Summary
Background
On April 25, 2017, a cluster of unexplained illnesses and deaths associated with a funeral was reported in Sinoe County, Liberia. Molecular testing identified Neisseria meningitidis serogroup C (NmC) in specimens from patients. We describe the epidemiological investigation of this cluster and metagenomic characterisation of the outbreak strain.
Methods
We collected epidemiological data from the field investigation and medical records review. Confirmed, probable, and suspected cases were defined on the basis of molecular testing and signs or symptoms of meningococcal disease. Metagenomic sequences from patient specimens were compared with 141 meningococcal isolate genomes to determine strain lineage.
Findings
28 meningococcal disease cases were identified, with dates of symptom onset from April 21 to April 30, 2017: 13 confirmed, three probable, and 12 suspected. 13 patients died. Six (21%) patients reported fever and 23 (82%) reported gastrointestinal symptoms. The attack rate for confirmed and probable cases among funeral attendees was 10%. Metagenomic sequences from six patient specimens were similar to a sequence type (ST) 10217 (clonal complex [CC] 10217) isolate genome from Niger, 2015. Multilocus sequencing identified five of seven alleles from one specimen that matched ST-9367, which is represented in the PubMLST database by one carriage isolate from Burkina Faso, in 2011, and belongs to CC10217.
Interpretation
This outbreak featured high attack and case fatality rates. Clinical presentation was broadly consistent with previous meningococcal disease outbreaks, but predominance of gastrointestinal symptoms was unusual compared with previous African meningitis epidemics. The outbreak strain was genetically similar to NmC CC10217, which caused meningococcal disease outbreaks in Niger and Nigeria. CC10217 had previously been identified only in the African meningitis belt.
Introduction
Meningococcal disease, caused by Neisseria meningitidis, is life-threatening and often presents as meningitis, with symptoms including fever, headache, stiff neck, and altered mental status. N meningitidis can also cause bloodstream infection, or meningococcaemia, which manifests with non-specific symptoms such as fever, vomiting, fatigue, and diarrhoea; petechiae or, in later stages, a characteristic purpuric rash can also be present. Meningococcal disease progresses rapidly, with 9–12% of meningitis cases and up to 40% of meningococcaemia cases being fatal despite prompt antibiotic treatment.1
Globally, the highest incidence of meningococcal disease is in the meningitis belt in sub-Saharan Africa, which stretches from Senegal to Ethiopia and has annual seasonal meningitis outbreaks, with large-scale epidemics occurring every 8–12 years.2 N meningitidis serogroup A, one of 12 meningococcal serogroups, caused most of the outbreaks within the meningitis belt countries before 2010.3,4 With introduction of the meningococcal serogroup A conjugate vaccine in 2010, serogroup A outbreaks have been nearly eliminated, and N meningitidis serogroups C, W, and X have become the predominant causes of meningococcal disease.3,4
Meningococcal strains are further classified into sequence types (STs) and clonal complexes (CCs) by use of multilocus sequence typing (MLST). Between 2010 and 2013, the common CCs encountered in Africa were CC11 (mainly associated with serogroup W) and CC181 (serogroup X).4,5 In 2013, a novel N meningitidis serogroup C (NmC) CC10217 strain emerged in Nigeria, causing the first reported outbreak of NmC in the meningitis belt since 1979.3 Since then, Nigeria and Niger have had multiple meningitis outbreaks due to CC10217, including epidemics of close to 10 000 suspected cases in Niger in 2015.6,7 Further expansion of this epidemic-associated CC is a substantial public health concern, particularly in light of global shortages of serogroup C-containing vaccines for outbreak response in Africa.
On April 25, 2017, a cluster of 14 unexplained illnesses and eight deaths associated with a funeral was reported in Liberia.8 Liberia is not located in the meningitis belt, but borders two meningitis belt countries, Guinea and Côte d’lvoire. By May 7, 31 cases with 13 deaths were reported. The initial differential diagnosis was broad and included Ebola virus disease, because Liberia had been affected by the 2014–15 Ebola virus epidemic. However, molecular testing of four specimens from patients at the US Centers for Disease Control and Prevention (CDC) headquarters (Atlanta, GA, USA) identified NmC as the causative agent of the illness cluster. In this outbreak report, we describe the epidemiological and laboratory findings from this cluster of initially unexplained illnesses in Liberia and the metagenomic analysis done to identify the cause of the outbreak.
Methods
Field investigation
The unexplained cluster of illnesses and deaths was initially detected through community event-based surveillance, in which community workers identify signs and symptoms of priority diseases or unusual events. After notification of the cluster by Liberia’s Sinoe County Health Team on April 25, 2017, a county-led field investigation and response was immediately launched with support from the National Rapid Response Team and epidemiologists from the CDC, WHO, and the African Field Epidemiology Network, from April 26 to May 27.8
An outbreak case was defined as a person who had visited or lived in Sinoe County (population 102 391; appendix) and presented with two or more symptoms, including headache, vomiting, mental confusion, and weakness, with illness onset on or after April 10, 2017. Patients and their families were interviewed by surveillance officers by use of a questionnaire to gather demographic information, food and environmental exposures, clinical features and timing of illness, and contacts with or knowledge of any other people who were or had been recently ill. Clinical information was also extracted from medical records. Active case finding was done in the community and health facilities to identify additional cases and their close contacts at Sinoe County clinics and the county’s sole hospital, which served as the referral health facility. Epidemiological links among cases were identified on the basis of contacts reported in the case investigation form or from patient and family member interviews. Epidemiological investigation forms, medical records, and notes from the field investigators were reconciled to generate the final dataset.
