Abstract
In our previous study, we identified differences in the levels of CDH2 and vascular endothelial growth factor (VEGF) between effective and ineffective clones of human umbilical cord blood (hUCB) mesenchymal stem cells (MSCs), with regard to the infarcted rat myocardium. In this study, we compared gene expression profiles between the effective and ineffective clones and identified that endothelin-1 (EDN1) is enriched in the effective clone. In the mechanistic analyses, EDN1 significantly increased expression of CDH2 and VEGF through endothelin receptor A (EDNRA), which was prevented by EDNRA blocker, BQ123. To decipher how EDN1 induced gene expression of CDH2, we performed a promoter activity assay and identified GATA2 and MZF1 as inducers of CDH2. EDN1 significantly enhanced the promoter activity of the CDH2 gene, which was obliterated by the deletion or point mutation at GATA2 or MZF1 binding sequence. Next, therapeutic efficacy of EDN1-priming of hUCB-MSCs was tested in a rat myocardial infarction (MI) model. EDN1-primed MSCs were superior to naive MSCs at 8 weeks after MI in improving myocardial contractility (p < 0.05), reducing fibrosis area (p < 0.05), increasing engraftment efficiency (p < 0.05), and improving capillary density (p < 0.05). In conclusion, EDN1 induces CDH2 and VEGF expression in hUCB-MSCs, leading to the improved therapeutic efficacy in rat MI, suggesting that EDN1 is a potential priming agent for MSCs in regenerative medicine.
Keywords: endothelin-1, human umbilical cord blood mesenchymal stem cell, therapeutic potency
Graphical Abstract
Introduction
Recently, human-umbilical-cord-blood-derived mesenchymal stem cells (hUCB-MSCs) have received attention for their potential application in allogeneic cell therapy.1 Protocols for isolating and expanding hUCB-MSCs have been successfully established, and hUCB-MSCs can be isolated from relatively young and healthy donors. However, despite the use of a standard isolation and expansion protocol, hUCB-MSCs from different donors vary in therapeutic efficacy when tested in a rat model of myocardial infarction (MI), suggesting that hUCB-MSCs are limited by individual variation.2
In a previous study, we compared the therapeutic efficacy of hUCB-MSCs obtained from different human donors in a rat MI model.2 We found that CDH2 is a critical marker for distinguishing the effective batch (M02 in the previous study) of hUCB-MSCs from the ineffective one (M01 in the previous study). In hUCB-MSCs, higher levels of CDH2 expression correspond to a higher expression of vascular endothelial growth factor (VEGF) and greater therapeutic efficacy; thus, it is important to ensure the presence of high levels of CDH2 and VEGF in hUCB-MSCs to be used for cell therapy.2 The ideal strategy to prime MSCs for elevating levels of CDH2 and VEGF would be small molecule, such as a peptide, which would be useful as a biomaterial component rather than artificial means, such as genetic manipulation.
In this study, we used the cDNA array data (M01 versus M02) from our previous report to identify a priming factor. From among the genes with greater than three-fold high expression in the effective batch (M02), endothelin-1 (EDN1) was identified. To examine whether EDN1 could be used as priming agent, we investigated its ability to increase the expression of CDH2 and VEGF in the ineffective batch, M01.
Mature EDN1 is a secreted peptide that comprises 21 amino acids and a potent vasoconstrictor.3 Although EDN1 is implicated in a wide variety of physiological and pathological processes in some organs, its action is unclear in others. EDN1 produces pleiotropic effects, including angiogenesis and enhanced viability of tumor cells.4, 5, 6 Both angiogenesis and cell viability are important determinants of the efficacy of stem cell therapy,7, 8 suggesting that EDN1 may be an effective priming agent for hUCB-MSCs. Moreover, to our knowledge, the potential for EDN1 to augment the therapeutic potency of human MSCs has not been reported. Therefore, in this study, we evaluated the possibility and potency of EDN1 as a priming agent for hUCB-MSCs using a rat MI model and its underlying mechanisms.
Results
Screening of Priming Factor to Improve the Regenerative Capability of hUCB-MSCs
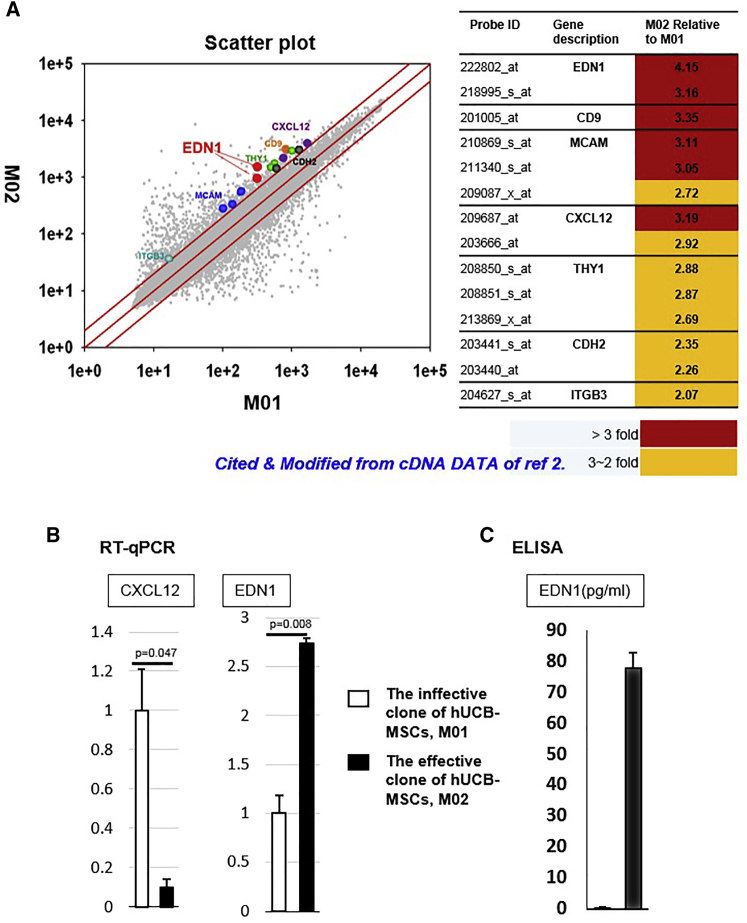
To identify factors that upregulate the therapeutic signature of hUCB-MSCs, we compared the cDNA array data obtained in our previous report from the effective batch with that from the ineffective one (M02 versus M01, respectively; Figure 1A).2 We identified seven genes related to surface and secretory factor with above two-fold higher expression in the effective batch (Figure 1A). In a previous study, we screened five surface-related genes: CD9; ITGB3; THY1; MCAM; and CDH2. Next, the RNA and protein levels of them were evaluated by qRT-PCR, fluorescence-activated cell sorting, and western blot. From these processes, CDH2 was selected to be a signature molecule for optimal hUCB-MSCs.2 However, in this study, we chose the secretory factors, EDN1 and C-X-C motif chemokine ligand 12 (CXCL12), which are feasible for application as priming factors. In the confirmation experiment using real-time PCR, expression of EDN1 mRNA was significantly higher (p = 0.008) in the effective batch, M02, whereas CXCL12 was not (Figure 1B). Identical results were obtained using ELISA (Figure 1C). Based on these results, we selected EDN1 as a potent priming factor to enhance the regenerative capability of ineffective hUCB-MSCs, M01.
Figure 1.
