Abstract
Technological developments in both the collection and analysis of molecular genetic data over the past few years have provided new opportunities for an improved understanding of the global response to pathogen exposure. Such developments are particularly dramatic for scientists studying the pig, where tools to measure the expression of tens of thousands of transcripts, as well as unprecedented data on the porcine genome sequence, have combined to expand our abilities to elucidate the porcine immune system. In this review, we describe these recent developments in the context of our work using primarily microar-rays to explore gene expression changes during infection of pigs by Salmonella. Thus while the focus is not a comprehensive review of all possible approaches, we provide links and information on both the tools we use as well as alternatives commonly available for transcriptomic data collection and analysis of porcine immune responses. Through this review, we expect readers will gain an appreciation for the necessary steps to plan, conduct, analyze and interpret the data from transcriptomic analyses directly applicable to their research interests.
Keywords: Porcine, Transcript profiling, Immune response, Salmonella, Microarray
1. Introduction
Host-pathogen interactions have been studied at the molecular, cellular, tissue and organismal levels for many years (see, for example, the set of reviews in Current Opinion in Immunology, 2007, Vol. 19). At the transcriptomic level, a meta-analysis of microarray data detecting the immune response to many types of pathogens in different human cell types has shown that a core set of ~500 genes are expressed in response to viruses, bacteria~(including Salmonella) and immune stimulants such as LPS (Jenner and Young, 2005). Because of the ubiquity and ability of Salmonella spp. to infect many important vertebrate species, both animal models (Santos et al., 2001; van der Sar et al., 2003) and cell culture systems (see below) have been used to understand Salmonella interactions with its host. Early immune responses are thought to be critical in resistance to Salmonella (Wick, 2004), and a large number of genes have been implicated in the host response to Salmonella (Detweiler et al., 2001; Rodenburg et al., 2007). Many studies have used lipopolysaccaride (LPS) as a model of the transcriptomic response to bacteria (Wells et al., 2003; Bliss et al., 2005; Wurfel et al., 2005; Beck et al., 2006). A direct comparison of the transcriptomic response to LPS versus Salmonella showed significant similarity in both the genes responding and the magnitude of the response (Rosenberger et al., 2000).
The cytokine protein and RNA responses to Salmonella have also been studied in the pig; most parameters and genes involved appear to be very similar to those in mouse (Dvorak et al., 2006). Initial interactions of Salmonella with the gut tissue have been studied in cell culture (Veldhuizen et al., 2006; Skjolaas et al., 2007), but also in explants of Peyer’s patch tissue (Hyland et al., 2006). In the latter study, IL1B and IL8 RNA (but not TNF) were found to increase after exposure to Salmonella enterica serovar Choleraesuis (SC) for 2 h. In IPEC-J2 cells, an in vitro model of porcine jejunal intestine, S. enterica serovar Typhimurium (ST) exposure for 1.5 h increased RNA for TNF, IL8 and CCL20, but induction of these cytokines was not observed in cells exposed to SC (Skjolaas et al., 2007). Using a separate porcine epithelial cell line, exposure to ST for 24 h induced RNA expression for the beta-defensin gene pBD-2 (Veldhuizen et al., 2006).
Immune responses of different types of porcine cells isolated from whole blood have also been studied. The in vitro response of pig peripheral blood mononuclear cells (PBMC) to Salmonella or mitogen treatment was characterized by increased IL2, IL4, and IFNG RNA, with no effect on IL10 expression, while bacterial F4 fimbrae increased expression of IFNG RNA only (Verfaillie et al., 2001). This group also found cytokine protein levels generally correlated well with RNA expression for the first 24 h of exposure.
Raymond and Wilkie (2004) investigated porcine T cell responses to stimulated dendritic cells (DCs), finding that the T cell response profile depended on how the DC was stimulated and the cytokine milieu during stimulation. They also evaluated monocyte and DC responses to specific pathogen-associated molecular pattern (PAMP) molecules. Treatment with LPS induced expression of TLR4 and T helper 1 (IFNG, IL12p35), T helper 2 (IL13) and regulatory T (IL10) cell response pathways. Cell-specific responses were observed for several of these genes; MHC Class II expression was greater after LPS stimulation in monocytes whereas B7 RNA increased in both cell types (Raymond and Wilkie, 2005). In CD14+ cells isolated from pig spleen, LPS pre-treatment was shown to decrease TNF and IL8, but not IL1B. However, gene expression in response to re-application of LPS indicated that pig monocytes undergo a similar LPS tolerance response (Cagiola et al., 2006) to that reported for murine macrophages.
The RNA response of several Toll-like receptor and chemoattractant genes to S. enterica serovar Choleraesuis (SC) and S. Typhimurium (ST)inoculationof pigs has been reported for a number of tissues (Burkey et al., 2007; Wang et al., 2007, 2008b). Relative to uninfected controls, quantitative real-time PCR (QPCR) analysis showed ST infection greatly increased IL8 expression in MLN and decreased expression of MIF RNA in colon, while in SC infected animals, TLR9 and MIF were decreased in colon and MIF and OPN were decreased in MLN (Burkey et al., 2007). Global transcriptional responses to pathogenic infections in the pig have been reported using microarrays (Afonso et al., 2004; Ledger et al., 2004; Li et al., 2004; Miller and Fox, 2004; Moser et al., 2004; Niewold et al., 2005; Zhao et al., 2006; Uthe et al., 2006; Uthe et al., 2007; Wang et al., 2007, 2008b; Tuggle et al., 2008) and multiple-gene and larger scale QPCR methods (Raymond and Wilkie, 2004; Baltes and Gerlach, 2004; Royaee et al., 2004; Dawson et al., 2005).
These studies (reviewed in Tuggle et al., 2007) have begun to identify immune genes involved in the host’s response to different pathogen infections. Taken together they have contributed to a better understanding of molecular pathways relating to health and disease in pigs.
This review centers on our approaches to use such transcriptomic data to unravel important pathways controlling the porcine response to Salmonella. We will not discuss recent publications on the host transcriptomic response to viruses (Bates et al., 2008; Flori et al., 2008a,b; Durand et al., 2009; Fernandes et al., 2009; Shi et al., 2009; Li et al., 2010; Tomas et al., 2010), to mycobacteria (Galindo et al., 2009), to Actinobacillus pleuropneumoniae (Hedegaard et al., 2007; Moser et al., 2008), or to Toxo-plasma gondii (Okomo-Adhiambo et al., 2006) infections or to non-infectious stimuli or other contrasts (Dvorak et al., 2006; Chowdhury et al., 2007; Nino-Soto et al., 2008a,b; Ponsuksili et al., 2008; Wang et al., 2008a). However, these authors used similar approaches to the broadly applicable methods discussed below. We will also not describe methods to measure miRNAs in tissues of immunological importance, although recent reports have identified miRNAs that are important in the immune response in other species (Pedersen and David, 2008; Bi et al., 2009). A number of reports have described miRNA identification in several porcine tissues focusing on reproduction or muscle development, although single papers have described isolation of swine miRNAs from intestine (Sharbati et al., 2010), or miRNAs potentially interacting with swine influenza virus (He et al., 2009).
2. Steps in producing and exploring transcriptomic data on immune response
2.1. Experimental design and tool choice—what question do you want to ask?
The most important first step in transcriptomic analyses (and in all experiments in fact) is to determine the question to address. In many transcriptome studies, the question is broadly exploratory, along the lines of “what are the genes and pathways that respond to the pathogen of interest in this tissue or cell type?” If so, then tools and processes that capture accurate and sensitive information on the largest numbers of transcripts for the lowest cost are optimal. Because of the lack of available space and complexity of these decisions for each lab, we cannot describe all possibilities but will briefly review the main choices for technologies in this area.