Laboratory confirmation of the outbreak and molecular characterisation of specimens
Haematology and blood chemistry testing were done at local laboratories, whereas Ebola and Lassa virus testing were done at the National Reference Laboratory in Liberia by reverse transcriptase PCR. Food samples were screened for toxins at the University of Natural Resources and Life Sciences (Vienna, Austria) by use of a high performance liquid chromatography-tandem-mass spectrometry-based multi-mycotoxin method.9 A forensic pathologist did two autopsies.
Testing for metals10 and organophosphates,11 as well as multipathogen screening with a quantitative PCR-based assay called TaqMan array card (TAC), which simultaneously screens for 23 respiratory pathogens, was done at the CDC. We used tier 1 and tier 2 TAC assays12 with a modified bead beating protocol to screen specimens for multiple disease causes. Direct real-time PCR (rtPCR) was done in specimens sent to the CDC and in additional specimens in Liberia to confirm the pathogen and serogroup.13
We selected specimens with abundant N meningitidis DNA for molecular characterisation. We used Sanger sequencing14 for MLST analysis and metagenomic sequencing for strain similarity analyses.13 For meta-genomic analysis, DNA was sequenced on an MiSeq system (Illumina, San Diego, CA, USA), and N meningitidis reads were identified with k-SLAM15 (version 1.0; appendix). The metagenomic N meningitidis reads were mapped to the FAM18 N meningitidis reference genome16 and compared with a genome alignment of 141 diverse N meningitidis isolates from the CDC isolate collection; the genome alignment of these isolates was also generated by mapping reads to the FAM18 reference genome. We calculated similarity between the metagenomic reads and each of the 141 isolate genomes at polymorphic positions, defined as positions in the core genome alignment where the consensus base call differed among the isolate genomes. The accuracy of similarity estimates calculated by this method was assessed by subsampling from whole genome sequence data (appendix).
Refined case definitions
The outbreak case definition was refined after laboratory testing confirmed the causative agent of the cluster. A confirmed case of meningococcal disease was defined as detection of NmC by rtPCR in any specimen from an outbreak case. A probable case was defined as the presence of purpura, detection of N meningitidis that was non-groupable in a sterile site specimen, or sudden death accompanied by at least three symptoms of meningococcaemia or meningitis, including documented fever, vomiting, diarrhoea, cold extremities, joint pain, muscle pain, chest pain, abdominal pain, neck stiffness, altered consciousness, headache, and convulsions. A suspected case was defined as the presence of three or more of these symptoms, but without meeting the confirmed or probable case definition. An outbreak case that did not meet the suspected case definition was defined as not having meningococcal disease; these cases were excluded from the analysis.
Statistical methods
We compared characteristics of confirmed and probable cases with those of suspected cases with χ2 test or Fisher’s exact test. Differences in median were tested with the Mann-Whitney test. A p value less than 0·05 was considered statistically significant. We calculated an attack rate for meningococcal disease by dividing the number of confirmed and probable meningococcal disease cases among people who attended the funeral by the estimated number of funeral attendees. Analyses were done with Epi Info (version 7·2) and SAS (version 9.4).
The full investigation was determined by the CDC to be a public health disease control activity and did not require review by an institutional review board.
Role of the funding source
The sponsor of the study had no role in design of the investigation, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the investigation and had final responsibility for the decision to submit for publication.
Results
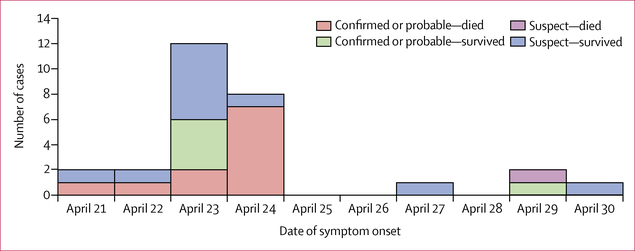
Approximately 150 people attended the funeral events (funeral, wake, and repast) on April 21–22, 2017. 31 disease cases were identified as part of the cluster by use of the outbreak case definition: 29 had attended the funeral, one was a close contact of a funeral attendee, and one had a meal with a driver who travelled from Sinoe County. No other suspected meningitis cases were reported in Sinoe County before or during the funeral events. 28 of 31 cases were classified as cases of meningococcal disease: 13 were classified as confirmed cases, three as probable, and 12 as suspected. The remaining three initially identified cases were not cases of meningococcal disease. Among the 28 meningococcal disease cases, most were reported from Sinoe County, about half were female, and half were aged 10–19 years (table 1). No meningococcal disease cases were reported in children younger than 10 years. By contrast, among the 61 other funeral attendees for whom age was available, five (8%) were younger than 10 years, 11 (18%) were aged 10–19 years, six (10%) were aged 20–29 years, 11 (18%) were aged 30–39 years, nine (15%) were aged 40–49 years, and 19 (31%) were 50 years or older. No patients reported recent travel outside of Liberia. The dates of symptom onset ranged from April 21 to April 30, 2017, with 20 (71%) patients reporting onset on April 23–24 (figure 1). Although 16 cases were reclassified as confirmed and probable, 15 of these had attended the funeral, resulting in an attack rate of 10% among approximately 150 funeral attendees.
