Abstract
Introduction:
Mesenchymal stem cells (MSCs) possess endogenous reparative properties and may serve as an exogenous therapeutic intervention in patients with chronic kidney disease. Cardiovascular risk factors clustering in the metabolic syndrome (MetS) might adversely affect cellular properties. To test the hypothesis that Mets interferes with MSC characteristics, we performed comprehensive comparison of the mRNA, microRNA, and protein content of MSCs isolated from Lean and MetS pigs.
Methods:
Domestic pigs were fed a 16-week Lean or MetS diet (n=4 each). Expression profiles of co-existing microRNAs, mRNAs, and proteins were obtained by high-throughput sequencing and liquid chromatography-mass spectrometry. TargetScan and ComiR were used to predict target genes of differentially expressed microRNAs, and DAVID 6.7 for functional annotation analysis to rank primary gene ontology categories for the microRNA target genes, mRNAs, and proteins.
Results:
Differential expression analysis revealed 12 microRNAs upregulated in MetS-MSCs compared to Lean-MSCs (fold change>1.4, p<0.05), which target 7,728 genes, whereas 33 mRNAs and 78 proteins were downregulated (fold change<0.7, p<0.05). Integrated analysis showed that targets of those microRNAs upregulated in MetS-MSCs overlap with at least half of mRNAs and proteins dysregulated in those cells. Functional analysis of overlapping mRNAs and proteins suggest that they are primarily involved in mitochondria, inflammation and transcription. MetS-MSCs also exhibited increased nuclear translocation of nuclear factor kappa-B, associated with increased SA-β-Galactosidase and decreased cytochrome-c oxidase-IV activity.
Conclusion:
MetS alters the transcriptome and proteome of swine adipose tissue-derived MSCs particularly genes involved in mitochondria, inflammation and transcription regulation. These alterations might limit the reparative function of endogenous MSC and their use as an exogenous regenerative therapy.
Keywords: mesenchymal stem cells, microRNA, sequencing, proteomics, senescence
Introduction
Chronic kidney disease (CKD) encompasses a group of disorders characterized by the gradual loss of kidney function. The overall prevalence of CKD worldwide is estimated to be 200 million people (1), and it is associated with substantial economic and social burden (2). Importantly, as CKD progresses to end-stage renal disease (ESRD), rates of morbidity and mortality increase significantly. Therefore, there is a pressing need for more effective strategies to attenuate its progression to ESRD.
Mesenchymal stem cells (MSCs) are mesoderm-derived multipotent stromal cells that have high self-renewal and multi-lineage differentiation potential, as well as immunomodulatory and anti-inflammatory capacity to repair damaged tissues. MSC exist in most organs and represent an important source of resident stem cells that can be recruited for local tissue repair. Furthermore, they can be harvested and delivered exogenously to augment tissue repair. They can be obtained from several sources, including hematopoietic bone marrow (3), adipose tissue (4), and placenta (5, 6). Importantly, MSCs can differentiate into many kidney cell types, constituting an attractive therapeutic option for repairing damaged kidneys (6, 7).
We have previously shown in swine reno-vascular disease that intra-renal delivery of MSCs with or without renal revascularization improves renal hemodynamics and function, and decreased inflammation, apoptosis, oxidative stress, microvascular loss, and fibrosis in the post-stenotic kidney (8, 9). Furthermore, we have recently shown that intra-renal delivery of MSCs in patients with renovascular disease improved blood flow, cortical and medullary perfusion, glomerular filtration rate, and tissue oxygenation in the post-stenotic kidney (10), underscoring the safety and efficacy of this intervention to ameliorate renal injury and dysfunction in human subjects.
