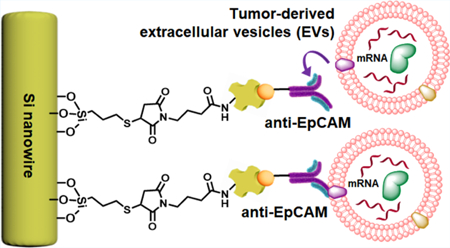
Abstract
Tumor-derived extracellular vesicles (EVs) present in bodily fluids are emerging liquid biopsy markers for non-invasive cancer diagnosis and treatment monitoring. Because the majority of EVs in circulation are not of tumor origin, it is critical to develop new platforms capable of enriching tumor-derived EVs from the blood. Herein, we introduce a biostructure-inspired NanoVilli Chip, capable of highly efficient and reproducible immunoaffinity capture of tumor-derived EVs from blood plasma samples. Anti-EpCAM-grafted silicon nanowire arrays were engineered to mimic the distinctive structures of intestinal microvilli, dramatically increasing surface area and enhancing tumor-derived EV capture. RNA in the captured EVs can be recovered for downstream molecular analyses by reverse transcription Droplet Digital PCR. We demonstrate that this assay can be applied to monitor the dynamic changes of ROS1 rearrangements and epidermal growth factor receptor T790M mutations that predict treatment responses and disease progression in non-small cell lung cancer patients.
Keywords: nanosubstrates, microfluidics, extracellular vesicles, ROS1 rearrangements, EGFR T790M mutation, non-small cell lung cancer
Graphical Abstract
INTRODUCTION
Extracellular vesicles1 (EVs) are a heterogeneous group of lipid bilayer-enclosed particles that play a crucial role in intercellular communication by transporting biomolecular cargo, including DNA, RNA, proteins, and lipids.2,3 EVs are classified into three categories according to their size and their biogenesis pathway of origin: (i) exosomes (30–150 nm);4,5 (ii) microvesicles (100–1000 nm);6 and (iii) apoptotic bodies (500–4000 nm).7 EVs are actively secreted by all cell types in the human body and can be found in a variety of body fluids. Oncogenic transformation often leads to increased EV production by tumor cells, resulting in increased levels of tumor-derived EVs in patients’ blood.8,9 Compared to well-studied circulating tumor cells (CTCs), which are challenging to detect until metastatic progression, tumor-derived EVs are present in circulation at relatively early stages of disease. These cancer-specific EVs can be collected from plasma or serum at any time over the course of treatment. Consequently, tumor-derived EVs are emerging candidates for liquid biopsy approaches10–12 for implementing non-invasive cancer diagnosis, prognosis, and treatment monitoring across all disease stages.
Since the biomolecular contents of tumor-derived EVs mirror those of the parental tumor cells, performing molecular characterization on tumor-derived EVs could provide key insights into the molecular mechanisms governing oncogenesis and disease progression. Most importantly, the fragile biomolecular contents inside individual EVs (e.g., tumor-specific RNA) are protected by the EV’s lipid bilayer, guaranteeing their availability for downstream molecular analysis. Recent studies have demonstrated the feasibility of detecting cancer driver mutations using mRNA extracted from enriched tumor-derived EVs in different solid tumors, for example, KRAS mutations in pancreatic cancer13 and epidermal growth factor receptor (EGFR) vIII mutation in glioblastoma.14 Performing mutational analyses using EV-derived mRNA results in improved sensitivity and better correlation with patients’ clinical outcomes over cell-free DNA (cfDNA)-based approaches.15,16 Moreover, well-preserved RNA in tumor-derived EVs is ideal for detecting gene rearrangements, as they have variable breakpoints and different fusion partners.
Since tumor-derived EVs constitute only a minor portion of the total number of EVs in circulation, the enrichment of tumor-derived EVs represents a considerable technical challenge. Conventional methods, such as ultracentrifugation,17–20 filtration,21,22 precipitation,23 and size-based microfluidic enrichment,24–29 can isolate entire populations of EVs from peripheral blood samples based on their physical properties (i.e., size and/ or density). However, these approaches are incapable of discriminating tumor-derived EVs from non-tumor-derived EVs. More recent research efforts have explored the application of immunoaffinity-based capture techniques for enriching tumor-derived EVs in different solid tumors.17,18,27 For example, pancreatic cancer-derived exosomes can be captured selectively using anti-GPC1-coated beads and isolated via flow cytometry,13 and the enrichment of glioblastoma-derived exosomes has been demonstrated in herringbone microfluidic devices (i.e., EVHB-Chip) with EGFRvIII antibodies used as the capture agent.14 To characterize and/or to quantify the trace amount of mRNA extracted from the enriched tumor-derived EVs, highly sensitive mRNA profiling technologies, for example, next-generation sequencing and Droplet Digital PCR (ddPCR), were adopted for downstream detection purposes.
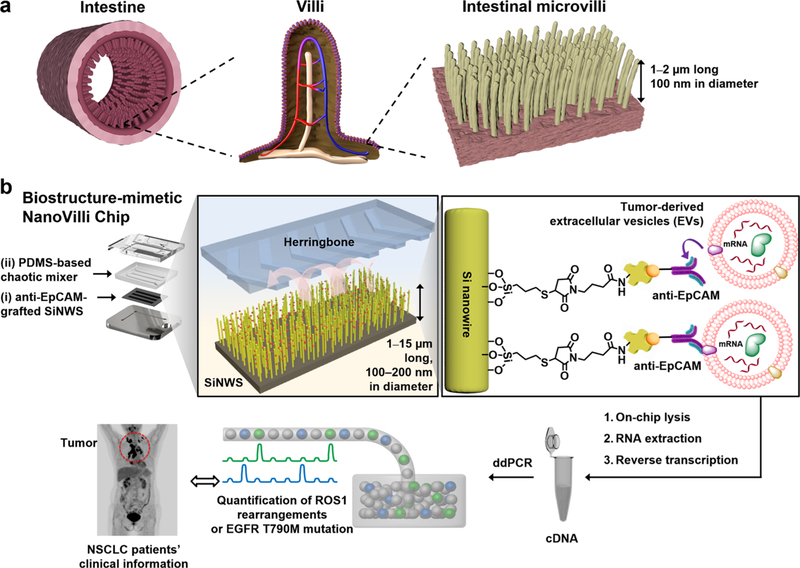
Inspired by the distinctive structures of intestinal microvilli (Figure 1a), which are densely packed on intestinal walls to increase mucosal surface area for enhanced absorption, we developed the NanoVilli Chips. These biostructure-inspired chips have antibody-grafted silicon (Si) nanowire arrays that are engineered in a densely packed manner to achieve efficient and reproducible immunoaffinity capture of tumor-derived EVs from blood plasma samples. A NanoVilli Chip is composed of two integral components (Figure 1b), that is, (i) an anti-epithelial cell adhesion molecule (EpCAM)-grafted Si nanowire substrate (SiNWS) and (ii) a superimposed polydimethylsiloxane (PDMS)-based chaotic mixer with a serpentine micro-channel, in which herringbone micropatterns introduce helical flow to facilitate direct physical contact between anti-EpCAM-grafted SiNWS and tumor-derived EVs in plasma. When a plasma sample containing tumor-derived EVs is run through a NanoVilli Chip, the integration of the anti-EpCAM-grafted SiNWS and the PDMS-based chaotic mixer leads to enhanced capture of tumor-derived EVs. Our SiNWS is optimized for interacting with nanoscale targets. In our previously described NanoVelcro chips,30–32 which utilize a similar device configuration, Velcro-like topographic interactions between nano-structured substrates and nanoscale cellular surface components immobilize CTCs on top of the SiNWS. Whereas microscale CTCs (diameters = 6–20 μm) can only interact with the uppermost portion of the SiNWS, free-floating nanoscale EVs (diameters = 30–1000 nm) can interact with both upper and deeper portions of the SiNWS (which are spaced 200–400 nm apart). EVs above 300 nm in diameter are primarily captured on the tips of the SiNWS, whereas EVs with sizes ranging between 30 and 300 nm are captured on both tips and sidewalls of the SiNWS. We designed NanoVilli Chips with longer Si nanowires (lengths = 10–15 μm) to increase functional surface area, enabling more efficient enrichment of tumor-derived EVs at both the tips and the sidewalls of individual Si nanowires. After capturing tumor-derived EVs on NanoVilli Chips, RNA recovered from the EVs can be evaluated with a Qubit 3.0 Fluorometer in combination with the Qubit RNA HS Assay and subjected to downstream analysis by reverse transcription Droplet Digital PCR (RT-ddPCR). We explored the clinical utility of NanoVilli Chips by applying this workflow to detect driver gene alterations in non-small cell lung cancer (NSCLC) quantitatively (e.g., ROS1 rearrangements or EGFR, T790M mutation). To understand how the embedded Si nanowires in NanoVilli Chips contribute to the highly efficient capture of tumor-derived EVs, scanning electron microscopy (SEM), transmission electron microscopy (TEM), and fluorescence microscopy were employed (Figure 2) to characterize the interactions between anti-EpCAM-grafted SiNWS and tumor-derived EVs. These observations were further validated with those obtained via computational simulation (see Supporting Information). Moreover, we progressively optimized the performance of NanoVilli Chips by systematically varying device operating conditions and configurations (e.g., flow rates, Si nanowire lengths, and anti-EpCAM concentrations). We evaluated these data to identify experimental conditions that enable efficient and reproducible enrichment of tumor-derived EVs from both artificial plasma samples and blood plasma samples obtained from NSCLC patients. The combined use of NanoVilli Chips and RT-ddPCR offers a new type of EV-based mRNA assay for quantitatively detecting and monitoring targetable oncogenic gene alterations in NSCLC patients.
