Abstract
Borna disease virus 1 (BoDV-1) is neurotropic prototype of Bornaviruses causing neurological diseases and maintaining persistent infection in brain cells of mammalian species. Long non-coding RNA (lncRNA) is transcript of more than 200 nucleotides without protein-coding function regulating various biological processes as proliferation, apoptosis, cell migration and viral infection. However, regulatory of lncRNAs in BoDV-1 infection remains unknown. To identify differential expression profiles and predict functions of lncRNA in BoDV-1 infection, microarray data showed that 3528 lncRNAs and 2661 lncRNAs were differentially expressed in Strain V and Hu-H1 BoDV-infected groups compared with control groups, respectively. Gene Ontology (GO) and pathway analysis suggested that differential lncRNAs may be involved in regulation of metabolic, biological regulation, cellular process, endocytosis, viral infections and cell adhesion processes, cancer in both BoDV-infected strains. ENSMUST00000128469 was found down-regulated in both BoDV-infected groups compared with control groups consistent with microarray (p < 0.05). ceRNA analysis indicated possible interaction networks as ENSMUST00000128469/miR-22-5p, miR-206-3p, miR-302b-5p, miR-302c-3p, miR-1a-3p/Igf1. Igf1 was found up-regulated in both BoDV-infected groups compared with control groups (p < 0.05). Possible functions of predicted target mRNAs and miRNAs of ENSMUST00000128469 were involved in cell proliferation, transcriptional misregulation and proteoglycan pathways enriched in cancer. lncRNA may be involved in regulation of Hu-H1 inhibited cell proliferation and promoted apoptosis through NF-kB, JNK/MAPK signaling, BCL2 and CDK6/E2F1 pathways different from Strain V. Possible interaction networks as ENSMUST00000128469/miR-22-5p, miR-206-3p, miR-302b-5p, miR-302c-3p, miR-1a-3p/Igf1 may involve in regulation of cell proliferation, apoptosis, and cancer.
Keywords: Borna disease virus, ceRNA, Infection, lncRNA, Mouse cortical neurons
Introduction
Bornaviruses, belonging to Mononegavirales Family Bornaviridae, as non-segmented negative single-stranded RNA viruses, now includes 8 species and 16 viruses.1, 2 Borna disease virus 1 (BoDV-1), a part of the species mammalian 1 bornavirus, is a highly neurotropic prototype of Bornaviruses and infects a variety of mammalian species including horses, sheep, rabbits, mice, rats, guinea pigs, dogs, cattle and primates.3, 4 Some people reported the presence of increased BoDV antibodies in blood serum samples, BoDV-RNA in peripheral blood samples and the presence of BoDV antigen in autopsy brain of patients with mental illness to support the possibility of human infection with BoDV.5 While, others thought that there was no connection between them.6 Therefore, the potential impact of human infection for mental disorders seemed discordant. Recently BoDV-1 was identified as the cause of deadly human encephalitis after organ transplantation,7, 8 which largely upgraded the significance of this infection in terms of human health and disease. BoDV-1, known to replicate and transcribe in the cell nucleus9 of brain cells as neurons and astrocytes,10 causing a wide range of neurological diseases including deficits in learning and social behavior ranging from immune-mediated disease to behavioral changes without inflammation,11 maintains strictly neurological and persistent infection. BoDV Strain V strain and BoDV Hu-H1 strain are two different strains of BoDV-1. BoDV Strain V strain obtained from Germany sick horses was passaged to rabbits and various cells. While BoDV Hu-H1 strain was isolated from the white blood cells of a bipolar patient.12 Our previous study found that function of target genes of differential miRNA in BoDV Hu-H1 strain infection in rat hippocampus was related to ‘IGF1mTOR’ and ‘IGF1’ pathways.13 BoDV infection provides a good model for studying relationship between persistent infection of viruses and development of chronic neurological symptoms, which may have impact on public health.14 However, underlying mechanisms of BoDV pathogenesis are unclear.
RNA structure provides biological functions for ribozyme and riboswitches, while most ncRNAs function as RNA-protein complexes, including ribosomes, snRNP, snoRNP, telomerase, microRNAs and lncRNAs.15 Long non-coding RNA (lncRNA) belongs to novel isomeric ncRNAs and is a transcript of more than 200 nucleotides without protein coding function. Recently, many studies shown that lncRNAs have multiple functions in various biological processes, such as proliferation, apoptosis, dysregulated cell migration in different diseases.15, 16, 17 Although more and more new functions of lncRNA have been discovered in many specific diseases or biological processes, a large number of lncRNA are still functionally uncharacteristic.18, 19 In recent years, with the application of high-throughput sequencing, a great number of lncRNAs related to viral infection have been discovered and intensively studied. Several lncRNAs have been described to affect cellular responses to viruses such as influenza virus, human immunodeficiency virus type 1, Kaposi'ssarcoma-associated herpesvirus and hepatitis B virus.20, 21, 22, 23 Analysis of interaction between virus and lncRNA will provide information about potential mechanisms of lncRNA acting in various steps of viral infection.
However, regulatory elements of lncRNAs in biological pathway and cellular processes of BoDV infection remain largely unknown. The aim of this study was to isolate lncRNA from Strain V and Hu-H1 BoDV infected and uninfected mouse cortical neurons to identify differential lncRNA expression profiles. Possible mechanism analysis was then performed to study on differential lncRNA and their target genes.
Materials and methods
Ethics statement
The Ethics Committee of Chongqing Medical University approved the study. All experiments were conducted in accordance with Chinese laws on the use of laboratory animals.
Virus and cell culture
The viral strains were kindly provided by Professor Hanns Ludwig of the Free University of Berlin, Germany. Preparation of standard BoDV strain solution and BoDV titration were performed as described previously.24 Each virus titer was approximately 2 × 105 focus forming units per milliliter (FFU/ml).