Screening for a Factor to Enhance the Regenerative Capability of hUCB-MSCs
(A) Scheme of comparison of the different gene expression profiles of the effective hUCB-MSCs, M02, and the ineffective ones, M01. Scatterplot of whole gene expression profiles of M02 and M01 from our previous report.2 Screened seven genes represented according to expression level of M02 relative to M01 are shown. First, surface and secretory molecules were selected and their statistically significant differences in expression were identified (p ≤ 0.05). Among them, we selected secretory factors, EDN1and CXCL12, for application as priming factors. (B) Expression of EDN1 and CXCL12 mRNA were confirmed using qRT-PCR. EDN1 mRNA showed significantly higher expression in the effective clone of hUCB-MSCs than in the ineffective one. The levels of CXCL12 mRNA did not change significantly. (C) ELISA showed higher EDN1 secretion from the effective clone.
EDN1 Is a Potent Stimulant of CDH2 and VEGF Expression
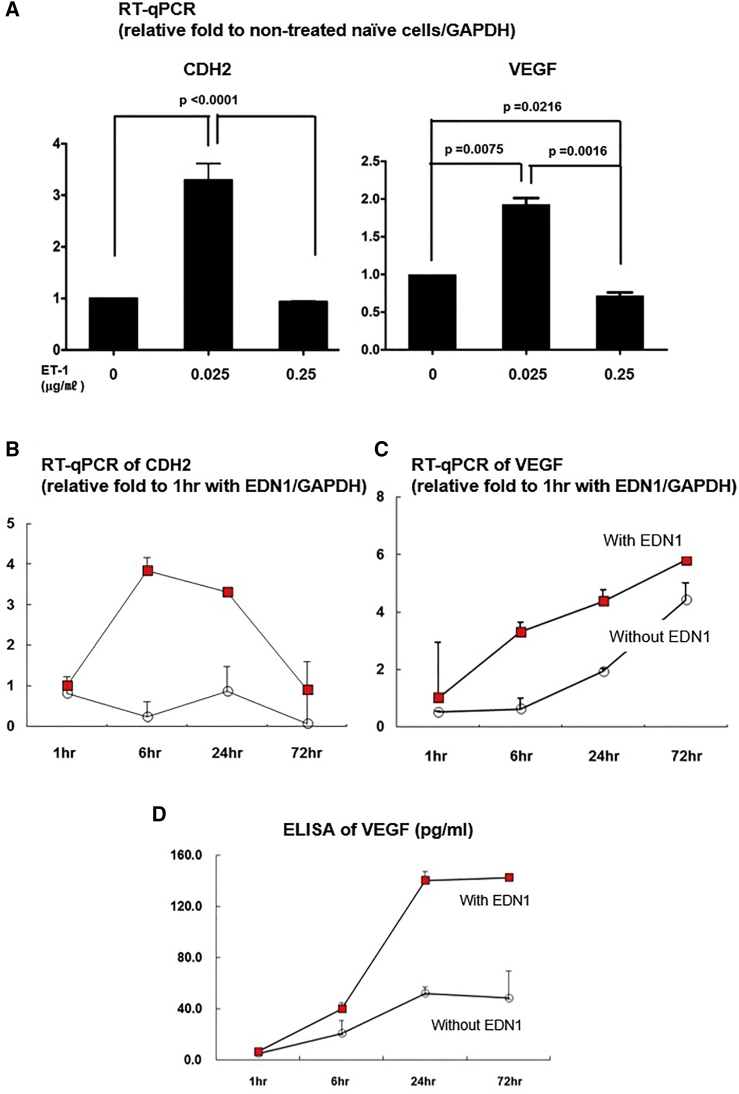
Treatment with low concentrations of a given factor is preferable for clinical applications. Therefore, to determine the optimal EDN1 concentration, hUCB-MSCs were treated for 24 h at a previously reported concentration (0.25 μg/mL) and a 10-fold lower concentration (0.025 μg/mL).9, 10 Treatment of hUCB-MSCs with 0.025 μg/mL EDN1 for 24 h resulted in a significant induction of CDH2 and VEGF, both of which are important for MSCs’ biologic effect. The dose of 0.25 μg/mL EDN1 had no significant effect (Figure 2A). Then, to optimize treatment time, we incubated the M01 hUCB-MSCs with 0.025 μg/mL EDN1 for 1, 6, 24, and 72 h and evaluated the expression of CDH2 and VEGF. Expression of CDH2 mRNA was highest at 6 h (Figure 2B), whereas that of VEGF mRNA and protein continued increasing until 24 and 72 h, respectively (Figures 2C and 2D). On the base of these results, we selected 0.025 μg/mL EDN1 for 24 h as the optimal condition of EDN1 priming to augment therapeutic efficacy via CDH2 and VEGF.
Figure 2.
EDN1 Upregulates Expression of CDH2 and VEGF
(A) qRT-PCR to determine the levels of CDH2 and VEGF mRNA. Transcription of CDH2 and VEGF increased with 0.025 μg/mL EDN1 compared with 0 and 0.25 μg/mL EDN1. (B–D) Optimization of EDN1 treatment time. (B) qRT-PCR to determine the levels of CDH2 is shown. Expression of CDH2 mRNA peaked at 6 h. (C) qRT-PCR to determine the levels of VEGF is shown. VEGF expression continues to increase from 1 to 72 h following EDN1 treatment. (D) ELISA to determine the levels of VEGF protein is shown. Secreted VEGF increased from 6 h and peaked at 24∼72 h. Taken together, the optimal condition of EDN1 treatment of cells is 0.025 μg/mL for 24 h.