Table 1:
Characteristics of 28 meningococcal disease cases overall and by case status
| Overall (n=28) | Confirmed and probable (n=16) | Suspected (n=12) | |
|---|---|---|---|
| Case status | |||
| Confirmed | 13 (46%) | 13 (81%) | 0 (0%) |
| Probable | 3 (11%) | 3 (19%) | 0 (0%) |
| Suspected | 12 (43%) | 0 (0%) | 12 (100%) |
| Sex | |||
| Female | 15 (54%) | 11 (69%) | 4 (33%) |
| Male | 13 (46%) | 5 (31%) | 8 (67%) |
| County of residence | |||
| Sinoe | 24 (86%) | 14 (88%) | 10 (83%) |
| Montserrado | 2 (7%) | 2 (13%) | 0 (0%) |
| Grand Bassa | 2 (7%) | 0 (0%) | 2 (17%) |
| Age (years) | |||
| 10–19 | 14 (50%) | 10 (63%) | 4 (33%) |
| 20–29 | 7 (25%) | 3 (19%) | 4 (33%) |
| 30–39 | 0 (0%) | 0 (0%) | 0 (0%) |
| 40–49 | 3 (11%) | 0 (0%) | 3 (25%) |
| ≥50 | 4 (14%) | 3 (19%) | 1 (8%) |
| Attended ≥1 funeral events (funeral, wake, and repast) | 26 (93%) | 15 (94%) | 11 (92%) |
| Signs and symptoms | |||
| Weakness | 26 (93%) | 15 (94%) | 11 (92%) |
| Abdominal pain | 22 (79%) | 11 (69%) | 11 (92%) |
| Any other body pain | 8 (29%) | 5 (31%) | 3 (25%) |
| Headache | 24 (86%) | 13 (81%) | 11 (92%) |
| Any gastrointestinal symptoms | 23 (82%) | 12 (75%) | 11 (92%) |
| Vomiting | 20 (71%) | 10 (63%) | 10 (83%) |
| Diarrhoea | 16 (57%) | 8 (50%) | 8 (67%) |
| Any neurological signs | 18 (64%) | 11 (69%) | 7 (58%) |
| Confusion | 15 (54%) | 9 (56%) | 6 (50%) |
| Convulsions | 4 (14%) | 4 (25%) | 0 (0%) |
| Hallucinations | 4 (14%) | 3 (19%) | 1 (8%) |
| Any other neurological signs | 6 (21%) | 4 (25%) | 2 (17%) |
| Any respiratory signs | 7 (25%) | 4 (25%) | 3 (25%) |
| Documented fever | 6 (21%) | 3 (19%) | 3 (25%) |
| Purpura | 5 (18%) | 5 (31%) | 0 (0%) |
| Cold extremities | 2 (7%) | 2 (13%) | 0 (0%) |
| Hospitalisation | |||
| Admitted | 22 (79%) | 12 (75%) | 10 (83%) |
| Dead upon arrival at hospital | 4 (14%) | 4 (25%) | 0 (0%) |
| Community* | 2 (7%) | 0 (0%) | 2 (17%) |
| Received antibiotics | 19 (68%) | 10 (63%) | 9 (75%) |
| Died | 13 (46%) | 12 (75%) | 1 (8%) |
| Median time between date of onset and hospital admission†, in days (IQR) | 2 (1–4) | 1–5 (1–2) | 6–5 (3–9) |
| Median timing between date of onset and death, in days (IQR) | 1 (1–2) | 1 (1–2) | 4 (4–4) |
Data are n (%), unless stated otherwise.
Patients did not present to a hospital.
Hospital admission includes patients who were dead upon arrival at the hospital.
Figure 1: Cases of meningococcal disease by date of onset.
Data are from 28 cases. Of the 31 outbreak cases, three were considered not cases of meningococcal disease and were excluded.
Predominant signs and symptoms of disease were weakness, headache, gastrointestinal symptoms (23 [82%] of 28 reported vomiting or diarrhoea), and abdominal pain; documented fever (based on temperature measurement) was uncommon (table 1). The clinical presentation was broadly similar when comparing confirmed and probable cases with suspected cases. However, purpura, convulsions, and cold extremities were reported only among confirmed and probable cases (table 1). Purpura was part of the probable case definition, whereas convulsions are consistent with late stages of meningitis and cold extremities with late stages of meningococcaemia. Autopsies done on two fatal confirmed cases that occurred in Montserrado County had findings consistent with meningococcal disease.
All meningococcal disease cases were admitted to a hospital or dead upon arrival at the hospital (table 1), with the exception of two suspected cases, who refused to be hospitalised. Additionally, the median time between date of onset and hospital admission (including deaths upon arrival at hospital) was significantly lower in confirmed and probable cases than in suspected cases (p=0·0002; table 1). Among all 28 cases, 13 (46%) patients died. The case fatality rate of confirmed and probable cases was significantly higher than that of suspected cases (p=0·0001; table 1). Of the 13 fatal cases, four patients were dead on arrival at the hospital and did not receive antibiotics. The remaining nine patients were admitted to a hospital, and seven received antibiotics. Furthermore, of the 17 patients who had blood, plasma, or cardiac blood specimens collected, ten were bacteraemic (seven died), and seven were not bacteraemic (all seven survived).