Cells obtained from mouse cortical neurons of C57 BL/6J mice (day 1 after birth) were mixed and then distributed to a poly-L-lysine-coated six-well plates at a density of 1.5 × 106 cells/well of the 35 wells. After removed the medium, cells were infected with 0.5 focus forming units of multiple infection (MOI) for 2 h. BoDV infection was carried out by adding a cell-released virus prepared as previously described to the culture medium.24 The control groups were added to the virus-free buffer as parallel control experiment. Then, excess virus was removed and the neurons were washed again in the neurobasal medium. Thereafter, all cells were cultured for 12 days in a humidified incubator (5% CO2, 37 °C). During this time, half of the medium was changed every three days.
Immunofluorescence
Our previous studies showed that BoDV P40-positive neurons could be detected almost 100% by day 12 in BoDV-infected neurons.25 Immunofluorescence assays were then applied on day 12 post infection. The percentage of neurons was determined by observing randomly selected cells in three independent experiments. Standard immunofluorescence was performed as described previously.25 Briefly, both BoDV-infected and control neurons were incubated in 6-well plates then permeabilized. After blocked and incubation, neuron-specific markers MAP-2 and anti-BoDV-specific p40 antigen primary monoclonal antibody were performed.26 After incubated with the second antibody and counterstained with DAPI, immunofluorescence was detected using an inverted fluorescence microscope (Nikon, Tokyo, Japan).
Microarray and data analysis
Total RNA from each sample was extracted using TRIzol and quantified using a NanoDrop 1000 spectrophotometer. RNA integrity was assessed using standard denaturing agarose gel electrophoresis.
Microarray Arraystar Mouse lncRNA Array v2.0 was designed to analyze lncRNA and protein-encoding RNA in mouse genome. A total of 31,423 lncRNAs were collected from authoritative data sources, including RefSeq, UCSC Knowngenes, Ensembl and other related literature. Microarray analysis was carried out by KangChen Bio-tech, Shanghai, PR China. For each microarray study, RNA from every 3 mouse cortical neuron samples from control and two BoDV strain infected groups was pooled and used for hybridization.
Data analysis Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using GeneSpring GX v11.5.1 software package (Agilent Technologies). Then lncRNA and mRNA with current or edge markers (“All Targets Value”) were selected for further data analysis. Differentially expressed lncRNA and mRNA between three groups were identified by fold change filtration. Hierarchical Clustering was performed using Agilent GeneSpring GX software (version 11.5.1).
LncRNA-mRNA association analysis
To reveal interaction between antisense lncRNA and mRNA, software RNAplex27 (http://www.tbi.univie.ac.at/RNA/RNAplex.1.html) was used to predict complementary correlation of antisense lncRNA and mRNA. lncRNA in less than 10 kb up/down stream may be cis-regulators. The cis-target genes are then subjected to enrichment analysis of GO and KEGG pathway. We analyzed correlation between lncRNAs and protein-coding genes to determine target genes of lncRNAs.
qRT-PCR
Total RNA was reverse transcribed using the ReverTra Ace qPCR Kit according to manufacturer's instructions. Expression levels of five differentially expressed lncRNAs both in two BoDV strains selected from the top 70 up-regulated and top 70 down-regulated lncRNAs were verified by quantitative real-time reverse transcription (qRT)—PCR using SYBR Green assays. mRNAs as ceRNA network target genes of lncRNA chosen from the five lncRNAs described above were verified by qRT-PCR using SYBR Green assays. GAPDH was used as an internal control. Tables S1 and S2 show the primer sequences. Gene expression levels were analyzed with the 2−ΔΔCT method.
ceRNA analyses
The co-expression network was illustrated using Cytoscape (v3.4.0). Analysis was carried out by KangChen Bio-tech, Shanghai, China, searching for potential miRNA responses elements on the sequences of lncRNA and mRNA. The miRNA binding sites were predicted by miRcode (http://www.mircode.org/) and miRNA-mRNA interactions were predicted by Targetscan (http://www.targetscan.org/).
Data analysis
Data were expressed as mean ± SD of three independent experiments with three biological replicates. Statistical analysis was performed using SPSS V.17.0 (SPSS, Chicago, USA). Fold change and Student‵s t test were used to analyze the statistical significance of microarray and RT-PCR results. P value < 0.05 (two-tailed) was considered statistically significant.
Results
Verification of BoDV infection
BoDV-p40 could be detected in all RNA samples from BoDV-infected mouse cortical neurons, but not detected in control groups. Immunofluorescence results showed that neuron purity was over 90% and neurons in Strain V and Hu-H1groups were infected with Strains V and Hu-H1 BoDV, respectively.
LncRNA is abnormally expressed in infected mouse cortical neurons compared with the control groups
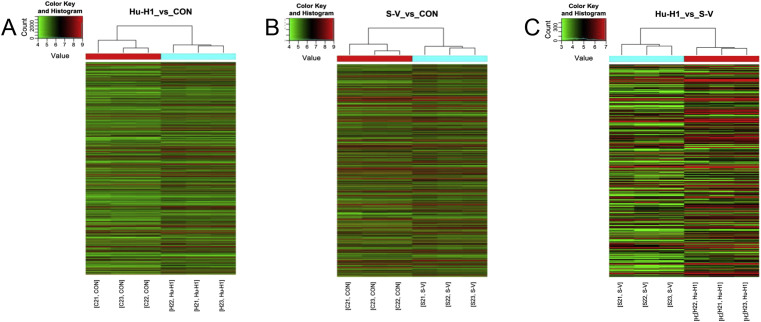
Microarray analysis was used to determine lncRNA expression profiles of mouse cortical neuron samples from control and different strains of BoDV-infected groups as Strain V and Hu-H1 (Fig. 1A–C). Using a pool of samples consisting of more than 31,423 lncRNAs, we evaluated that a total of 2661 lncRNAs were differentially (fold change FC > 2, P < 0.05) expressed in Hu-H1 BoDV-infected groups compared to control groups with 1545 lncRNAs up-regulated (>2 fold) and 1116 lncRNAs down-regulated. A total of 3528 lncRNAs expressed in Strain V BoDV-infected groups were differentially (FC > 2, P < 0.05) compared to control groups with 1649 lncRNAs up-regulated (>2 fold) and 1879 lncRNAs down-regulated. A total of 379 lncRNAs were differentially (FC > 2, P < 0.05) expressed in Hu-H1 BoDV-infected groups compared to Strain V BoDV-infected groups with 278 lncRNAs up-regulated (>2 fold) and 101 lncRNAs down-regulated (Table S3). Detailed informations, including top 70 up-regulated and top 70 down-regulated lncRNAs in Hu-H1 and Strain V BoDV-infected groups compared to control groups, were provided in Tables S4 and S5.