EDN1 Induces CDH2 and VEGF through EDNRA
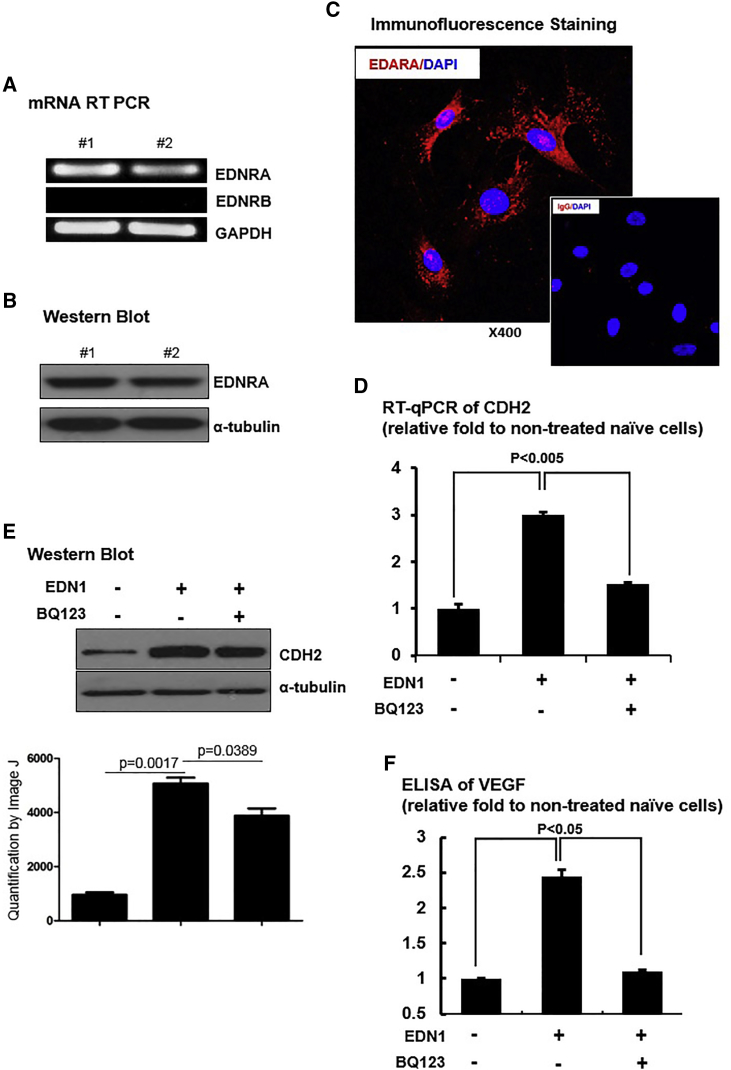
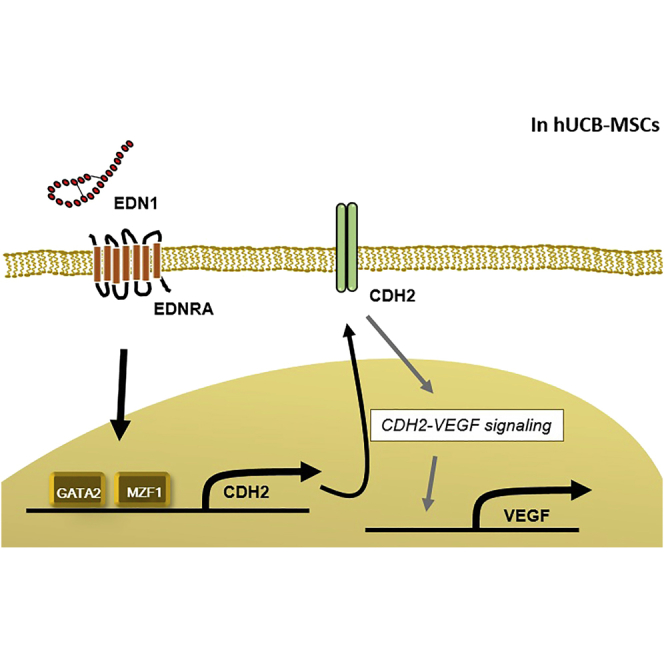
Two types of EDN1 receptors exist, endothelin receptor type A (EDNRA) and B (EDNRB), whose distribution and actions vary among different types of cells or organs. Several studies report that EDNRA and EDNRB have distinct effects through dissimilar downstream signaling pathways.11, 12, 13, 14 To identify the underlying mechanism of the effects of EDN1 on hUCB-MSCs, we first identified the EDN1 receptor expressed in hUCB-MSCs. hUCB-MSCs showed high levels of EDNRA mRNA but no expression of EDNRB mRNA (Figure 3A). Protein expression of EDNRA was also confirmed by western blot analysis and immunofluorescence staining (Figures 3B and 3C). To test whether the EDN1 priming effect is mediated through EDNRA, we treated hUCB-MSCs with a specific EDNRA blocker, BQ123, before priming with EDN1. MSCs treated with EDN1 increased CDH2 mRNA and protein expressions, which were repressed by BQ123 (Figures 3D and 3E). Secretion of VEGF also showed a response similar to that of CDH2 expression, induced by EDN1 and declined by EDNRA blocking (Figure 3F). These results suggest that EDN1 upregulates CDH2 and VEGF via mainly EDNRA in hUCB-MSCs.
Figure 3.
EDN1 Induced CDH2 and VEGF Levels through EDNRA
Expression of EDN1 receptors was assessed at the mRNA and protein levels in ineffective hUCB-MSCs. Only EDNRA was expressed in ineffective hUCB-MSCs. (A) RT-PCR to determine EDNRA and EDNRB mRNA is shown. (B) Western blot of EDNRA is shown. (C) Immunofluorescence staining of EDNRA is shown. (D) qRT-PCR to determine the levels of CDH2 mRNA is shown. (E) Western blot to determine the levels of CDH2 protein is shown. An EDNRA blocker, BQ123, suppressed upregulation of CDH2 mRNA and protein under EDN1 treatment. (F) ELISA to determine the levels of VEGF protein is shown. BQ123 reduced levels of secreted VEGF following treatment with EDN1. Overall, EDNRA mediates induction of the CDH2 and VEGF by EDN1.
EDN1 Induces CDH2 Expression via Transcription Factors, GATA2 and MZF1
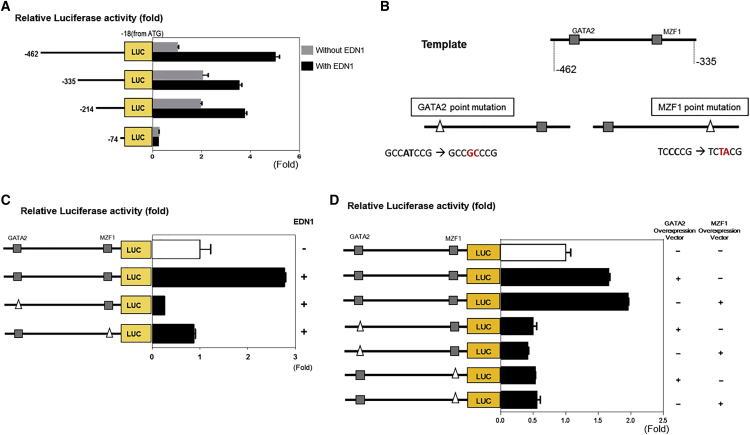
To identify the transcription factors regulating CDH2 induction by EDN1, we performed a luciferase reporter assay with a control DNA fragment of CDH2 promoter region comprising bases −462 to −18 from the ATG as well as with a series of deletion fragments (bases −335 to −18, −214 to −18, and −74 to −18).15 EDN1 significantly induced the promoter activity of the control fragment, which was retarded by deletion of the region from −462 to −335 bp (Figure 4A). Deletion from −214 to −74 bp decreased promoter activity in both the control and EDN1 treatments. These results indicate that the region between −462 and −335 bp includes the binding site of putative transcription factors that are induced by EDN1. DNA sequences from −462 to −335 bp were analyzed using the TFSEARCH V1.3 database, and putative GATA2 and MZF1 binding regions were identified. To evaluate the transcriptional activity of these sites, we prepared point mutants (Figure 4B). A point mutation in either the GATA2 (ΔGATA2) or MZF1 (ΔMZF1) binding site of the fragment significantly blocked the EDN1 effect on promoter activity (Figure 4C). To determine the biological relevance of the transcriptional activity of GATA2 and MZF1 in this region, we used GATA2 and MZF1 expression vectors (Figure 4D). The promoter activity of the −462- to −335-bp region was increased with co-expression of GATA2 or MZF1, whereas no response was observed in each mutant form, even in the presence of co-expression of GATA2 or MZF1, respectively (Figure 4E). Interestingly, luciferase activity was not turned on by MZF1 transfection in the ΔGATA2 mutant (ΔGATA2-MZF1), even with normal MZF1 site. Similarly, it was not turned on by GATA2 transfection in the ΔMZF1 mutant (ΔMZF1-GATA2), even with normal GATA2 site (Figure 4E). These results suggest that GATA2 and MZF1 in combination upregulate the transcriptional activity of the CDH2 promoter, and functional interaction between the two factors may be important.
Figure 4.
