ABSTRACT
Melting snowfields in polar and alpine regions often exhibit a red and orange colouration caused by microalgae. The diversity of these organisms is still poorly understood. We applied a polyphasic approach using three molecular markers and light and electron microscopy to investigate spherical cysts sampled from alpine mountains in Europe, North America and South America as well as from both polar regions. Molecular analyses revealed the presence of a single independent lineage within the Chlamydomonadales. The genus Sanguina is described, with Sanguina nivaloides as its type. It is distinguishable from other red cysts forming alga by the number of cell wall layers, cell size, cell surface morphology and habitat preference. Sanguina nivaloides is a diverse species containing a total of 18 haplotypes according to nuclear ribosomal DNA internal transcribed spacer 2, with low nucleotide divergence (≤3.5%). Based on molecular data we demonstrate that it has a cosmopolitan distribution with an absence of geographical structuring, indicating an effective dispersal strategy with the cysts being transported all around the globe, including trans-equatorially. Additionally, Sanguina aurantia is described, with small spherical orange cysts often clustered by means of mucilaginous sheaths, and causing orange blooms in snow in subarctic and Arctic regions.
Keywords: snow algae, red snow, haplotype network, Chlamydomonas nivalis
Red and orange spherical cysts causing snow colouration across several continents were investigated with regards to their geographic distribution, ecology, ultrastructure and phylogeny; the cosmopolitan distribution of a new independent lineage Sanguina within the Chlamydomonadales was molecularly confirmed.
INTRODUCTION
Several algal species are reported to cause the macroscopic phenomenon of orange and red to pinkish snow, the latter often referred to as ‘watermelon snow’ or ‘blood snow’ when occurring as mass developments, including Chlamydomonas nivalis (Wille 1903; Kol 1968; Czygan 1970; Fjerdingstad et al. 1974; Kawecka 1981; Gradinger and Nürnberg 1996; Leya 2004), Chloromonas nivalis (Remias et al. 2010; Matsuzaki, Nozaki and Kawachi 2018), Cr. brevispina (Hoham, Roemer and Mullet 1979), Cr. polyptera (Remias et al. 2013), Cr. rubroleosa (Ling and Seppelt 1993), Cr. nivalis subsp. tatrae (Procházková et al. 2018a), Chlorosarcina antarctica (Ling 2002), Smithsonimonas abbotii (Kol 1942), Chlainomonas kolii (Novis 2002) and Chlainomonas rubra (Hoham 1974).
One of the first field samples of red snow was collected during the Northern Expedition under Captain Ross in August 1818 to Baffin Bay (75°54' N 67°15' W) between northern Greenland and Canada (Bauer 1819) and brought back to England to be investigated. Red snow has been reported from almost all polar and alpine regions (Kol 1968; Marchant 1982; Mataloni and Tesolín 1997; Thomas and Broady 1997). The first phylogenetic studies that identified a separate clade comprising solely red spherical cysts were based on 18S rRNA gene data (Leya 2004; Remias et al. 2010, 2013), but a deeper molecular identification based on nuclear ribosomal DNA internal transcribed spacer 2 (ITS2) sequence data so far has been missing for this taxon except for a recent study by Segawa et al. (2018) investigating samples from both poles. Red spherical cysts have also been the focus of a few metagenomic studies carried out in the USA (Brown, Ungerer and Jumpponen 2016), Japan (Terashima et al. 2017), the Arctic (Lutz et al. 2016; Segawa et al. 2018) and the Antarctic (Segawa et al. 2018). However, detailed investigations of red snow from mid-latitude mountain ranges like those in Europe are lacking.
Though modern microscopes are much better equipped than 200 years ago, a persistent problem is that the red spherical cysts abundantly found in melting snow simply do not reveal enough intracellular details that can be used for a deep morphological description and thus, identification. The unicellular, thick-walled cyst stages [‘cysts’ = unicellular life cycle stage protected from unfavourable environmental conditions with a wall, which was built exogenously or endogenously according to Ettl and Gärtner (2014)] dominate snow surfaces during summer. The characteristic dark blood-red pigment astaxanthin is accumulated in similarly large quantities only in Haematococcus lacustris, Chlainomonas rubra and some others (Remias, Lütz-Meindl and Lütz 2005). In many cases astaxanthin covers the cysts' chloroplast structure almost entirely, making light microscopy hardly illuminating. A first revision of Chlamydomonas by Pröschold et al. (2001) revealed how important chloroplast morphology is for consistency with the phylogeny of the Chlamydomonadaceae. Many snow algal species are held as live cultures in culture collections (e.g. CCCryo, UTEX, CCAP), but none of these ‘green’ species ever formed a single phylogenetic clade together with red spherical cysts from red snow. Instead, all 18S rRNA gene phylogenetic studies have revealed a single clade comprising immotile spherical red cysts (Leya 2004; Leya, Müller, Ling et al.2004; Remias et al. 2010, 2013). This ‘red snow’ clade is clearly separated from other clades of Chlamydomonas, Chloromonas, Lobochlamys, Oogamochlamys, Haematococcus, Carteria, etc. Most likely, green, flagellated isolates from red snow field samples often were misinterpreted as having resulted from red spherical cysts or even vice versa; e.g. Novakovskaya et al. (2018) thought to have observed such a phenomenon when linking green isolates of Cr. reticulata from snow samples to red cysts in the same sample. The lack of knowledge about other life cycle stages, whether vegetative (e.g. green actively dividing cells with a well-visible chloroplast) or sexual (e.g. gametes), and the lack of sufficient molecular data from red spherical cysts, have been the most hindering in identifying their true taxonomy.
In this study we used a polyphasic approach to study field material composed of spherical red and orange cysts sampled from alpine habitats in Europe, North America, South America and both polar regions. Our aims were (i) to reveal their molecular diversity based on ITS2 rDNA and rbcL gene sequences, (ii) to describe their morphological and ultrastructural cellular details using light, transmission and scanning electron microscopy, and (iii) to characterise their habitat conditions (pH, conductivity, elevation). Furthermore, we wanted (iv) to describe the geographic distribution of haplotypes detected by establishing a haplotype network, and (v) to test the existence of spatial patterns (Mantel test, sPCA). Based on the results, a new genus-level taxonomy of the examined species is proposed with a description of two new species: the new genus Sanguina Leya, Procházková et Nedbalová with Sanguina nivaloides Procházková, Leya et Nedbalová and Sanguina aurantia Leya, Procházková et Nedbalová.
MATERIALS AND METHODS
Sampling localities
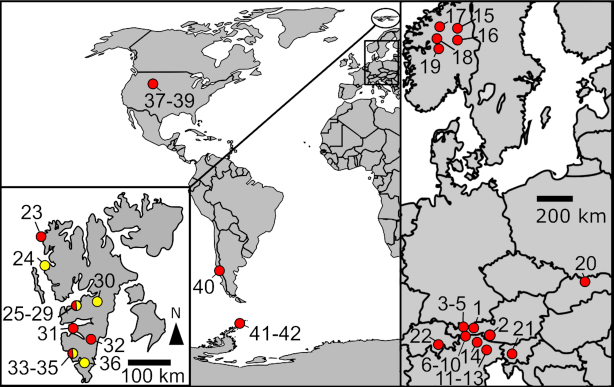
Patches of clearly visible red- or orange-coloured snow caused by spherical red or orange cells were collected from 42 localities in melting seasons between the years 1999 and 2018 (Fig. 1, Supplementary Tables 1 and 2): 22 samples originated from alpine sites in continental Europe, three from sites in North America and one from South America. Moreover, 14 samples were collected at polar sites on the island of Spitsbergen (Svalbard archipelago) and two from polar sites on the Antarctic Peninsula.
Figure 1.
Sampling locations of Sanguina nivaloides (red circles), Sanguina aurantia (yellow circles) and regions where both species were sampled (red-yellow circles) in continental Europe (right), on the archipelago of Svalbard (left), in North and South America (middle) and in Antarctica (middle). Numbering corresponds to the sample origin from the following countries: Austria (1–10; 1-Haferkarlespitze, 2-High Tauern, 3–5 Kühtai, 6–10 Ötztal Alps), Italy (11–14; 11–13 Dolomites, 14-Sarntal Alps), Norway (15–19), Slovakia (20-High Tatra Mountains), Slovenia (21-Julien Alps), Switzerland (22-Urner Alps), Svalbard (Norway) (23–36; 23-Amsterdamøya, 24-Ny Ålesund, 25–29 Longyearbyen, 30-Raggfjellet, 31-Midterhuken, 32-Doktorbreen, 33–35 Hornsund, 36-Chomjakovbreen), Colorado (USA) (37–39 Rocky Mountains), Argentina (40-Patagonia), Antarctica (41–42 Antarctic Peninsula). Habitat description of sampling localities including geographical data are shown in Supplementary Table 1.
To ensure the selection of ‘clean’ (mono-specific) bloom spots and to avoid a taxonomically diverse sample, a field microscope was used to check the sample in the field before collection. In the lab, samples were further thoroughly examined using laboratory light microscopy (see below) prior to molecular analyses. If the sample appeared to be too diverse, it was discarded. In addition, Alanagreh et al. (2017) reported that the dominant haplotype was usually evident from the Sanger sequencing, similar as in cloning approach.
Measurements of pH and conductivity and snow water equivalent (referred to as ‘snow water content’ in the paper cited) were carried out according to Procházková et al. (2018a).
In addition we were also able to investigate red cysts collected in 1851, namely from the desiccated sample ‘MB_ES_1781c’, further labelled ‘Rother Schnee Crimsen [sic] Cliff Beverly 1851, 76°3'N.B. 1781c’ obtained from the Ehrenberg Collection at the Museum für Naturkunde in Berlin. This historical material was collected during the Grinnel Arctic Expedition (1850–1851) under Lieut. De Haven of the U.S. American Marines from red snow on scarlet-coloured cliffs (Crimson Cliffs) in Baffin Bay on the Greenland coast (Beverly Greenland), though a bit further north of the location, from where the Ross type sample of 1818 originated (75°54' N 67°15' W; Ross 1820). Unfortunately, the latter material was not traceable and could not be included in this study.
Light and electron microscopy
Light microscopy was performed using an Olympus BX43 (Olympus Corporation, Japan) equipped with a DXM 1200F digital camera (Nikon, USA). Microphotographs were further processed using the QuickPHOTO Camera 3.0 software (Promicra, Czech Republic). The same software or cell∧P software (version 3.4, Olympus Soft Imaging Solutions GmbH, Germany) was used to measure cell sizes. Preparation of samples for scanning and transmission electron microscopy (SEM and TEM) followed the protocol described in Procházková et al. (2018a). The diameters of around 40 cysts were measured for each field sample. Measurements of red cysts from 1851 from the desiccated sample ‘MB_ES_1781c’ were also included.
DNA extraction and PCR
DNA isolation was carried out using the DNeasy Plant Mini Kit (Qiagen, Germany) as described in Procházková et al. (2018a). If less than 20 mg wet biomass was available, DNA was extracted using the Instagene Matrix (Bio-Rad Laboratories, USA) according to Remias et al. (2016). The 18S small subunit ribosomal RNA gene (18S rRNA gene), internal transcribed spacer region 2 (ITS2 rDNA) and ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene regions were amplified from DNA by polymerase chain reaction (PCR) using the primers listed in Supplementary Table 3. Primer pairs involved in the PCR of 18S were P2/P4, 18F2/18R2, FC/RF and FA/RD. In order to amplify ITS2 we used ITS1/ITS4, ITS5/ITS4, SSU/LSU, Al1500af/LR3 and TW81/AB28. For PCR of rbcL, we used the primer pairs rbcL1F/rbcL7R, rbcL1F/rbcL4R, rbcL10F/rbcL7R, rbcL8F/rbcL7R, Snow-F3/Snow-R12 and Snow-F0/Snow-R12. The amplification reactions for these markers are described in Procházková et al. (2018a). PCR products were purified and sequenced using an Applied Biosystems automated sequencer (ABI 3730xl) at Macrogen (Netherlands). Chromatograms of all newly obtained sequences were carefully examined using the program FinchTV 1.4.0 (Geospiza, USA). Only sequences showing distinct single peaks in the electropherogram were used. The nuclear rDNA ITS2 region was identified using a web interface for the ITS2 database showing the position of the 26S motif (http://its2.bioapps.biozentrum.uniwuerzburg.de/cgi-bin/index.pl?annotator; Koetschan et al. 2010). The sequences obtained were submitted to the NIH genetic sequence database GenBank at NCBI (accession numbers are shown in Supplementary Tables 4–6).
Molecular analyses
Our 18S rRNA gene alignment contained 96 sequences (1544 bp) and the rbcL matrix consisted of 62 sequences (954 bp); members of Chaetophora-clade were selected as the outgroup. The best-fit nucleotide substitution model was estimated by jModeltest 2.0.1 (Posada 2008). Based on the Akaike Information Criterion (AIC), the TIM2+I+G model was selected for the 18S rRNA gene. Three partitions were set for rbcL gene sequences and the following substitution models were applied: GTR+I+G (1st codon position), TIM3+I (2nd codon position) and GTR+I+G (3rd codon position). The 18S rRNA gene and rbcL phylogenetic trees were inferred by Bayesian Inference (BI) and Maximum Likelihood (ML) according to Nedbalová et al. (2017), with the minor modification that Markov Chain Monte Carlo runs were carried out for three million generations in BI. Convergence of the two cold chains was checked by the average standard deviation of split frequencies (0.000014 and 0.001613 for the 18S rRNA gene and rbcL dataset, respectively). Bootstrap analyses were performed by maximum likelihood and Bayesian posterior probabilities were calculated as described by Nedbalová et al. (2017).
The 42 sequences of ITS2 rDNA obtained during this study were used as the main dataset which was split into a red snow dataset (35 sequences) and an orange snow dataset (seven sequences). The extended dataset additionally included selected published sequences, which were assigned to the investigated species based on a pairwise comparison using BLAST. The newly generated ITS2 sequences of our red snow sample RS_0015–2010 (accession number MK728599) and orange snow sample RS_0017–2010 (accession number MK728634) were used as a query. Samples of these additional sequences originated from Colorado and Washington states (USA) (NCBI accession codes KX063716.1-KX063729.1 and KX063743.1-KX063753.1; Brown, Ungerer and Jumpponen 2016), and from Alaska (AB902998.1), Greenland (AB902971.1), the Austrian Alps (GU117577.1; Remias et al. 2010) Japan (AB903028.1) and Tadjikistan (AB902973.1, AB903025.1). The individual sequences of specimens with ambiguous positions (one or two ambiguous bases out of 205 bp in these specimens from the main dataset: redCol, 4HT, 2SLOV, 4D, 9D; one or eight ambiguous bases out of 205 bp in specimens of the extended dataset: AL1, JP1) were resolved using the PHASE algorithm implemented in DnaSP v.6.0. (Librado and Rozas 2009). PHASE assumes Hardy-Weinberg equilibrium and uses a coalescent-based Bayesian method to infer haplotypes (Librado and Rozas 2009). This means the PHASE algorithm can successfully infer haplotypes if a sufficiently high number of homozygotic sequences are used. All samples were resolved with a strong support. Haplotypes were delineated at the 100% similarity threshold (i.e. two sequences belong to the same haplotype if they are identical). Only different haplotypes were used in the ITS2 secondary structure modelling, whereas all haplotypes were used in the ITS2 haplotype network analysis. For the ITS2 haplotype network of a species causing red snow the main dataset and extended dataset consisted of 40 and 59 sequences, respectively. For the ITS2 haplotype network of a species causing orange snow the main dataset and extended dataset consisted of seven and 21 sequences, respectively.