Patients with renal disease are often exposed to cardiovascular risk factors (11–13), including the metabolic syndrome (MetS), a constellation of obesity, hypertension, hyperlipidemia, and insulin resistance that frequently coexists with renal disease (14, 15). We have previously probed in pigs with MetS alterations in specific pathways, involving either mRNA or microRNA regulating insulin signaling, mitochondrial function, or inflammation (16–19), in adipose tissue-derived MSC. However, how the overall transcriptome and proteome of adipose tissue-derived MSCs in MetS compares to that in Lean-MSCs and which pathways are primarily affected remains unknown. We hypothesized that MetS induces a congruent alteration in the gene, protein, and microRNA profile of these MSCs. To test this, we took advantage of our established swine model of MetS. In addition, we took advantage of high-throughput mRNA and microRNA sequencing and liquid chromatography-mass spectrometry (LC-MS) to compare the gene, protein, and microRNA profile between Lean- and MetS- MSCs.
Materials and Methods
Animals and Protocol
The Mayo Clinic Animal Care and Use Committee approved our study. Three-month-old female domestic pigs were randomized into Lean and MetS groups (n=4 each) for 16 weeks. MetS pigs were fed a high-cholesterol/carbohydrate diet (15) (5B4L, protein 16.1%, ether extract fat 43.0%, and carbohydrates 40.8%, Purina Test Diet, Richmond, IN) and Lean pigs a standard chow (13% protein, 2% fat, 6% fiber, Purina Animal Nutrition LLC, MN) for the duration of the study.
Systemic Measurements
Body weight and blood pressure were recorded after 16 weeks of diet. Total cholesterol, low-density lipoprotein (LDL), and triglyceride levels were measured by enzymatic methods and insulin resistance by homeostasis model-assessment of insulin resistance (HOMA-IR) (20). Animals were then euthanized with a lethal intravenous dose of sodium pentobarbital (100 mg/kg IV, Fatal Plus®, Vortech Pharmaceuticals, Dearborn, MI), and subcutaneous abdominal adipose tissue collected for MSC isolation.
MSC isolation, characterization, and culture
MSCs are plastic-adherent when maintained in standard culture conditions, express classic MSC surface markers, and differentiate into osteoblasts, adipocytes, and chondroblasts in vitro (21). Isolation of primary MSCs from abdominal fat (5–10g) was performed using collagenase-H. Cells were cultured for 3 weeks in advanced MEM medium (Gibco/Invitrogen, Carlsbad CA) supplemented with platelet lysate (PLTmax, Mill Creek Life Sciences, Rochester, MN), and used at the third passage, as previously shown (8, 22, 23). Characterization of MSCs was confirmed in vitro by positive immuno-fluorescent staining and flow cytometry for CD44, CD73, CD90, and CD105 (24), in accordance with our observations in human MSCs (21, 22).
mRNA-sequencing analysis
RNA sequencing was analyzed as described previously (22). Sequencing RNA libraries were prepared according to the manufacturer’s protocol (TruSeq RNA Sample Prep Kit v2, Illumina, San Diego, USA) and loaded onto flow cells (8–10pM) to generate cluster densities of 700,000/mm2 following the standard protocol for the Illumina cBot and cBot Paired-end cluster kit version-3. Cells were sequenced on an Illumina HiSeq 2000 using TruSeq SBS kit version 3 and HCS v2.0.12 data collection software, and data analyzed using the MAPRSeq v.1.2.1 system and the Bioinformatics Core standard tool, which includes alignment with TopHat 2.0.6 (25, 26) and gene counts with the feature Counts software (25–27). mRNA-Seq data were analyzed using CAP-miRSeq v1.1 (28) and normalization and differential expression analysis performed using edgeR 2.6.2 (29). Gene expression was normalized to 1 million reads and corrected for gene length (reads per kilobasepair per million mapped reads, RPKM). mRNAs with fold-change >1.4 and p values<0.05 were considered to be upregulated in MetS- compared to Lean-MSCs, whereas those with fold-change <0.7 and p<0.05 were considered downregulated. Functional annotation clustering analysis was performed using DAVID6.7 database (http://david.abcc.ncifcrf.gov/) to obtain a ranking of primary gene ontology categories for the upregulated and downregulated mRNAs.
