Key Points
The bat MHC I exhibits distinctive structural characteristics.
Significant differences exist in the peptidome between inside the cell and outside the cell.
A cluster of uniquely inserted residues may help promote high-affinity peptide binding.
Abstract
Bats are natural reservoir hosts, harboring more than 100 viruses, some of which are lethal to humans. The asymptomatic coexistence with viruses is thought to be connected to the unique immune system of bats. MHC class I (MHC I) presentation is closely related to cytotoxic lymphocyte immunity, which plays an important role in viral resistance. To investigate the characteristics of MHC I presentation in bats, the crystal structures of peptide–MHC I complexes of Pteropus alecto, Ptal-N*01:01/HEV-1 (DFANTFLP) and Ptal-N*01:01/HEV-2 (DYINTNLVP), and two related mutants, Ptal-N*01:01/HEV-1PΩL (DFANTFLL) and Ptal-N*01:01ΔMDL/HEV-1, were determined. Through structural analysis, we found that Ptal-N*01:01 had a multi-Ala–assembled pocket B and a flexible hydrophobic pocket F, which could accommodate variable anchor residues and allow Ptal-N*01:01 to bind numerous peptides. Three sequential amino acids, Met, Asp, and Leu, absent from the α1 domain of the H chain in other mammals, were present in this domain in the bat. Upon deleting these amino acids and determining the structure in p/Ptal-N*01:01ΔMDL/HEV-1, we found they helped form an extra salt-bridge chain between the H chain and the N-terminal aspartic acid of the peptide. By introducing an MHC I random peptide library for de novo liquid chromatography–tandem mass spectrometry analysis, we found that this insertion module, present in all types of bats, can promote MHC I presentation of peptides with high affinity during the peptide exchange process. This study will help us better understand how bat MHC I presents high-affinity peptides from an extensive binding peptidome and provides a foundation to understand the cellular immunity of bats.
Introduction
The bat is an important reservoir host of zoonotic viruses (1–3). To date, over 1000 bat subspecies, accounting for 25% of all mammal species, have been recognized (4, 5). The origin of bats can be traced to 52.5 million years ago (6). As the only type of mammal that can fly continuously, bats can carry and spread viruses easily (7). Surprisingly, bats exhibit no visible symptoms of viral diseases but carry viruses that are lethal to humans and other mammals, such as Hendra virus (HEV) (8, 9), Nipah virus (10, 11), severe acute respiratory syndrome–coronavirus (CoV) (12–14), Middle East respiratory syndrome–CoV (15–17), and Ebola virus (18). Therefore, bats have led to tremendous threats and panics all over the world. The Australian black flying fox (Pteropus alecto), a giant bat species in Australia, Papua New Guinea, and Indonesia, has attracted much attention since the discovery of its relationship with the emergence of human fatal zoonotic viruses (19).
Recently, several studies have been carried out to explain the immunologic mechanisms by which bats coexist with viruses (20–28). Sequencing of the whole genomes of P. alecto and Myotis davidii revealed unexpected concentrations of positively selected genes in the DNA damage checkpoint and NF-κB pathways that are related to the expansion and contraction of important gene families (20). Subsequent research reported that these bats hosted contracted type I IFN loci and could constitutively express IFN-α in bat-derived cells, avoiding viral infection (21). This finding provided vital evidence that a special immune system exists in bats and helped to decipher how they withstood viral contamination during early invasion. However, a thorough understanding of the nature of bat immunity remains lacking, especially regarding the role of adaptive immunity (i.e., cell-mediated immunity in clearing endogenous Ag via CTLs activated by MHC class I [MHC I] bound with specific epitopes).
The first information on bat adaptive cellular immunity might date to 1979, when it was reported that under the stimulation of PHA, a type of T cell mitogen, the generation of lymphocytes in bats was delayed (∼2 d) relative to that in mice (22). Since then, several studies have reproduced this phenomenon (23–25). Subsequently, high-quality whole-genome sequencing unexpectedly revealed 12 MHC I loci located both within and outside the canonical MHC loci that interact with the expansion of type I IFNs and a cluster of differentiation 159a (CD159a and NKG2) genes and lead to a higher activation threshold of NK cells (26). This finding implied an important and unique role of MHC I in bats during both Ag presentation and the related CTL-mediated immunity against viral infection. Moreover, a recent study reported the functional characterization of bat MHC I molecules (P. alecto MHC I allele [Ptal]–N*01:01), which revealed that bat MHC I can present HEV-derived epitopes in HEV-infected cells (27, 28). In this study, several bat MHC I–binding partners, including calnexin, calreticulin, protein disulfide isomerase A3, tapasin, TAP1, and TAP2, were identified. Through polypeptide identification by mass spectrometry (MS), the preference of pocket B and pocket F of Ptal-N*01:01 for hydrophobic amino acids was discovered. Notably, P1-D, the aspartic acid as the first amino acid of the N terminus of the binding polypeptide, was presented at >10-fold the expected frequency (27). In addition, the proline at the C terminus of the polypeptide (PΩ-P) was reported as the second preferred amino acid. However, because no studies on the structure of the bat MHC I molecule have been performed, it remains unclear how these residues participate in the process of viral antigenic peptide presentation, including whether this expanded MHC I loci strengthens the CTL-mediated immune response in bats.
Because of the distinctive characteristics of bat cellular immunity and the pivotal role of MHC I in Ag presentation, the structural biological study of MHC I presentation in bats will help clarify how the CTL-mediated immune response functions during viral infection in bats. In this study, crystal structures of peptide–MHC I (p/MHC I) complexes of P. alecto, Ptal-N*01:01/HEV-1 (DFANTFLP) and Ptal-N*01:01/HEV-2 (DYINTNLVP), and two related mutants, Ptal-N*01:01/HEV-1PΩL (DFANTFLL) and p/Ptal-N*01:01ΔMDL/HEV-1, were determined. Through these structures, we illuminated the special features of bat MHC I and the structural basis of the presentation of abundant peptides from lethal viruses. More interestingly, we found that relative to the α1 domain of the H chain (HC) in other mammals, 3 aa (MDL52–54) have been added to that of bats; these help form an additional salt-bridge chain and contribute to the maintenance of peptide self-stability. By using a random peptide library in de novo liquid chromatography–tandem MS (RPLD-MS) analysis, we found an absence of P1-D preference in the peptides presented by Ptal-N*01:01 without the cell components, which was in contrast to the peptidome eluted from the cell surface (27). This result implied that this salt-bridge chain was closely related to the P1-D choice in the cell. The pulling force engendered from the additional ionic bond confers peptides with N-terminal aspartic acids high affinity with bat MHC I, which may be selected during the process of peptide exchange in the cell environment. Our results elucidated the structural basis of the special bat MHC I presentation and will help us better understand bat antiviral cellular immunity as well as how bats coexist with lethal viruses.
Materials and Methods
Peptide synthesis
The peptides used in this experiment are listed in Supplemental Table I. HEV-1 and HEV-2, the two epitope peptides, were reported previously (27, 28). EBOLAGP1_483 was predicted by NetMHCpan 4.0 with the highest score (http://www.cbs.dtu.dk/services/NetMHCpan/) (score = 0.46) (29). Two synthetic random peptide libraries, Ran_9Xsplitted (XXXXXXXXX, where X is a random amino acid other than cysteine) and HEV-1PΩX (DFANTFLX), were synthesized separately to ensure randomness (30). The peptides were synthesized, purified to >90% using reverse-phase HPLC (SciLight Biotechnology and Top-Biotechnology), stored as lyophilized powder at −80°C, and dissolved in DMSO before use.
Protein preparation
With information from the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP) (31) and TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (32), the extracellular region without signal peptide of bat MHC I (termed Ptal-N*01:01) HC (residues 1–279) (GenBank no. AMD11115.1) and β-2-microglobulin (β2m) (residues 1–98) (GenBank no. XP_006920478.1) were codon optimized and synthesized (GENEWIZ, Beijing). Then, they were built into pET-21a (Novagen) and named pET-21a-PtalH and pET-21a-PtalB, respectively (33). The constructed plasmids were transformed into Escherichia coli strain BL21 (DE3). After prokaryotic expression and purification as inclusion bodies, as described previously (33), the purified proteins were dissolved in 6 M guanidine hydrochloride at a final concentration of 30 mg/ml and stored at −20°C.
Assembly of p/Ptal-N*01:01 complexes
At a molar ratio of P. alecto MHC I HC (Ptal-N*01:01): P. alecto MHC I L chain (Ptal-β2m): peptide (HEV-1, HEV-2, and HEV-1PΩL) = 1: 1: 3, inclusion bodies were renatured using a previously described gradual dilution method in refolding buffer composed of 100 mM Tris (pH 8), 400 mM l-arginine, 2 mM EDTA, 5 mM reduced glutathione, and 0.5 mM glutathione disulfide for 24 h at 4°C (33). Then, the product was concentrated with a 10-kDa filter followed by column chromatography (Superdex 200 16/60 column and Resource Q anion exchange column, GE Healthcare) to obtain the purified p/Ptal-N*01:01 complexes (33).