While we focus on the use of microarray technology to collect transcriptomic data in this review, it is by far not the only method, and other technologies such as Differential Display (DD), Suppression Subtractive Hybridization (SSH), and Serial Analysis of Gene Expression (SAGE) have been used to identify differentially expressed (DE) genes during the immune response in pigs (Tuggle et al., 2007 and references therein). These latter technologies, especially DD and SSH, require significant wet lab analyses, as many different combination of primers are required to survey the transcriptome significantly. The popularity of microarrays is due primarily for their breadth of coverage and relative sensitivity and simplicity over these other methods to generate global RNA profiles. More recent technologies such as RNA Seq (Wang et al., 2009) have not yet been reported in porcine immunogenomics, but a number of groups are developing such data. It is anticipated that RNA-Seq and similar methods based on the new sequencing technologies will replace microarrays in the future, especially for initial screening experiments, due to their anticipated lower cost and broader representation of the transcriptome.
In pigs, there are several options for collecting transcriptomic data and the best approach depends on the level of molecular and bioinformatic expertise available to the lab. If such expertise is minimal, then data collection using a fee-for-service approach is probably most appropriate. One such option is the use of the Affymetrix Porcine GeneChip®, which requires only RNA preparation in one’s laboratory; the RNA is then provided to a dedicated service facility which many universities and research institutes have available. Expression data is provided by the Facility ready for statistical analysis as described below. If more “wet-lab” expertise is available, data collection can be less expensive, especially for a large project, through the use of custom oligonucleotide arrays in one’s laboratory. cDNA arrays have been replaced by such oligonucleotide arrays, as synthesis costs have dropped significantly and algorithms to minimize cross-hybridization have improved. Thus two major issues with cDNA arrays, the need for highly accurate clone and PCR product tracking during array production, and the concern of cross-hybridization to common domains within multiple cDNAs, are significantly diminished for oligonucleotide arrays, which can also be created for less expense, at least on a per array basis, than purchased arrays. However, in any such cost comparison it is important to consider the labor and materials costs not only on the array production but also the target labeling, hybridization and data acquisition, so that all costs are recognized during the decision-making process. Finally, there are hybrid approaches, where several companies sell arrays for use in individual laboratories or selected universities print arrays for use in labs nationwide, as is done for several of the swine long oligo arrays. In pigs, sets of long (70-mer) oligonucleotides have been designed and validated for transcriptomics research in the past few years (Zhao et al., 2005; Steibel et al., 2009). Most recently, an oligonucleotide array, the Pigoligoarray, with functional annotation for 16,225 of the 18,524 porcine-specific oligonucleotides has been evaluated (Steibel et al., 2009). For the 4 tissues examined, the array was found to be useful for accurate measurement of gene expression on a global scale. In the work described below, we used the Porcine GeneChip®, from Affymetrix, which has 23,937 probe sets with a total of 19,253 annotations currently (see Section 2.3; Couture et al., 2009).
2.2. Statistical analysis of microarray data
There are many excellent reviews of the various approaches to statistical analysis of microarray data (Quackenbush, 2001; Quackenbush, 2002; Roberts, 2008). The MicroArray Quality Control (MAQC) project reviewed major aspects of microarray data analysis in a special issue of Nature Biotechnology (MAQC Consortium, 2006). The following is only a short general synopsis of this topic and focuses on our specific approaches to assess expression response using transcriptomics. We do want to emphasize that any experimental design should incorporate as much biological replication as possible, while eliminating technical replication that had been thought to be important early in the field but is no longer deemed important for microarray analysis. The earliest statistical analysesof microarrays depended on the experimental design of comparing two differently labeled (Cy3, Cy5) samples hybridized to the same array, thus many aspects of the technique that lead to nuisance variation are diminished (Schena et al., 1995). Such work compared levels of Cy3 and Cy5 expression, setting an ad hoc x-fold difference in expression as the criteria for declaring a gene as differentially expressed (DE). In many experiments the design was a comparison of test samples to the same control sample (the reference design).
However, an analytical approach based only on a fold change filter is simplistic and an insensitive method to find all differential expression. Experimental designs and data analyses have become statistically more rigorous; details can be found in the reviews listed above. In the work below, we describe our analysis of Affymetrix-based data for which company-provided software is used to produce an estimate of expression for the transcript in question. First, a unique Affymetrix algorithm combines the hybridization signals for a set of probes to estimate the signal for each transcript. The signal for each probeset across the Genechip is normalized. Data normalization is a significant field in its own right, and the type of normalization used depends on the specifics of the microarray technology as well as the kinds of questions to be answered (Quackenbush, 2002). Here we summarize normalization, carried out in the following experiments, as the method to adjust raw individual hybridization signals within a microarray experiment so that results across biological replicates can be combined. Such normalizations often involve an adjustment based on the overall level of hybridization across the microarray, although many refinements including local background measurements and other methods can be applied (Quackenbush, 2002). An ANOVA model is then used to compare responses across time points or treatments. As this method performs many thousands of statistical significance tests at the same time, it is important to correct for such multiple testing by estimating the false discovery rate (FDR) (Storey and Tibshirani, 2003). The FDR q-value provides an estimate of the likelihood that the members of a list of differentially expressed are incorrectly predicted to be DE; commonly used q-values are 0.01–0.1, which indicate that no more than 1–10% of the genes are false discoveries; i.e., the higher the q value the more likely the gene is NOT differentially expressed.
2.3. Bioinformatics analysis of microarray data
2.3.1. Microarray element annotation
Once the genes that differentially respond to infection, treatments, or other variables, have been identified, we can proceed to explore specific biological questions of interest. As shown in Fig. 1, several immediate analyses can be envisioned. Because all these studies focus on pig gene expression, it is important to remember that most of the available tools rely on the transfer of gene function or annotations to the array elements in the target species (in our case, pig) from closely related and more widely studied species, e.g., human or mouse, through sequence comparison. If it is found that a pig sequence is sufficiently similar to an annotated gene sequence in another species, comparative analysis can be used to predict the identity and function(s) of the porcine transcript based on the annotations associated with the matching sequences in other species. Thus, it is critical to obtain the most up to date and comprehensive annotation of the gene sequences on the microarray.
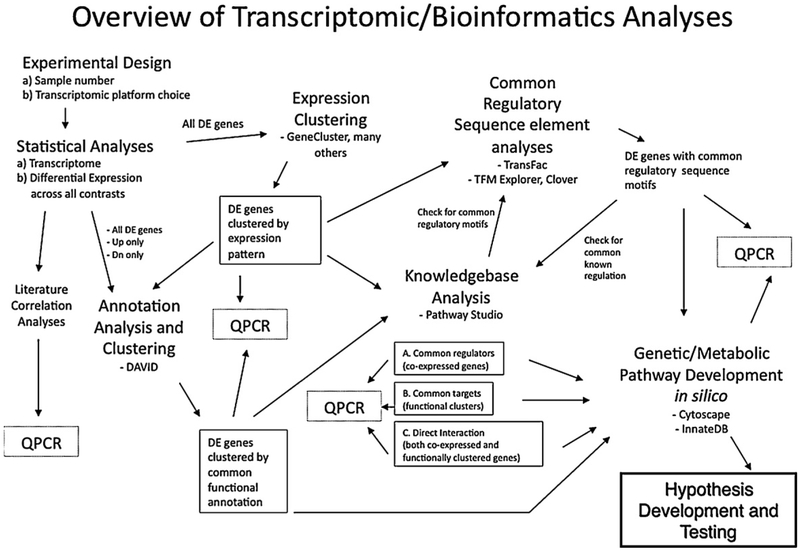
Fig. 1.