The ITS2 rDNA sequences were folded using the Mfold server at http://mfold.rna.albany.edu/?q5mfold (Zuker 2003). The secondary structure model that was consistent with the following specific features of the nuclear rDNA ITS2 was selected, namely four helices and a U–U mismatch in helix II (Coleman 2007). The ITS2 sequences were aligned on the basis of the sequence-structure analysis (Schultz and Wolf 2009) using the 4SALE tool (Seibel et al. 2006, 2008). At this step a manual validation and correction of the sequence alignment and secondary structure was necessary. The alignments of the main and the extended dataset included 213 bp and both were used in the consensus secondary structure modelling and haplotype network. The secondary structure of the nuclear rDNA ITS2 of specimen RS_0015–2010 (accession number MK728599) was drawn using Visualization Applet for RNA (VARNA) version 3.9 (Darty, Denise and Ponty 2009). The consensus secondary structure was edited using Inkscape software version 0.91 (Free Software Foundation Inc., USA). The number of compensatory base changes (CBC; a double-sided base change of a nucleotide pair in a helix, retaining the secondary structure) between haplotypes was evaluated using the 4SALE tool. The nuclear network for ITS2 was constructed by TCS software version 1.21 (Clement, Posada and Crandall 2000) using statistic parsimony under a 95% connection limit. Then we ran a second analysis, reducing the connection limit to 91% in an attempt to infer connections among sub-networks. Gaps were considered the fifth phylogenetic character (Vukić, Ulqini and Šanda 2017). The final edit of the haplotype network was again carried out in Inkscape.
Geographic relationships
To investigate the spatial genetic structure of the red snow alga studied, we summarised genotype allele frequency data of the variable ITS2 rDNA marker by performing a spatial principal component analysis (sPCA). A principal component analysis (PCA) summarises multivariate genetic information into a few linearly uncorrelated variables, the principal components (Jombart, Pontier and Dufour 2009). The PCA method was recently modified to include spatial information to investigate the part of the genetic variation that is or is not spatially structured, the sPCA (Jombart et al. 2008). The sPCA analyses a matrix of relative allele frequencies, which contains genotypes in rows and alleles in columns. Geographic coordinates of sampling points are used as input data for the creation of a list of weights derived from a connection network (function ‘chooseCN’). Our sPCAs were performed with the adegenet package in R software (Jombart 2008), and using the function ‘spca’ and bar plots of the sPCA eigenvalues were produced showing the amount of genetic diversity (as measured by the multivariate method being used) represented by each principal component. To test for the existence of global and/or local structure, we performed two Monte Carlo multivariate tests (Jombart et al. 2008). The outputs are represented as figures showing two histograms (one for the global test and one for the local test) of permuted test statistics, and the reference distribution of these statistics is obtained by randomly permuting the rows of the genotype datamatrix. Spatial PCA was carried out for two versions of the datasets of the red spherical cysts: (i) Europe, America, Antarctica and Svalbard, and (ii) Europe. To assess the significance of the correlation between ITS2-based genetic and geographic distance matrices of the investigated species causing red snow, we also performed a classical Mantel test (‘mantel.randtest’) using the package adegenet in the R software. Pairwise Euclidean distances for both distance matrices were computed using the function ‘dist’. Haplotype diversity of the species studied within and among mountain ranges (European Alps, Tatra Mountains, continental Norway, Svalbard, Rocky Mountains, Patagonia) and polar regions (Svalbard, Antarctica) were evaluated using the program MEGA (version 7.0.14); genetic distances between ITS2 sequences were generated using both uncorrected (p-distance) and corrected pairwise distance [Kimura two-parameter (K2P) distance]. The number of base differences, uncorrected (p-distance) and corrected pairwise distance (K2P) between 18 ITS2 haplotypes of red cysts and two ITS2 haplotypes of orange cysts were evaluated with MEGA as well.
RESULTS
Habitat conditions
The species causing red snow in this study, Sanguina nivaloides, occurred on temporary (alpine meadows, polar rocks, permafrost) or persistent (glaciers) snowfields in both alpine and polar regions (Supplementary Fig. 1). This species was hardly found at mountain sites below the timber line (e.g. sample ‘subalpine’). It was observed occasionally in massive amounts on open icy glacier surfaces, though as long as there is a snow layer on the glacier, this habitat is also occupied. The species causing orange snow in this study, Sanguina aurantia, occurred at similar habitats on Svalbard only. At localities where these species are found once, they appear consistently year after year, forming reddish or orange blooms in summer, producing the well-known macroscopic phenomenon of red snow or blood snow due to the dominating red pigment astaxanthin in its cysts. Colour shades can vary from orange (orange spherical cysts) and pink to dark cherry and blood red (red spherical cysts) (Supplementary Fig. 2). These localities are usually well-defined in their extent and can cover areas from a few square metres up to several hundreds of square metres. Over summer and with melting of the snow cover algal populations are slowly exposed on the snow surface, and are often washed down from snow fields in alpine regions, resulting in intensely coloured streaks on the otherwise white snow. If a snow field is not persistent over the summer months, the cysts can end up on permafrost or rocky soil, and then covered by fresh snow at the end of the summer. There they are a ‘seed bank’ serving as an inoculum for the next melting season when the newly encountered habitat is suitable. In addition, they may be dispersed to other locations by strong winds or storms, which are e.g. typical for autumn in the polar region around Svalbard. Snow fields with blooms of Sanguina nivaloides can be almost flat or be part of a steep slope, with any variation in between. Blooms have been observed at sites close to alpine pathways and on pastures, as well as at polar sites far away from any signs of civilisation, e.g. on remote inland glaciers on Spitsbergen. Also, organic nutrient input was not a decisive factor, as we found red snow both far away and also close to bird colonies. Red snowfields were located at elevations between 13 and 585 m a.s.l. in polar regions, and between 1914 and 3731 m a.s.l. in alpine regions (Supplementary Table 1). The orange snowfields were located between 10 and 500 m a.s.l. in polar regions (Supplementary Table 2). In snowfields with S. nivaloides or S. aurantia, blooms had low electrical conductivities (3–26 µS cm−1, occasionally up to 84, but usually well below 100 µS cm−1) and pH values between slightly acidic (pH 4.7) and neutral (pH 7.1) (Supplementary Tables 1 and 2). Snow temperatures were naturally stable near 0°C. The water equivalent of a red blooming snowfield in the Austrian Alps amounted to 51.6 ± 1.6% (sample WP123). Snow field inclination and its cardinal direction towards the sun are thought to account for decisive differences in incident solar radiation and temperature, but neither were correlated with the occurrence of red snow.
Cytological observations
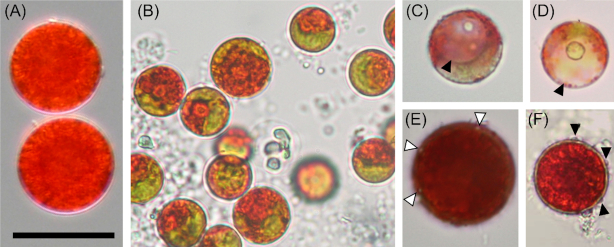
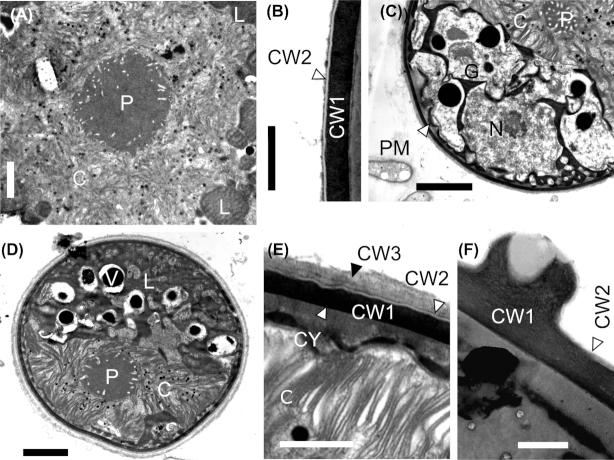
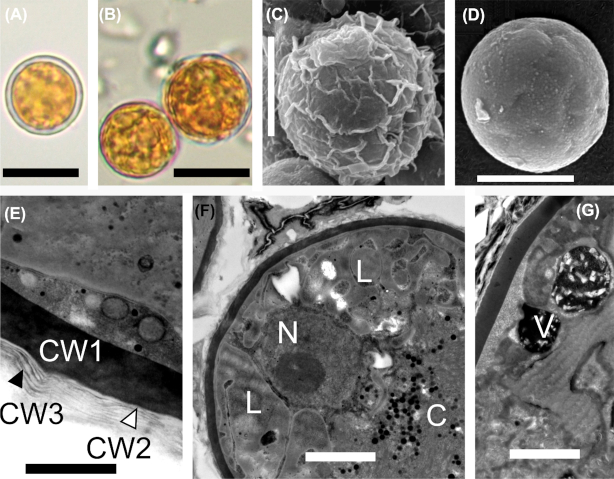
Light microscopy (Fig. 2) and electron microscopy (SEM: Fig. 3, TEM: Fig. 4) showed that field samples of Sanguina nivaloides consisted of red spherical cells exhibiting different accumulation levels of secondary carotenoids in abundant lipid droplets, which in mature cysts fully masked the chloroplasts making any detailed structure unrecognisable by light microscopy (Fig. 2A) and thus unusable for identification. A single parietal chloroplast and one or two large, unpigmented vacuoles were observed in young cells in their early stage of development only (Fig. 2B–D and Fig. 4C and D). The typical mature red cyst was smooth-walled (Figs 2A and 3A), but in two cases the red snow also consisted of populations of cysts with nipples (Figs 2E, 3B and 4F). A few scattered cysts with short blunt nibs (Fig. 3C) or pronounced papillae (Figs 2F and 3D) or cells carrying uniformly distributed small smooth papillae (Fig. 3E) were noticed in a population of smooth-walled cells . Sanguina aurantia field samples (Fig. 5) consisted of smaller spherical cysts with a striking orange-coloured cell content, the cells had an outer distant cell layer or the distant layer was absent. A few scattered cells of ‘ruby’-like cysts (the term ‘ruby’ is frequently used for this type of cyst as the dark red pigmented cyst content and the wall structure appear like a ruby encased in mother-of-pearl) were found in two red snow samples (Supplementary Fig. 3A-F, marked with an asterisk in Supplementary Table 4). We are certain that no sequences of those ruby cysts were used in any of the analyses in this study, as they would have been greatly outnumbered by the dominating smooth-walled cysts, even assuming they were equally amenable to DNA extraction in the first place. As far as the intracellular ultrastructure of our samples is concerned, mature cysts had a naked pyrenoid in a single chloroplast, which is forced into a central position by surrounding lipid globules (Fig. 4A) and cell walls consisted of two layers (Fig. 4B). Early stages of red cysts differed in the presence of a clearly visible single parietal chloroplast (Fig. 4C and D) and a three-layered cell wall, the outermost being fuzzy (Fig. 4E). Orange-coloured cysts of Sanguina aurantia differed from red cysts of Sanguina nivaloides in the structure of the second layer of their cell wall, being multilayered and slightly undulating on the surface (Fig. 5C and E). This layer most likely reflects the maturity of the cysts, as orange-coloured cysts of the same cell diameter but with smooth surfaces also existed in this population of Sanguina aurantia (Fig. 5D). Cytosolic electron-dense vacuoles filled with a crystalline structure were observed in both red cysts of Sanguina nivaloides and in orange-coloured cysts of Sanguina aurantia (Fig. 5G).
Figure 2.
Light microphotographs of Sanguina nivaloides. (A) Typical mature red cyst with smooth cell wall (holotype specimen CCCryo RS_0015–2010), abundant lipid droplets entirely masking the cell interior and structure of the chloroplast. (B-D) Young red cysts in early stage of development, (B) showing visible parts of a single parietal chloroplast (paratype sample WP123), (C-D) depicting the large unpigmented vacuole (arrowhead) (samples: Sva4 and Saddle). (E-F) Mature red cysts of less frequent haplotypes causing red blooms, (E) visualises nipples on the cell surface (arrowheads, paratype specimen CCCryo RS_0003–2004). (F) shows pronounced papillae on cell surface (arrowheads, sample 4D). Scale bar 20 µm.
Figure 3.
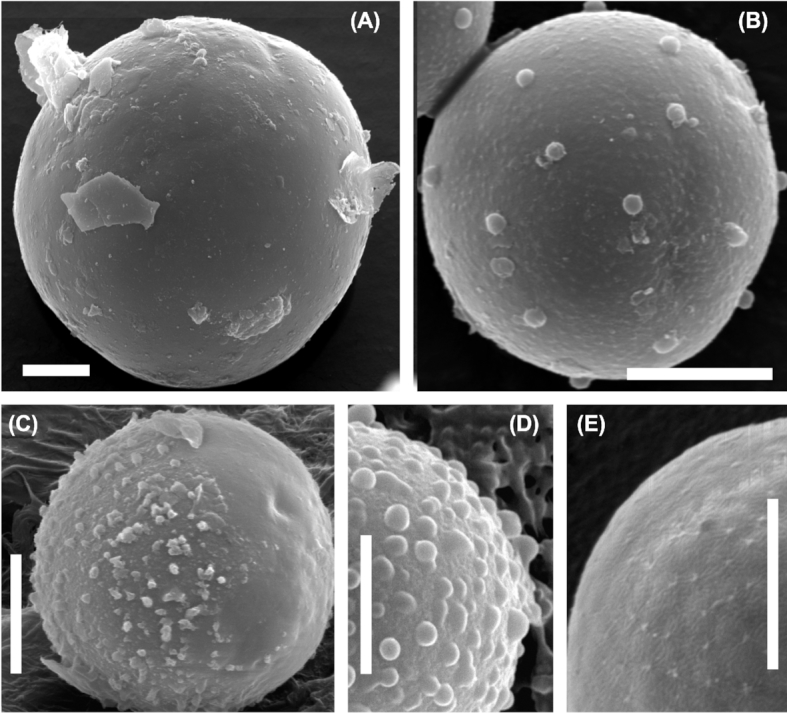
Scanning electron microphotographs of Sanguina nivaloides. (A) Typical mature red cyst with smooth cell wall (holotype specimen RS_0015–2010). (B-D) Mature red cysts of less frequent haplotypes causing blooms, (B) showing nipples on cell surface (paratype specimen RS_0003–2004), (C) depicts the smooth surface with short blunt nibs, which may represent the compressed remnants of the presumably transient primary cell wall, this cell wall layer may decompose over time (note: on the left side of the cell 'papillae' are present, on the right side they are absent, specimen RS_0011), (D) shows the pronounced papillae on the cell surface (sample 4D). (E) Scattered cells with small papillae on their cell surfaces in a population of smooth-walled cells (sample redCol). Scale bars 5 µm.
