genetic main effect and an interaction effect, and it may thus help identify interaction effects for which the 1-df test lacks sufficient power (27).
Table 1.
Distribution (Number) of Participants by Ancestry in a Genome-Wide Meta-Analysis of Gene-Alcohol Interaction and Lipid Levels, 2017a
| Analysis Stage | Ancestry Group | Meta-Analysis | ||||
|---|---|---|---|---|---|---|
| European | African | Asian | Hispanic | Brazilian | ||
| 1 | 89,893 | 20,989 | 12,450 | 3,994 | 0 | 127,326 |
| 2b | 136,986 | 4,475 | 108,431 | 13,714 | 3,652 | 267,258 |
| Totalc | 226,879 | 25,464 | 120,881 | 17,708 | 3,652 | 394,584 |
Abbreviation: df, degrees of freedom.
a For each lipid trait, association analyses were performed accounting for 2 alcohol consumption status variables: “current drinker” and “regular drinker.” For each ancestry group, study-specific results were combined to perform the 1-df test for an interaction effect and the 2-df joint test of the genetic main effect and interaction with drinking exposure. Persons from 5 ancestry groups were included: European, African, Asian, Hispanic, and Brazilian.
b Variants selected for follow-up at P ≤ 1 × 10−6 using a 1-df interaction test and a joint 2-df interaction test.
c Variants found to be significant at P ≤ 5 × 10−8 using a joint 2-df interaction test or a 1-df interaction test.
Overview of participating studies
This analysis covered the period July 2014–November 2017 and included men and women aged 18–80 years from 5 ancestry groups: European, African, Asian, Hispanic, and Brazilian. Investigators in each study obtained informed consent from participants and approval from the appropriate institutional review boards. Although the participating studies are based on different study designs and populations, all of them have data on lipid levels, alcohol consumption, and genotypes across the genome. In total, the analysis comprised 394,584 individuals.
Stage 1 studies
Stage 1 included 89,893 European-ancestry participants, 20,989 African-ancestry participants, 12,450 Asian-ancestry participants, and 3,994 Hispanic-ancestry participants, for an overall total of 127,326 individuals from 45 studies (see Web Table 1, available at https://academic.oup.com/aje), namely: the Age, Gene/Environment Susceptibility (AGES)-Reykjavik Study (1967—Reykjavik, Iceland), the Atherosclerosis Risk in Communities (ARIC) Study (1987–1989—Washington County, Maryland; Forsyth County, North Carolina; Jackson, Mississippi; and Minneapolis, Minnesota), the Coronary Artery Risk Development in Young Adults (CARDIA) Study (1985–1986—Birmingham, Alabama; Chicago, Illinois; Minneapolis, Minnesota; and Oakland, California), the Cardiovascular Health Study (CHS) (1989–1990 and 1992–1993—Forsyth County, North Carolina; Sacramento County, California; Washington County, Maryland; and Pittsburgh, Pennsylvania), the CROATIA-Korcula Study (2007—Korcula, Croatia), the CROATIA-Vis Study (2003–2004—Vis, Croatia), the Erasmus Rucphen Family (ERF) Study (2002–2005—Rotterdam, the Netherlands), the Family Heart Study (FamHS) (1992–1995—Salt Lake City, Utah; Forsyth County, North Carolina; Minneapolis, Minnesota; and Framingham, Massachusetts), the Framingham Heart Study (1948—Framingham, Massachusetts), the Genetic Epidemiology Network of Arteriopathy (GENOA) Study (1995–2000—Rochester, Minnesota and Jackson, Mississippi), the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) Study (2003–2005—provinces of Hebei, Henan, Shandong, Shaanxi, and Jiangsu, China), the Generation Scotland: Scottish Family Health Study (GS_SFHS) (2006–2011—Scotland, United Kingdom), the Health, Aging and Body Composition (HABC) Study (1997–1998—Pittsburgh, Pennsylvania and Memphis, Tennessee), the Healthy Aging in Neighborhoods of Diversity Across the Life Span (HANDLS) Study (2004–2009—Baltimore, Maryland), the Health, Risk Factors, Exercise Training and Genetics (HERITAGE) Study (1995–2000—Arizona; Indiana; Minnesota; Texas; and Quebec, Canada), the Howard University Family Study (HUFS) (2001–2008—Washington, DC), the Hypertension Genetic Epidemiology Network (HyperGEN) Study (1996–1999—Birmingham, Alabama; Salt Lake City, Utah; Forsyth County, North Carolina; Minneapolis, Minnesota; and Framingham, Massachusetts), the Jackson Heart Study (JHS) (2000–2004—Jackson, Mississippi), the Multi-Ethnic Study of Atherosclerosis (MESA) (2000–2002—Los Angeles, California; St. Paul, Minnesota; Chicago, Illinois; Winston-Salem, North Carolina; Baltimore, Maryland; and New York, New York), the Netherlands Epidemiology of Obesity (NEO) Study (2008–2012—Leiden, the Netherlands), Rotterdam Study 1 (RS1) (1990—Rotterdam, the Netherlands), Rotterdam Study 2 (RS2) (2000–2001—Rotterdam, the Netherlands), Rotterdam Study 3 (RS3) (2006–2008—Rotterdam, the Netherlands), the Singapore Chinese Eye Study (SCES) (2009–2011—Singapore), the Singapore Chinese Health Study–Coronary Heart Disease Study (SCHS-CHD) (1993–1998—Singapore), the Singapore Malay Eye Study (SiMES) (2004–2006—Singapore), the Singapore Indian Eye Study (SINDI) (2007–2009—Singapore), the Singapore 2 (SP2) Study (SP2-1M and SP2-610) (1982–1998—Singapore), the Women’s Genome Health Study (WGHS) (1992–1995—United States), and the Women’s Health Initiative (WHI) (WHI Genomics and Randomized Trials Network (WHI_GARNET) and WHI Memory Study (WHI_WHIMS); 1993–1998—United States).
Stage 2 studies
Stage 2 included 136,986 European-ancestry, 4,475 African-ancestry, 108,431 Asian-ancestry, 13,714 Hispanic-ancestry, and 3,652 Brazilian-ancestry participants, for an overall total of 267,258 individuals from the following 66 studies (Web Table 2): the 1982 Pelotas Birth Cohort Study (1982—Pelotas, Brazil), the African American Diabetes Heart Study (AA-DHS) (1998–2005—Winston-Salem, North Carolina), the Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT) (1998–2000—Denmark, Finland, Ireland, Norway, Sweden, and the United Kingdom), the Baependi Heart Study (2010—Baependi, Brazil), the BioBank Japan (BBJ) Project (2003–2008—Japan), the Beijing Eye Study (BES-Omniexpress) (2001—Beijing, China), the British Genetics of Hypertension (BRIGHT) Study (1995—United Kingdom), the Cardio-metabolic Genome Epidemiology Network Amagasaki Study (CAGE-Amagasaki) (2002–2003—Amagasaki, Japan), the Data From the Epidemiological Study on the Insulin Resistance Syndrome (DESIR) Study (1994–1996—France), the Dongfeng-Tongji (DFTJ) Cohort Study (2008—Shiyan City, China), the Diabetes Heart Study (DHS) (1998–2005—Winston-Salem, North Carolina), the Dose Responses to Exercise Training (DR’s EXTRA) Study (2005–2006—Kuopio, Finland), the Estonian Genome Center of the University of Tartu (EGCUT) Study (2002–2010—Estonia), the European Prospective Investigation Into Cancer and Nutrition (EPIC) (1992–1997—France, Italy, Spain, the United Kingdom, the Netherlands, Germany, Sweden, Denmark, Norway, and Greece), the Fenland Study (Fenland-GWAS and Fenland-Omics) (1950–1975—Cambridgeshire, England, United Kingdom), the Finland-United States Investigation of NIDDM Genetics (FUSION) Study (1994—Finland), the Genetic Studies of Atherosclerosis Risk (GeneSTAR) Study (1983–2006—Baltimore, Maryland), the Gene × Lifestyle Interactions and Complex Traits Involved in Elevated Disease Risk (GLACIER) Study (1985–2004—Sweden), the Genetic Regulation of Arterial Pressure of Humans in the Community (GRAPHIC) Study (2003–2005—Leicestershire, England, United Kingdom), the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) (2008–2011—Chicago, Illinois; Miami, Florida; New York, New York; and San Diego, California), the Health and Retirement Study (HRS) (2006–2010—United States), the Hypertension Genetic Epidemiology Network (HyperGEN)-Axiom Study (which used the Axiom Genome-Wide ASI 1 Array Plate; Thermo Fisher Scientific, Waltham, Massachusetts) (1996–1999—Birmingham, Alabama; Salt Lake City, Utah; Forsyth County, North Carolina; Minneapolis, Minnesota; and Framingham, Massachusetts), the Italian Network Genetic Isolates—Carlantino (INGI-CARL) Study (2005–2006—Carlantino, Italy), the Italian Network Genetic Isolates—Friuli-Venezia Giulia (INGI-FVG) Study (2013—Friuli-Venezia Giulia, Italy), the EPIC-InterAct Case-Cohort Study (InterAct) (1991–2007—France, Italy, Spain, the United Kingdom, the Netherlands, Germany, Sweden, and Denmark), the Insulin Resistance Atherosclerosis Study (IRAS; IRAS Cohort Study and IRAS Family Study) (1999–2005—San Antonio, Texas and San Luis Valley, Colorado), the Cooperative Health Research in the Augsburg Region S3 (KORA_S3) Study (1994–1995—Augsburg, Germany), the Cooperative Health Research in the Augsburg Region S4 (KORA_S4) Study (1991–2001—Augsburg, Germany), the Lothian Birth Cohort 1936 (LBC1936) Study (2004–2007—Lothian, Scotland, United Kingdom), the LifeLines Cohort Study (2006–2013—the Netherlands), the London Life Sciences Prospective Population (LOLIPOP) Study (2003–2007—London, England, United Kingdom), the Long Life Family Study (LLFS) (2006–2009—Boston, Massachusetts; New York, New York; Pittsburgh, Pennsylvania; and Denmark), the Kingston Gene-by-Environment (Loyola GxE) Study (1994–1995—Kingston, Jamaica), the Spanish Town (Loyola SPT) Study (1994–1995—Kingston, Jamaica), the Metabolic Syndrome in Men (METSIM) Study (2005–2010—Kuopio, Finland), the Netherlands Study of Depression and Anxiety (NESDA) (2004–2007—the Netherlands), the Obesity in Adults (OBA) Study (2005—France), the Prevention of Renal and Vascular End Stage Disease (PREVEND) Study (1997–1998—Groningen, the Netherlands), the Precocious Coronary Artery Disease (PROCARDIS) Study (2004–2008—United Kingdom, Italy, Sweden, and Germany), the Ragama Health Study (RHS) (2007—Ragama, Sri Lanka), the Stockholm Heart Epidemiology Program (SHEEP) (1992–1994—Stockholm County, Sweden), the Study of Health in Pomerania (SHIP) (participants from the baseline examination (SHIP-0) (1997–2001—Greifswald, Stralsund, and Anklam, Germany) and participants from a new sample for a new cohort drawn from the same area (SHIP-Trend) (2008–2012—Greifswald, Stralsund, and Anklam, Germany)), the Shanghai Women’s Health Study/Shanghai Men’s Health Study (SWHS/SMHS) (1997–2000—Shanghai, China), the TwinGene Project (data from the Swedish Twin Registry; 2004–2008—Sweden), and the Cardiovascular Risk in Young Finns Study (YFS) (1980—Finland).
Calculation of variance
An additional study not included in stage 1 or stage 2 was used to determine the variance explained by variants at known and new loci: the Airwave Health Monitoring Study (2004–2015—England, Scotland, and Wales, United Kingdom).
Phenotype and lifestyle variables
Three lipid traits were analyzed separately: HDL-C (mg/dL), LDL-C (mg/dL), and TG (mg/dL). HDL-C and TG were directly assayed, while LDL-C was either directly assayed or estimated using the Friedewald equation: LDL-C = TC − HDL-C − (TG/5) (28). Only fasting samples (≥8 hours) were used to assay TG, and the Friedewald equation was only used in samples with fasting TG concentrations less than or equal to 400 mg/dL. LDL-C values were adjusted for use of statins (Web Appendix). HDL-C and TG values were natural log-transformed prior to analyses.
Alcohol consumption was assessed using 2 dichotomized alcohol consumption variables: “current drinking” status, defined as any recurrent drinking behavior, and “regular drinking” status, defined as the subset of current drinkers who consumed at least 2 drinks per week. Because the standard pure ethanol content in 1 alcoholic drink may vary among countries, for this study a standard drink was defined to contain approximately 13 g of pure ethanol, and this measure was used to standardize the definitions across studies.
Genotyping and imputation
Information on genotyping and imputation for each of the stage 1 and stage 2 studies is presented in Web Table 3 and Web Table 4, respectively. For imputation, most studies used the 1000 Genomes Project Phase I Integrated Release Version 3 Haplotypes (2010–2011 data freeze; March 14, 2012, haplotypes), which contain haplotypes for 1,092 individuals from multiple ancestry groups (29).
Study-specific analysis
Study-specific regression analyses were performed for each variant, using models containing the genetic variant, the alcohol consumption variable (current drinking status or regular drinking status), and their interaction. Variants were coded according to the additive model, so that the β coefficient represents the effect size per copy of the coded allele. These regressions were adjusted for age, sex, ancestry-informative principal components, and study-specific variables where appropriate (such as center for multicenter studies). Information on the principal components and study-specific variables adjusted for in each study-specific analysis is provided in Web Tables 3 and 4.
In stage 1, investigators in each study performed genome-wide association analyses within each ancestry group and provided the CHARGE Consortium with information on the estimated genetic main effect, the estimated interaction effect, and a robust estimate of the corresponding covariance matrix. In stage 2, investigators in each study performed analyses only for the selected variants identified in stage 1. Study-specific association analyses were performed using various software programs (Web Appendix and Web Tables 3 and 4). Extensive quality control using the R (R Foundation for Statistical Computing, Vienna, Austria) package “EasyQC” was performed for all study-specific GWAS results, as described in the Web Appendix (30).
Meta-analysis
We implemented METAL software to meta-analyze the genetic main and interaction effects jointly using the 2-df approach of Manning et al. (27) and Willer et al. (31) and to meta-analyze the interaction coefficients alone using inverse-variance–weighted meta-analysis (1-df test). For each meta-analysis, results were obtained from Wald tests, performed using genetic main-effect estimates, interaction effect estimates, and robust estimates of the corresponding covariance matrix.
In stage 1, ancestry-specific meta-analyses were performed for each of the 12 analyses (3 lipids × 2 alcohol consumption exposures × 2 tests). Genomic control correction was applied twice (32), first to the study-specific GWAS results (Web Table 5) and then to the ancestry-specific meta-analysis results. The results from each ancestry group were then combined in a transancestry meta-analysis.