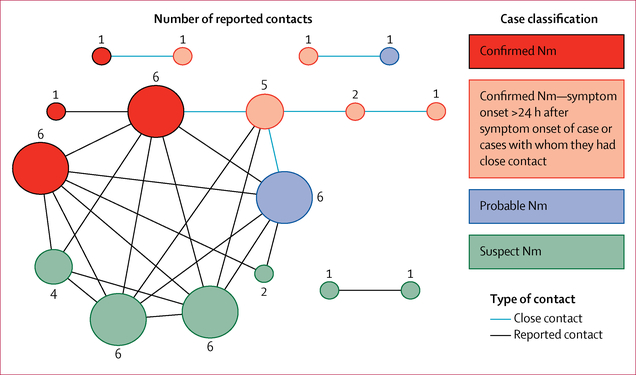
Among the 28 meningococcal disease cases, 17 (61%) patients reported contacts with one or more patients (figure 2). Five confirmed cases had close contact with one or more patients who had earlier onsets; all five patients reported symptom onset more than 24 h (1–5 days) after symptom onset of the patient, or patients, with whom they had close contact.
Figure 2: Reported contacts among cases.
17 cases with any reported contact with other cases are shown. Each circle represents a case; a line between circles indicates contact between cases with the weight of the line illustrating the type of contact. Circles are coloured on the basis of case classification, and size of circle is based on number of additional cases with whom contact was reported. Nm=Neisseria meningitidis.
At the start of the outbreak, samples were not collected systematically for all cases because the cause and scope of the outbreak was unknown. Haematology tests, chemistry tests, or both were done on specimens from 12 cases; no consistent abnormalities were identified to guide determination of outbreak cause. All specimens tested for Ebola and Lassa virus by PCR (n=23) were negative. Toxicology testing of urine (n=3) and food (n=10) specimens did not detect sufficient amounts of any toxin to explain the outbreak.
63 clinical specimens from 24 of 31 cases, including 24 specimens from the two autopsies, were available for testing. Six specimens from four cases were tested by TAC, and N meningitidis was detected in all six specimens. We did N meningitidis serogroup-specific rtPCR on the same six specimens and one additional oral swab specimen—from one of the same four cases—and confirmed that all specimens were positive for NmC. Analyses with rtPCR detected N meningitidis in 40 (63%) of 63 specimens; 39 of these 40 specimens tested positive for NmC (98%) and one for non-groupable N meningitidis (2%), which was negative for invasive serogroups A, B, C, W, X, and Y. Overall, we detected N meningitidis in 14 of 24 cases with available specimens, of which 11 cases had positive specimens from sterile sites. Of the 13 patients who died, 11 had specimens tested; all were positive for NmC (table 2).
Table 2:
Confirmatory results for specimens collected from 22 of 28 patients*
| Sterile site specimens |
Non-sterile site specimens |
Outcome | |||
|---|---|---|---|---|---|
| Specimen types | Organism detected | Specimen types | Organism detected | ||
| 1 | Cardiac blood† | Neisseria meningitidis serogroup C | Oral swab† | N meningitidis serogroup C | Dead |
| 2 | Plasma† | N meningitidis serogroup C | Oral swab | N meningitidis serogroup C | Dead |
| 3 | NA | NA | Oral swab | N meningitidis serogroup C | Dead |
| 4 | Cardiac blood† | N meningitidis serogroup C | Oral swab† | N meningitidis serogroup C | Dead |
| 5 | Plasma | N meningitidis serogroup C | NA | NA | Dead |
| 6 | Plasma | No N meningitidis DNA detected | Urine | No N meningitidis DNA detected | Alive |
| 7 | Various tissues and fluids†‡ | N meningitidis serogroup C | Urine | N meningitidis serogroup C | Dead |
| 8 | Various tissues and fluids‡ | N meningitidis serogroup C | NA | NA | Dead |
| 9 | Plasma | N meningitidis serogroup C | Urine | N meningitidis serogroup C | Dead |
| 10 | Plasma | No N meningitidis DNA detected | Urine | No N meningitidis DNA detected | Alive |
| 11 | Plasma | N meningitidis serogroup C | NA | NA | Alive |
| 12 | Plasma | N meningitidis serogroup C | Urine | N meningitidis serogroup C | Alive |
| 13 | Plasma | No N meningitidis DNA detected | NA | NA | Alive |
| 14 | Blood | N meningitidis serogroup C | Oral swab | N meningitidis serogroup C | Dead |
| 15 | NA | NA | Urine | No N meningitidis DNA detected | Alive |
| 16 | Blood, CSF | No N meningitidis DNA detected | Urine | No N meningitidis DNA detected | Alive |
| 17 | CSF, plasma | No N meningitidis DNA detected | NA | NA | Alive |
| 18 | Plasma | No N meningitidis DNA detected | Urine | No N meningitidis DNA detected | Alive |
| 19 | NA | NA | Oral swab | N meningitidis serogroup C | Dead |
| 20 | NA | NA | Oral swab | N meningitidis serogroup C | Dead |
| 21 | Blood | N meningitidis NG | NA | NA | Alive |
| 22 | Skin lesion, blood | No N meningitidis DNA detected | NA | NA | Alive |
NA=Not applicable; no specimen was received. Cerebrospinal fluid (CSF).
Two additional outbreak cases from the initially identified 31 cases had plasma specimens tested; both specimens were negative for N meningitidis DNA and both patients were classified as not having meningococcal disease.
Specimens initially screened positive for N meningitidis with the TaqMan Array Card assay.