Preparation of p/Ptal-N*01:01ΔMDL
Using overlapped PCR, pET-21a-PtalH without residues 52–54 was obtained and named pET-21a-MDL (primers used for mutation were as follows: 5′-TGCACCGTGGGTTGAGCAGC-3′ and 5′-GCTGCTCAACCCACGGTGCA-3′). The recombinant protein in the inclusion bodies was further purified by the method described above. The product was termed Ptal-N*01:01ΔMDL and was refolded with β2m and the peptide. The complex was then further purified by gel filtration and anion exchange chromatography as described previously (33).
Liquid chromatography–tandem MS
The proteins and binding peptides, p/Ptal-N*01:01, were mixed with 0.2 N acetic acid and placed in a 90°C water bath for 5 min for peptide elution. They were then concentrated with a 3-kDa filter to collect the peptides. The samples were desalted using C18 tips built in house as follows: first, 200 μl of methanol was used to revitalize the desalting tips, and then the tips were equilibrated with 200 μl of 0.1% (v/v) trifluoroacetic acid (TFA). The peptide was washed twice with 200 μl of 0.1% (v/v) TFA and then eluted with 200 μl of a solution of 0.1% (v/v) TFA and 75% (v/v) acetonitrile. These eluted peptides were lyophilized and stored at −80°C. To separate these peptides, the EasyNano LC 1000 system (San Jose, California, Thermo Fisher Scientific) was used. The desalted peptides were first loaded on a trap column (5-μm pore size, 150-μm i.d. × 3-cm length, 120 Å) constructed in house and then separated on a custom-made C18 column (3-μm pore size, 75-μm i.d. × 15-cm length, 100 Å) constructed in house with a flow rate of 450 μl/min. A 60-min linear gradient was performed as follows: 3% B (0.1% formic acid in acetonitrile [v/v]/97% A (0.1% formic acid in H2O [v/v]) to 6% B in 8 min, 6% B to 22% B in 37 min, 22% B to 35% B in 8 min, 35% B to 100% B in 2 min, and 100% B for 5 min. The acquisition of MS data was carried out by a Q Exactive HF (Bremen, Thermo Fisher Scientific) in data-dependent acquisition mode, and the top 20 precursors by intensity from mass range m/z 300 to 1800 were sequentially fragmented with higher energy collisional dissociation, normalized collision energy 27. The dynamic exclusion time was 20 s. Automatic gain control for MS1 and MS2 was set to 3e6 and 1e, and the resolution for MS1 and MS2 was set to 120 and 30K.
De novo analysis and peptide scoring
Based on the spectrum information, the software resolved each of the nonapeptides by probability and derived the peptide with the highest probability from each spectrum. For the identified polypeptides, adjusted by the detection threshold, sequence length = 9, and score ≥ 50 (34, 35). First, to determine the restricted motif of p/Ptal-N*01:01 presentation, the SD (σ) and average value () as well as the coefficient of variation (Vs) were calculated as follows (36):
Using this method, the likelihood of each amino acid occurring at positions P1 to PΩ of the peptides was estimated. A peptide position with a higher Vs was predicted to be a restricted position in the presentation. Second, according to the weighting probability (based on the position-probability matrix) of every type of amino acid located at every single locus of nine (37, 38), the elements and specific amino acids at the predicted restricted motif (given the number of restricted motifs and numbered from 1) were valued () as follows:
where is the number of nonapeptides obtained from the MS experiment and is the set of symbols in the alphabet, given , with representing the amino acid species at specific positions with amino acid abbreviations. is the set of symbols in the alphabet, and is an indicator function where is valued as 1 when and 0 otherwise. After each amino acid type at the position of the restricted motif is valued, the target peptides were scored (S) by the added values of specific amino acids at their predicted restricted motifs (specific is represented as ) as follows:
Crystallization and data collection
Two epitope peptides, HEV-1 (DFANTFLP), derived from HEV (NC_001906) phosphoprotein P residues 481–488, and HEV-2 (DYINTNLVP), derived from HEV fusion protein residues 177–185, and an artificial nonapeptide, HEV-1PΩL (DFANTFLL), were assembled and crystallized with Ptal-N*01:01 and Ptal-β2m. The HEV-1 epitope was assembled and crystallized with p/Ptal-N*01:01ΔMDL complexed with β2m. All of the crystals were first screened by a crystallization screen kit (Hampton Research) by the sitting-drop method. Purified protein complexes were diluted by molecular sieve buffer to 4 and 8 μg/μl and then mixed with crystallization screen kit buffers at a volume ratio of 1:1. p/Ptal-N*01:01/HEV-1, p/Ptal-N*01:01/HEV-2, and p/Ptal-N*01:01/HEV-1PΩL grew well as relatively thin slices in 0.2 M lithium sulfate monohydrate, 0.1 M BIS-TRIS pH 6.5, and 25% w/v polyethylene glycol 3350. p/Ptal-N*01:01ΔMDL/HEV-1 grew well as relatively thick slices in 0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dehydrate pH 5.6, and 30% w/v polyethylene glycol 4000. After sitting-drop crystallization, the crystal growth conditions for the complexes were optimized by the hanging-drop method. Diffraction data were collected at Beamline BL17U (wavelength, 0.97892 Å) of the Shanghai Synchrotron Radiation Facility with the help of an R-AXIS IV++ imaging plate detector. The obtained diffraction data were indexed and integrated by iMosflm (39); then, scaling and merging were performed through CCP4i suite (40).
Structure determination, refinement, and data analysis
For all the scaled and merged diffraction data of p/Ptal-N*01:01/HEV-1, p/Ptal-N*01:01/HEV-2, p/Ptal-N*01:01/HEV-1PΩL, and p/Ptal-N*01:01ΔMDL/HEV-1, the phases were calculated after the structure was determined by MOLREP (41) and Phaser (42) [using the Molecular Replacement Method (43)] in CCP4i to obtain the model and coordinates. After correction of the maps in coot (44) and several rounds of refinement in Refmac5 (45), phenix.refine (46) was used for further refinement. Finally, MolProbity (47) tools were used in Phenix for model quality assessment (48) (Table I). PyMOL (Schrödinger, LLC) and Chimera (University of California–San Francisco) were used for structure-correlation diagram analysis and plotting (49). Sequence alignment and phylogenetic tree construction were performed by Clustal Ω and Jalview (50, 51).
Table I. X-ray diffraction data collection and refinement statistics.
| Ptal-N*01:01/HEV-1 | Ptal-N*01:01/HEV-2 | Ptal-N*01:01/HEV-1PΩL | Ptal-N*01:01ΔMDL/HEV-1 | |
|---|---|---|---|---|
| Data collectiona | ||||
| Space group | C2221 | C2221 | C2221 | C2221 |
| Cell parameters (Å) | 72.66, 86.93, 135.54 | 72.126, 87.871, 133.99 | 72.876, 86.44, 135.12 | 73.343, 80.198, 129.54 |
| Resolution | 135.54–1.93 | 50.00–1.97 | 135.12–1.93 | 64.77–2.70 |
| Rmergeb | 0.110 (0.603) | 0.213 (0.614) | 0.285 (0.628) | 0.431 (0.845) |
| Average I/σ | 16.0 | 2.6 | 19.0 | 5.8 |
| Completeness (%) | 99.8 (100.0) | 75.0 (37.5) | 100.0 (100.0) | 99.5 (98.7) |
| N (observed) | 466, 731 | 118, 661 | 203, 807 | 68, 066 |
| N (unique) | 23, 392 | 12, 065 | 14, 538 | 9, 390 |
| Refinea | ||||
| Resolution | 14.90–2.20 | 28.71–2.70 | 67.56–2.54 | 64.77–2.90 |
| Rwork/Rfreec | 0.1844/0.2300 | 0.2153/0.2925 | 0.2178/0.2811 | 0.2129/0.2932 |
| RMSD (bonds) | 0.01 | 0.00 | 0.00 | 0.00 |
| RMSD (angles) | 0.91 | 0.65 | 0.66 | 0.53 |
| Average B-factor | 49.90 | 47.77 | 37.38 | 22.50 |
| Ramachandran outliers (%) | 0.79 | 0.79 | 0.53 | 0.80 |
| Ramachandran favored (%) | 95.51 | 96.58 | 94.20 | 95.48 |
| Rotamer outliers | 1.2 | 0.3 | 0.6 | 0.0 |
| C-β outliers | 0 | 0 | 0 | 0 |
| Clashscore | 6.89 | 8.31 | 8.52 | 5.92 |
Numbers in parentheses indicate the highest resolution shell.
Rmerge =, where I is the integrated intensity of the reflection.
R =. Rfree is the R factor for a subset (5%) of reflections that was selected prior to refinement calculations and not included in the refinement.