Proteome analysis
For LC-MS proteomic analysis, MSCs were solubilized and lysed, and protein samples denatured by incubation at 85°C for 10min (30, 31). Aliquots were resolubilized in reducing sample buffer and samples electrophoresed in 4–20% TGX Ready gels at 200V for 30min. Gel sections were digested with trypsin (31) and peptides extracted and transferred onto a 35cmx100μm PicoFrit column 9 (NewObjective), self-packed with Agilent Poroshell 120S 2.7μm EC-C18 stationary phase, using a Dionex UltiMate 3000 RSLC LC system (Thermo-Fisher Scientific, Waltham, MA). Peptides were separated and eluting peptides analyzed using a QExactive mass spectrometer (Thermo-Fisher Scientific, Waltham, MA). We then used label-free peptide MS1 intensity-based methods to identify differentially expressed proteins between MSCs and MaxQuant 1.5.1 software to assess Data quality, and reversed protein sequences were appended to the database for estimating protein identification false discovery rates (FDRs). Protein group intensities of each sample were log2 transformed (31), normalized, and modeled using a Gaussian-linked generalized linear model. Data was normalized by protein loading, and differential p-values FDR-corrected (32) . Proteins with fold change >1.4 and p<0.05 were considered upregulated in MetS- vs. Lean-MSCs, whereas those with fold-change <0.7 and p<0.05 were considered downregulated. Proteins dysregulated in MetS- compared to Lean- MSCs were classified by their molecular function and cellular localization (33). Functional annotation analysis was performed using DAVID6.7 database to identify ontology categories for differentially expressed proteins.
microRNA-sequencing analysis
High-throughput microRNA sequencing and functional analysis was performed (23) utilizing CAP-miRSeq-v1.1 workflow (26, 28). FASTQs and aligned BAMS were generated, and raw and normalized mature microRNA expression counts transferred to a excel file. We utilized EdgeR 2.62 to identify differential expressed of microRNAs between Lean- and MetS-MSCs (29, 34) microRNAs with fold-change >2 and p<0.05 were considered upregulated in MetS-MSCs, and those with fold-change <2 and p<0.05 downregulated. We used TargetScan 7.1 (http://www.targetscan.org/vert_71) and ComiR (http://lagavulin.ccbb.pitt.edu/comir/index.php) to predict target genes of microRNAs upregulated and downregulated in MetS-MSCs versus Lean-MSCs. Functional annotation clustering analysis of microRNA target genes was performed using DAVID6.7.
Integrated analysis
To visualize mRNAs and proteins downregulated corresponding to target genes of microRNA upregulated in MetS-MSCs, three-way Venn diagrams were constructed using VENNY 2.1, (http://bioinfogp.cnb.csic.es/tools/venny/). Conversely, three-way Venn diagrams were used to visualize microRNA target genes downregulated in MetS-MSCs and corresponding mRNAs and proteins upregulated in MetS-MSCs. Individual gene information of overlapping genes was obtained using Gene cards (http://www.genecards.org/).
MSC functional studies
To explore the functional implications of these findings, we compared representative elements of inflammation, senescence, and mitochondrial function between Lean- and MetS-MSCs. Nuclear translocation of the pro-inflammatory transcription factor nuclear factor (NF)-kB was evaluated by immunofluorescent staining (abcam, 1:200, Cambridge, MA, USA). Nuclear DNA was stained with 4’,6’-diamino-2-phenylindole (DAPI), Thermo Fisher Scientific, Waltham, MA. Double positive (NFkB/DAPI) areas were quantified using a computer-aided image analysis program (ZEN® 2012 blue edition; Carl Zeiss SMT, Oberkochen, Germany), and the results from all fields were averaged. MSCs senescence was evaluated by SA-β-Galactosidase (SA-ß-GAL) activity using a Cellular Senescence Activity Assay kit (Enzo Life Sciences, Farmingdale, NY, USA) following the vendor’s protocol (35). The percentage of the positively stained area was quantified using ZEN®. In addition, mitochondria were isolated from MSCs using the MITO-ISO kit (Catalog #8268, ScienCell, Carlsbad, California) (36), and cytochrome-c oxidase (COX)-IV activity assessed by fluorometric methods (Abnova Cat#KA3950, Caltag Med system, UK).