Overview of transcriptomic/bioinformatics analyses. A schematic view of approaches our collaborative group has developed to analyze porcine transcriptomic data. See text in Sections 2.3.1–2.3.7 for details.
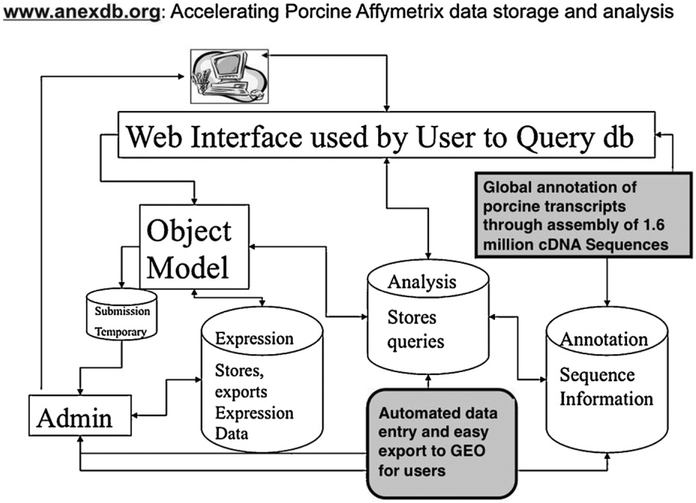
We have recently assembled all available cDNA sequences to create an Iowa Porcine Assembly (IPA) which is a set of ~140,000 consensus porcine sequences and ~105,000 singletons for all known pig mRNAs (Fig. 2; Couture et al., 2009). We have annotated these sequences using sequence similarity of the porcine consensus sequence to those annotations reported for human, mouse, rat, and bovine sequences in GenBank. These IPA sequences were then used to annotate the Affymetrix Genechip sequence elements; this resulted in excellent annotation coverage (∼ 80%) of the Affymetrix elements. We are now applying our IPA annotation to the Pigoligoarray evaluated by Steibel and colleagues so that information in the future can be more easily integrated across platforms (Couture et al., unpublished data). These annotations will be updated when the draft porcine genome sequence is released, which is expected in late 2010. These annotations are available at www.anexdb.org, where a download file provides up-to-date annotations of the major expression platforms for the pig. In addition, the AnexDB.org website has been organized to assist in transcriptome analyses; we have created data storage, analysis and GEO submission tools (Fig. 2). This includes tools to store the numerical data as well as sample preparation, hybridization protocols and data collection details. Such data can be kept completely private in the database until publication. Most journals now require that all publications reporting microarray data must make available the Minimal Information About a Microarray Experiment (MIAME) information. At AnexDB.org, we provide tools that automate much of this analysis pipeline, including generation of the correctly formatted ‘soft’ file containing all MIAME information required for NCBI-GEO submission of microarray data.
Fig. 2.
Schematic diagram of the major parts of the ANEXdb.org website and database for porcine transcriptomic data storage and analysis. See text in Section 2.3.1 for details on the main functions of this bioinformatic resource.
2.3.2. Embedding the microarray results into current literature
A first step for transcriptomic experiments is to compare the list of DE genes with the current literature. This primary step can help integrate the microarray data with known responses at porcine immune response genes. This can be done by individually checking genes for available functional investigations by other groups in PubMed. Using annotation information for the gene symbol of the tran script of interest, a researcher can easily access relevant genome, cDNA and functional information that has been integrated at a number of databases. The most comprehensive of these genome browsers are those available at the NCBI web portal (www.ncbi.nlm.nih.gov) and the EMBL web portal (www.ensembl.org/index.html). Particularly for the porcine genomics community, the latter site has excellent resources for the analysis of the pig genome (see http://www.ensembl.org/Sus scrofa/Info/Index). Our group has found the Online Mendelian Inheritance in Man (OMIM) database (www.ncbi.nlm.nih.gov/OMIM) and the Online Mendelian Inheritance in Animals (OMIA) (www.ncbi.nlm.nih.gov/omia) to be especially useful as a starting point to learn about specific genes. OMIM contains extensive information on the structure, function, and phenotypes of known mutations in human genes and their counterparts in model organisms. However, the real value of microarray analyses is the power of measuring gene expression of so many genes that the responses of important pathways and networks can be recognized and measured, as described below.
2.3.3. Function clustering and analysis using Gene Ontology and other gene annotation databases
Once individual genes in a list of differentially responsive transcripts have been compared to available literature, a new type of exploratory analysis of these genes can be performed. An important question to ask of the data is: are there known functions or other attributes—annotations—for genes in this list that are over-represented compared to a background list of genes? In other words, what are the enriched biological ‘signatures’ or ‘clues’ hidden in this list that can help one understand the immune response represented by the list? The most widely used set of annotations are those provided by the Gene Ontology (GO) Consortium, which applies a set of descriptive terms from a defined vocabulary to genes for which some functional data is available. Terms covering three descriptive areas are available: Biological Process, Molecular Function, and Cellular Component.
To determine all annotations for a set of genes, there are a number of software tools that are available. These are proprietary packages (such as Ingenuity or GeneSpring), software available as downloadable local programs (such as GoMiner; http://discover.nci.nih.gov/gominer/index.jsp), as well as free web-based tools, such as the Database for Annotation, Visualization and Integrated Discovery (DAVID) created by NIAID scientists (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003). While DAVID provides a number of different analytical tools, we have primarily used DAVID to annotate lists of genes in whole blood responding to Salmonella infection (Huang et al., manuscript in preparation) as well as a number of our other projects (Lkhagvadorj et al., 2009; Lkhagvadorj et al., 2010). In the DAVID on-line tool, one uploads a list of genes to analyze as well as a background gene list. The DAVID tool can calculate the frequency of GO terms associated with all genes in the uploaded list, and calculates those terms that are over-represented relative to the background. Over-representation of other gene annotations, such as KEGG pathways, Protein Information Resource (PIR) information, etc., can also be calculated depending on user specifications; a recent detailed description of current DAVID functionality is available (Huang da et al., 2009). We have also developed our own specific list of GO terms related to specific immune pathways and functions (a GO-Slim) by using OBO-edit and used it to develop a better understanding of the immune-response specific pathways in gene lists responsive to Salmonella infections (Wang et al., 2007, 2008b).
A comment on the selection of the ‘background’ list of genes is warranted. The default for this list in DAVID is the human genome; this may be inappropriate for any porcine gene list, but especially for those lists created from microarray platforms for which there is either incomplete genome coverage or non-random selection of elements representing transcripts on the array. The modified Fisher’s Exact test calculations used by DAVID to determine over-representation—finding a higher frequency of terms in a specified list as compared to the background—depends on the assumption that the background is a set of genes that has a chance to be included in the differentially expressed list. For example, if many genes with a specific term such as “immune response” are present at 10% in a list of genes responding to LPS in macrophage cells, but present at much lower levels in a list of all human genes, this would be returned as an over-represented term. However, it is likely that many of the genes in a human genome background list, contributing to the overall frequencies of GO terms in that list, were not expressed in immune cells and could not be in the DE list. Thus it is more appropriate to use the largest set of genes that could have been in the DE list: the transcriptome for the tissue or cell type under study. Therefore we define our transcriptome, and thus our background list, as all those genes that show at least one hybridization signal above background across our entire set of Affymetrix chips for the tissue of interest. By using GO enrichment approaches such as these, we have shown that annotations for genes up-regulated in mesenteric lymph node from animals infected with SC are enriched in apoptosis, innate immune response and defense response terms, while annotations for down-regulated genes in these tissues are enriched in cell adhesion and calcium ion binding terms (Wang et al., 2008b).