GATA2 and MZF1 Are Transcription Factors of CDH2 Induction by EDN1
(A) Luciferase reporter assay to determine activity of the CDH2 promoter region (the −462- to −18-bp fragment from the ATG) and fragments of the promoter sequence (bases −335 to −18, −214 to −18, and −74 to −18). EDN1 significantly enhanced the promoter activity (−462- to −18-bp fragment; p < 0.05). Deletion of the sequence between −462 and −335 bp retarded this effect (−462 to −18 bp versus −335 to −18; p < 0.05). (B) The sequence from −462 to −335 bp contained one GATA2 and one MZF1 binding sequence. Putative GATA2 and MZF1 binding regions were identified using the TFSEARCH database v1.3, and point mutagenesis was carried out. (C) Luciferase reporter assay to determine the promoter activity of the −462- to −335-bp fragment with and without point mutations. The promoter activity of the −462- to −335-bp region was increased by EDN1 treatment (p < 0.05) but was obliterated by point mutations at either GATA2 or MZF1 site (p < 0.05). (D) Luciferase reporter assay to determine the promoter activity of the −462- to −335-bp fragment with and without point mutations in cells overexpressing GATA2 or MZF1 is shown. The promoter activity of the −462- to −335-bp region was increased by co-expression of GATA2 or MZF1 (p < 0.05). Reduced activity was observed in each point mutant (ΔGATA2-MZF1 or ΔMZF1-GATA2; no significant difference).
EDN1 Treatment Enhances the Therapeutic Efficacy of hUCB-MSCs
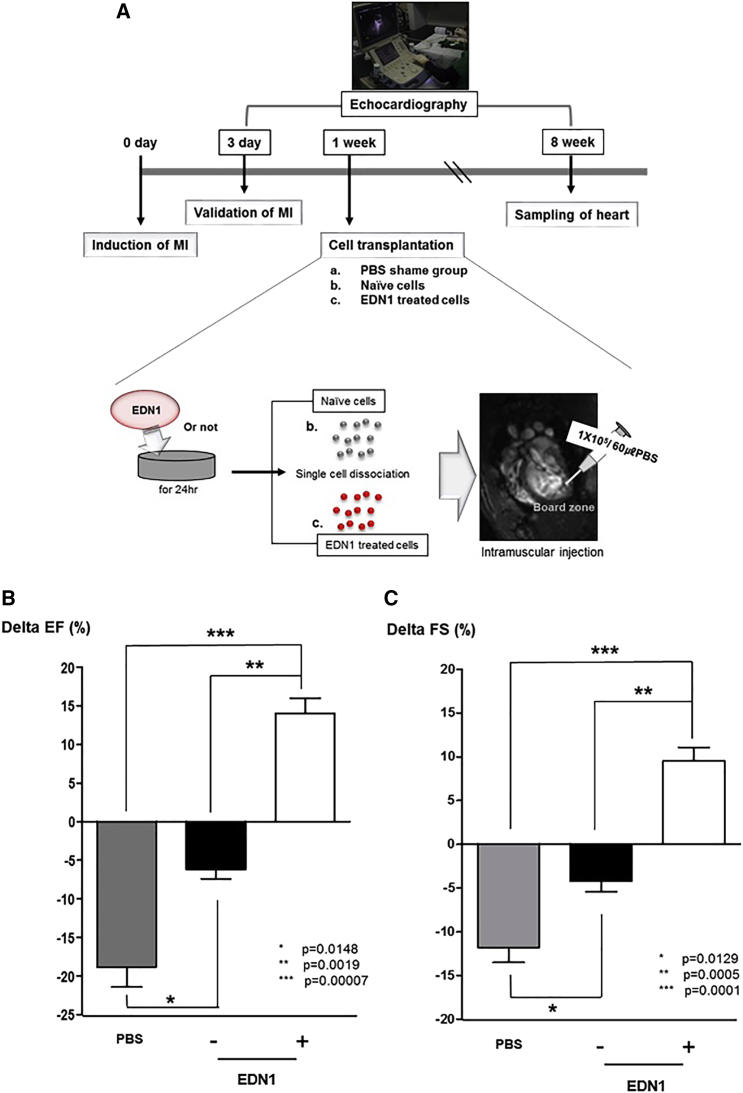
In order to confirm the priming effect of EDN1 on the therapeutic efficacy of hUCB-MSCs, we implanted 1 × 105 hUCB-MSCs to the infarcted myocardium of rats at 1 week after MI (Figure 5A). 8 weeks after induction of MI, the left ventricle ejection fraction (EF) of control rats treated with PBS was significantly decreased (ΔEF = −18.81% ± 2.60% in PBS; Figure 5B). Such a reduction in EF was remarkably prevented by transplantation of naive hUCB-MSCs (ΔEF = −6.18% ± 1.20% in naive MSCs; Figure 5B). However, the implantation of EDN1-treated hUCB-MSC increased EF compared with the baseline EF, 3 days after MI (ΔEF = 14.02% ± 2.00% in EDN1-primed MSCs; Figure 5B). There were significant differences in the change of EF among the three groups (Figure 5B; p = 0.015 for PBS versus naive MSCs, p = 0.002 for naive MSCs versus EDN1-primed MSCs, and p = 0.0001 for PBS versus EDN1-primed MSCs). Similar results were obtained regarding left ventricle fractional shortening (FS), indicating that EDN1 group had significantly improved FS compared to the control MSCs- or PBS-injected group (ΔFS = −11.78% ± 1.72% in PBS, −4.20% ± 1.23% in naive MSCs, and 9.52% ± 1.57% in EDN1-primed MSCs; p = 0.013 for PBS versus naive MSCs, p = 0.001 for naive MSCs versus EDN1, and p = 0.0001 for PBS versus EDN1; Figure 5C).
Figure 5.
Transplantation of EDN1-Primed hUCB-MSCs Improved Myocardial Regeneration in a Rat Myocardial Infarction (MI) Model
(A) Scheme for cell transplantation in MI. EDN1-primed and naive ineffective hUCB-MSCs were transplanted in infarcted heart at 1 week after induction of MI. (B) 8 weeks after induction of MI, delta EF in the EDN1-treated hUCB-MSC group was improved significantly compared to that in the naive hUCB-MSCs or PBS-injected groups (p < 0.05). (C) 8 weeks after induction of MI, delta FS was significantly improved in the EDN1 group compared to the control MSCs or PBS-injected groups (p < 0.05). *p = 0.01, PBS versus naive MSCs; **p = 0.001, naive MSCs versus EDN1-primed MSCs; and ***p = 0.00007, PBS versus EDN1-primed MSCs in delta EF. *p = 0.01, PBS versus naive MSCs; **p = 0.0005, naive MSCs versus EDN1-primed MSCs; and ***p = 0.0001, PBS versus EDN1 primed MSCs in delta FS (n = 7 in each group).