Figure 1.
A-C Entire hierarchical clusterings of differentially expressed lncRNAs among Hu-H1, Strain V (group-S-V) BoDV-infected mouse cortical neuron and control groups (group-CON), respectively; up- and down-regulated genes are colored in red and green.
GO and pathway analysis for differentially expressed lncRNAs
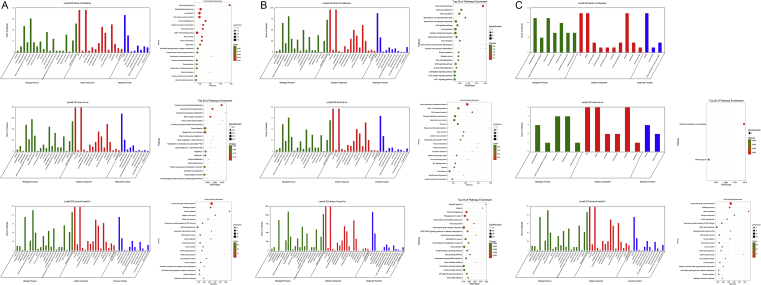
It has been shown that lncRNA mediates expression of adjacent and distant genes by antisense-, cis- or trans-acting regulation, respectively. A total of 206 antisense-target genes, 232 cis-target genes and 2023 trans-target genes were further identified from Ensembl data source for GO and pathway analysis. Through GO analysis, we found that differentially expressed lncRNAs in Hu-H1 BoDV infection were mainly enriched in biological regulation, cellular process, single-organism process, cell and cell part, organelle, binding, membrane part, as well as sphingolipid signaling pathway, axon guidance, cell adhesion molecules, cGMP-PKG signaling pathway, focal adhesion, Epstein–Barr virus infection, neuroactive ligand–receptor interaction, GABAergic synapse, cAMP signaling pathway through pathway analysis (Fig. 2A).
Figure 2.
A-C Enrichment analysis of pathways and GO terms for differentially expressed lncRNAs in Hu-H1 vs control groups, S-V vs control groups and Hu-H1 vs S-V groups, respectively.
We found that differentially expressed lncRNAs in Strain V BoDV infection were mainly enriched for biological regulation, cellular process, single-organism process, cell and cell part, organelle, binding, membrane part, as well as natural killer cell mediated cytotoxicity, axon guidance, ubiquitin mediated proteolysis, protein processing in endoplasmic reticulum, adrenergic signaling in cardiomyocytes, proteoglycans in cancer, cGMP-PKG signaling pathway, focal adhesion, Epstein–Barr virus infection, chemokine signaling pathway, neuroactive ligand–receptor interaction through pathway analysis (Fig. 2B).
We found that differentially expressed lncRNAs in Hu-H1 and Strain V BoDV infection were mainly enriched for biological regulation, cellular process, single-organism process, cell and cell part, organelle, binding, membrane, as well as cAMP-PKG signaling pathway, GABAergic synapse, neuroactive ligand–receptor interaction through pathway analysis (Fig. 2C).
qRT-PCR validation of lncRNA
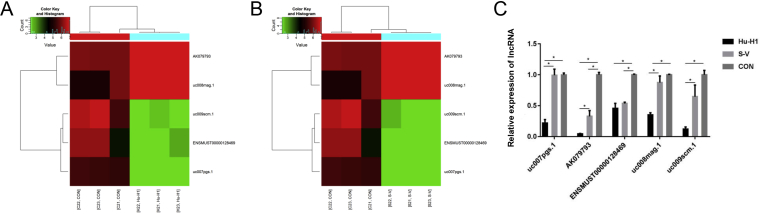
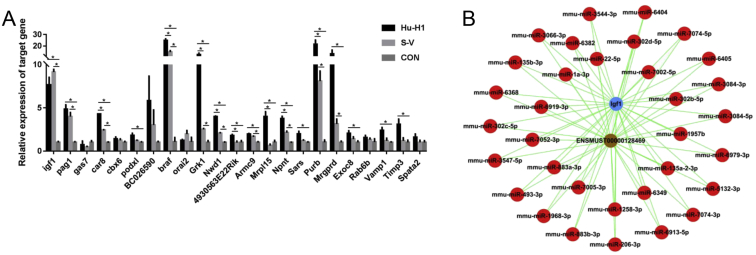
Five differentially expressed lncRNAs from top 70 up-regulated and top 70 down-regulated lncRNAs in Hu-H1 and Strain V BoDV-infected groups which had the same change in both Hu-H1 and Strain V BoDV-infected groups were selected (Fig. 3A and B) and analyzed for their expression levels in BoDV Strain V, Hu-H1 infected and control groups via qRT-PCR. ENSMUST00000128469 was found down-regulated both in Strain V and Hu-H1 BoDV infection groups compared with control groups consistent with array (p < 0.05). uc007 pgs.1, uc008mag.1 and uc009scm.1 in Hu-H1 BoDV groups were found both down-regulated consistent with array (p < 0.05), while expressed levels of uc007 pgs.1, uc008mag.1, uc009scm.1 in Strain V BoDV groups and AK079793 in both Hu-H1 and Strain V BoDV groups were found not consistent with array (Fig. 3C).
Figure 3.