Figure 4.
Transmission electron microphotographs of Sanguina nivaloides. (A, B) Mature red cyst (holotype specimen RS_0015–2010), (A) shows the centrally located plastid with a naked pyrenoid, surrounded by abundant lipid droplets. (B) depicts the electron-dense thick cell wall covered with a very thin transparent layer. (C-E) Young red cyst (paratype sample WP123), (C) shows the large cytoplasmatic-free area (arrowhead) between the cytoplasmatic membrane and the cell wall, the nucleus and the parietal plastid, as well as parts of the Golgi aparatus, no lipid bodies visible, (D) depicts the parietal plastid with a naked pyrenoid, lipid bodies and vacuoles, (E) shows a detail of the cell wall consisting of three layers: the innermost—thick electron dense (CW1), the middle one—very thin electron transparent (CW2; trilaminar sheath) and the fuzzy upper layer (CW3; presumably the primary cell wall). (F) A detail of a cell wall nipple of less frequent haplotypes (paratype specimen RS 0003–2004). C = chloroplast, CY = cytoplasm, CW1 = cell wall 1, CW2 = cell wall 2, G = Golgi stacks, L = lipid globules, N = nucleus, P = pyrenoid, PM = plasmatic membrane, V = vacuole. Scale bars = 2 µm for (A, C, D). Scale bars = 0.5 µm for (B, E, F).
Figure 5.
Light and electron micrographs of Sanguina aurantia (holotype specimen RS_0017–2010), (A) depicts the distant outer cell layer. (B) shows that the distant cell layer is absent. (C-D) outermost layer is wrinkled and when it is decomposed, the smooth cell wall below may appear. (E) shows details of the cell wall with the electron-dense layer (CW1) being covered by several more or less undulating thin transparent layers (CW2, CW3). (F) shows a cross-section of the cyst showing the nucleus and the centrally compressed chloroplast. (G) depicts the cytosolic electron-dense vacuoles filled with crystalline structures. C = chloroplast, CW1 = cell wall 1, CW2 = cell wall 2, CW = cell wall 3, L = lipid globules, N = nucleus, V = vacuole. Scale bars = 10 µm for (A, B), 5 µm for (C, D), 0.5 µm for (E) and 2 µm for (F, G).
Size ranges of the orange-coloured cysts and red cysts overlapped, with 6.2–15.5 µm and 7.3–39.0 µm, respectively. The variability of cell size ranges in field samples of each haplotype of both snow algal species (for description see below) is shown in Supplementary Fig. 4. The material from Baffin Bay (sample ‘MB_ES_1781c’ from the Ehrenberg Collection at the Museum für Naturkunde in Berlin) contained cells in the size range typical for red cysts (Fig. 6).
Figure 6.
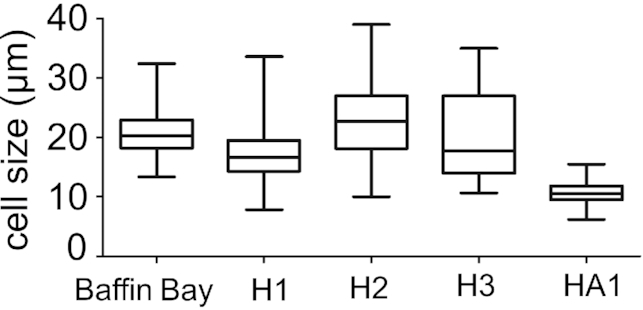
Cell size ranges of cysts of the most common haplotypes of Sanguina nivaloides (H1 and H2 = red cysts with smooth cell wall, H3 = red cysts with nipples) and Sanguina aurantia (HA1 = small orange cysts) in the dataset studied compared with cell sizes from the Crimson Cliffs in Baffin Bay on Greenland (specimen MB_ES_1781c) (Ehrenberg 1851; Leya 2008).
Molecular identification of genus Sanguina and its closest relatives
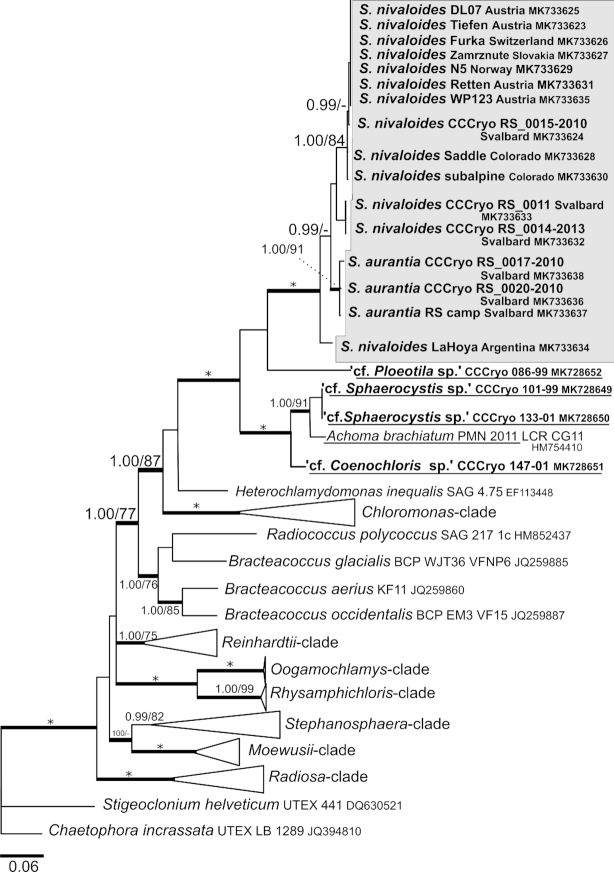
According to our 18S rRNA gene and rbcL phylogenies (Figs 7 and 8), the genus Sanguina forms a monophyletic lineage within the Chlamydomonadaceae containing samples from nearly all regions of the Earth (Greenland, North America, South America, Africa, Europe, Asia, Svalbard and Antarctica). The closest relatives to the genus Sanguina are organisms found on glaciers or glacier-associated environments, such as the strain CCCryo 086–99 (cf. Ploeotila sp.) from a glacier surface or uncultured eukaryote clones from glacial debris in Alaska and periglacial environments on the top of Mount Kilimanjaro in Tanzania. Slightly distant relatives represent strains from the Arctic permafrost in close vicinity to glaciers and snow fields, e.g. CCCryo 101–99 and CCCryo 133–01 (both cf. Sphaerocystis sp.) or Arctic snow fields, e.g. CCCryo 147–01 (cf. Coenochloris sp.), and from soil such as ‘uncultured Haematococcaceae clones’ isolated from an aspen rhizosphere and Gloeocystis sp. from desert soil in Washington State. Phylogeny based on rbcL data (Fig. 8) showed an increased molecular variability in comparison with 18S rRNA gene data among specimens of the genus Sanguina because of the degeneracy of the genetic code, which accounts for the existence of synonymous mutations. Interestingly, the small orange cysts of Sanguina aurantia formed a small subclade within the red cyst samples of Sanguina nivaloides. The rbcL phylogeny confirmed the positions of cf. Sphaerocystis and cf. Coenochloris as slightly distant relatives to Sanguina nivaloides. Additionally, it revealed that these genera form closely related lineages (Fig. 8) to Achoma brachiatum isolated from an alpine herbfield soil in New Zealand (Novis and Visnovsky 2012). Whether any of them could be assigned to the genus Achoma is discussed further below, but remains to be investigated. To sum up, the monophyly of the Sanguina-clade and the phylogenetic positions of its closest known culturable relatives were confirmed for both markers (Supplementary Figs 5 and 6). Moreover, our phylogenetic analyses demonstrate that the positions of strains SAG 26.86 of Cr. typhlos (= UTEX 1969 = CCCryo 122–00), and Cr. reticulata SAG 29.83 (= UTEX 1970 = CCCryo 213–05) in the Chloromonas-clade based on 18S rRNA gene and rbcL phylogenetic trees clearly differed from the position of S. nivaloides (Supplementary Figs 5 and 6). These strains were isolated from snow samples and were formerly believed to cause red snow, and thus, they originally were (mis)identified as Cd. nivalis and Cd. yellowstonensis (Sutton 1970), respectively. The former is still listed under two different names, as Cr. typhlos (SAG) and Cd. augustae (UTEX), as is the latter, Cr. reticulata (SAG) and Cr. clathrata (UTEX); in both cases the designations at SAG are correct. Moreover, strain CCAP 11/128 (= UTEX 2824 = CPCC 528 = CCCryo 214–05; accession number HQ404886.1) is listed as Cd. nivalis at UTEX (and others), but in our phylogeny proved to be Cr. typhlos based on the comparison of its ITS2 sequences (pairwise blast search), with 100% identity and no CBC for this marker with CCCryo 122–00 of Cr. typhlos (= SAG 26.86 = UTEX 1969 = CCAP 11/51B = IAM C-263 = NIES 2226 = IU 1969 [original deposition]; accession number HQ404869.1).
Figure 7.
18S ribosomal RNA gene-based Bayesian phylogenetic tree. The Sanguina-clade with Sanguina nivalis and Sanguina aurantia is highlighted in a grey box. Posterior probabilities (≥0.95) and bootstrap values from maximum likelihood analyses (≥75%) are shown. Full statistical support (1.00/100) is marked with an asterisk. Thick branches represent nodes receiving the highest posterior probability support (1.00). Newly obtained sequences are in bold. Accession numbers, strain or field sample codes are indicated after each species name. Species names of the closest species that are available as laboratory cultures are underlined. Abbreviation: unc, uncultured.
Figure 8.
rbcL gene-based Bayesian phylogenetic tree. The Sanguina (S.)-clade with Sanguina nivaloides and Sanguina aurantia is marked in a grey box. Posterior probabilities (≥0.95) and bootstrap values from maximum likelihood analyses (≥75%) are shown. Full statistical support (1.00/100) is marked with an asterisk. Thick branches represent nodes receiving the highest posterior probability support (1.00). Newly obtained sequences are in bold. Accession numbers, strain or field sample codes are indicated after each species name. Species names of the closest species that are available as laboratory cultures are underlined.
The CBC species concept applied to Sanguina nivaloides and Sanguina aurantia
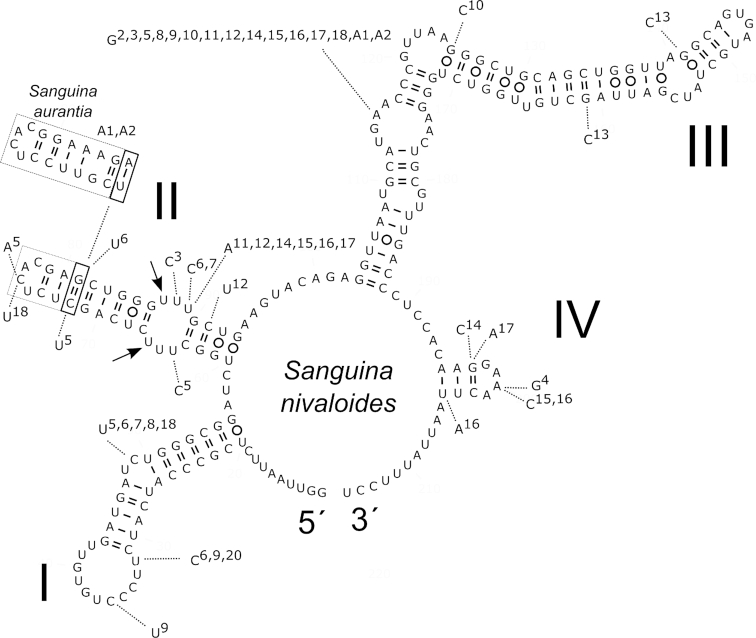
The network analysis of field samples of Sanguina nivaloides from Europe, North America, South America and Antarctica (35 specimens) applying a 95% connection limit resulted in a total of 18 ITS2 haplotypes interconnected in one network (Fig. 9). Haplotypes differed by one up to seven nucleotide changes out of 205 base pairs (Table 1). The network analysis of Sanguina aurantia field samples from Svalbard (seven specimens) resulted in two ITS2 haplotypes differing by one nucleotide change out of 213 bp. Based on the comparison of ITS2 rDNA secondary structures, one CBC was found between Sanguina nivaloides (the red cysts) and Sanguina aurantia (the small orange cysts) at the 13th position in helix II (Fig. 10). However, there is no consensus as to whether the CBC detected at this position can be used for a species separation within the Chlorophyceae. In other words, some authors treat this position as a variable one (e.g. Demchenko et al. 2012), others as a conservative one (e.g. Caisová, Marin and Melkonian 2013). Most recently, CBCs in the 5´ end of helix III were only treated as relevant for evaluating species boundaries between closely related taxa (Segawa et al. 2018). Sequence identities between red cysts and small orange cysts were ≥95.1%, whereas sequence identities within red cysts were ≥96.5% (Supplementary Table 7), suggesting that both types of cysts investigated in this study are very closely related. Among the red cysts of Sanguina nivaloides no CBC was found in the conserved parts of this marker structure, suggesting that according to the CBC species concept as proposed by Coleman (2000), all our samples of red snow from Europe, North America, South America and Antarctica were caused by one species. The presence of the one CBC and the number of base differences between ITS2 haplotypes of Sanguina nivaloides (red cysts) and Sanguina aurantia (orange cysts) are summarised in Table 1. Interestingly, two haplotypes of red cysts (H4 and H5) lack the CBC at the 13th position in helix II in their ITS2 secondary structure when compared to small orange cysts (Table 1). When comparing three molecular markers, the lowest intraspecific variability between S. nivaloides and S. aurantia haplotypes was detected in 18S (≤0.2%), was higher in rbcL (≤4.4%) and the highest was in ITS2 (≤4.9%) (Supplementary Tables 7–9).
Figure 9.