Results
The overall structure of p/Ptal-N*01:01
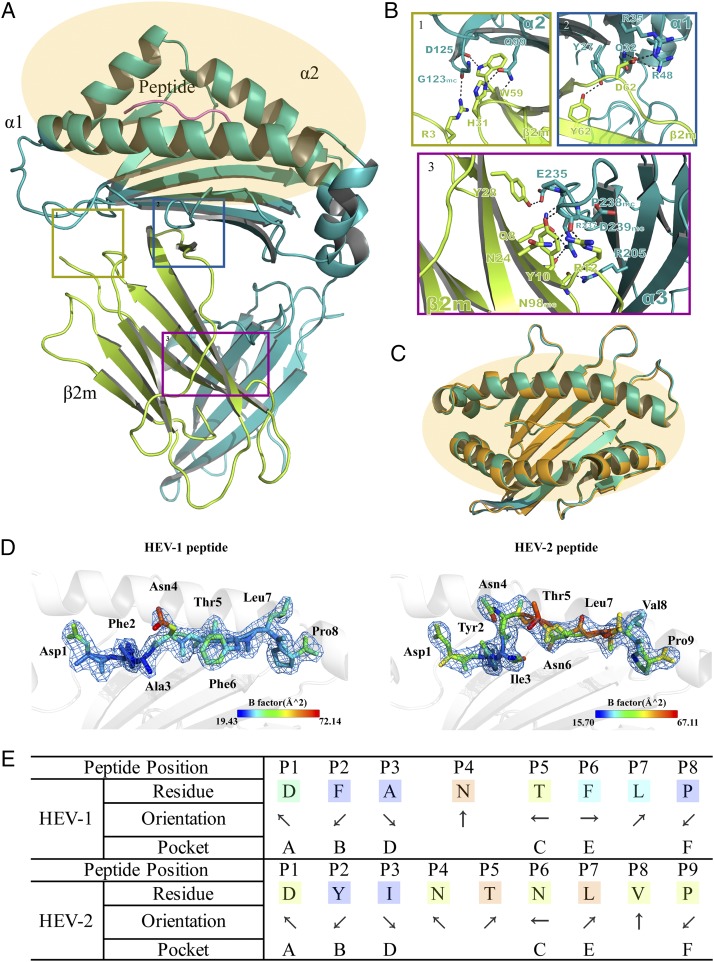
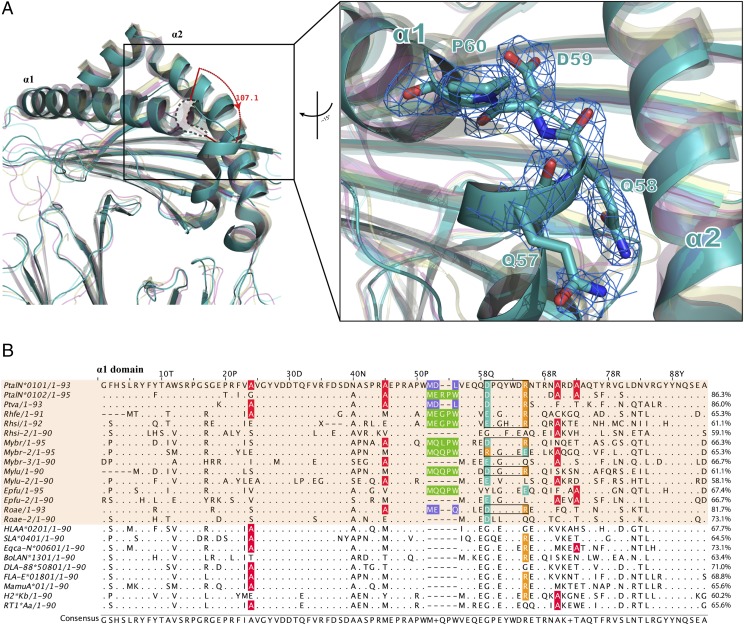
The crystal structures of Ptal-N*01:01 presenting the HEV-derived epitope octapeptide and nonapeptide, termed HEV-1 (DFANTFLP) and HEV-2 (DYINTNLVP), were determined to resolutions of 2.2 and 2.7 Å, respectively, through molecular replacement using a 1.5-Å coordinate from HLA-B*3508 (Protein Data Bank identification [PDB ID]: 3VFN) as an initial phasing model. The two crystals were crystallized in the C2221 orthorhombic space group with no other asymmetric units (Table I). The topological structures of the p/Ptal-N*01:01/HEV-1 and p/Ptal-N*01:01/HEV-2 complexes shared similar features (Fig. 1A, 1C) analogous to those of the known mammalian complexes, with the α1 and α2 domains consisting of antiparallel β-sheets spanned by two long α-helixes supported by the α3 domain and β2m (52). The L chains (β2m) and HC were linked by hydrogens and Van der Waals forces that are conserved in mammals, which are also the same in Ptal-N*01:01, mainly consisting of 17 H-bonds and forming the contact area of ∼1500.3 Å2. The H-bonds between p/Ptal-N*01:01 HC and β2m are shown in Fig. 1B. The Ag-binding groove (ABG) is formed by the α1 and α2 domains to bind polypeptide ligands derived from viruses to help activate CTL-mediated immunity during virus infection (52). The HEV-1 and HEV-2 epitope peptides are both located in the ABG (Fig. 1C, 1D). The difference in length between the HEV-1 and HEV-2 epitope peptides resulted in conformational differences in the polypeptide, primarily at the P4 and P5 positions. Residues P3–P6 of HEV-1 were more rooted in the ABG, and the transition between P3–P6 was more dramatic in HEV-2 (Fig. 1C). Generally, P4 of HEV-1 corresponded to P4 and P5 of HEV-2. Other positions of both HEV-1 and HEV-2 shared similar anchoring sites and orientations (Fig. 1E). Compared with the known MHC I molecular structures, the most significant trait of p/Ptal-N*01:01 was at the helix of the α1 domain, which α-helix was flipped at ∼107.1° in the left upper corner of the α1 domain (Fig. 2A). Through sequence alignment, this type of folding in p/Ptal-N*01:01 was triggered by the pushing force from the 3 aa MDL52–54, as shown in Fig. 2B. Similar to MDL52–54, this type of residue insertion module was common among all published bat subspecies MHC I molecule sequences. As shown in Fig. 2B, the three residue insertions were found in P. vampyrus, P. alecto, and Rousettus aegyptiacus (three megabats) and 5 aa insertions throughout the other bat subspecies.
FIGURE 1.
Overview of p/Ptal-N*01:01/HEV-1 and binding peptide comparison with p/Ptal-N*01:01/HEV-2. (A) The HC of p/Ptal-N*01:01/HEV-1 is shown in teal, composed of three domains named α1, α2, and α3. The L chain, β2m, is shown in yellow. The peptide HEV-1 (DFANTFLP) is shown in violet. The ABG is highlighted in orange. (B) The H-bonds between β2m and the HC (from 1 to 3 showing the interactions in domains α2, α1, and α3, respectively). The H-bonds are represented by black dotted lines. The corresponding residues are labeled with amino acid abbreviations and primary sequence numbers. Mc indicates the main chain of the corresponding amino acids. Nitrogen atoms are shown in blue and oxygen atoms in red. (C) Comparison of the binding clefts of p/Ptal-N*01:01/HEV-1 (octapeptide) in teal and p/Ptal-N*01:01/HEV-2 (nonapeptide) in bright orange. With p/Ptal-N*01:01/HEV-1 as the reference, the RMSD between the HC was 0.436 Å, between β2m was 0.292 Å, and between the ligands was 1.814 Å. (D) The peptide comparison between HEV-1 and HEV-2 binding with p/Ptal-N*01:01 shown as a cartoon representation. Corresponding amino acids are labeled and colored by B-factor. (E) The orientation and location of HEV-1 and HEV-2 binding in the pocket of p/Ptal-N*01:01. The arrows pointing upward or downward represent the direction of the amino acid side chains toward the TCR or the MHC I base, respectively. The arrows pointing left or right represent the amino acid side chains facing the α1 domain or the α2 domain, respectively.
FIGURE 2.
Structural and sequence alignment. (A) Structural comparison among p/Ptal-N*01:01/HEV-1 (teal), bony fish MHC I (PDB ID: 5H5Z, magenta), chicken MHC I p/BF2*04:01 (PDB ID: 4EOR, yellow), and human MHC I p/HLA-A*0201 (PDB ID: 3I6K, cyan). The magnified inset on the right suggests that the p/Ptal-N*01:01’s ABG in the α1 domain is different from the reported MHC I structures of other representative species. At the upper right corner of the proximal end in the figure, there was an α helix flipped ∼107.1° compared with those in other MHC I molecules. The (2Fo-Fc) map of these structurally differential amino acids (shown as teal sticks) is shown on the right. All the structures are shown as cartoon representations. p/Ptal-N*01:01/HEV-1 has been highlighted with no transparency, whereas other MHC I molecules are shown with 50% transparency. (B) Protein primary sequence alignment of MHC I α1 domains in different selected representative mammals. Bat MHC I molecules are shown in a transparent orange background. Alanine in pocket B is colored red. The inserted 3 or 5 aa are boxed and colored in purple or green, respectively. Special salt bridges formed between Asp59 and Arg65 are boxed in black, acidic amino acids are colored teal, and basic amino acids are colored orange. With p/Ptal-N*01:01 as the reference; the similarities of other sequences are marked at the end of each sequence.
