Abstract
Background
IQCJ-SCHIP1 antisense RNA 1 (IQCJ-SCHIP1-AS1) was a functional novel long non-coding RNA (lncRNA) revealed by our previous expression profile. In this study, we aim to investigate its clinical relevance and biological significance in colorectal cancer (CRC).
Methods
We measured the expression levels of IQCJ-SCHIP1-AS1 in 86 paired CRC tissues using quantitative RT-PCR assay, and then analyzed its association with patient prognoses. Moreover, gain-of-function and loss-of-function studies were performed to examine the biological functions of IQCJ-SCHIP1-AS1. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene set enrichment analysis (GSEA) were used to elucidate potential mechanisms of IQCJ-SCHIP1-AS1 in CRC.
Results
More than 2-fold decreased expression of IQCJ-SCHIP1-AS1 was found in half of CRC tissues (53.5%, 46/86). IQCJ-SCHIP1-AS1 down-regulation was correlated with poor differentiation (P=0.025), advanced depth of tumor (P=0.022), lymphatic invasion (P=0.010), advanced tumor stage (P=0.006), and poor prognosis (P=0.0027) in CRC patients. The Cox proportional hazards model demonstrated that IQCJ-SCHIP1-AS1 expression was an independent prognostic factor for CRC (HR =0.247, 95% CI: 0.081–0.752, P=0.014). Moreover, knockdown of IQCJ-SCHIP1-AS1 promoted CRC cell proliferation through increasing cell cycle progression and impairing cell apoptosis. Additionally, bioinformatics analysis showed that differential expression genes in IQCJ-SCHIP1-AS1-depleted CRC cells were enriched in the pathways of cell cycle, DNA replication, and p53.
Conclusions
Our results demonstrate that IQCJ-SCHIP1-AS1 has an indicative tumor suppressor role and appears to be a potential prognostic factor in CRC for the first time.
Keywords: Long non-coding RNA (lncRNA), IQCJ-SCHIP1 antisense RNA 1 (IQCJ-SCHIP1-AS1), biomarker, colorectal cancer (CRC)
Introduction
Colorectal cancer (CRC) is the third deadly cancer worldwide, and its incidence increases with age (1). Despite the fact advances in multimodality therapy have been made in recent decades, the recurrence and mortality rate of CRC did not substantially decrease (2-4). The detailed molecular mechanisms related to CRC tumorigenesis and progression remain poorly understood (5,6). Therefore, it is urgent to identify novel biomarkers with prognostic and/or therapeutic values in CRC to further unveil the mechanisms involved in CRC occurrence and development.
Long non-coding RNAs (lncRNAs) are a set of large transcripts (more than 200 nucleotides in length) with limited protein-coding potential (7,8). Emerging evidences have suggested that lncRNA is an important class of regulators, which involves in various biological processes, such as cellular development, immune regulation, especially tumorigenesis and progression (9,10). Notably, accumulating studies reveal that dysregulation of lncRNAs are associated with tumorigenesis and progression in many types of cancer (11-13). For example, LncSox4 interacting with Stat3 and their complex directly binds to the Sox4 promoter region to initiate the transcription of Sox4, which promotes the self-renewal of liver tumor-initiating cells (14). LncRNA PCAT1 directly binds to FKBP51, leading to activation of AKT and NF-κB signalings in castration-resistant prostate cancer (15). We previously identified several lncRNAs (including FEZF1-AS1, UCA1, LINC00152, and SNHG6), which exert tumor-promoting functions in CRC (16-19).
In this study, we identified a novel lncRNA, IQCJ-SCHIP1 antisense RNA 1 (IQCJ-SCHIP1-AS1), which located in 3q25.33 in human genome. It was aberrantly expressed in CRC tissues according to our previous findings (18). The expression and functional role of IQCJ-SCHIP1-AS1 had not been reported in human diseases. We compared the expression of IQCJ-SCHIP1-AS1 in 86 paired CRC tissues and their adjacent noncancerous tissues (NCTs). The results indicated that decreased IQCJ-SCHIP1-AS1 expression was positively correlated with tumor progression and poor prognosis in CRC patients. IQCJ-SCHIP1-AS1 could significantly inhibit CRC cell proliferation though blocking cell cycle and inducing cell apoptosis. In addition, bioinformatics analyses predicted that IQCJ-SCHIP1-AS1 was involved in cell cycle, DNA replication and p53 signaling pathways. These results indicated that IQCJ-SCHIP1-AS1 could be a potential prognostic factor and tumor suppressor for CRC.
Methods
Cell lines and cell culture
The CRC cell lines, HT29, LoVo, HCT116, and SW480, were purchased from the American Type Culture Collection (ATCC). All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Hyclone, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) at 37 °C in a humidified atmosphere of 5% CO2.
Patients and tissue samples
A total of 86 paired CRC tissues and adjacent NCTs used in this study were collected from the Affiliated Hospital of Jiangnan University. Our study was approved by the Clinical Research Ethics Committees of Affiliated Hospital of Jiangnan University, and all specimens were collected with informed consent. After tumor resection, tissue specimens were immediately snap-frozen in liquid nitrogen and then stored at −80 °C for total RNA extraction. The detailed information of patients was listed in Table 1.