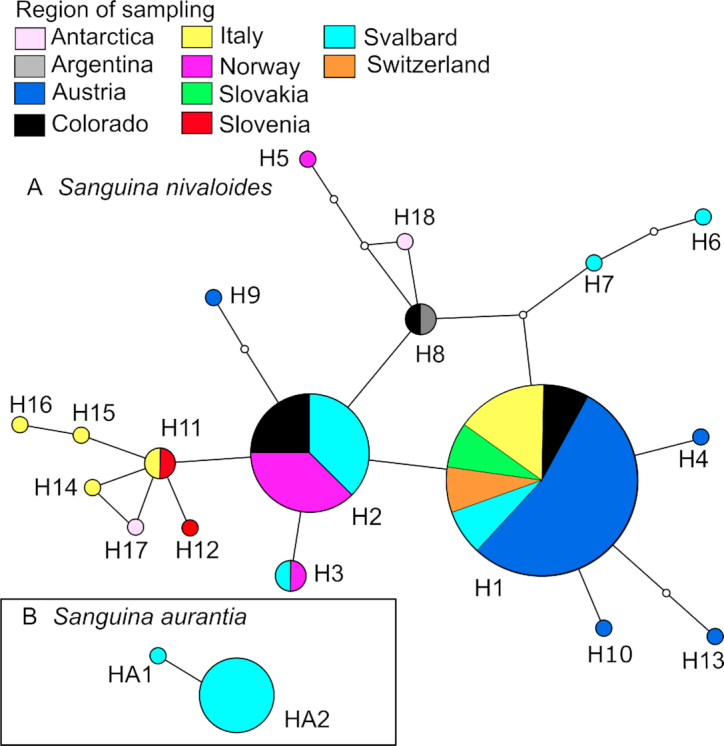
Geographic distribution of ITS2 rDNA haplotypes detected in this study: (A) Sanguina nivaloides, (B) Sanguina aurantia. Each haplotype network was constructed by a statistical parsimony method with a 95% connection limit. The Sanguina nivaloides network includes haplotypes from polar regions (Antarctica, Svalbard) and alpine regions in Europe (Austria, Italy, Norway, Slovakia, Slovenia, Switzerland), North America (Colorado) and South America (Argentina). The Sanguina aurantia network includes haplotypes from Svalbard. Each ball represents a haplotype (i.e. in this study's field samples, which have identical ITS2 sequences). The size of the ball is proportional to the number of specimens sampled, which belong to that specific haplotype. The colour represents a country of sampling. Lines connect each haplotype with its most similar relative. Open dots represent mutation steps between haplotypes, one dot indicates a change of one base pair.
Table 1.
Amount of compensatory base changes in the ITS2 secondary structure (lower left) and the number of base differences/number of indels (each of two bp length) (upper right; e.g. ‘3/4’ means that 3 differences and 4 indels were found) between 18 haplotypes of Sanguina nivaloides (H1-H18) and two haplotypes of Sanguina aurantia (HA1, HA2).
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H13 | H14 | H15 | H16 | H17 | H18 | HA1 | HA2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | 1 | 2 | 1 | 5 | 4 | 2 | 2 | 3 | 1 | 2 | 3 | 2 | 3 | 3 | 4 | 3 | 3 | 3/4 | 4/4 | |
| H2 | 0 | 1 | 2 | 4 | 5 | 3 | 1 | 2 | 2 | 1 | 2 | 3 | 2 | 2 | 3 | 2 | 2 | 2/4 | 3/4 | |
| H3 | 0 | 0 | 3 | 5 | 6 | 4 | 2 | 3 | 3 | 2 | 3 | 4 | 3 | 3 | 4 | 3 | 3 | 3/4 | 4/4 | |
| H4 | 0 | 0 | 0 | 6 | 5 | 3 | 3 | 4 | 2 | 3 | 4 | 3 | 4 | 3 | 4 | 4 | 4 | 4/4 | 5/4 | |
| H5 | 0 | 0 | 0 | 0 | 7 | 5 | 3 | 6 | 6 | 5 | 6 | 7 | 6 | 6 | 7 | 3 | 6 | 4/4 | 5/4 | |
| H6 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 5 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 5 | 6 | 6/4 | 5/4 | |
| H7 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 5 | 3 | 3 | 4 | 4 | 4 | 4 | 5 | 3 | 4 | 5/4 | 6/4 | |
| H8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 2 | 3 | 4 | 3 | 3 | 4 | 1 | 3 | 3/4 | 4/4 | |
| H9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 4 | 5 | 4 | 4 | 5 | 4 | 4 | 4/4 | 3/4 | |
| H10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 3 | 4 | 4 | 5 | 4 | 4 | 4/4 | 5/4 | |
| H11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 1 | 1 | 2 | 3 | 1 | 3/4 | 4/4 | |
| H12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 2 | 2 | 3 | 4 | 2 | 4/4 | 5/4 | |
| H13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 | 6 | 5 | 5 | 5/4 | 6/4 | |
| H14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 4 | 1 | 4/4 | 5/4 | |
| H15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 2 | 4/4 | 5/4 | |
| H16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 3 | 5/4 | 6/4 | |
| H17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4/4 | 5/4 | |
| H18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4/4 | 5/4 | |
| HA1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| HA2 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
Figure 10.
Comparison of the secondary structure of the ITS2 rDNA transcripts of 18 haplotypes of Sanguina nivaloides (H1-H18) and two haplotypes of Sanguina aurantia (HA1, HA2). Helices are labelled with Latin numbers (I-IV). Nucleotide differences of all haplotypes are shown outside the structure and are linked by dotted lines. Numbers 2–18, A1 and A2 indicate in which haplotype the difference was detected when compared to haplotype number 1 of Sanguina nivaloides (represented by the holotype specimen CCCryo RS_0015–2010, accession number MK728599). One compensatory base change between Sanguina nivaloides and Sanguina aurantia is indicated by the bold rectangle. The dashed rectangle at the end of helix II is showing an expansion segment (length is not conserved). Note: the U–U mismatch in helix II (arrows). The 5´ and 3´ ends of the ITS2 transcript are indicated.
The geographic distribution of ITS2 haplotypes
For Sanguina nivaloides (red cysts), the most frequent haplotype of this study (H1) was found in the High Tatra Mountains (Slovakia), the Swiss Alps (Switzerland), the High Tauern (Austria), Tyrol (Austria), the Sarntal Alps (Italy), the Dolomites (Italy), the Rocky Mountains (Colorado, USA) and on Spitsbergen (Svalbard, Norway). The second most abundant haplotype (H2) occurred on several sites in continental Norway and on Spitsbergen as well as in the Rocky Mountains (Colorado, USA). A smooth cell surface was typical for most of the haplotypes of red cysts, whereas pronounced nipples were found only in a single haplotype (H3) recorded from Italy and Svalbard. Small papillae were rare and only found in scattered cells in three specimens with smooth-walled cysts (Italy—‘1D’, Switzerland—‘Furka’, Slovakia—‘Zamrznute’) belonging to the most common haplotype (H1). Sanguina aurantia (small orange cysts) were represented here by two haplotypes (HA1 and HA2), reported from Svalbard and phylogenetically already attributed to a clade called ‘Roter Schnee’ (Leya 2004). The Austrian Alps were shown to be rich in haplotype diversity of Sanguina nivaloides, with the presence of at least five (H1, H4, H9, H10 and H13). The haplotype network of this species indicates the presence of shared haplotypes among European mountain ranges, between alpine and polar habitats (e.g. Austria versus Svalbard) and between the continents investigated (Europe, North America and South America). Interestingly, the small orange cysts of Sanguina aurantia were not restricted only to the island of Spitsbergen in Svalbard as was implied from preliminary conclusions, but its haplotypes HA2, HA3 and HA4 were found at sampling sites in Colorado and Washington (USA), as shown in Supplementary Fig. 7. The extended version of the haplotype network of S. nivaloides detected four further haplotypes for this species (sequence JP1 was phased into sequences belonging to H1 and H22, the latter was connected to the ITS2 haplotype network when the TCS connection limit was lowered to 91%), which confirmed its cosmopolitan distribution e.g. in Japan, Tajikistan, Greenland and Alaska. However, for these sequences published prior to this study, no scanning electron nor light microscopy pictures of cells were available for comparison.
Spatial structuring of Sanguina nivaloides metapopulations
The sPCA revealed positive and negative eigenvalues for both two S. nivaloides datasets, indicating global (positive values) and local (negative values) patterns. However, the tests on global and local structures were not significant (P > 0.05, Supplementary Table 10), so S. nivaloides showed an absence of a distinct global spatial genetic structure and no genetic differentiation between European regions (Supplementary Fig. 8). In other words, we did not find any phylogeographic signal in the distribution of S. nivaloides. Moreover, the Mantel test on pairwise geographic and genetic matrices showed no significant correlation (Supplementary Table 11). Genetic distances of Sanguina nivaloides ITS2 sequences within and between mountain ranges (European Alps, High Tatra Mountains, continental Norway, Rocky Mountains, Patagonia) and polar regions (Svalbard, Antarctica) were very similar (Table 2). These findings suggest a high dispersal capacity of the cysts, which could promote a continuous gene flow over long distances (on scales of different continents!). At the same time, a successful independent evolution of haplotypes in separate mountain ranges and polar regions might also be taking place as inferred from the genetic distances observed within these groups. That both scenarios are possible for Sanguina nivaloides is visualised by the geographical distribution of ITS2 haplotypes in its network (Fig. 9): an identical haplotype occurred in several regions, and a particular region was inhabited by a few different haplotypes.
Table 2.
Mean uncorrected (p-distance; %) and corrected pairwise genetic distance (Kimura-2-parameter K2P; %) between Sanguina nivaloides field samples from different regions based on the ITS2 rDNA nucleotide sequence below and above the diagonal. Mean uncorrected and corrected pairwise genetic distances are shown in the first column. The presence of n.c. (not calculated) in the results denotes cases in which it was not possible to estimate evolutionary distances (single sequence from the region).
| Mean intraspecific distance | Svalbard | European Alps | Norway | Tatra Mountains | Rocky Mountains | Patagonia | Antarctica | |
|---|---|---|---|---|---|---|---|---|
| Svalbard | 1.30 | 1.20 | 1.20 | 0.80 | 0.90 | 1.00 | 1.60 | |
| European Alps | 1.00 | 1.20 | 1.20 | 0.60 | 0.80 | 1.20 | 1.60 | |
| Norway | 1.00 | 1.20 | 1.20 | 1.00 | 0.70 | 0.80 | 1.40 | |
| Tatra Mountains | n.c. | 0.80 | 0.60 | 1.00 | 0.50 | 1.00 | 1.50 | |
| Rocky Mountains | 0.50 | 0.90 | 0.80 | 0.70 | 0.50 | 0.50 | 1.10 | |
| Patagonia | n.c. | 1.00 | 1.20 | 0.80 | 1.00 | 0.50 | 1.00 | |
| Antarctica | 2.00 | 1.50 | 1.50 | 1.30 | 1.50 | 1.10 | 1.00 |
Taxonomic treatment
Sanguina Leya, Procházková et Nedbalová, gen. nov.
Type of the name of the genus: Sanguina nivaloides Procházková, Leya et Nedbalová, sp. nov.
Registration: http://phycobank.org/100627
Initially, we attempted to reinstate one of the old genus names typified by Uredo nivalis F.A.Bauer 1819—Protococcus C.Agardh, 1824 or Sphaerella Sommerfelt, 1824—but both genera names have to be rejected, in the former case versus Chlamydomonas Ehrenberg, 1833 (nom. et orth. cons.) (original publication: Ehrenberg 1834), in the latter versus Chlorococcum Meneghini 1842 (original publication: Meneghini 1843) (nomen cons.) (Farr and Zijlstra 1996+). The complicated historic synonymy and the fact that it is not possible to prove that Uredo nivalis F.A.Bauer 1819 is conspecific with our recent findings are the reasons why the newly established clade is described by us as a new genus and species.
Etymology: The genus name Sanguina refers to the striking blood-red colouration of snow caused by the bloom of its cysts.
Description: Cysts (it is unresolved whether these are of asexual or sexual origin) are spherical, contain one nucleus, a central chloroplast with one naked pyrenoid, and abundant peripheral lipid globules containing red-coloured carotenoids. According to the maturity of cysts and their astaxanthin to chlorophyll-a ratios, cysts can appear in different colour shades as orange-peel orange or blood red. As the secondary carotenoids are stored in lipid globules of different sizes the cytoplasm can appear speckled to homogeneous. The surface of the cysts can appear smooth, with blunt (nipples, papillae) or sharp (nibs) protrusions. In mature cysts, carotenoids mask other cell organelles beyond recognition. In young cysts a single parietal chloroplast is clearly visible. At the light microscopy level one cell wall layer can be distinguished in mature cysts and two layers in young cysts. The outermost layer in young cysts is most probably the primary cell wall, which later is shed during cyst maturation exposing the developing secondary cell wall. Vegetative stages are unknown, and no culturable strains exist.The genus is restricted in its habitat to snowfields and glacier-based snowfields in polar and alpine regions.
Diagnosis: The genus Sanguina differs from other genera within the Chlamydomonadaceae [i.e. the genus Chlamydomonas sensu Prӧschold et al. (2001)] in its phylogeny, as this species forms an independent lineage in phylogenetic trees based on 18S rRNA gene (Fig. 7) or rbcL data (Fig. 8) [characteristic 18S rRNA gene and rbcL sequences for holotype specimen CCCryo RS 0015–2010: accession numbers JQ790560 (new resequencing of this field material in the course of this study resulted in the identical sequence) and MK733624, respectively]. Moreover, Sanguina is distinguishable from the genus Chlamydomonas in its clear habitat preference. The former genus develops cysts rich in astaxanthin, which causes the striking red or orange colouration of snow in alpine and polar regions. The morphology of vegetative stages of Sanguina is unknown because no culturable, proliferating strain exists, thus hampering a comparison of such life cycle stages to those of the genus Chlamydomonas. Regarding its fully spherical, red cysts, Sanguina differs from mature cysts of Chlainomonas sp. in its cell wall arrangement and plastid organisation: the latter has multilayered cell walls including an opaque layer, plastids are located close to the cell wall (parietal) and mean cyst sizes are larger in diameter (Procházková et al. 2018b). Chlainomonas sp. also differs in the blood-red, but ovoid quadriflagellate cell stages that are quite distinct from the immobile spherical cysts of Sanguina. Moreover, Chlainomonas sp. prefers water-logged habitats, and thus usually causes the red colouration of snow-slush on frozen lakes. Sanguina, on the other hand, is rather found in terrestrial habitats such as snow-covered rocks or soil and permafrost. If Sanguina is found close to water-logged localities, it tends to dwell on the dry parts of snow. Smithsonimonas abbotii (Kol 1942), a snow alga described from red snow above the timber line in Alaska, might be closely related to the genus Chlainomonas. It differs from Sanguina in its cells being surrounded by a bell-shaped cell wall, which is smooth when young and becomes progressively warty when older. Large quantities of astaxanthin are also accumulated in the resistant red cysts (aplanospores) of the genus Haematococcus (Allewaert et al. 2015; Mazumdar et al. 2018), which differs, however, in its 18S rRNA gene and rbcL sequences.