The variants that were at least suggestively associated with lipid levels (P < 1 × 10−6) in any of the stage 1 interaction analyses were pursued for stage 2 analysis. In stage 2, we used the same approaches as in stage 1 to perform ancestry-specific and transancestry meta-analyses. Finally, ancestry-specific and transancestry meta-analyses were performed to combine stage 1 results with stage 2 results. Variants with P values less than 5 × 10−8 for either the 2-df joint test of genetic main and interaction effects or the 1-df test of interaction effects were considered genome-wide-significant. False discovery rate (FDR) q values were calculated using the Benjamini and Hochberg method (33) implemented with the “p.adjust” function in R, correcting for the number of tests performed in stage 1. FDR q values less than 0.05 thus indicate a <5% FDR even after considering the multiple testing introduced by performing genome-wide analyses on multiple outcomes using multiple models. An independent locus was defined as the ±1 mega–base-pair region surrounding an index variant. For each locus, the closest genes were determined on the basis of proximity to the index variant. For loci with intergenic index variants, we included the closest gene in each direction.
Additional analyses
The percentages of variance in HDL-C, LDL-C, and TG levels explained by all previously known and novel variants were evaluated in 10 studies on multiple ancestry groups (Web Appendix). The HaploReg (34), RegulomeDB (35), and Genotype-Tissue Expression (GTEx) (36) software packages were used to annotate variants at significant loci. We also used Data-driven Expression Prioritized Integration for Complex Traits (DEPICT) software (37) to prioritize genes at the loci associated with lipid levels in the combined analysis of stages 1 and 2. More details on gene prioritization using DEPICT can be found in the Web Appendix.
Lastly, we examined the associations of index variants at the 147 significant loci with coronary artery disease and myocardial infarction using publicly available summary association results from a large GWAS of these phenotypes performed by the Coronary Artery Disease Genome-Wide Replication and Meta-Analysis (CARDIoGRAM) plus Coronary Artery Disease (C4D) Genetics Consortium (CARDIoGRAMplusC4D Consortium) (38).
RESULTS
Descriptive statistics for the studies participating in stage 1 of the analysis are shown in Web Table 1: 56.1% of stage 1 participants were current drinkers, and 39.9% were regular drinkers. The stage 1 genome-wide analyses identified 25,115 variants in 495 independent loci that were at least suggestively associated (P < 1 × 10−6) with HDL-C, LDL-C, or TG using either the 1-df test of the interaction or the 2-df test that jointly assessed genetic main and interaction effects. The 1-df interaction test identified 356 suggestively associated variants, while the 2-df joint test identified an additional 24,759 variants. Manhattan and quantile-quantile plots are shown in Web Figure 1 and Web Figure 2, respectively.
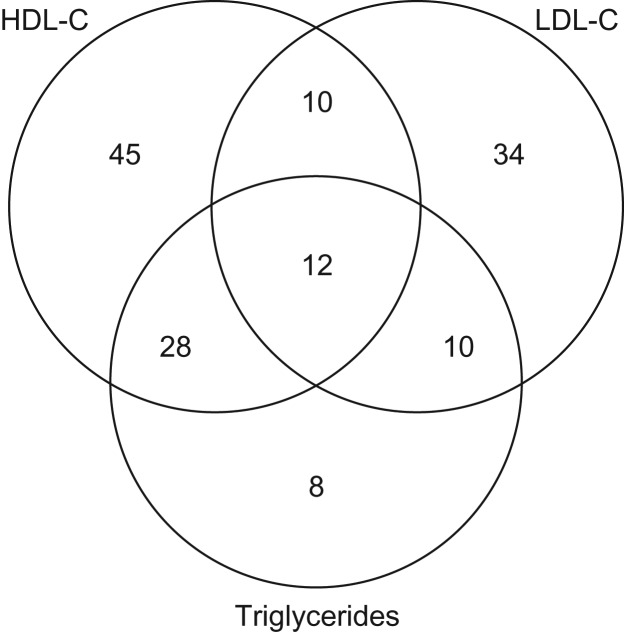
The 25,115 variants were then evaluated in stage 2. Descriptive statistics for the studies participating in stage 2 are shown in Web Table 2: 58.5% of stage 2 participants were current drinkers, and 41.0% were regular drinkers. The combined analysis of stage 1 and stage 2 findings identified 22,590 variants at 147 independent loci with genome-wide significance (P < 5 × 10−8; Web Table 6). All genome-wide-significant associations were identified through the 2-df joint tests of main and interaction effects. There were no genome-wide-significant 1-df interaction associations in the combined analysis of stage 1 and stage 2. At genome-wide significance, 95 of the 147 loci were associated with HDL-C, 66 were associated with LDL-C, and 58 were associated with TG. Of the 147 loci, 60 loci were associated with more than 1 lipid trait, as shown in a Venn diagram in Figure 1.
Figure 1.
Distribution of genome-wide-significant associations at 147 genetic loci identified as being associated with 3 lipid traits (high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides) in a genome-wide meta-analysis of gene-alcohol interaction and lipid levels, 2017.
Novel loci
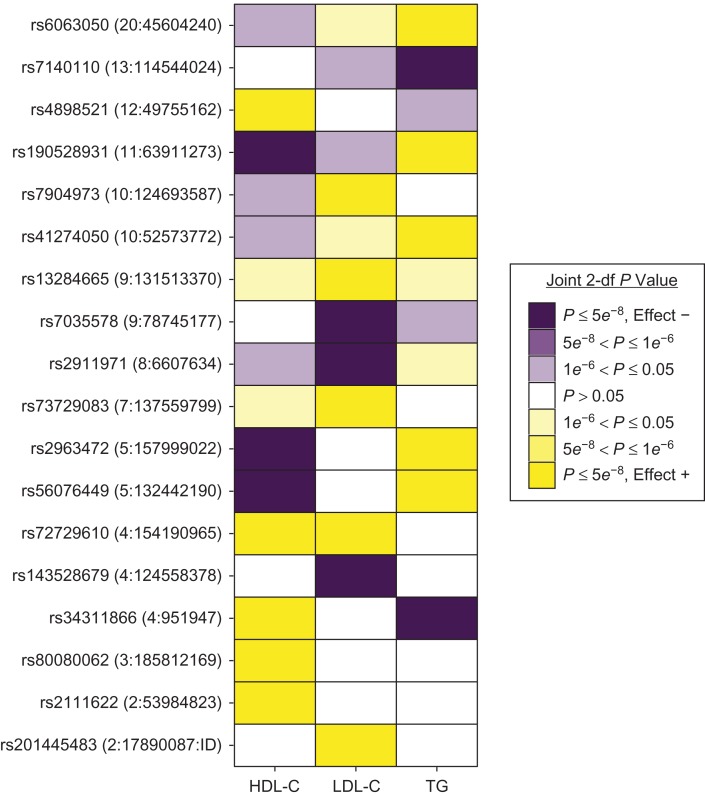
Of the 147 identified genome-wide-significant loci, 18 are novel lipid loci that have not been previously identified by other association studies for HDL-C, LDL-C, TG, or total cholesterol (Table 2 and Web Figure 3) (2–8). A concurrent genetic association study of exonic variants also identified 4 of these 18 novel loci (39), as indicated in Table 2. Eight of the novel loci involved HDL-C, 8 involved LDL-C, and 7 involved TG, as shown in the heat map in Figure 2. The most significant associations at each of the 18 novel loci all had FDR q values less than 0.05 (Table 2), indicating that they were unlikely to be false-positive findings introduced by multiple testing. As shown in forest plots (Web Figure 4), the 2-df associations at the novel loci were predominantly driven by genetic main effects, with a smaller contribution from interaction effects. Furthermore, of the 18 index variants, 15 had at least suggestively significant (P < 1 × 10−6) genetic main effects in stage 1 (Web Appendix and Web Table 7). None of the associations at the 18 novel loci displayed heterogeneity across ancestry groups (Table 2).
Table 2.
Novel Loci Discovered in a Genome-Wide Meta-Analysis of Gene-Alcohol Interaction and Lipid Levels (Combined Analysis of Stages 1 and 2) Using a 2–Degrees-of-Freedom Model That Jointly Tested Main and Interaction Effects, 2017
| rsID | Chromosome No.:Position | Allelesa | Frequencyb | Closest Gene(s) | Main Effectc | Interaction Effectc | Joint P Valuec | Joint FDR q Valuec | Interaction P Valued | Heterogeneity P Valuee | Most Significant 2-df Model |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs190528931f | 11:63911273 | A/C | 0.04 | MACROD1 | 0.0109 | −0.0023 | 1.9 × 10−16 | 3.6 × 10−11 | 0.32 | 0.96 | METAg—HDL-C—CURDRINKh |
| rs7904973f | 10:124693587 | T/G | 0.55 | C10orf88 | 0.9200 | −0.1500 | 1.9 × 10−15 | 3.5 × 10−10 | 0.38 | 0.89 | META—LDL-C—CURDRINK |
| rs73729083 | 7:137559799 | C/T | 0.91 | CREB3L2 | 4.0100 | 0.6500 | 8.2 × 10−15 | 1.4 × 10−9 | 0.57 | 0.22 | META—LDL-C—CURDRINK |
| rs80080062 | 3:185812169 | G/C | 0.87 | ETV5 | 0.0061 | 0.0031 | 1.1 × 10−12 | 1.7 × 10−7 | 0.38 | 0.85 | META—HDL-C—REGDRINKi |
| rs7140110 | 13:114544024 | C/T | 0.73 | GAS6-AS1 | −0.0100 | −0.0040 | 3.4 × 10−12 | 5.1 × 10−7 | 0.19 | 0.42 | META—TG—CURDRINK |
| rs34311866 | 4:951947 | C/T | 0.83 | TMEM175 | −0.0200 | 0.0040 | 1.5 × 10−11 | 2.1 × 10−6 | 0.42 | 0.90 | EUR—TG—CURDRINK |
| rs2911971 | 8:6607634 | G/C | 0.34 | AGPAT5 | −0.7500 | 0.0100 | 7.5 × 10−11 | 1.1 × 10−5 | 0.53 | 0.49 | META—LDL-C—CURDRINK |
| rs56076449 | 5:132442190 | G/T | 0.79 | HSPA4/FSTL4 | 0.0130 | −0.0020 | 9.3 × 10−11 | 1.3 × 10−5 | 0.80 | 0.80 | META—TG—REGDRINK |
| rs41274050f | 10:52573772 | T/C | 0.01 | A1CF | 0.1080 | −0.0310 | 9.6 × 10−10 | 1.3 × 10−4 | 0.62 | 1.00 | EUR—TG—REGDRINK |
| rs7035578 | 9:78745177 | A/G | 0.15 | PCSK5 | −1.2700 | 0.0800 | 1.2 × 10−9 | 1.6 × 10−4 | 0.70 | 0.82 | EUR—LDL-C—CURDRINK |
| rs201445483 | 2:17890087 | I/D | 0.83 | SMC6 | 1.4300 | 0.6800 | 4.7 × 10−9 | 6.0 × 10−4 | 0.17 | 0.46 | META—LDL-C—CURDRINK |
| rs72729610 | 4:154190965 | G/A | 0.86 | TRIM2 | 0.0075 | −0.0036 | 5.6 × 10−9 | 7.2 × 10−4 | 0.08 | 0.26 | META—HDL-C—REGDRINK |
| rs143528679 | 4:124558378 | G/A | 0.10 | SPRY1/LINC01091 | −1.2000 | −5.6300 | 6.3 × 10−9 | 8.0 × 10−4 | 6.4 × 10−4 | 0.10 | AFR—LDL-C—CURDRINK |
| rs2111622f | 2:53984823 | G/A | 0.77 | ASB3/GPR75-ASB3 | 0.0008 | −0.0072 | 7.9 × 10−9 | 9.9 × 10−4 | 0.01 | 0.12 | EUR—HDL-C—CURDRINK |
| rs13284665 | 9:131513370 | G/A | 0.88 | ZER1 | 1.9900 | −0.8900 | 1.1 × 10−8 | 1.3 × 10−3 | 0.35 | 0.89 | EUR—LDL-C—CURDRINK |
| rs4898521 | 12:49755162 | A/T | 0.95 | DNAJC22/SPATS2 | 0.0179 | −0.0107 | 1.3 × 10−8 | 1.7 × 10−3 | 0.06 | 1.00 | EUR—HDL-C—REGDRINK |
| rs6063050 | 20:45604240 | C/T | 0.75 | EYA2 | 0.0110 | 0.0000 | 2.9 × 10−8 | 3.6 × 10−3 | 0.30 | 0.39 | META—TG—CURDRINK |
| rs2963472 | 5:157999022 | A/G | 0.21 | LOC101927697/EBF1 | 0.0140 | −0.0020 | 3.5 × 10−8 | 4.2 × 10−3 | 0.96 | 0.23 | EUR—TG—REGDRINK |
Abbreviations: A1CF, APOBEC1 complementation factor gene; ADP, adenosine diphosphate; AFR, African ancestry; AGPAT5, 1-acylglycerol-3-phosphate O-acyltransferase 5 gene; APOBEC1, apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1; AS1, antisense RNA 1 gene; ASB3, ankyrin repeat and SOCS box containing 3 gene; cAMP, cyclic adenosine 3',5'-monophosphate; C10orf88, chromosome 10 open reading frame 88 gene; CREB3L2, cAMP responsive element binding protein 3 like 2 gene; CURDRINK, current drinkers; df, degrees of freedom; DNAJC22, DnaJ heat shock protein family (Hsp40) member C22 gene; EBF, early B cell factor; EBF1, EBF transcription factor 1 gene; ETS, E26 transformation-specific; ETV5, ETS variant 5 gene; EUR, European ancestry; EYA, eyes absent; EYA2, EYA transcriptional coactivator and phosphatase 2 gene; FDR, false discovery rate; FSTL4, follistatin like 4 gene; GAS6, growth arrest specific 6 gene; GPR75, G protein-coupled receptor 75 gene; HDL-C, high-density lipoprotein cholesterol; Hsp, heat shock protein; HSPA4, heat shock protein family a (Hsp70) member 4 gene; LDL-C, low-density lipoprotein cholesterol; LINC01091, long intergenic non-protein coding RNA 1091 gene; LOC101927697, uncharacterized locus 101927697; MACROD1, macro domain-containing protein 1 gene; META, meta-analysis; PCSK5, proprotein convertase subtilisin/kexin type 5 gene; REGDRINK, regular drinkers; rsID, reference SNP identifier; RTK, receptor tyrosine kinase; SMC6, structural maintenance of chromosomes 6 gene; SNP, single nucleotide polymorphism; SOCS, suppressor of cytokine signaling; SPATS2, spermatogenesis associated serine rich 2 gene; SPRY1, sprouty RTK signaling antagonist 1 gene; TG, triglycerides; TMEM175, transmembrane protein 175 gene; TRIM2, tripartite motif containing 2 gene; ZER1, Zyg-11 related cell cycle regulator gene.
a Coded allele/noncoded allele.
b Frequency of the coded allele.
c These estimates pertain to the 2-df joint test of main and interaction effects.
d These P values pertain to 1-df tests of interaction effects.
e Significance of the stage 1 heterogeneity across ancestry groups in the most significant 2-df model.
f These loci were also discovered by Liu et al. (39) in a concurrent association study focused on exonic variants.
g Trans-ancestry meta-analysis.
h Alcohol consumption categorized into drinkers and nondrinkers.
i Alcohol consumption categorized into regular drinkers and non–regular drinkers.