Table 1. Correlation of IQCJ-SCHIP1-AS1 expression with clinicopathologic features in CRC.
| Characteristics | n | Low expression (%) | High expression (%) | χ2 | P value |
|---|---|---|---|---|---|
| Ages (years) | 0.977 | 0.323 | |||
| ≥60 | 50 | 29 (58.0) | 21 (42.0) | ||
| <60 | 36 | 17 (47.2) | 19 (52.8) | ||
| Gender | 0.698 | 0.403 | |||
| Female | 45 | 26 (57.8) | 19 (42.2) | ||
| Male | 41 | 20 (48.8) | 21 (51.2) | ||
| Tumor size (cm) | 0.197 | 0.264 | |||
| ≥5 | 54 | 26 (48.1) | 28 (51.9) | ||
| <5 | 32 | 20 (62.5) | 12 (37.5) | ||
| Location | 1.485 | 0.223 | |||
| Rectum | 37 | 17 (45.9) | 20 (54.1) | ||
| Colon | 49 | 29 (59.2) | 20 (40.8) | ||
| Differentiation | 4.993 | 0.025 | |||
| Poorly | 74 | 36 (48.6) | 38 (51.4) | ||
| Well+ moderately | 12 | 10 (83.3) | 2 (16.7) | ||
| Depth of tumor | 5.254 | 0.022 | |||
| T3 + T4 | 15 | 4 (26.7) | 11 (73.3) | ||
| T1 + T2 | 71 | 42 (59.2) | 29 (40.8) | ||
| Lymphatic invasion | 6.589 | 0.010 | |||
| Yes | 41 | 16 (39.0) | 25 (61.0) | ||
| No | 45 | 30 (66.7) | 15 (33.3) | ||
| Tumor stage | 7.684 | 0.006 | |||
| III + IV | 40 | 15 (37.5) | 25 (62.5) | ||
| I + II | 46 | 31 (67.4) | 15 (32.6) |
IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer.
RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted from tissues and cells with TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s instructions. Total RNA was then reversed transcribed for complementary DNA (cDNA) synthesis using HiFiScript cDNA Synthesis Kit (CWBIO, China). Quantitative RT-PCR (qRT-PCR) was performed using UltraSYBR Mixture (With ROX) (CWBIO), in a total volume of 16 µl containing cDNA, 2× UltraSYBR Mixture (With ROX), 0.2 µM of each primer and ddH2O. The primers used for qRT-PCR were listed in Table S1. β-actin was used as the internal control.
Table S1. Sequences of primer pairs for quantitative real-time PCR.
| Gene | Forward primer sequence (5'/3') | Reverse primer sequence (5'/3') |
|---|---|---|
| IQCJ-SCHIP1-AS1 | AGTCATCCTTCTGCGTGCATCA | TGCTCCTGTATTTGAATGGGTTGAT |
| GADD45A | AGCAGAAGACCGAAAGGATG | AGGCACAACACCACGTTATC |
| MCM2 | ATGGCGGAATCATCGGAATCC | GGTGAGGGCATCAGTACGC |
| FEN1 | ATGACATCAAGAGCTACTTTGGC | GGCGAACAGCAATCAGGAACT |
| SKP2 | TTAGTCGGGAGAACTTTCCAGGTG | AGTCACGTCTGGGTGCAGATTT |
| E2F1 | CATCAGTACCTGGCCGAGAG | TGGTGGTCAGATTCAGTGAGG |
| CCNB1 | GCACTTTCCTCCTTCTCA | CGATGTGGCATACTTGTT |
| β-actin | GCTCTCTGCTCCTCCTGTTC | ACGACCAAATCCGTTGACTC |
Transfection
The sequence of IQCJ-SCHIP1-AS1 (NR_121669) was cloned into pLenti-EF1a-EGFP-F2A-Puro-CMV-MCS vector to construct over-expression plasmid by GENEWIZ (China). HT29 and SW480 cells with relatively low expression of IQCJ-SCHIP1-AS1 were transfected with the IQCJ-SCHIP1-AS1 over-expression vector or control vector; IQCJ-SCHIP1-AS1 si-RNAs and scrambled control si-RNAs were transfected into HCT116 and LoVo cells with relatively high expression of IQCJ-SCHIP1-AS1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The si-RNAs of IQCJ-SCHIP1-AS1 (sense: 5’-AACCTGGTCTCCATAATAdTdT-3’) were purchased from RiboBio (China). All cells were seeded into six-well plate and harvested after 48 hours to test transfection efficiencies by qRT-PCR.
CCK8 assay and colony formation assay
Cell Counting Kit 8 (CCK8, Dojindo, Japan) and colony formation assay were used to evaluate cell proliferation ability. For CCK8 assay, cells were seeded in a 96-well plate for 4 days, and the absorbance was measured at 450 nm once a day according to the manufacturer’s instructions. For the colony formation assay, cells (1,000) were plated in 6-well plate and cultured in DMED containing 10% FBS at 37 °C for 2 weeks. Colonies were then fixed in methanol, and stained with 0.1% crystal violet (Beyotime, China) for 30 min before counted.
Cell cycle and apoptosis analysis
HCT116 and LoVo cells were harvested 48 h after transfection. For cell cycle analysis, cells were fixed in chilled 75% ethanol at 4 °C overnight, and subsequently re-suspended in 800 µL of phosphate-buffered saline. Next, the fixed cells were stained with Propidium Iodide (PI, CWBIO) according to the instruction for detecting cell cycle progress, and then assessed by flow cytometer system (BD Biosciences, USA) to analyze the percent of cell numbers in different phase of cell cycle. For cell apoptosis analysis, transfected cells were harvested by Trypsin-EDTA Solution (Beyotime). Double staining with 7-amino-actinomycin D (7-AAD) and Annexin V-phycoerythrin (PE) was performed using the Annexin V-PE/7-AAD Apoptosis Detection Kit (CWBIO) according to the manufacturer’s recommendations. Then, the cells were analyzed by flow cytometer system to estimate the percentage of cell apoptosis.
RNA-seq assay and bioinformatics analysis
RNA-seq assay was performed to measure the mRNA expression profile of IQCJ-SCHIP1-AS1-silenced CRC cells at GENEWIZ using HiSeq3000 (Illumina). The clusterProfiler R package (http://bioconductor.org/pack-ages/release/bioc/html/clusterProfiler.html) was used to identify and visualize the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched of differential expression genes (DEGs) (20). Gene set enrichment analysis (GSEA) was launched to identify DEGs of statistical difference by using GSEA v3 software (http://www.brodinstitute.org/gsea/index.jsp) (21).