Sanguina nivaloides Procházková, Leya et Nedbalová, sp. nov.
? = Uredo nivalis F.A.Bauer, Microscopical observation on the red snow.— The Quarterly Journal of Literature, Science and The Arts, London: 222–229 (incl. plate VI), Figs 1-7. 1819
? = Chlamydomonas nivalis (F.A.Bauer) Wille, Algologische Notizen IX-XIV. Nyt Magazin for Naturvidenskaberne 41: 103, 126, plate III, Figs 44, 45. 1903.
Neither of the original field samples of Uredo nivalis from 1818 and of Chlamydomonas nivalis from Wille are traceable, therefore these names are included in the synonymy with a question mark.
Nomenclatural remarks: Though we clearly do not attempt to synonymise Cd. nivalis with S. nivaloides, simply because there is no conclusive way to do so, for completeness we want to give an overview of the nomenclatural complexity relating to Cd. nivalis. With regard to the type of material sampled in Baffin Bay in 1818, three homotypic synonyms are listed on the website of AlgaeBase (Guiry and Guiry 2018): Uredo nivalis F.A.Bauer 1819, Protococcus nivalis C.Agardh 1824 and Sphaerella nivalis (F.A.Bauer) Sommerfelt 1824. In this context Uredo nivalis represents the lectotype and basionym, still reflecting that the (colourless) globules were regarded as a fungus. The genus Uredo was typified by Persoon (1801) with its type species U. betae and thus is conserved against Cd. nivalis. Wrangel (1823) described Lepraria kermesina, the lectotype species of Protococcus, and he considered this lichen to be the organism causing the red snow. L. kermesina was then listed as a synonym of Protococcus nivalis. Haematococcus Flotow, 1844 was also listed as a synonym, but it was finally decided to exclude both, Lepraria kermesina and Haematococcus, from the synonymy (Andersen 2018). With regard to Protococcus C.Agardh, 1824 and Sphaerella Sommerfelt, 1824—both genus names have to be rejected, in the former case versus Chlamydomonas Ehrenberg, 1833 (original publication: Ehrenberg 1834) (nom. et orth. cons.), in the latter versus Chlorococcum Meneghini, 1842 (original publication: Meneghini 1843) (nomen cons.) (Farr and Zijlstra 1996+). In the Index Nominum Genericorum and the literature, further synonyms for Cd. nivalis are listed. In her extensive and outstanding monography on the biology and limnology of snow and ice, Kol (1968) cites these synonyms, which are given here for completeness as well (some citations have been corrected, as far as possible, as wrong volume numbers or pages were cited originally): Palmella nivalis Hooker 1825, p. 184; Coccochloris nivalis Sprengel (ex parte) 1827, p. 373; Coccophysium nivale Link 1833, p. 342; Gloiococcus grevillei Shuttleworth 1840a, p. 54 and Shuttleworth 1840b, p. 405 resp.; Disceraea nivalis Vogt 1844, p. 217 with plate 1: Figs 1-5 (as ‘Discerea nivalis’); Hysginum nivale Perty 1852, p. 95; Chlamydococcus nivalis (F.A.Bauer) A.Braun nach F.Cohn 1861, p. 54, Haematococcus nivalis (F.A.Bauer) C.Agardh 1828-1835, Icon. Alg., Haematococcus nivalis Flotow 1844. Further information on the synonymy and references on world-wide findings of Cd. nivalis also can be found in Kol (1968).
Holotype: Specimen CCCryo RS_0015–2010 of field sample 004/10 consisting of cysts in a desiccated (non-viable) state in the herbarium of the Botanic Garden and Botanical Museum Berlin-Dahlem (herbarium acronym: B), Freie Universität Berlin, Königin-Luise-Straße 6–8, 14 195 Berlin, Germany, no. B 40 00 43192 [http://herbarium.bgbm.org/object/B400043192, represented by our Figs 2A and 3A.
Isotypes: (1) desiccated, under nitrogen, in the Micropalaeo Collection, Museum für Naturkunde, Leibniz Institute for Evolution and Biodiversity, Science, Invalidenstraße 43, 10 115 Berlin, Germany, no. ECO103, (2) frozen at -80°C at DNA-bank in the herbarium of the Botanic Garden and Botanical Museum Berlin-Dahlem (herbarium acronym: B), no. B GT 00 24094 [http://herbarium.bgbm.org/object/BGT0024094], (3) frozen at -150°C (in triplicate) in the SAG biobank at the Department Experimental Phycology and Culture Collection of Algae at the Goettingen University (EPSAG), Nikolausberger Weg 18, 37 073 Goettingen, Germany, nos. Z000696411, Z000696414, Z000696417, (4) frozen at -150°C (in triplicate) in the CCCryo biobank at the Fraunhofer Institute for Immunology & Cell Therapy, Branch Bioanalytics and Bioprocesses IZI-BB, Am Muehlenberg 13, 144 760 Postdam, Germany, nos. 2–07-02–14, 2–07-02–15, 2–07-02–16, and (5) desiccated (in triplicate) in the CCCryo biobank, nos. D2–07-02–17, D2–07-02–18, D2–07-02–19.
Type locality: a steep snowfield surrounded by moss vegetation and rocks, stretching down northwest to sea level from Mount Midterhuken (760 m a.s.l.), southwest of Mariasundet between Bellsund and Van Mijenfjorden, Nathorst Land, Svalbard, Norway (77°39'44.298''N 14°48'58.903''E; altitude: 15 m a.s.l., specimen CCCryo RS 0015–2010, collected on 4 August 2010 by Thomas Leya & Guntram Weithoff as collectors' sample number 004/10).
Paratypes: (1) Specimen CCCryo RS_0003–2004 frozen at -150°C in the CCCryo biobank at Fraunhofer IZI-BB, no. 2–07-02–21 (Figs 2E, 3B and 4F), (2) specimen WP123 frozen at -150°C in the CCCryo biobank at Fraunhofer IZI-BB, no. 2–07-02–22 (Figs 2B and 4C-E).
Paratype localities: CCCryo RS_0003–2004: northeastern part of Doktorbreen, Nathorst Land, Svalbard, Norway (77°34'0.001''N 16°53'59.999''E; altitude: 430 m a.s.l., collected on 6 August 2004 by Thomas Leya, sample number 013–01/04); WP123: Schwarzmoos, Kühtai, Stockach, Tyrol, Austria (47°13'35.0‘N 11°01'03.1’E; altitude: 2352 m a.s.l., collected on 25 May 2017 by Lenka Procházková & Daniel Remias, sample number WP123).
Registration: http://phycobank.org/100628
Etymology: the species epithet refers to the habitat of this species (snow).
Description: Usually smooth-walled cysts (Figs 2A and 3A), which can also be ornamented more or less regularly with nipples (Figs 2E and 3B), blunt nibs, or small or pronounced papillae. Cysts of size 7.8–39 μm in diameter, causing red snow. The astaxanthin to chlorophyll-a ratio of the red cysts ranges between 20:1 and 34:1 depending on the level of cyst maturity (Bidigare et al. 1993; Müller et al. 1998; Remias, Lütz-Meindl and Lütz 2005). At the light microscopy level usually only one cell wall layer is distinguishable in mature cysts (Fig. 2A) (using TEM two layers are distinguishable, Fig. 4B) and two cell wall layers for young cysts in early stages of development (Fig. 2B) (using TEM three layers are visible, Fig. 4E). The detailed structure of the cell wall is usually visible in TEM only; it contains a very thin trilaminar sheath formed below the transient primary cell wall during morphogenesis of the secondary cell wall. In early cyst stages a parietal chloroplast is clearly visible in LM, as are a large unpigmented vacuole and a naked pyrenoid (Fig. 2B). The temporary third outermost cell wall in TEM is fuzzy (Fig. 4E). Vegetative stages are unknown, as no culturable strains exist. Sanguina nivaloides is an extremophilic alga living in melting snowfields in polar and alpine regions.
Diagnosis:In the stage of mature cysts S. nivaloides has a single chloroplast located in the central position surrounding one single nucleus; (nearly) its entire cytoplasm appears dark red due to the secondary carotenoid astaxanthin. It differs from aplanospores of the genus Haematococcus, which have their chloroplasts localised in the interspace between oil droplets, resulting in chloroplasts with a net-like appearance (Wayama et al. 2013). It differs from Bracteacoccus engadinensis, which also accumulates astaxanthin in its multinuclear coenoblasts, but only in the central area, and its chloroplast is also composed of several (4–10) polygonal discs (Ettl and Gärtner 2014). It differs from Chlainomonas rubra, Chlainomonas kolii and Chlainomonas sp. quadriflagellates in its spherical cell shape (the latter have an ovoid cell shape) and its cell wall organisation (in C. rubra the protoplast is surrounded by a massive sheath of gelatinous material, C. kolii has a net-like outer envelope, C. sp. has a partially thickened cell wall) and in the absence of flagellar grooves in their cysts; persistant flagellar grooves were observed in the thick-walled cysts for C. kolii and C. rubra at least during cyst formation (Hoham 1974; Novis et al. 2008). Most importantly, S. nivaloides differs genetically in its 18S rRNA gene and rbcL sequences from red aplanospores of the genus Haematococcus such as Haematoccoccus lacustris (epitype, strain SAG 34–1b; 18S: AF159369.1) (Hepperle et al. 1998; Nakada and Ota 2016) and Haematococcus alpinus (strain: LCR-CC-261f, 18S: MF093734, rbcL: MF093733) (Mazumdar et al. 2018). Sanguina nivaloides differs from Sanguina aurantia in the macroscopic coloration of snowfields (the latter causes orange snow), cell size ranges (the latter is usually smaller, up to 16 µm), pigmentation (the latter has one magnitude lower level of astaxanthin to chlorophyll–a ratio in mature cysts), distribution (the latter was found in the Arctic and subarctic regions in North America to date) and physical association between cells (Sanguina nivaloides has cells that are almost always solitary, whereas Sanguina aurantia is often clustered by means of mucilaginous sheaths surrounding the cells). The genus Sanguina forms a monophyletic clade, and S. nivaloides is unique in its ITS2 rDNA sequence. For the holotype specimen CCCryo RS 0015–2010 the ITS2 rDNA sequence is characteristic (accession number MK728599).
Distribution: melting snow in Europe, Asia, Africa, Antarctica, the Arctic, North America, South America, New Zealand and Australia.
Sanguina aurantia Leya, Procházková et Nedbalová, sp. nov.
Holotype: specimen RS_0017–2010 of field sample 005/10–1b consisting of cysts in desiccated (non-viable) state in the herbarium of the Botanic Garden and Botanical Museum Berlin-Dahlem (herbarium acronym: B), Freie Universität Berlin, Königin-Luise-Straße 6–8, 14 195 Berlin, Germany, no. B 40 00 43197 [ http://herbarium.bgbm.org/object/B400043197], represented by our Fig. 5B and D.
Isotype: frozen at -80°C at DNA-bank in the herbarium of the Botanic Garden and Botanical Museum Berlin-Dahlem (herbarium acronym: B), no. B GT 00 38001 [http://herbarium.bgbm.org/object/BGT0038001].
Type locality: snowfield at the south foot of Fugleberget, Isbjörnhamna, Hornsund, Wedel Jarlsberg Land, Svalbard, Norway (77°0'42.156''N 15°32'13.2''E; altitude: 55 m a.s.l., collected on 6 August 2010 by Thomas Leya & Guntram Weithoff, sample number 005/10–1b).
Registration: http://phycobank.org/101398
Etymology: the species epithet refers to the intracellular colouration of the mature cysts (orange).
Description: Usually smooth-walled cysts (Figs 5B), sometimes the outermost layer is wrinkled and when it is decomposed, the smooth cell wall below may appear. Cysts of size 6.2–15.5 µm, causing orange snow. The astaxanthin to chlorophyll ratio of the orange cysts ranges between 0.8:1 and 2.6:1 (Müller et al. 1998). At the light microscopy level usually only one cell wall layer is distinguishable in mature cysts (Fig. 5B) (using TEM two layers are distinguishable), in some cases also a distant cell wall layer is present (Fig. 5A; in TEM three layers are visible and the outermost third layer might be multilayered, Fig. 5E). The detailed structure of the cell wall is usually visible in TEM only; it contains a very thin trilaminar sheath formed below the transient primary cell wall during morphogenesis of the secondary cell wall. Vegetative stages are unknown, as no culturable strains exist.
Diagnosis: Sanguina aurantia differs from Sanguina nivaloides in the macroscopic orange colouration of snowfields (the latter causes red snow), cell size ranges (the latter is usually larger, up to 40 µm), pigmentation (the latter has a higher level of astaxanthin to chlorophyll-a ratio in mature cysts), distribution (the latter was shown to be cosmopolitan) and physical association between cells (Sanguina aurantia is often clustered by means of a mucilaginous sheath surrounding the cells, whereas Sanguina nivaloides has cells that are almost always solitary). For the holotype specimen CCCryo RS_0017–2010 the ITS2 rDNA sequence is characteristic (accession number MK728634).
Distribution: in melting snow in subpolar and polar regions, so far the known locations are only in the northern hemisphere (North America, the Arctic).
DISCUSSION
The first collections of red snow date back to British expeditions to the far north looking for a navigable North-West passage (N.N. 1818; Bauer 1819; Ross 1819; Bauer 1820; Ross 1820). In our study many samples of red snow cysts from around the world have yielded sequences that fell in the new clade Sanguina, so one is tempted to assume that the historical sample of 1818 also probably represents the red snow-causing species S. nivaloides; however, this cannot be proven yet. A genetic comparison was not possible as the historic sample of 1818 could not be located in museums or herbaria in either London or Berlin. Intact DNA could also not be isolated from the similar sample collected during the Grinnell Arctic Expedition at Beverly Greenland [76°3' N in 1851 (Ehrenberg 1851; Leya 2008) a bit further north of the original location of 1818–1819 at the Crimson Cliffs (75°54' North, 67°15' West)]. Thus, we do not attempt to synonymise S. nivaloides with Cd. nivalis. In this study we have suggested that all our investigated red and orange spherical cysts belong to Sanguina nivaloides and Sanguina aurantia, respectively. We showed that the genus Sanguina forms one independent lineage in the Chlamydomonadaceae. The conservative nuclear marker 18S was 100% identical for these two species. Strains CCCryo 086–99, CCCryo 101–99, 133–01 and 147–01 represent the most closely related and culturable isolates/species and appear to be distributed into several lineages that are sister genera to the clade of Sanguina, some of them possibly representing the genus Achoma (Novis and Visnovsky 2012). The revised genus Chloromonas (Pröschold et al. 2001) so far contains the majority of other culturable snow algal species involved in coloured snow, but is robustly separated from the Sanguina-clade.