Figure 1.
Inspired by the distinctive structures of intestinal microvilli (a), which are densely packed on the intestinal walls to increase their intestinal mucosal surface areas for enhanced absorption, we designed and tested a biostructure-inspired NanoVilli Chip (b) featuring densely packed anti-epithelial cell adhesion molecule (anti-EpCAM)-grafted silicon (Si) nanowire arrays to achieve highly efficient and reproducible immunoaffinity capture of tumor-derived EVs. A NanoVilli Chip is composed of (i) an anti-EpCAM-grafted Si nanowire substrate (SiNWS) and (ii) a superimposed PDMS-based chaotic mixer. Captured tumor-derived EVs were lysed in the device to release EV-derived RNA, which was extracted for downstream analysis via RT-ddPCR. We utilize this workflow to detect gene alterations such as ROS1 rearrangements or EGFR T790M mutations in NSCLC quantitatively.
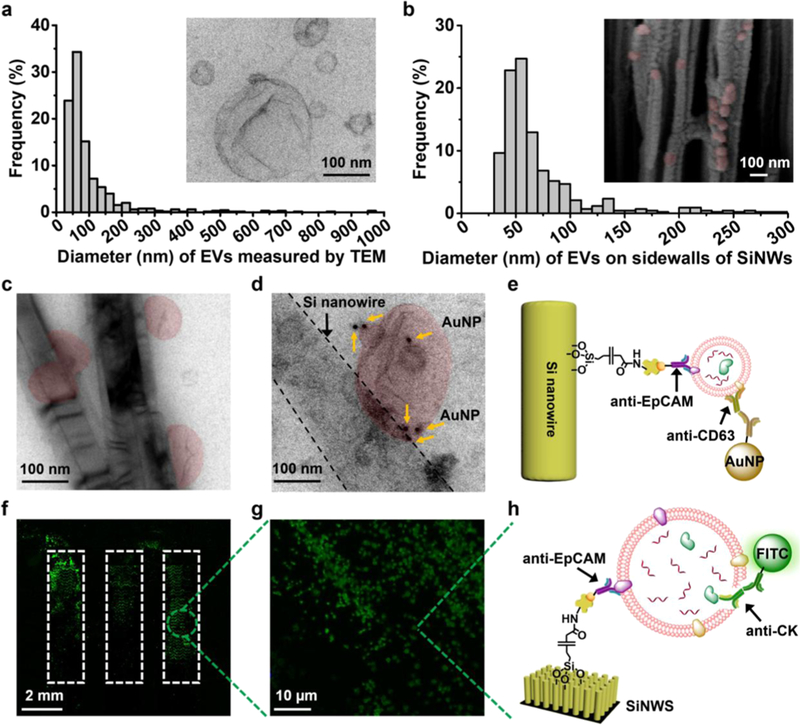
Figure 2.
Characterization of tumor-derived EVs in solution and on anti-epithelial cell adhesion molecule (anti-EpCAM)-grafted silicon nanowire substrates (SiNWSs) in NanoVilli Chips. (a) Size distribution (n = 653, diameters = 30–1000 nm) of HCC78-derived EVs, measured by TEM. Inset: a representative TEM image (scale bar, 100 nm) of HCC78-derived EVs. (b) Size distribution of HCC78-derived EVs (n = 425, diameters = 30–300 nm) immunoaffinity-captured on the sidewalls of Si nanowires (SiNWs) measured by SEM. Inset: a representative cross-sectional SEM image (scale bar, 100 nm) of a device with immobilized HCC78-derived EVs (colored in red). (c) TEM image of HCC78-derived EVs (colored in red) immobilized on the sidewalls of Si nanowires. Scale bar, 100 nm. (d) Immunogold staining by anti-CD63 was employed to verify the identity of EVs captured on Si nanowires. (e) Schematic illustrating the immobilization of 10 nm gold nanoparticles via anti-CD63 onto a tumor-derived EV attached to the sidewall of a Si nanowire by anti-EpCAM. (f,g) Fluorescence microscopy images confirming the capture of HCC78-derived EVs immobilized on the SiNWS using an antibody targeting to the epithelial tumor marker CK. (h) Schematic depicting how anti-EpCAM and anti-CK were used for EV capture and immunostaining of CK, respectively.
There has recently been a major strategic shift in the clinical management of NSCLC. Following initial tissue-based histo-logical classification schemes, NSCLC has been further classified based on molecular phenotype (e.g., ALK/ROS1 rearrangements33,34 and EGFR mutations35) in order to guide the implementation of effective targeted therapeutic strategies employing tyrosine kinase inhibitors (TKIs). Considering the profound risk associated with invasive tissue-based diagnostic approaches, clinicians increasingly prefer non-invasive diagnostic solutions36 for both initial diagnosis and longitudinal monitoring of disease progression.37 NanoVilli Chips were developed to harvest tumor-derived EVs to enable non-invasive characterization of tumors. We demonstrate the feasibility of quantifying the dynamic changes in both ROS1 rearrangements and the EGFR T790M mutations from tumor-derived EVs in NSCLC patients and correlate these data with patient outcomes measured by radiographic imaging, which is the current gold standard for evaluating the therapeutic response of solid tumors clinically.
RESULTS AND DISCUSSION
Fabrication of NanoVilli Chips.