Tissues include liver, brain, skin, kidney, and lung; fluids include cardiac blood, CSF, vitreous humor, blood, lung, and plasma.
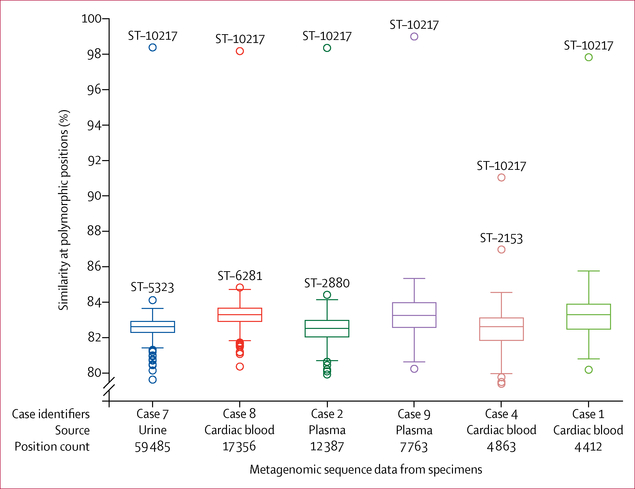
We selected eight specimens from six cases, including cardiac blood, plasma, blood, urine, and vitreous humour fluid, with abundant N meningitidis DNA for molecular characterisation. All eight specimens were analysed by metagenomic sequencing, and one blood specimen was tested by MLST analysis. Metagenomic sequencing identified 340–4374 N meningitidis DNA fragments in each specimen. Specimens from patient one (blood) and patient three (plasma) were excluded from further analysis because of insufficient reads. For each patient, N meningitidis reads, from the specimen with most reads, were compared with a reference collection of 141 diverse N meningitidis genomes representing 140 sequence types, with more than 0·5% genetic variation between each pair of genomes (appendix). N meningitidis DNA sequence similarity between the references and the specimens ranged from 79% (3860 of 4863) to 99% (7704 of 7763) when assessed at all polymorphic positions where reads for a given specimen were mapped (86 739 total positions, range 4412–59 485 positions per specimen; figure 3). For all specimens tested, N meningitidis DNA sequences were most similar to the genome of an ST-10217 NmC isolate obtained during the 2015 epidemic in Niger.7 N meningitidis DNA sequences from five specimens had more than 98% (>4351 of 4412) similarity to the ST-10217 genome, and one specimen (case 4, figure 3) had 91% (4449 of 4863) similarity. By contrast, N meningitidis DNA sequences had less than 87% (<4229 of 4863) similarity to any other genome in the reference collection.
Figure 3: Metagenomic analysis from six non-oral specimens with high abundance of Neisseria meningitidis DNA.
Similarity of 141 N meningitidis isolate genomes to the N meningitidis sequences identified in specimens from each of six cases. Metagenomic datasets are labelled with the case identifier, the source of the specimen, and the number of polymorphic positions within the alignment of 141 isolate genomes for which the specimen sequences had a single base call. The percentage similarity to each of the 141 isolate genomes is plotted for each specimen; a boxplot shows the first, second, and third quartiles, and error bars cover points beyond the first or third quartile, up to 1·5 times the IQR. Outliers above the whiskers are labelled with the sequence type (ST) of the isolate.
Only one specimen was successfully analysed by MLST, with five of seven MLST genes identified. In PubMLST, the only sequence type that contains the five alleles identified in the patient specimen is ST-9367, which was identified from a non-groupable carriage isolate obtained from Burkina Faso in 2011. ST-9367 whole genome sequencing data were not available for comparison by the metagenomics method. Four of five alleles identified in the outbreak specimen were also present in ST-10217. Both ST-10217 and ST-9367 belong to CC10217 and are similar, with MLST profiles that differ by only one nucleotide in fumC (appendix).
Discussion
During and after the 2014–15 Ebola epidemic, the Liberian Ministry of Health and international partners made substantial investments in strengthening Liberia’s surveillance and laboratory capacity for outbreak response.17,18 These efforts enabled prompt detection and notification of this cluster, rapid rule-out of Ebola and Lassa viruses, and immediate implementation of infection control and contact tracing procedures during this outbreak. Although the predominance of gastrointestinal symptoms prompted initial concerns about toxic exposure, thorough laboratory analyses and indepth case investigations led to diagnosis and confirmation of meningococcal disease. After confirmation of the outbreak cause on May 16, 2017, the administration of antibiotic chemoprophylaxis (ciprofloxacin) was attempted to funeral attendees and contacts of cases, by use of Liberia’s contact tracing infrastructure. Because of the absence of ongoing meningococcal transmission over several weeks, vaccination was not implemented.