This study is the first record of ITS2 sequence data of spherical cysts forming red snow from Argentina and many mid-latitude alpine sites in Europe. Moreover, our data will increase the knowledge about this taxon due to several new findings on Svalbard. The ITS2 marker was previously used in this context only in studies of snow samples from the USA (Brown, Ungerer and Jumpponen 2016), Greenland, Alaska, Japan and Tajikistan (referenced in accession numbers only, see Materials and Methods) and also most recently from Antarctica and Svalbard (Segawa et al. 2018). The set of different molecular data used in this study has proved to provide valuable insights into the relationship of this taxonomically complex algal group. Haplotype networks have been used to demonstrate the intraspecific diversity (i.e. within a species) (Škaloud et al. 2015). Based on our results, S. nivaloides represents a diverse species containing 18 different ITS2 genotypes and S. aurantia has two ITS2 genotypes. According to the metagenomic study of Brown, Ungerer and Jumpponen (2016) in Colorado and Washington (USA), sequences of this locus of cysts in blooming snowfields were distributed into 30 haplotypes classified as ‘Coenochloris’ and 27 haplotypes termed as ‘Chlamydomonas’. Further samplings in other mountain regions might reveal additional unique genotypes as shown by Segawa et al. (2018). Similarly, 15 haplotypes of ITS2 rDNA were resolved for the common ‘blood rain alga’ Haematococcus lacustris (referred to as H. pluvialis in the paper cited) in temperate Europe (Allewaert et al. 2015). Intraspecific diversity within the widespread lichen photobiont Asterochloris lobophora in Europe, America and Asia accounted for 18 haplotypes of ITS2 rDNA, while half that amount of intraspecific diversity was found for other Asterochloris species (Škaloud et al. 2015).
Although the overall mean distance among the ITS2 red cysts haplotypes of S. nivaloides was rather low [p-distance of 1.7%; this value was similarly low for Asterochloris lineages described by Škaloud et al. (2015)], a divergence between haplotypes can be observed with regard to the red cysts of Sanguina nivaloides and orange cysts of S. aurantia in their variable ITS2 marker (sequence differences up to 5%), which might indicate a currently ongoing genetic diversification. Such a process was described for the soil alga Bracteacoccus bullatus by Fučíková and Lewis (2012). Further detailed pigment studies, physiological experiments with cysts, sequencing of further highly variable molecular markers (Škaloud et al. 2015) and cultivation attempts, possibly using strategies so far unconsidered, are clearly necessary to bring a richer understanding of morphological and molecular variability within the genus Sanguina.
When comparing red snow samples of Baffin Bay (Greenland) from 1851 with our recent samples, this showed that cell sizes corresponded well to the cell size ranges determined in this study (Fig. 6). Spherical red globules as well as green globules 'mostly with a small stalk and fluffy attachment, being the thallus or the root and leaf layer’ were reported from this sample [quotation translated into English from Ehrenberg (1851)]. While we could identify the former in the desiccated sample, the latter were not visible. However, we have also occasionally observed such stalks in recent samples [Fig. 2b in Leya (2008)]. We assume that they are fungal mycelia, which are not rare in field samples of red snow (Brown, Olson and Jumpponen 2015).
Cysts of Sanguina nivaloides are usually smooth-walled, and other specific variations in cell surface structure (e.g. cysts with nipples) may either reflect a level in the process of cyst maturation reached at the time of sampling or a causality between intraspecific variability and morphology (as e.g. all samples of H3 are expected to be represented by cysts with nipples, which is the case for both specimens of H3 in this study). To test this hypothesis, more specimens of the same morphologically different haplotype are needed (i.e. for H7, H14). Additionally, the observation of populations of smooth-walled cysts with the rare presence of cells with small papillae in two of our samples is in good correspondence with the findings reported from red snow in North America by Brown, Ungerer and Jumpponen (2016): they point out that very low haplotype diversity exists within the same algal bloom sampled: each algal patch was dominated by only one haplotype, with a very minor proportion of subdominant haplotypes. In the past, blooms of other snow algal taxa producing red cysts were often wrongly assigned to ‘Chlamydomonas nivalis’, e.g. most often Chlainomonas was not recognised [see Figs 23 and 24 in Stein and Amundsen (1967), in Kawecka (1981), and in Fig. 4 in Kobayashi and Fukushima (1952)]. The first comprehensive cell ultrastructure and secondary pigment profile comparisons between cysts of Chlainomonas and ‘Chlamydomonas nivalis’ were performed by Remias et al. (2016).
In our study, cell sizes of S. nivaloides red cysts correspond to the findings of Mosser et al. (1977) and Remias et al. (2005). The sizes of S. aurantia orange-coloured cysts from our study are in the range reported from Arctic pack ice floes (Gradinger and Nürnberg 1996) and from Spitsbergen (Müller et al. 1998; Leya 2004; Stibal et al. 2007). We assume that cell size can be highly plastic during their seasonal development, which would also explain the variable cell sizes among the haplotypes of red cysts we found, since our locations were sampled at different times during the snow melting season and were also varying in aspects related to their topographical properties (snowfield's slope angle, cardinal orientation of the site to the sun, elevation) and snow texture. Interestingly, the largest mean cell sizes were observed in samples collected either late in the season (e.g. samples ‘2RON’, ‘NOR4–1’) or on glacier-based snowfields (e.g. samples '4HT', 'Foxfona', 'Retten'). Indeed, cell sizes may also reflect culture conditions as in the lichen photobiont Asteriochloris (Škaloud et al. 2015). An alternative hypothesis could be that cell sizes of S. nivaloides and S. aurantia haplotypes are genetically determined with some variability. Interestingly, we observed narrow cell size variations when comparing genetically very uniform small orange cysts of S. aurantia from the different locations on Svalbard (two haplotypes only). More observations of orange cysts of S. aurantia from other geographic regions are needed to test this hypothesis, e.g. a bloom in North America was genetically identical for ITS2 (accession number KX063716; Brown, Ungerer and Jumpponen 2016) to S. aurantia orange cysts sampled in Svalbard (this study), but for the former no morphology data were collected. Interestingly, some haplotypes of S. nivaloides red cysts [i.e. H9 (‘WP123'), H18 (‘Esperanza-8')] were as small as S. aurantia orange cysts. Considering this fact, the sole parameter of cell size only should not be used for distinguishing between S. nivaloides and S. aurantia by light microscopy. Other aspects of mature cysts also should be considered such as cell wall morphology, intracellular pigmentation reflecting the astaxanthin to chlorophyll-a ratio and the macroscopic colour of the blooming snowfield.
Due to their small size (Wilkinson et al. 2012) and ability to resist drought and freezing stress, the cysts of S. nivaloides and S. aurantia are suitable propagules for long distance dispersal by prevailing air streams (Brown, Larson and Bold 1964; Schlichting Jr. 1969), birds (e.g. polar birds migrating between the two polar regions, such as the Arctic tern Sterna paradisaea) and ocean currents (Gillespie et al. 2012). Our data showed a cosmopolitan distribution of S. nivaloides in alpine and polar snowfields in both hemispheres, which supports the theory of a trans-equatorial dispersal of microbes (Hodač et al. 2016). No population structure was detected when analysing the ITS2 rDNA data, as there was no phylogeographic signal. Metagenomic analyses have shown red pigmented snow algae to be cosmopolitans based on the analysis of partial sequences of the 18S rRNA gene (Lutz et al. 2016) as well as of the ITS2 rDNA (Segawa et al. 2018). The latter study also detected endemic snow algal phylotypes being distributed in one of the two polar regions. Antarctica was regarded for a long time as a continent in biological isolation until large amounts of vascular plant pollen were collected on Signy Island (South Orkney Islands), and a parallel observation of the daily synoptic weather patterns revealed that roughly 1.5 times per year a corridor of strong winds develops between Tierra del Fuego (Argentina) and Antarctica (Marshall 1996). This explains our finding that the South American sample of S. nivaloides (‘La Hoya’, H8) was nearly identical in its ITS2 rDNA sequence with the haplotype from maritime Antarctica (H18) and with the most common haplotype in Europe (H1). The distribution of these small microorganisms could be controlled by selection due to local environmental factors as well as dispersal limitations. Moreover, a strong founder effect from only few propagules and intense kin competition is assumed to take place (Brown, Ungerer and Jumpponen 2016).
Concerning local environmental factors at the moment of sampling of encysted cells, the usually slightly acidic pH and low conductivity of snow patches with algal blooms resembled those of pristine, white snow presumably free of any algae. However, Brown and Jumpponen (2019) showed that snow algae are seemingly omnipresent, though in low abundances, at the surface of melting subpolar and alpine ‘white snow’ that are void of a visible algal bloom. Nitrate, ammonium and phosphate were not limiting nutrients for Arctic algal communities in a study by Spijkerman et al. (2012). On the other hand, atmospheric trace gases were recently recognised to play a role in supporting primary production in oligotrophic habitats in polar regions (Ji et al. 2017). None of these findings conclusively explains the patchy occurrence of red snow, which often can be observed in the same pattern year after year at the same location. Spijkerman et al. (2012) suggested that the highly patchy distribution of algal blooms in snow could be partly a result of small-scale topographical and geological characteristics influencing snowfield persistence or disappearance due to enhanced or reduced snow melt or snow accumulation. Similarly, inputs of mineral impurities can result in significantly earlier snow melt (Di Mauro et al. 2018). The slower the melting, the more prolonged is the period that is available for a snow algal population to develop. This is realised in alpine regions at higher elevations (above 1900 m a.s.l. in this study). With regards to the availability of liquid water, the algal cells seem to thrive during the growth season in the thin but persistent aqueous reservoir that by the laws of physics remains liquid around and between snow and ice crystals (Jones et al. 2001). To find a conclusive answer about which factors on a small scale actually determine the development of snow blooms such as those of S. nivaloides and S. aurantia or whether the cysts are just randomly dispersed there, a time series sampling would be necessary to observe the development of a blooming snow field while concurrently measuring various parameters.
Red spherical cysts from the European Alps morphologically identical to S. nivaloides cysts were shown to tolerate short periods of elevated UV-B radiation (Remias et al. 2010) due to the massive production of secondary carotenoids, which act mainly as shading pigments, but also as very powerful radical scavengers. This is especially true of ketocarotenoid astaxanthin, which absorbs energy in the near UV-range. Sanguinanivaloides shows high photophysiological plasticity with a highly efficient photosystem under low irradiation and no photoinhibition up to 2000 μmol m−2 s−1 (Procházková et al. 2018b). Additionally, inorganic rock dust particles covering mature cysts seem to provide additional shading protection (Lutz-Meindl and Lutz 2006). Sanguina nivaloides cysts are required to survive over summer when snow has melted, and provided they still contain chlorophyll and are not photoinhibited, the photosynthetic apparatus should continue to function (Remias, Lütz-Meindl and Lütz 2005). After total snow melt, cysts most likely ‘oversummer’ the rest of the year, generally in a dried state, on rocks, permafrost or moss-covered soil, all being subject to drastic diurnal temperature fluctuations; non-vegetated soils in particular can experience temperature changes of 30°C within 24 h (Ley, Williams and Schmidt 2004). Therefore, S. nivaloides cysts can be regarded as being eurythermic and desiccation-resistant. The question remains whether the vegetative stages (gametes or other green stages if they exist) are dependent on cold temperatures, which would characterise them as cryophiles.
Key triggers to a successful S. nivaloides cultivation have not been elucidated yet. Indeed, many green proliferating strains have been isolated from snowfields, but molecular methods and phylogeny have always shown that these isolates are taxa other than S. nivaloides. A well-known example of such a misinterpretation was strain UTEX 1969, which for years was listed as Chlamydomonas nivalis and frequently used in studies addressing cold adaptation, just as other strains erroneously were assigned to this taxon (Lu et al. 2012a,b, 2016). When, in 1990, H. Ettl and U.G. Schlösser (pers. comm.) re-identified many UTEX, CCAP and SAG strains of Chlamydomonas, they found that many were incorrectly named—among them also Cd. nivalis strains, which were first assigned to Chlamydomonas augustae, but later partly renamed again on the basis of the phylogenetic analyses as Chloromonas typhlos (Matsuzaki, Hara and Nozaki 2012) or Cr. reticulata (CCALA 753; CCALA 754). At present, it must be accepted that we do not have any living strain on hand to enable the studies on the life cycle of S. nivaloides. All diagrams describing the life cycle of this species are thus just hypothetical without any empirical proof (e.g. in terms of molecular data) (Müller, Leya and Fuhr 2001; Sattler et al. 2010; Remias 2012). Any conclusions can be made only very cautiously and indirectly based on the knowledge of life cycles of more distantly related species. The sexual life cycle of several Chlamydomonadacean species usually include the formation of zygotes into zygospores with a thick cell wall, which represents the dormant stage in the life cycle (Žárský, Kalina and Sulek 1985). After the formation and growth of the secondary cell wall, the primary cell wall of the zygospore is shed from the surface (VanWinkle‐Swift and Rickoll 2008) and the ornamented structure of the mature zygote becomes apparent (Pröschold et al. 2001). We observed a similar cell wall organisation in the ruby cysts type, which have been found so far on the Northern hemisphere in continental Norway (sample ʿN4’ and ʿNOR4–1’ in this study), in Svalbard [sample RS_0011 in this study and in Müller et al. (1998)], on Iceland (D. Remias, pers. comm.), on the western coast of Canada (L. Quarmby, pers. comm.), in Washington State in the USA (K. Thomas, pers. comm.), Oceania (P. Novis, pers. comm.), as well as in the southern hemisphere on the Antarctic King George Island (T. Leya, pers. comm.). Wille (1903) was referring to this distinctive cellular morphology in his study of Cd. nivalis from Djupvatshytten (Norway) (plate III, Figs 44 and 45). Their identity needs to be revealed by single cell sequencing. The first insights into their 18S rRNA gene phylogeny suggest that the ruby-type cysts might in fact be members of the Chloromonas-clade (Segawa et al. 2018).
