Abstract
The biological mechanisms driving associations between alcohol consumption and chronic diseases might include epigenetic modification of DNA methylation. We explored the hypothesis that alcohol consumption is associated with methylation in an epigenome-wide association study of blood and normal breast tissue DNA. Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, California) array data on blood DNA methylation was examined in a discovery set of 2,878 non-Hispanic white women from the Sister Study (United States, 2004–2015) who provided detailed questionnaire information on lifetime alcohol use. Robust linear regression modeling was used to identify significant associations (false discovery rate of Q < 0.05) between the number of alcoholic drinks per week and DNA methylation at 5,458 cytosine-phosphate-guanine (CpG) sites. Associations were replicated (P < 0.05) for 677 CpGs in an independent set of 187 blood DNA samples from the Sister Study and for 628 CpGs in an independent set of 171 normal breast DNA samples; 1,207 CpGs were replicated in either blood or normal breast, with 98 CpGs replicated in both tissues. Individual gene effects were notable for phosphoglycerate dehydrogenase (PGHDH), peptidyl-prolyl cis-trans isomerase (PPIF), solute carrier 15 (SLC15), solute carrier family 43 member 1 (SLC43A1), and solute carrier family 7 member 11 (SLC7A11). We also found that high alcohol consumption was associated with significantly lower global methylation as measured by the average of CpGs on the entire array.
Keywords: alcohol consumption, breast cancer, DNA methylation, epigenome-wide association study
Alcohol consumption is an important risk factor for multiple chronic diseases including many cancers (1–5). Alcohol likely influences disease risk through a wide variety of biological mechanisms that might include epigenetic alterations. Studies have reported associations between DNA methylation in brain or buccal tissue and chronic alcohol use or alcohol use disorder (6–9), and a large pooled cohort analysis identified multiple potential DNA methylation markers of heavy alcohol consumption in blood (10). Women appear to be more vulnerable to some negative health consequences of alcohol at relatively low levels of consumption (11). The risk of breast cancer increases even with a moderate level of drinking, equivalent to the consumption of 1 alcoholic beverage a day or 7 drinks per week. These moderate levels fall within the “low-risk” category for alcohol use disorder in nutritional recommendations for women’s alcohol consumption (12, 13).
Identification of DNA methylation patterns linked to alcohol consumption could contribute to our understanding of how alcohol consumption influences women’s risk of chronic diseases, particularly breast cancer. Here, we identified alcohol-associated methylation sites using blood DNA samples from a large nationwide cohort of women and replicate a subset of those sites in independent DNA samples from both blood and normal breast tissue.
METHODS
Study population
The Sister Study is a prospective cohort study of 50,884 women in the United States with a family history of breast cancer who were free of breast cancer themselves (14). To be eligible for the study, women had to have a sister with breast cancer but be free of breast cancer at the time of study enrollment and blood collection. Participants provided detailed exposure and health-related information in a computer-assisted telephone interview and provided a blood sample during a home study visit. They complete annual, biennial, and triennial updates on cancer and various other lifestyle, exposure, and health factors (14–17).
Breast tissue methylation data came from participants in the Normal Breast Study (18). In that study, 479 women undergoing various types of breast surgery between 2009 and 2013 at the University of North Carolina were enrolled, provided demographic and health information via telephone interview, and donated breast tissue samples during surgery. The study included 399 women with breast cancer and 75 women without malignant disease; 171 participants were previously selected for DNA methylation analysis (18).
Alcohol exposure classification
Computer-assisted telephone interviews conducted at study baseline were used to gather information on current alcohol consumption along with detailed information on past alcohol consumption habits across a woman’s lifetime. Our analysis focused on recent alcohol use; our main exposure of interest was the self-reported average number of alcoholic beverages consumed per week over the 12 months prior to the baseline interview. An alcoholic beverage is defined as a 12-ounce bottle or can of beer, a 5-ounce glass of wine, 1 wine cooler, 1 shot of liquor, or 1 mixed drink or cocktail. Alcohol consumption is analyzed as a continuous variable. We also report women’s self-reported drinking habits, in which they classify themselves as one of the following: never drinker, social past drinker, social current drinker, regular past drinker, or regular current drinker.
Alcohol consumption information in the Normal Breast Study was less detailed than in the Sister Study cohort, so alcohol consumption for this cohort was defined as an ordinal categorical variable with the following values: never drank alcohol, former drinker of alcohol, current occasional/light drinker of alcohol, current moderate/heavy drinker of alcohol.
DNA methylation assays
We previously generated array-based DNA methylation data sets for several projects within the Sister Study cohort. Our discovery set consists of women who were included in a prospective case-cohort study to identify DNA methylation markers associated with subsequent breast cancer risk (19). All blood samples were collected before any of the study participants had been diagnosed with breast cancer. Participant selection has been previously described (14): Briefly, a random subsample of cohort participants were selected, as were all additional participants who reported a diagnosis of invasive breast cancer or ductal carcinoma in situ during study follow-up as of March 2015. Participants were excluded from the sample if they reported an ethnicity other than non-Hispanic white or if they had no available blood sample. These exclusions resulted in a discovery set sample of 2,878 women. This subset of participants includes 1,262 women from the cohort who remained cancer-free as of the most recent study follow-up and 1,616 women who were free of breast cancer at the time of their blood draw but who developed breast cancer during the study follow-up period.
Genomic DNA was extracted using an automated system in the Molecular Genetics Core Facility at the National Institute of Environmental Health Sciences or using DNAQuik at BioServe Biotechnologies Ltd. (Beltsville, MD), and methylation arrays were analyzed at the National Institutes of Health Center for Inherited Disease Research (see Web Appendix 1, available at https://academic.oup.com/aje).
Replication set 1: replication in blood DNA
The first replication set comprised 187 white non-Hispanic Sister Study participants between the ages of 40–59 years who were part of a previous nested case-control study of prenatal exposure and DNA methylation (20). Participants in the replication set do not overlap with the discovery set but have the same data available on alcohol use and blood DNA methylation (20).
Replication set 2: replication in normal breast tissue DNA
We used DNA methylation array data from fresh-frozen normal breast tissue to examine alcohol-related methylation in a tissue other than blood. Methylation data from breast tissue samples come from women enrolled in the Normal Breast Study (18); processing details are in Web Appendix 1.