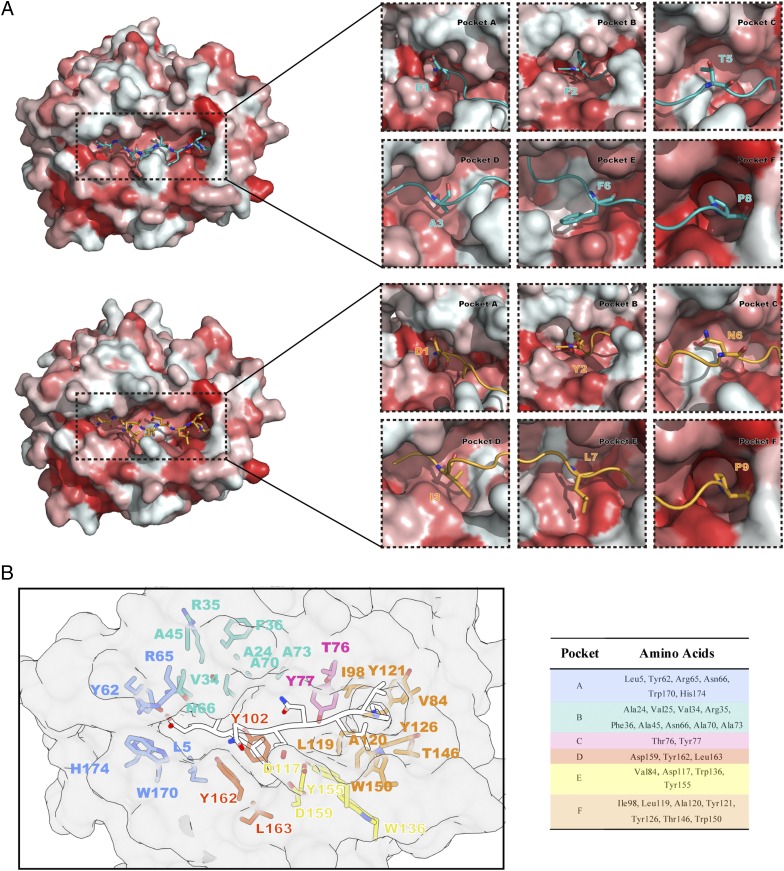
The high adaptability of ABG for binding peptides
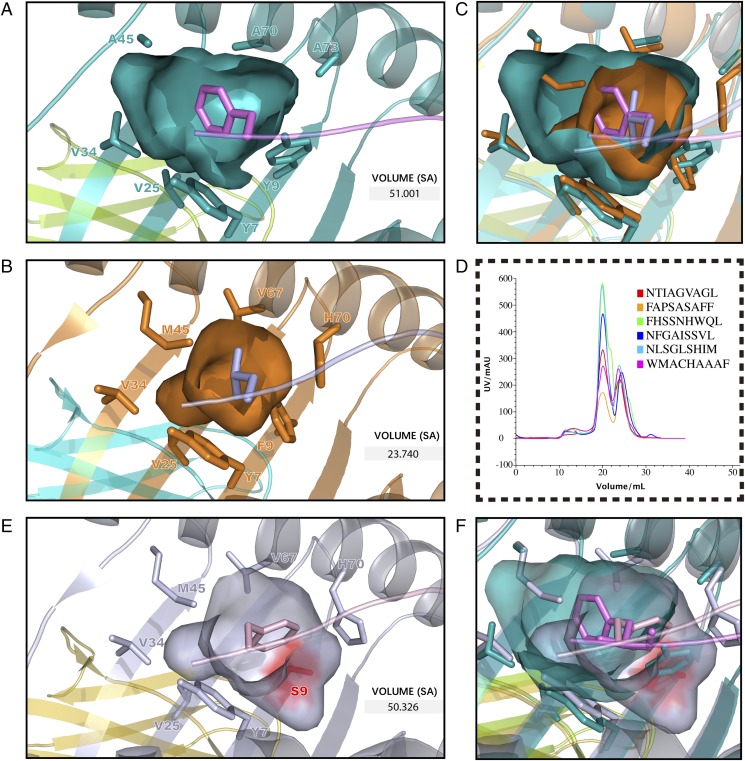
The ABG of p/Ptal-N*01:01 was decided by six pockets named pockets A through F. Structural alignment between p/Ptal-N*01:01/HEV-1 and p/Ptal-N*01:01/HEV-2 (root-mean-square deviation [RMSD] of HC = 0.436 Å) revealed a minor difference in the pocket surface shape [shown in terms of the Eisenberg hydrophobicity scale (53)] regardless of whether the binding peptide was an octapeptide or a nonapeptide (Fig. 3A). The ABG residue composition of p/Ptal-N*01:01 is shown in Fig. 3B. In the structure of p/Ptal-N*01:01, P2 and PΩ fit deeply into pocket B and pocket F, respectively, which were important for peptide binding. Pocket B possessed a multi-Ala assembly at positions 24, 45, 70, and 73, respectively, making pocket B more spacious (Fig. 4A). In the reported p/MHC I structures, most of the human MHC I molecules, such as HLA-A*0201, which was in allele group HLA-A*02 and globally common, have a comparatively large pocket B (23.74 Å3), as shown in Fig. 4B. However, the multi-Ala assembly caused pocket B of p/Ptal-N*01:01 to possess a volume of 51.00 Å3, which was significantly larger than that of HLA-A*0201 (Fig. 4A–C). We also compared pocket B of Ptal-N*01:01 with HLA-A*2402, which also has a preference for amino acids with a bulky side chain at P2. It is worth noting that HLA-A*2402 has a similar B pocket composition with HLA-A*0201, including Met45, which influenced the size as shown in Fig. 4B. However, Ser9 of HLA-A*2402 provides a large space for the side chain at P2 and guides it from the other side. Compared with pocket B of HLA-A*2402, Ptal-N*01:01 uses a different approach (Fig. 4E, 4F). In the peptide refolding experiment, histidine, threonine, phenylalanine, leucine, tyrosine, and alanine (amino acids with different traits at the P2 position) could help p/Ptal-N*01:01 refold well (Fig. 4D). This behavior illustrated that pocket B of p/Ptal-N*01:01 could hold multifarious hydrophobic or mildly hydrophilic amino acids, even with a remarkable differential in the side-chain spatial conformation.
FIGURE 3.
Pocket view and surface comparison between p/Ptal-N*01:01/HEV-1 and p/Ptal-N*01:01/HEV-2. (A) On the left is the model of Ptal-N*01:01/HEV-1 (up) and Ptal-N*01:01/HEV-2 (down), surface colored according to the Eisenberg hydrophobicity scale. Hydrophilic amino acids to hydrophobic amino acids are colored in a gradient from white to red. On the right is a comparison between six pockets of p/Ptal-N*01:01/HEV-1 and p/Ptal-N*01:01/HEV-2. The pockets and corresponding amino acids are labeled. The peptides of p/Ptal-N*01:01/HEV-1 (teal) and p/Ptal-N*01:01/HEV-2 (bright orange) are shown in cartoon and stick representation. (B) The pocket residues are colored and labeled with corresponding amino acid abbreviations and sequence numbers, as shown in the table on the right, and peptide HEV-2 is shown in cartoon representation with 50% transparency in white.
FIGURE 4.
The relatively large pocket B of p/Ptal-N*01:01. (A) Pocket B of p/Ptal-N*01:01/HEV-1 (teal) is shown as cavities and pockets. (B) Pocket B of p/HLA-A*0201 (PDB ID: 3I6K, bright orange) is shown as cavities and pockets. (C) Structure comparison of pocket B between Ptal-N*01:01 and HLA-A*0201. (D) AKTA molecular sieve purification results of in vitro peptide binding assay of p/Ptal-N*01:01. The abscissa shows the peak volume (ml), whereas the ordinate represents the intensity of UV (mAU). (E) Pocket B of p/HLA-A*2402 (PDB ID: 3I6L, blue white) is shown as cavities and pockets. Ser9 is colored in red for observation. (F) Structure comparison of pocket B between Ptal-N*01:01 and HLA-A*2402. The pockets are shown as surfaces composed of amino acids, which are shown as sticks. The predicted Richards solvent-accessible (SA) surface volumes are labeled.