Statistical analyses
Statistical analyses were performed by the analysis software SPSS 23.0 software (IBM, USA) or R software (version 3.5.1). The GraphPad prism 6.0 software and R software were utilized to make diagrams. The most appropriate cutoff values for IQCJ-SCHIP1-AS1 were obtained by generating receiver operating characteristic (ROC) curves. The Pearson χ2 test or Fisher’s exact tests was used to analyze the association between the expression of IQCJ-SCHIP1-AS1 and clinicopathologic factors. Kaplan-Meier survival analysis and the log-rank test were used to determine differences in survival rates. Survival data were also evaluated using Cox regression model. Student’s t test was used to determine significant differences of other continuous data. P values less than 0.05 were considered as statistically significant.
Results
Levels of IQCJ-SCHIP1-AS1 expression are decreased in CRC tissues
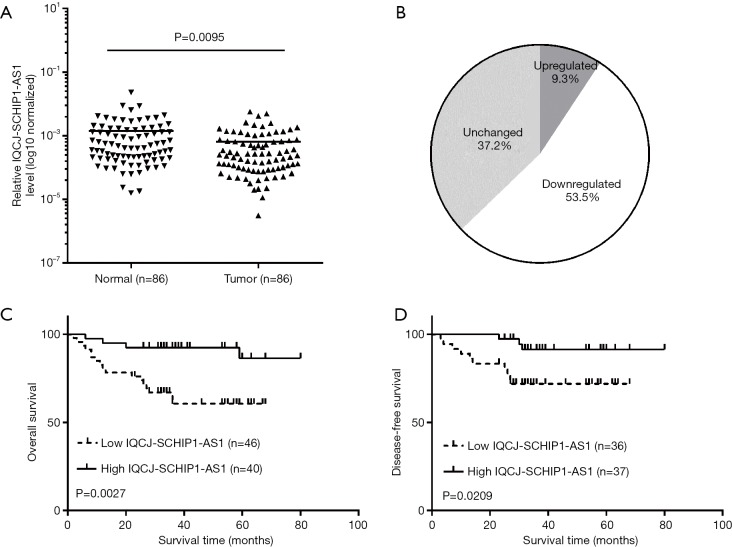
In order to investigate the expression of IQCJ-SCHIP1-AS1 in CRC, we analyzed the expression of IQCJ-SCHIP1-AS1 in 86 pairs CRC tissues and their matched NCTs. We found that IQCJ-SCHIP1-AS1 expression was significantly decreased in CRC tissues compared with paired NCTs (P=0.0095, Figure 1A), and half of CRC tissues (53.5%, 46 of 86) showed more than 2-fold down-regulation of IQCJ-SCHIP1-AS1 compared to paired NCTs (Figure 1B).
Figure 1.
IQCJ-SCHIP1-AS1 is down-regulated and predicts poor prognosis in CRC. (A) IQCJ-SCHIP1-AS1 expression levels were analysed in 86 paired CRC and adjacent NCTs by qRT-PCR; (B) IQCJ-SCHIP1-AS1 was significantly down-regulated in 53.5% of CRC tissues compared with NCTs; (C,D) Kaplan-Meier overall survival curves and disease-free survival curves for CRC patients were stratified according to relative IQCJ-SCHIP1-AS1 expression in CRC. IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer; NCT, noncancerous tissue; qRT-PCR, quantitative RT-PCR.
Low expression of IQCJ-SCHIP1-AS1 correlates with the advanced tumor stage of CRC patients
To study the correlation between IQCJ-SCHIP1-AS1 expression and clinicopathologic characteristics, the 86 CRC patients were classified into a relatively high (n=40) versus low group (n=46) using a cut-off value selected by the ROC curve (Figure S1; Table S2). As summarized in Table 1, decreased expression of IQCJ-SCHIP1-AS1 was significantly associated with poor differentiation (poorly vs. well+moderately; P=0.025), advanced depth of tumor (T3 + T4 vs. T1 + T2; P=0.022), lymphatic invasion (yes vs. no; P=0.010), and advanced tumor stage (III + IV vs. I + II; P=0.006). However, IQCJ-SCHIP1-AS1 expression was not associated with other clinicopathologic factors.
Figure S1.
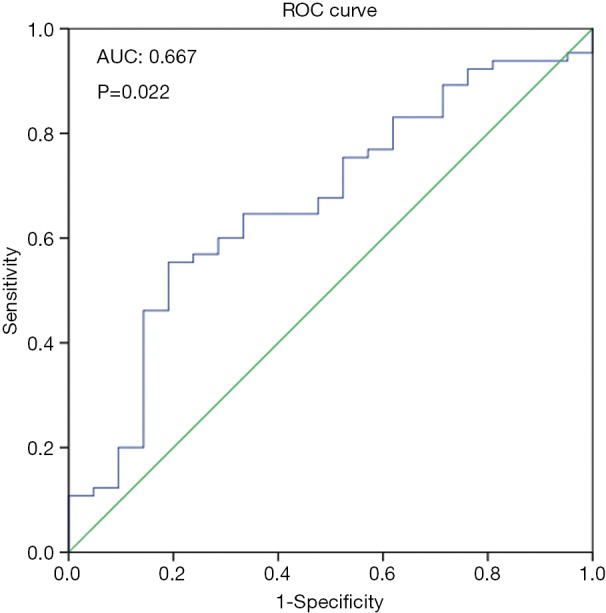
ROC curve built to classify the expression of IQCJ-SCHIP1-AS1 into a relatively high group versus a relatively low group on the prognosis of 86 patients with CRC. IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer.