In the past several years, multiple NmC outbreaks in the meningitis belt have been associated with CC10217, including a 2013 outbreak in Nigeria6 and large-scale outbreaks in Niger and Nigeria in 2015.7 Our molecular analysis identified the Liberia outbreak strain as being most similar to strains from CC10217. Although we were unable to completely define the sequence type of the Liberia outbreak strain, the detection of an outbreak with an NmC strain that is a member of CC10217 is concerning in a region where NmC is emerging and the population is unlikely to have immunity to NmC. The introduction of a strain from an epidemic-associated CC might have contributed to the unusually high attack rate among funeral attendees observed in this outbreak.19 The spread of this strain highlights the potential for global transmission of meningococcal strains from the African meningitis belt, as shown by the worldwide meningococcal disease outbreak after the Hajj, in 2000.20
In CC10217 meningococcal disease outbreaks of the past decade, reported symptoms have been unremarkable;21 however, this Liberia outbreak featured a predominance of gastrointestinal symptoms. Although gastrointestinal symptoms are consistent with meningococcal disease, cases with primarily gastrointestinal symptoms have been considered rare.2,22 However, gastrointestinal symptoms have been described as a more common symptom of meningococcal disease, particularly among NmW ST-11 cases in the UK23 and Chile.24 Notably, among NmW cases with gastrointestinal symptoms in the UK, 71% (five of seven) died within 24 h of presentation,23 similar to the rapid disease progression observed in this outbreak. Similarly, diarrhoea was significantly associated with death in the NmW ST-11 outbreak in Chile.24 The potential human or bacteriological factors contributing to the emergence of meningococcal disease with predominantly gastrointestinal symptoms are unclear.
In addition to the high prevalence of gastrointestinal symptoms, the tight temporal clustering of cases, low prevalence of documented fever, and rapid disease progression initially led to suspicions of a toxic cause for the outbreak. However, these features are also consistent with meningococcal disease. The temporal clustering of cases, mimicking a point source exposure, has been observed in some previous meningococcal disease outbreaks.25,26 Although fever is typically among the most common meningococcal disease symptoms, absence of documented fever was noted in 40% (24 of 60) of cases in a 2012 NmW ST-11 outbreak in Chile24 and has been observed in septic patients.27 This observation might account for the absence of documented fever among the patients in this outbreak who died on arrival at the hospital or within hours of presenting to care. Given the rapidity with which meningococcal disease can progress to overwhelming sepsis, the absence of fever does not rule out meningococcal disease, particularly when purpura or other symptoms characteristic of meningococcal disease are also noted.
Although meningitis has historically been the primary presentation of meningococcal disease in the meningitis belt, the majority of patients in this outbreak presented with meningococcaemia, including five patients with purpura. Along with late presentation to care, this difference in reported disease presentations might help to explain the very high case fatality rate in this outbreak compared with previous reports of cases in the meningitis belt.21 Although climatic or other differences between Liberia and countries in the meningitis belt could affect the predominant presentation of meningococcal disease in these areas, there are few data on the prevalence of meningococcaemia in the meningitis belt, because blood is not routinely drawn or tested for meningococcal detection. However, NmC infection in other areas of the world frequently presents as meningococcaemia (CDC, unpublished data).28 With the emergence of NmC as the predominant cause of meningococcal disease in the African meningitis belt, meningococcaemia, particularly with purpura, must be recognised by clinicians and public health professionals in the region as a potential presentation of meningococcal disease.
The spread of NmC CC10217 to Liberia and throughout the meningitis belt shows that the emergence, expansion, and spread of a novel meningococcal clone continues to have the potential to cause devastating outbreaks. Although NmA epidemics have been largely eliminated after the introduction of meningococcal serogroup A conjugate vaccine, the NmC outbreaks within and outside the meningitis belt highlight the need to address global shortages of serogroup C-containing vaccines to ensure an adequate supply for future outbreak responses. Sustained investment in global health security to strengthen prevention, detection, and response to infectious disease threats—as epitomised by Liberia’s successful improvements in disease surveillance, laboratory systems, workforce development, and emergency operations after the Ebola virus epidemic— might also provide benefits in tracking and controlling the spread of CC10217 and related strains. Continued strengthening of public health surveillance and laboratory capacity in Africa, as well as networking with reference laboratories, will also facilitate outbreak detection and implementation of appropriate response measures.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for English-language articles published from database inception to Dec 1, 2017, regarding outbreaks of meningococcal disease with search terms including “Neisseria meningitidis”, “Neisseria meningitidis serogroup C”, “meningococcal”, “Africa”, “epidemic”, “outbreak”, and “meningitis”. We reviewed these articles and others citing or cited by the articles retrieved. Outbreaks of N meningitidis serogroup C (NmC) clonal complex (CC) 10217 were first reported in Nigeria in 2013, and in Niger in 2015, both of which lie in the meningitis belt. Few meningococcal disease outbreaks have been previously reported in Africa outside of the meningitis belt. Although gastrointestinal symptoms are consistent with meningococcal disease, they have rarely been identified as predominant presenting symptoms. However, in the past several years, they have been recognised as a more common symptom of meningococcal disease.
Added value of this study
We describe the epidemiological investigation of an unusual meningococcal disease outbreak in Liberia and the metagenomic characterisation of the NmC outbreak strain. Of the 31 cases identified as part of the initial cluster of unknown illness, 13 were confirmed meningococcal disease cases and three were probable. Most patients presented with predominantly gastrointestinal symptoms and only six had documented fever; however, five had purpura. Metagenomic analysis revealed that the outbreak strain was genetically similar to the NmC CC10217 strain that caused the meningococcal disease epidemics in Nigeria in 2013, and in Niger in 2015. Gastrointestinal symptoms or absence of fever should not be considered to rule out meningococcal disease, particularly when purpura is present. This NmC outbreak shows that the emergence, expansion, and spread of a novel meningococcal clone continues to have the potential to cause devastating outbreaks.