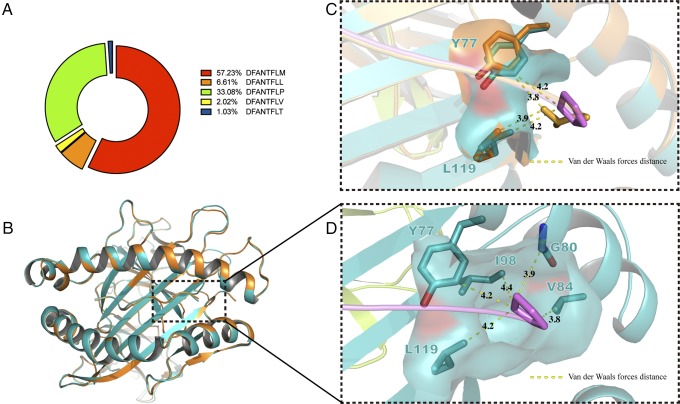
Another essential part of MHC I was pocket F, which formed conserved H-bonds between -COOH of PΩ and Tyr87. Pocket F of p/Ptal*N-01:01 was previously reported to dominantly enwrap proline at PΩ (27). The proline preference at PΩ was rarely reported, indicating that pocket F of p/Ptal-N*01:01 is uncommon and characteristic. First, the detailed pocket F binding feature was reverified quantitatively and accurately. p/Ptal-N*01:01 was refolded with split-synthesized DFANTFLX (X equal to any of the 20 aa, excluding cystine). After the peptides in the p/Ptal-N*01:01 complex were eluted and collected, they were sampled by liquid chromatography–tandem MS (LC-MS/MS) to retrieve DFANTFLX. The peptide chromatographic peak areas detected were extracted to compare with the relative content of each, and the result is shown in Fig. 5A. DFANTFLM represented a relatively large proportion, followed by DFANTFLP and DFANTFLL, which further verified the proline preference of the F pocket. To explain this phenomenon, Ptal-N*01:01 HC was refolded with β2m and DFANTFLL (HEV-1PΩL). After purification, we resolved the crystal structure of p/Ptal-N*01:01/HEV-1PΩL. In the structure of p/Ptal-N*01:01/HEV-1, proline at PΩ interacted with Ile98, Tyr77, Leu119, Gly80, and Val84 through Van der Waals forces, surrounding itself in a very hydrophobic environment. Gly80 played an important role in the formation of Van der Waals forces between PΩ proline and pocket F, which was the only amino acid without a side chain. If this residue was mutated to any other amino acid, the force network that interacts with proline would be destroyed by extending the distance between the side chain of amino acid at position 80 and the PΩ proline to ∼2.5 Å (<3.0 Å). Compared with the binding peptide in p/Ptal-N*01:01/HEV-1, the main chain of HEV-1PΩL showed no visible change (RMSD = 0.130 Å) (Fig. 5B). However, we found that Leu119 was slightly deflected, resulting in a suitable distance between Leu119 and PΩ maintained at 3.9 Å ∼4.2 Å (Fig. 5C, 5D). This deflection worked as a soft paw to keep the hydrophobic PΩ residues with different types of side chains attached to pocket F. These properties allowed pocket F to accommodate hydrophobic amino acids even if the side chains themselves were not suitable for anchoring, as in the case of proline. The essential and featured pocket B and pocket F together provided p/Ptal-N*01:01 with the potential for high adaptability for peptide binding.
FIGURE 5.
The flexible pocket F of p/Ptal-N*01:01. (A) MS results for DFANTFLX refolded with p/Ptal-N*01:01. (B) The structural alignment between p/Ptal-N*01:01/HEV-1 (teal) and p/Ptal-N*01:01/HEV-1PΩL (bright orange). With p/Ptal-N*01:01/HEV-1 as a reference, the HC RMSD was 0.379 Å, the β2m RMSD was 0.201 Å, and the ligand RMSD was 0.130 Å. (C) The flexible pocket F. (D) The Van der Waals forces around the PΩ proline in Ptal-N*01:01/HEV-1. The corresponding amino acids are shown as sticks. PΩ proline and PΩ leucine are colored violet and bright orange, respectively, and the corresponding distance is indicated by yellow dashed lines and labeled with the length in angstroms.
The salt-bridge chain reinforces the interaction between pocket A and P1-D
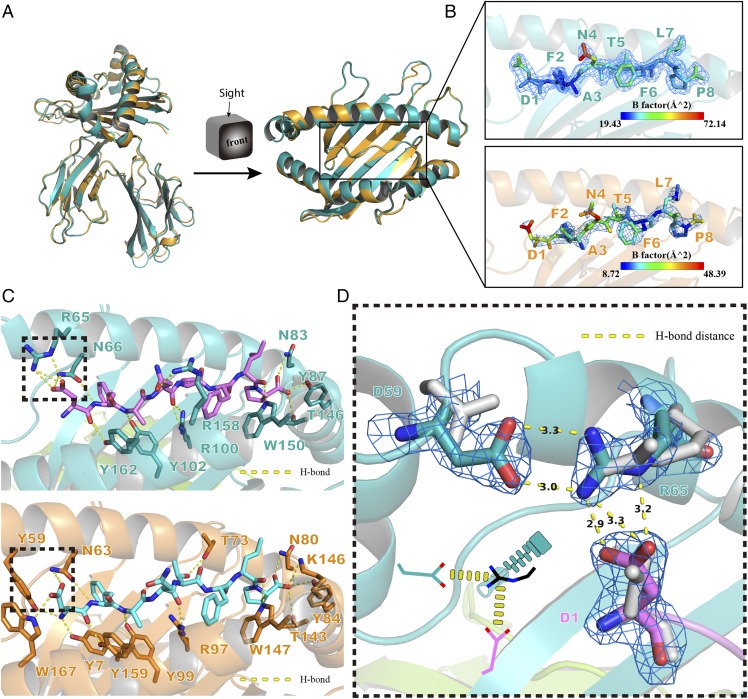
There were three additional residues, MDL52–54, at the α1 domain of p/Ptal-N*01:01. To elucidate the function, they were deleted from Ptal-N*01:01 HC. After the preparation and purification of this mutant HC refolded with HEV-1 peptide and β2m, the complex was obtained and termed p/Ptal-N*01:01ΔMDL/HEV-1. The crystal structure of p/Ptal-N*01:01ΔMDL/HEV-1 was resolved and is shown in Fig. 6A. Although the peptide is bound in p/Ptal-N*01:01ΔMDL, the structure appeared unchanged compared with that of p/Ptal-N*01:01 (RMSD of atomic positions = 0.393 Å). However, from the Debye–Waller factor (B-factor), the peptide bound to p/Ptal-N*01:01ΔMDL was obviously unstable compared with p/Ptal-N*01:01, especially at P1 (Fig. 6B). The H-bonds between the peptide and the ABG were analyzed, and Arg65 of p/Ptal-N*01:01 was found to form a salt bridge with P1-D of the peptide. However, p/Ptal-N*01:01ΔMDL has lost this salt bridge (Fig. 6C). In addition, Asp59 in p/Ptal-N*01:01 formed another salt bridge with Arg65, which fixed the complex. The first salt bridge between Asp59 and Arg65 (the left arm) and the second salt bridge between Arg65 and P1-D (the right arm) formed a salt-bridge chain that served as a zipper, pulling the α1 domain and peptide together tightly; this feature was absent from the structure of p/Ptal-N*01:01ΔMDL (Fig. 6D). This salt-bridge chain was probably related to the previously reported P1-D preference (27).
FIGURE 6.
The special salt-bridge chain in p/Ptal-N*01:01. (A) Front and top view of structural alignment between p/Ptal-N*01:01/HEV-1 (teal) and p/Ptal-N*01:01ΔMDL/HEV-1 (bright orange). Using Ptal-N*01:01/HEV-1 as the reference, the HC RMSD was 1.081 Å, the β2m RMSD was 0.446 Å, and the ligand RMSD was 0.393 Å. (B) The side view of ligands bound in MHC I of Ptal-N*01:01/HEV-1 (teal) and p/Ptal-N*01:01ΔMDL/HEV-1 (bright orange). Ligands are colored by B-factor and enveloped in the (2Fo-Fc) electron density. (C) H-bond interactions between peptide and ABG predicted in p/Ptal-N*01:01 (up) and p/Ptal-N*01:01ΔMDL (down). Binding groove composition residues were labeled with amino acid abbreviations, and H-bonds are shown as yellow dotted lines. (D) The special salt-bridge chain formed in p/Ptal-N*01:01/HEV-1 shown as sticks enveloped in (2Fo-Fc) electron density compared with p/Ptal-N*01:01ΔMDL/HEV-1 in gray. The lower left corner shows a sketch map of this special salt-bridge chain.