Statistical analysis
Methylation data preprocessing and quality control for all data sets were completed using the ENmix package in R (R Foundation for Statistical Computing, Vienna, Austria) (21). Methylation preprocessing and quality control methods are described in detail in Web Appendix 1. After quality control exclusions, there were 2,775 participant samples included in the discovery analysis. β values were logit-transformed into M values for statistical analysis.
Multivariable robust linear regression analysis was performed to identify associations between cytosine-phosphate-guanine (CpG) site M values and the number of alcoholic beverages consumed per week. We adjusted for the following variables in the robust linear regression model: age at blood draw, breast cancer case status, smoking status, body mass index, and surrogate variables derived from array nonnegative control probe data. All association tests were adjusted for the proportions of white blood cell types, estimated using a method described by Houseman et al. (22, 23). To correct for multiple testing of the approximately 450,000 CpG sites included on the array, we estimate the false discovery rate (FDR) using the Q-value method (24). CpGs with significance values smaller than the FDR Q (<0.05) are considered differentially methylated with alcohol consumption. All analyses were repeated in the replication sets; a CpG site passing the FDR in the discovery set was considered replicated if its association test in the replication set produced a nominal P value of < 0.05 with the same direction of association.
For replication in normal breast tissue, associations between CpG site methylation values and the participant’s alcohol consumption level were tested using a reference-free method (25) to control for possible differences in cell-type composition among samples. Models also adjusted for age at breast surgery, menopausal status, and race.
We conducted a sensitivity analysis limited to women who did not go on to develop breast cancer in the follow-up period to ensure that the identified methylation differences were not due solely to early, as-yet-undiagnosed breast cancer (n = 1,262).
Pathway analysis
We performed Ingenuity Pathway Analysis (QIAGEN Sciences, Germantown, Maryland) to identify pathways enriched for alcohol-related differentially methylated CpGs. CpGs were mapped to the nearest genes based on the Illumina annotation files for the HumanMethylation450 BeadChip (Illumina Inc., San Diego, California). Permutation P values were calculated based on 10,000 permutations of P values for array CpGs.
Functional enrichment analysis
We used publicly available Encyclopedia of DNA Elements (ENCODE) ChIP-seq data (www.encodeproject.org) for transcription-factor binding sites and histone modification sites to perform functional enrichment analysis by comparing the set of differentially methylated CpGs with all CpGs on the array that were included in the analysis. We performed χ2 tests to determine enrichment; a Bonferroni-adjusted P value of 0.05 was used as the significance threshold.
Array-wide methylation averages
To determine whether alcohol consumption was associated with an overall increase or decrease in methylation across the genomic regions covered by the array, for each woman we calculated the mean β value of all CpG sites on the array (n = 413,652 sites) and for specific subsets of CpGs based on genomic context. Subgroup analyses were performed using mean values for CpGs with the following annotations: gene bodies, within 1,500 base pairs (bp) of transcription start sites, within 200 base pairs of transcription start sites, 5′ untranslated region (5′UTR), 3′untranslated region (3′UTR), and “other.” We performed robust linear regression analysis to determine associations between the number of alcoholic beverages consumed per week and an individual’s mean DNA methylation value overall and for subgroups of CpGs based on genomic context.
DNA methylation and gene expression in breast tumor
We performed conditional logistic regression of publicly available 450K DNA methylation data for 175 paired breast tumor and adjacent normal tissues samples from the Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) to identify differentially methylated CpG sites between tumor and normal tissue (FDR Q < 0.05). To identify CpG sites whose methylation was significantly associated with gene expression in breast cancer, we used Illumina annotation to map CpGs to individual genes; we then used paired methylation and expression data from 868 TCGA breast cancer tissue samples to identify CpGs with significant (Bonferroni-corrected P < 0.05) Spearman correlations between DNA methylation and gene expression.
RESULTS
Sample characteristics
Basic characteristics of Sister Study participants with Illumina Infinium Human450 methylation array data are summarized in Table 1. The average age of participants was 57.8 (standard deviation, 8.8) years in the discovery set and 50.5 (standard deviation, 4.8) years in the blood replication set. A large portion of women in both the discovery set and replication set consumed alcohol, with 76.4% of women in the discovery set and 81% of the replication set self-reporting that they are current regular drinkers. The participants in the discovery set report an average of 3.1 alcoholic drinks consumed per week in the previous 12 months. The average number of drinks consumed per week in the replication set was 3.2.
Table 1.
Demographic Information for the Discovery Set Participants and Blood Replication Set Participants With Blood Methylation Data Meeting Quality Thresholds, Sister Study, United States, 2004–2015
| Demographic Information | Discovery Set (n = 2,775) | Blood Replication Set (n = 187) | ||
|---|---|---|---|---|
| No. | % | No. | % | |
| Body mass indexa | ||||
| Missing | 2 | 0.1 | 0 | 0.0 |
| <18.5 | 27 | 1.0 | 1 | 0.5 |
| 18.5–24.9 | 1,047 | 37.7 | 91 | 48.7 |
| 25.0–29.9 | 900 | 32.4 | 49 | 26.2 |
| 30.0–34.5 | 493 | 17.8 | 27 | 14.4 |
| 35.0–39.5 | 180 | 6.5 | 11 | 5.9 |
| ≥40 | 126 | 4.5 | 8 | 4.3 |
| Menopausal status | ||||
| Missing | 1 | 0.0 | 86 | 46.0 |
| Premenopausal | 874 | 31.5 | 55 | 29.4 |
| Postmenopausal | 1,900 | 68.5 | 46 | 24.6 |
| Ever pregnant | ||||
| Missing | 1 | 0.0 | 0 | 0.0 |
| No | 350 | 12.6 | 36 | 19.3 |
| Yes | 2,424 | 87.4 | 151 | 80.7 |
| Used hormone replacement therapy | ||||
| Unknown | 7 | 0.3 | 1 | 0.5 |
| No | 1,401 | 50.5 | 122 | 65.2 |
| Yes | 1,367 | 49.3 | 64 | 34.2 |
| Smoking status | ||||
| Never smoker | 1,447 | 52.1 | 109 | 58.3 |
| Former smoker | 1,125 | 40.5 | 67 | 35.8 |
| Current smoker | 203 | 7.3 | 11 | 5.9 |
| Alcohol use | ||||
| Never drinker | 80 | 2.9 | 3 | 1.6 |
| Social past drinker | 149 | 5.4 | 11 | 5.9 |
| Social current drinker | 209 | 7.5 | 10 | 5.3 |
| Regular past drinker | 218 | 7.9 | 10 | 5.3 |
| Regular current drinker | 2,119 | 76.4 | 153 | 81.8 |
| No. of drinks consumed per week in the previous 12 monthsb | 3.1 (4.7) | 3.2 (4.5) | ||
a Body mass index calculated as weight (kg)/height (m)2.
b Values are expressed as mean (standard deviation).