2.3.4. Expression clustering to find genes with similar transcriptomic response to infection or stimulus
Depending on the type of experiment, a second valuable global analysis approach is to identify groups of genes that respond similarly to infection or immune stimulation at the RNA expression level. Several methods are available to ‘cluster’ genes by expression pattern across experimental samples. Such clustering methods (Belacel et al., 2006) can be broadly classified into (a) hierarchical methods or model based methods e.g., probabilistic mixture models (Medvedovic and Sivaganesan, 2002), and (b) model-free methods e.g., spectral clustering (von Luxburg, 2007). In each of these categories, clustering algorithms can generate different types of clusters e.g., non-overlapping clusters of data points or hierarchical organization of clusters. Each class of methods has its own advantages and disadvantages, requiring care in the choice of specific methods in specific settings (Belacel et al., 2006); we have primarily relied on hierarchical clustering, which work well for time course data such as response after infection. Clustering techniques can be particularly powerful for a time course experiment, as not only can expression pattern gene clusters be identified, but inferences can be made as to cause and effect during the immune response. Further, combining expression clustering with GO annotation of specific clusters can be very illustrative. One can find enriched functions that may identify specific pathways activated (or repressed) at specific times, allowing inference of multi-stage gene-gene interactions. For example, a seminal paper in the use of systems biology tools in immunology showed that expression clustering could identify murine regulatory pathways controlling the response to LPS (Gilchrist et al., 2006). This group showed that specific transcription factors (TF) in a cluster of up-regulated genes early in LPS treatment of mouse macrophage cells in culture controlled the expression of other sets of genes that clustered together with a maximal response later than the TF-dominated early cluster.
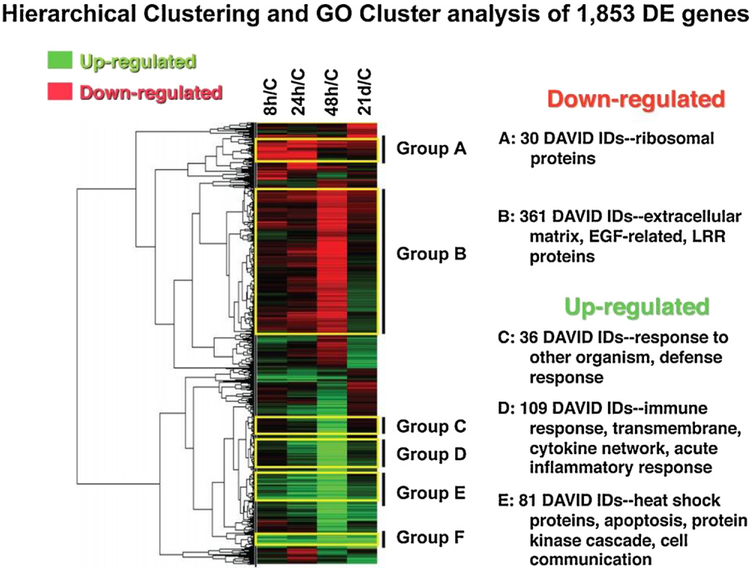
We have used GeneCluster software to identify sets of genes and their functions responding similarly to infection in mesenteric lymph nodes of SC inoculated pigs (Wang et al., 2008b). As illustrated in Fig. 3 using TreeView software, we detect large clusters of genes down-regulated (Groups A–B) or up-regulated (Groups C–F) at one or more time points during infection. Group A genes, down-regulated for the first 24 h post infection, are over-represented for ribosomal annotations, while Group B genes, only down-regulated by 48 h, are enriched for extracellular matrix proteins (Fig. 3).
Fig. 3.
An example of using hierarchical clustering of gene expression patterns to find co-expressed clusters of genes and the general functions represented by such gene clusters.
Adapted from Wang et al. (2008b).
2.3.5. Using promoter sequences of co-expressed genes to find common regulatory motifs
Genes with a similar response to an immune stimulus are co-expressed, and may be co-regulated; i.e., there may be a common regulatory factor controlling this co-expression response. To look for evidence of a common regulatory factor controlling multiple genes in a list, one approach is to search in the promoter DNA of such genes for over-represented sequence motifs known to mediate TF action. At the time of writing, the porcine genome community was close to completion of a full draft sequence for the pig, but the location of promoters near porcine genes has not yet been established. Due to lack of knowledge of promoter sequences for the pig, we developed perl scripts to obtain the orthologous human promoter sequences for porcine genes shown to be differentially up-regulated due to SC infection. Within these human promoters, we searched for TF motifs using the TransFac database (http://www.gene-regulation.com/pub/databases.html), as well as both TFM Explorer (Defrance and Touzet, 2006) and Clover (Frith et al., 2004) packages to determine over-abundance of motifs in the Group E set of genes (Fig. 3) that were up-regulated by 8–24 h post-inoculation (Wang et al., 2008b). We identified 95% of known NFkB-regulated genes in the Group E gene list, as well as 51 genes that have not previously been shown to be bound by NFkB. Similar results with slightly lower percentages were obtained for a set of genesup-regulated only by 48 hpi. These porcine genes that were co-expressed with many known NFkB target genes (activated early in infection) and with NFkB motifs near their orthologous human promoters are proposed to be previously unrecognized members of an NFkB-dependent regulatory pathway responding to SC infection (Wang et al., 2008b).
2.3.6. Using ‘Knowledgebase’ text-mining tools to efficiently mine the available literature.
Once you have a list of genes that have some common attribute such as a co-expression pattern or common function, searching the voluminous primary literature to find commonalities among gene list members is of interest but an extremely time-consuming task. We have found value in using a text-mining software tool, such as Pathway Studio (Ariadne Genomics, Inc.), which accelerates the process of such literature searches, and can provide new insight as well. This software accepts a list of genes/proteins and searches a proprietary database that holds published information on relationships between genes, between proteins, as well as gene-protein, small-molecule-gene interactions, etc. It is possible to select a specific type of relationship to search for, such as “direct-regulator of”, and specify only those genes/proteins that regulate two or more genes in a list. This would be an example of a search for a “common regulator” of the genes in the list. We recently used Pathway Studio to find the known targets of the NFkB regulatory complex in a list of genes up-regulated by SC early in infection (8–48 h post infection: Wang et al., 2008b). This information was then used to inform further exploratory analyses such as those described above for gene regulatory networks. While we could have found such information by searching many published articles, the software was able to dramatically cut down the time required for such searches, while providing a level of comprehensiveness and a repeatable methodology to the search. A number of other Pathway Studiorelationship filters, such as “common target” (to look for common functions of the gene list), or relationships within the gene list (to look at molecular or regulatory interactions among list members) are available. Furthermore, the software can create publication-quality figures depicting such relationships (Wang et al., 2008b).