The CBC species concept was successfully applied for several algal groups, e.g. snow-dwelling Chloromonas species (Matsuzaki et al. 2019). This concept can be used also in a metagenomic study: Segawa et al. (2018) showed that their ‘Chlamydomonas’ snow group B (conspecific with S. nivaloides) differs by one CBC in helix III only compared with the closely related ‘Chlamydomonas’ snow group A (see their Supplementary Figures S2 and S8). On the other hand, it was shown that the CBCs are not diagnostic in some cases at the species level and that even genera, families and orders of green algae can lack CBCs in such regions (Caisova, Marin and Melkonian 2011; Škaloud and Rindi 2013). Therefore, the presence and number of CBCs are probably direct consequences of the accumulation of mutations during the evolutionary process, simply reflecting the genetic distance among organisms. In our study, two field samples of Sanguina nivaloides (‘NOR4–1’ and ‘RS_0014–2013’) lack any CBC in their secondary structure when compared with Sanguina aurantia. Both two newly described species are closely related: they have identical 18S rRNA gene and more than 95% identical ITS2 (one CBC found). In the rbcL phylogeny, S. aurantia has been placed within Sanguina nivaloides field samples; this might be linked to unusual gene substitutions in rbcL, which was reported to result in artefacts in the phylogeny (Nozaki, Onishi and Morita 2002). Species boundaries between S. nivaloides, S. aurantia and other related species should be estimated by combination of substantial morphological, ecological and genetic difference (investigation of several molecular markers, the level of ITS2 rDNA sequence similarity should be also taken into account). S. aurantia has uniquely different ITS2 haplotypes to S. nivaloides, orange spherical cysts and red spherical cysts are clearly distinguishable, both microscopically and macroscopically, and the area of their geographic distribution is only partly overlapping. No strain for these species is available yet. According to our 18S rRNA gene and rbcL phylogenies, the closest known and culturable relatives of Sanguina nivaloides are strains CCCryo 133–01/101–99 (currently assigned to cf. Sphaerocystis sp.), strain CCCryo 147–01 (cf. Coenochloris sp.) and CCCryo 086a-99 (cf. Ploeotila sp.). Some of these relatives are known for their production of astaxanthin (Leya et al. 2009), though in less dominating amounts than in S. nivaloides, but none of them has ever been reported to cause striking snow colouring. They have been isolated from different habitats on Svalbard such as a moss and a snowfield as well as from a glacier. These strains may rather be regarded as permafrost algae than snow algae. Demchenko (2013) investigated the above strains and described them all as having coccoid cells. When studying their life cycles he observed zooid formation in strains CCCryo 101–99 and CCCryo 086b-99. Motile stages so far have not been described for the only known species of Ploeotila (P. ramosa T.Mrozinska-Webb) and since he also found slightly different characteristics in the other two strains in comparison to Sphaerocystis or Coenochloris, their attribution to these genera remains unclear (hence the ‘cf.’). Our rbcL phylogeny (Fig. 7) confirmed that these tentative genera form closely related lineages to Achoma brachiatum isolated from an alpine herbfield soil in New Zealand (Novis and Visnovsky 2012). However, as outlined above, CCCryo 101–99 and 133–01 clearly differ from strain CCCryo 147–01, the former two producing zoids and considerable amounts of secondary carotenoids and the latter lacking such cell stages and ability. Which of them actually might have to be assigned to the genus Achoma remains to be investigated. The three strains CCCryo 086a-99, 101–99 and 147–01 further markedly differ in the following aspects: cells are single or organised in clusters of tetrads in CCCryo 133–01, in microscopic colonies in an extracellular polymeric substance (EPS) in CCCryo 147–01, and in cells or a group of cells sitting on a mucilaginous stalk in CCCryo 086a-99. Chloroplasts are parietal (CCCryo 133–01, CCCryo 086a-99) and pot-like (CCCryo 147–01), and a pyrenoid is present. Yet S. nivaloides shares several features with the above mentioned three CCCryo strains: its origin from cold habitats, its parietal chloroplast with a pyrenoid (at least observable in young cysts) and its prevailing morphologic organisation as a coccoid life form.
Our study has shown that sequencing of the field-collected cysts was a suitable strategy and also the only option to decipher their phylogenetic position, as establishing actively growing isolates from them has yet to be successful. This newly described genus Sanguina with two closely related species S. nivaloides and S. aurantia represents a single monophyletic lineage, independent from other Chlamydomonadacean algae. Using molecular methods we showed that S. nivaloides has a cosmopolitan distribution in polar and alpine regions, having detected the same ITS2 haplotypes on several continents. It is possible to distinguish the cysts of S. nivaloides from red-coloured cysts of other algae based on the cell size, number of cell walls, plastid organisation and habitat preference. Further studies could involve single-cell sequencing methods, e.g. with a further focus on the ruby-type cysts. The genetic variability below species level might be detected using a microsatellite-based approach (Nagai et al. 2007) developed from single cell genomics (Muramoto et al. 2010). Additionally, sampling at known locations early in the season might help to discover vegetative stages of this species, enabling us to study this interesting alga in much more detail using viable cultures/isolates. The molecular identity of other red coccoid species from snow reported by Kol (1968) such as Chlamydomonas antarcticus (red cysts with a broad mucilage layer) or Trochiscia spp. (red cysts with spikes) might also yet be revealed.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Petr Sklenář, Marie Bulínová, Adéla Moravcová, Eva Hejduková, Martin Pusztai (all: Charles University, Prague, Czech Republic) and Daniel Remias (University of Applied Sciences Upper Austria, Wells, Austria) for field sampling. Our sincere thanks to Jasna Vukić (Charles University, Prague, Czech Republic) for her hints concerning haplotype network reconstruction. We are immensely grateful to Wolf-Henning Kusber (Botanischer Garten und Botanisches Museum Berlin, Germany) for his comments and great help with the taxonomic treatment. We also thank David Lazarus (Museum für Naturkunde, Berlin, Germany) for letting us have some of the historical red snow field sample of 1851. TL is specifically thankful to Günter R. Fuhr for having made numerous scientific voyages to Svalbard and the Antarctic and many invaluable experiences in the Arctic possible. We thank the reviewers, explicitly Phil Novis, for their constructive comments.
FUNDING
This work was supported by the Czech Science Foundation (GACR) project [18–02634S to LP and LN] and by the institutional long-term research plan RVO67985939 of the Institute of Botany of the Czech Academy of Sciences.
Conflicts of interest. None declared.
REFERENCES
- Agardh CA. Systema Algarum. Lundae. 1824;XI-XXI:13–4. [Google Scholar]
- Agardh CA. Icones Algarum Europaerum. Leipzig: Leopold Voss, 1828–35. [Google Scholar]
- anagreh L, Pegg C, Harikumar A et al.. Assessing intragenomic variation of the internal transcribed spacer two: Adapting the Illumina metagenomics protocol. PLoS One. 2017;12:e0181491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allewaert CC, Vanormelingen P, Pröschold T et al.. Species diversity in European Haematococcus pluvialis (Chlorophyceae, Volvocales). Phycologia. 2015;54:583–98. [Google Scholar]
- Andersen RA. Report of the nomenclature committee for algae: 19. Taxon. 2018;67:1029–30. [Google Scholar]
- Bauer F. Microscopical observation on the red snow. Quart J Sci Arts. 1819;7:222–9.(incl. plate VI). [Google Scholar]
- Bauer F. Red snow of Baffin's Bay. Amer J Sci Arts. 1820;2:356. [Google Scholar]
- Bidigare RR, Ondrusek ME, Kennicutt MC II et al.. Evidence for a photoprotective function for secondary carotenoids of snow algae. J Phycol. 1993;29:427–34. [Google Scholar]
- Brown RM, Larson DA, Bold HC. Airborne algae: Their abundance and heterogenity. Science. 1964;143:583–4. [DOI] [PubMed] [Google Scholar]
- Brown SP, Jumpponen A. Microbial ecology of snow reveals taxa-specific biogeographical structure. Microb Ecol. 2019;77:946–58. [DOI] [PubMed] [Google Scholar]
- Brown SP, Olson BJSC, Jumpponen A. Fungi and algae co-occur in snow: An issue of shared habitat or algal facilitation of heterotrophs?. Arctic, Antarctic, and Alpine Research. 2015;47:729–49. [Google Scholar]
- Brown SP, Ungerer MC, Jumpponen A. A community of clones: Snow algae are diverse communities of spatially structured clones. Int J Plant Sci. 2016;177:432–9. [Google Scholar]
- Caisová L, Marin B, Melkonian M. A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol Biol. 2011;11:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caisová L, Marin B, Melkonian M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist. 2013;164:482–96. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–9. [DOI] [PubMed] [Google Scholar]
- Cohn F, Chlamydococcus nivalis (F.A.Bauer) A. Braun. Rabenhorst: Algae Exsiccatae, 1861, 1141. [Google Scholar]
- Coleman AW. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist. 2000;151:1–9. [DOI] [PubMed] [Google Scholar]
- Coleman AW. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007;35:3322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czygan F-C. Blutregen und Blutschnee: Stickstoffmangel-Zellen von Haematococcus pluvialis und Chlamydomonas nivalis. Arch Mikrobiol. 1970;74:69–76. [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko E. Morphology, ecophysiology and molecular phylogeny of marine and psychrophilic green flagellates belonging to Microglena and their adaptation to changing ecological conditions. DAAD Reports. 2013;322:1–15. [Google Scholar]
- Demchenko E, Mikhailyuk T, Coleman AW et al.. Generic and species concepts in Microglena (previously the Chlamydomonas monadina group) revised using an integrative approach. Eur J Phycol. 2012;47:264–90. [Google Scholar]
- Di Mauro B, Garzonio R, Rossini M et al.. Saharan dust events in the European Alps: Role in snowmelt and geochemical characterization. The Cryosphere. 2019; 13:1147–65. [Google Scholar]
- Ehrenberg CG. Dritter Beitrag zur Erkenntnis großer Organisation in der Richtung des kleinsten Raumes. Physikalische Abhandlungen der Koeniglichen Akademie der Wissenschaften zu Berlin. 1834;1833:145–336. [Google Scholar]
- Ehrenberg CG, König. Preuss. Akad. Wiss. Berlin; Über eine frische Probe der die Crimson Cliffs scharlachroth färbenden Substanz aus der Baffins Bai und das sie begleitende kleinste Leben. Bericht über die zur Bekanntmachung geeigneten Verhandl. 1851:741–4. [Google Scholar]
- Ettl H, Gärtner G. Syllabus der Boden-, Luft- und Flechtenalgen. Springer, 2014. [Google Scholar]
- Farr ER, Zijlstra G. Index nominum genericorum (Plantarum), 1996+. http://botany.si.edu/ing/(18 March 2019, date last accessed).
- Fjerdingstad E, Kemp K, Fjerdingstad E et al.. Chemical analyses of red “snow” from East-Greenland with remarks on Chlamydomonas nivalis (Bau.) Wille. Arch Hydrobiol. 1974;73:70–83. [Google Scholar]
- Flotow J. Über Haematococcus pluvialis. Nov Act Leop Carol. 1844;20:413–606. [Google Scholar]
- Fučíková K, Lewis LA. Intersection of Chlorella, Muriella and Bracteacoccus: Resurrecting the genus Chromochloris Kol et Chodat (Chlorophyceae, Chlorophyta). Fottea. 2012;12:83–93. [Google Scholar]
- Gillespie RG, Baldwin BG, Waters JM et al.. Long-distance dispersal: A framework for hypothesis testing. Trends Ecol Evol. 2012;27:47–56. [DOI] [PubMed] [Google Scholar]
- Gradinger R, Nürnberg D. Snow algal communities on arctic pack ice floes dominated by Chlamydomonas nivalis (Bauer) Wille. Proc NIPR Symp Polar Biol. 1996;9:35–43. [Google Scholar]
- Guiry MD, Guiry GM. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway, 2018. http://www.algaebase.org (14 June 2018, date last accessed).
- Hepperle D, Nozaki H, Hohenberger S et al.. Phylogenetic position of the Phacotaceae within the Chlamydophyceae as revealed by analysis of 18s rDNA and rbcL sequences. J Mol Evol. 1998;47:420–30. [DOI] [PubMed] [Google Scholar]
- Hodač L, Hallmann C, Spitzer K et al.. Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol Ecol. 2016;92:fiw122. [DOI] [PubMed] [Google Scholar]
- Hoham RW. New findings in the life history of the snow alga, Chlainomonas rubra (Stein et Brooke) comb. nov. (Chlorophyta, Volvocales). Syesis. 1974;7:239–47. [Google Scholar]
- Hoham RW, Roemer SC, Mullet JE. The life history and ecology of the snow alga Chloromonas brevispina comb. nov. (Chlorophyta, Volvocales). Phycologia. 1979;18:55–70. [Google Scholar]
- Hooker JD. 36. Red snow. Edinb J Sci. 1825;2:184. [Google Scholar]
- Ji M, Greening C, Vanwonterghem I et al.. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature. 2017;552:400–3. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–5. [DOI] [PubMed] [Google Scholar]
- Jombart T, Pontier D, Dufour AB. Genetic markers in the playground of multivariate analysis. Heredity. 2009;102:330–41. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Dufour AB et al.. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity. 2008;101:92–103. [DOI] [PubMed] [Google Scholar]
- Jones HG, Pomeroy JW, Walker DA et al.(eds.) Snow ecology. An interdisciplinary examination of snow-covered ecosystems. Cambridge: Cambridge University Press, 2001; 378. [Google Scholar]
- Kawecka B. Biology and ecology of snow algae 2. Formation of aplanospores in Chlamydomonas nivalis (Bauer) Wille (Chlorophyta, Volvocales). Acta Hydrobiol. 1981;23:211–5. [Google Scholar]
- Kobayashi Y, Fukushima H. On the red and green snow newly found in Japan II. Bot Mag Tokyo. 1952;65:128–36. [Google Scholar]
- Koetschan C, Förster F, Keller A et al.. The ITS2 Database III-sequences and structures for phylogeny. Nucleic Acids Res. 2010;38:D275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol E. The snow and ice algae of Alaska. Smithsonian Miscellaneous Collection. 1942;101:1–36. [Google Scholar]
- Kol E. Kryobiologie: Biologie und Limnologie des Schnees und Eises, I. Kryovegetation. Die Binnengewässer Einzeldarstellungen aus der Limnologie und ihren Nachbargebieten Volume XXIV volume 24. Stuttgart: E. Schweizerbart'sche Verlagsbuchhandlung (Nägele u. Obermiller), 1968.