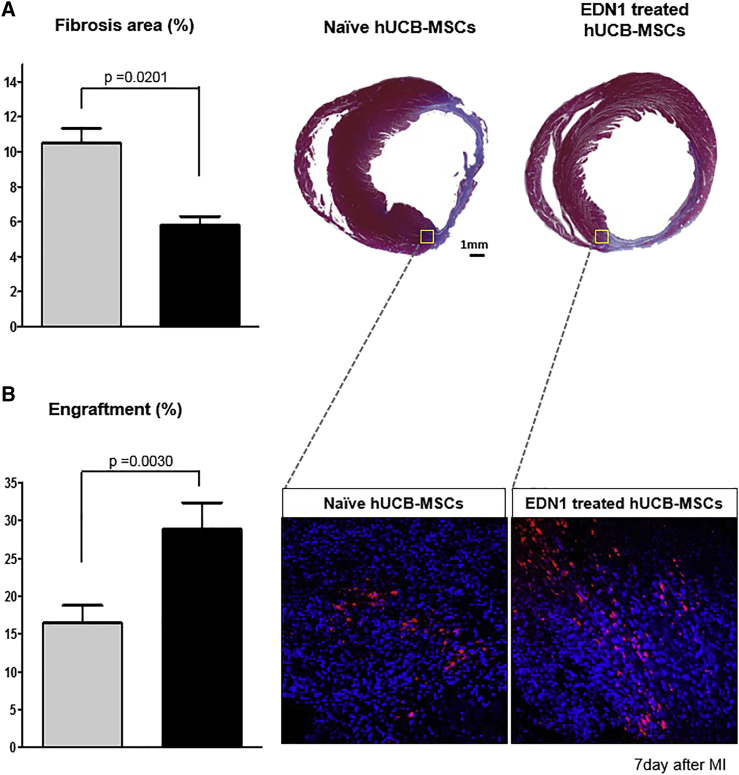
In histological analysis of the heart, Masson’s trichrome staining showed a significantly smaller fibrosis area in the EDN1-primed MSCs group than that observed in the naive MSCs group (5.81% ± 0.49% versus 10.50% ± 0.84%; p = 0.02; Figure 6A). In the analysis of engraftment efficiency, the implanted DiI-labeled hUCB-MSCs were detected more frequently in the EDN1-primed MSCs group than in the naive MSCs group (28.81% ± 3.56% versus 16.44% ± 2.37%; p = 0.003; Figure 6B). These results suggest that EDN1 priming augments hUCB-MSCs viability and engraftment in the infarcted myocardium.
Figure 6.
EDN1-Primed hUCB-MSCs Survive Longer after Implantation and Reduce Infarct Scar in the Rat MI Model
(A) Masson’s trichrome staining of infarcted rat hearts and quantitative analysis of the fibrosis area (blue). The fibrosis area was significantly reduced in the EDN1-treated hUCB-MSC group compared with the naive hUCB-MSCs group (p = 0.02). (B) Analysis of the engraftment using DiI labeling of transplanted hUCB-MSCs is shown. The intensity of DiI-labeled cells was higher after transplantation of EDN1-treated hUCB-MSCs than after transplantation of naive hUCB-MSCs (p = 0.003).
Discussion
EDN1 Is Not Only Vasoactive Peptide but Also Regulator of Tumor or Stem Cells
In the earlier era of research, EDN1 was established as a key molecule regulating vascular constriction and cardiovascular hypertrophy. Later, EDN1 was reported to affect tumor cell proliferation and survival.4 Furthermore, EDN1 was reported to affect cell differentiation, tissue generation, and epithelial-mesenchymal transition (EMT)16 as well as angiogenesis6 and lymphangiogenesis.17 We hypothesized that effective factors in tumor cells like EDN1 could also be effective in stem cells because of the examples of FGF2 and activin that promote angiogenesis and proliferation in tumor cells18 as well as control stemness and differentiation of human cells.19, 20 Therefore, we selected the small peptide EDN1 as a priming agent for MSCs. Moreover, small peptides have the benefits of cost effectiveness, immunity, and biological specificity compared to large-size proteins or therapeutic genes.21 Our results herein show that EDN1 improves the therapeutic efficacy of hUCB-MSCs. Therefore, EDN1 is a “double-edged sword” that shows completely different results depending on the context where it is applied.
GATA2 and MZF1 Are Key Transcription Factors for Induction of CDH2 by EDN1
GATA2 and MZF1 are transcription factors that contain zinc finger motifs on their DNA binding domain.22, 23 In this study, we established that GATA2 and MZF1 played a significant role in CDH2 induction by EDN1. Although GATA2 and MZF1 could interact with several binding sequences in the putative CDH2 promoter region, we found that one binding sequence for each factor in the region from −462 to −335 bp was essential for mediating the priming effect of EDN1 on CDH2 expression. The specific role of GATA2 and MZF1 binding element in the region from −462 to −335 bp in mediating the EDN1 priming effect was well confirmed by two separate experiments: reduction of promoter activity by point mutation in the binding elements and induction of promoter activity by overexpression of GATA2 and MZF1 transcription factors.
Consequently, binding of GATA2 and MZF1 to this promoter sequence was the mechanism by which EDN1 increased CDH2, leading to VEGF secretion and the enhanced therapeutic efficacy of hUCB-MSCs in MI.
Feasibility of Application of EDN1 as a Priming Agent for hUCB-MSCs
Other methods for priming hUCB-MSCs to induce the expression of CDH2 and VEGF without EDN1 treatment would be overexpression of either CDH2 or VEGF. However, these methods require genetic manipulation or viral vectors that may not be safe in clinical applications. Therefore, priming hUCB-MSCs using EDN1 is advantageous because it is performed without genetic manipulation or viral vectors. Furthermore, priming can be simply achieved using EDN1 at low concentrations and over a relatively short treatment time.
In conclusion, EDN1 could be a promising priming agent for improving therapeutic efficacy of human MSCs in cell therapy. We identified a new mechanism by which EDN1 could regulate the biology of MSCs; it activates GATA2 and MZF1 transcription factors to increase CDH2 expression and VEGF secretion, leading to augmentation of the therapeutic efficacy of MSCs. In this context, our results may be helpful for future studies on the use of EDN1 in clinical research and trials.
Materials and Methods
cDNA Microarray
Total RNAs from hUCB-MSCs (M01 or M02) were isolated using TRIzol (Invitrogen) and the RNeasy Mini kit (QIAGEN). The purity and integrity of the total RNA were checked by Bioanalyzer (Agilent). Probe synthesis for total RNA, hybridization, detection, and scanning were performed according to the manufacturer’s protocols (Affymetrix). Briefly, cDNA was synthesized from total RNA using the One-Cycle cDNA Synthesis Kit. After clean up with a Sample Cleanup Module, double-strand cDNA was used for in vitro transcription (IVT). cDNA was transcribed using the GeneChip IVT Labeling Kit, in the presence of biotin-labeled CTP and UTP. After clean up with a Sample Cleanup Module, labeled cRNA was fragmented in fragmentation buffer. Fragmented cRNA was hybridized to the Affymetrix Human Genome U133 Plus 2.0 Array. After hybridization, the arrays were washed in a GeneChip Fluidics Station 450 with wash buffer and were stained with a streptavidin-phycoerythrin complex. Staining intensities were determined with a GeneChip scanner 3000 using GCOS Affymetrix software. As a normalization process, RMA (robust multi-array average) normalization was applied to remove systematic variance. By applying RMA, the raw intensity values are background corrected, log2 transformed, and then quantile normalized. We identified genes that were differentially expressed using t test (p < 0.05) among triplicated experiments.