The nanostructures-embedded substrates (i.e., SiNWS) were fabricated via photolithography, followed by silver (Ag) nanoparticle-templated wet etching38 to generate vertically aligned nanowire arrays on a Si wafer.39 This fabrication process confers precise control over the diameters (100–200 nm), lengths (1–2 or 10–15 μm), and spacings (200–400 nm) of the Si nanowires (confirmed by SEM, Figure S1), resulting in large surface areas that enable enhanced immunoaffinity capture of tumor-derived EVs. A 4-step modification process was designed for the preparation of anti-EpCAM-grafted SiNWS (Figure S2). Chaotic mixers were prepared by thermally curing PDMS pre-polymer (Sylgard 184) on a Si-based replicate mold (master wafer). On the mold, the herringbone patterns were fabricated by inductively coupled plasma-reactive ion etching (ICP-RIE). Compared to the SU-8 photolithographically deposited patterns used previously,40 the ICP-RIE fabricated patterns on Si are much more durable over time with repeated usage. We altered the chaotic mixing behavior in our devices based on findings reported by Sheng et al.,41 where the spacings of herringbone patterns and the microchannel heights/widths/lengths (70 μm × 2 mm × 60 mm) were configured to optimize physical contact between anti-EpCAM-grafted SiNWS and tumor-derived EVs in plasma. Prior to EV capture studies, a custom-designed chip holder was employed to couple the PDMS-based chaotic mixers onto anti-EpCAM-grafted SiNWS to complete the chip assembly (Figure S3). This chip holder also serves as an interface with syringe/ syringe pumps used for handling plasma samples and reagents.
Characterization of EV Captured in NanoVilli Chips.
To study the function and performance of NanoVilli Chips, tumor-derived EVs were purified by ultracentrifugation from serum-free culture media of HCC78 NSCLC cells which harbor the SLC34A2-ROS1 rearrangement. These HCC78-derived EVs were first characterized by both dynamic light scattering (DLS, Figure S4) and TEM. The inset in Figure 2a shows a typical TEM image of the HCC78-derived EVs after uranyl acetate negative staining. These EVs exhibited cup- or spherical-shaped morphologies with sizes ranging between 30 and 1000 nm. As a model system for testing NanoVilli Chips, artificial plasma samples were prepared by spiking aliquoted 10 μL HCC78-derived EVs into 90 μL freshly isolated healthy donor (HD) blood plasma. After EV capture, the NanoVilli Chips were disassembled to remove the PDMS-based chaotic mixers. To prepare samples for SEM imaging, the SiNWS underwent paraformaldehyde (PFA) fixation, ethanol dehydration, and vacuum sputter coating with gold. The SiNWS were then cut to expose the cross sections of the Si nanowire arrays. The inset in Figure 2b shows a cross-sectional SEM image of Si nanowires with HCC78-derived EVs (diameters = 30–300 nm) adhered along the sidewalls of the nanowires. For samples characterized by TEM, Si nanowires with immobilized EVs were mechanically detached from the underlying substrate. The detached Si nanowires were collected and transferred onto TEM grids. The immobilized EVs along the sidewalls of Si nanowires range between 30 and 300 nm in diameter (Figure 2c). Additionally, both SEM and TEM images showed that EVs with diameters greater than 300 nm were immobilized on the tips of SiNWS (Figure S5a–d), which is expected, given that these EVs are too large to fit into the spacings (200–400 nm) between the Si nanowires. In comparison, negligible amounts of EVs were captured in control experiments where the anti-EpCAM capture agent was absent (Figure S6). To confirm the identity of EVs, immunogold staining via anti-CD63 (an EV surface marker) was employed to label EVs with 10 nm gold nanoparticles before and after anti-EpCAM-based immunoaffinity capture onto Si nanowires, respectively. The TEM images showed that both precapture (Figure S7) and postcapture (Figure 2d,e) EVs could be decorated with 10 nm gold nanoparticles via anti-CD63.
The HCC78-derived EVs also express cytokeratin (CK) due to their epithelial origin, which enables immunohistochemical characterization of tumor-derived EVs immobilized on the SiNWS (Figure 2f–h) via fluorescence microscopy (Nikon, 90i). As shown in Figure 2f,g, CK-positive EVs trapped on the tips of SiNWS were visualized by fluorescence microscopy. The actual size distribution of these EVs was determined by SEM (Figure S5b,d).
EV-Derived RNA Assay Using NanoVilli Chips.
To optimize EV-capture performance for NanoVilli Chips, we systematically examined different experimental parameters, including flow rates, Si nanowire lengths, and anti-EpCAM concentrations. In each study, a 100 μL artificial plasma sample was run through a NanoVilli Chip. Subsequently, a TRIzol solution (Zymo Research, USA) was introduced into the device to lyse the captured EVs. The resulting lysate was subjected to RNA extraction using a Direct-zolRNA MicroPrep Kit (Zymo Research, USA), followed by treatment with DNase I to remove residual DNA. The extracted EV-derived RNA was then evaluated and quantified using a Qubit 3.0 Fluorometer in combination with the Qubit RNA HS Assay. The amount of the extract EV-derived RNA is denoted as RNACap−EV. In parallel, 90 μL healthy-donor plasma samples were analyzed via the same workflow, where we denote the systems’ RNA background as RNAbg. To determine the EV-capture efficiencies of NanoVilli Chips, we also quantified RNA directly extracted from aliquoted 10 μL HCC78-derived EVs (that were not passed through a NanoVilli Chip), labeled as RNAori−EV. The EV-capture performance of NanoVilli Chips was assessed by calculating the RNA recovery rate using the following equation:
| (1) |
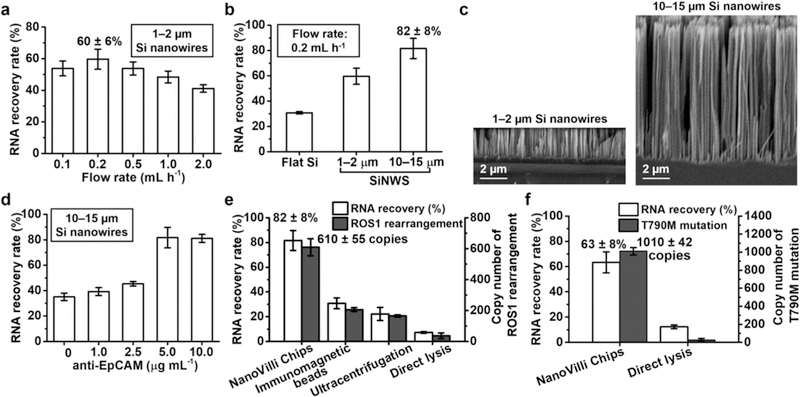
To study how flow rate affects EV-capture performance, 100 μL artificial plasma samples were injected into NanoVilli Chips (Si nanowire lengths = 1–2 μm) at flow rates of 0.1, 0.2, 0.5, 1.0, and 2.0 mL h−1. A flow rate of 0.2 mL h−1 resulted in an optimal RNA recovery rate of 60 ± 6% (Figure 3a). We subsequently used this flow rate to investigate the relationship between Si nanowire length and EV-capture performance (Figure 3b). NanoVilli Chips with long Si nanowires (lengths = 10–15 μm) exhibited an 82 ± 8% RNA recovery rate, which was significantly higher than 60 ± 6 and 31 ± 1% observed for the devices with shorter Si nanowires (lengths = 1–2 μm) and flat Si substrates, respectively. The lengths of the embedded Si nanowires NanoVilli Chips were verified by SEM (Figure 3c). Additionally, SEM was also used to characterize how EVs (n = 500, diameters = 30–300 nm) distributed along the depth of Si nanowires (Figure S9a). The results in Figure S9b showed that 53.4, 20.4, 20.4, 5.8, and 0% of EVs were immobilized at depths of 0–1, 1–2, 2–5, 5–9, and 9–10 μm from the top of Si nanowires, respectively. This demonstrates that the increased surface area of the longer Si nanowires improves immunoaffinity capture of tumor-derived EVs such that a total length of 10 μm is sufficient.
Figure 3.