Figure 2.
Significance and direction of effects of index genetic variants identified at 18 novel loci for 3 lipid traits (high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG)) in a genome-wide meta-analysis of gene-alcohol interaction and lipid levels, 2017. For each combination of index variant and lipid trait, the effect direction of and P value for the most significant association is shown. For example, the rs11:63911273 variant was most significantly associated with HDL-C in the transancestry meta-analysis, using the “current drinker” alcohol consumption variable. Shades of purple and yellow represent negative and positive directions of effect, respectively, while associations of either direction with a P value greater than 0.05 are white. Effect +: the direction of effect is greater than or equal to zero; Effect –: the direction of effect is less than zero. df, degrees of freedom; ID, identification; rs, reference SNP; SNP, single nucleotide polymorphism.
Known loci
The remaining 129 of the 147 significant loci had been identified in previous GWAS of lipid traits (Web Table 6) (2–8). This is a subset of all known lipid loci: Web Table 8 shows the significance of 314 reported index variants in all 236 known lipid loci among all 2-df joint tests and 1-df interaction tests of the combined analysis of stage 1 and stage 2, or stage 1 alone for variants not meeting the stage 2 inclusion criteria (2–8). Considering only the 314 known variants, no 1-df interactions were significant in the European, African, or transancestry meta-analyses (P < 8.8 × 10−6, corresponding to 0.05/(314 variants × 3 lipid traits × 2 alcohol consumption variables × 3 ancestry groups)).
Additional analyses
The percentages of variance in LDL-C, HDL-C, and TG concentrations explained by various loci were calculated in individual studies of multiple ancestry groups. Across ancestry groups, the mean variance explained by known lipid loci was 9.1% for HDL-C, 10.4% for LDL-C, and 7.5% for TG. The total percentage of additional variance explained by the 18 novel loci, including both genetic main effects and interaction effects, was 0.2 for HDL-C, 0.3 for LDL-C, and 0.4 for TG. Ancestry-specific and study-specific estimates are shown in Web Table 9.
Functional annotations using HaploReg (34) and RegulomeDB (35) for variants at the 147 loci that were associated in the combined analysis of stages 1 and 2 are presented in Web Table 10, and associations of these variants with gene expression levels from the GTEx database (36) in a variety of tissues are shown in Web Table 11. A total of 443 variants were associated with gene expression levels, of which 27 variants were indicated by RegulomeDB as having strong evidence for an association with enhancer function.
Our gene prioritization analyses with DEPICT highlighted (FDR q values <5%) 165 genes at HDL-C-associated loci, 110 genes at LDL-C-associated loci, and 87 genes at TG-associated loci (Web Tables 11–14). Thus, at some loci, multiple potential causal genes were prioritized. DEPICT identified 656, 877, and 497 reconstituted gene sets that were significantly enriched (FDR q values <5%) for genes at HDL-C, LDL-C, and TG loci, respectively (Web Table 15). This large number of processes and enriched gene sets underscores the complex genetic, biological, and physiological mechanisms underlying lipid traits. Among the most significantly enriched gene sets were processes related to “total amount of body fat” and “abnormal liver morphology.” Finally, DEPICT revealed that genes at associated HDL-C, LDL-C, or TG loci were significantly enriched (FDR q values <5%) for association with gene expression in 23 tissues, 14 cell types, and 12 physiological systems (Web Table 16). We found a compelling enrichment of genes acting in hepatocytes and liver processes at associated loci for each of the 3 traits and of genes acting in adipose tissues for HDL-C and TG loci (Web Table 16, Web Figure 5).
Fourteen index variants at known lipid loci were associated with coronary artery disease with P values less than 1.7 × 10−4 (0.05/(147 variants × 2 disease outcomes)), and 11 of these were also associated with myocardial infarction (Web Table 17) (38). None of the index variants at novel loci were significantly associated with these clinical endpoints.
DISCUSSION
We performed a GWAS of lipid levels incorporating interactions with alcohol consumption and identified 147 genome-wide-significant lipid loci, of which 18 are novel.
Despite the large sample of 394,584 individuals, which is comparable to sample sizes in other successful genetic interaction studies (40, 41), genome-wide-significant interactions were not found in the present study. Gene-alcohol interactions also do not appear to have contributed substantially to the discovery of the 18 novel loci, given that the genetic main effect of index variants at 15 of the 18 novel loci passed the stage 1 suggestive significance threshold. Below, we highlight 3 of the novel loci that harbor especially promising candidate genes with putative roles in lipid metabolism based on existing evidence from cellular and experimental models.
One of the newly identified associations for LDL-C maps to the proprotein convertase subtilisin/kexin type 5 gene (PCSK5), a member of the same gene family as proprotein convertase subtilisin/kexin type 9 (PCSK9), which is targeted by new drugs that successfully lower LDL-C levels (42, 43). Several independent lines of evidence support the involvement of PCSK5 in the regulation of lipid levels. First, in a candidate gene study, Iatan et al. (44) found that variants in PCSK5 were associated with levels of HDL-C. Additionally, in vitro studies of cell lines show that proprotein convertase subtilisin/kexin type 5 (PCSK5) inactivates endothelial lipase directly through cleavage and that it may also inactivate endothelial lipase and lipoprotein lipase indirectly through activation of angiopoietin-like protein 3 (45). Endothelial lipase, lipoprotein lipase, and angiopoietin-like protein 3 have all been robustly implicated in the regulation of lipid levels, probably with primary roles in the metabolism of HDL-C and TG (3, 46–48). In our study, the PCSK5 locus was associated at genome-wide significance only with LDL-C levels; it was associated at nominal significance (P < 0.05) with TG levels.
One novel locus for TG mapped to the apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1) complementation factor gene (A1CF), which encodes APOBEC1 complementation factor. Liu et al. (39) also identified the same index variant (rs41274050) in association with TG in a concurrent study. They showed that introducing the minor allele of rs41274050 in mice led to increased TG levels, confirming the functional role of this missense variant in the regulation of TG levels (39). APOBEC1 complementation factor forms an enzymatic complex with APOBEC1 and deaminates apolipoprotein B mRNA (49). This site-specific deamination of cytidine to uridine results in the production of the apolipoprotein B 48 isoform as opposed to the apolipoprotein B 100 isoform (49). The apolipoprotein B 48 isoform is critical in the assembly and secretion of chylomicrons, which mainly carry dietary-derived TG (50). Interestingly, a recent GWAS carried out among persons of East Asian ancestry identified the apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1) locus as being associated with HDL-C levels (5)—an association that we confirmed in our analysis (index variant: 12:7725904:ID). At nominal significance, both of the index variants near A1CF and APOBEC1 were associated with all 3 lipid traits (P < 0.05). Given the role of apolipoprotein B 100 in atherosclerosis, promoting the synthesis of apolipoprotein B 48 instead of apolipoprotein B 100 may represent a possible therapeutic strategy for the prevention of cardiovascular disease (51). Neither the index variant at A1CF nor APOBEC1 was significantly associated with coronary artery disease or myocardial infarction in the largest GWAS of these outcomes. However, further studies are needed to characterize their role in cardiovascular disease, given our multiancestry design and the European-driven design of the available GWAS data on cardiovascular disease outcomes (Web Table 17).
Variants closest to the mono-ADP ribosylhydrolase 1 (macro domain-containing protein 1) gene (MACROD1) were associated with HDL-C levels and TG levels. This locus was also reported in the concurrent study by Liu et al. (39), although the index variant in their study was located in the phospholipase C beta 3 (PLCB3) gene, around 120 kilo–base pairs away from the index variant in the present study. We found that variants at this locus were associated with expression levels of another nearby gene, the vascular endothelial growth factor B gene (VEGFB), in adipose and heart tissue (Web Table 11). Vascular endothelial growth factor B (VEGF-B) is reportedly involved in endothelial fatty acid transport, with Vegfb−/− mice showing less accumulation of lipids in muscle, heart, and brown adipose tissue but a greater uptake of fatty acids in white adipose tissue and higher body weight (52). Additionally, inhibition of the vascular endothelial growth factor B (Vegfb) protein in a mouse model of type 2 diabetes resulted in an improved glycemic profile, as well as a reduction of dyslipidemia (53). Mice lacking the Vegfb protein had lower levels of TG and LDL-C accompanied by higher levels of HDL-C. Subsequent studies in other mouse models did not corroborate these findings: Dijkstra et al. (54) found in an independent strain of mice that knocking out the mouse vascular endothelial growth factor B gene (Vegfb) had no effect on TG and total cholesterol levels, while Rubciuc et al. (55) and Rafii and Carmeliet (56) reported that transduction of the human VEGFB gene into mice led to increased vascularity in adipose tissue and an improved lipid profile. Our results provide insight into the effects of VEGF-B in humans to complement the divergent reports from rodent studies. The A allele of index variant rs190528931 is associated with decreased expression of VEGFB in adipose and heart tissue, decreased levels of HDL-C, and increased levels of TG. Additionally, rs190528931 was also associated with nominally significant increased levels of LDL-C (P < 0.05). Hence, evidence from our study suggests that inhibition of VEGF-B does not improve lipid profile but instead promotes dyslipidemia.
The strengths of this study include the large sample size, the diverse ancestral composition of the sample, and the use of a dense reference panel for genotype imputation (57). A limitation of this study is the imbalance in ancestry groups between stage 1 and stage 2. Persons of African ancestry were well-represented in stage 1 but underrepresented in stage 2. In contrast, persons of Asian and Hispanic ancestry were relatively underrepresented in stage 1 as compared with stage 2. A more balanced division of participants across stages 1 and 2 may have led to the identification of additional loci. Additionally, alcohol consumption may be underreported in both self-administered questionnaires and interviews, leading to a loss of statistical power due to misclassification (58). Similarly, the classification of alcohol consumption into categories such as “regular drinkers” and “current drinkers” may have reduced power relative to treating it as a fully quantitative variable (59). Nevertheless, the use of such categories was necessary for harmonizing data from 111 studies with heterogeneous measurement of alcohol consumption. It is plausible that more comprehensive characterization of alcohol consumption could reveal interactions that were missed in our study.
In conclusion, we identified 18 novel loci that were significantly associated with lipid traits, and these include several loci with genes (PCSK5, VEGFB, and A1CF) that have a putative role in lipid metabolism based on existing evidence from cellular and experimental models. The associations identified in this study appear to be driven by genetic main effects, and it remains uncertain whether alcohol consumption modifies the association of genetic variants with lipid levels.
Supplementary Material
ACKNOWLEDGMENTS
Authors: Paul S. de Vries, Michael R. Brown, Amy R. Bentley, Yun J. Sung, Thomas W. Winkler, Ioanna Ntalla, Karen Schwander, Aldi T. Kraja, Xiuqing Guo, Nora Franceschini, Ching-Yu Cheng, Xueling Sim, Dina Vojinovic, Jennifer E. Huffman, Solomon K. Musani, Changwei Li, Mary F. Feitosa, Melissa A. Richard, Raymond Noordam, Hugues Aschard, Traci M. Bartz, Lawrence F. Bielak, Xuan Deng, Rajkumar Dorajoo, Kurt K. Lohman, Alisa K. Manning, Tuomo Rankinen, Albert V. Smith, Salman M. Tajuddin, Evangelos Evangelou, Mariaelisa Graff, Maris Alver, Mathilde Boissel, Jin Fang Chai, Xu Chen, Jasmin Divers, Ilaria Gandin, Chuan Gao, Anuj Goel, Yanick Hagemeijer, Sarah E. Harris, Fernando P. Hartwig, Meian He, Andrea R. V. R. Horimoto, Fang-Chi Hsu, Anne U. Jackson, Anuradhani Kasturiratne, Pirjo Komulainen, Brigitte Kühnel, Federica Laguzzi, Joseph H. Lee, Jian’an Luan, Leo-Pekka Lyytikäinen, Nana Matoba, Ilja M. Nolte, Maik Pietzner, Muhammad Riaz, M. Abdullah Said, Robert A. Scott, Tamar Sofer, Alena Stančáková, Fumihiko Takeuchi, Bamidele O. Tayo, Peter J. van der Most, Tibor V. Varga, Yajuan Wang, Erin B. Ware, Wanqing Wen, Lisa R. Yanek, Weihua Zhang, Jing Hua Zhao, Saima Afaq, Najaf Amin, Marzyeh Amini, Dan E. Arking, Tin Aung, Christie Ballantyne, Eric Boerwinkle, Ulrich Broeckel, Archie Campbell, Mickaël Canouil, Sabanayagam Charumathi, Yii-Der Ida Chen, John M. Connell, Ulf de Faire, Lisa de las Fuentes, Renée de Mutsert, H. Janaka de Silva, Jingzhong Ding, Anna F. Dominiczak, Qing Duan, Charles B. Eaton, Ruben N. Eppinga, Jessica D. Faul, Virginia Fisher, Terrence Forrester, Oscar H. Franco, Yechiel Friedlander, Mohsen Ghanbari, Franco Giulianini, Hans J. Grabe, Megan L. Grove, C. Charles Gu, Tamara B. Harris, Sami Heikkinen, Chew-Kiat Heng, Makoto Hirata, James E. Hixson, Barbara V. Howard, M. Arfan Ikram, the InterAct Consortium, David R. Jacobs, Jr., Craig Johnson, Jost Bruno Jonas, Candace M. Kammerer, Tomohiro Katsuya, Chiea Chuen Khor, Tuomas O. Kilpeläinen, Woon-Puay Koh, Heikki A. Koistinen, Ivana Kolcic, Charles Kooperberg, Jose E. Krieger, Steve B. Kritchevsky, Michiaki Kubo, Johanna Kuusisto, Timo A. Lakka, Carl D. Langefeld, Claudia Langenberg, Lenore J. Launer, Benjamin Lehne, Rozenn N. Lemaitre, Yize Li, Jingjing Liang, Jianjun Liu, Kiang Liu, Marie Loh, Tin Louie, Reedik Mägi, Ani W. Manichaikul, Colin A. McKenzie, Thomas Meitinger, Andres Metspalu, Yuri Milaneschi, Lili Milani, Karen L. Mohlke, Thomas H. Mosley, Jr., Kenneth J. Mukamal, Mike A. Nalls, Matthias Nauck, Christopher P. Nelson, Nona Sotoodehnia, Jeff R. O’Connell, Nicholette D. Palmer, Raha Pazoki, Nancy L. Pedersen, Annette Peters, Patricia A. Peyser, Ozren Polasek, Neil Poulter, Leslie J. Raffel, Olli T. Raitakari, Alex P. Reiner, Treva K. Rice, Stephen S. Rich, Antonietta Robino, Jennifer G. Robinson, Lynda M. Rose, Igor Rudan, Carsten O. Schmidt, Pamela J. Schreiner, William R. Scott, Peter Sever, Yuan Shi, Stephen Sidney, Mario Sims, Blair H. Smith, Jennifer A. Smith, Harold Snieder, John M. Starr, Konstantin Strauch, Nicholas Tan, Kent D. Taylor, Yik Ying Teo, Yih Chung Tham, André G. Uitterlinden, Diana van Heemst, Dragana Vuckovic, Melanie Waldenberger, Lihua Wang, Yujie Wang, Zhe Wang, Wen Bin Wei, Christine Williams, Gregory Wilson, Sr., Mary K. Wojczynski, Jie Yao, Bing Yu, Caizheng Yu, Jian-Min Yuan, Wei Zhao, Alan B. Zonderman, Diane M. Becker, Michael Boehnke, Donald W. Bowden, John C. Chambers, Ian J. Deary, Tõnu Esko, Martin Farrall, Paul W. Franks, Barry I. Freedman, Philippe Froguel, Paolo Gasparini, Christian Gieger, Bernardo L. Horta, Yoichiro Kamatani, Norihiro Kato, Jaspal S. Kooner, Markku Laakso, Karin Leander, Terho Lehtimäki, the LifeLines Cohort Study Group, Patrik K. E. Magnusson, Brenda Penninx, Alexandre C. Pereira, Rainer Rauramaa, Nilesh J. Samani, James Scott, Xiao-Ou Shu, Pim van der Harst, Lynne E. Wagenknecht, Ya Xing Wang, Nicholas J. Wareham, Hugh Watkins, David R. Weir, Ananda R. Wickremasinghe, Wei Zheng, Paul Elliott, Kari E. North, Claude Bouchard, Michele K. Evans, Vilmundur Gudnason, Ching-Ti Liu, Yongmei Liu, Bruce M. Psaty, Paul M. Ridker, Rob M. van Dam, Sharon L. R. Kardia, Xiaofeng Zhu, Charles N. Rotimi, Dennis O. Mook-Kanamori, Myriam Fornage, Tanika N. Kelly, Ervin R. Fox, Caroline Hayward, Cornelia M. van Duijn, E. Shyong Tai, Tien Yin Wong, Jingmin Liu, Jerome I. Rotter, W. James Gauderman, Michael A. Province, Patricia B. Munroe, Kenneth Rice, Daniel I. Chasman, L. Adrienne Cupples, Dabeeru C. Rao*, and Alanna C. Morrison*.