Methylation sites related to alcohol consumption
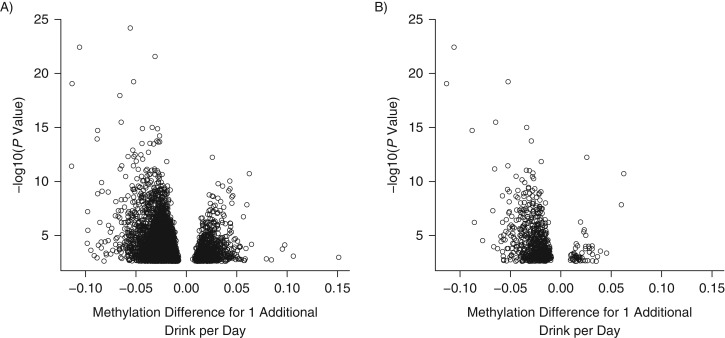
Based on the DNA methylation data for the 128 duplicates, we noticed a large proportion of CpGs on the array having within-sample variation similar to the between-sample variation. To identify more reliable CpG markers, we restricted our analysis to the 144,655 CpGs with intraclass correlation coefficients of >0.5 based on CpGFilter (26). We performed association analysis for each of these CpGs and found that DNA methylation levels for 5,460 of them were significantly (FDR Q < 0.05) correlated with the number of alcoholic drinks women reported consuming per week over the previous 12 months. With use of the genomic control method (27), 1,433 CpGs were significant at an FDR Q value of 0.05, and 78 of the CpGs reached an adjusted P value of 1 × 10−6 (Web Figure 1); 68 of the 5,460 identified CpGs passed strict Bonferroni correction (P < 3.5 × 10−7).
Using data from the replication set of blood DNA samples from 187 women, 677 of 5,460 CpG sites were confirmed (P ≤ 0.05) to have an association between blood DNA methylation and number of drinks per week (Web Table 1), including 23 of the 68 CpGs passing the Bonferroni correction threshold. Fourteen of these CpGs replicated at a Bonferroni correction threshold. Volcano plots of effect size versus log-transformed P values demonstrate that both the initial and replicated set of CpGs show similar patterns: Nearly 4-fold more CpGs have significant methylation loss than gain (Figure 1). This asymmetry in direction of effect persisted even after stratifying CpGs by average population methylation level into low-, medium-, and high-methylation sets (Web Figure 2). The replicated CpG associations with the smallest Q values are presented in Table 2; 2 of the top hits (cg14476101 and cg16246545) are approximately 50 base pairs apart in the first intron of the phosphoglycerate dehydrogenase gene (PHGDH). The dose-response plot at cg14476101 shows progressively decreased methylation with a higher number of drinks per week (P < 0.001) (Figure 2). Other genes with notable methylation differences included peptidyl-prolyl cis-trans isomerase (PPIF), solute carrier 15 (SLC15), solute carrier family 43 member 1 (SLC43A1), and solute carrier family 7 member 11 (SLC7A11).
Figure 1.
Volcano plots of associations between blood DNA cytosine-phosphate-guanine (CpG) methylation and alcohol consumption, Sister Study, United States, 2004–2015. A) Effect size (methylation change in percent from 1 additional drink/day) on x-axis versus log-transformed P value on y-axis for 5,460 CpG sites showing association with alcohol at FDR Q of <0.05. B) 667 CpGs replicated in independent blood sample DNAs. In both panels, the number of CpGs showing lower methylation among those who consumed alcohol are much more numerous than the number of CpGs with higher methylation.
Table 2.
Top Replicated Cytosine-Phosphate-Guanine Site Associations Between Blood DNA Methylation Levels and Number of Drinks per Week in the Past Year, Sister Study, United States, 2004–2015
| CpG Site | Gene | Chromosome | Discovery Set (n = 2,775) | Replication Set (n = 187) | |||
|---|---|---|---|---|---|---|---|
| % Methylated | Point Estimatea | Q Value | Point Estimatea | P Value | |||
| cg14476101 | PHGDH | 1 | 74.9 | −0.015 | 2 × 10−18 | −0.027 | 5 × 10−11 |
| cg11376147 | SLC43A1 | 11 | 12 | −0.007 | 2 × 10−15 | −0.012 | 2 × 10−4 |
| cg06690548 | SLC7A11 | 4 | 95.4 | −0.016 | 2 × 10−15 | −0.019 | 7 × 10−5 |
| cg16246545 | PHGDH | 1 | 55 | −0.009 | 5 × 10−12 | −0.017 | 1 × 10−6 |
| cg27519140 | RPP21 | 6 | 88.7 | −0.005 | 1 × 10−11 | −0.005 | 0.04 |
| cg00422488 | ANP32B | 9 | 2.7 | −0.013 | 2 × 10−11 | −0.011 | 3 × 10−3 |
| cg05288253 | ANKRD11 | 16 | 70.2 | −0.004 | 1 × 10−10 | −0.003 | 0.03 |
| cg13740985 | PSAT1 | 9 | 33.5 | 0.004 | 2 × 10−9 | 0.003 | 0.02 |
| cg11440375 | DKFZP686I152 | 6 | 81.1 | −0.003 | 5 × 10−9 | −0.003 | 0.03 |
| cg02711608 | SLC1A5 | 19 | 18.1 | −0.008 | 1 × 10−8 | −0.005 | 0.04 |
| cg27637303 | N/A | 2 | 71.3 | −0.009 | 2 × 10−8 | −0.018 | 3 × 10−6 |
| cg16260349 | TSPAN14 | 10 | 65.7 | −0.005 | 3 × 10−8 | −0.006 | 5 × 10−3 |
| cg17466510 | N/A | 1 | 77.2 | −0.004 | 3 × 10−8 | −0.01 | 8 × 10−4 |
| cg18763536 | ETV6 | 12 | 34.6 | −0.004 | 4 × 10−8 | −0.007 | 3 × 10−4 |
| cg22994830 | PRKAR1B | 7 | 25.2 | 0.009 | 5 × 10−8 | 0.01 | 7 × 10−3 |
| cg03217115 | N/A | 2 | 61.9 | −0.006 | 8 × 10−8 | −0.013 | 4 × 10−6 |
| cg06983052 | LRRC8D | 1 | 69.5 | −0.004 | 8 × 10−8 | −0.01 | 5 × 10−3 |
Abbreviations: ANKRD11, ankyrin repeat domain 11; ANP32B, acidic leucine-rich nuclear phosphoprotein 32 family member b; cAMP, cyclic adenosine monophosphate; CpG, cytosine-phosphate-guanine; DKFZP686I152, hypothetical protein DKFZP686I152; ETV6, ETS variant 6; LRRC8D, leucine-rich repeat containing 8 VRAC subunit D; N/A, not applicable; PHGDH, phosphoglycerate dehydrogenase; PRKAR1B, protein kinase cAMP-dependent type 1 regulatory subunit β; PSAT1, phosphoserine aminotransferase 1; RPP21, ribonuclease p subunit 21; SLC1A5, solute carrier family 1 member 5; SLC43A1, solute carrier family 43 member 1; SLC7A11, solute carrier family 7 member 11; TSPAN14, tetrespanin 14.