The differential preference for P1-D in or out of the cell environment
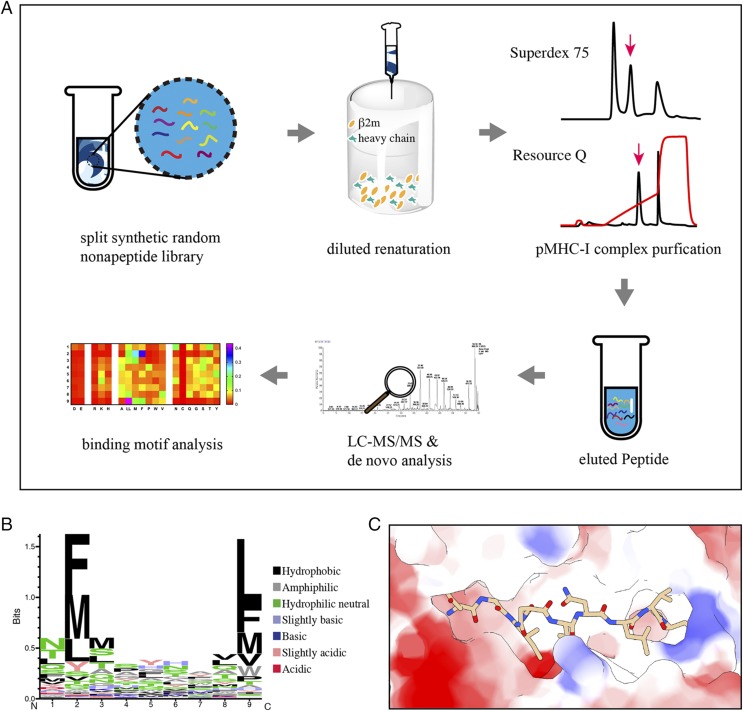
To investigate the presentation of p/Ptal-N*01:01 without cellular components, the split-synthesized random nonapeptide library was renatured with Ptal-N*01:01 HC and β2m. After size-exclusion chromatography followed by ion exchange chromatography, the enriched random peptides were eluted from the p/Ptal-N*01:01 complex and loaded for de novo peptide sequencing via LC-MS/MS, termed RPLD-MS (Fig. 7A). Finally, 2264 profoundly convinced nonapeptides were detected. To verify the accuracy of the MS result, two polypeptides (DENOVOMDM and DENOVOFHS) were chosen that did not wholly conform to the predicted motif but were still detected and had the capacity to assemble with Ptal-N*01:01 HC and β2m. The results are shown in Supplemental Table I. According to the weighted probability (position-probability matrix) (37, 38) of every type of amino acid located at each of the nine loci, we scored each of the amino acids as described in Materials and Methods and then depicted the resulting logo by a heat map. The Seq2logo (http://www.cbs.dtu.dk/biotools/Seq2Logo-2.1/) with the Shannon presentation type is presented (Fig. 7B) (54, 55). From P1 to PΩ, the Vs is 0.92, 1.78, 0.90, 0.60, 0.62, 0.63, 0.47, 0.73, and 1.81, which suggests marked restrictive features mainly in P2 and PΩ, with hydrophobic amino acids such as phenylalanine, methionine, and leucine. All of the peptides used in this experiment were scored and are listed in Supplemental Table I; the results corresponded to the in vitro renaturation results. The mass spectral results were further verified. To clarify the peptide-binding capacity of p/Ptal-N*01:01 more intuitively, the selected virus-derived proteins were scanned at the motif-compliant level based on the scoring system referred to in the Materials and Methods with every 9 aa as a scan step unit. The results further suggested that p/Ptal-N*01:01 had high adaptive potential to bind abundant peptides (Supplemental Fig. 1). Similar presentation characteristics to those revealed from the in-cell peptide MS results were observed, including the P2 and PΩ binding components composed mainly of hydrophobic amino acids (Fig. 7B). The electrostatic potential was calculated and colored according to Coulomb law (49) (Fig. 7C). An obvious negative electrostatic potential of ABG was shown, meaning that acidic amino acids do not easily adhere. Notably, the proportion of aspartic acid at P1 was ∼3.5% to <5.1% (expected frequency, the average weighting of amino acids at P1) and differed significantly from previously reported values (∼60%, >10-fold of expected frequency). This difference illustrated that differences in some mechanisms in or out of the cellular environment have caused differences in the aspartic acid priority in pocket A. Additionally, with respect to the previously reported experiment (27) as well as our results, the preference of proline at PΩ is shown as positively correlated with the preference of P1-D both in and out of the cell (Figs. 5A, 7C). In Fig. 6A, the Ptal-N*01:01–bound polypeptide shifted to the left by ∼0.4 Å in the horizontal direction compared with the mutant, which may indicate that P1-D is pulled to the left through interactions with Arg65. As mentioned before, Gly80 plays an important role in the proline preference in pocket F, and this pulling force will move the polypeptide farther away from Gly80 and make the peptide with a PΩ-P binding more stable, which also explains why the PΩ-P preference can be reduced after P1-D no longer has a preference (Fig. 8).
FIGURE 7.
Results of LC-MS/MS de novo analysis. (A) Schematic view of RPLD-MS flow and results. Gray arrows indicate the direction of the process. The split synthetic random nonapeptide library was renatured with β2m and MHC I HC using the diluted renaturation method. After purification through molecular sieve chromatography and ion exchange chromatography, the purified p/MHC I complex (the peak indicated by the pink arrow) solution was concentrated, and the bound peptides were eluted. Then, the eluted peptides were sampled for LC-MS/MS and de novo analysis. Finally, we obtained the binding motif shown as a heat map of the amino acid (shown as abbreviation) preference distribution at positions 1 ∼9. (B) Sequence logo showing the amino acid weighting probabilities at every position of the presented peptide in the type of Shannon. (C) The ABG was colored by Coulomb law with peptide (HEV-2) bound.
FIGURE 8.
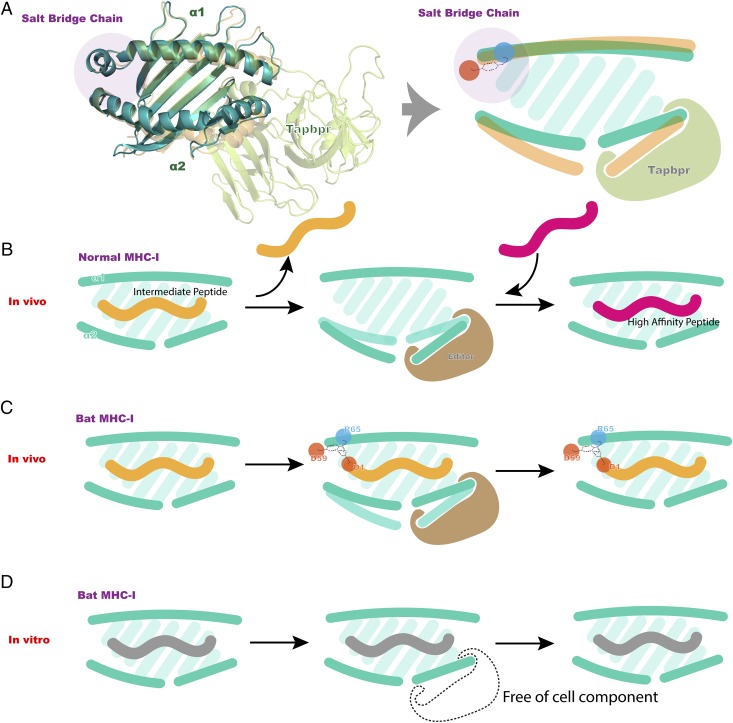
Pattern diagram of bat MHC I peptide presentation. (A) Predicted model of the p/MHC I/Tapbpr complex in 60% transparency compared with p/Ptal-N*01:01/HEV-1 ABG and its analog on the right. (B) Schematic of normal peptide editing and replacement process in vivo. (C) Schematic of the salt-bridge chain in bat MHC I potentially influencing peptide editing and replacement processes in vivo. (D) Schematic of salt-bridge chains potentially influencing peptide editing and replacement processes in vitro. The salt-bridge chain is highlighted in purple circles, whereas the acid and basic amino acids are highlighted in red and blue circles, respectively.
Structural details of bat MHC I
To clarify the MHC I molecular characteristics in bat species that differ from those of other mammals, a full-length sequence alignment was carried out (Supplemental Fig. 2A), including annotations of the salt-bridge chain, ABG pockets, and predicted residues interacted with TCR and CD8αα, illustrated in Supplemental Fig. 2B. The maximum likelihood phylogenetic tree analysis (Supplemental Fig. 2C) indicated that bat MHC I shared an evolutionary branch with other mammals. This finding demonstrated that the bat MHC I molecule has evolved as an immune molecule with distinct differences among species. The constructed superposition model of p/Ptal-N*01:01/HEV-1 (as well as the modeled HC of Ptal-N*01:02, Rhinolophus ferrumequinum and R. sinicus) complexed with TCR and the CD8αα complex revealed no differences (data not shown). Especially for MHC I/CD8αα, the amino acid sequences were highly conserved compared with those in other mammals. The results of the structural conservation analysis of the six pockets are shown in Supplemental Fig. 3. Some characteristic amino acids could provide clues to clarify the way peptides bind the bat MHC I molecule’s ABG. Pocket B was thought to be a binding anchor pocket for most mammals and to encapsulate the side chain at P2, improving the peptide binding. Notably, Ala45 and Ala73 in the B pocket of bat MHC I were very distinct and rarely observed in other mammals. Through structural analysis, we found that they could significantly influence the B pocket size and provide a larger B pocket than in most other MHC I molecules. This large pocket B would facilitate the binding of suitable anchoring residues. In vitro binding experiments showed that pocket B can tolerate various amino acid types. The prevalence of Ala45 in bats suggested that bat MHC I pocket B was generally larger and was beneficial for enwrapping peptide side chains.