A-B Hierarchical clusterings of five differentially expressed lncRNAs selected from the top 70 up- and down-regulated lncRNAs among Hu-H1, Strain V BoDV-infected mouse cortical neuron and control groups, respectively; up- and down-regulated genes are colored in red and green. C Validation of five lncRNAs using qRT-PCR comparing Hu-H1, strain V BoDV-infected mouse cortical neuron groups and control groups.
Establishment of the lncRNA/miRNA/mRNA gene co-expression network
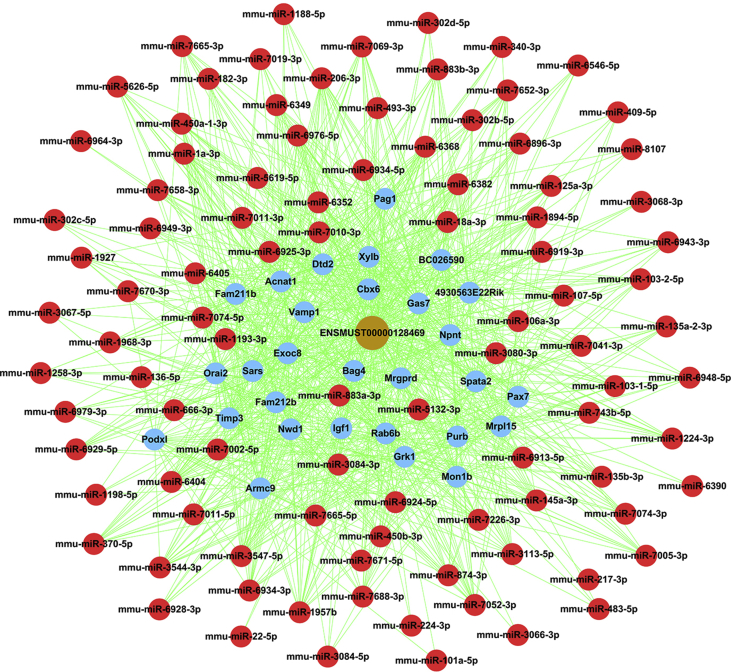
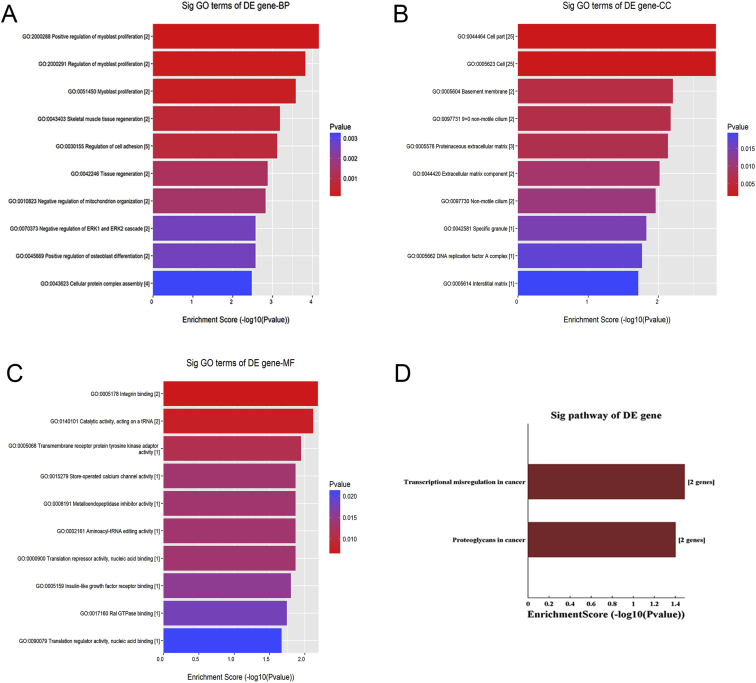
To investigate molecular mechanism of lncRNA, based on ceRNA analysis, we identified lncRNA/miRNA/mRNA interaction network of ENSMUST00000128469. ceRNA network included lncRNA ENSMUST00000128469, 96 miRNAs and 31 mRNAs (Fig. 4). Predicted target mRNAs were mainly enriched for regulation of myoblast proliferation, skeletal muscle tissue regeneration, regulation of cell adhesion, negative regulation of ERK1 and ERK2 cascade, cell and cell part, basement membrane, non-motile cilium, proteinaceous extracellular matrix, extracellular matrix component, integrin binding, catalytic activity, acting on a tRNA, transmembrane receptor protein tyrosine kinase adaptor activity, store operated calcium channel activity, metalloendopeptidase inhibitor activity, as well as transcriptional misregulation and proteoglycans in cancer through pathway analysis (Fig. 5). Then, we further identified several promising networks of lncRNA/miRNA/mRNA interactions. mRNAs as ceRNA network target genes of ENSMUST00000128469 were selected and analyzed for their expression levels in BoDV Strain V and Hu-H1 infection groups and control groups via qRT-PCR. Igf1, Pag1, Car8, BC026590, Braf, Grk1, Nwd1, Npnt, Purb, Mrgprd were found to be up-regulated both in Strain V and Hu-H1 BoDV-infected groups compared with control groups (p < 0.05) (Fig. 6A). Our previous study found that function of target genes of differential miRNA in BoDV infection in rat hippocampus was related to ‘IGF1mTOR’ and ‘IGF1’ pathways.13 We focused on Igf1 and found possible lncRNA/miRNA/mRNA interactions as ENSMUST00000128469/miR-22-5p, miR-206-3p, miR-302b-5p, miR-302c-3p, miR-1a-3p/Igf1 (Fig. 6B).
Figure 4.
ceRNA analysis indicated potential lncRNA/miRNA/mRNA interactions of ENSMUST00000128469. Red nodes mean miRNAs, gray node means ENSMUST00000128469, blue nodes mean mRNAs.
Figure 5.