a Association point estimates are presented as the difference in average percent methylation associated with an increase of 1 drink per day (7 drinks per week).
Figure 2.
Distribution of methylation values in the discovery set (Sister Study, United States, 2004–2015) for site cg14476101 in the phosphoglycerate dehydrogenase gene (PHGDH), categorized by number of alcoholic drinks consumed per week.
To determine whether associations were driven by women with the highest alcohol consumption, we performed sensitivity analysis using only women who reported consuming less than 15 drinks per week (n = 2,301). Although some CpGs no longer passed the FDR test, a large proportion of CpGs in both analyses remained statistically significant, indicating that the differential methylation continues to be evident even after excluding women with the highest levels of alcohol consumption.
Results limited to women who did not go on to develop breast cancer were also highly correlated with the results from the main analysis, indicating that associations between alcohol consumption and DNA methylation were not due solely to early breast cancer (Web Figure 3).
We specifically examined 144 CpG sites previously reported to be associated with high alcohol consumption (10); 104 of those CpGs were associated with alcohol use in our discovery set at P < 0.05, with 77 of the sites passing the FDR Q threshold of <0.05.
Functional enrichment and pathway analysis
To assess potentially shared transcription factor characteristics of alcohol-associated CpGs sites with increased or decreased methylation, we considered each group separately, using as a referent the characteristics of the 144,655 CpGs with intraclass correlation coefficients of >0.5 on the array (Web Table 2). Sites with decreased methylation were significantly enriched for transcription factor binding sites found at active enhancers with approximately 2-fold enrichment for RNA polymerase II subunit A (POLR2A) (P = 5.7 × 10−40) and metallothionein-like protein 3A (MT3A) (P = 3.5 × 10−28). In contrast, sites with increased methylation had 2-fold enrichment of binding sites for runt-related transcription factor 3 (RUNX3) (P = 1.7 × 10−10) and E74-like ETS transcription factor 1 (ELF1) (P = 2.1 × 10−9).
Although sites with increased and decreased methylation had different profiles for histone modifications, both were consistent with transcriptional activation (Web Table 3). Sites with decreased methylation were enriched for histone modifications characteristic of enhancer or activator regions including H3K4me1 (P = 1.1 × 10−259) and H3K79me2 (P = 6.8 × 10−248), while sites with decreased methylation showed highest enrichment for H3K36me3 (P = 9.7 × 10−27), a histone modification found in the body of actively transcribed genes.
Pathway analysis of the 677 replicated CpG sites identified 23 significantly (P < 0.01) enriched pathways, including the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling and hereditary breast cancer signaling (Table 3 and Web Table 4).
Table 3.
Top Results of Ingenuity Pathway Enrichment Analysis for the 677 Replicated Cytosine-Phosphate-Guanine Sites Associated With Alcohol Consumption, Sister Study, United States, 2004–2015
| Pathway | Permutation P Value |
|---|---|
| GM-CSF signaling | 0.0003 |
| Integrin signaling | 0.0005 |
| Role of macrophages fibroblasts and endothelial cells in rheumatoid arthritis | 0.0006 |
| Chronic myeloid leukemia signaling | 0.0007 |
| Germ cell–Sertoli cell junction signaling | 0.002 |
| Growth hormone signaling | 0.002 |
| NRF2-mediated oxidative stress response | 0.002 |
| Pancreatic-adenocarcinoma signaling | 0.003 |
| Virus entry via endocytic pathways | 0.003 |
| Hereditary breast cancer signaling | 0.004 |
| IGF-1 signaling | 0.004 |
| Paxillin signaling | 0.005 |
| FAK signaling | 0.006 |
| Glucocorticoid receptor signaling | 0.006 |
| MSP-RON signaling pathway | 0.007 |
| T-cell receptor signaling | 0.007 |
| Cardiac hypertrophy signaling | 0.007 |
| PKC-θ signaling in T lymphocytes | 0.007 |
| ErbB2-ErbB3 signaling | 0.008 |
| Regulation of eIF4 and p70S6K signaling | 0.009 |
| Leukocyte extravasation signaling | 0.009 |
| Telomerase signaling | 0.01 |
| Role of JAK1 and JAK3 in γc cytokine signaling | 0.01 |
Abbreviations: eIF4, eukaryotic initiation factor-4; ErbB2-ErbB3, epidermal growth factor receptor tyrosine kinase B2 and B3; FAK, focal adhesion kinase; GM-CSF, granulocyte-macrophage colony-stimulating factor; IGF-1, insulin-like growth factor 1; JAK, ribosomal protein S6 kinase (p70S6K), Janus kinase; MSP-RON, macrophage-stimulating protein-protein tyrosine kinase/receptor d’origine nantais; NRF2, nuclear factor erythroid 2-related factor; p70S6K, ribosomal protein S6 kinase; PKC-θ, protein kinase C-θ.