Optimization of NanoVilli Chips for immunoaffinity capture of tumor-derived EVs using artificial plasma samples. (a) EV-capture performance of NanoVilli Chips (Si nanowires lengths = 1–2 μm) was studied at flow rates of 0.1, 0.2, 0.5, 1.0, and 2.0 mL h−1. (b) EV-capture performance observed for three different control groups: flat Si substrates, short Si nanowires (lengths = 1–2 μm), and long Si nanowires (lengths = 10–15 μm). (c) SEM images (scale bar, 2 μm) of the two NanoVilli Chips with different lengths of embedded Si nanowires (1–2 vs 10–15 μm). (d) EV-capture performance observed for NanoVilli Chips with Si nanowires (lengths = 10–15 μm) coated by biotinylated anti-epithelial cell adhesion molecule (anti-EpCAM) at concentrations of 0, 1.0, 2.5, 5.0, and 10.0 μg mL−1. (e) RNA recovery rates and copy numbers of ROS1 rearrangements observed for NanoVilli Chips, immunomagnetic beads and ultracentrifugation. As a control, the artificial plasma samples were directly subjected to RT-ddPCR analysis to demonstrate the necessity of conducting EV enrichment. (f) General applicability of NanoVilli Chips was validated using different artificial plasma samples containing EVs purified from NCI-H1975 NSCLC cells harboring EGFR T790M mutation.
To validate the results of the EV distribution observed by SEM, a computational simulation was conducted (see Supporting Information). The well-known laminar boundary layer effect42 dominates fluid behavior at the surface of microfluidic channels. These laminar boundary layers were estimated to be about 1.3 μm thick (Figure S9c). Therefore, the flow velocity near the top of Si nanowire matrix is very slow and the EV diffusion into the Si nanowire matrix is primarily attributed to Brownian motion of EVs. A dissipative particle dynamics (DPD) simulation43 was used to study the EV capture process by the Si nanowire matrix when the EVs diffuse from the top to the bottom of Si nanowire matrix (Figure S9d). As shown in Figure S9e, 52.1, 25.0, 14.6, 8.3, and 0% of EVs were located at depths of 0–1, 1–2, 2–5, 5–9, and 9–10 μm from the top of Si nanowires, respectively. An empirical function with the exponential form was used to describe the EV distribution probability profiles along the depth of Si nanowire. The results derived from the experiment and DPD simulation were very close.
We next attempted to reduce the consumption of our EV-capture agent (anti-EpCAM) without compromising the EV-capture performance at the optimal flow rate and nanowire configurations identified earlier. Five different biotinylated anti-EpCAM concentrations (i.e., 0.5, 1.0, 2.5, 5.0, and 10.0 μg mL−1) were tested for SiNWS modification. At concentrations <5.0 μg mL−1, EV-capture performance was reduced dramatically (Figure 3d). When the concentration was higher than 5.0 μg mL−1, the EV-capture performance did not improve, indicating that 5.0 μg mL−1 is sufficient to capture EVs in NanoVilli Chips. EV capture capacity along the channel was tested by segmentally quantifying RNA recovery rates of the three channels (Figure S10b). The results indicated that 90% of the EVs were captured in the f irst channel of the NanoVilli Chips.
With the optimal EV-capture conditions identified, we compared the performances of NanoVilli Chips with two commonly used EV enrichment methods (i.e., immunomagnetic beads18 and ultracentrifugation17) using the artificial plasma samples. Since HCC78 NSCLC cells harbor-specific SLC34A2-ROS1 rearrangement, these artificial plasma samples can be used to validate the feasibility of detecting ROS1 rearrangement in the EVs captured by NanoVilli Chips. In parallel with the RNA quantification, matching RNA samples obtained from the three EV enrichment methods were subjected to the RT-ddPCR assay to quantify the ROS1 rearrangement copy number (Figure S11). The results summarized in Figure 3e indicate that NanoVilli Chips exhibited a superior RNA recovery rate of 82 ± 8% compared to the 31 ± 4 and 22 ± 5% observed for immunomagnetic beads and ultracentrifugation, respectively. We observed consistent performance in detecting ROS1 rearrangements (610 ± 55, 206 ± 12, and 165 ± 8 copies) when comparing NanoVilli Chips, immunomagnetic beads, and ultracentrifugation. For control purposes, the artificial plasma samples were directly processed and subjected to the RT-ddPCR assays. The resultant low RNA recovery rate (7 ± 1%) may due to the RNase and proteins in the background plasma had a negative effect on the RNA quality during direct lysing process, highlighting the necessity of EV enrichment for reliable EV-based RNA analysis. Finally, we validated the general applicability of NanoVilli Chips for enriching NSCLC-derived EVs using different artificial plasma samples containing EVs purified from NCI-H1975 cells (harboring EGFR T790M point mutation). As shown in Figure 3f, an EV-capture efficiency of 63 ± 8% was measured with the NanoVilli Chip, which is significantly higher than the 12 ± 2% observed following a direct lysis method. Using RT-ddPCR, 1010 ± 42 copies of EGFR T790M mutation were detected in enriched EV-derived RNA (whereas 27 ± 17 copies of EGFR T790M mutation were observed for the direct lysis method). Overall, the optimized conditions developed for NanoVilli Chips enabled efficient purification of tumor-derived EVs from artificial plasma samples with capture efficiencies ranging from 63 to 82% in a period of 30 min.
Non-Invasive Detection of Gene Alterations in NSCLC Patients.
We operated the NanoVilli Chips at the optimal conditions identified in our initial studies to enrich tumor-derived EVs from NSCLC patient blood plasma samples. A cohort of 13 NSCLC patients seven harboring a clinically confirmed CD74-ROS1 rearrangement (treatment naiv̈e, stages III−IV) and six with an acquired EGFR T790M mutation (resistant to the prior EGFR-TKI treatment, i.e., gefitinib or erlotinib, stages III−IV)—were recruited for this feasibility study (Table 1). Control studies were performed in parallel on nine HDs. In each study, 200 μL samples of processed plasma were run through a NanoVilli Chip. For the seven ROS1-rearranged NSCLC patients, 18–468 copies of the CD74-ROS1 rearrangement were detected in the NanoVilli Chip-enriched EVs at diagnosis. For the six EGFR T790M-mutated NSCLC patients, 0–225 copies of the acquired EGFR T790M mutation were detected at their time of disease relapse. In the control studies, all of the nine HDs were negative for both ROS1 rearrangement and EGFR T790M mutation (Table 1).
Table 1.
Clinical Characteristics of (NSCLC, Adenocarcinoma) Patients and HD Enrolled in This Study
| Patient No. | Gender | Age (Y) | Smoking history (Y) | Tumor grade | Clinical stage | Gene status (Tissue)b | Copy numbers ofT790M mutation/ROSl rearrangement in EVsa |
|---|---|---|---|---|---|---|---|
| R01 | Female | 62 | None | 3 | IIIB | CD74-ROS1 | 18 |
| R02 | Male | 41 | None | 2 | hib | CD74-ROS1 | 27 |
| R03 | Male | 61 | 35 | 2 | IV | CD74-ROS1 | 54 |
| R04 | Male | 34 | None | 3 | TV | CD74-ROS1 | 54 |
| R05 | Male | 61 | 35 | 2 | IV | CD74-ROS1 | 99 |
| R06 | Male | 34 | None | 3 | IV | CD74-ROS1 | 396 |
| R07-1 | Male | 32 | None | 3 | IV | CD74-ROS1 | 324 |
| R07-2 | Male | 32 | None | 3 | IV | N/A | 0 |
| R07-3 | Male | 32 | None | 3 | IV | N/A | 468 |
| E01 | Female | 55 | None | 3 | IV | T790M | 36 |
| E02 | Female | 62 | None | 3 | TV | T790M | 36 |
| E03 | Male | 66 | None | 3 | IV | T790M | 90 |
| E04-1 | Male | 53 | None | 3 | IV | N/A | 0 |
| E04-2 | Male | 53 | None | 3 | IV | T790M | 225 |
| E04-3 | Male | 53 | None | 3 | IV | N/A | 9 |
| E05 | Male | 62 | None | 2 | IV | T790M | 72 |
| E06 | Male | 61 | None | 2 | IV | T790M | 81 |
| HD01 | Male | 30 | None | N/A | N/A | N/A | 0 |
| HD02 | Male | 26 | None | N/A | N/A | N/A | 0 |
| HD03 | Male | 29 | None | N/A | N/A | N/A | 0 |
| HD04 | Male | 46 | None | N/A | N/A | N/A | 0 |
| HD05 | Female | 36 | None | N/A | N/A | N/A | 0 |
| HD06 | Male | 32 | None | N/A | N/A | N/A | 0 |
| HD07 | Male | 56 | None | N/A | N/A | N/A | 0 |
| HD08 | Female | 58 | None | N/A | N/A | N/A | 0 |
| HD09 | Male | 60 | None | N/A | N/A | N/A | 0 |
Per 0.2 mL plasma.