Enrichment analysis of pathways and GO terms for predicted target mRNAs of ceRNA analysis of ENSMUST00000128469. A-C: GO analysis according to biological process, cellular component, and molecular function, respectively. D: Pathway analysis is a functional analysis that involves mapping genes to KEGG pathways. The lower the P-value, more significant the pathway (The recommended P-value cut-off is 0.05).
Figure 6.
A: Validation of mRNA by ceRNA analysis of ENSMUST00000128469 using qRT-PCR comparing Hu-H1, strain V BoDV-infected mouse cortical neuron and control groups. B: ceRNA analysis indicated potential lncRNA/miRNA/mRNA interactions of ENSMUST00000128469/miRNA/Igf1. Red nodes mean miRNAs, gray node means ENSMUST00000128469, blue nodes mean mRNAs.
Discussion
Our study is the first study to perform a genome-wide analysis of differentially expressed lncRNA in different infection of BoDV strains. Further analysis of lncRNAs and their target genes provided new insights into the mechanisms of BoDV infection.
Since little is known about function of these lncRNAs, through GO and pathway analysis we found that lncRNA target genes in both strain V and Hu-H1 strains were enriched in metabolic, biological regulations, cellular process, endocytosis, cancer, viral infections and cell adhesion. Previous studies showed that two BoDV strains alter metabolic pathways as amino acids and energy metabolism.28 BoDV surface glycoprotein (G) directed BoDV entry cell through receptor-mediated endocytosis.29 BoDV infection may reduce expression of vascular cell adhesion molecule 1 leading to maintenance of persistent infection.30 The germline cells that borna virus retained and integrated sequence into host genome can be genetically involved in cancer formation as endogenous viral elements and act as tumor suppressors.31 Homo sapiens EBLNs-1 (hsEBLNs-1) as one of endogenous bornavirus-like N-elements integrated in the germline of humans and human ancestors, retains a long open reading frame (ORF) that encodes 366 amino acids, comparable with the full-length BoDV N protein.32 Sofuku et al showed that hsEBLN-1 may have anti-tumor effects most probably through functioning as lncRNA downregulation of COMMD3 expression potentiating NF-kB pathway.33 In addition, BoDV N-protein possessed a NF-kB inhibitory sequence,34 working as an antagonist of hsEBLN-1.
However, Our team's past studies found that Hu-H1 inhibited cell proliferation and promoted apoptosis of human oligodendrocytes, while Strain V was the opposite.35 Hu-H1 strain had anti-proliferative and no apoptosis effect, while Strain V caused increase in apoptosis of SH-SH5Y cells during initial infection.36 Our studies of miRNAs in the hippocampi of neonatal rats infected with BoDV Hu-H1 revealed that BoDV Hu-H1 induced downregulation miR-126 may involved in inhibition of NF-kB leading to induction of apoptosis, downregulation miR-200b could involved in microglia activation regulated through the JNK/MAPK signaling and downregulation miR-449a to influence cell cycle regulation, proliferation and apoptosis through the JNK/MAPK signaling and BCL2 and CDK6/E2F1 pathways.13 That may be why BoDV Hu-H1 infection may have different mechanism from BoDV Strain V infection. BoDV as the only RNA virus known to persist in the nucleus of infected cells inhibited histone acetylation by BoDV phosphoprotein interference with histone acetyltransferase activity accompanied by regulation of viral replication and ensuring long-term maintenance in infected cells, which was found both by our team in BoDV Hu-H1 infection in human oligodendroglia cells37, 38 and BoDV Giessen strain He/80 infection in rat primary cortical neurons.39, 40 This revealed the perfect adaptation of this “old” virus to its host, which may contribute to neuronal persistence and limit cell damage. Therefore, lncRNA may be involved in regulation of above BoDV-infected process, and have different roles in different BoDV-infected in cell cycle regulation, proliferation and apoptosis.
Currently, ceRNA analysis is new method for predicting function of lncRNA. Our study is also first using ceRNA analysis to indicate potential lncRNA/miRNA/mRNA interactions of lncRNA-ENSMUST00000128469 in BoDV-infection as ENSMUST00000128469/miR-22-5p, miR-206-3p, miR-302b-5p, miR-302c-3p, miR-1a-3p/Igf1, suggesting its potential role as a ceRNA that may regulate pathogenesis of BoDV. Of course, additional functional studies in vivo and vitro are needed.
Our study found that Igf1 may be involved in neuronal disease caused by strain V and Hu-H1 BoDV infection. Igf1 signaling pathway controlled growth of most animal species after birth,41 as major regulator of aging process from rodents to humans.42 The major signaling pathway for IGF-1R-mediated apoptosis protection is dependent on phosphatidylinositol 3-kinase, Akt/protein kinase B, and the phosphorylation and inactivation of BAD, a member of the Bcl-2 family of proteins, which may explain the remarkable efficacy of IGF-1R in protecting cells from apoptosis.43 Activation of Igf1 was involved in regulation of various cellular functions of PI3K/Akt/mTOR pathway as cell proliferation, differentiation, apoptosis, and associated with cancers as leukemia44 rectal cancer,45 and esophageal adenocarcinoma activated by ERK1/2 mitotic pattern.46 Igf1 mediated inhibition of NF-κB activation in microglia.47 BoDV directly interfered with NF-κB pathway.34 Igf1 may be involved in pathogenesis of BoDV-infected with BoDV N-protein as mentioned above by acting on NF-κB IGF-1, the most potent natural activator of anti-apoptotic Akt/protein kinase b (PKB), was a target gene of ENSMUST00000128469. miR-22-5p affected expression of breast cancer and acute myeloid leukemia.48 miR-206-3p inhibited Akt phosphorylation and inactivated PI3K/Akt signaling pathway.49 miR-206 was involved in Alzheimer's disease by inhibiting expression of brain-derived neurotrophic factor (BDNF),50 which induced mutation triggering eliminated by BoDV.51 miR-302b-3p had tumor suppressor effect in gastric cancer and reduced AKT phosphorylation.52 miR-302C-3p inhibited both migration and invasion of MHCC97H cells in vitro and AKT-mediated epithelial–mesenchymal transition in HCC cells.53 miR-1a-3p regulated Akt/mTOR/S6K signaling pathway.54 According to the Ensemble Transcript ID, ENSMUST00000128469 could be found as synonym with the serine/threonine protein kinase PAK6 (mouse). PAK6, an effector kinase of the Rho family GTPase Chp/RhoV, predominantly expressed in the brain, positively regulated by androgen receptor, may also be involved in stress-signaling stimulated by p38-MAP kinase.55 Expression of PAK6 was targeted by miR-429 in colon cancer cells,56 by miR-328 and miR-23a in prostate cancer cells involved in cell growth and migration.57, 58 PAK6 was recognized as a therapeutic candidate gene for epileptic encephalopathy.59 Combined deletion of Pak5 and Pak6 genes could lead to a significant impairment of mobility, memory, and learning.60 This was consistent with prediction of target mRNA and miRNA by ceRNA analysis of ENSMUST00000128469, and transcriptional misregulation and proteoglycan pathways enriched in cancer in our study.