Table S2. Test result variable(s): expression value of IQCJ-SCHIP1-AS1 in CRC tumor tissue.
| Positive if greater than or equal toa | Sensitivity | 1 − specificity | Youden index |
|---|---|---|---|
| 0.0002706132192b | 0.554 | 0.190 | 0.363 |
| 0.000275166794250 | 0.538 | 0.190 | 0.348 |
| 0.000279573272350 | 0.523 | 0.190 | 0.333 |
| 0.000261215506800 | 0.569 | 0.238 | 0.331 |
| 0.000357864807250 | 0.462 | 0.143 | 0.319 |
| 0.000284081892200 | 0.508 | 0.190 | 0.317 |
| 0.000268740864400 | 0.554 | 0.238 | 0.316 |
| 0.000223552414600 | 0.600 | 0.286 | 0.314 |
| 0.000179304902450 | 0.646 | 0.333 | 0.313 |
| 0.000397934274500 | 0.446 | 0.143 | 0.303 |
| 0.000297215779000 | 0.492 | 0.190 | 0.302 |
| 0.000237221398850 | 0.585 | 0.286 | 0.299 |
| 0.000187824833900 | 0.631 | 0.333 | 0.297 |
| 0.000432997280450 | 0.431 | 0.143 | 0.288 |
| 0.000319611268150 | 0.477 | 0.190 | 0.286 |
| 0.000247384204700 | 0.569 | 0.286 | 0.284 |
| 0.000200400462350 | 0.615 | 0.333 | 0.282 |
| 0.000470405154400 | 0.415 | 0.143 | 0.273 |
| 0.000339413934550 | 0.462 | 0.190 | 0.271 |
| 0.000211660922300 | 0.600 | 0.333 | 0.267 |
| 0.000167003481100 | 0.646 | 0.381 | 0.265 |
| 0.000504282473050 | 0.400 | 0.143 | 0.257 |
| 0.000519320734850 | 0.385 | 0.143 | 0.242 |
| 0.000116129045700 | 0.754 | 0.524 | 0.230 |
| 0.000546701530850 | 0.369 | 0.143 | 0.226 |
| 0.000157993472250 | 0.646 | 0.429 | 0.218 |
| 0.000120836923000 | 0.738 | 0.524 | 0.215 |
| 0.000084983706850 | 0.831 | 0.619 | 0.212 |
| 0.000599425183600 | 0.354 | 0.143 | 0.211 |
| 0.000136875799750 | 0.677 | 0.476 | 0.201 |
| 0.000125722068700 | 0.723 | 0.524 | 0.199 |
| 0.000101200325450 | 0.769 | 0.571 | 0.198 |
| 0.000086379608950 | 0.815 | 0.619 | 0.196 |
| 0.000643203219600 | 0.338 | 0.143 | 0.196 |
| 0.000144815153800 | 0.662 | 0.476 | 0.185 |
| 0.000127925763300 | 0.708 | 0.524 | 0.184 |
| 0.000108435572750 | 0.754 | 0.571 | 0.182 |
| 0.000089581161400 | 0.800 | 0.619 | 0.181 |
| 0.000654610173400 | 0.323 | 0.143 | 0.180 |
| 0.000058887613250 | 0.892 | 0.714 | 0.178 |
| 0.000152161242200 | 0.646 | 0.476 | 0.170 |
| 0.000130793908000 | 0.692 | 0.524 | 0.168 |
| 0.000093031432900 | 0.785 | 0.619 | 0.166 |
| 0.000668629857300 | 0.308 | 0.143 | 0.165 |
| 0.000080674026100 | 0.831 | 0.667 | 0.164 |
| 0.000048338710750 | 0.923 | 0.762 | 0.161 |
| 0.000132474490100 | 0.677 | 0.524 | 0.153 |
| 0.000097546861050 | 0.769 | 0.619 | 0.150 |
| 0.000691641816350 | 0.292 | 0.143 | 0.149 |
| 0.000070327245450 | 0.862 | 0.714 | 0.147 |
| 0.000049409671750 | 0.908 | 0.762 | 0.146 |
| 0.000727965220000 | 0.277 | 0.143 | 0.134 |
| 0.000050833468900 | 0.892 | 0.762 | 0.130 |
| 0.000044345755000 | 0.938 | 0.810 | 0.129 |
| 0.000759048853050 | 0.262 | 0.143 | 0.119 |
| 0.000075729581300 | 0.831 | 0.714 | 0.116 |
| 0.000046834674200 | 0.923 | 0.810 | 0.114 |
| 0.001903120220800 | 0.108 | 0.000 | 0.108 |
| 0.001222834202400 | 0.200 | 0.095 | 0.105 |
| 0.000890296538600 | 0.246 | 0.143 | 0.103 |
| 0.002064414891300 | 0.092 | 0.000 | 0.092 |
| 0.001299808700400 | 0.185 | 0.095 | 0.089 |
| 0.001050241645900 | 0.231 | 0.143 | 0.088 |
| 0.000041457995650 | 0.938 | 0.857 | 0.081 |
| 0.002335693929550 | 0.077 | 0.000 | 0.077 |
| 0.001622908901600 | 0.123 | 0.048 | 0.075 |
| 0.001331503741500 | 0.169 | 0.095 | 0.074 |
| 0.001106094991250 | 0.215 | 0.143 | 0.073 |
| 0.002547419134750 | 0.062 | 0.000 | 0.062 |
| 0.001762966827000 | 0.108 | 0.048 | 0.060 |
| 0.001359698150000 | 0.154 | 0.095 | 0.059 |
| 0.001137913129150 | 0.200 | 0.143 | 0.057 |
| 0.003369201772200 | 0.046 | 0.000 | 0.046 |
| 0.001371560930750 | 0.138 | 0.095 | 0.043 |
| 0.000035849223150 | 0.938 | 0.905 | 0.034 |
| 0.004564488852250 | 0.031 | 0.000 | 0.031 |
| 0.001475000768000 | 0.123 | 0.095 | 0.028 |
| 0.005372266489100 | 0.015 | 0.000 | 0.015 |
| 0.000021989212750 | 0.954 | 0.952 | 0.001 |
| 0.000000000000000 | 1.000 | 1.000 | 0.000 |
| 1.000000000000000 | 0.000 | 0.000 | 0.000 |
| 0.000027221775000 | 0.938 | 0.952 | −0.014 |
| 0.000007205870200 | 0.985 | 1.000 | −0.015 |
| 0.000015531637100 | 0.969 | 1.000 | −0.031 |
| 0.000020543586600 | 0.954 | 1.000 | −0.046 |
aThe smallest cutoff value is the minimum observed test value minus 1, and the largest cutoff value is the maximum observed test value plus 1. All the other cutoff values are the averages of two consecutive ordered observed test values. bThe largest sum of the sensitivity plus specificity minus 1 is the largest Youden index, which of the corresponding expression value is the cut-off value. Therefore, the cut-off value is 0.0002706132192. IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer.