Author affiliations: Human Genetics Center, Department of Epidemiology, Human Genetics, and Environmental Sciences, School of Public Health, University of Texas Health Science Center at Houston, Houston, Texas (Paul S. de Vries, Michael R. Brown, Megan L. Grove, Zhe Wang, Bing Yu, Myriam Fornage, Alanna C. Morrison); Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland (Amy R. Bentley, Charles N. Rotimi); Division of Biostatistics, School of Medicine, Washington University, St. Louis, Missouri (Yun J. Sung, Karen Schwander, Lisa de las Fuentes, C. Charles Gu, Yize Li, Treva K. Rice, Dabeeru C. Rao); Department of Genetic Epidemiology, Faculty of Medicine, University of Regensburg, Regensburg, Germany (Thomas W. Winkler); Clinical Pharmacology Centre, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom (Ioanna Ntalla, Patricia B. Munroe); Division of Statistical Genomics, Department of Genetics, School of Medicine, Washington University, St. Louis, Missouri (Aldi T. Kraja, Mary F. Feitosa, Lihua Wang, Christine Williams, Mary K. Wojczynski, Michael A. Province); Genomic Outcomes, Pediatrics, Institute for Translational Genomics and Population Sciences, LA BioMed, Harbor-UCLA Medical Center, Torrance, California (Xiuqing Guo, Yii-Der Ida Chen, Kent D. Taylor, Jie Yao, Jerome I. Rotter); Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Nora Franceschini, Mariaelisa Graff, Yujie Wang, Kari E. North); Singapore Eye Research Institute, Singapore National Eye Centre, Singapore, Republic of Singapore (Ching-Yu Cheng, Tin Aung, Sabanayagam Charumathi, Yuan Shi, Nicholas Tan, Yih Chung Tham, Tien Yin Wong); Centre for Quantitative Medicine, Academic Medicine Research Institute, Ophthalmology and Visual Sciences Academic Clinical Program, Duke-NUS Medical School, Singapore, Republic of Singapore (Ching-Yu Cheng, Sabanayagam Charumathi); Department of Ophthalmology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Republic of Singapore (Ching-Yu Cheng, Tin Aung, Nicholas Tan, Tien Yin Wong); Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore, Republic of Singapore (Xueling Sim, Jin Fang Chai, Woon-Puay Koh, Yik Ying Teo, Rob M. van Dam, E. Shyong Tai); Department of Epidemiology, Erasmus University Medical Center, Rotterdam, the Netherlands (Dina Vojinovic, Najaf Amin, Oscar H. Franco, Mohsen Ghanbari, M. Arfan Ikram, André G. Uitterlinden, Cornelia M. van Duijn); MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, United Kingdom (Jennifer E. Huffman, Caroline Hayward); Jackson Heart Study, Department of Medicine, University of Mississippi Medical Center, Jackson, Mississippi (Solomon K. Musani, Mario Sims); Department of Epidemiology and Biostatistics, College of Public Health, University of Georgia at Athens, Athens, Georgia (Changwei Li); Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, Texas (Melissa A. Richard, Myriam Fornage); Department of Internal Medicine and Department of Gerontology and Geriatrics, Leiden University Medical Center, Leiden, the Netherlands (Raymond Noordam, Diana van Heemst); Department of Epidemiology, T.H. Chan School of Public Health, Harvard University, Boston, Massachusetts (Hugues Aschard); Centre de Bioinformatique, Biostatistique et Biologie Intégrative, Institut Pasteur, Paris, France (Hugues Aschard); Cardiovascular Health Research Unit, Department of Biostatistics, School of Public Health, University of Washington, Seattle, Washington (Traci M. Bartz, Rozenn N. Lemaitre, Nona Sotoodehnia, Bruce M. Psaty); Cardiovascular Health Research Unit, Department of Medicine, School of Medicine, University of Washington, Seattle, Washington (Traci M. Bartz, Rozenn N. Lemaitre, Nona Sotoodehnia, Bruce M. Psaty); Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan (Lawrence F. Bielak, Patricia A. Peyser, Jennifer A. Smith, Wei Zhao, Sharon L. R. Kardia); Department of Biostatistics, School of Public Health, Boston University, Boston, Massachusetts (Xuan Deng, Virginia Fisher, Ching-Ti Liu, L. Adrienne Cupples); Genome Institute of Singapore, Agency for Science Technology and Research, Singapore, Republic of Singapore (Rajkumar Dorajoo, Chiea Chuen Khor, Jianjun Liu, Yik Ying Teo); Division of Public Health Sciences, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Kurt K. Lohman, Jasmin Divers, Fang-Chi Hsu, Carl D. Langefeld, Lynne E. Wagenknecht); Division of Biostatistical Sciences, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Kurt K. Lohman, Jasmin Divers, Fang-Chi Hsu, Carl D. Langefeld); Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, Boston, Massachusetts (Alisa K. Manning); Department of Medicine, Harvard Medical School, Boston, Massachusetts (Alisa K. Manning, Tamar Sofer); Human Genomics Laboratory, Pennington Biomedical Research Center, Baton Rouge, Louisiana (Tuomo Rankinen, Claude Bouchard); Icelandic Heart Association, Kopavogur, Iceland (Albert V. Smith, Vilmundur Gudnason); Faculty of Medicine, University of Iceland, Reykjavik, Iceland (Albert V. Smith, Vilmundur Gudnason); Health Disparities Research Section, Laboratory of Epidemiology and Population Sciences, National Institute on Aging, National Institutes of Health, Baltimore, Maryland (Salman M. Tajuddin, Michele K. Evans); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, United Kingdom (Evangelos Evangelou, Weihua Zhang, Saima Afaq, Benjamin Lehne, Marie Loh, Raha Pazoki, William R. Scott, John C. Chambers, Paul Elliott); Department of Hygiene and Epidemiology, University of Ioannina Medical School, Ioannina, Greece (Evangelos Evangelou); Estonian Genome Center, University of Tartu, Tartu, Estonia (Maris Alver, Reedik Mägi, Andres Metspalu, Lili Milani, Tõnu Esko); Centre National de la Recherche Scientifique Unité Mixte de Recherche 8199, European Genomic Institute for Diabetes, Institut Pasteur de Lille, University of Lille, Lille, France (Mathilde Boissel, Mickaël Canouil, Philippe Froguel); Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden (Xu Chen, Nancy L. Pedersen, Patrik K. E. Magnusson); Department of Medical, Surgical, and Health Sciences, University of Trieste, Trieste, Italy (Ilaria Gandin, Dragana Vuckovic, Paolo Gasparini); Molecular Genetics and Genomics Program, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Chuan Gao); Division of Cardiovascular Medicine, Radcliffe Department of Medicine, University of Oxford, Oxford, United Kingdom (Anuj Goel, Martin Farrall, Hugh Watkins); Wellcome Centre for Human Genetics, University of Oxford, Oxford, United Kingdom (Anuj Goel, Martin Farrall, Hugh Watkins); Department of Cardiology, Faculty of Medical Sciences, University of Groningen and University Medical Center Groningen, Groningen, the Netherlands (Yanick Hagemeijer, M. Abdullah Said, Ruben N. Eppinga, Pim van der Harst); Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh, United Kingdom (Sarah E. Harris, John M. Starr, Ian J. Deary); Medical Genetics Section, University of Edinburgh Centre for Genomic and Experimental Medicine and MRC Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, United Kingdom (Sarah E. Harris); Postgraduate Programme in Epidemiology, Federal University of Pelotas, Pelotas, Brazil (Fernando P. Hartwig, Bernardo L. Horta); MRC Integrative Epidemiology Unit, University of Bristol, Bristol, United Kingdom (Fernando P. Hartwig); Department of Occupational and Environmental Health and State Key Laboratory of Environmental Health (Incubation), Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (Meian He, Caizheng Yu); Laboratory of Genetics and Molecular Cardiology, Heart Institute, University of São Paulo Medical School, São Paulo, Brazil (Andrea R. V. R. Horimoto, Jose E. Krieger, Alexandre C. Pereira); Department of Biostatistics and Center for Statistical Genetics, School of Public Health, University of Michigan, Ann Arbor, Michigan (Anne U. Jackson, Michael Boehnke); Department of Public Health, Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka (Anuradhani Kasturiratne, Ananda R. Wickremasinghe); Foundation for Research in Health Exercise and Nutrition, Kuopio Research Institute of Exercise Medicine, Kuopio, Finland (Pirjo Komulainen, Timo A. Lakka, Rainer Rauramaa); Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (Brigitte Kühnel, Melanie Waldenberger, Christian Gieger); Institute of Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (Brigitte Kühnel, Annette Peters, Melanie Waldenberger); Unit of Cardiovascular Epidemiology, Institute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden (Federica Laguzzi, Ulf de Faire, Karin Leander); Sergievsky Center and Taub Institute, Columbia University Medical Center, New York, New York (Joseph H. Lee); MRC Epidemiology Unit, University of Cambridge, Cambridge, United Kingdom (Jian’an Luan, Robert A. Scott, Jing Hua Zhao, Claudia Langenberg, Nicholas J. Wareham); Department of Clinical Chemistry, Fimlab Laboratories, Tampere, Finland (Leo-Pekka Lyytikäinen, Terho Lehtimäki); Department of Clinical Chemistry and Finnish Cardiovascular Research Center–Tampere, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland (Leo-Pekka Lyytikäinen, Terho Lehtimäki); Laboratory for Statistical Analysis, Center for Integrative Medical Sciences, Kokuritsu Kenkyu Kaihatsu Hojin Rikagaku Kenkyusho, Yokohama, Japan (Nana Matoba, Yoichiro Kamatani, Michiaki Kubo); Department of Epidemiology, Faculty of Medical Sciences, University of Groningen and University Medical Center Groningen, Groningen, the Netherlands (Ilja M. Nolte, Peter J. van der Most, Marzyeh Amini, Harold Snieder); Institute of Clinical Chemistry and Laboratory Medicine, University Medicine Greifswald, Greifswald, Germany (Maik Pietzner, Matthias Nauck); German Centre for Cardiovascular Research, Partner Site Greifswald, Greifswald, Germany (Maik Pietzner, Matthias Nauck); Department of Cardiovascular Sciences, University of Leicester, Leicester, United Kingdom (Muhammad Riaz, Christopher P. Nelson, Nilesh J. Samani); NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, United Kingdom (Muhammad Riaz, Christopher P. Nelson, Nilesh J. Samani); Division of Sleep and Circadian Disorders, Brigham and Women’s Hospital, Boston, Massachusetts (Tamar Sofer); Institute of Clinical Medicine, School of Medicine, University of Eastern Finland, Kuopio, Finland (Alena Stančáková, Sami Heikkinen, Johanna Kuusisto, Markku Laakso); Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan (Fumihiko Takeuchi, Norihiro Kato); Department of Public Health Sciences, Stritch School of Medicine, Loyola University Chicago, Maywood, Illinois (Bamidele O. Tayo); Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University Diabetes Centre, Skåne University Hospital, Malmö, Sweden (Tibor V. Varga, Paul W. Franks); Department of Population and Quantitative Health and Sciences, School of Medicine, Case Western Reserve University, Cleveland, Ohio (Yajuan Wang, Jingjing Liang, Xiaofeng Zhu); Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, Michigan (Erin B. Ware, Jessica D. Faul, Jennifer A. Smith, David R. Weir); Division of Epidemiology, Department of Medicine, School of Medicine, Vanderbilt University, Nashville, Tennessee (Wanqing Wen, Xiao-Ou Shu, Wei Zheng); Division of General Internal Medicine, Department of Medicine, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Lisa R. Yanek, Diane M. Becker); Department of Cardiology, Ealing Hospital, Middlesex, United Kingdom (Weihua Zhang, John C. Chambers, Jaspal S. Kooner); McKusick-Nathans Institute of Genetic Medicine, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Dan E. Arking); Ophthalmology and Visual Sciences Academic Clinical Program, Duke-NUS Medical School, National University of Singapore, Singapore, Republic of Singapore (Tin Aung, Tien Yin Wong); Section of Cardiovascular Research, Baylor College of Medicine, Houston, Texas (Christie Ballantyne); Houston Methodist Debakey Heart and Vascular Center, Houston, Texas (Christie Ballantyne); Department of Epidemiology, Human Genetics, and Environmental Sciences, School of Public Health, University of Texas, Houston, Texas (Eric Boerwinkle, James E. Hixson); Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas (Eric Boerwinkle); Section of Genomic Pediatrics, Department of Pediatrics, Medicine and Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin (Ulrich Broeckel); Centre for Genomic and Experimental Medicine, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, United Kingdom (Archie Campbell); Ninewells Hospital and Medical School, University of Dundee, Dundee, United Kingdom (John M. Connell); Cardiovascular Division, Department of Medicine, School of Medicine, Washington University, St. Louis, Missouri (Lisa de las Fuentes); Clinical Epidemiology, Leiden University Medical Center, Leiden, Netherlands (Renée de Mutsert, Dennis O. Mook-Kanamori); Department of Medicine, Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka (H. Janaka de Silva); Center on Diabetes, Obesity, and Metabolism, Section of Gerontology and Geriatric Medicine, Division of Public Health Sciences, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Jingzhong Ding); Institute of Cardiovascular and Medical Sciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom (Anna F. Dominiczak); Department of Genetics, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Qing Duan, Karen L. Mohlke); Department of Family Medicine and Epidemiology, Alpert Medical School of Brown University, Providence, Rhode Island (Charles B. Eaton); Tropical Metabolism Research Unit, Tropical Medicine Research Institute, University of the West