N/A: not available.
Dynamic Monitoring of Gene Alterations over the Course of Treatment Intervention.
We then investigated the feasibility of combining tumor-derived EV enrichment by NanoVilli Chips and RT-ddPCR to monitor dynamic changes in disease course during treatment. Serial blood draws were obtained from patient R07 with the ROS1 rearrangement before and after crizotinib treatment. The copy numbers of rearranged ROS1 are plotted in Figure 4a. Matching serial computed tomography (CT) images depict the lesions in the patient’s chest on days 0, 30, and 75 post-crizotinib treatment. The patient was found to have a partial response on day 30 but unfortunately relapsed after day 75 (Figure 4b). The patient died of untreatable tumor burden on day 78. Dynamic changes in ROS1 copy numbers were consistent with CT imaging and clinical outcomes, suggesting that this EV-based mRNA assay may serve as a complementary diagnostic tool for monitoring treatment outcomes in NSCLC patients with ROS1 rearrangements. In Figure 4c, we track patient E04 over a period of 279 days. Blood was collected serially at three time points: day 0, when the patient had responded to a first-generation EGFR-TKI (i.e., gefitinib); day 133, when the patient acquired resistance; and day 279, when the patient had a partial response after 146 days of treatment with the third-generation EGFR-TKI osimertinib, which targets the acquired EGFR T790M mutation. The emergence of the acquired EGFR T790M mutation (copy number increased from 0 to 225) between days 0 and 133 indicated resistance to the initial therapy, suggesting a possible timepoint for switching from gefitinib to osimertinib. At 146 days post-osimertinib treatment, the EGFR T790M copy numbers decreased from 225 to 9, consistent with the tumor shrinkage observed via CT imaging (Figure 4d). The US Food and Drug Administration has approved osimertinib for the treatment of patients with an acquired EGFR T790M mutation who progress during prior EGFR-TKIs treatments.44 Confirmation of the EGFR T790M mutation by tissue re-biopsy is required for treatment selection after relapse from prior EGFR-TKIs treatments. These results suggest our NanoVilli Chip-based tumor-derived EV capturing platform is compatible with detecting both gene rearrangements (i.e., ROS1 rearrangements) and gene mutations (i.e., EGFR T790M mutation) for monitoring early treatment response and guiding the selection of alternative therapies non-invasively.
Figure 4.
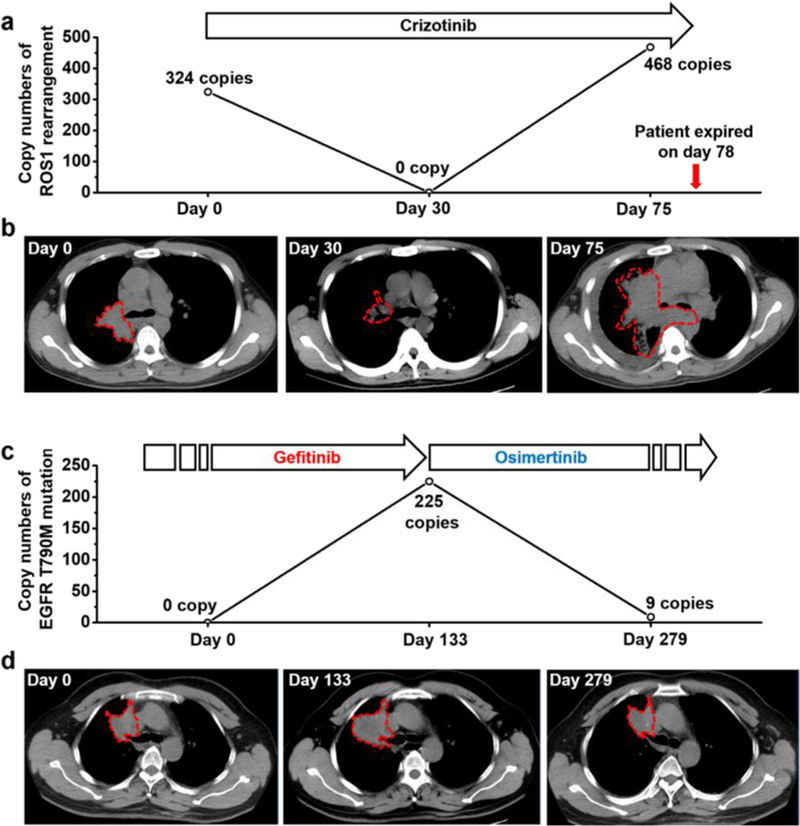
NanoVilli Chips combined with RT-ddPCR analysis can be used to detect and to monitor ROS1 rearrangements or acquired EGFR T790M mutation in tumor-derived EVs purified from NSCLC patients’ blood. (a) Dynamic change (0–75 days) of the ROS1 rearrangements observed for patient R07 with CD74-ROS1 rearrangement before and after crizotinib treatment. (b) Chest CT scans taken at days 0, 30, 75 post-crizotinib treatment. (c) Dynamic changes (0–279 days) of EGFR T790M mutation observed for patient E04 before and after osimertinib treatment. (d) Chest CT images were taken at day 0 (following response to gefitinib treatment), day 133 (disease relapse), and day 279 (post-treatment with osimertinib).
CONCLUSIONS AND PROSPECTS
We have successfully developed and demonstrated a bio-inspired device capable of highly efficient and reproducible immunoaffinity capture of tumor-derived EVs from blood plasma samples. The anti-EpCAM-grafted Si nanowire arrays that comprise these NanoVilli Chips mimic the distinctive structures of intestinal microvilli, providing dramatically increased surface area for capturing tumor-derived EVs. A PDMS-based microfluidic chaotic mixer is used to establish direct physical contact between tumor-derived EVs and anti-EpCAM-grafted SiNWS, further enhancing the EV-capture performance. We studied the influence of flow rate, length of Si nanowires, and anti-EpCAM concentrations to identify conditions that yield optimal EV-capture performance. When operated at these optimal conditions, NanoVilli Chips enable highly efficient, reproducible, and rapid (30 min) enrichment of tumor-derived EVs from both artificial plasma samples as well as plasma samples isolated from NSCLC patients. By coupling NanoVilli Chips with a downstream RT-ddPCR, we have developed a new type of EV-based mRNA assay for quantitatively detecting and monitoring targetable oncogenic gene alterations. In clinically relevant applications, tumor-derived EVs captured on NanoVilli Chips can provide critical diagnostic information as a source for detecting specific oncogenic gene alterations that correlate with treatment responses and disease progression to inform the clinical management of NSCLC patients.
EXPERIMENTAL SECTION
Fabrication of the Anti-EpCAM-Grafted Silicon Nanowire Substrate.