Low expression of IQCJ-SCHIP1-AS1 predicts poor prognosis in CRC patients
The prognostic value of IQCJ-SCHIP1-AS1 was further examined in these 86 CRC patients. The results indicated that patients with low expression of IQCJ-SCHIP1-AS1 had a significantly shorter overall survival time than those with high expression of IQCJ-SCHIP1-AS1 (median survival time: 34.5 vs. 42 months; number of deaths: 21 of 86; log rank =9.024, P=0.0027, Figure 1C). There are 17 and 4 deaths in patients with low and high expression of IQCJ-SCHIP1-AS1, respectively. Kaplan–Meier survival analysis also revealed that low expression of IQCJ-SCHIP1-AS1 partially predicted the disease-free survival (median survival time: 34 vs. 36 months; number of relapse: 13 of 73; log rank =5.338, P=0.0209, Figure 1D). There are 10 and 3 cases of recurrence of CRC in patients with low and high expression of IQCJ-SCHIP1-AS1, respectively. Additionally, the multivariate Cox regression analysis was performed to determine the prognostic significance of IQCJ-SCHIP1-AS1. The results suggested that IQCJ-SCHIP1-AS1 expression could serve as an independent prognostic factor in CRC patients (HR =0.247, 95% CI: 0.081–0.752, P=0.014, Table 2).
Table 2. Univariate regression analyses of parameters associated with prognosis of CRC patients.
| Characteristics | Subset | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| Age | ≥60 vs. <60 years | 1.512 (0.641–3.565) | 0.345 | |||
| Gender | Male vs. female | 0.650 (0.269–1.569) | 0.338 | |||
| Tumor size | ≥5 vs. <5 cm | 1.526 (0.648–3.596) | 0.334 | |||
| Location | Colon vs. rectum | 1.302 (0.540–3.142) | 0.557 | |||
| Differentiation | Poorly vs. well+ moderately | 2.983 (1.154–7.711) | 0.024 | 2.157 (0.821–5.669) | 0.119 | |
| Depth of tumor | T3 + T4 vs. T1 + T2 | 4.892 (0.656–36.466) | 0.121 | |||
| Lymphatic invasion | Yes vs. no | 11.035 (2.566–47.462) | 0.001 | |||
| Tumor stage | III + IV vs. I + II | 10.468 (2.434–45.028) | 0.002 | |||
| IQCJ-SCHIP1-AS1 | High vs. low | 0.218 (0.073–0.650) | 0.006 | 0.247 (0.081–0.752) | 0.014 | |
HR, hazard ratio; CI, confidence interval; IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer.
IQCJ-SCHIP1-AS1 inhibits CRC cell proliferation
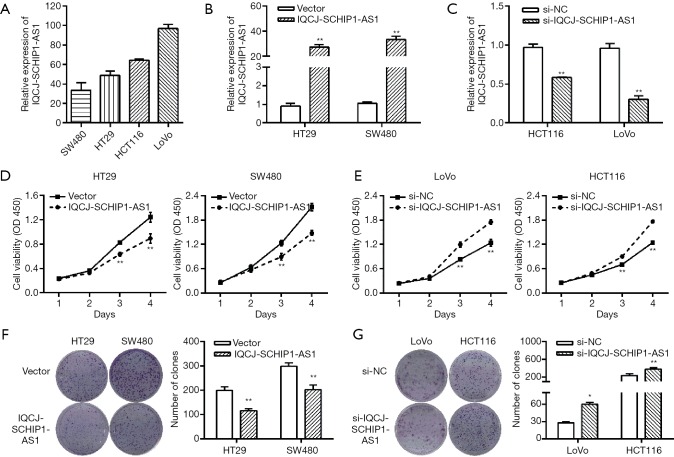
To explore the effects of IQCJ-SCHIP1-AS1 on cell functions, we first investigated the expression levels of IQCJ-SCHIP1-AS1 in CRC cell lines (HCT116, SW480, HT29, and LoVo) by qRT-PCR (Figure 2A). Then, we overexpressed IQCJ-SCHIP1-AS1 in both SW480 and HT29 cells, which express relatively low levels of endogenous IQCJ-SCHIP1-AS1 (Figure 2B). Meanwhile, we successfully knocked down IQCJ-SCHIP1-AS1 expression in HCT116 and LoVo cells, which express relatively high levels of endogenous IQCJ-SCHIP1-AS1 (Figure 2C). A CCK8 assay was performed to investigate the effect of IQCJ-SCHIP1-AS1 on CRC cell proliferation. Our results revealed that overexpression of IQCJ-SCHIP1-AS1 dramatically decreased cell proliferation (Figure 2D), whereas silence of IQCJ-SCHIP1-AS1 expression strikingly promoted cell growth (Figure 2E). Colony formation assay is also consistent with previous shown proliferation data, which IQCJ-SCHIP1-AS1 overexpression inhibited the colony formation abilities of HT29 and SW480 cells (Figure 2F). On the other hand, IQCJ-SCHIP1-AS1 knockdown enhanced the colony formation abilities of CRC cells (Figure 2G). Collectively, these data demonstrated that IQCJ-SCHIP1-AS1 could inhibit CRC cell proliferation.