Indies, Mona, Jamaica (Terrence Forrester, Colin A. McKenzie); Institute of Social and Preventive Medicine, University of Bern, Bern, Switzerland (Oscar H. Franco); Unit of Epidemiology, Braun School of Public Health, Hebrew University-Hadassah Medical Center, Jerusalem, Israel (Yechiel Friedlander); Department of Genetics, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran (Mohsen Ghanbari); Division of Preventive Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Franco Giulainini, Lynda M. Rose, Paul M. Ridker, Daniel I. Chasman); Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald, Germany (Hans J. Grabe); Laboratory of Epidemiology and Population Sciences, National Institute on Aging, National Institutes of Health, Bethesda, Maryland (Tamara B. Harris, Lenore J. Launer); Institute of Biomedicine, School of Medicine, University of Eastern Finland, Kuopio Campus, Kuopio, Finland (Sami Heikkinen, Timo A. Lakka); Department of Paediatrics, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Republic of Singapore (Chew-Kiat Heng); Khoo Teck Puat–National University Children’s Medical Institute, National University Health System, Singapore, Republic of Singapore (Chew-Kiat Heng); Laboratory of Genome Technology, Human Genome Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan (Makoto Hirata); MedStar Health Research Institute, Hyattsville, Maryland (Barbara V. Howard); Center for Clinical and Translational Sciences and Department of Medicine, School of Medicine, Georgetown University, Washington, DC (Barbara V. Howard); Center for Clinical and Translational Sciences and Department of Medicine, College of Medicine, Howard University, Washington, DC (Barbara V. Howard); Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, Rotterdam, the Netherlands (M. Arfan Ikram); Department of Neurology, Erasmus University Medical Center, Rotterdam, the Netherlands (M. Arfan Ikram); Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, Minnesota (David R. Jacobs, Jr., Pamela J. Schreiner); Collaborative Health Studies Coordinating Center, University of Washington, Seattle, Washington (Craig Johnson); Department of Ophthalmology, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany (Jost Bruno Jonas); Beijing Institute of Ophthalmology, Beijing Ophthalmology and Visual Science Key Laboratory, Beijing Tongren Eye Center, Capital Medical University, Beijing, China (Jost Bruno Jonas); Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania (Candace M. Kammerer); Department of Clinical Gene Therapy, Graduate School of Medicine, Osaka University, Suita, Japan (Tomohiro Katsuya); Department of Geriatric and General Medicine, Graduate School of Medicine, Osaka University, Suita, Japan (Tomohiro Katsuya); Department of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Republic of Singapore (Chiea Chuen Khor); Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark (Tuomas O. Kilpeläinen); Department of Environmental Medicine and Public Health, Icahn School of Medicine at Mount Sinai, New York, New York (Tuomas O. Kilpeläinen); Programme in Health Services and Systems Research, Duke-NUS Medical School, Singapore, Republic of Singapore (Woon-Puay Koh, E. Shyong Tai); Department of Health, National Institute for Health and Welfare, Helsinki, Finland (Heikki A. Koistinen); Department of Medicine, Faculty of Medicine, University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland (Heikki A. Koistinen); Minerva Foundation Institute for Medical Research, Helsinki, Finland (Heikki A. Koistinen); Department of Public Health, School of Medicine, University of Split, Split, Croatia (Ivan Kolcic, Ozren Polasek); Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington (Charles Kooperberg, Alex P. Reiner); Department of Epidemiology, School of Public Health, University of Washington, Seattle, Washington (Charles Kooperberg, Alex P. Reiner); Sticht Center for Healthy Aging and Alzheimer’s Prevention, Department of Internal Medicine, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Steve B. Kritchevsky); Department of Clinical Physiology and Nuclear Medicine, Kuopio University Hospital, Kuopio, Finland (Timo A. Lakka); Division of Epidemiology, Department of Preventive Medicine, Feinberg School of Medicine, Northwestern University, Chicago, Illinois (Kiang Liu); Translational Laboratory in Genetic Medicine, Agency for Science, Technology and Research, Singapore, Republic of Singapore (Marie Loh); Department of Biostatistics, School of Public Health, University of Washington, Seattle, Washington (Tin Louie, Kenneth Rice); Biostatistics Section, Center for Public Health Genomics, School of Medicine, University of Virginia, Charlottesville, Virginia (Ani W. Manichaikul); Institute of Human Genetics, German Research Center for Environmental Health, Neuherberg, Germany (Thomas Meitinger); Institute of Human Genetics, Technische Universität München, Munich, Germany (Thomas Meitinger); Department of Psychiatry, Amsterdam Neuroscience and Amsterdam Public Health Research Institute, Amsterdam University Medical Center, Amsterdam, the Netherlands (Yuri Milaneschi, Brenda Penninx); Division of Geriatrics, School of Medicine, University of Mississippi Medical Center, Jackson, Mississippi (Thomas H. Mosley, Jr.); Division of General Medicine and Primary Care, Department of Medicine, Beth Israel Deaconess Medical Center, Boston, Massachusetts (Kenneth J. Mukamal); Data Tecnica International, Glen Echo, Maryland (Mike A. Nalls); Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, Maryland (Mike A. Nalls); Division of Endocrinology, Diabetes, and Nutrition, School of Medicine, University of Maryland, Baltimore, Maryland (Jeff R. O’Connell); Program for Personalized and Genomic Medicine, School of Medicine, University of Maryland, Baltimore, Maryland (Jeff R. O’Connell); Department of Biochemistry, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Nicholette D. Palmer, Donald W. Bowden); German Centre for Cardiovascular Research, Neuherberg, Germany (Annette Peters); Psychiatric Hospital “Sveti Ivan,” Zagreb, Croatia (Ozren Polasek); Gen-Info Ltd., Zagreb, Croatia (Ozren Polasek); International Centre for Circulatory Health, School of Public Health, Imperial College London, London, United Kingdom (Neil Poulter); Division of Genetic and Genomic Medicine, Department of Pediatrics, School of Medicine, University of California, Irvine, Irvine, California (Leslie J. Raffel); Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland (Olli T. Raitakari); Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland (Olli T. Raitakari); Center for Public Health Genomics, School of Medicine, University of Virginia, Charlottesville, Virginia (Stephen S. Rich); Institute for Maternal and Child Health–IRCCS “Burlo Garofolo,” Trieste, Italy (Antonietta Robino, Paolo Gasparini); Department of Epidemiology, College of Public Health, University of Iowa, Iowa City, Iowa (Jennifer G. Robinson); Department of Internal Medicine, Carver College of Medicine, University of Iowa, Iowa City, Iowa (Jennifer G. Robinson); Centre for Global Health Research, Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, United Kingdom (Igor Rudan); Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany (Carsten O. Schmidt); National Heart and Lung Institute, Imperial College London, London, United Kingdom (William R. Scott, Peter Sever, Jaspal S. Kooner, James Scott); Division of Research, Kaiser Permanente Northern California, Oakland, California (Stephen Sidney); Division of Population Health and Genomics, Ninewells Hospital and Medical School, University of Dundee, Dundee, United Kingdom (Blair H. Smith); Alzheimer Scotland Dementia Research Centre, University of Edinburgh, Edinburgh, United Kingdom (John M. Starr); Institute of Genetic Epidemiology, German Research Center for Environmental Health, Neuherberg, Germany (Konstantin Strauch); Department of Genetic Epidemiology, Institute for Medical Informatics, Biometry and Epidemiology and Faculty of Medicine, Ludwig Maximilian University, Munich, Germany (Konstantin Strauch); Life Sciences Institute, National University of Singapore, Singapore, Republic of Singapore (Yik Ying Teo); Graduate School for Integrative Science and Engineering, National University of Singapore, Singapore, Republic of Singapore (Yik Ying Teo); Department of Statistics and Applied Probability, Faculty of Science, National University of Singapore, Singapore, Republic of Singapore (Yik Ying Teo); Department of Internal Medicine, Erasmus University Medical Center, Rotterdam, the Netherlands (André G. Uitterlinden); Beijing Tongren Eye Center, Beijing Tongren Hospital, Capital Medical University, Beijing, China (Wen Bin Wei); Jackson Heart Study, School of Public Health, Jackson State University, Jackson, Mississippi (Gregory Wilson, Sr.); Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania (Jian-Min Yuan); Division of Cancer Control and Population Sciences, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Jian-Min Yuan); Behavioral Epidemiology Section, Laboratory of Epidemiology and Population Sciences, National Institute on Aging, National Institutes of Health, Baltimore, Maryland (Alan B. Zonderman); Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Republic of Singapore (John C. Chambers); Imperial College Healthcare NHS Trust, London, United Kingdom (John C. Chambers, Jaspal S. Kooner); MRC-PHE Centre for Environment and Health, Imperial College London, London, United Kingdom (John C. Chambers, Paul Elliott); NIHR Imperial College Biomedical Research Centre, Imperial College London, London, United Kingdom (John C. Chambers, Paul Elliott, Jaspal S. Kooner); Department of Psychology, Faculty of Medicine, University of Edinburgh, Edinburgh, United Kingdom (Ian J. Deary); Broad Institute of the Massachusetts Institute of Technology and Harvard University, Boston, Massachusetts (Tõnu Esko); Department of Nutrition, T. H. Chan School of Public Health, Harvard University, Boston, Massachusetts (Paul W. Franks, Rob M. van Dam); Department of Public Health and Clinical Medicine, Faculty of Medicine, Umeå University, Umeå, Sweden (Paul W. Franks); Oxford Centre for Diabetes, Endocrinology and Metabolism, Radcliffe Department of Medicine, Medical Sciences Division, University of Oxford, Oxford, United Kingdom (Paul W. Franks); Section of Nephrology, Department of Internal Medicine, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Barry I. Freedman); Department of Genomics of Common Disease, Imperial College London, London, United Kingdom (Philippe Froguel); German Center for Diabetes Research, Neuherberg, Germany (Christian Gieger); Department of Genetics, Faculty of Medical Sciences, University of Groningen and University Medical Center Groningen, Groningen, the Netherlands (Pim van der Harst); Beijing Institute of Ophthalmology, Beijing Tongren Eye Center, Beijing Ophthalmology and Visual Science Key Laboratory, Beijing Tongren Hospital, Capital Medical University, Beijing, China (Ya Xing Wang); Health Data Research UK, Imperial College London, London, United Kingdom (Paul Elliott); UK Dementia Research Institute, Imperial College London, London, United Kingdom (Paul Elliott); Carolina Center of Genome Sciences, University of North Carolina, Chapel Hill, North Carolina (Kari E. North); Section of Public Health Sciences, Department of Epidemiology and Prevention, Division of Public Health Sciences, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, North Carolina (Yongmei Liu); Department of Medicine, School of Medicine, University of Washington, Seattle, Washington (Bruce M. Psaty); Departments of Epidemiology and Health Services, School of Public Health, University of Washington, Seattle, Washington (Bruce M. Psaty); Kaiser Permanente Washington, Health Research Institute, Seattle, Washington (Bruce M. Psaty); Department of Medicine, Harvard Medical School, Boston, Massachusetts (Paul M. Ridker, Daniel I. Chasman); Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Republic of Singapore (Jianjun Liu, Rob M. van Dam, E. Shyong Tai); Department of Public Health and Primary Care, Leiden University Medical Center, Leiden, the Netherlands (Dennis O. Mook-Kanamori); Department of Epidemiology, School of Public Health and Tropical Medicine, Tulane University, New Orleans, Louisiana (Tanika N. Kelly); Division of Cardiology, Department of Medicine, University of Mississippi Medical Center, Jackson, Mississippi (Ervin R. Fox); Women’s Health Initiative Clinical Coordinating Center, Fred Hutchinson Cancer Research Center, Seattle, Washington (Jingmin Liu); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California (W. James Gauderman); NIHR Barts Cardiovascular Biomedical Research Unit, Queen Mary University of London, London, United Kingdom (Patricia B. Munroe); and Framingham Heart Study, Framingham, Massachusetts (L. Adrienne Cupples).
P.S.d.V., M.R.B., A.R.B., Y.J.S., T.W.W., and I.N. contributed equally to this article as junior authors. P.B.M., K.R., D.I.C., L.A.C., D.C.R., and A.C.M. contributed equally to this article as senior authors.
This work was supported by grant R01HL118305 from the National Heart, Lung, and Blood Institute (NHLBI), US National Institutes of Health (NIH). Infrastructure for the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium was supported in part by NHLBI grant R01HL105756. American Heart Association grant 17POST33350042 made it possible for P.S.d.V. to lead this project. Study-specific funding sources are provided below, starting with stage 1 studies and followed by stage 2 studies.