Average methylation across the array
To investigate whether the decrease in methylation was widespread across the genome, we considered the association between alcohol consumption and the average methylation across all CpGs sites on the array, as well as for CpGs grouped by genomic context. Increased alcohol consumption was associated with significantly lower average methylation across the array as a whole, and this association was still evident after parsing CpGs by genomic context (Table 4).
Table 4.
Associations Between Total Average Methylation Across All Cytosine-Phosphate-Guanine Sites on the Array and Cytosine-Phosphate-Guanine Site Subgroups With Current Alcohol Consumption Level, Sister Study, United States, 2004–2015
| Gene Region | Methylation Difference in %a | |
|---|---|---|
| Difference | P Value | |
| All locations | −7.5 × 10−4 | 0.01 |
| Gene body | −8.8 × 10−4 | 0.01 |
| Other | −7.3 × 10−4 | 0.09 |
| TSS1500b | −1.1 × 10−3 | 0.008 |
| TSS200c | −1.8 × 10−6 | 0.66 |
| 5′UTRd | −1.2 × 10−3 | 0.0003 |
| 3′UTRe | −8.4 × 10−4 | 0.05 |
Abbreviations: TSS, transcription start site; UTR, untranslated region.
aDifference associated with 1 additional alcoholic drink per week.
b Within 1,500 base pairs of a transcription start site.
c Within 200 base pairs of a transcription start site.
d The 5′ untranslated region.
e The 3′ untranslated region.
Alcohol effects also seen in normal breast tissue
To determine whether the alcohol effects identified in blood DNA were also present in normal breast tissue DNA, we examined Illumina Infinium450 methylation data from normal breast tissue DNA samples available from 175 women in the Normal Breast Study. Of these women, 171 had provided self-reports of their alcohol consumption; 31 women reported never consuming alcohol, 18 were former drinkers, and 122 reported that they were current drinkers. Of the current drinkers, 67 women were “occasional current drinkers” (drink currently, but not every week), and 55 women were “moderate/heavy drinkers” (drink at least once a week). Of the 5,458 CpG sites identified in our discovery set, 628 CpG sites were replicated in normal breast tissue, and 98 CpGs from the discovery set replicated in both the blood and breast tissue sets (Table 5).
Table 5.
Topa Cytosine-Phosphate-Guanine Sites With Associations Between Alcohol Consumption and DNA Methylation That Replicate in Both Blood and in Normal Breast Tissue Samples, Sister Study (2004–2015), Normal Breast Study (2009–2013), and the Cancer Genome Atlas (2012–2016), United States
| Probe | Gene | Blood Discovery Set Methylation Coefficient | P Value | Breast Tissue Methylation Coefficient | P Value |
|---|---|---|---|---|---|
| cg02711608 | SLC1A5 | −0.007 | 4 × 10−12 | −0.07 | 0.02 |
| cg06180389 | CD53 | −0.003 | 5 × 10−11 | −0.07 | 0.002 |
| cg19566658 | TRIP6 | −0.004 | 4 × 10−10 | −0.08 | 8 × 10−5 |
| cg19939077 | PPIF | −0.007 | 9 × 10−10 | −0.06 | 0.006 |
| cg19695041 | TACC1 | −0.003 | 1 × 10−9 | −0.04 | 0.03 |
| cg06745030 | B3GNT7 | −0.004 | 2 × 10−9 | −0.08 | 0.003 |
| cg02583484 | HNRNPA1P10 | −0.006 | 2 × 10−9 | −0.08 | 0.001 |
| cg16290996 | SNORD75 | −0.007 | 8 × 10−9 | −0.09 | 0.015 |
| cg11701312 | MIR4754 | −0.003 | 2 × 10−8 | −0.06 | 0.009 |
| cg03523740 | TXLNA | −0.003 | 2 × 10−8 | −0.05 | 0.004 |
| cg02118194 | MYPOP | −0.004 | 3 × 10−8 | −0.05 | 0.0009 |
| cg13105234 | N/A | −0.005 | 1 × 10−7 | −0.14 | 0.0001 |
| cg17593384 | AFF1 | −0.003 | 2 × 10−7 | −0.07 | 0.03 |
| cg17962756 | N/A | −0.004 | 2 × 10−7 | −0.07 | 0.02 |
| cg00326958 | HNRNPF | −0.008 | 6 × 10−7 | −0.05 | 0.03 |
| cg10143507 | N/A | −0.005 | 8 × 10−7 | −0.05 | 0.001 |
| cg06414073 | GAS2L3 | −0.003 | 1 × 10−6 | −0.05 | 0.02 |
| cg27479634 | ATP5G2 | −0.005 | 1 × 10−6 | −0.06 | 0.003 |
| cg23202985 | N/A | −0.002 | 1 × 10−6 | −0.12 | 4 × 10−7 |
| cg22548088 | MLLT1 | −0.002 | 1 × 10−6 | −0.04 | 0.02 |
| cg08841048 | ZNF428 | −0.005 | 2 × 10−6 | −0.07 | 0.03 |
| cg23598378 | C6orf132 | 0.003 | 3 × 10−6 | 0.09 | 7 × 10−5 |
| cg09325711 | RALA | −0.007 | 4 × 10−6 | −0.10 | 0.02 |
| cg03756485 | GNPTAB | −0.003 | 5 × 10−6 | −0.12 | 3 × 10−5 |
Abbreviations: AFF1, AF4/FMR2 family member 1; ATP5G2, adenosine triphosphate membrane subunit c2; B3GNT7, β-1,3-N-acetylglucosaminyltransferase 2; C6orf132, chromosome 6 open reading frame 132; CD53, CD53 molecule; GAS2L3, growth arrest specific protein 2-like 3; GNPTAB, N-acetylglucosamine-1-phosphate transferase subunits α and β; HNRNPA1P10, heterogeneous nuclear ribonucleoprotein A1 pseudogene 10; HNRNPF, heterogenous nuclear ribonucleoprotein F; MIR4754, microRNA 4,754; MLLT1, myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog) (translocated to 1); MYPOP, Myb related transcription factor, partner of profiling; N/A, not applicable; PPIF, peptidyl-prolyl cis-trans isomerase; RALA, RAS-like proto-oncogene a; SLC1A5, solute carrier family 1 member 5; SNORD75, small nucleoloar RNA c/d box 75; TACC1, transforming acidic coiled-coil containing protein 1; TRIP6, thyroid receptor-interacting protein 6; TXLNA, taxilin α; ZNF428, zinc finger protein 428.
a Blood association P ≤ 5 × 10−6.