First, thiol groups were introduced onto SiNWS by exposure to (3-mercaptopropyl)trimethoxysilane (MPS, 211.4 mg, 200 μL, Sigma-Aldrich, USA) vapor at room temperature for 45 min. The SiNWS were rinsed with ethanol three times to wash off unbound reagents. Second, freshly prepared MPS−SiNWS were incubated with the N-maleimidobutyryl-oxysuccinimide ester (GMBS, 0.25 mM in DMSO, Sigma-Aldrich, USA) solution for 30 min to attach GMBS on the surface of SiNWS. Third, GMBS−SiNWS were reacted with streptavidin (SA, 10 μg mL−1, Thermo Fisher Scientific, USA) solution at room temperature for 30 min to immobilize SA. The obtained SA−SiNWS were rinsed with 1× phosphate-buffered saline (PBS, pH 7.4, Thermo Fisher Scientific) to remove excess SA. Fourth, to graft anti-EpCAM onto the SA−SiNWS, biotinylated anti-EpCAM (Abcam, USA) at concentrations of 1.0, 2.5, or 5.0 μg mL−1 in PBS (100 μL) was incubated on the SA−SiNWS for 30 min at room temperature. After rinsing off the unbounded biotinylated anti-EpCAM, the anti-EpCAM-grafted SiNWS were blocked with 5% bovine serum albumin (BSA, Thermo Fisher Scientific) solution for 30 min. The total inner volume of three microfluidic channels in a NanoVilli Chip was 20 μL.
Culture of NSCLC Cell Lines.
NSCLC cell lines including HCC78 and NCI-H1975 were obtained from the American Type Culture Collection and regularly tested and found negative for mycoplasma contamination. These NSCLC cells were cultured in RPMI-1640 growth medium (Thermo Fisher Scientific, USA) with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific), 1% (v/v) GlutaMAX-I (Thermo Fisher Scientific), and penicillin−streptomycin (100 U mL−1, Thermo Fisher Scientific) in a humidified incubator with 5% CO2 at 37 °C.
Preparation and Isolation of NSCLC Cell-Derived EVs.
Both HCC78 and H1975 NSCLC cells were grown in 18 Nunc EasYDish dishes (145 cm2, Thermo Fisher Scientific) for 3 days. The cells were then cultured in serum-free medium (Thermo Fisher Scientific) for 24–48 h. Thereafter, the culture medium was collected for centrifugation at 300g (4 °C) for 10 min to remove cells and cell debris. The supernatants were transferred to new Falcon 50 mL Conical Centrifuge Tubes (Thermo Fisher Scientific) and centrifuged at 2800 g (4 °C) for 10 min to eliminate remaining cellular debris and large particles. The supernatants were carefully transferred into Ultra-Clear Tubes (38.5 mL, Beckman Coulter, Inc., USA), followed by ultracentrifugation using an Optima L-100 XP Ultracentrifuge (Beckman Coulter, Inc, USA) at 100 000g (4 °C) for 70 min. After removing the supernatant, EV pellets at the bottom of the tubes were resuspended into 400 μL of PBS (Thermo Fisher Scientific) and were stored at −80 °C for future use.
Preparation of Artificial Plasma Samples Containing NSCLC Cell-Derived EVs.
The plasma was isolated from the blood samples of HDs with approval from the UCLA Institutional Review Board (IRB, #00000173). Artificial plasma samples (each had a total volume of 100 μL) were prepared by spiking 10 μL of NSCLC cell-derived EVs (see above) into 90 μL of healthy-donor plasma.
Capture of Tumor-Derived EVs on NanoVilli Chips.
Prior to the injection of artificial plasma samples, 200 μL of PBS was introduced into a NanoVilli Chip via an automated digital fluidic handler at a flow rate of 0.5 mL h−1 to test for leaks. Next, 100 μL of artificial plasma or blood plasma containing tumor-derived EVs was introduced into the NanoVilli Chip at an optimal flow rate of 0.2 mL h−1. For the optimization of flow rates, replicates of 100 μL of artificial plasma samples were introduced into NanoVilli Chips at flow rates of 0.2, 0.5, 1.0, and 2.0 mL h−1, respectively.
Characterization of the Embedded Silicon Nanowires and Captured EVs by SEM.
To characterize the Si nanowires embedded in the SiNWS, we cut the SiNWS to expose the cross sections of the silicon nanowire arrays. The broken SiNWS was placed on the SEM sample holder for SEM imaging (ZEISS Supra 40VP SEM at an accelerating voltage of 10 keV). For SEM characterization of EVs captured on Si nanowires, the SiNWS were separated from the NanoVilli Chip after capturing EVs from 100 μL of artificial plasma samples. The EVs immobilized on SiNWS were fixed in 4% PFA for 1 h. The samples were dehydrated by sequential immersion in 30, 50, 75, 85, 95, and 100% ethanol solutions for 10 min per solution. After overnight lyophilization, sputter-coating with gold was performed at room temperature. The morphology of EVs immobilized on Si nanowires was observed using a ZEISS Supra 40VP SEM at an accelerating voltage of 10 keV.
TEM Characterization of HCC78-Derived EVs.
The HCC78-derived EVs in solution or captured by the Si nanowires were fixed in 4% PFA for 30 min prior to morphological characterization and determining the size distribution of tumor-derived EVs via TEM. Afterward, the EV samples were deposited onto 200-mesh formvar and carbon coated copper grids and incubated for 5 min. After wiping off the excess sample, the grids were treated with 2% uranyl acetate for 10 min and then washed three times with deionized water. Grids were dried for TEM imaging by JEM1200-EX (JEOL USA Inc.) at 80 kV. To verify the identity of EVs in solution and captured on Si nanowires, immunogold staining by anti-CD63 was employed for TEM imaging. Fixed EVs in solution or captured on Si nanowires were applied to 200-mesh formvar and carbon-coated nickel grids and incubated for 5 min before being wiped off from the grids. Then, the grids were incubated in a blocking solution (0.4% BSA in PBS) for 30 min and then rinsed three times using deionized water. Thereafter, the grids were incubated with mouse anti-CD63 (Abcam, USA, positive control) or with blocking solution (negative control) for 1 h. After being rinsed three times with deionized water, the grids were incubated with goat anti-mouse IgG H&L 10 nm gold (Abcam, USA) for 1 h. After again being rinsed three times using deionized water, the grids were negatively stained using 2% uranyl acetate and then dried for TEM imaging using a JEM1200-EX (JEOL USA Inc.) at 80 kV.
Immunostaining by Anti-CK and Fluorescence Characterization of Tumor-Derived EVs Immobilized on Silicon Nanowire Substrates.
Tumor-derived EVs immobilized on SiNWS were fixed with 4% PFA for 10 min, followed by incubation with 0.1% Triton X 100 in PBS for 10 min at room temperature. Then, they were incubated with a PBS solution containing Pan-CK antibody [Abcam, USA, 1:100 (v/v)] and Normal Donkey serum (Jackson Immuno-Research, USA, 2%) at 4 °C overnight. After being washed with PBS three times, the tumor-derived EVs captured on SiNWS were further incubated with Donkey anti-Rabbit IgG (H + L) [Alexa Fluor 488, Thermo Fisher Scientific, USA, 1:500 (v/v)] for 1 h. After washing off the excess reagent, the tumor-derived EVs immobilized on SiNWS were characterized using a fluorescence microscope (Nikon 90i, exposure time = 200 ms).
Extraction of RNA from Tumor-Derived EVs Captured on NanoVilli Chips.
To extract RNA from tumor-derived EVs captured on NanoVilli Chips, we performed on-chip lysis of EVs by introducing 600 μL of TRIzol solution (Zymo Research, USA) and 600 μL of anhydrous ethanol (Sigma-Aldrich) sequentially through the NanoVilli Chip. The effluent solution was collected in a 2.0 mL RNase-free Eppendorf tube at the same time. Then, RNA was purified using a Direct-zolRNA MicroPrep Kit (Zymo Research). The enzyme DNase I was used to digest DNA for 15 min to make sure that cfDNA was not analyzed in the measurements. The RNA was dissolved in DNase/ RNase-free water and then measured with a Qubit 3.0 Fluorometer (Thermo Fisher Scientific) in combination with the Qubit RNA HS Assay (Thermo Fisher Scientific) using the manufacturer’s protocol.