Culture and Treatment of hUCB-MSCs
hUCB-MSCs (Medipost) were cultured in α-minimum essential medium (α-MEM) (Gibco-BRL) containing 10% fetal bovine serum (FBS) (Gibco-BRL), 10,000 units/mL penicillin G sodium, and 10,000 μg/mL streptomycin sulfate (InvivoGen, San Diego, CA, USA). We used hUCB-MSCs from the seventh or eighth passage in this study. For mechanistic analysis, hUCB-MSCs were treated with 0.025 or 0.25 μg/mL EDN1 (Sigma, St. Louis, MO, USA) for 1, 6, 24, and 72 h and 10 nM EDNRA blocker BQ123 (Sigma) for 1 h before treatment with 0.025 μg/mL EDN1 for 24 h. To study the therapeutic efficacy, hUCB-MSCs were treated with 0.025 μg/mL EDN1 for 24 h and then prepared as a single-cell suspension for the in vivo experiments.
qRT-PCR Analysis
Total RNA from the cultured hUCB-MSCs was extracted using the RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions. The cDNA was synthesized from approximately 1 mg of total RNA using the PrimeScript 1st strand cDNA Synthesis kit (Takara, Tokyo, Japan) and subjected to PCR amplification with specific primers. The reactions were performed using the ABI Prism 7000 sequence detection system (Applied Biosystems) and SYBR Green I dye (Sigma). GAPDH was simultaneously run as a control and used for normalization. Control wells without cDNA were included as negative controls. Each test sample was run in triplicate. The results are reported as the relative expression after normalization of the transcript amount to the endogenous control using the 2−ΔΔCT method.24 Threshold cycle (CT) indicates the fractional cycle number at which the amount of amplified target reaches a fixed threshold. The qRT-PCR primer sequences were as follows: EDN1, forward: 5′-gctcgtccctgatggataaa-3′, reverse: 5′-ttcctgcttggcaaaaattc-3′; EDNRA, forward: 5′-cttgcccttggagaccttatct-3′, reverse: 5′-aacactaagagcgcagaggttg-3′; EDNRB, forward: 5′-gataatgacgccacccactaag-3′, reverse: 5′-gaacacaaggcaggacacaac-3′; CDH2, forward: 5′-gacaatgcccctcaagtgtt-3′, reverse: 5′-ccattaagccgagtgatggt-3′; VEGF, forward: 5′-gggcagaatcatcacgaagt-3′, reverse: 5′-tggtgatgttggactcctca-3′; and GAPDH, forward: 5′-tgtgaggaggggagattca-3′, reverse: 5′-caacgaatttggctacagca-3′.
Western Blot Analysis
Lysis buffer containing 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1% NP 40, 0.1% SDS, and 0.5% deoxycholate and Protease Inhibitor Cocktail (Roche, Indianapolis, IN, USA) was used to prepare all cell protein lysates. Total protein lysates (25–30 μg) were electrophoresed in SDS–polyacrylamide gel and immunoblotted with an anti-EDNRA antibody (1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or an anti-CDH2 antibody (1:1,000; Santa Cruz Biotechnology). A primary antibody against α-tubulin (1:5,000; Sigma) was used as an internal control.
ELISA Assay
Supernatant were collected at 1, 6, 24, and 72 h from monolayer cultured hUCB-MSCs to analysis the levels of VEGF or at 24 h for analyzing EDN1 levels. The levels of secreted VEGF and EDN1 were measured by VEGF ELISA (R&D Systems, Minneapolis, MN, USA) or Endothelin ELISA (Invitrogen) according to the manufacturer’s protocols. The absorbance at 450 nm was measured using ELISA reader (VERSAmax; AccuScan Instruments, Columbus, OH, USA).
Luciferase Assay and Site-Directed Mutagenesis
CDH2 promoter activity was determined using fragments encoding the human CDH2 5′ promoter in pGL3basic (fragments: −462 to +18, −335 to −18, −214 to −18, and −74 to −18 bp). A QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA) was used to achieve point mutations of specific DNA sequences for investigation of GATA2 and MZF1 binding to the CDH2 promoter region. The following mutation primers were used: GATA2 binding site mutation sequence: 5′-gctcttggggagcgccgcccgctccacttccacc-3′; MZF1 binding site mutation sequence: 5′-tccaccggccaaggtctacgccgctgcatccctc-3′. The identities of the constructs and mutants were examined by sequencing.
Transfection was performed using Metafectene-Pro (Biontex, Planegg, Germany) in α-MEM without FBS.
To assay the promoter activity in the presence of transcription factors, pGL3 plasmids containing CDH2 promoter fragment (100 ng of each and 10 ng Renilla) were cotransfected with 500 ng of GATA2 (pCMV-GATA2; OriGene) or MZF1 (pCMV-MZF1; OriGene) expression vectors. Before vector transfection, we validated vector expression by qRT-PCR.
Luciferase analysis was performed following the procedure recommended by the company with GLOMAX system 20/20 (Promega).
Immunofluorescence Staining
Sections of rat infarcted myocardium were de-paraffinized and subjected to antigen retrieval with citrate buffer (Dako, Glostrup, Denmark). Blocking was conducted with 0.1% BSA and 0.01% Triton X-100 in PBS. Subsequently, sections were incubated overnight at 4°C with lectin from Bandeiraea simplicifolia tetramethylrhodamine isothiocyanate (TRITC)-conjugated antibody (1:100; Sigma-Aldrich). After three washes, tissue sections were mounted using a fluorescent mounting medium with DAPI (IHC World, Woodstock, NY). To calculate the fibrosis area of infarcted regions, sections were stained with H&E and Masson’s trichrome. The SABIA software program (Metoosoft, Seoul, Korea) was used for quantitative analysis of the fibrosis area following Masson’s trichrome staining. The engraftment percentage of DiI-stained injected cells was observed 7 days after cell transplantation in the heart and calculated by ImageJ (https://imagej.nih.gov/ij/).
Rat MI Model
All animal experimental procedures were performed using protocols approved by the “Guide for the Care and Use of Laboratory Animals” announced by the NIH, USA, and by the Experimental Animal Committee of Clinical Research Institute, Seoul National University Hospital, Seoul, Korea.
Male Sprague-Dawley rats (8–10 weeks old; 240–290 g) were used in our experiments. Rats were anesthetized with 50 mg/kg Zoletil (Virbac, Carros, France) and Rompun (Bayer, Leverkusen, Germany) by intraperitoneal injection and then fixed in the supine position. A left thoracotomy was conducted in the fourth intercostal space, and the pericardium was opened. Initiation of MI was performed by making a 3- or 4-cm incision along the left side of the sternum. Ligation of the main trunk of the left coronary arteries was performed with 6-0 silk sutures. After ligation of the proximal left artery descending (LAD), an irreversible pale area was demarcated on the surface of the middle and apical portion of the left ventricle (LV), and the chest was then closed in layers.
To validate the MI state in rats, an echocardiographic study (Vivid I; GE Healthcare) was performed 4 days before and 7 weeks after hUCB-MSC transplantation using an 11.5-MHz transducer. LV end-diastolic and end-systolic dimensions (LVEDDs and LVESDs, respectively) were measured according to the leading-edge method of the American Society of Echocardiography. To avoid investigators’ bias, the operator for echocardiography involved in the experiment was blinded to the assigned treatment. The LVFS was calculated as 100 × (LVEDD − LVESD)/LVEDD. The LVEF was automatically calculated by the built-in software.2, 7, 25
hUCB-MSC Transplantation
Rats were randomly assigned into groups 2 days after MI induction, a PBS-injected control group and hUCB-MSC injected groups, anesthetized, and ventilated; the chests were opened using the protocol described for the initial surgery. We implanted the minimal effective dose (1 × 105; 60 μL) of cells from each hUCB-MSC group, as determined from our previous study.2 hUCB-MSCs were injected into three equidistant peri-infarct zones.
To quantify transplanted cells, hUCB-MSCs were stained with Cell Tracker CM-DiI (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocol before transplantation.
Statistical Analysis
GraphPad Prism 5 (GraphPad, La Jolla, CA, USA) was used to perform statistical analyses. Results are expressed as the mean ± SEM. t test for two samples of values, ANOVA for more comparison between more than two samples of values. p < 0.05 was regarded as statistically significant.