Alcohol-related CpG sites and changes in breast cancer
To determine whether the 98 CpG sites associated with alcohol consumption in both blood and normal breast tissue also showed aberrant DNA methylation in breast cancer, we analyzed publicly available data from TCGA. Seven of the 98 CpG sites (in genes for cyclin g-associated kinase (GAK), integrin subunit α 6 (ITGA6), actin-binding LIM protein family member 1 (ABLIM1), transmembrane protein 129 (TMEM129), RAS-like proto-oncogene A (RALA), SH3 domain-containing 21 (SH3D21), and PPIF) were significantly differentially methylated in tumor versus normal comparisons, and all 7 sites were correlated with gene expression (Table 6). At all 7 sites, the direction of methylation change associated with alcohol consumption coincided with the direction in tumor tissue.
Table 6.
Cytosine-Phosphate-Guanine Sites Differentially Methylated in Both Blood and Normal Breast Tissue With Increasing Alcohol Consumption That Also are Differentially Methylated in Breast Tumor Compared With Normal Tissue and Are Associated With Gene-Expression Levels in Tumor, Sister Study, United States, 2004–2015
| Probe | Gene | Blood Methylation Direction With Higher Alcohol Consumption | Blood P Value | Normal Breast Methylation Direction With Higher Alcohol Consumption | Normal Breast P Value | Breast Tumor Methylation Relative to Normal Breast | Breast Tumor P Value | Correlation Between Methylation and Gene Expression in Breast Tumor | Correlation P Value |
|---|---|---|---|---|---|---|---|---|---|
| cg03915012 | GAK | ↑ | 0.0002 | ↑ | 2 × 10−5 | ↑ | 0.0001 | 0.147 | 1 × 10−5 |
| cg06262280 | ITGA6 | ↓ | 2 × 10−5 | ↓ | 0.003 | ↓ | 0.02 | 0.133 | 8 × 10−5 |
| cg07978738 | ABLIM1 | ↑ | 0.002 | ↑ | 0.04 | ↑ | 0.0002 | −0.214 | 2.0E-10 |
| cg08629884 | EM129 | ↓ | 0.0004 | ↓ | 0.0003 | ↓ | 3 × 10−5 | −0.086 | 0.01 |
| cg09325711 | RALA | ↓ | 4 × 10−6 | ↓ | 0.02 | ↓ | 4 × 10−5 | −0.110 | 0.001 |
| cg17710804 | SH3D21 | ↓ | 0.0004 | ↓ | 0.0001 | ↓ | 3 × 10−5 | −0.269 | 9 × 10−16 |
| cg19939077 | PPIF | ↓ | 9 × 10−10 | ↓ | 0.007 | ↓ | 2 × 10−6 | −0.252 | 5 × 10−14 |
Abbreviations: ABLIM1, actin binding LIM protein 1; EM129, transmembrane protein 129; GAK, cyclin g associated kinase; ITGA6, integrin subunit α 6; PPIF, peptidyl-prolyl cis-trans isomerase; RALA, RAS-like proto-oncogene; SH3D21, SRC homology 3 domain containing 21.
DISCUSSION
We performed an epigenome-wide association study of alcohol consumption and DNA methylation using participants from the Sister Study and the Normal Breast Study. Using a discovery set of 2,878 women, we identified 5,458 CpGs associated with alcohol; of those, 1,207 CpGs replicated in independent DNA samples of blood or normal breast tissue, with 98 CpGs replicating in both tissues. Of the CpGs replicated in blood, 93% showed evidence of an association between lower methylation and higher alcohol consumption, and of those replicated in breast tissue, 63% showed evidence of an association between lower methylation and higher alcohol consumption. Alcohol-associated CpGs were significantly more likely to coincide with histone modifications and transcription factors found at actively transcribed genes, suggesting that these methylation changes might affect transcription levels.
As assessed by the average methylation of all CpGs on the array, there was evidence that alcohol consumption was associated with significantly lower global methylation. Significantly lower methylation was also evident when CpGs on the array were parsed by genomic location/CpG density into 6 different categories. Decreased methylation is consistent with effects reported in animal models: Acetaldehyde, a metabolite of alcohol, has been shown to inhibit the function of DNA methyltransferases (DNMTs) in vitro (28), and alcohol decreases DNMT mRNA levels in rats (29), with chronic in utero exposure leading to global decreases in DNA methylation (28). DNMTs are responsible for maintaining existing DNA methylation patterns and de novo methylation. In humans DNMT3b mRNA expression levels are lower in individuals with chronic alcoholism (30). Alcohol also impairs donation of methyl groups to DNMTs via inhibition of S-adenosyl-L-methionine (SAMe) synthesis (31–33).
Alcohol consumption is associated with increased risk of several cancers, including breast cancer (3, 5). In our study, 2 of the alcohol-related CpGs (cg14476101 and cg16246545) with the strongest associations are approximately 50 base pairs apart in the first intron of the PHGDH, a gene that has been shown to modulate cancer cell proliferation (34, 35). PHGDH can shift glucose metabolism to produce high levels of serine (36) and support tumor growth (37). Increased expression of PHGDH is observed in many tumors types (38–43) and is associated with poor prognosis in several cancers (44–46), and ectopic expression of PHGDH in mammary epithelial cells leads to phenotypic changes that could predispose cells to malignant transformation (36). Using TCGA data we found that in breast tumor tissue, increased methylation at cg14476101 and cg16246545 is strongly associated with increasing expression of the PHGDH mRNA, providing a possible link between alcohol consumption and breast carcinogenesis.