2.3.7. Immune network analysis in silico—toward systems understanding of immune response
A higher level analysis that can integrate several of the above datasets is systems biology (Klipp et al., 2005; Bruggeman and Westerhoff, 2007; Gardy et al., 2009). For example, using a network analysis approach, an immunologist could use visualization tools such as those available in Cytoscape (www.cytoscape.org/) or InnateDB (Lynn et al., 2008; www.innatedb.org), to take datasets from many gene expression experiments and, combined with other data such as protein-protein interaction data, use correlations among datasets to draw network diagrams that illustrate the connections between genes (Gardy et al., 2009; Zak and Aderem, 2009). A network diagram, where genes are the nodes and the relationship between any two genes are the edges drawn between those nodes, can help visualize important genes in the immune process; such important genes may have much higher number of connections than on average and are called “hubs” in the network. As well, networks are often drawn such that the edge lengths are related to the correlation values used to create the network; for example, two genes with very similar patterns of expression across many treatments would have a very short edge length connecting the two nodes. Sets of similarly responding genes across different conditions would cluster in the network and show a high level of connections in a small three-dimensional space. Such approaches can be powerful methods to uncover cryptic networks; the larger and more integrated the datasets, the more powerful the method becomes. Systematic collection of specific immune cell type transcriptomes (Hyatt et al., 2006), and a network analysis of these data and immune response data is being used in human and mouse immunology to move toward a “systems biology” understanding of innate immune responses (Heng et al., 2008; Gardy et al., 2009; Zak and Aderem, 2009). While not used as of yet in livestock transcriptomics, we predict broader use of systems biology approaches in animal species as relevant and useful data accumulate.
2.3.8. Hypothesis generation and testing
The analyses (as described above in Sections 2.3.1–2.3.7) can guide hypothesis generation, an important outcome of microarray experiments (Fig. 2). There are many possible paths through the above tools to come to a hypothesis to be tested; it is also possible to develop a hypothesis based on a single such analysis. As discussed above, we found in Wang et al. (2008b) that many genes that were up-regulated in the first 8–24 h post SC infection are known to be involved in the innate inflammatory response, and about ~25% were also previously shown to be regulated by NFkB. We then showed that the majority of the Group E genes do have NFkB regulatory motifs in the promoters of their human orthologs (Wang et al., 2008b). Thus we hypothesize that many of the remaining Group E genes, co-expressed with known NFkB target genes, may in fact be regulated by NFkB; most of these genes were not known to be NFkB targets during infection. We are currently testing a number of these genes for their response to LPS in culture and the dependency of this response on NFkB signaling (Couture et al., data not shown).
3. Use of transcriptomics to find genes associated with quantitative disease resistance traits
In the last few years, RNA profiling has been used to investigate not only the common RNA response to infection across biological replicates, but also the variation in response to LPS treatment among individuals in a population or across inbred strains (Wells et al., 2003; Wurfel et al., 2005; Beck et al., 2006). Different inbred mouse strains showed significant differences in pathway response to LPS challenge (Wells et al., 2003). In an attempt to understand modifiers of human innate response differences to LPS in whole blood ex vivo, Wurfel et al. (2005) tested blood from 102 donors, incubating the blood with LPS and measuring levels of 7 cytokines released. They then selected high (n = 3) and low (n = 3) LPS responders (in terms of cytokines released) and profiled whole blood RNA using a human Affymetrix array. This approach enabled them to identify 80 LPS-responsive genes as well as 36 genes differentially expressed between high and low responders before stimulation.
Recently, we have initiated work to identify genes differentially expressed between pigs with different infection outcomes. We infected 40 individual pigs with ST, and measured numbers of shed bacteria up to 21 days post-inoculation (dpi, Uthe et al., 2009). Four pigs showed shedding only up to 7 dpi, with relatively low numbers of bacteria shed (low shedder phenotype, LS). On the other hand, six pigs shed continuously throughout the test period (Persistent Shedder phenotype, PS). In these 10 animals, we have profiled using Affymetrix technology the peripheral blood RNA before infection (t0) and at the early inflammatory stage (48 hpi, t2) when all animals are shedding bacteria (Huang et al., manuscript in preparation). Interestingly, we find significant numbers of differentially expressed genes in whole blood between LS and PS animals at t2, and many genes also show differential expression during infection in both shedding types (Table 1). Less that 5% of these expression differences are correlated with the numbers of different cell types as measured in complete blood counts (CBCs), indicating that differential expression is not due to changes in cell populations (data not shown). To characterize pathways and functions associated with these phenotypes, we selected genes up-regulated or down-regulated in either Low or Persistent shedders and annotated these genes to identify Gene Ontology functions over- or under-represented in these gene lists (Table 2). This analysis found striking results. First, blood from animals with a PS phenotype has increased intracellular-oriented responses and decreased extracellular-oriented responses (data not shown). Second, the blood of LS phenotype animals had a nearly opposite response; with increased expression of gene annotated in extracellular signaling pathways (Huang et al., manuscript in preparation).
Table 1.
Number of Affymetrix Porcine GeneChip® probesets showing significant differences in expression in whole blood for contrasts shown.
| Criteria | Shed by time | Shed (at t0) | Shed (at t2) | Time (in LS) | Time (in PS) |
|---|---|---|---|---|---|
| q < 0.05 | 1442 | 0 | 243 | 171 | 3379 |
| q < 0.10 | 3308 | 0 | 1313 | 837 | 4992 |
t2: 48 h post inoculation LS: low shedder PS: persistent shedder. q value: number of genes with False discovery controlled at 5% or 10%.
Table 2.
Gene ontology annotation of infection response genes shows distinct differences in persistent shedding and low shedding animals.