Author Contributions
Conceptualization: E.J.L. and H.-S.K. Methodology: E.J.L., I.H., G.-H.K., D.M., S.Y.K., I.-C.H., and P.J.M. Data analysis: E.J.L., S.-Y.L., and H.-S.K. Writing of the original manuscript: E.J.L., I.W., and G.-H.K. Review and editing of the manuscript: E.J.L. and H.-S.K.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by a grant of the Korea Health Technology R&D Project Strategic Center of Cell and Bio Therapy (grant number: HI17C2085) and Korea Research-Driven Hospital (grant number: HI14C1277) through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (MHW), Republic of Korea. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chang S.A., Lee E.J., Kang H.J., Zhang S.Y., Kim J.H., Li L., Youn S.W., Lee C.S., Kim K.H., Won J.Y. Impact of myocardial infarct proteins and oscillating pressure on the differentiation of mesenchymal stem cells: effect of acute myocardial infarction on stem cell differentiation. Stem Cells. 2008;26:1901–1912. doi: 10.1634/stemcells.2007-0708. [DOI] [PubMed] [Google Scholar]; Chang, S.A., Lee, E.J., Kang, H.J., Zhang, S.Y., Kim, J.H., Li, L., Youn, S.W., Lee, C.S., Kim, K.H., Won, J.Y., et al. (2008). Impact of myocardial infarct proteins and oscillating pressure on the differentiation of mesenchymal stem cells: effect of acute myocardial infarction on stem cell differentiation. Stem Cells 26, 1901-1912. [DOI] [PubMed]
- 2.Lee E.J., Choi E.-K., Kang S.K., Kim G.-H., Park J.Y., Kang H.-J., Lee S.-W., Kim K.-H., Kwon J.S., Lee K.H. N-cadherin determines individual variations in the therapeutic efficacy of human umbilical cord blood-derived mesenchymal stem cells in a rat model of myocardial infarction. Mol. Ther. 2012;20:155–167. doi: 10.1038/mt.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, E.J., Choi, E.-K., Kang, S.K., Kim, G.-H., Park, J.Y., Kang, H.-J., Lee, S.-W., Kim, K.-H., Kwon, J.S., Lee, K.H., et al. (2012). N-cadherin determines individual variations in the therapeutic efficacy of human umbilical cord blood-derived mesenchymal stem cells in a rat model of myocardial infarction. Mol. Ther. 20, 155-167. [DOI] [PMC free article] [PubMed]
- 3.Yanagisawa M., Masaki T. Molecular biology and biochemistry of the endothelins. Trends Pharmacol. Sci. 1989;10:374–378. doi: 10.1016/0165-6147(89)90011-4. [DOI] [PubMed] [Google Scholar]; Yanagisawa, M., and Masaki, T. (1989). Molecular biology and biochemistry of the endothelins. Trends Pharmacol. Sci. 10, 374-378. [DOI] [PubMed]
- 4.Del Bufalo D., Di Castro V., Biroccio A., Varmi M., Salani D., Rosanò L., Trisciuoglio D., Spinella F., Bagnato A. Endothelin-1 protects ovarian carcinoma cells against paclitaxel-induced apoptosis: requirement for Akt activation. Mol. Pharmacol. 2002;61:524–532. doi: 10.1124/mol.61.3.524. [DOI] [PubMed] [Google Scholar]; Del Bufalo, D., Di Castro, V., Biroccio, A., Varmi, M., Salani, D., Rosano, L., Trisciuoglio, D., Spinella, F., and Bagnato, A. (2002). Endothelin-1 protects ovarian carcinoma cells against paclitaxel-induced apoptosis: requirement for Akt activation. Mol. Pharmacol. 61, 524-532. [DOI] [PubMed]
- 5.Nelson J.B., Udan M.S., Guruli G., Pflug B.R. Endothelin-1 inhibits apoptosis in prostate cancer. Neoplasia. 2005;7:631–637. doi: 10.1593/neo.04787. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nelson, J.B., Udan, M.S., Guruli, G., and Pflug, B.R. (2005). Endothelin-1 inhibits apoptosis in prostate cancer. Neoplasia 7, 631-637. [DOI] [PMC free article] [PubMed]
- 6.Wu M.H., Huang C.Y., Lin J.A., Wang S.W., Peng C.Y., Cheng H.C., Tang C.H. Endothelin-1 promotes vascular endothelial growth factor-dependent angiogenesis in human chondrosarcoma cells. Oncogene. 2014;33:1725–1735. doi: 10.1038/onc.2013.109. [DOI] [PubMed] [Google Scholar]; Wu, M.H., Huang, C.Y., Lin, J.A., Wang, S.W., Peng, C.Y., Cheng, H.C., and Tang, C.H. (2014). Endothelin-1 promotes vascular endothelial growth factor-dependent angiogenesis in human chondrosarcoma cells. Oncogene 33, 1725-1735. [DOI] [PubMed]
- 7.Lee E.J., Park S.J., Kang S.K., Kim G.H., Kang H.J., Lee S.W., Jeon H.B., Kim H.S. Spherical bullet formation via E-cadherin promotes therapeutic potency of mesenchymal stem cells derived from human umbilical cord blood for myocardial infarction. Mol. Ther. 2012;20:1424–1433. doi: 10.1038/mt.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, E.J., Park, S.J., Kang, S.K., Kim, G.H., Kang, H.J., Lee, S.W., Jeon, H.B., and Kim, H.S. (2012). Spherical bullet formation via E-cadherin promotes therapeutic potency of mesenchymal stem cells derived from human umbilical cord blood for myocardial infarction. Mol. Ther. 20, 1424-1433. [DOI] [PMC free article] [PubMed]
- 8.Lee E.J., Hwang I., Lee J.Y., Park J.N., Kim K.C., Kim G.H., Kang C.M., Kim I., Lee S.Y., Kim H.S. Hepatocyte growth factor improves the therapeutic efficacy of human bone marrow mesenchymal stem cells via RAD51. Mol. Ther. 2018;26:845–859. doi: 10.1016/j.ymthe.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, E.J., Hwang, I., Lee, J.Y., Park, J.N., Kim, K.C., Kim, G.H., Kang, C.M., Kim, I., Lee, S.Y., and Kim, H.S. (2018). Hepatocyte growth factor improves the therapeutic efficacy of human bone marrow mesenchymal stem cells via RAD51. Mol. Ther. 26, 845-859. [DOI] [PMC free article] [PubMed]
- 9.Morini M., Astigiano S., Gitton Y., Emionite L., Mirisola V., Levi G., Barbieri O. Mutually exclusive expression of DLX2 and DLX5/6 is associated with the metastatic potential of the human breast cancer cell line MDA-MB-231. BMC Cancer. 2010;10:649. doi: 10.1186/1471-2407-10-649. [DOI] [PMC free article] [PubMed] [Google Scholar]; Morini, M., Astigiano, S., Gitton, Y., Emionite, L., Mirisola, V., Levi, G., and Barbieri, O. (2010). Mutually exclusive expression of DLX2 and DLX5/6 is associated with the metastatic potential of the human breast cancer cell line MDA-MB-231. BMC Cancer 10, 649. [DOI] [PMC free article] [PubMed]
- 10.Spinella F., Caprara V., Di Castro V., Rosanò L., Cianfrocca R., Natali P.G., Bagnato A. Endothelin-1 induces the transactivation of vascular endothelial growth factor receptor-3 and modulates cell migration and vasculogenic mimicry in melanoma cells. J. Mol. Med. (Berl.) 2013;91:395–405. doi: 10.1007/s00109-012-0956-2. [DOI] [PubMed] [Google Scholar]; Spinella, F., Caprara, V., Di Castro, V., Rosano, L., Cianfrocca, R., Natali, P.G., and Bagnato, A. (2013). Endothelin-1 induces the transactivation of vascular endothelial growth factor receptor-3 and modulates cell migration and vasculogenic mimicry in melanoma cells. J. Mol. Med. (Berl.) 91, 395-405. [DOI] [PubMed]