Similarly, cg02711608 in the solute carrier family 1 member 5 (SLC1A5) gene had decreased methylation with increasing alcohol consumption in both blood and in breast tissue. SLC1A5 mediates glutamine uptake that is important for rapid proliferation of neoplastic cells (47). Many tumor types, including colorectal cancers and triple-negative breast tumors (48–51), express high levels of SLC1A5 and conversely, in vitro inhibition of SLC1A5 has been shown to slow tumor cell proliferation (52–55).
Of the 98 CpG sites that replicated in both blood and breast tissue, 7 (cg03915012 in cyclin g-associated kinase (GAK), cg06262280 in integrin subunit α 6 (ITGA6), cg07978738 in actin-binding LIM protein family member 1 (ABLIM1), cg08629884 in transmembrane protein 129 (TMEM129), cg09325711 in RAS-like proto-oncogene A (RALA), cg17710804 in SH3 domain-containing 21 (SH3D21), and cg19939077 in PPIF) had significant methylation changes in breast tumors in the same direction seen with higher levels of alcohol consumption. Although there is little literature on the role of these genes in cancer, in tamoxifen-treated estrogen receptor–positive breast cancer, increased PPIF expression has been associated with increased risk of future metastasis (56).
Strengths and limitations
Our data are cross-sectional and thus we cannot determine whether the observed methylation differences are caused by alcohol consumption. We also cannot determine whether the associations we describe persist if women stop drinking alcohol. We restricted our analysis to non-Hispanic white women, so these associations might not be evident in other ethnic groups. However, this study benefits from large sample sizes and multiple replication sets in both blood and breast tissue. We also leveraged multiple existing databases to provide genomic context for the identified differentially methylated CpG sites.
Conclusion
This study utilized a large discovery set and multiple replication sets in both blood and normal breast tissue to identify associations between higher alcohol consumption and methylation at 98 CpG sites. Individual alcohol-CpG methylation associations were particularly notable for sites in PGHDH, PPIF, SLC15, SLC43A1, and SLC7A11. Many of the sites we identified in blood are reported in a recent study of methylation and alcohol consumption (10), supporting the hypothesis that these associations are robust and reproducible across different study populations. Seven of the CpG sites with replicated differential methylation in normal breast tissue related to alcohol consumption also showed differential methylation in breast tumors compared with normal tissue. These sites show the same directional change as with alcohol consumption; given the strong link between heavier alcohol consumption and risk of breast cancer development, it is possible that methylation of these sites is related to breast cancer tumorigenesis and might be worthwhile targets for future study.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Epidemiology Branch, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina (Lauren E. Wilson, Zongli Xu, Sophia Harlid, Alexandra J. White, Dale P. Sandler, Jack A. Taylor); Department of Population Health Sciences, Duke University School of Medicine, Durham, North Carolina (Lauren E. Wilson); Department of Radiation Sciences, Oncology, Umea University, Umea, Sweden (Sophia Harlid); and Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Melissa A. Troester).
This work was supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences (grants Z01 ES049033, Z01 ES049032, and Z01 ES044005).
Conflict of interest: none declared.
Abbreviations
- CpG
cytosine-phosphate-guanine
- DNMT
DNA methyltransferase
- FDR
false discovery rate
- PHGDH
phosphoglycerate dehydrogenase gene
- PPIF
peptidyl-prolyl cis-trans isomerase gene
- SLC15
solute carrier 15 gene
- SLC43A1
solute carrier family 43 member 1 gene
- SLC7A11
solute carrier family 7 member 11 gene
- TCGA
the Cancer Genome Atlas
REFERENCES
- 1. Rehm J, Mathers C, Popova S, et al. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373(9682):2223–2233. [DOI] [PubMed] [Google Scholar]
- 2. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans Alcohol consumption and ethyl carbamate. IARC Monogr Eval Carcinog Risks Hum. 2010;96:3–1383. [PMC free article] [PubMed] [Google Scholar]
- 3. Corrao G, Bagnardi V, Zambon A, et al. A meta-analysis of alcohol consumption and the risk of 15 diseases. Prev Med. 2004;38(5):613–619. [DOI] [PubMed] [Google Scholar]
- 4. Williams LA, Olshan AF, Hong CC, et al. Alcohol intake and breast cancer risk in African American women from the AMBER Consortium. Cancer Epidemiol Biomarkers Prev. 2017;26(5):787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mostofsky E, Mukamal KJ, Giovannucci EL, et al. Key findings on alcohol consumption and a variety of health outcomes from the Nurses’ Health Study. Am J Public Health. 2016;106(9):1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hagerty SL, Bidwell LC, Harlaar N, et al. An exploratory association study of alcohol use disorder and DNA methylation. Alcohol Clin Exp Res. 2016;40(8):1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim DS, Kim YH, Lee WK, et al. Effect of alcohol consumption on peripheral blood Alu methylation in Korean men. Biomarkers. 2016:21(3):243–248. [DOI] [PubMed] [Google Scholar]
- 8. Zhang H, Herman AI, Kranzler HR, et al. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013;37(suppl 1):E108–E115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Philibert RA, Penaluna B, White T, et al. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9(9):1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu C, Marioni RE, Hedman AK, et al. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry. 2018;23(2):422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. National Institutes of Health (NIH), Office of Research on Women’s Health, Office of the Director, and the National Institute on Alcohol Abuse and Alcoholism Alcohol: A Women’s Health Issue. 2nd ed Washington, DC: National Institute on Alcohol Abuse and Alcoholism; 2015. [Google Scholar]
- 12. Scoccianti C, Lauby-Secretan B, Bello PY, et al. Female breast cancer and alcohol consumption: a review of the literature. Am J Prev Med. 2014;46(3 suppl 1):S16–S25. [DOI] [PubMed] [Google Scholar]
- 13. Department of Health and Human Services, Office of Disease Prevention and Health Promotion 2015–2020 Dietary Guidelines for Americans. https://health.gov/dietaryguidelines/2015/. Updated 2015. Accessed December 12, 2017.