Therefore, we could presume that inhibition of NF-kB1 through N-protein, thereby escaping innate immunity of the host, is one key of BoDV virus-driven mechanism. While inhibition of NF-kB through down-regulation of miR-126 has been induced promotion of apoptosis which the natural strain Hu-H1 did but the more adapted strain V did not. From the host side, a possible antagonist may be hsEBLN-1 via downregulation of the COMMD3 gene and a strong protection against apoptosis mediated by IGF-1 through multiple mechanisms. Another important support to establish virus persistence works through epigenetic signaling via BoDV P-protein involved in decreasing histone acetylation. All processes above may lncRNA especially ENSMUST00000128469 involve in.
In summary, although our study was preliminary and lacked additional functional experiments, it provided comprehensive understanding of lncRNA involved in different BoDV-infected strains and helped elucidate molecular mechanisms behind this infection. ceRNA analysis was revealed that ENSMUST00000128469/miR-22-5p, miR-206-3p, miR-302b-5p, miR-302c-3p, miR-1a-3p/Igf1 network may have most potential for further research.
Conclusions
lncRNA may play important and different roles in infection of different BoDV strains. GO and pathway analyses indicated that lncRNA may be involved in regulation of metabolic, biological regulation, cellular process, endocytosis, viral infections and cell adhesion processes, cancer in both BoDV-infected strains. lncRNA target genes may have different regulatory roles in different BoDV-infected strains in cell cycle regulation, proliferation and apoptosis. lncRNA may be involved in regulation of Hu-H1 inhibited cell proliferation and promoted apoptosis through NF-kB, JNK/MAPK signaling, BCL2 and CDK6/E2F1 pathways different from Strain V. Possible interaction networks as ENSMUST00000128469/miR-22-5p, miR-206-3p, miR-302b-5p, miR-302c-3p, miR-1a-3p/Igf1 indicated by ceRNA analysis may involve in regulation of cell proliferation, apoptosis, and cancer.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
We thank Professor Hanns Ludwig, Berlin Free University, Germany, and Dr. Liv Bode, Robert Koch Institute, Germany, for providing BoDV strain Hu-H1 and Strain V. This work was supported by the National Key Research and Development Program of China, China (Grant No. YFA0505700) and the Natural Science Foundation of China, China (Grant No.81601207).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2019.04.002.
Contributor Information
Lin Sun, Email: tilamisu789456123@126.com.
Yujie Guo, Email: g1240725344@163.com.
Peng He, Email: hepeng000@sina.com.
Xiaoyan Xu, Email: xxy2922@163.com.
Xiong Zhang, Email: 1059859279@qq.com.
Haiyang Wang, Email: wanghaiyang6939@163.com.
Tian Tang, Email: tangtian2333@163.com.
Wei Zhou, Email: 431125a22ha.cdb@sina.cn.
Ping Xu, Email: xuping527@vip.sina.com.
Peng Xie, Email: xiepeng@cqmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lipkin W.I., Briese T. Bornaviridae. In: Knipe D., Howley P., Griffin D., editors. 5th ed. vol. II. Lippincott Williams and Wilkins; Philadelphia PA: 2007. pp. 1829–1851. (Fields Virology). [Google Scholar]
- 2.Amarasinghe G.K., Bao Y., Basler C.F. Taxonomy of the order Mononegavirales: update 2017. Arch Virol. 2017;162(8):2493–2504. doi: 10.1007/s00705-017-3311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinnunen P.M., Palva A., Vaheri A., Vapalahti O. Epidemiology and host spectrum of Borna disease virus infections. J Gen Virol. 2013;94(Pt 2):247–262. doi: 10.1099/vir.0.046961-0. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Wang X., Zhan Q. Evidence for natural Borna disease virus infection in healthy domestic animals in three areas of western China. Arch Virol. 2014;159(8):1941–1949. doi: 10.1007/s00705-013-1971-5. [DOI] [PubMed] [Google Scholar]
- 5.Bode L., Ludwig H. Borna disease virus infection, a human mental-health risk. Clin Microbiol Rev. 2003;16(3):534–545. doi: 10.1128/CMR.16.3.534-545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikuta K., Ibrahim M.S., Kobayashi T., Tomonaga K. Borna disease virus and infection in humans. Front Biosci. 2002;7:d470–d495. doi: 10.2741/A789. [DOI] [PubMed] [Google Scholar]
- 7.Schlottau K., Forth L., Angstwurm K. Fatal encephalitic borna disease virus 1 in solid-organ transplant recipients. N Engl J Med. 2018;379(14):1377–1379. doi: 10.1056/NEJMc1803115. [DOI] [PubMed] [Google Scholar]
- 8.Korn K., Coras R., Bobinger T. Fatal encephalitis associated with borna disease virus 1. N Engl J Med. 2018;379(14):1375–1377. doi: 10.1056/NEJMc1800724. [DOI] [PubMed] [Google Scholar]
- 9.Briese T., de la Torre J.C., Lewis A., Ludwig H., Lipkin W.I. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc Natl Acad Sci U S A. 1992;89(23):11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennartz F., Bayer K., Czerwonka N. Surface glycoprotein of Borna disease virus mediates virus spread from cell to cell. Cell Microbiol. 2016;18(3):340–354. doi: 10.1111/cmi.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pletnikov M.V., Moran T.H., Carbone K.M. Borna disease virus infection of the neonatal rat: developmental brain injury model of autism spectrum disorders. Front Biosci – J Vis Lit. 2002;7:d593–d607. doi: 10.2741/A797. [DOI] [PubMed] [Google Scholar]
- 12.Bode L., Durrwald R., Rantam F.A., Ferszt R., Ludwig H. First isolates of infectious human Borna disease virus from patients with mood disorders. Mol Psychiatr. 1996;1(3):200–212. [PubMed] [Google Scholar]