- 11.Schneider M.P., Boesen E.I., Pollock D.M. Contrasting actions of endothelin ET(A) and ET(B) receptors in cardiovascular disease. Annu. Rev. Pharmacol. Toxicol. 2007;47:731–759. doi: 10.1146/annurev.pharmtox.47.120505.105134. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schneider, M.P., Boesen, E.I., and Pollock, D.M. (2007). Contrasting actions of endothelin ET(A) and ET(B) receptors in cardiovascular disease. Annu. Rev. Pharmacol. Toxicol. 47, 731-759. [DOI] [PMC free article] [PubMed]
- 12.Mazzuca M.Q., Khalil R.A. Vascular endothelin receptor type B: structure, function and dysregulation in vascular disease. Biochem. Pharmacol. 2012;84:147–162. doi: 10.1016/j.bcp.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mazzuca, M.Q., and Khalil, R.A. (2012). Vascular endothelin receptor type B: structure, function and dysregulation in vascular disease. Biochem. Pharmacol. 84, 147-162. [DOI] [PMC free article] [PubMed]
- 13.Ivy D., Wilson N. Tale of 2 endothelin receptor antagonists in Eisenmenger syndrome. Circulation. 2019;139:64–66. doi: 10.1161/CIRCULATIONAHA.118.037171. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ivy, D., and Wilson, N. (2019). Tale of 2 endothelin receptor antagonists in Eisenmenger syndrome. Circulation 139, 64-66. [DOI] [PMC free article] [PubMed]
- 14.Martynyuk T.V., Nakonechnikov S.N., Chazova I.Y. New horizons for the use of the second generation of endothelin receptor antagonist macitentan in patients with pulmonary hypertension. Ter. Arkh. 2018;90:72–80. doi: 10.26442/terarkh201890472-80. [DOI] [PubMed] [Google Scholar]; Martynyuk, T.V., Nakonechnikov, S.N., and Chazova, I.Y. (2018). New horizons for the use of the second generation of endothelin receptor antagonist macitentan in patients with pulmonary hypertension. Ter. Arkh. 90, 72-80. [DOI] [PubMed]
- 15.Le Mée S., Fromigué O., Marie P.J. Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Exp. Cell Res. 2005;302:129–142. doi: 10.1016/j.yexcr.2004.08.028. [DOI] [PubMed] [Google Scholar]; Le Mee, S., Fromigue, O., and Marie, P.J. (2005). Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Exp. Cell Res. 302, 129-142. [DOI] [PubMed]
- 16.Jamal S., Schneider R.J. UV-induction of keratinocyte endothelin-1 downregulates E-cadherin in melanocytes and melanoma cells. J. Clin. Invest. 2002;110:443–452. doi: 10.1172/JCI13729. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jamal, S., and Schneider, R.J. (2002). UV-induction of keratinocyte endothelin-1 downregulates E-cadherin in melanocytes and melanoma cells. J. Clin. Invest. 110, 443-452. [DOI] [PMC free article] [PubMed]
- 17.Spinella F., Garrafa E., Di Castro V., Rosanò L., Nicotra M.R., Caruso A., Natali P.G., Bagnato A. Endothelin-1 stimulates lymphatic endothelial cells and lymphatic vessels to grow and invade. Cancer Res. 2009;69:2669–2676. doi: 10.1158/0008-5472.CAN-08-1879. [DOI] [PubMed] [Google Scholar]; Spinella, F., Garrafa, E., Di Castro, V., Rosano, L., Nicotra, M.R., Caruso, A., Natali, P.G., and Bagnato, A. (2009). Endothelin-1 stimulates lymphatic endothelial cells and lymphatic vessels to grow and invade. Cancer Res. 69, 2669-2676. [DOI] [PubMed]
- 18.Haley E.M., Kim Y. The role of basic fibroblast growth factor in glioblastoma multiforme and glioblastoma stem cells and in their in vitro culture. Cancer Lett. 2014;346:1–5. doi: 10.1016/j.canlet.2013.12.003. [DOI] [PubMed] [Google Scholar]; Haley, E.M., and Kim, Y. (2014). The role of basic fibroblast growth factor in glioblastoma multiforme and glioblastoma stem cells and in their in vitro culture. Cancer Lett. 346, 1-5. [DOI] [PubMed]
- 19.Yu P., Pan G., Yu J., Thomson J.A. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu, P., Pan, G., Yu, J., and Thomson, J.A. (2011). FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 8, 326-334. [DOI] [PMC free article] [PubMed]
- 20.Guo S., Mao X., He F., Liu H., Ming L. Activin A supplement in the hESCs culture enhances the endoderm differentiation efficiency. Cell Biol. Int. 2014;38:849–856. doi: 10.1002/cbin.10274. [DOI] [PubMed] [Google Scholar]; Guo, S., Mao, X., He, F., Liu, H., and Ming, L. (2014). Activin A supplement in the hESCs culture enhances the endoderm differentiation efficiency. Cell Biol. Int. 38, 849-856. [DOI] [PubMed]
- 21.Collier J.H., Segura T. Evolving the use of peptides as components of biomaterials. Biomaterials. 2011;32:4198–4204. doi: 10.1016/j.biomaterials.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; Collier, J.H., and Segura, T. (2011). Evolving the use of peptides as components of biomaterials. Biomaterials 32, 4198-4204. [DOI] [PMC free article] [PubMed]
- 22.Lee M.E., Temizer D.H., Clifford J.A., Quertermous T. Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J. Biol. Chem. 1991;266:16188–16192. [PubMed] [Google Scholar]; Lee, M.E., Temizer, D.H., Clifford, J.A., and Quertermous, T. (1991). Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J. Biol. Chem. 266, 16188-16192. [PubMed]
- 23.Hromas R., Collins S.J., Hickstein D., Raskind W., Deaven L.L., O’Hara P., Hagen F.S., Kaushansky K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J. Biol. Chem. 1991;266:14183–14187. [PubMed] [Google Scholar]; Hromas, R., Collins, S.J., Hickstein, D., Raskind, W., Deaven, L.L., O’Hara, P., Hagen, F.S., and Kaushansky, K. (1991). A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J. Biol. Chem. 266, 14183-14187. [PubMed]
- 24.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]; Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402-408. [DOI] [PubMed]
- 25.Lee S.W., Kang H.J., Lee J.Y., Youn S.W., Won J.Y., Kim J.H., Lee H.C., Lee E.J., Oh S.I., Oh B.H. Oscillating pressure treatment upregulates connexin43 expression in skeletal myoblasts and enhances therapeutic efficacy for myocardial infarction. Cell Transplant. 2009;18:1123–1135. doi: 10.3727/096368909X12483162196809. [DOI] [PubMed] [Google Scholar]; Lee, S.W., Kang, H.J., Lee, J.Y., Youn, S.W., Won, J.Y., Kim, J.H., Lee, H.C., Lee, E.J., Oh, S.I., Oh, B.H., et al. (2009). Oscillating pressure treatment upregulates connexin43 expression in skeletal myoblasts and enhances therapeutic efficacy for myocardial infarction. Cell Transplant. 18, 1123-1135. [DOI] [PubMed]