- 14. Sandler DP, Hodgson ME, Deming-Halverson SL, et al. The Sister Study cohort: baseline methods and participant characteristics. Environ Health Perspect. 2017;125(12):127003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ekenga CC, Parks CG, Sandler DP. Chemical exposures in the workplace and breast cancer risk: a prospective cohort study. Int J Cancer. 2015;137(7):1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson LE, D’Aloisio AA, Sandler DP, et al. Long-term use of calcium channel blocking drugs and breast cancer risk in a prospective cohort of US and Puerto Rican women. Breast Cancer Res. 2016;18:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. White AJ, DeRoo LA, Weinberg CR, et al. Lifetime alcohol intake, binge drinking behaviors, and breast cancer risk. Am J Epidemiol. 2017;186(5):541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hair BY, Xu Z, Kirk EL, et al. Body mass index associated with genome-wide methylation in breast tissue. Breast Cancer Res Treat. 2015;151(2):453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Brien KM, Sandler DP, Xu Z, et al. Vitamin D, DNA methylation, and breast cancer. Breast Cancer Res. 2018;20:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harlid S, Xu Z, Panduri V, et al. In utero exposure to diethylstilbestrol and blood DNA methylation in women ages 40–59 years from the sister study. PLoS One. 2015;10(3):e0118757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu Z, Niu L, Li L, et al. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44(3):e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Accomando WP, Wiencke JK, Houseman EA, et al. Decreased NK cells in patients with head and neck cancer determined in archival DNA. Clin Cancer Res. 2012;18(22):6147–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J, Just AC, Schwartz J, et al. CpGFilter: model-based CpG probe filtering with replicates for epigenome-wide association studies. Bioinformatics. 2016;32(3):469–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60(3):155–166. [DOI] [PubMed] [Google Scholar]
- 28. Garro AJ, McBeth DL, Lima V, et al. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15(3):395–398. [DOI] [PubMed] [Google Scholar]
- 29. Bielawski DM, Abel EL. The effect of administering ethanol as single vs. divided doses on blood alcohol levels in the rat. Neurotoxicol Teratol. 2002;24(4):559–562. [DOI] [PubMed] [Google Scholar]
- 30. Bönsch D, Lenz B, Fiszer R, et al. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm (Vienna). 2006;113(9):1299–1304. [DOI] [PubMed] [Google Scholar]
- 31. Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7(8):599–612. [DOI] [PubMed] [Google Scholar]
- 32. Martínez-Chantar ML, García-Trevijano ER, Latasa MU, et al. Importance of a deficiency in S-adenosyl-L-methionine synthesis in the pathogenesis of liver injury. Am J Clin Nutr. 2002;76(5):1177S–1182S. [DOI] [PubMed] [Google Scholar]
- 33. Torres L, Ávila MA, Carretero MV, et al. Liver-specific methionine adenosyltransferase MAT1A gene expression is associated with a specific pattern of promoter methylation and histone acetylation: implications for MAT1A silencing during transformation. FASEB J. 2000;14(1):95–102. [DOI] [PubMed] [Google Scholar]
- 34. Polet F, Corbet C, Pinto A, et al. Reducing the serine availability complements the inhibition of the glutamine metabolism to block leukemia cell growth. Oncotarget. 2016;7(2):1765–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samanta D, Park Y, Andrabi SA, et al. PHGDH expression is required for mitochondrial redox homeostasis, breast cancer stem cell maintenance, and lung metastasis. Cancer Res. 2016;76(15):4430–4442. [DOI] [PubMed] [Google Scholar]
- 36. Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mullarky E, Mattaini KR, Vander Heiden MG, et al. PHGDH amplification and altered glucose metabolism in human melanoma. Pigment Cell Melanoma Res. 2011;24(6):1112–1115. [DOI] [PubMed] [Google Scholar]
- 39. Yoon S, Kim JG, Seo AN, et al. Clinical implication of serine metabolism-associated enzymes in colon cancer. Oncology. 2015;89(6):351–359. [DOI] [PubMed] [Google Scholar]
- 40. Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pollari S, Käkönen SM, Edgren H, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125(2):421–430. [DOI] [PubMed] [Google Scholar]
- 42. Liu J, Guo S, Li Q, et al. Phosphoglycerate dehydrogenase induces glioma cells proliferation and invasion by stabilizing forkhead box M1. J Neurooncol. 2013;111(3):245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jing Z, Heng W, Xia L, et al. Downregulation of phosphoglycerate dehydrogenase inhibits proliferation and enhances cisplatin sensitivity in cervical adenocarcinoma cells by regulating Bcl-2 and caspase-3. Cancer Biol Ther. 2015;16(4):541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jia XQ, Zhang S, Zhu HJ, et al. Increased expression of PHGDH and prognostic significance in colorectal cancer. Transl Oncol. 2016;9(3):191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu J, Ma J, Wang X, et al. High expression of PHGDH predicts poor prognosis in non-small cell lung cancer. Transl Oncol. 2016;9(6):592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xian Y, Zhang S, Wang X, et al. Phosphoglycerate dehydrogenase is a novel predictor for poor prognosis in gastric cancer. Onco Targets Ther. 2016;9:5553–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Geldermalsen M, Wang Q, Nagarajah R, et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35(24):3201–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang F, Zhao Y, Zhao J, et al. Upregulated SLC1A5 promotes cell growth and survival in colorectal cancer. Int J Clin Exp Pathol. 2014;7(9):6006–6014. [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y, Yang L, An H, et al. High expression of Solute Carrier Family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma. Sci Rep. 2015;5:16954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin Cancer Biol. 2005;15(4):254–266. [DOI] [PubMed] [Google Scholar]
- 52. Wang Q, Hardie RA, Hoy AJ, et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J Pathol. 2015;236(3):278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hassanein M, Hoeksema MD, Shiota M, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res. 2013;19(3):560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hassanein M, Qian J, Hoeksema MD, et al. Targeting SLC1a5-mediated glutamine dependence in non-small cell lung cancer. Int J Cancer. 2015;137(7):1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Willems L, Jacque N, Jacquel A, et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood. 2013;122(20):3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Madhavan S, Gusev Y, Singh S, et al. ERRγ target genes are poor prognostic factors in Tamoxifen-treated breast cancer. J Exp Clin Cancer Res. 2015;34:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.