Notably, Asn66 in other mammals is usually a glutamic acid, which might create charge repulsion with Asp at P1 (33). Residue 66 in bats was mostly N/S/Q, side chains without marked charge properties, which might contribute to the stability of the charged amino acids at P1 and to the formation of a salt-bridge chain. Conserved salt-bridge chains have been found across almost all bats, which is related to the bat’s special additional amino acids: MDL/MEQ, MER(G)PW/MEGPW, and MQQ(L)PW. This insertion is expected to cause subsequent amino acid anteversion relative to the direction toward the ligand and push Asp59 close to Arg65, resulting in salt bridges between the two side chains. After the Arg65 was fixed by this interaction, aspartic acid at P1 would also form the other salt bridge with Arg65. To the best of our knowledge, this kind of salt-bridge chain was never previously reported in MHC I, and as in the sequence alignment, this finding is not isolated among bats. The HC of Ptal-N*01:02, R. sinicus, and R. ferrumequinum were modeled (data not shown). They could also potentially form salt-bridge chains between Asp/Glu/Arg59 and Arg65 or Glu65. There were also bat MHC I molecules without this type of insertion (as shown in the structure of p/Ptal-N*01:01ΔMDL, no amino acids inserted would break the salt-bridge chain) and ones in which the insertion would not help form a salt-bridge chain; however, these cases were unusual.
Discussion
Recent research has verified that bats harbor a significantly higher proportion of zoonotic viruses than do all other mammalian orders, resulting in widespread concern (56). It is crucial to find clues as to why bats can cope with virulent infections, such as Ebola, HEV, severe acute respiratory syndrome–CoV, and Middle East respiratory syndrome–CoV, that are often lethal to other mammals. Previous reports, such as reports of bats’ multiple MHC I loci and the capacity of bat MHC I to present HEV-derived epitopes in the cell (26, 27), suggest that bat MHC I plays a special role in antiviral immunity and is closely associated with antiviral defense. To better understand and explain the presentation features of bat MHC I, the crystal structure of p/Ptal-N*01:01 was determined. From the structure, we found that the large pocket B and the flexible pocket F worked as the restricted positions for residue anchoring, providing p/Ptal-N*01:01 with high adaptability for peptide binding. By sequence alignment of MHC I in bat species, we found that amino acids closely related to B pocket size, such as Ala45 and Ala73, are ubiquitous in bats compared with other mammals. Even if not all of the binding peptides could contribute to highly stable p/MHC I complexes, they would help MHC I to exist as an intermediate situation in vivo, waiting for replacement with higher-affinity peptides. Only a small fraction of the binding peptides with certain affinities and specific motifs could dock at the cell membrane TCRs. The high adaptability of the peptide and the unique characteristics of the MHC I gene distribution in the bat, with locations both inside and outside the canonical MHC locus, might facilitate the activation of cell-mediated immunity (57).
Compared with the sequence of the reported MHC I structures, there were three additional MDL52–54 in the α1 domain of Ptal-N*01:01, which is a common phenomenon in bats. By analyzing the structural differences in the α1 region between p/Ptal-N*01:01/HEV-1 and its mutant, p/Ptal-N*01:01ΔMDL/HEV-1, we found that this type of insertion was involved in the formation of a salt-bridge chain consisting of one salt bridge between Asp59 and Arg65 (the left arm) and another salt bridge between Arg65 and P1-D (the right arm) characterizing the specificity of pocket A. A recent study demonstrated that pocket A of p/Ptal-N*01:01 had a significant preference (60%) for P1-D in the cell (27). However, the side chain of the P1 residue consistently stretched out of pocket A, which can be expected to generally prevent the P1 pocket from showing apparent restrictions (58). We suspected that this phenomenon might be related to the impact of the salt-bridge chain during peptide exchange. To prove our speculation, RPLD-MS was performed to decipher the in vitro presentation characteristics of p/Ptal-N*01:01 in complete isolation from the cell environment. The aspartic acid at P1 of the binding peptide accounted for only 3.5% of the binding preference, showing that the high-level preference for aspartic acid at P1 disappeared. Furthermore, the in vitro peptide-binding assays demonstrated that P1 was not a significantly restrictive position (Supplemental Table I).
We speculate that the different P1 residue situations in vivo and in vitro are related to the unique salt bridge formed by MDL52–54. The model diagram of our experimental results is shown in Fig. 8. In vivo, a type of low-affinity peptide to a high-affinity peptide exchange called catalyzed proofreading can occur (Fig. 8A, 8B). The unloaded MHC I structure complexed with Tapbpr was reported previously (59). Using the reported structure as a template (PDB ID: 5OPI), the Ptal-N*01:01/Tapbpr complex was modeled (default multiple target mode, https://swissmodel.expasy.org/) with the amino acid sequence of Ptal-N*01:01 HC (GenBank: AMD11115.1) and the Tapasin-related protein of P. alecto (GenBank: ELK10383.1). Afterwards, the unique conformation around the salt-bridge chain was further polished in coot based on the resolved map of p/Ptal-N*01:01/HEV-1. We observed that during peptide exchange, the α2 domain of the MHC I HC would be squeezed open by the inserted Tapbpr loop, whereas the α1 domain would remain relatively unchanged. During peptide processing and exchange, because of the relative stability of the α1 domain, a novel salt bridge in the bat MHC I molecule would maintain attachment to the Asp at P1 of the peptide and influence the polypeptide replacement process (Fig. 8B). The MDL52–54 insertion led to tight binding of P1-D. During the proofreading process, P1-D enabled peptides with a certain affinity to survive the exchange (Fig. 8C). Thus, the preference for aspartic acid at P1 in the cell increased. In our experiment, renaturation was performed under conditions isolated from cellular components and deficient in proofreading-related chaperones. High-ratio peptides would remain bound and not exchange, leading to the disappearance of the P1-D preference (Fig. 8D).
A split synthetic random peptide library for identifying ligand-binding activity (30) has been applied to the screening of MHC I ligands. The results indicated the existence of a similar motif to naturally processed and presented peptides, demonstrating that MHC I complexes refolded in E. coli retain their peptide-binding specificity (60). Recently, de novo LC-MS/MS has rapidly developed and is widely used in peptide sequencing. De novo LC-MS/MS can execute sequencing directly from MS/MS and does not depend on databases, as it can use the fragment ion information from MS/MS in an unbiased manner (61). It completely avoids the large problems of low identification rates (low sensitivity) and high false-discovery rates (low accuracy) of existing database search tools originating from the static search space (62). In this study, we combined the use of a synthetic RPLD-MS and constructed a work flow for rapid and efficient identification of MHC I binding motifs outside the cell environment. Based on our comparison of the peptidome identified with and without cellular components and previous findings, we speculate that this salt-bridge chain might lead to the preference for Asp at the N terminus of the peptide and increase its binding stability during peptide exchange. In this scenario, when the P1 of the binding peptide is Asp, the salt bridge is expected to prevent it from detaching from the ABG during reciprocating peptide exchange. The presentation of Asp in the cell at the P1 position might increase the overall affinity of MHC I–binding polypeptides and improve cellular immunity against viral infections.
In conclusion, through the structural study of p/Ptal-N*01:01, we have described the detailed presentation characteristics of bat MHC I. The large multi-Ala–assembled pocket B and flexible pocket F allow Ptal-N*01:01 to accommodate diverse peptides, and the novel salt-bridge chain near pocket A might promote the presentation of high-affinity peptides by Ptal-N*01:01 after binding peptide exchange in vivo. The identified features are common among bats and might facilitate the activation of efficient cell-mediated immunity. The present study provides insight into the cellular immunity of bats and the mechanisms underlying the ability of bats to survive many viruses that are lethal in other mammals.
Supplementary Material
Acknowledgments
The authors thank everyone who assisted in this work.
This work was supported by the National Natural Science Foundation of China (Grant 31572493), the Beijing Municipal Natural Science Foundation (Grant 6182029), and the National Natural Science Foundation of China (Grant 31572653).
The coordinates and structural factors generated in this study have been submitted to the Protein Data Bank (https://deposit-pdbj.wwpdb.org/deposition/) under accession numbers 6ILC, 6ILE, 6ILF, and 6ILG.
The online version of this article contains supplemental material.