- 13.Zhao M., Sun L., Chen S. Borna disease virus infection impacts microRNAs associated with nervous system development, cell differentiation, proliferation and apoptosis in the hippocampi of neonatal rats. Mol Med Rep. 2015;12(3):3697–3703. doi: 10.3892/mmr.2015.3828. [DOI] [PubMed] [Google Scholar]
- 14.Scordel C., Huttin A., Cochet-Bernoin M. Borna disease virus phosphoprotein impairs the developmental program controlling neurogenesis and reduces human GABAergic neurogenesis. PLoS Pathog. 2015;11(4):e1004859. doi: 10.1371/journal.ppat.1004859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 17.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Kerin T., Ramanathan A., Rivas K., Grepo N., Coetzee G.A., Campbell D.B. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4(128) doi: 10.1126/scitranslmed.3003479. 128ra140. [DOI] [PubMed] [Google Scholar]
- 19.Guil S., Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19(11):1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 20.Landeras-Bueno S., Ortin J. Regulation of influenza virus infection by long non-coding RNAs. Virus Res. 2016;212:78–84. doi: 10.1016/j.virusres.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Lazar D.C., Morris K.V., Saayman S.M. The emerging role of long non-coding RNAs in HIV infection. Virus Res. 2016;212:114–126. doi: 10.1016/j.virusres.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad N.K. New insights into the expression and functions of the Kaposi's sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res. 2016;212:53–63. doi: 10.1016/j.virusres.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyo B., Nicholson S.A., Arbuthnot P.B. The role of long non-coding RNAs in hepatitis B virus-related hepatocellular carcinoma. Virus Res. 2016;212:103–113. doi: 10.1016/j.virusres.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 24.Huang R., Gao H., Zhang L. Borna disease virus infection perturbs energy metabolites and amino acids in cultured human oligodendroglia cells. PLoS One. 2012;7(9):e44665. doi: 10.1371/journal.pone.0044665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Liu S., Zhang L. Real-time qPCR identifies suitable reference genes for Borna disease virus-infected rat cortical neurons. Int J Mol Sci. 2014;15(12):21825–21839. doi: 10.3390/ijms151221825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig H., Furuya K., Bode L., Klein N., Durrwald R., Lee D.S. Biology and neurobiology of Borna disease viruses (BDV), defined by antibodies, neutralizability and their pathogenic potential. Arch Virol Suppl. 1993;7:111–133. doi: 10.1007/978-3-7091-9300-6_10. [DOI] [PubMed] [Google Scholar]
- 27.Tafer H., Hofacker I.L. RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24(22):2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 28.Liu S., Bode L., Zhang L. GC-MS-Based metabonomic profiling displayed differing effects of borna disease virus natural strain Hu-H1 and laboratory strain V infection in rat cortical neurons. Int J Mol Sci. 2015;16(8):19347–19368. doi: 10.3390/ijms160819347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemente R., Sisman E., Aza-Blanc P., de la Torre J.C. Identification of host factors involved in borna disease virus cell entry through a small interfering RNA functional genetic screen. J Virol. 2010;84(7):3562–3575. doi: 10.1128/JVI.02274-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtaki N., Kamitani W., Watanabe Y. Downregulation of an astrocyte-derived inflammatory protein, S100B, reduces vascular inflammatory responses in brains persistently infected with Borna disease virus. J Virol. 2007;81(11):5940–5948. doi: 10.1128/JVI.02137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda T. Potential links between hepadnavirus and bornavirus sequences in the host genome and cancer. Front Microbiol. 2017;8:2537. doi: 10.3389/fmicb.2017.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horie M., Honda T., Suzuki Y. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463(7277):84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sofuku K., Parrish N.F., Honda T., Tomonaga K. Transcription profiling demonstrates epigenetic control of non-retroviral RNA virus-derived elements in the human genome. Cell Rep. 2015;12(10):1548–1554. doi: 10.1016/j.celrep.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Makino A., Fujino K., Parrish N.F., Honda T., Tomonaga K. Borna disease virus possesses an NF-kB inhibitory sequence in the nucleoprotein gene. Sci Rep. 2015;5:8696. doi: 10.1038/srep08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D., Lei Y., Deng J. Human but not laboratory borna disease virus inhibits proliferation and induces apoptosis in human oligodendrocytes in vitro. PLoS One. 2013;8(6):e66623. doi: 10.1371/journal.pone.0066623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H., Zhang H., Li D. Different inhibitory effects on the proliferation and apoptosis of human and laboratory Borna disease virusinfected human neuroblastoma SHSY5Y cells in vitro. Mol Med Rep. 2018;17(1):925–931. doi: 10.3892/mmr.2017.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Zhao L., Yang Y. Human borna disease virus infection impacts host proteome and histone lysine acetylation in human oligodendroglia cells. Virology. 2014;464–465:196–205. doi: 10.1016/j.virol.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonnaud E.M., Szelechowski M., Betourne A. Borna disease virus phosphoprotein modulates epigenetic signaling in neurons to control viral replication. J Virol. 