| Persistent shedders (PS) |
| Generally: intracellular response genes |
| ↑ Response to biotic stimulus, immune resp. |
| ↑ Proteasome, endopeptidase activity |
| ↑ Protein catabolism |
| ↑ Protein kinase cascade, reg. of NFkB cascade |
| ↑ Programmed cell death, apoptosis |
| ↑ Immunoglobulin domain |
| ↑ Vacuole/lysozyme/lytic vacuole |
| ↑ TOLL receptor signaling, NFkB/ILlR sig. |
| ↑ Multiple sclerosis/diabetes/arthritis |
| ↑ SH2 domain |
| ↑ Pleckstrin |
| ↑ Asthma/lupus-Genetic Assoc |
| ↑ CHOLERA-Genetic Assoc |
| ↓ Signal peptide |
| ↓ Intrinsic to plasma membrane |
| ↓ Signal transducer, receptor activity |
| ↓ Extracellular matrix |
| ↓ Morphogenesis, organ morphogenesis |
| ↓ Cell-cell signaling |
| ↓ Fibrinogen alpha/beta/gamma |
| ↓ Ion channel activity |
| Low shedders (LS) |
| Generally: extracellular response genes |
| ↑ Signal peptide, cell communication, receptor |
| ↑ Response to biotic stimulus, immune resp. |
| ↑ Integral to plasma membrane |
| ↑ Extracellular matrix |
| ↑ Cell-cell signaling, structural component |
| ↑ Carbohydrate/heparin binding |
| ↑ Angiogenesis |
| ↑ Tissue/organ development/remodeling |
| ↑ Fibronectin type III |
| ↑ Organ morphogenesis |
| ↓ Cell cycle, M phase |
| ↓ Nucleosome, nucleus/nuclear protein |
| ↓ Nucleic acid binding, reg. of biol. process |
| ↓ Microtubule organizing center |
| ↓ RNA localization, RNA splicing |
| ↓ Response to DNA damage stimulus |
| ↓ Chromosome segregation |
| ↓ Cellular processes—phys., metabolic |
| ↓ Zinc finger, ion binding |
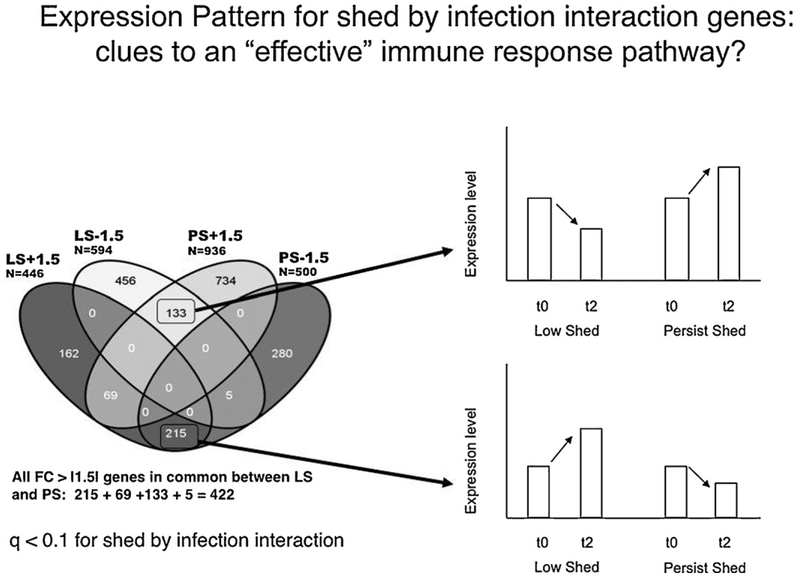
Of special interest is the large number of genes that show significant shed by time interaction, indicating the response to infection (time variable) depends on which phenotypic class is examined (Table 1). To find the genes that show high differences between the pheno-type classes, genes with significant shed x time interaction (q-value < 0.1) were sorted by Fold Change (FC) for time (response to infection), separately for LS and for PS animals. Four lists were prepared (FC > +1.5 and FC < −1.5 for each phenotype) and these lists were examined for overlap (Fig. 4). While it is expected that genes with significant shed by time interactions would show different expression between classes, the large numbers of genes with opposite expression patterns (totaling 348 genes; Fig. 4) clearly shows that the immune pathways measured in blood are strikingly different between these two phenotypes. We believe that these genes may indicate pathways controlling a more effective immune response to Salmonella infection, as their expression pattern correlates with bacterial load as measured by fecal shedding, and we plan to carefully study these genes for their roles in controlling variation in disease phenotypes during bacterial infection.
Fig. 4.
Genes with significant shed x infection interaction may be clues as to an effective immune response pathway. For example, nearly 50% of genes (215 of 448) showing high up-regulation response to infection in low shedders (LS +1.5) are in common with high down-regulated genes in Persistent shedding animals (PS −1.5). The response of such genes to infection is dependent on the class of animal (LS or PS) in which they are present. The global function of such genes may be useful in understanding variation in immune response to Salmonella.
4. Summary and future
Clearly the porcine immune response community is just starting to use the tools of trancriptomics and boinformatics to unravel the intricacies of host-pathogen interactions. If we can look to human and mouse studies as a guide, there are tremendous advances in store for researchers using such genomic approaches to study and manipulate porcine immunology and immunogenetics. With the increased use of high-throughput sequencing approaches, researchers will have access to whole-genome datasets with little to no technological limitations in the biological interpretation of the data. The limitations will be only in the imaginations of the scientists to design the optimal experiments to take advantage of these truly extraordinary opportunities for advancing molecular, cellular, and physiological knowledge and to turn such knowledge into understanding and practical application.
Footnotes
Conflict of interest statement
The authors declare there are no conflicts of interest to be disclosed. Funding of several aspects of the work described in this manuscript performed in the authors’ laboratories came from the USDA-NRICGP, Iowa State University Center for Integrated Animal Genomics, USDA-Food Safety Consortium, USDA-ARS, and the National Pork Board. None of these study sponsors had a role in writing or submission of this manuscript.
References
- Afonso CL, Piccone ME, Zaffuto KM, Neilan J, Kutish GF, Lu Z, Balinsky CA, Gibb TR, Bean TJ, Zsak L, Rock DL, 2004. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J. Virol 78, 1858–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N, Gerlach GF, 2004. Identification of genes transcribed by Actinobacillus pleuropneumoniae in necrotic porcine lung tissue by using selective capture of transcribed sequences. Infect. Immun 72, 6711–6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JS, Petry DB, Eudy J, Bough L, Johnson RK, 2008. Differential expression in lung and bronchial lymph node of pigs with high and low responses to infection with porcine reproductive and respiratory syndrome virus. J. Anim. Sci 86, 3279–3289. [DOI] [PubMed] [Google Scholar]
- Beck GC, Rafat N, Brinkkoetter P, Hanusch C, Schulte J, Haak M, van Ackern K, van der Woude FJ, Yard BA, 2006. Heterogeneity in lipopolysaccharide responsiveness of endothelial cells identified by gene expression profiling: role of transcription factors. Clin. Exp. Immunol 143, 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belacel N, Wang Q, Cuperlovic-Culf M, 2006. Clustering methods for microarray gene expression data. OMICS 10, 507–531. [DOI] [PubMed] [Google Scholar]
- Bi Y, Liu G, Yang R, 2009. MicroRNAs: novel regulators during the immune response. J. Cell. Physiol 218, 467–472. [DOI] [PubMed] [Google Scholar]
- Bliss TW, Dohms JE, Emara MG, Keeler CL Jr., 2005. Gene expression profiling of avian macrophage activation. Vet. Immunol. Immunopathol 105, 289–299. [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV, 2007. The nature of systems biology.Trends Microbiol. 15, 45–50. [DOI] [PubMed] [Google Scholar]
- Burkey TE, Skjolaas KA, Dritz SS, Minton JE, 2007. Expression of Toll-like receptors, interleukin 8, macrophage migration inhibitory factor, and osteopontin in tissues from pigs challenged with Salmonella enterica serovar Typhimurium or serovar Choleraesuis. Vet. Immunol. Immunopathol 115, 309–319. [DOI] [PubMed] [Google Scholar]
- Cagiola M, Giulio S, Miriam M, Katia F, Paola P, Macri A, Pasquali P, 2006. In vitro down regulation of proinflammatory cytokines induced by LPS tolerance in pig CD14+ cells. Vet. Immunol. Immunopathol 112, 316–320. [DOI] [PubMed] [Google Scholar]
- Chowdhury SR, King DE, Willing BP, Band MR, Beever JE, Lane AB, Loor JJ, Marini JC, Rund LA, Schook LB, Van Kessel AG, Gaskins HR, 2007. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture O, Callenberg K, Koul N, Pandit S, Younes R, Hu ZL, Dekkers J, Reecy J, Honavar V, Tuggle C, 2009. ANEXdb: an integrated animal ANnotation and microarray EXpression database. Mamm. Genome 20, 768–777. [DOI] [PubMed] [Google Scholar]