- ABG
- Ag-binding groove
- CoV
- coronavirus
- HC
- H chain
- HEV
- Hendra virus
- LC-MS/MS
- liquid chromatography–tandem MS
- β2m
- β-2-microglobulin
- MHC I
- MHC class I
- MS
- mass spectrometry
- PDB ID
- Protein Data Bank identification
- p/MHC I
- peptide–MHC I
- PΩ-P
- proline at the C terminus of the polypeptide
- Ptal
- P. alecto MHC I allele
- RMSD
- root-mean-square deviation
- RPLD-MS
- random peptide library in de novo liquid chromatography–tandem MS
- TFA
- trifluoroacetic acid
- Vs
- coefficient of variation.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Pawan J. L. 1959. Rabies in the vampire bat of Trinidad, with special reference to the clinical course and the latency of infection. Caribb. Med. J. 21: 137–156. [PubMed] [Google Scholar]
- 2.Pawan J. L. 1959. The transmission of paralytic rabies in Trinidad by the vampire bat (Desmodus rotundus murinus Wagner. Caribb. Med. J. 21: 110–136. [PubMed] [Google Scholar]
- 3.Zhou P., Fan H., Lan T., Yang X. L., Shi W. F., Zhang W., Zhu Y., Zhang Y. W., Xie Q. M., Mani S., et al. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones K. E., Bininda-Emonds O. R., Gittleman J. L. 2005. Bats, clocks, and rocks: diversification patterns in Chiroptera. Evolution 59: 2243–2255. [PubMed] [Google Scholar]
- 5.Turmelle A. S., Olival K. J. 2009. Correlates of viral richness in bats (order Chiroptera). Ecohealth 6: 522–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons N. B., Seymour K. L., Habersetzer J., Gunnell G. F. 2008. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature 451: 818–821. [DOI] [PubMed] [Google Scholar]
- 7.Wang L. F., Walker P. J., Poon L. L. 2011. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr. Opin. Virol. 1: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halpin K., Young P. L., Field H. E., Mackenzie J. S. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81: 1927–1932. [DOI] [PubMed] [Google Scholar]
- 9.Young P. L., Halpin K., Selleck P. W., Field H., Gravel J. L., Kelly M. A., Mackenzie J. S. 1996. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg. Infect. Dis. 2: 239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua K. B., Koh C. L., Hooi P. S., Wee K. F., Khong J. H., Chua B. H., Chan Y. P., Lim M. E., Lam S. K. 2002. Isolation of Nipah virus from Malaysian island flying-foxes. Microbes Infect. 4: 145–151. [DOI] [PubMed] [Google Scholar]
- 11.Rahman S. A., Hassan S. S., Olival K. J., Mohamed M., Chang L. Y., Hassan L., Saad N. M., Shohaimi S. A., Mamat Z. C., Naim M. S., et al. Henipavirus Ecology Research Group 2010 Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg. Infect. Dis. 16: 1990–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau S. K., Woo P. C., Li K. S., Huang Y., Tsoi H. W., Wong B. H., Wong S. S., Leung S. Y., Chan K. H., Yuen K. Y. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102: 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J. H., Wang H., Crameri G., Hu Z., Zhang H., et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310: 676–679. [DOI] [PubMed] [Google Scholar]
- 14.Ge X. Y., Li J. L., Yang X. L., Chmura A. A., Zhu G., Epstein J. H., Mazet J. K., Hu B., Zhang W., Peng C., et al. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller M. A., Raj V. S., Muth D., Meyer B., Kallies S., Smits S. L., Wollny R., Bestebroer T. M., Specht S., Suliman T., et al. 2012. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio 3: e00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Boheemen S., de Graaf M., Lauber C., Bestebroer T. M., Raj V. S., Zaki A. M., Osterhaus A. D., Haagmans B. L., Gorbalenya A. E., Snijder E. J., Fouchier R. A. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3: e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., et al. 2014. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 16: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leroy E. M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Délicat A., Paweska J. T., Gonzalez J. P., Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438: 575–576. [DOI] [PubMed] [Google Scholar]
- 19.Walsh M. G., Wiethoelter A., Haseeb M. A. 2017. The impact of human population pressure on flying fox niches and the potential consequences for Hendra virus spillover. Sci. Rep. 7: 8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K. A., Fang X., Wynne J. W., Xiong Z., Baker M. L., Zhao W., et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339: 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P., Tachedjian M., Wynne J. W., Boyd V., Cui J., Smith I., Cowled C., Ng J. H., Mok L., Michalski W. P., et al. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. USA 113: 2696–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurray D. N., Thomas M. E. 1979. Cell-mediated immunity in two species of bats. J. Mammal. 60: 576–581. [Google Scholar]
- 23.Paul B. N., Chakravarty A. K. 1986. In vitro analysis of delayed immune response in a bat, Pteropus giganteus: process of con-A mediated activation. Dev. Comp. Immunol. 10: 55–67. [DOI] [PubMed] [Google Scholar]
- 24.Chakravarty A. K., Paul B. N. 1987. Analysis of suppressor factor in delayed immune responses of a bat, Pteropus giganteus. Dev. Comp. Immunol. 11: 649–660. [DOI] [PubMed] [Google Scholar]
- 25.Paul B. N., Chakravarty A. K. 1987. Phytohaemagglutinin mediated activation of bat (Pteropus giganteus) lymphocytes. Indian J. Exp. Biol. 25: 1–4. [PubMed] [Google Scholar]
- 26.Pavlovich S. S., Lovett S. P., Koroleva G., Guito J. C., Arnold C. E., Nagle E. R., Kulcsar K., Lee A., Thibaud-Nissen F., Hume A. J., et al. 2018. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell 173: 1098–1110.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wynne J. W., Woon A. P., Dudek N. L., Croft N. P., Ng J. H., Baker M. L., Wang L. F., Purcell A. W. 2016. Characterization of the antigen processing machinery and endogenous peptide presentation of a bat MHC class I molecule. J. Immunol. 196: 4468–4476. [DOI] [PubMed] [Google Scholar]
- 28.Ng J. H., Tachedjian M., Deakin J., Wynne J. W., Cui J., Haring V., Broz I., Chen H., Belov K., Wang L. F., Baker M. L. 2016. Evolution and comparative analysis of the bat MHC-I region. Sci. Rep. 6: 21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. 2017. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199: 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam K. S., Salmon S. E., Hersh E. M., Hruby V. J., Kazmierski W. M., Knapp R. J. 1991. A new type of synthetic peptide library for identifying ligand-binding activity. [Published erratum appears in 1992 Nature 358: 434.]; [Published erratum appears in 1992 Nature 360: 768.] Nature 354: 82–84. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen H. 2017. Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611: 59–73. [DOI] [PubMed] [Google Scholar]
- 32.Möller S., Croning M. D., Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17: 646–653. [DOI] [PubMed] [Google Scholar]
- 33.Liang R., Sun Y., Liu Y., Wang J., Wu Y., Li Z., Ma L., Zhang N., Zhang L., Wei X., et al. 2018. Major histocompatibility complex class I (FLA-E*01801) molecular structure in domestic cats demonstrates species-specific characteristics in presenting viral antigen peptides. J. Virol. 92: e01631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi H., Chen H., He K., Wu L., Yang B., Sun R. X., Liu J., Zeng W. F., Song C. Q., He S. M., Dong M. Q. 2013. pNovo+: de novo peptide sequencing using complementary HCD and ETD tandem mass spectra. J. Proteome Res. 12: 615–625. [DOI] [PubMed] [Google Scholar]
- 35.Muth T., Weilnböck L., Rapp E., Huber C. G., Martens L., Vaudel M., Barsnes H. 2014. DeNovoGUI: an open source graphical user interface for de novo sequencing of tandem mass spectra. J. Proteome Res. 13: 1143–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caron E., Kowalewski D. J., Chiek Koh C., Sturm T., Schuster H., Aebersold R. 2015. Analysis of major histocompatibility complex (MHC) immunopeptidomes using mass spectrometry. Mol. Cell. Proteomics 14: 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202. [DOI] [PubMed] [Google Scholar]
- 38.Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. 1982. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 10: 2997–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- 41.Vagin A., Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66: 22–25. [DOI] [PubMed] [Google Scholar]
- 42.McCoy A. J. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossmann M. G. 1990. The molecular replacement method. Acta Crystallogr. A 46: 73–82. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Lohkamp B., Scott W. G., Cowtan K. 2010. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., et al. 2018. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27: 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 50.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. 2009. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329: 506–512. [DOI] [PubMed] [Google Scholar]
- 53.Eisenberg D., Schwarz E., Komaromy M., Wall R. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179: 125–142. [DOI] [PubMed] [Google Scholar]
- 54.Thomsen M. C., Nielsen M. 2012. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40(Web Server Issue): W281–W287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27: 379–423, 623–656. [Google Scholar]
- 56.Olival K. J., Hosseini P. R., Zambrana-Torrelio C., Ross N., Bogich T. L., Daszak P. 2017. Host and viral traits predict zoonotic spillover from mammals. [Published erratum appears in 2017 Nature 548: 612.] Nature 546: 646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koch M., Camp S., Collen T., Avila D., Salomonsen J., Wallny H. J., van Hateren A., Hunt L., Jacob J. P., Johnston F., et al. 2007. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27: 885–899. [DOI] [PubMed] [Google Scholar]
- 58.Rammensee H. G., Friede T., Stevanoviíc S. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41: 178–228. [DOI] [PubMed] [Google Scholar]
- 59.Thomas C., Tampé R. 2017. Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing. Science 358: 1060–1064. [DOI] [PubMed] [Google Scholar]
- 60.Davenport M. P., Smith K. J., Barouch D., Reid S. W., Bodnar W. M., Willis A. C., Hunt D. F., Hill A. V. 1997. HLA class I binding motifs derived from random peptide libraries differ at the COOH terminus from those of eluted peptides. J. Exp. Med. 185: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muth T., Hartkopf F., Vaudel M., Renard B. Y. 2018. A potential golden age to come-current tools, recent use cases, and future avenues for de novo sequencing in proteomics. Proteomics 18: e1700150. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., Ma B. 2012. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics 11: M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.