Quantification of ROS1 Rearrangements or EGFR T790M Mutation from EV-Derived mRNA by RT-ddPCR.
EV-derived mRNA was reverse-transcribed to cDNA using a Maxima H Minus Reverse Transcriptase Kit (Thermo Fisher Scientific). The EV-derived mRNA was added into a reaction solution containing 1× RT Buffer, dNTPs (0.5 mM), Random Hexamer (8 μM), Maxima H Minus Reverse Transcriptase (6.5 U μL−1), and RNase inhibitor (1 U μL−1). The reaction was run at 55 °C for 30 min and then 85 °C for 5 min. The cDNA generated from EV-derived mRNA was detected by the PrimePCR ddPCR Expert Design Assay Kit (dHsaEXD73338942, ROS1 rearrangements) or PrimePCR ddPCR Mutation Assay Kit (dHsaCP2000020, EGFR T790M mutation, Bio-Rad, USA) according to the manufacturer’s instructions. For ddPCR, droplets were generated within a DG8 Cartridge which was preloaded with sample (20 μL) and droplet generation oil (70 μL) for each sample. All droplets were transferred into a 96-well plate accordingly and sealed with a PX1 PCR Plate Sealer. A programmed Thermal Cycler was set at 96 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 60 °C for 60 s, and finally 98 °C for 10 min. The droplets containing amplicons were quantified with a QX200 Droplet Reader using the QuantaSoft software package.
Collection of Blood Plasma Samples from NSCLC Patients and HDs.
Blood samples were collected from 12 NSCLC patients in Guangdong Provincial Hospital of Traditional Chinese Medicine and 9 HDs at UCLA in accordance with the Institutional Review Board (IRB). We consecutively enrolled 6 NSCLC (stages III and IV) patients with known ROS1 rearrangements45 from October 2016 to June 2017 and 6 NSCLC patients with known EGFR T790M mutation from January 2018 to June 2018. Blood samples were centrifuged at 300g for 5 min and then 2000g for 5 min at 4 °C. Plasma was collected and stored I at −80 °C. For each blood plasma sample, 200 μL of plasma was directly run through a NanoVilli Chip.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (R33 CA17 4562, U01CA198900, R01CA21835 6, R21CA216807, and R21CA235340). J.D. gratefully acknowledges financial support from the China Scholarship Council (201706010064).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.9b01406.
Fabrication of SiNWS; chemical modification to prepare anti-EpCAM-grafted SiNWS; methods for PDMS chaotic mixers; photograph and schematic of the NanoVilli device; size characterization of HCC78-derived EVs by DLS; electronic microscopy and fluorescence characterization of EV-captured SiNWS; computational simulation analysis; negative control of cytokeratin fluorescence staining; quantification of gene alterations from EV-derived RNA by RT-ddPCR; isolation of tumor-derived EVs by magnetic beads; and collection of blood plasma samples from NSCLC patients (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Shah R; Patel T; Freedman JE Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med 2018, 379, 958–966. [DOI] [PubMed] [Google Scholar]
- (2).Raposo G; Stoorvogel W Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol 2013, 200, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Revenfeld ALS; Bæk R; Nielsen MH; Stensballe A; Varming K; Jørgensen M Diagnostic and Prognostic Potential of Extracellular Vesicles in Peripheral Blood. Clin. Ther 2014, 36, 830–846. [DOI] [PubMed] [Google Scholar]
- (4).Garcia-Romero N; Esteban-Rubio S; Rackov G; Carrión-Navarro J; Belda-Iniesta C; Ayuso-Sacido A Extracellular Vesicles Compartment in Liquid Biopsies: Clinical Application. Mol. Aspects Med 2018, 60, 27–37. [DOI] [PubMed] [Google Scholar]
- (5).Cocucci E; Meldolesi J Ectosomes and Exosomes: Shedding the Confusion between Extracellular Vesicles. Trends Cell Biol 2015, 25, 364–372. [DOI] [PubMed] [Google Scholar]
- (6).Corso G; Mager I; Lee Y; Gorgens A; Bultema J; Giebel B; Wood MJA; Nordin JZ; Andaloussi SE Reproducible and Scalable Purification of Extracellular Vesicles Using Combined Bind-Elute and Size Exclusion Chromatography. Sci. Rep 2017, 7, 11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Akers JC; Gonda D; Kim R; Carter BS; Chen CC Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J. Neuro-Oncol 2013, 113, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Balaj L; Lessard R; Dai L; Cho Y-J; Pomeroy SL; Breakefield XO; Skog J Tumour Microvesicles Contain Retro-transposon Elements and Amplified Oncogene Sequences. Nat. Commun 2011, 2, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Eldh M; Bagge RO; Lässer C; Svanvik J; Sjöstrand M; Mattsson J; Lindnér P; Choi D-S; Gho YS; Lötvall J Microrna in Exosomes Isolated Directly from the Liver Circulation in Patients with Metastatic Uveal Melanoma. BMC Cancer 2014, 14, 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).King HW; Michael MZ; Gleadle JM Hypoxic Enhancement of Exosome Release by Breast Cancer Cells. BMC Cancer 2012, 12, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gercel-Taylor C; Atay S; Tullis RH; Kesimer M; Taylor DD Nanoparticle Analysis of Circulating Cell-Derived Vesicles in Ovarian Cancer Patients. Anal. Biochem 2012, 428, 44–53. [DOI] [PubMed] [Google Scholar]
- (12).Taylor DD; Gercel-Taylor C The Origin, Function, and Diagnostic Potential of RNA within Extracellular Vesicles Present in Human Biological Fluids. Front. Genet 2013, 4, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Melo SA; Luecke LB; Kahlert C; Fernandez AF; Gammon ST; Kaye J; LeBleu VS; Mittendorf EA; Weitz J; Rahbari N; Reissfelder C; Pilarsky C; Fraga MF; Piwnica-Worms D; Kalluri R Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 2015, 523, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Reategui E; van der Vos KE; Lai CP; Zeinali M; Atai NA; Aldikacti B; Floyd FP Jr.; Khankhel AH; Thapar V; Hochberg FH; Sequist LV; Nahed BV; Carter BS; Toner M; Balaj L; Ting DT; Breakefield XO; Stott SL Engineered Nanointerfaces for Microfluidic Isolation and Molecular Profiling of Tumor-Specific Extracellular Vesicles. Nat. Commun 2018, 9, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Möhrmann L; Huang HJ; Hong DS; Tsimberidou AM; Fu S; Piha-Paul SA; Subbiah V; Karp DD; Naing A; Krug A; Enderle D; Priewasser T; Noerholm M; Eitan E; Coticchia C; Stoll G; Jordan L-M; Eng C; Kopetz ES; Skog J; Meric-Bernstam F; Janku F Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin. Cancer Res 2018, 24, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Castellanos-Rizaldos E; Grimm DG; Tadigotla V; Hurley J; Healy J; Neal PL; Sher M; Venkatesan R; Karlovich C; Raponi M; Krug A; Noerholm M; Tannous J; Tannous BA; Raez LE; Skog JK Exosome-Based Detection ofEGFRT790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin. Cancer Res 2018, 24, 2944–2950. [DOI] [PubMed] [Google Scholar]