Stage 1 studies—Age, Gene/Environment Susceptibility (AGES)-Reykjavik Study: The AGES-Reykjavik Study was funded by NIH contract N01-AG012100; the Intramural Research Program of the NIH, National Institute on Aging (NIA); Intramural Research Program award ZIAEY000401 from the National Eye Institute, NIH; award IAA Y2-DC_1004-02 from the Division of Scientific Programs, National Institute on Deafness and Other Communication Disorders, NIH; the Icelandic Heart Association (Hjartavernd); and the Icelandic Parliament (Althingi). The study was approved by the Icelandic National Bioethics Committee. Atherosclerosis Risk in Communities (ARIC) Study: The ARIC Study was carried out as a collaborative study supported by NHLBI contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C; NHLBI grants R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute, NIH, contract U01HG004402; and NIH contract HHSN268200625226C. Infrastructure for ARIC was partly supported by grant UL1RR025005, a component of the NIH Roadmap for Medical Research. Baependi Heart Study: The Baependi Heart Study was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (grant 2013/17368-0), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, and the Hospital Samaritano Society (grant 25000.180.664/2011-35), through the Brazilian Ministry of Health’s Support Program for Institutional Development of the Unified Health System. Cardiovascular Health Study: The Cardiovascular Health Study was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, and N01HC85086 and NHLBI grants U01HL080295, R01HL085251, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and R01HL130114, with an additional contribution from the National Institute of Neurological Disorders and Stroke, NIH. Additional support was provided through grant R01AG023629 from the NIA. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute grant UL1TR000124, and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Diabetes Research Center grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Coronary Artery Risk Development in Young Adults (CARDIA) Study: The CARDIA Study was conducted and supported by the NHLBI in collaboration with the University of Alabama at Birmingham (grants HHSN268201300025C and HHSN268201300026C), Northwestern University (grant HHSN268201300027C), the University of Minnesota (grant HHSN268201300028C), the Kaiser Foundation Research Institute (grant HHSN268201300029C), and the Johns Hopkins School of Medicine (grant HHSN268200900041C). The CARDIA Study was also partially supported by the Intramural Research Program of the NIH, NIA. Genotyping was funded as part of the NHLBI Candidate-Gene Association Resource (grant N01-HC-65226) and the National Human Genome Research Institute’s Gene, Environment Association Studies (GENEVA) Consortium (grants U01-HG004729, U01-HG04424, and U01-HG004446). CROATIA-Korcula Study: The CROATIA-Korcula Study was funded by the Medical Research Council (United Kingdom), the Croatian Ministry of Science, Education and Sports (grant 216-1080315-0302), the European Commission Sixth Framework Programme EUROSPAN Project (contract LSHG-CT-2006-018947), and the Croatian Science Foundation (grant 8875). Single nucleotide polymorphism genotyping for the CROATIA-Korcula cohort was performed by the German Research Center for Environmental Health (Helmholtz Zentrum München), Neuherberg, Germany. CROATIA-Vis Study: The CROATIA-Vis Study was funded by the Medical Research Council, the Croatian Ministry of Science, Education and Sports (grant 216-1080315-0302), the European Commission Sixth Framework Programme EUROSPAN Project (contract LSHG-CT-2006-018947), and the Croatian Science Foundation (grant 8875). Single nucleotide polymorphism genotyping for the CROATIA-Vis cohort was performed in the core genotyping laboratory of the Wellcome Trust Clinical Research Facility (Western General Hospital, Edinburgh, United Kingdom). Erasmus Rucphen Family (ERF) Study: The ERF Study, as a part of the European Special Populations Research Network (EUROSPAN), was supported by European Commission Sixth Framework Programme Specific Targeted Research Projects grant 018947 (grant LSHG-CT-2006-01947) and also received funding from the European Commission Seventh Framework Programme (grant FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 with the European Commission under the Fifth Framework Programme (“Quality of Life and Management of Living Resources”; grant QLG2-CT-2002-01254). The ERF Study was further supported by the European Network for Genetic and Genomic Epidemiology (ENGAGE) Consortium and the Centre for Medical Systems Biology. High-throughput analysis of the ERF data was supported by a joint grant from the Netherlands Organisation for Scientific Research and the Russian Foundation for Basic Research (grant 047.017.043). The ERF Study was further supported by a grant from the Netherlands Organisation for Health Research and Development (ZonMw) (project 91111025). Family Heart Study: The Family Heart Study was funded by grants R01HL118305 and R01HL117078 from the NHLBI and grants 5R01DK07568102 and 5R01DK089256 from the NIDDK, NIH. Framingham Heart Study: The Framingham Heart Study was supported by the NHLBI and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) Project. This work was partially supported by the NHLBI (contracts N01-HC-25195 and HHSN268201500001I) and a contract with Affymetrix, Inc. (Santa Clara, California) for genotyping services (contract N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II), funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The research was partially supported by grant R01-DK089256 from the NIDDK (Principal Investigators: Ingrid B. Borecki, L. Adrienne Cupples, and Kari North). Genetic Epidemiology Network of Arteriopathy (GENOA) Study: Support for the GENOA Study was provided by the NHLBI (grants HL119443, HL118305, HL054464, HL054457, HL054481, HL071917, and HL087660). Genotyping was performed at the Mayo Clinic (Rochester, Minnesota) (Stephen T. Turner, Mariza de Andrade, Julie Cunningham). Genetic Epidemiology Network of Salt Sensitivity (GenSalt) Study: The GenSalt Study was supported by research grants (grants U01HL072507, R01HL087263, and R01HL090682) from the NHLBI. Healthy Aging in Neighborhoods of Diversity Across the Life Span (HANDLS): The HANDLS Study was supported by the Intramural Research Program of the NIH, NIA and the National Center on Minority Health and Health Disparities (project Z01-AG000513). Data analyses for the HANDLS Study utilized the high-performance computational resources of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov; http://hpc.nih.gov). Health, Aging, and Body Composition (HABC) Study: The HABC Study was funded by the NIA. The research was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The genome-wide association study (GWAS) was funded by NIA grant 1R01AG032098-01A1 to the Division of Public Health Sciences, Wake Forest School of Medicine, Wake Forest University, and genotyping services were provided by the Center for Inherited Disease Research (Baltimore, Maryland). The Center for Inherited Disease Research is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract HHSN268200782096C). The research was supported in part by the Intramural Research Program of the NIH, NIA. Health, Risk Factors, Exercise Training and Genetics (HERITAGE) Study: The HERITAGE Study was supported by NHLBI grant HL-45670. Howard University Family Study: The Howard University Family Study was supported by NIH grant S06GM008016-320107 to Charles N. Rotimi and NIH grant S06GM008016-380111 to Adebowale Adeyemo. Participant enrollment was carried out at the Howard University General Clinical Research Center, supported by NIH grant 2M01RR010284. The research was supported in part by the Intramural Research Program of the NIH, Center for Research on Genomics and Global Health. The Center for Research on Genomics and Global Health was supported by the National Human Genome Research Institute, the NIDDK, the Center for Information Technology, and the Office of the Director of the NIH (grant Z01HG200362). Genotyping support was provided by the Coriell Institute for Medical Research. Hypertension Genetic Epidemiology Network (HyperGEN) Study: The HyperGEN Study was funded by the following cooperative agreements (U10 agreements) with the NHLBI: HL54471, HL54472, HL54473, HL54495, HL54496, HL54497, HL54509, HL54515, and 2 R01 HL55673-12. The study involves the following centers—the University of Utah (Network Coordinating Center, Field Center, and Molecular Genetics Laboratory), the University of Alabama at Birmingham (Field Center and Echo Coordinating and Analysis Center), the Medical College of Wisconsin (Echo Genotyping Laboratory), Boston University (Field Center), the University of Minnesota (Field Center and Biochemistry Laboratory), the University of North Carolina (Field Center), Washington University (Data Coordinating Center), Weil Cornell Medical College (Echo Reading Center), and the NHLBI. Generation Scotland: Scottish Family Health Study (GS_SFHS): The GS_SFHS received core support from the Chief Scientist Office of the Scottish Government Health and Social Care Directorates (grant CZD/16/6) and the Scottish Funding Council (grant HR03006). Genotyping of the GS_SFHS samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility (Western General Hospital, Edinburgh, United Kingdom) and was funded by the Medical Research Council and the Wellcome Trust (Strategic Award “Stratifying Resilience and Depression Longitudinally” (STRADL) reference no. 104036/Z/14/Z). Jackson Heart Study: The Jackson Heart Study was supported by contracts HSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, and HHSN268201300050C from the National Institute on Minority Health and Health Disparities, NIH. Multi-Ethnic Study of Atherosclerosis (MESA): MESA was supported by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, and N01-HC-95169, by NHLBI grant HL071205, and by grant UL1-DR-001079 from the National Center for Research Resources, NIH. Funding for SHARe genotyping in MESA was provided by NHLBI contract N02-HL-6-4278. This publication was partially developed under Science to Achieve Results (STAR) research assistance agreement RD831697 (MESA Air), awarded by the US Environmental Protection Agency. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute grant UL1TR001881, and NIDDK Diabetes Research Center grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Netherlands Epidemiology of Obesity (NEO) Study: Genotyping in the NEO Study was supported by the Centre National de Génotypage (Paris, France), headed by Jean-Francois Deleuze. The NEO Study was supported by the participating departments, Division 2, and the Board of Directors of Leiden University Medical Center and by Leiden University (research profile area: Vascular and Regenerative Medicine). Dennis Mook-Kanamori was supported by the NWO (ZonMw-Veni grant 916.14.023). 1982 Pelotas Birth Cohort Study: The 1982 Pelotas Birth Cohort Study was conducted by the Postgraduate Program in Epidemiology at Universidade Federal de Pelotas (Pelotas, Brazil) with the collaboration of the Brazilian Public Health Association. From 2004 to 2013, the Wellcome Trust supported the study. The International Development Research Center, the World Health Organization, the Overseas Development Administration, the European Union, the National Support Program for Centers of Excellence, the Brazilian National Research Council, and the Brazilian Ministry of Health supported previous phases of the study. Genotyping of 1982 Pelotas Birth Cohort Study participants was supported by the Department of Science and Technology (Ministry of Health, Brazil), the National Fund for Scientific and Technological Development (Ministry of Science and Technology, Brazil), Funding of Studies and Projects (Ministry of Science and Technology, Brazil), and Coordination of Improvement of Higher Education Personnel (Ministry of Education, Brazil). Rotterdam Study: The Rotterdam Study was funded by Erasmus Medical Center (Erasmus MC) and Erasmus University (Rotterdam, the Netherlands), the Netherlands Organisation for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture and Science, the Ministry of Health, Welfare and Sports, the European Commission (Directorate-General for Science Research and Development (DG XII)), and the Municipality of Rotterdam. Generation and management of GWAS genotype data for the Rotterdam Study was conducted at the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC. The GWAS data sets were supported by the NWO (grants 175.010.2005.011 and 911-03-012); the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC; the Research Institute for Diseases in the Elderly (RIDE2 grant 014-93-015); the Netherlands Genomics Initiative/NWO; and the Netherlands Consortium for Healthy Aging (project 050-060-810). Singapore Chinese Eye Study, Singapore Malay Eye Study, and Singapore Indian Eye Study: The Singapore Malay Eye Study, the Singapore Indian Eye Study, and the Singapore Chinese Eye Study were supported by the National Medical Research Council (Singapore) (grants 0796/2003, 1176/2008, 1149/2008, STAR/0003/2008, 1249/2010, CG/SERI/2010, CIRG/1371/2013, and CIRG/1417/2015) and the Biomedical Research Council (Singapore) (grants 08/1/35/19/550 and 09/1/35/19/616). Ching-Yu Cheng was supported by an award from the National Medical Research Council (grant CSA/033/2012). The Singapore Tissue Network and the Genome Institute of Singapore (Agency for Science, Technology and Research, Singapore) provided services for tissue archival and genotyping, respectively. Singapore Chinese Health Study–Coronary Heart Disease Study: The Singapore Chinese Health Study was supported by the NIH (grants RO1 CA144034 and UM1 CA182876); the Coronary Heart Disease Study, a nested case-control study of myocardial infarction, was supported by the National Medical Research Council (Singapore) (grant 1270/2010); and genotyping was supported by the Hebrew University of Jerusalem–Campus for Research Excellence and Technological Enterprise (HUJ-CREATE) Programme of the National Research Foundation (Singapore) (project 370062002). Singapore 2 (SP2) Prospective Study Program: The SP2 Study (including the SP2-1M and SP2-610 subsets) was supported by individual research grants and clinician scientist awards from the National Medical Research Council (Singapore) and the Biomedical Research Council (Singapore). Women’s Genome Health Study: The Women’s Genome Health Study was supported by the NHLBI (grants HL043851 and HL080467) and the National Cancer Institute, NIH (grants CA047988 and UM1CA182913), with collaborative scientific support and funding for genotyping provided by Amgen, Inc. (Thousand Oaks, California). Women’s Health Initiative: The Women’s Health Initiative was funded by the NHLBI through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