- Dawson HD, Beshah E, Nishi S, Solano-Aguilar G, Morimoto M, Zhao A, Madden KB, Ledbetter TK, Dubey JP, Shea-Donohue T, Lunney JK, Urban JF Jr., 2005. Localized multigene expression patterns support an evolving Th1/Th2-like paradigm in response to infections with Toxoplasma gondii and Ascaris suum. Infect. Immun. 73, 1116– 1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defrance M, Touzet H, 2006. Predicting transcription factor binding sites using local over-representation and comparative genomics. BMC Bioinform. 7, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA, 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- Detweiler CS, Cunanan DB, Falkow S, 2001. Host microarray analysis reveals a role for the Salmonella response regulator phoP in human macrophage cell death. Proc Natl. Acad. Sci. U.S.A. 98, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand SV, Hulst MM, de Wit AA, Mastebroek L, Loeffen WL, 2009. Activation and modulation of antiviral and apoptotic genes in pigs infected with classical swine fever viruses of high, moderate or low virulence. Arch. Virol 154, 1417–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak CM, Hirsch GN, Hyland KA, Hendrickson JA, Thompson BS, Rutherford MS, Murtaugh MP, 2006. Genomic dissection of mucosal immunobiology in the porcine small intestine. Physiol. Genomics 28, 5–14. [DOI] [PubMed] [Google Scholar]
- Fernandes LT, Tomas A, Bensaid A, Perez-Enciso M, Sibila M, Sanchez A, Segales J, 2009. Exploratory study on the transcriptional profile of pigs subclinically infected with porcine circovirus type 2. Anim. Biotechnol 20, 96–109. [DOI] [PubMed] [Google Scholar]
- Flori L, Rogel-Gaillard C, Cochet M, Lemonnier G, Hugot K, Chardon P, Robin S, Lefevre F, 2008a. Transcriptomic analysis of the dialogue between Pseudorabies virus and porcine epithelial cells during infection. BMC Genomics 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flori L, Rogel-Gaillard C, Mariani V, Lemonnier G, Cochet M, Hugot K, Chardon P, Robin S, Lefevre F, 2008b. A combined transcriptomic approach to analyse the dialogue between pseudorabies virus and porcine cells. Dev. Biol. (Basel) 132, 99–104. [DOI] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z, 2004. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 32, 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo RC, Ayoubi P, Naranjo V, Gortazar C, Kocan KM, de la Fuente J, 2009. Gene expression profiles of European wild boar naturally infected with Mycobacterium bovis. Vet. Immunol. Immunopathol 129, 119–125. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Lynn DJ, Brinkman FS, Hancock RE, 2009. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 30, 249–262. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A, 2006. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178. [DOI] [PubMed] [Google Scholar]
- He T, Feng G, Chen H, Wang L, Wang Y, 2009. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation 4, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard J, Skovgaard K, Mortensen S, Sorensen P, Jensen TK, Hornshoj H, Bendixen C, Heegaard PM, 2007. Molecular characterisation of the early response in pigs to experimental infection with Actinobacillus pleuropneumoniae using cDNA microarrays. Acta Vet. Scand 49, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW, Immunological Genome Project Consortium, 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, Goldrath A, Nguyen L, Luckey J, Yamagata T, Herman A, Jacobs J, Mathis D, Benoist C, 2006. Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol 7, 686–691. [DOI] [PubMed] [Google Scholar]
- Hyland KA, Brown DR, Murtaugh MP, 2006. Salmonella enter-ica serovar Choleraesuis infection of the porcine jejunal Peyer’s patch rapidly induces IL-1beta and IL-8 expression. Vet. Immunol. Immunopathol 109, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Young RA, 2005. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol 3, 281–294. [DOI] [PubMed] [Google Scholar]
- Klipp E, Herwig R, Kowald A, Wierling C, Lehrach H, 2005. Systems Biology in Practice. Wiley-VCH. [Google Scholar]
- Ledger TN, Pinton P, Bourges D, Roumi P, Salmon H, Oswald IP, 2004. Development of a macroarray to specifically analyze immuno- logical gene expression in swine. Clin. Diagn. Lab. Immunol 11, 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu YJ, Feng L, Cai XB, Tang HB, Sun SK, Zhang HY, Liang JJ, Luo TR, 2010. Global transcriptional profiles in peripheral blood mononuclear cell during classical swine fever virus infection. Virus Res. 148, 60–70. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang J, Block ER, Patel JM, 2004. Nitric oxide-modulated marker gene expression of signal transduction pathways in lung endothelial cells. Nitric Oxide 11, 290–297. [DOI] [PubMed] [Google Scholar]
- Lkhagvadorj S, Qu L, Cai W, Couture OP, Barb CR, Hausman GJ, Nettleton D, Anderson LL, Dekkers JC, Tuggle CK, 2009. Microarray gene expression profiles of fasting induced changes in liver and adi-pose tissues of pigs expressing the melanocortin-4 receptor D298N variant. Physiol. Genomics 38, 98–111. [DOI] [PubMed] [Google Scholar]
- Lkhagvadorj S, Qu L, Cai W, Couture OP, Barb CR, Hausman GJ, Nettleton D, Anderson LL, Dekkers JC, Tuggle CK, 2010. Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Regul. Integr. Comp. Physiol 298, R494–507. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, Lo R, Naseer M, Que J, Yau M, Acab M, Tulpan D, Whiteside MD, Chikatamarla A, Mah B, Munzner T, Hokamp K, Hancock RE, Brinkman FS, 2008. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol 4, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium MAQC, 2006. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol 24, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic M, Sivaganesan S, 2002. Bayesian infinite mixture model based clustering of gene expression profiles. Bioinformatics 18, 1194–1206. [DOI] [PubMed] [Google Scholar]
- Miller LC, Fox JM, 2004. Apoptosis and porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol 102, 131–142. [DOI] [PubMed] [Google Scholar]
- Moser RJ, Reverter A, Kerr CA, Beh KJ, Lehnert SA, 2004. A mixed-model approach for the analysis of cDNA microarray gene expression data from extreme-performing pigs after infection with Actinobacillus pleuropneumoniae. J. Anim. Sci 82, 1261–1271. [DOI] [PubMed] [Google Scholar]
- Moser RJ, Reverter A, Lehnert SA, 2008. Gene expression profiling of porcine peripheral blood leukocytes after infection with Actinobacillus pleuropneumoniae. Vet. Immunol. Immunopathol 121, 260–274. [DOI] [PubMed] [Google Scholar]
- Niewold TA, Kerstens HH, van der Meulen J, Smits MA, Hulst MM,2005. Development of a porcine small intestinal cDNA micro-array: characterization and functional analysis of the response to enterotoxigenic E. coli. Vet. Immunol. Immunopathol 105, 317–329. [DOI] [PubMed] [Google Scholar]
- Nino-Soto MI, Jozani RJ, Bridle B, Mallard BA, 2008a. Analysis of gene expression patterns by microarray hybridization in blood mononu-clear cells of SLA-DRB1 defined Canadian Yorkshire pigs. BMC Res. Notes 1, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino-Soto MI, Jozani RJ, Bridle B, Mallard BA, 2008b. CDNA microarray analysis of gene expression patterns in blood mononuclear cells of SLA-DRB1-defined Yorkshire pigs. Dev. Biol. (Basel) 132, 321–325. [DOI] [PubMed] [Google Scholar]