Figure 2.
IQCJ-SCHIP1-AS1 inhibits CRC cell proliferation in vitro. (A) The expression of IQCJ-SCHIP1-AS1 in CRC cell lines were measured by qRT-PCR. (B,C) SW480 and HT29 cells (B) were transfected with IQCJ-SCHIP1-AS1 overexpression vector and control vector; HCT116 and LoVo cells (C) were transfected with IQCJ-SCHIP1-AS1 si-RNAs. (D,E) Effects of IQCJ-SCHIP1-AS1 overexpression and knockdown on cell growth were detected by CCK8 assay. (F,G) Effects of IQCJ-SCHIP1-AS1 on colony formation were performed in CRC cells. *, P<0.05; **, P<0.01. IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer; qRT-PCR, quantitative RT-PCR.
IQCJ-SCHIP1-AS1 blocks CRC cell cycle progression and promotes cell apoptosis
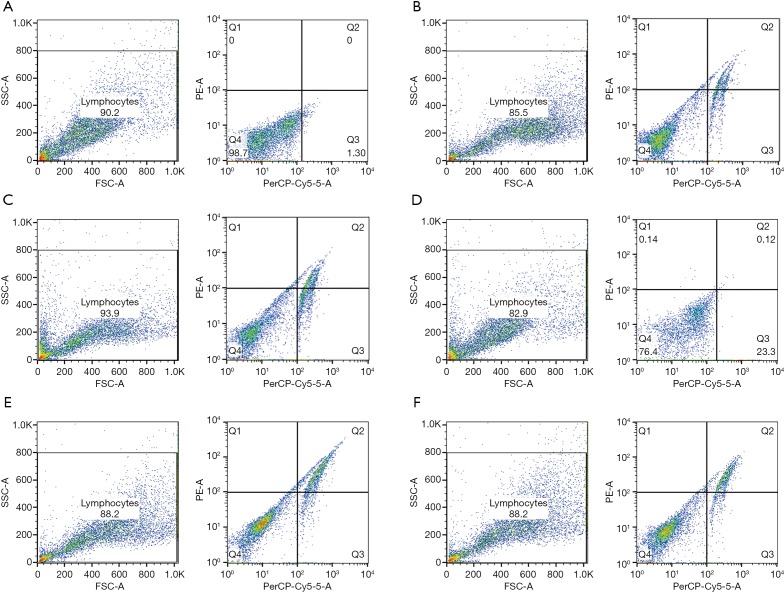
To further determine the effects of IQCJ-SCHIP1-AS1 on CRC growth, cell cycle distribution and apoptosis were analyzed using flow cytometry in IQCJ-SCHIP1-AS1-depleted or overexpressing CRC cells. The results revealed that cell cycle was arrested in G0/G1 phase in the IQCJ-SCHIP1-AS1 overexpressing HT29 and SW480 cells (Figure 3A). IQCJ-SCHIP1-AS1 depletion significantly promoted cell cycle progression in HCT116 and LoVo cells (Figure 3B). IQCJ-SCHIP1-AS1 overexpression enhanced apoptosis, whereas IQCJ-SCHIP1-AS1 knockdown inhibited apoptosis in CRC cells (Figures 3C,D,S2). These data demonstrated that IQCJ-SCHIP1-AS1 could inhibit CRC cell proliferation through blocking cell cycle progression and enhancing apoptosis. These investigations suggest that IQCJ-SCHIP1-AS1 could function as a tumor suppressor in CRC.
Figure 3.
Downregulation of IQCJ-SCHIP1-AS1 promotes cell cycle progression and inhibits cell apoptosis of CRC. (A,B) Flow cytometry assays were performed to analyze the cell cycle progression. The bar chart represented the percentage of cells in G0/G1, S, or G2/M phase. (C,D) Flow cytometry assays were performed to analyze the cell apoptosis. *, P<0.05; **, P<0.01. IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer.
Figure S2.
Effects of IQCJ-SCHIP1-AS1 on CRC cell apoptosis. (A) The crossgate was delimited according to the boundary of the negative control in HT29; (B and C) Flow cytometry assays were performed in HT29 (B: vector; C: IQCJ-SCHIP1-AS1); (D) The crossgate was delimited according to the boundary of the negative control in LoVo; (E and F) Flow cytometry assays were performed in LoVo (E: si-NC; F: si-IQCJ-SCHIP1-AS1). IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer.