Implications of all the available evidence
With the emergence of serogroup C as an important cause of meningitis outbreaks within and outside the African meningitis belt, clinicians and public health professionals should be aware that the disease might present as either meningitis or meningococcaemia, and symptoms of meningococcaemia, particularly purpura, should be recognised. Continued strengthening of public health surveillance and laboratory capacity in Africa, as well as networking with reference laboratories, is crucial to ensure that outbreaks are rapidly detected and appropriate response measures are implemented. The NmC outbreaks in the past several years, within and outside the meningitis belt, highlight the need to address global shortages of serogroup C-containing vaccines to ensure an adequate supply for future outbreak response.
Acknowledgments
This work was part of a public health response and supported by the US Global Health Security funds. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention.
Funding US Global Health Security.
Footnotes
Declaration of interests
We declare no competing interests.
The Liberian Meningococcal Disease Outbreak Response Team
Y Konway, S Q Wiah, V Doedeh, U Bao, G Senneh, L Gorwor, P Gonotee, T Paasewe, G Tamatai, J Yarkeh, S Smith, A Brima-Davis, G Dauda, T Monger, L W Gornor-Pewu, S Lombeh (Sinoe County Health Team, Liberia); H W Wilson, M Korvayan, N Dovillie, R Jetoh, F Taweh, Y V Walker, P Hardy, M Freeman (National Public Health Institute of Liberia, Paynesville, Liberia); G George, G Kerwillain, S Toe, E Ghartey, L Larway, D Gweh (Liberian Ministry of Health, Paynesville, Liberia); D Allen, S Friesen, G Gwesa, C Kinkade, M Reed (CDC Liberia Country Office, Paynesville, Liberia); A Chang, J George, J Schier, J Thomas (National Center for Environment Health) and M H Diaz, T Jenkins, B E Mahon, S E Schmink, S J Joseph, J L Waller, J Whaley, J M Winchell (Division of Bacterial Diseases) and R R Arthur, S Fuller, K Christian, J T Redd (Division of Global Health Protection; US Centers for Disease Control and Prevention, Atlanta, GA, USA); K D M Yealue II, J Naiene (WHO, Geneva, Switzerland); J A Frimpong, M Amo-Addae, O Stephen (African Field Epidemiology Network, Kampala, Uganda); A V Gottberg (National Institute for Communicable Diseases, Johannesburg, South Africa); M Taha (Institut Pasteur, Paris, France).
Contributor Information
Catherine H Bozio, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
Jeni Vuong, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
E Kainne Dokubo, Division of Global Health Protection, Center for Global Health, US Centers for Disease Control and Prevention, Monrovia, Liberia.
Mosoka P Fallah, National Public Health Institute of Liberia, Monrovia, Liberia.
Lucy A McNamara, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
Caelin C Potts, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
John Doedeh, Liberia Ministry of Health, Monrovia, Liberia.
Miatta Gbanya, Liberia Ministry of Health, Monrovia, Liberia.
Adam C Retchless, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
Jaymin C Patel, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
Thomas A Clark, Division of Reproductive Health, National Center for Chronic Diseases Prevention and Health Promotion, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
Henry Kohar, National Public Health Institute of Liberia, Monrovia, Liberia.
Thomas Nagbe, National Public Health Institute of Liberia, Monrovia, Liberia.
Peter Clement, World Health Organization—Liberia, Monrovia, Liberia.
Victoria Katawera, World Health Organization—Liberia, Monrovia, Liberia.
Nuha Mahmoud, World Health Organization—Liberia, Monrovia, Liberia.
Harouna M Djingarey, World Health Organization—AFRO, Brazzaville, Republic of Congo.
Anne Perrocheau, World Health Organization, Geneva, Switzerland.
Dhamari Naidoo, World Health Organization, Geneva, Switzerland.
Mardia Stone, World Health Organization—Liberia, Monrovia, Liberia.
Roseline N George, National Public Health Institute of Liberia, Monrovia, Liberia.
Desmond Williams, Division of Global Health Protection, Center for Global Health, US Centers for Disease Control and Prevention, Monrovia, Liberia.
Alex Gasasira, World Health Organization—Liberia, Monrovia, Liberia.
Tolbert Nyenswah, National Public Health Institute of Liberia, Monrovia, Liberia.
Xin Wang, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
LeAnne M Fox, Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, US Centers for Disease Control and Prevention, Atlanta, GA, USA.