- Okomo-Adhiambo M, Beattie C, Rink A, 2006. cDNA microarray analysis of host-pathogen interactions in a porcine in vitro model for Toxoplasma gondii infection. Infect. Immun 74, 4254–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen I, David M, 2008. MicroRNAs in the immune response. Cytokine 43, 391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsuksili S, Murani E, Wimmers K, 2008. Porcine genome-wide gene expression in response to tetanus toxoid vaccine. Dev. Biol. (Basel) 132, 185–195. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, 2001. Computational analysis of microarray data. Nat.Rev. Genet 2, 418–427. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, 2002. Microarray data normalization and transformation.Nat. Genet 32 (Suppl.), 496–501. [DOI] [PubMed] [Google Scholar]
- Raymond CR, Wilkie BN, 2004. Th-1/Th-2 type cytokine profiles of pig T-cells cultured with antigen-treated monocyte-derived dendritic cells. Vaccine 22, 1016–1023. [DOI] [PubMed] [Google Scholar]
- Raymond CR, Wilkie BN, 2005. Toll-like receptor, MHC II. B7 and cytokine expression by porcine monocytes and monocyte-derived dendritic cells in response to microbial pathogen-associated molecular patterns. Vet. Immunol. Immunopathol 107, 235–247. [DOI] [PubMed] [Google Scholar]
- Roberts PC, 2008. Gene expression microarray data analysis demystified. Biotechnol. Annu. Rev 14, 29–61. [DOI] [PubMed] [Google Scholar]
- Rodenburg W, Bovee-Oudenhoven IM, Kramer E, van der Meer R,Keijer J, 2007. Gene expression response of the rat small intestine following oral Salmonella infection. Physiol. Genomics 30, 123–133. [DOI] [PubMed] [Google Scholar]
- Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB, 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol 164, 5894–5904. [DOI] [PubMed] [Google Scholar]
- Royaee AR, Husmann RJ, Dawson HD, Calzada-Nova G, Schnitzlein WM, Zuckermann FA, Lunney JK, 2004. Deciphering the involvement of innate immune factors in the development of the host response to PRRSV vaccination. Vet. Immunol. Immunopathol 102, 199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ, 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3, 1335–1344. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO, 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Sharbati S, Friedländer MR, Sharbati J, Hoeke L, Chen W, Keller A, Stähler PF, Rajewsky N, Einspanier R, 2010. Deciphering the porcine intestinal microRNA transcriptome. BMC Genomics 11, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Sun J, Guo H, Tu C, 2009. Genomic expression profiling of peripheral blood leukocytes of pigs infected with highly virulent classical swine fever virus strain Shimen. J. Gen. Virol 90, 1670–1680. [DOI] [PubMed] [Google Scholar]
- Skjolaas KA, Burkey TE, Dritz SS, Minton JE, 2007. Effects of Salmonella enterica serovar Typhimurium, or serovar Choleraesuis. Lactobacillus reuteri and Bacillus licheniformis on chemokine and cytokine expression in the swine jejunal epithelial cell line, IPEC-J2. Vet. Immunol. Immunopathol 115, 299–308. [DOI] [PubMed] [Google Scholar]
- Steibel JP, Wysocki M, Lunney JK, Ramos AM, Hu ZL, Rothschild MF, Ernst CW, 2009. Assessment of the swine protein-annotated oligonucleotide microarray. Anim. Genet [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas A, Fernandes LT, Sanchez A, Segales J, 2010. Time course differential gene expression in response to porcine circovirus type 2 subclinical infection. Vet. Res 41, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuggle CK, Wang Y, Couture O, 2007. Advances in swine transcriptomics. Int. J. Biol. Sci 3, 132–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuggle CK, Wang YF, Couture OP, Qu L, Uthe JJ, Kuhar D, Lunney JK, Nettleton D, Dekkers JC, Bearson SM, 2008. Computational integration of structural and functional genomics data across species to develop information on the porcine inflammatory gene regulatory pathway. Dev. Biol. (Basel) 132, 105–113. [DOI] [PubMed] [Google Scholar]
- Uthe JJ, Royaee A, Lunney JK, Stabel TJ, Zhao SH, Tuggle CK, Bearson SM, 2007. Porcine differential gene expression in response to Salmonella enterica serovars Choleraesuis and Typhimurium. Mol. Immunol 44, 2900–2914. [DOI] [PubMed] [Google Scholar]
- Uthe JJ, Stabel TJ, Zhao SH, Tuggle CK, Bearson SM, 2006. Analysis of porcine differential gene expression following challenge with Salmonella enterica serovar Choleraesuis using suppression subtractive hybridization. Vet. Microbiol 114, 60–71. [DOI] [PubMed] [Google Scholar]
- Uthe JJ, Wang Y, Qu L, Nettleton D, Tuggle CK, Bearson SM, 2009. Correlating blood immune parameters and a CCT7 genetic variant with the shedding of Salmonella enterica serovar Typhimurium in swine. Vet. Microbiol 135, 384–388. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Musters RJ, van Eeden FJ, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W, 2003. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 5, 601–611. [DOI] [PubMed] [Google Scholar]
- Veldhuizen EJ, Hendriks HG, Hogenkamp A, van Dijk A, Gaastra W, Tooten PC, Haagsman HP, 2006. Differential regulation of porcine beta-defensins 1 and 2 upon Salmonella infection in the intestinal epithelial cell line IPI-2I. Vet. Immunol. Immunopathol 114, 94– 102. [DOI] [PubMed] [Google Scholar]
- Verfaillie T, Cox E, To LT, Vanrompay D, Bouchaut H, Buys N, Goddeeris BM, 2001. Comparative analysis of porcine cytokine production by mRNA and protein detection. Vet. Immunol. Immunopathol 81, 97–112. [DOI] [PubMed] [Google Scholar]
- von Luxburg U, 2007. A tutorial on spectral clustering. Stat. Comput 17, 395–416. [Google Scholar]
- Wang J, Chen L, Li P, Li X, Zhou H, Wang F, Li D, Yin Y, Wu G, 2008a. Gene expression is altered in piglet small intestine by weaning and dietary glutamine supplementation. J. Nutr 138, 1025– 1032. [DOI] [PubMed] [Google Scholar]
- Wang Y, Couture OP, Qu L, Uthe JJ, Bearson SM, Kuhar D, Lunney JK, Nettleton D, Dekkers JC, Tuggle CK, 2008b. Analysis of porcine transcriptional response to Salmonella enterica serovar Choleraesuis suggests novel targets of NFkappaB are activated in the mesenteric lymph node. BMC Genomics 9, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qu L, Uthe JJ, Bearson SM, Kuhar D, Lunney JK, Couture OP, Nettleton D, Dekkers JC, Tuggle CK, 2007. Global transcriptional response of porcine mesenteric lymph nodes to Salmonella enterica serovar Typhimurium. Genomics 90, 72–84. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M, 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CA, Ravasi T, Faulkner GJ, Carninci P, Okazaki Y, Hayashizaki Y, Sweet M, Wainwright BJ, Hume DA, 2003. Genetic control of the innate immune response. BMC Immunol. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick MJ, 2004. Living in the danger zone: innate immunity to Salmonella. Curr. Opin. Microbiol 7, 51–57. [DOI] [PubMed] [Google Scholar]
- Wurfel MM, Park WY, Radella F, Ruzinski J, Sandstrom A, Strout J, Bumgarner RE, Martin TR, 2005. Identification of high and low responders to lipopolysaccharide in normal subjects: an unbiased approach to identify modulators of innate immunity. J. Immunol 175, 2570–2578. [DOI] [PubMed] [Google Scholar]
- Zak DE, Aderem A, 2009. Systems biology of innate immunity. Immunol. Rev 227, 264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SH, Recknor J, Lunney JK, Nettleton D, Kuhar D, Orley S, Tuggle CK, 2005. Validation of a first generation of porcine long oligonucleotide microarray for transcriptional profiling in the pig. Genomics 86, 618–625. [DOI] [PubMed] [Google Scholar]
- Zhao SH, Kuhar D, Lunney JK, Dawson H, Guidry C, Uthe JJ, Bearson SM, Recknor J, Nettleton D, Tuggle CK, 2006. Gene expression profiling in Salmonella Choleraesuis-infected porcine lung using a long oligonucleotide microarray. Mamm. Genome 17, 777–789. [DOI] [PubMed] [Google Scholar]