Functional and pathway analysis of IQCJ-SCHIP1-AS1 in CRC
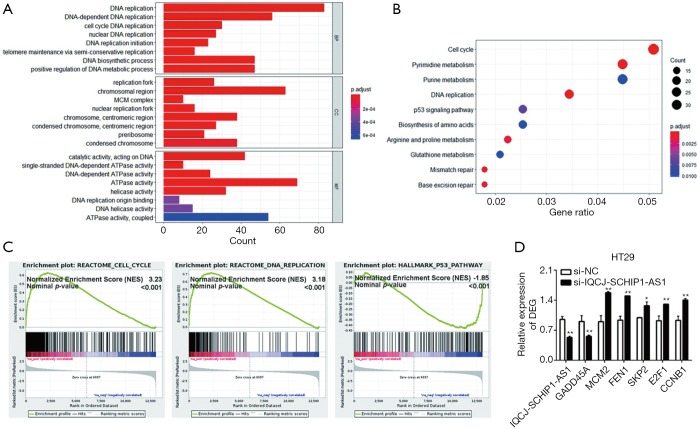
To explore the underlying mechanisms of IQCJ-SCHIP1-AS1 in CRC, the gene expression profiling in IQCJ-SCHIP1-AS1-depleted HT29 cells has been studied. Bioinformatics analyses were performed based on the top 1,502 DEGs in IQCJ-SCHIP1-AS1-silenced cells compared with the control cells. GO terms of the top related DEGs were mainly involved in several DNA replication-related biological processes (Figure 4A). Besides, both KEGG analysis and GSEA showed that the top related DEGs mainly involved in cell cycle, DNA replication and p53 signaling pathways that have strong correlation with tumorigenesis and progression (Figure 4B,C). Several key DEGs in these pathways, including cell cycle (SKP2, E2F1, CCNB1), DNA replication (MCM2, FEN1) and p53 signaling pathways (GADD45A), were selected and validated further by qRT-PCR. The results confirmed our results of RNA-seq analysis (Figure 4D). In conclusion, we observed that these genes potentially regulated by IQCJ-SCHIP1-AS1 mainly concentrated in cell proliferation related pathways, which were consistent with the results of functional assays.
Figure 4.
Functional and pathway analysis of IQCJ-SCHIP1-AS1 in CRC. (A,B) GO annotations (A) and KEGG pathway (B) of the top DEGs potentially regulated by IQCJ-SCHIP1-AS1 were analysed in R software. (C) GSEA of the top DEGs was performed in GSEA v3 software. (D) Several key DEGs in these significantly related pathways were validated by qRT-PCR. *, P<0.05; **, P<0.01. IQCJ-SCHIP1-AS1, IQCJ-SCHIP1 antisense RNA 1; CRC, colorectal cancer; qRT-PCR, quantitative RT-PCR; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genome; DEG, differential expression gene.
Conclusions
Recently, increasing evidences indicate that large numbers of lncRNAs were found to be aberrantly expressed and involved in the occurrence and development of human cancers (22-24). Such lncRNAs function as oncogenes and tumor suppressors and their aberrant expression is often associated with the prognosis of patients, which renders lncRNAs as the valuable prognostic biomarkers. In the present study, we provided the first evidence that IQCJ-SCHIP1-AS1 was downregulated in CRC, and was associated with poor prognosis. Functional assays revealed the proliferation-inhibiting effects of IQCJ-SCHIP1-AS1 in CRC cells. All these data indicated that IQCJ-SCHIP1-AS1 may function as a novel tumor suppressor in CRC.
Previous studies have showed the key regulatory roles of lncRNA in tumors. For example, lncRNA MALAT1 is reported to be overexpressed in variety of tumor types, including CRC, and identified to be associated with the short overall survival rate of CRC patients (25). Lnc-Spry1 is downregulated and mediates TGF-β-induced epithelial–mesenchymal transition by transcriptional and post-transcriptional regulatory mechanisms (26). High lnc-ZNF180-2 expression is associated with poor outcome in clear cell renal cell carcinoma (ccRCC) patients and might act as a biomarker to predict the survival of ccRCC patients (27). Our previous study demonstrated that LINC00152 functions as an oncogene to promote cell proliferation, metastasis and chemoresistance of CRC cells by regulating miR-139-5p/Notch1 axis (17). We also showed that FEZF1-AS1 promotes tumor proliferation and metastasis by regulating PKM2 signaling (18). In addition, we revealed that UCA1 enhances cell proliferation and drug resistance in CRC cells though inhibiting miR-204-5p, a well-known tumor suppressor (16,28).
The expression and function of IQCJ-SCHIP1-AS1 have not been illustrated so far. The present study further measured the expression of IQCJ-SCHIP1-AS1 in an expanded CRC cohort and revealed that IQCJ-SCHIP1-AS1 was down-regulated in more than half of CRC tissues compared with their corresponding NCTs. Downregulation of IQCJ-SCHIP1-AS1 was positively correlated with the advanced tumor stage and poor survival, which suggests that it may be a promising prognostic factor for CRC. In the present study, we also found that IQCJ-SCHIP1-AS1 could inhibit cell proliferation through blocking cell cycle progression and promoting cell apoptosis in CRC, which further suggested the tumor suppressive roles of IQCJ-SCHIP1-AS1 in CRC. Moreover, GO and KEGG analyses and GSEA also revealed that these DEGs potentially regulated by IQCJ-SCHIP1-AS1 mainly concentrated in cell cycle, DNA replication and p53 signaling pathways, which suggests that IOCJ-SCHIP1-AS1 may inhibit cell proliferation through these pathways. It is well known that cell cycle and DNA replication are closely related with cell proliferation. Many studies also have reported that p53 acts as a tumor suppressor in CRC and inhibits tumor formation (29,30). Therefore, we proposed that IQCJ-SCHIP1-AS1 can affect CRC cell proliferation through these pathways of cell cycle, DNA replication and p53. Further studies are required to clarify the molecular mechanisms or signaling pathways that IQCJ-SCHIP1-AS1 mediated cell proliferation.
In conclusion, we identified a novel lncRNA, IQCJ-SCHIP1-AS1, that was down-regulated in CRC and predicted poor prognosis. In addition, we demonstrated, for the first time, the proliferation-suppressing functions of IQCJ-SCHIP1-AS1 in CRC. The present work suggests that IQCJ-SCHIP1-AS1 is a potential therapeutic target and prognostic factor for CRC.