Stage 2 studies—African American Diabetes Heart Study: The African American Diabetes Heart Study was supported by NIH grants R01 DK071891 and R01 HL092301 and the General Clinical Research Center of Wake Forest School of Medicine, Wake Forest University (grant M01-RR-07122). Airwave Health Monitoring Study: The Airwave Health Monitoring Study was funded by the United Kingdom Home Office (grant 780-TETRA), with additional support from the National Institute for Health Research (NIHR), the Imperial College Healthcare NHS Trust, and the Imperial College Biomedical Research Centre. The study received ethical approval from the National Health Service Multicentre Research Ethics Committee. This work used computing resources provided by the MRC-funded UK Medical Bioinformatics Partnership Programme (grant MR/L01632X/1). Paul Elliott received support from the Medical Research Council and Public Health England for the MRC-PHE Centre for Environment and Health (grant MR/L01341X/1) and from the NIHR Health Protection Research Unit in Health Impact of Environmental Hazards (grant HPRU-2012-10141). Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT): ASCOT and the collection of samples for the ASCOT DNA repository were supported by Pfizer, Inc. (New York, New York); by the Servier Research Group (Paris, France); and by Leo Laboratories (Copenhagen, Denmark). Genotyping was funded by the Centre National de Génotypage, the Medical Research Council, and the NIHR. This work forms part of the research program of the NIHR Cardiovascular Biomedical Research Unit (NIHR Barts Biomedical Research Centre) at Barts and The London School of Medicine and Dentistry, Queen Mary University of London. Practicia B. Munroe received support from the NIHR Barts Biomedical Research Centre at Barts and The London School of Medicine and Dentistry, Queen Mary University of London. Beijing Eye Study: The Beijing Eye Study was supported by the National Key Laboratory Fund, Beijing, China. BioBank Japan Project: The BioBank Japan Project was supported by the Japan Agency for Medical Research and Development and by the Japanese Ministry of Education, Culture, Sports, Sciences and Technology. British Genetics of Hypertension (BRIGHT) Study: The BRIGHT Study was supported by the Medical Research Council (grant G9521010D) and the British Heart Foundation (grant PG/02/128) and forms part of the research program of the NIHR Cardiovascular Biomedical Unit (NIHR Barts Biomedical Research Centre) at Barts and The London School of Medicine and Dentistry, Queen Mary University of London. Cardio-metabolic Genome Epidemiology Network Amagasaki (CAGE-Amagasaki) Study: The Cardio-metabolic Genome Epidemiology Network studies were supported by Core Research for Evolutional Science and Technology (CREST) grants from the Japan Science and Technology Agency; the Program for Promotion of Fundamental Studies in Health Sciences, National Institute of Biomedical Innovation (Japan); and a grant from the National Center for Global Health and Medicine (Japan). Cooperative Health Research in the Augsburg Region (KORA) Study: The KORA Study was initiated and financed by the German Research Center for Environmental Health (Helmholtz Zentrum München), which is supported by the German Federal Ministry of Education and Research and the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences, Ludwig Maximilians University, as part of the LMUinnovativ projects. Data From the Epidemiological Study on the Insulin Resistance Syndrome (DESIR) Study: The DESIR Study Group comprises investigators from Institut National de la Santé et de la Recherche Médicale (INSERM) Unité 1018 (Paris: B. Balkau, P. Ducimetière, and E. Eschwège), INSERM Unité 367 (Paris: F. Alhenc-Gelas), Centre Hospitalier Universitaire d’Angers (A. Girault), Bichat Hospital (Paris: F. Fumeron, M. Marre, and R. Roussel), Centre Hospitalier Universitaire de Rennes (F. Bonnet), Centre National de la Recherche Scientifique Unité Mixte de Recherche 8199 (Lille: A. Bonnefond and P. Froguel), Medical Examination Services (Alençon, Angers, Blois, Caen, Chartres, Chateauroux, Cholet, Le Mans, Orléans, and Tours), the Research Institute for General Medicine (J. Cogneau), the general practitioners of the region, and the Cross-Regional Institute for Health (C. Born, E. Caces, M. Cailleau, N. Copin, J. G. Moreau, F. Rakotozafy, J. Tichet, and S. Vol). The DESIR Study was supported by INSERM contracts with the Caisse Nationale d’Assurance Maladie des Travailleurs Salariés (Paris, France), Eli Lilly and Company (Indianapolis, Indiana), Novartis Pharmaceuticals Corporation (East Hanover, New Jersey), and Sanofi-Aventis France S.A. (Paris, France), as well as by INSERM (Réseaux en Santé Publique, Interactions Entre les Déterminants de la Santé, Cohortes Santé Très Grande Infrastructure de Recherche 2008), the Association Diabète Risque Vasculaire, the Fédération Française de Cardiologie, La Fondation de France, the Association de Langue Française pour l’Etude du Diabète et des Maladies Métabolique (ALFEDIAM), the Office National Interprofessionnel des Vins (ONIVINS), the Société Francophone du Diabète, Ardix Medical (Suresnes, France), Bayer Diagnostics (Tarrytown, New York), Becton Dickinson (Franklin Lakes, New Jersey), Cardionics, Inc. (Webster, Texas), Merck Santé S.A.S. (Lyon, France), Novo Nordisk (Bagsværd, Denmark), Laboratoires Pierre Fabre (Paris, France), Roche, Inc. (Basel, Switzerland), and Topcon Corporation (Tokyo, Japan). Dongfeng-Tongji Cohort Study: The Dongfeng-Tongji Cohort Study was supported by grants from the National Basic Research Program (grant 2011CB503800), the Programme of Introducing Talents of Discipline, the National Natural Science Foundation of China (grants NSFC-81473051, NSFC-81522040, and NSFC-81230069), and the Program for the New Century Excellent Talents in University (grant NCET-11-0169). Diabetes Heart Study: The Diabetes Heart Study was supported by the NIH (grants HL67348 and HL092301). Dose Responses to Exercise Training (DR’s EXTRA) Study: The DR’s EXTRA Study was supported by grants from the Ministry of Education and Culture of Finland (grants 722 and 627; 2004–2010); the Academy of Finland (grants 102318, 104943, 123885, and 211119); European Commission Sixth Framework Programme Integrated Project (EXGENESIS) grant LSHM-CT-2004-005272; the City of Kuopio (Kupio, Finland); the Juho Vainio Foundation; the Finnish Diabetes Association; the Finnish Foundation for Cardiovascular Research; Kuopio University Hospital; the Päivikki and Sakari Sohlberg Foundation; and the Social Insurance Institution of Finland. Estonian Genome Center of the University of Tartu (EGCUT): The EGCUT Study is part of the Estonian Biobank and was supported by European Union H2020 grants 692145, 676550, and 654248; Estonian Research Council grant IUT20-60; the Nordic Information for Action eScience Center; the European Institute for Innovation and Technology (EIT Health); the NIH (grant 2R01DK075787-06A1); and the European Union, through the European Regional Development Fund (Center of Excellence for Genomics and Translational Medicine (GenTransMed) project 2014-2020.4.01.15-0012). European Prospective Investigation Into Cancer and Nutrition (EPIC): The EPIC Study was funded by Cancer Research UK, the British Heart Foundation, the Medical Research Council, the Ministry of Agriculture, Fisheries and Food (United Kingdom), and the Europe Against Cancer Programme of the Commission of the European Communities. European Investigation Into Cancer and Nutrition–InterAct Case-Cohort Study (EPIC-InterAct): The EPIC-InterAct Study received funding from the European Union (Integrated Project LSHM-CT-2006-037197 in the European Commission Sixth Framework Programme). Fenland Study: The Fenland Study was funded by the Wellcome Trust and the Medical Research Council (grants MC_U106179471 and MC_UU_12015/1). Finland-United States Investigation of NIDDM Genetics (FUSION): The FUSION Study was supported by NIH grants DK093757, DK072193, DK062370, and ZIA-HG000024. Genotyping was conducted at the Genetic Resources Core Facility at the Johns Hopkins Institute of Genetic Medicine. Gene × Lifestyle Interactions and Complex Traits Involved in Elevated Disease Risk (GLACIER) Study: The GLACIER Study was supported by Novo Nordisk, the Swedish Research Council, the Swedish Heart-Lung Foundation, the European Research Council, and the Skåne Health Authority through grants to Paul W. Franks. Genetic Regulation of Arterial Pressure of Humans in the Community (GRAPHIC) Study: The GRAPHIC Study was funded by the British Heart Foundation (grant BHF/RG/2000004). This work falls under the portfolio of research supported by the NIHR Leicester Cardiovascular Biomedical Research Unit. Christopher P. Nelson and Nilesh J. Samani were supported by the British Heart Foundation, and Nilesh J. Samani is an NIHR Senior Investigator. Genetic Studies of Atherosclerosis Risk (GeneSTAR): GeneSTAR was supported by grants from the NHLBI (grants HL49762, HL59684, HL58625, HL071025, U01 HL72518, HL087698, HL092165, HL099747, and K23HL105897), the National Institute of Nursing Research, NIH (grant NR0224103), and the National Institute of Neurological Disorders and Stroke, NIH (grant NS062059) and by grants from the National Center for Research Resources to the Johns Hopkins General Clinical Research Center (grant M01-RR000052) and the Johns Hopkins Institute for Clinical and Translational Research (grant UL1 RR 025005). Hispanic Community Health Study/Study of Latinos (HCHS/SOL): The HCHS/SOL baseline examination was supported by contracts from the NHLBI to the University of North Carolina (grant N01-HC65233), the University of Miami (grant N01-HC65234), Albert Einstein College of Medicine (grant N01-HC65235), Northwestern University (grant N01-HC65236), and San Diego State University (grant N01-HC65237). The National Institute on Minority Health and Health Disparities, the National Institute on Deafness and Other Communication Disorders, the National Institute of Dental and Craniofacial Research, the NIDDK, the National Institute of Neurological Disorders and Stroke, and the NIH Office of Dietary Supplements additionally contributed funding to HCHS/SOL. The Genetic Analysis Center at the University of Washington was supported by NHLBI and National Institute of Dental and Craniofacial Research contracts (contracts HHSN268201300005C AM03 and MOD03). Additional analysis support was provided by NIDDK grants 1R01DK101855-01 and 13GRNT16490017 from the American Heart Association. Genotyping was also supported by National Center for Advancing Translational Sciences grant UL1TR000124 and NIDDK grant DK063491 to the Southern California Diabetes Endocrinology Research Center. HCHS/SOL was also supported in part by the Intramural Research Program of the NIH, NIDDK (contract HHSB268201200054C) and Illumina, Inc. (San Diego, California). Health and Retirement Study: The Health and Retirement Study was supported by the NIA (grants U01AG009740 and R03 AG046389). Genotyping was funded separately by the NIA (grants RC2 AG036495 and RC4 AG039029). Genotyping for the Health and Retirement Study was conducted by the NIH Center for Inherited Disease Research at Johns Hopkins University. Genotyping quality control and final preparation of the data were performed by the Genetics Coordinating Center at the University of Washington. Hypertension Genetic Epidemiology Network (HyperGEN)-Axiom Study: The HyperGEN-Axiom Study was supported by the NIH (NHLBI grant HL086718). Italian Network Genetic Isolates (INGI-CARL) Study: The INGI-CARL Study was partially supported by Regione Friuli-Venezia Giulia (grant L.26.2008) and the Italian Ministry of Health (grant GR-2011-02349604). Italian Network Genetic Isolates (INGI-FVG) Study: The INGI-FVG Study was partially supported by Regione Friuli-Venezia Giulia (grant L.26.2008) and the Italian Ministry of Health (grant GR-2011-02349604). Insulin Resistance Atherosclerosis Study (IRAS): The IRAS was supported by the NHLBI (grants HL047887, HL047889, HL047890, and HL47902). The IRAS Family Study was supported by the NHLBI (grants HL060944, HL061019, and HL060919). Genotyping for IRAS was supported by the Genetics Underlying Diabetes in Hispanics (GUARDIAN) Consortium with grant support from the NIDDK (grant DK085175) and in part by NIH grants UL1TR000124 (Clinical and Translational Science Institute) and DK063491 (Diabetes Research Center). Kingston Gene-by-Environment (Loyola GxE) Study: The Loyola GxE Study, a subset of the International Collaborative Study of Hypertension in Blacks, was supported by NIH grant R01HL53353. Lothian Birth Cohort 1936: Lothian Birth Cohort 1936 was supported by Age UK (the Disconnected Mind Project). Genotyping was funded by the Biotechnology and Biological Sciences Research Council (grant BB/F019394/1). The work was undertaken by the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross-council Lifelong Health and Wellbeing Initiative (grant MR/K026992/1). Funding was also received from the Medical Research Council. LifeLines Cohort Study: The LifeLines Cohort Study and generation and management of GWAS genotype data for the LifeLines Cohort Study were supported by the Netherlands Organisation for Scientific Research (grant 175.010.2007.006); the Dutch government’s Economic Structure Enhancing Fund; the Netherlands Ministry of Economic Affairs; the Netherlands Ministry of Education, Culture and Science; the Netherlands Ministry of Health, Welfare and Sport; the Northern Netherlands Collaboration of Provinces; the Province of Groningen; University Medical Center Groningen; the University of Groningen; the Dutch Kidney Foundation; and the Dutch Diabetes Research Foundation. London Life Sciences Prospective Population (LOLIPOP) Study: The LOLIPOP Study was supported by the NIHR Comprehensive Biomedical Research Centre Imperial College Healthcare NHS Trust, the British Heart Foundation (grant SP/04/002), the Medical Research Council (grants G0601966 and G0700931), the Wellcome Trust (grant 084723/Z/08/Z), the NIHR (grant RP-PG-0407-10371), the European Commission Seventh Framework Programme (EpiMigrant grant 279143), and Action on Hearing Loss (grant G51). Long Life Family Study: The Long Life Family Study was supported by NIA grant U01AG023746. Metabolic Syndrome in Men (METSIM) Study: The METSIM Study was supported by the Academy of Finland (contract 124243), the Finnish Heart Foundation, the Finnish Diabetes Foundation, the Center for Advancement of Technology (Tekes) (contract 1510/31/06), the Commission of the European Communities (grant HEALTH-F2-2007 201681), and the NIH (grants DK093757, DK072193, DK062370, and ZIA-HG000024). Genotyping was conducted in the Genetic Resources Core Facility at the Johns Hopkins Institute of Genetic Medicine. Netherlands Study of Depression and Anxiety (NESDA): The infrastructure for the NESDA Study was funded through the Geestkracht Program of the NWO (ZonMw grant 10-000-1002) and matching funds from participating universities and mental health-care organizations. Genotyping in NESDA was funded by the Genetic Association Information Network of the Foundation for the NIH. Statistical analyses were carried out using the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Organisation for Scientific Research (NWO grant 480-05-003), along with a supplement from the Dutch Brain Foundation. Obesity in Adults (OBA) Study: In the OBA Study, obese French adults were recruited by the laboratory of Professor Philippe Froguel (“Integrated Genomics and Metabolic Diseases Modeling”; Centre National de la Recherche Scientifique Unité Mixte de Recherche 8199, Université de Lille 2, and Institut Pasteur de Lille). Prevention of Renal and Vascular End Stage Disease (PREVEND) Study: Genetic analysis in the PREVEND Study was supported by the Dutch Kidney Foundation (grant E033), the European Union project GENECURE (European Commission Sixth Framework Programme grant LSHM-CT-2006-037697), the NIH (grant 2R01LM010098), and the Netherlands Organisation for Health Research and Development (NWO-Groot grant 175.010.2007.006, NWO Veni grant 916.761.70, and ZonMw grant 90.700.441). Precocious Coronary Artery Disease (PROCARDIS) Study: The PROCARDIS Study was supported by the European Commission Sixth Framework Program (grant LSHM-CT-2007-037273), AstraZeneca plc (Cambridge, United Kingdom), the British Heart Foundation, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Swedish Heart-Lung Foundation, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular Program of Karolinska Institutet, the Foundation for Strategic Research, and the Stockholm County Council (grant 560283). Martin Farrall and Hugh Watkins received a Wellcome Trust core award (award 090532/Z/09/Z) and support from the BHF Centre of Research Excellence. Anuj Goel and Hugh Watkins received support from the European Commission Seventh Framework Programme (2007–2013) under grant agreement HEALTH-F2-2013-601456 (CVGenes@Target), and Anuj Goel received support from the Wellcome Trust Institutional Strategic Support Fund. Ragama Health Study: The Ragama Health Study was supported by a grant from the National Center for Global Health and Medicine. Spanish Town (Loyola SPT) Study: The Loyola SPT Study, a subset of the International Collaborative Study of Hypertension in Blacks, was supported by NIH grant R01HL53353. Stockholm Heart Epidemiology Program (SHEEP) Study: The SHEEP Study was supported by grants from the Swedish Research Council for Health, Working Life and Welfare, the Stockholm County Council, the Swedish Research Council, the Swedish Heart and Lung Foundation, and the Cardiovascular Programme at Karolinska Institutet. Study of Health in Pomerania (SHIP): The SHIP Study is part of the Community Medicine Research Net of the University of Greifswald (Greifswald, Germany), which was funded by the Federal Ministry of Education and Research (grants 01ZZ9603, 01ZZ0103, and 01ZZ0403), the German Ministry of Cultural Affairs, and the Social Ministry of the Federal State of Mecklenburg-West Pomerania, as well as the network “Greifswald Approach to Individualized Medicine (GANI_MED),” funded by the Federal Ministry of Education and Research (grant 03IS2061A). The generation of genome-wide genotype data was supported by the Federal Ministry of Education and Research (grant 03ZIK012) and a joint grant from Siemens Healthcare (Erlangen, Germany) and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the Center of Knowledge Interchange program of Siemens AG (Berlin, Germany). Shanghai Women’s Health Study/Shanghai Men’s Health Study: The Shanghai Women’s Health Study and the Shanghai Men’s Health Study were supported by research grants UM1CA182910 and UM1CA173640, respectively, from the National Cancer Institute. TwinGene Project: The TwinGene Project received funding from the Swedish Research Council (grant M-2005-1112), GenomEUtwin (grants EU/QLRT-2001-01254 and QLG2-CT-2002-01254), the NIH (grant DK U01-066134), the Swedish Foundation for Strategic Research, and the Heart and Lung Foundation (grant 20070481). The TwinGene Project is part of the Swedish Twin Registry, which is managed by the Karolinska Institutet and receives funding through the Swedish Research Council (grant 2017–00641). Cardiovascular Risk in Young Finns Study: The Cardiovascular Risk in Young Finns Study was financially supported by the Academy of Finland (grants 286284, 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi)); the Social Insurance Institution of Finland; the Kuopio, Tampere and Turku University Hospital Medical Funds (grant X51001); the Juho Vainio Foundation; the Paavo Nurmi Foundation; the Finnish Foundation for Cardiovascular Research; the Finnish Cultural Foundation; the Tampere Tuberculosis Foundation; the Emil Aaltonen Foundation; the Yrjö Jahnsson Foundation; the Signe and Ane Gyllenberg Foundation; and the Diabetes Research Foundation of the Finnish Diabetes Association.