2015;89(11):5996–6008. doi: 10.1128/JVI.00454-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams B.L., Hornig M., Yaddanapudi K., Lipkin W.I. Hippocampal poly(ADP-Ribose) polymerase 1 and caspase 3 activation in neonatal bornavirus infection. J Virol. 2008;82(4):1748–1758. doi: 10.1128/JVI.02014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poenisch M., Burger N., Staeheli P., Bauer G., Schneider U. Protein X of Borna disease virus inhibits apoptosis and promotes viral persistence in the central nervous systems of newborn-infected rats. J Virol. 2009;83(9):4297–4307. doi: 10.1128/JVI.02321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poinsot P., Schwarzer M., Peretti N., Leulier F. The emerging connections between IGF1, the intestinal microbiome, Lactobacillus strains and bone growth. J Mol Endocrinol. 2018;61(1):T103–T113. doi: 10.1530/JME-17-0292. [DOI] [PubMed] [Google Scholar]
- 42.Gubbi S., Quipildor G.F., Barzilai N., Huffman D.M., Milman S. 40 YEARS of IGF1: IGF1: the Jekyll and Hyde of the aging brain. J Mol Endocrinol. 2018;61(1):T171–T185. doi: 10.1530/JME-18-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peruzzi F., Prisco M., Dews M. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol Cell Biol. 1999;19(10):7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullighan C.G. IGF1 brings growing pains for T-ALL LSCs. Cell Stem Cell. 2018;23(5):632–633. doi: 10.1016/j.stem.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J., Cao J., Zhou L. MiR-1260b inhibitor enhances the chemosensitivity of colorectal cancer cells to fluorouracil by targeting PDCD4/IGF1. Oncol Lett. 2018;16(4):5131–5139. doi: 10.3892/ol.2018.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo W., Pencina K.M., Gagliano-Juca T. Effects of an ActRIIB.Fc ligand trap on cardiac function in simian immunodeficiency virus-infected male rhesus macaques. J Endocr Soc. 2018;2(8):817–831. doi: 10.1210/js.2018-00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu H., Lin H., Duan W. Intrathecal injection of scAAV9-hIGF1 prolongs the survival of ALS model mice by inhibiting the NF-kB pathway. Neuroscience. 2018;381:1–10. doi: 10.1016/j.neuroscience.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Yao H., Sun P., Duan M. microRNA-22 can regulate expression of the long non-coding RNA MEG3 in acute myeloid leukemia. Oncotarget. 2017;8(39):65211–65217. doi: 10.18632/oncotarget.18059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang R., Ma F., Li W., Ouyang S., Liu Z., Wu J. miR-206-3p inhibits 3T3-L1 cell adipogenesis via the c-Met/PI3K/Akt pathway. Int J Mol Sci. 2017;18(7) doi: 10.3390/ijms18071510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C.N., Wang Y.J., Wang H. The anti-dementia effects of donepezil involve miR-206-3p in the Hippocampus and cortex. Biol Pharm Bull. 2017;40(4):465–472. doi: 10.1248/bpb.b16-00898. [DOI] [PubMed] [Google Scholar]
- 51.Hans A., Bajramovic J.J., Syan S. Persistent, noncytolytic infection of neurons by Borna disease virus interferes with ERK 1/2 signaling and abrogates BDNF-induced synaptogenesis. FASEB J. 2004;18(7):863–865. doi: 10.1096/fj.03-0764fje. [DOI] [PubMed] [Google Scholar]
- 52.Guo B., Zhao Z., Wang Z. MicroRNA-302b-3p suppresses cell proliferation through AKT pathway by targeting IGF-1R in human gastric cancer. Cell Physiol Biochem. 2017;42(4):1701–1711. doi: 10.1159/000479419. [DOI] [PubMed] [Google Scholar]
- 53.Yang L., Guo Y., Liu X. The tumor suppressive miR-302c-3p inhibits migration and invasion of hepatocellular carcinoma cells by targeting TRAF4. J Cancer. 2018;9(15):2693–2701. doi: 10.7150/jca.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Ma J., Qiu W. Guanidinoacetic acid regulates myogenic differentiation and muscle growth through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K signaling pathway. Int J Mol Sci. 2018;19(9) doi: 10.3390/ijms19092837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar R., Sanawar R., Li X., Li F. Structure, biochemistry, and biology of PAK kinases. Gene. 2017;605:20–31. doi: 10.1016/j.gene.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian X., Wei Z., Wang J., Liu P., Qin Y., Zhong M. MicroRNA-429 inhibits the migration and invasion of colon cancer cells by targeting PAK6/cofilin signaling. Oncol Rep. 2015;34(2):707–714. doi: 10.3892/or.2015.4039. [DOI] [PubMed] [Google Scholar]
- 57.Liu C., Zhang L., Huang Y. MicroRNA328 directly targets p21activated protein kinase 6 inhibiting prostate cancer proliferation and enhancing docetaxel sensitivity. Mol Med Rep. 2015;12(5):7389–7395. doi: 10.3892/mmr.2015.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai S., Chen R., Li X. Downregulation of microRNA-23a suppresses prostate cancer metastasis by targeting the PAK6-LIMK1 signaling pathway. Oncotarget. 2015;6(6):3904–3917. doi: 10.18632/oncotarget.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliver K.L., Lukic V., Freytag S., Scheffer I.E., Berkovic S.F., Bahlo M. In silico prioritization based on coexpression can aid epileptic encephalopathy gene discovery. Neurol Genet. 2016;2(1):e51. doi: 10.1212/NXG.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furnari M.A., Jobes M.L., Nekrasova T., Minden A., Wagner G.C. Functional deficits in PAK5, PAK6 and PAK5/PAK6 knockout mice. PLoS One. 2013;8(4):e61321. doi: 10.1371/journal.pone.0061321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.