References
- 1.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease. N Engl J Med 2001; 344: 1378–88. [DOI] [PubMed] [Google Scholar]
- 2.Greenwood B Manson lecture. Meningococcal meningitis in Africa. Trans R Soc Trop Med Hyg 1999; 93: 341–53. [DOI] [PubMed] [Google Scholar]
- 3.Funk A, Uadiale K, Kamau C, Caugant DA, Ango U, Greig J. Sequential outbreaks due to a new strain of Neisseria meningitidis serogroup C in northern Nigeria, 2013–14. PLoS Curr 2014; 6: ecurrents.outbreaks.b50c2aaf1032b3ccade0fca0b63ee518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trotter CL, Lingani C, Fernandez K, et al. Impact of MenAfriVac in nine countries of the African meningitis belt, 2010–15: an analysis of surveillance data. Lancet Infect Dis 2017; 17: 867–72. [DOI] [PubMed] [Google Scholar]
- 5.WHO. Meningococcal disease in countries of the African meningitis, 2012—emerging needs and future perspectives. Wkly Epidemiol Rec 2013; 88: 129–36. [PubMed] [Google Scholar]
- 6.Chow J, Uadiale K, Bestman A, et al. Invasive meningococcal meningitis serogroup C outbreak in northwest Nigeria, 2015—third consecutive outbreak of a new strain. PLoS Curr 2016; 8: ecurrents.outbreaks.06d10b6b4e690917d8b0a04268906143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kretz CB, Retchless AC, Sidikou F, et al. Whole-genome characterization of epidemic Neisseria meningitidis serogroup C and resurgence of serogroup W, Niger, 2015. Emerg Infect Dis 2016; 22: 1762–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doedeh J, Frimpong JA, Yealue KDM 2nd, et al. Rapid field response to a cluster of illnesses and deaths—Sinoe County, Liberia, April–May, 2017. MMWR Morb Mortal Wkly Rep 2017; 66: 1140–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malachova A, Sulyok M, Beltran E, Berthiller F, Krska R. Optimization and validation of a quantitative liquid chromatography-tandem mass spectrometric method covering 295 bacterial and fungal metabolites including all regulated mycotoxins in four model food matrices. J Chromatogr A 2014; 1362: 145–56. [DOI] [PubMed] [Google Scholar]
- 10.Caldwell KLJH, Jarrett J, Jones RL. Inductively coupled plasma mass spectrometry to measure multiple toxic elements in urine in NHANES 1999–2000. Atomic Spectroscopy 2005; 26: 1–7. [Google Scholar]
- 11.Odetokun MS, Montesano MA, Weerasekera G, Whitehead RD Jr, Needham LL, Barr DB. Quantification of dialkylphosphate metabolites of organophosphorus insecticides in human urine using 96-well plate sample preparation and high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2010; 878: 2567–74. [DOI] [PubMed] [Google Scholar]
- 12.Diaz MH, Waller JL, Napoliello RA, et al. Optimization of multiple pathogen detection using the TaqMan array card: application for a population-based study of neonatal infection. PLoS One 2013; 8: e66183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vuong J, Collard JM, Whaley MJ, et al. Development of real-time PCR methods for the detection of bacterial meningitis pathogens without DNA extraction. PLoS One 2016; 11: e0147765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 1998; 95: 3140–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ainsworth D, Sternberg MJE, Raczy C, Butcher SA. k-SLAM: accurate and ultra-fast taxonomic classification and gene identification for large metagenomic data sets. Nucleic Acids Res 2017; 45: 1649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentley SD, Vernikos GS, Snyder LA, et al. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet 2007; 3: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zolia Y, Musa E, Wesseh CS, et al. Setting the scene for post-Ebola health system recovery and resilience in Liberia: lessons learned and the way forward. Health Syst Policy Res 2016; 4: 42. [Google Scholar]
- 18.Ministry of Health Liberia, WHO, US Centers for Disease Control and Prevention. National technical guidelines for integrated disease surveillance and response. 2016. http://moh.gov.lr/wp-content/uploads/2016/12/IDSRTechnical_Guidelines_20161128.pdf (accessed Dec 1, 2017).
- 19.Jackson LA, Schuchat A, Reeves MW, Wenger JD. Serogroup C meningococcal outbreaks in the United States. An emerging threat. JAMA 1995; 273: 383–89. [PubMed] [Google Scholar]
- 20.Aguilera JF, Perrocheau A, Meffre C, Hahne S, Group WW. Outbreak of serogroup W135 meningococcal disease after the Hajj pilgrimage, Europe, 2000. Emerg Infect Dis 2002; 8: 761–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coldiron ME, Salou H, Sidikou F, et al. Case-fatality rates and sequelae resulting from Neisseria meningitidis serogroup C epidemic, Niger, 2015. Emerg Infect Dis 2016; 22: 1827–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odegaard A Unusual manifestations of meningococcal infection: a review. NIPH Ann 1983; 6: 59–63. [PubMed] [Google Scholar]
- 23.Campbell H, Parikh SR, Borrow R, Kaczmarski E, Ramsay ME, Ladhani SN. Presentation with gastrointestinal symptoms and high case fatality associated with group W meningococcal disease (MenW) in teenagers, England, July 2015 to January 2016. Euro Surveill 2016; 21. [DOI] [PubMed] [Google Scholar]
- 24.Moreno G, Lopez D, Vergara N, Gallegos D, Advis MF, Loayza S. Clinical characterization of cases with meningococcal disease by W135 group in Chile, 2012. Rev Chilena Infectol 2013; 30: 350–60 (in Spanish). [DOI] [PubMed] [Google Scholar]
- 25.US Centers for Disease Control and Prevention. Outbreak of meningococcal disease associated with an elementary school—Oklahoma, March 2010. MMWR Morb Mortal Wkly Rep 2012; 61: 217–21. [PubMed] [Google Scholar]
- 26.Zangwill KM, Schuchat A, Riedo FX, et al. School-based clusters of meningococcal disease in the United States: descriptive epidemiology and a case-control analysis. JAMA 1997; 277: 389–95. [PubMed] [Google Scholar]
- 27.Rumbus Z, Matics R, Hegyi P, et al. Fever is associated with reduced, hypothermia with increased mortality in septic patients: a meta-analysis of clinical trials. PLoS One 2017; 12: e0170152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans-Jones LG, Whittle HC, Onyewotu II, Egler LJ, Greenwood BM. Comparative study of group A and group C meningococcal infection. Arch Dis Child 1977; 52: 320–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.