We thank the participants in all of the included studies for their important contributions. Similarly, we thank all of the people who have contributed to each of these studies, including volunteers, physicians, pharmacists, administrative staff, laboratory staff, recruitment center staff, and data managers, among others. We thank Matthew Brown for his help in tracking authorship information. Investigators in the Diabetes Heart Study and the African American Diabetes Heart Study acknowledge the study participants for their cooperation. The ASCOT investigators thank all ASCOT participants, physicians, nurses, and practices in the participating countries for their important contributions; in particular, they thank Clare Muckian and David Toomey for their help in DNA extraction, storage, and handling. We acknowledge the staff of Barts and The London Genome Centre for genotyping. The BRIGHT Study investigators are extremely grateful to all of the patients who participated in the study and to the BRIGHT nursing team. In relation to the Cardiovascular Risk in Young Finns Study, the expert technical assistance of Irina Lisinen in the statistical analyses is gratefully acknowledged. In relation to the CROATIA-Vis and CROATIA-Korcula studies, we acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to the University of Split and Zagreb Medical Schools, the Institute for Anthropological Research in Zagreb, and the Croatian Institute for Public Health. In relation to the EPIC Study, we thank staff from the technical, field epidemiology, and data functional group teams of the MRC Epidemiology Unit in Cambridge, United Kingdom, for carrying out sample preparation, DNA provision and quality control, genotyping, and data handling. We specifically thank Sarah Dawson for coordinating the sample provision for biomarker measurements, Abigail Britten for coordinating DNA sample provision and genotyping of candidate markers, Nicola Kerrison, Chris Gillson, and Abigail Britten for data provision and genotyping quality control, and Matt Sims for writing the technical laboratory specification for the intermediate pathway biomarker measurements and for overseeing the laboratory work. In relation to the ERF Study, we are grateful to P. Veraart for her help in genealogy, J. Vergeer for supervision of the laboratory work, P. Snijders for his help in data collection, and E. M. van Leeuwen for genetic imputation. We thank the Fenland Study Investigators, the Fenland Study Coordination Team, and the epidemiology field, data, and laboratory teams. We are grateful to the Scottish School of Primary Care for their help in subject recruitment for the GS_SFHS. In relation to the GLACIER Study, we thank the health professionals and data managers involved in the Västerbottens Intervention Project. We are also grateful to the staff of the Northern Sweden Biobank for preparing materials and to K. Enqvist and T. Johansson (Västerbottens County Council, Umeå, Sweden) for DNA preparation. We acknowledge the Jackson Heart Study team institutions (University of Mississippi Medical Center, Jackson State University, and Tougaloo College) and participants for their long-term commitment, which continues to improve our understanding of the genetic epidemiology of cardiovascular and other chronic diseases among African Americans. We acknowledge the services of the LifeLines Cohort Study staff, as well as the contributing research centers delivering data to LifeLines. We thank the MESA Coordinating Center, the MESA investigators, and the MESA study staff for their valuable contributions. We thank the NEO study group, Petra Noordijk, Pat van Beelen, and Ingeborg de Jonge for coordination, laboratory work, and data management in the NEO Study. In relation to the Rotterdam Study, we thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera, Marjolein Peters, and Carolina Medina-Gomez for their help in creating the GWAS database and Karol Estrada, Yurii Aulchenko, and Carolina Medina-Gomez for the creation and analysis of imputed data. We thank the Women’s Health Initiative investigators and staff for their dedication and for making the program possible.
The following persons are members of the LifeLines Cohort Study Group in the specified departments at the University of Groningen, University Medical Center Groningen, Groningen, the Netherlands: Department of Epidemiology (Behrooz Z. Alizadeh, H. Marike Boezen, and Harold Snieder); Department of Genetics (Lude Franke, Morris Swertz, and Cisca Wijmenga); Department of Cardiology (Pim van der Harst); Department of Internal Medicine, Division of Nephrology (Gerjan Navis); Department of Medical Biology (Marianne Rots); and Department of Endocrinology (Bruce H. R. Wolffenbuttel). A full list of Cardiovascular Health Study principal investigators and institutions can be found at CHS-NHLBI.org. A complete list of HyperGEN investigators is available at http://www.biostat.wustl.edu/hypergen/Acknowledge.html. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. A full list of Women’s Health Initiative investigators can be found at http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf.
This research was presented at the 67th Annual Meeting of the American Society of Human Genetics, Orlando, Florida, October 17–21, 2017.
Ethics approval for the GS_SFHS was given by the NHS Tayside committee on research ethics.
This article has not been formally reviewed by the US Environmental Protection Agency. The views expressed in this document are solely those of the authors, and the Environmental Protection Agency does not endorse any products or commercial services mentioned. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the NIH.
We declare no competing financial interests, except for the following. B.M.P. serves on the Data and Safety Monitoring Board of a clinical trial funded by the manufacturer (Zoll Lifecor Corporation, Pittsburgh, Pennsylvania) and on the Steering Committee of the Yale Open Data Access Project, funded by Johnson & Johnson (New Brunswick, New Jersey). O.H.F. received grants from Metagenics, Inc. (Aliso Viejo, California) on women’s health and epigenetics and from Nestlé S.A. (Vevey, Switzerland) on child health. M.A.N.’s participation was supported by a consulting contract between Data Tecnica International (Glen Echo, Maryland) and the NIA (Bethesda, Maryland). M.A.N. also consults for Illumina Inc. (San Diego, California), the Michael J. Fox Foundation for Parkinson’s Research (New York, New York), and UC Health (Oakland, California), among other organizations. N.P. has received financial support and consultancy fees from several pharmaceutical companies that manufacture either blood pressure-lowering or lipid-lowering agents or both (Sanofi (Guildford, United Kingdom), Amgen (Uxbridge, United Kingdom), Takeda (London, United Kingdom), Servier (Suresnes, France), and Pfizer (Tadwork, United Kingdom)) but holds no stock or shares in any such companies. P.S. has received research awards from Pfizer Inc. (New York, New York). J.B.J. serves as a consultant for the Mundipharma Company (Cambridge, United Kingdom), is a patent holder with Biocompatibles UK Ltd. (Franham, United Kingdom) (patent no. 20120263794; “Treatment of Eye Diseases Using Encapsulated Cells Encoding and Secreting Neuroprotective Factor and/or Anti-Angiogenic Factor”), and is a patent applicant with the University of Heidelberg (Heidelberg, Germany) (Europäische Patentanmeldung no. 15 000 771.4; “Agents for Use in the Therapeutic or Prophylactic Treatment of Myopia or Hyperopia”).
Abbreviations
- A1CF
APOBEC1 complementation factor gene
- APOBEC1
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
- APOBEC1
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 gene
- CHARGE
Cohorts for Heart and Aging Research in Genomic Epidemiology
- DEPICT
Data-driven Expression Prioritized Integration for Complex Traits
- df
degrees of freedom
- FDR
false discovery rate
- GWAS
genome-wide association study(ies)
- HDL-C
high-density lipoprotein cholesterol
- LDL-C
low-density lipoprotein cholesterol
- PCSK5
proprotein convertase subtilisin/kexin type 5
- PCSK5
proprotein convertase subtilisin/kexin type 5 gene
- PCSK9
proprotein convertase subtilisin/kexin type 9 gene
- TG
triglycerides
- VEGF-B
vascular endothelial growth factor B
- VEGFB
vascular endothelial growth factor B gene
REFERENCES
- 1. Roth GA, Johnson C, Abajobir A, et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990–2015. J Am Coll Cardiol. 2017;70(1):1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asselbergs FW, Guo Y, van Iperen EP, et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet. 2012;91(5):823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Global Lipids Genetics Consortium, Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peloso GM, Auer PL, Bis JC, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94(2):223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spracklen CN, Chen P, Kim YJ, et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum Mol Genet. 2017;26(9):1770–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Surakka I, Horikoshi M, Mägi R, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47(6):589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Leeuwen EM, Sabo A, Bis JC, et al. Meta-analysis of 49 549 individuals imputed with the 1000 Genomes Project reveals an exonic damaging variant in ANGPTL4 determining fasting TG levels. J Med Genet. 2016;53(7):441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muth ND, Laughlin GA, von Mühlen D, et al. High-density lipoprotein subclasses are a potential intermediary between alcohol intake and reduced risk of cardiovascular disease: the Rancho Bernardo Study. Br J Nutr. 2010;104(7):1034–1042. [DOI] [PubMed] [Google Scholar]
- 10. Gardner CD, Tribble DL, Young DR, et al. Associations of HDL, HDL2, and HDL3 cholesterol and apolipoproteins A-I and B with lifestyle factors in healthy women and men: the Stanford Five City Project. Prev Med. 2000;31(4):346–356. [DOI] [PubMed] [Google Scholar]
- 11. Wakabayashi I. Relationship between alcohol intake and lipid accumulation product in middle-aged men. Alcohol Alcohol. 2013;48(5):535–542. [DOI] [PubMed] [Google Scholar]
- 12. Klop B, do Rego AT, Cabezas MC. Alcohol and plasma triglycerides. Curr Opin Lipidol. 2013;24(4):321–326. [DOI] [PubMed] [Google Scholar]
- 13. Whitfield JB, Heath AC, Madden PA, et al. Metabolic and biochemical effects of low-to-moderate alcohol consumption. Alcohol Clin Exp Res. 2013;37(4):575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rifler JP, Lorcerie F, Durand P, et al. A moderate red wine intake improves blood lipid parameters and erythrocytes membrane fluidity in post myocardial infarct patients. Mol Nutr Food Res. 2012;56(2):345–351. [DOI] [PubMed] [Google Scholar]
- 15. Brinton EA. Effects of ethanol intake on lipoproteins. Curr Atheroscler Rep. 2012;14(2):108–114. [DOI] [PubMed] [Google Scholar]
- 16. Kechagias S, Zanjani S, Gjellan S, et al. Effects of moderate red wine consumption on liver fat and blood lipids: a prospective randomized study. Ann Med. 2011;43(7):545–554. [DOI] [PubMed] [Google Scholar]
- 17. Wakabayashi I. Associations of alcohol drinking and cigarette smoking with serum lipid levels in healthy middle-aged men. Alcohol Alcohol. 2008;43(3):274–280. [DOI] [PubMed] [Google Scholar]
- 18. Rakic V, Puddey IB, Dimmitt SB, et al. A controlled trial of the effects of pattern of alcohol intake on serum lipid levels in regular drinkers. Atherosclerosis. 1998;137(2):243–252. [DOI] [PubMed] [Google Scholar]
- 19. Brien SE, Ronksley PE, Turner BJ, et al. Effect of alcohol consumption on biological markers associated with risk of coronary heart disease: systematic review and meta-analysis of interventional studies. BMJ. 2011;342:d636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rimm EB, Williams P, Fosher K, et al. Moderate alcohol intake and lower risk of coronary heart disease: meta-analysis of effects on lipids and haemostatic factors. BMJ. 1999;319(7224):1523–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tabara Y, Ueshima H, Takashima N, et al. Mendelian randomization analysis in three Japanese populations supports a causal role of alcohol consumption in lowering low-density lipid cholesterol levels and particle numbers. Atherosclerosis. 2016;254:242–248. [DOI] [PubMed] [Google Scholar]
- 22. Vu KN, Ballantyne CM, Hoogeveen RC, et al. Causal role of alcohol consumption in an improved lipid profile: the Atherosclerosis Risk in Communities (ARIC) Study. PLoS One. 2016;11(2):e0148765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirk EP. Genes, environment, and the heart: putting the pieces together. Circ Cardiovasc Genet. 2017;10(3):e001764. [DOI] [PubMed] [Google Scholar]
- 24. Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao DC, Sung YJ, Winkler TW, et al. Multiancestry study of gene-lifestyle interactions for cardiovascular traits in 610 475 individuals from 124 cohorts: design and rationale. Circ Cardiovasc Genet. 2017;10(3):e001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. [DOI] [PubMed] [Google Scholar]
- 27. Manning AK, LaValley M, Liu CT, et al. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet Epidemiol. 2011;35(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 29. 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winkler TW, Day FR, Croteau-Chonka DC, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. [DOI] [PubMed] [Google Scholar]
- 33. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 34. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(database issue):D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pers TH, Karjalainen JM, Chan Y, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nikpay M, Goel A, Won HH, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu DJ, Peloso GM, Yu H, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49(12):1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Justice AE, Winkler TW, Feitosa MF, et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat Commun. 2017;8:14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winkler TW, Justice AE, Graff M, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11(10):e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lepor NE, Kereiakes DJ. The PCSK9 inhibitors: a novel therapeutic target enters clinical practice. Am Health Drug Benefits. 2015;8(9):483–488. [PMC free article] [PubMed] [Google Scholar]
- 43. Stein EA, Mellis S, Yancopoulos GD, et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med. 2012;366(12):1108–1118. [DOI] [PubMed] [Google Scholar]
- 44. Iatan I, Dastani Z, Do R, et al. Genetic variation at the proprotein convertase subtilisin/kexin type 5 gene modulates high-density lipoprotein cholesterol levels. Circ Cardiovasc Genet. 2009;2(5):467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin W, Wang X, Millar JS, et al. Hepatic proprotein convertases modulate HDL metabolism. Cell Metab. 2007;6(2):129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shimamura M, Matsuda M, Yasumo H, et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27(2):366–372. [DOI] [PubMed] [Google Scholar]
- 47. Jin W, Millar JS, Broedl U, et al. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J Clin Invest. 2003;111(3):357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsutsumi K. Lipoprotein lipase and atherosclerosis. Curr Vasc Pharmacol. 2003;1(1):11–17. [DOI] [PubMed] [Google Scholar]
- 49. Chester A, Scott J, Anant S, et al. RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochim Biophys Acta. 2000;1494(1-2):1–13. [DOI] [PubMed] [Google Scholar]
- 50. Kendrick JS, Chan L, Higgins JA. Superior role of apolipoprotein B48 over apolipoprotein B100 in chylomicron assembly and fat absorption: an investigation of apobec-1 knock-out and wild-type mice. Biochem J. 2001;356(3):821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khoo B, Krainer AR. Splicing therapeutics in SMN2 and APOB. Curr Opin Mol Ther. 2009;11(2):108–115. [PMC free article] [PubMed] [Google Scholar]
- 52. Hagberg CE, Falkevall A, Wang X, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464(7290):917–921. [DOI] [PubMed] [Google Scholar]
- 53. Hagberg CE, Mehlem A, Falkevall A, et al. Targeting VEGF-B as a novel treatment for insulin resistance and type 2 diabetes [letter]. Nature. 2012;490(7420):426–430. [DOI] [PubMed] [Google Scholar]
- 54. Dijkstra MH, Pirinen E, Huusko J, et al. Lack of cardiac and high-fat diet induced metabolic phenotypes in two independent strains of Vegf-b knockout mice. Sci Rep. 2014;4:6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robciuc MR, Kivelä R, Williams IM, et al. VEGFB/VEGFR1-induced expansion of adipose vasculature counteracts obesity and related metabolic complications. Cell Metab. 2016;23(4):712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rafii S, Carmeliet P. VEGF-B improves metabolic health through vascular pruning of fat. Cell Metab. 2016;23(4):571–573. [DOI] [PubMed] [Google Scholar]
- 57. de Vries PS, Sabater-Lleal M, Chasman DI, et al. Comparison of HapMap and 1000 Genomes reference panels in a large-scale genome-wide association study. PLoS One. 2017;12(1):e0167742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livingston M, Callinan S. Underreporting in alcohol surveys: whose drinking is underestimated? J Stud Alcohol Drugs. 2015;76(1):158–164. [PubMed] [Google Scholar]
- 59. Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332(7549):1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.