- (17).Tauro BJ; Greening DW; Mathias RA; Ji H; Mathivanan S; Scott AM; Simpson RJ Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human Colon Cancer Cell Line LIM1863-Derived Exosomes. Methods 2012, 56, 293–304. [DOI] [PubMed] [Google Scholar]
- (18).Kalra H; Adda CG; Liem M; Ang C-S; Mechler A; Simpson RJ; Hulett MD; Mathivanan S Comparative Proteomics Evaluation of Plasma Exosome Isolation Techniques and Assessment of the Stability of Exosomes in Normal Human Blood Plasma. Proteomics 2013, 13, 3354–3364. [DOI] [PubMed] [Google Scholar]
- (19).Lamparski HG; Metha-Damani A; Yao J-Y; Patel S; Hsu D-H; Ruegg C; Le Pecq J-B Production and Characterization of Clinical Grade Exosomes Derived from Dendritic Cells. J. Immunol. Methods 2002, 270, 211–226. [DOI] [PubMed] [Google Scholar]
- (20).Caradec J; Kharmate G; Hosseini-Beheshti E; Adomat H; Gleave M; Guns E Reproducibility and Efficiency of Serum-Derived Exosome Extraction Methods. Clin. Biochem 2014, 47, 1286–1292. [DOI] [PubMed] [Google Scholar]
- (21).Heinemann ML; Ilmer M; Silva LP; Hawke DH; Recio A; Vorontsova MA; Alt E; Vykoukal J Benchtop Isolation and Characterization of Functional Exosomes by Sequential Filtration. J. Chromatogr. A 2014, 1371, 125–135. [DOI] [PubMed] [Google Scholar]
- (22).Heinemann ML; Vykoukal J Sequential Filtration: A Gentle Method for the Isolation of Functional Extracellular Vesicles. In Extracellular Vesicles: Methods and Protocols; Kuo WP, Jia S, Eds.; Humana Press Inc: Totowa, 2017; Vol. 1660, pp 33–41. [DOI] [PubMed] [Google Scholar]
- (23).Rekker K; Saare M; Roost AM; Kubo A-L; Zarovni N; Chiesi A; Salumets A; Peters M Comparison of Serum Exosome Isolation Methods for Microrna Profiling. Clin. Biochem 2014, 47, 135–138. [DOI] [PubMed] [Google Scholar]
- (24).Davies RT; Kim J; Jang SC; Choi E-J; Gho YS; Park J Microfluidic Filtration System to Isolate Extracellular Vesicles from Blood. Lab Chip 2012, 12, 5202–5210. [DOI] [PubMed] [Google Scholar]
- (25).Kanwar SS; Dunlay CJ; Simeone DM; Nagrath S Microfluidic Device (Exochip) for On-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab Chip 2014, 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhao Z; Yang Y; Zeng Y; He M A Microfluidic Exosearch Chip for Multiplexed Exosome Detection Towards Blood-Based Ovarian Cancer Diagnosis. Lab Chip 2016, 16, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yang F; Liao X; Tian Y; Li G Exosome Separation Using Microfluidic Systems: Size-Based, Immunoaffinity-Based and Dynamic Methodologies. Biotechnol. J 2017, 12, 1600699. [DOI] [PubMed] [Google Scholar]
- (28).Wang Z; Wu H.-j.; Fine D; Schmulen J; Hu Y; Godin B; Zhang JXJ; Liu X Ciliated Micropillars for the Microfluidic-Based Isolation of Nanoscale Lipid Vesicles. Lab Chip 2013, 13, 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wunsch BH; Smith JT; Gifford SM; Wang C; Brink M; Bruce RL; Austin RH; Stolovitzky G; Astier Y Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids Down to 20 nm. Nat. Nanotechnol 2016, 11, 936–940. [DOI] [PubMed] [Google Scholar]
- (30).Lin M; Chen J-F; Lu Y-T; Zhang Y; Song J; Hou S; Ke Z; Tseng H-R Nanostructure Embedded Microchips for Detection, Isolation, and Characterization of Circulating Tumor Cells. Acc. Chem. Res 2014, 47, 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chen J-F; Zhu Y; Lu Y-T; Hodara E; Hou S; Agopian VG; Tomlinson JS; Posadas EM; Tseng H-R Clinical Applications of Nanovelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells. Theranostics 2016, 6, 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Jan YJ; Chen J-F; Zhu Y; Lu Y-T; Chen SH; Chung H; Smalley M; Huang Y-W; Dong J; Chen L-C; Yu H-H; Tomlinson JS; Hou S; Agopian VG; Posadas EM; Tseng H-R Nanovelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells. Adv. Drug Delivery Rev 2018, 125, 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kohno T; Nakaoku T; Tsuta K; Tsuchihara K; Matsumoto S; Yoh K; Goto K Beyond ALK-RET, ROS1 and Other Oncogene Fusions in Lung Cancer. Transl. Lung Cancer Res 2015, 4, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rothschild S Targeted Therapies in Non-Small Cell Lung Cancer-Beyond EGFR and ALK. Cancers 2015, 7, 930–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Maemondo M; Inoue A; Kobayashi K; Sugawara S; Oizumi S; Isobe H; Gemma A; Harada M; Yoshizawa H; Kinoshita I; Fujita Y; Okinaga S; Hirano H; Yoshimori K; Harada T; Ogura T; Ando M; Miyazawa H; Tanaka T; Saijo Y; Hagiwara K; Morita S; Nukiwa T Gefitinib or Chemotherapy for Non-Small-Cell Lung Cancer with Mutated EGFR. N. Engl. J. Med 2010, 362, 2380–2388. [DOI] [PubMed] [Google Scholar]
- (36).Crowley E; Di Nicolantonio F; Loupakis F; Bardelli A Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat. Rev. Clin. Oncol 2013, 10, 472–484. [DOI] [PubMed] [Google Scholar]
- (37).Myung JH; Park S.-j.; Wang AZ; Hong S Integration of Biomimicry and Nanotechnology for Significantly Improved Detection of Circulating Tumor Cells (CTCs). Adv. Drug Delivery Rev 2018, 125, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Peng K-Q; Yan Y-J; Gao S-P; Zhu J Synthesis of Large-Area Silicon Nanowire Arrays via Self-Assembling Nanoelectrochemistry. Adv. Mater 2002, 14, 1164–1167. [Google Scholar]
- (39).Lu Y-T; Zhao L; Shen Q; Garcia MA; Wu D; Hou S; Song M; Xu X; Ouyang W-H; Ouyang WW-L; Lichterman J; Luo Z; Xuan X; Huang J; Chung LWK; Rettig M; Tseng H-R; Shao C; Posadas EM Nanovelcro Chip for CTC Enumeration in Prostate Cancer Patients. Methods 2013, 64, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wang S; Liu K; Liu J; Yu ZT-F; Xu X; Zhao L; Lee T; Lee EK; Reiss J; Lee Y-K; Chung LWK; Huang J; Rettig M; Seligson D; Duraiswamy KN; Shen CK-F; Tseng H-R Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angew. Chem., Int. Ed 2011, 50, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sheng W; Ogunwobi OO; Chen T; Zhang J; George TJ; Liu C; Fan ZH Capture, Release and Culture of Circulating Tumor Cells from Pancreatic Cancer Patients Using an Enhanced Mixing Chip. Lab Chip 2014, 14, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kármán TV Über Laminare Und Turbulente Reibung. Z. Angew. Math. Mech 1921, 1, 233–252. [Google Scholar]
- (43).Groot RD; Warren PB Dissipative Particle Dynamics: Bridging the Gap between Atomistic and Mesoscopic Simulation. J. Chem. Phys 1997, 107, 4423–4435. [Google Scholar]
- (44).Wang S; Cang S; Liu D Third-Generation Inhibitors Targeting EGFR T790M Mutation in Advanced Non-Small Cell Lung Cancer. J. Hematol. Oncol 2016, 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wu W; Haderk F; Bivona T Non-Canonical Thinking for Targeting ALK-Fusion Onco-Proteins in Lung Cancer. Cancers 2017, 9, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.