- Ley R, Williams MW, Schmidt SK. Microbial population dynamics in an extreme environment: controlling factors in talus soils at 3750 m in the Colorado Rocky Mountains. Biogeochemistry. 2004;68:313–35. [Google Scholar]
- Leya T. Feldstudien und genetische Untersuchungen zur Kryophilie der Schneealgen Nordwestspitzbergens. Berichte aus der Biologie. Aachen: Shaker, 2004. [Google Scholar]
- Leya T. Die „Ross-Proben“ von den Crimson Cliffs: Probe „MB_ES_1781c“ aus der Ehrenberg Sammlung des Naturkundemuseums Berlin. Berlin: Fraunhofer IBMT, 2008, 6. [Google Scholar]
- Leya T, Müller T, Ling HU et al.. Snow algae from north-western Spitsbergen (Svalbard). Reports on Polar and Marine Research. 2004;492:46–54. [Google Scholar]
- Leya T, Rahn A, Lütz C et al.. Response of arctic snow and permafrost algae to high light and nitrogen stress by changes in pigment composition and applied aspects for biotechnology. FEMS Microbiol Ecol. 2009;67:432–43. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. [DOI] [PubMed] [Google Scholar]
- Ling HU. Snow algae of the Windmill Islands, continental Antarctica: Chlorosarcina antarctica comb. nov. (Chlorophyceae, Chlorophyta) from pink snow, with discussion of Chlorosarcina and allied genera. Phycologia. 2002;41:1–9. [Google Scholar]
- Ling HU, Seppelt RD. Snow algae of the Windmill Islands, continental Antarctica. 2. Chloromonas rubroleosa sp. nov. (Volvocales, Chlorophyta), an alga of red snow. Eur J Phycol. 1993;28:77–84. [Google Scholar]
- Link DHF. 45./2. Coccophysium nivale. Handbuch zur Erkennung der nutzbarsten und am häufigsten vorkommenden Gewächse Drittel Teil. Berlin: Haude und Spenersche Buchhandlung; (S.J. Joseephy), 1833, 341–2. [Google Scholar]
- Lu N, Wei D, Chen F et al.. Lipidomic profiling reveals lipid regulation in the snow alga Chlamydomonas nivalis in response to nitrate or phosphate deprivation. Process Biochem. 2012a;48:605–13. [Google Scholar]
- Lu N, Wei D, Jiang X-L et al.. Regulation of lipid metabolism in the snow alga Chlamydomonas nivalis in response to NaCl stress: An integrated analysis by cytomic and lipidomic approaches. Process Biochem. 2012b;47:1163–70. [Google Scholar]
- Lu N, Chen JH, Wei D et al.. Global metabolic regulation of the snow alga Chlamydomonas nivalis in response to nitrate or phosphate deprivation by a metabolome profile analysis. Int J Mol Sci. 2016;17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S, Anesio AM, Raiswell R et al.. The biogeography of red snow microbiomes and their role in melting arctic glaciers. Nat Commun. 2016;7:11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütz-Meindl U, Lütz C. Analysis of element accumulation in cell wall attached and intracellular particles of snow algae by EELS and ESI. Micron. 2006;37:452–8. [DOI] [PubMed] [Google Scholar]
- Marchant HJ. Snow algae from the Australian Snowy Mountains. Phycologia. 1982;21:178–84. [Google Scholar]
- Marshall WA. Biological particles over Antarctica. Nature. 1996;383:680. [Google Scholar]
- Mataloni G, Tesolín G. A preliminary survey of cryobiontic algal communities from Cierva Point (Antarctic Peninsula). Antarct Sci. 1997;9:250–8. [Google Scholar]
- Matsuzaki R, Hara Y, Nozaki H. A taxonomic revision of Chloromonas reticulata (Volvocales, Chlorophyceae), the type species of the genus Chloromonas, based on multigene phylogeny and comparative light and electron microscopy. Phycologia. 2012;51:74–85. [Google Scholar]
- Matsuzaki R, Nozaki H, Kawachi M. Taxonomic revision of Chloromonas nivalis (Volvocales, Chlorophyceae) strains, with the new description of two snow-inhabiting Chloromonas species. PLoS One. 2018;13:e0193603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki R, Nozaki H, Takeuchi N et al.. Taxonomic re-examination of “Chloromonas nivalis (Volvocales, Chlorophyceae) zygotes” from Japan and description of C. muramotoi sp. nov. PLoS One. 2019;14:e0210986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar N, Gopalakrishnan KK, Visnovsky G et al.. A novel alpine species of Haematococcus (Chlamydomonadales: Chlorophyta) from New Zealand. N Z J Bot. 2018;56:216–26. [Google Scholar]
- Muramoto K, Nakada T, Shitara T et al.. Re-examination of the snow algal species Chloromonas miwae (Fukushima) Muramoto et al., comb. nov. (Volvocales, Chlorophyceae) from Japan, based on molecular phylogeny and cultured material. Eur J Phycol. 2010;45:27–37. [Google Scholar]
- Meneghini G. Monographia Nostochinearum italicarum addito specimine de Rivulariis. Memorie della Reale Accademia delle Scienze di Torino, ser 2. 1843;5:1–143 pls I-XVII. [Google Scholar]
- Mosser JL, Mosser AG, Brock TD. Photosynthesis in the snow: The alga Chlamydomonas nivalis (Chlorophyceae). J Phycol. 1977;13:22–7. [Google Scholar]
- Müller T, Leya T, Fuhr G. Persistent snow algal fields in Spitsbergen: Field observations and a hypothesis about the annual cell circulation. Arct Antarct Alp Res. 2001;33:42–51. [Google Scholar]
- Müller T, Bleiß W, Martin C-D et al.. Snow algae from northwest Svalbard: their identification, distribution, pigment and nutrient content. Polar Biol. 1998;20:14–32. [Google Scholar]
- Nagai S, Lian C, Yamaguchi S et al.. Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. J Phycol. 2007;43:43–54. [Google Scholar]
- N.N Captain Sir John Ross has brought from Baffin's Bay a Quantity of Red Snow. London: London Times, 1818. [Google Scholar]
- Nakada T, Ota S. What is the correct name for the type of Haematococcus Flot. (Volvocales, Chlorophyceae)?. Taxon. 2016;65:343–8. [Google Scholar]
- Nedbalová L, Mihál M, Kvíderová J et al.. Identity, ecology and ecophysiology of planktic green algae dominating in ice-covered lakes on James Ross Island (northeastern Antarctic Peninsula). Extremophiles. 2017;21:187–200. [DOI] [PubMed] [Google Scholar]
- Novakovskaya IV, Patova EN, Boldina ON et al.. Molecular phylogenetic analyses, ecology and morphological characteristics of Chloromonas reticulata (Goroschankin) Gobi which causes red blooming of snow in the subpolar Urals. Cryptogam, Algol. 2018;39:199–213. [Google Scholar]
- Novis PM. Ecology of the snow alga Chlainomonas kolii (Chlamydomonadales, Chlorophyta) in New Zealand. Phycologia. 2002;41:280–92. [Google Scholar]
- Novis PM, Visnovsky G. Novel alpine algae from New Zealand: Chlorophyta. Phytotaxa. 2012;39:1–30. [Google Scholar]
- Novis PM, Hoham RW, Beer T et al.. Two snow species of the quadriflagellate green alga Chlainomonas (Chlorophyta, Volvocales): Ultrastructure and phylogenetic position within the Chloromonas clade. J Phycol. 2008;44:1001–12. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Onishi K, Morita E. Differences in pyrenoid morphology are correlated with differences in the rbcL genes of members of the Chloromonas lineage (Volvocales, Chlorophyceae). J Mol Evol. 2002;55:414–30. [DOI] [PubMed] [Google Scholar]
- Persoon CH, Göttingen; Synopsis Methodica Fungorum. 1801;1:214. [Google Scholar]
- Perty M. Zur Kenntnis kleinster Lebensformen. Bern. 1852:1–228. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. [DOI] [PubMed] [Google Scholar]
- Procházková L, Remias D, Řezanka T et al.. Chloromonas nivalis subsp. tatrae, subsp. nov. (Chlamydomonadales, Chlorophyta): re-examination of a snow alga from the High Tatra Mountains (Slovakia). Fottea. 2018a;18:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procházková L, Remias D, Holzinger A et al.. Ecophysiological and morphological comparison of two populations of Chlainomonas sp. (Chlorophyta) causing red snow on ice-covered lakes in the High Tatras and Austrian Alps. Eur J Phycol. 2018b;53:230–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Marin B, Schlösser UG et al.. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist. 2001;152:265–300. [DOI] [PubMed] [Google Scholar]
- Remias D. Cell Structure and Physiology of Alpine Snow and Ice Algae. In: Lütz C.(ed.) Plants in Alpine Regions: Cell Physiology of Adaptation and Survival Strategies. Wien: Springer, 2012, 175–85. [Google Scholar]
- Remias D, Lütz-Meindl U, Lütz C. Photosynthesis, pigments and ultrastructure of the alpine snow alga Chlamydomonas nivalis. Eur J Phycol. 2005;40:259–68. [Google Scholar]
- Remias D, Karsten U, Lütz C et al.. Physiological and morphological processes in the Alpine snow alga Chloromonas nivalis (Chlorophyceae) during cyst formation. Protoplasma. 2010;243:73–86. [DOI] [PubMed] [Google Scholar]
- Remias D, Wastian H, Lütz C et al.. Insights into the biology and phylogeny of Chloromonas polyptera (Chlorophyta), an alga causing orange snow in Maritime Antarctica. Antarct Sci. 2013;25:648–56. [Google Scholar]
- Remias D, Pichrtová M, Pangratz M et al.. Ecophysiology, secondary pigments and ultrastructure of Chlainomonas sp. (Chlorophyta) from the European Alps compared with Chlamydomonas nivalis forming red snow. FEMS Microbiol Ecol. 2016;92:fiw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A Voyage of Discovery, Made Under the order of the Admirality, in His Majesty's Ships Isabella and Alexander for the Purpose of Exploring Baffin's Bay, and Inquiring into the Probability of a North-West Passage. London: John Murray, 1819. [Google Scholar]
- Ross J.(ed.) Entdeckungsreise unter den Befehlen der Britischen Admiraltät mit den Königlichen Schiffen Isabella und Alexander um Baffins-Bay auszuforschen und die Möglichkeit einer nordwestlichen Durchfahrt zu Untersuchen. Leipzig: Friedrich Fleischer, 1820. [Google Scholar]
- Sattler B, Remias D, Lütz C et al.. Leben auf Schnee und Eis. In: Erschbamer B, Koch EM.(eds.) Glaziale und periglaziale Lebensräume im Raum Obergurgl. Innsbruck: Innsbruck University Press iup, 2010. [Google Scholar]
- Schlichting HE., Jr The importance of airborne algae and Protozoa. J Air Pollut Control Assoc. 1969;19:946–51. [DOI] [PubMed] [Google Scholar]
- Schultz J, Wolf M. ITS2 sequence-structure analysis in phylogenetics: A how-to manual for molecular systematics. Mol Phylogen Evol. 2009;52:520–3. [DOI] [PubMed] [Google Scholar]
- Segawa T, Matsuzaki R, Takeuchi N et al.. Bipolar dispersal of red-snow algae. Nat Commun. 2018;9:3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, Müller T, Dandekar T et al.. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res Notes. 2008;1:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, Müller T, Dandekar T et al.. 4SALE - A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics. 2006;7:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuttleworth RJ. Upon the colouring matter of red snow. The Edinburgh New Philosophical Journal. 1840a;29:54–64. (with 1 coloured plate). [Google Scholar]
- Shuttleworth RJ. Nouvelles observations sur la matière colorant de la neige rouge. 1840b;25, Geneva. [Google Scholar]
- Škaloud P, Rindi F. Ecological differentiation of cryptic species within an asexual protist morphospecies: a case study of filamentous green alga Klebsormidium (Streptophyta). J Eukaryot Microbiol. 2013;60:350–62. [DOI] [PubMed] [Google Scholar]
- Škaloud P, Steinová J, Řídká T et al.. Assembling the challenging puzzle of algal biodiversity: species delimitation within the genus Asterochloris (Trebouxiophyceae, Chlorophyta). J Phycol. 2015;51:507–27. [DOI] [PubMed] [Google Scholar]
- Sommerfelt SC. Om den røde Sne, eller Sphærella nivalis Sommerf., Uredo nivalis Auct. Mag Nat videnskab. 1824;4:249–53. [Google Scholar]
- Spijkerman E, Wacker A, Weithoff G et al.. Elemental and fatty acid composition of snow algae in Arctic habitats. Front Microbiol. 2012;3:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel C. Coccochloris nivalis Spreng. Syst Veg. 1827;4:373. [Google Scholar]
- Stein JR, Amundsen CC. Studies on snow algae and fungi from the front range of Colorado. Can J Bot. 1967;45:2033–45. [Google Scholar]
- Stibal M, Elster J, Šabacká M et al.. Seasonal and diel changes in photosynthetic activity of the snow alga Chlamydomonas nivalis (Chlorophyceae) from Svalbard determined by pulse amplitude modulation fluorometry. FEMS Microbiol Ecol. 2007;59:265–73. [DOI] [PubMed] [Google Scholar]
- Sutton EA. The physiology and life histories of selected cryophytes of the Pacific Northwest Department of Oceanography volume PhD. Corvallis, OR: Oregon State University, 1970, 98. [Google Scholar]
- Terashima M, Umezawa K, Mori S et al.. Microbial community analysis of colored snow from an alpine snowfield in northern Japan reveals the prevalence of betaproteobacteria with snow algae. Front Microbiol. 2017;8:1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WH, Broady PA. Distribution of coloured snow and associated algal genera in New Zealand. N Z J Bot. 1997;35:113–7. [Google Scholar]
- VanWinkle‐Swift KP, Rickoll WL. The zygospore wall of Chlamydomonas monoica (Chlorophyceae): morphogenesis and evidence for the presence of sporopollenin. J Phycol. 2008;33:655–65. [Google Scholar]
- Vogt C. Neige rouge. In: Desor E.(ed.) Excursions et séjours dans les glaciers et les hautes régions des Alpes, de M Agassiz et de ses compagnons de voyage. Neuchatel and Paris, 1844, 215–24.(16; pl. 1: fig. 1-5 (as 'Discerea nivalis‘). [Google Scholar]
- Vukić J, Ulqini D, Šanda R. Occurrence of Knipowitschia goerneri Ahnelt, 1991 (Gobiidae) in southern Albania confirmed with molecular tools. J Appl Ichthyol. 2017;33:284–90. [Google Scholar]
- Wayama M, Ota S, Matsuura H et al.. Three-dimensional ultrastructural study of oil and astaxanthin accumulation during encystment in the green alga Haematococcus pluvialis. PLoS One. 2013;8:e53618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DM, Koumoutsaris S, Mitchell EAD et al.. Modelling the effect of size on the aerial dispersal of microorganism. J Biogeogr. 2012;39:89–97. [Google Scholar]
- Wille N. Algologische Notizen IX-XIV. Nyt Magazin for Naturvidenskaberne. 1903;41:89–185. [Google Scholar]
- Wrangel FA. Microscopiska och Physiologiska Undersökningar rörande utvecklingen af Lepraria Kermesina och dess likhet med den sa kallade röda Snön. Tilläg till Anmärkningarne rörande Byssus Jolithus Linn. Kongl Vetenskaps-Akademiens Handlingar för år 1823, Stockholm. 1823;1:71–95. [Google Scholar]
- Žárský V, Kalina T, Sulek J. Notes on the sexual reproduction of Chlamydomonas geitleri Ettl. Arch Protistenkd. 1985;130:343–53. [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.