Acknowledgements
The authors thank for Dr. Chen Chen for critical reading of this article.
Funding: This work was supported by the National Natural Science Foundation of China (81672328, 81802469, 81772636 and 81802462), Natural Science Foundation of Jiangsu Province (BK20150004 and BK20180618), Fundamental Research Funds for the Central Universities (NOJUSRP51619B), National First-class Discipline Program of Food Science and Technology (JUFSTR20180101), Medical Key Professionals Program of Jiangsu Province (AF052141), Projects of the Wuxi Health and Family Planning Commission (CXTP003, YGZXZ1401 and Z201806) and Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1868).
Ethical Statement: The study was approved by the Clinical Research Ethics Committees of Affiliated Hospital of Jiangnan University and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 2.Franke AJ, Parekh H, Starr JS, et al. Total Neoadjuvant Therapy: A Shifting Paradigm in Locally Advanced Rectal Cancer Management. Clin Colorectal Cancer 2018;17:1-12. 10.1016/j.clcc.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 3.Jia H, Shen X, Guan Y, et al. Predicting the pathological response to neoadjuvant chemoradiation using untargeted metabolomics in locally advanced rectal cancer. Radiother Oncol 2018;128:548-56. 10.1016/j.radonc.2018.06.022 [DOI] [PubMed] [Google Scholar]
- 4.Du D, Su Z, Wang D, et al. Optimal Interval to Surgery After Neoadjuvant Chemoradiotherapy in Rectal Cancer: A Systematic Review and Meta-analysis. Clin Colorectal Cancer 2018;17:13-24. 10.1016/j.clcc.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 5.Aziz MA, Yousef Z, Saleh AM, et al. Towards personalized medicine of colorectal cancer. Crit Rev Oncol Hematol 2017;118:70-8. 10.1016/j.critrevonc.2017.08.007 [DOI] [PubMed] [Google Scholar]
- 6.Haraldsdottir S, Einarsdottir HM, Smaradottir A, et al. Colorectal cancer - review. Laeknabladid 2014;100:75-82. [DOI] [PubMed] [Google Scholar]
- 7.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics 2017;17:135-43. 10.1007/s10142-016-0524-x [DOI] [PubMed] [Google Scholar]
- 9.Karlic R, Ganesh S, Franke V, et al. Long non-coding RNA exchange during the oocyte-to-embryo transition in mice. DNA Res 2017;24:129-41. 10.1093/dnares/dsx008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 11.Xu YC, Liang CJ, Zhang DX, et al. LncSHRG promotes hepatocellular carcinoma progression by activating HES6. Oncotarget 2017;8:70630-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu P, Wang Y, Wu J, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun 2016;7:13608. 10.1038/ncomms13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Leng K, Li Z, et al. The prognostic potential and carcinogenesis of long non-coding RNA TUG1 in human cholangiocarcinoma. Oncotarget 2017;8:65823-35. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Chen ZZ, Huang L, Wu YH, et al. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun 2016;7:12598. 10.1038/ncomms12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang Z, Yu J, Sun L, et al. LncRNA PCAT1 activates AKT and NF-kappaB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKalpha complex. Nucleic Acids Res 2019. [Epub ahead of print]. 10.1093/nar/gkz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep 2016;6:23892. 10.1038/srep23892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bian Z, Zhang J, Li M, et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis 2017;6:395. 10.1038/s41389-017-0008-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian Z, Zhang J, Li M, et al. LncRNA-FEZF1-AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin Cancer Res 2018;24:4808-19. 10.1158/1078-0432.CCR-17-2967 [DOI] [PubMed] [Google Scholar]
- 19.Li M, Bian Z, Yao S, et al. Up-regulated expression of SNHG6 predicts poor prognosis in colorectal cancer. Pathol Res Pract 2018;214:784-9. 10.1016/j.prp.2017.12.014 [DOI] [PubMed] [Google Scholar]
- 20.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blondeau JJ, Deng M, Syring I, et al. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin Epigenetics 2015;7:10. 10.1186/s13148-015-0047-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo J, Xu L, Jiang Y, et al. Expression profile of long non-coding RNAs in colorectal cancer: A microarray analysis. Oncol Rep 2016;35:2035-44. 10.3892/or.2016.4606 [DOI] [PubMed] [Google Scholar]
- 24.Yao J, Huang JX, Lin M, et al. Microarray expression profile analysis of aberrant long non-coding RNAs in esophageal squamous cell carcinoma. Int J Oncol 2016;48:2543-57. 10.3892/ijo.2016.3457 [DOI] [PubMed] [Google Scholar]
- 25.Ji Q, Zhang L, Liu X, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer 2014;111:736-48. 10.1038/bjc.2014.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Mateo C, Torres B, Gutiérrez G, et al. Downregulation of Lnc-Spry1 mediates TGF-β-induced epithelial-mesenchymal transition by transcriptional and posttranscriptional regulatory mechanisms. Cell Death And Differentiation 2017;24:785. 10.1038/cdd.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellinger J, Alam J, Rothenburg J, et al. The long non-coding RNA lnc-ZNF180-2 is a prognostic biomarker in patients with clear cell renal cell carcinoma. American journal of cancer research 2015;5:2799-807. [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res 2014;20:6187-99. 10.1158/1078-0432.CCR-14-1030 [DOI] [PubMed] [Google Scholar]
- 29.Li H, Rokavec M, Jiang L, et al. Antagonistic Effects of p53 and HIF1A on microRNA-34a Regulation of PPP1R11 and STAT3 and Hypoxia-induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells. Gastroenterology 2017;153:505-20. 10.1053/j.gastro.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Yue X, Zhao Y, et al. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat Commun 2014;5:5218. 10.1038/ncomms6218 [DOI] [PMC free article] [PubMed] [Google Scholar]