Results
Systemic characteristics
Systemic characteristics of all animals at 16 weeks are shown in Table 1. MetS pigs compared to Lean pigs had higher body weight and blood pressure. Comparable fasting glucose levels were noted in MetS and Lean pigs, however insulin and HOMA-IR were higher in MetS versus Lean pigs. Total cholesterol, LDL, and triglyceride levels were higher in MetS compared to Lean pigs.
Table 1.
Systemic characteristic of Lean and metabolic syndrome (MetS) pigs (n=4 each)
| Parameter | Lean | MetS |
|---|---|---|
| Body Weight (Kg) | 74.5±11.1 | 91.7±2.2* |
| Mean blood pressure (mmHg) | 99.2±11.9 | 125.3±7.9* |
| Total cholesterol (mg/dl) | 84.2±6.8 | 425.3±84.7* |
| LDL cholesterol (mg/dl) | 32.8±6.7 | 359.3±156.3* |
| Triglycerides (mg/dl) | 8.1±1.3 | 18.5±5.4* |
| Fasting glucose (mg/dl) | 128.5±14.9 | 118.0±19.6 |
| Fasting insulin (μU/ml) | 0.4±0.1 | 0.7±0.1* |
| HOMA-IR score | 0.7±0.1 | 1.7±0.4* |
p≤0.05 vs. Lean. LDL: Low-density lipoprotein, HOMA-IR: Homeostasis model assessment of insulin resistance.
mRNAs
Of all annotated genes (n=10,413), mapping of RNA reads revealed 511 (4.9%) mRNAs that were upregulated in MetS-MSCs compared to Lean-MSCs (Figure 1A). Functional annotation clustering analysis showed that those genes are involved in calcium-dependent membrane targeting, regulation of inflammatory response, ribonucleotide biosynthetic process, and cell activation during immune response, and activation and adhesion. Contrarily, only 33 genes (0.31%) were downregulated in MetS-MSCs compared to Lean-MSCs (Figure 1B). The genes encode proteins involved in regulation of transcription, transmembrane region, glycoprotein, plasma membrane, and organelle lumen, zinc ion binding, and extracellular region.
Figure 1.
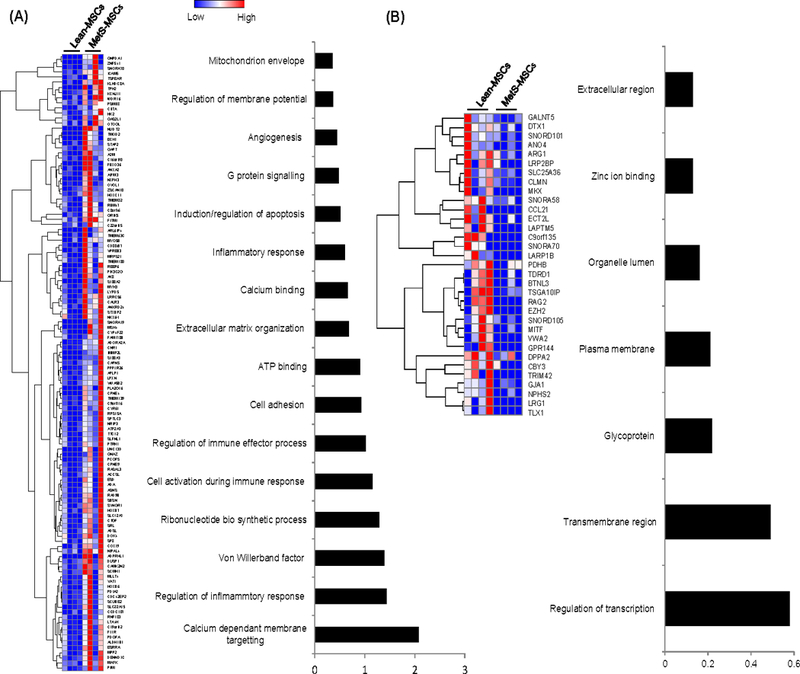
Comparison of mRNAs in MetS-MSCs and Lean-MSCs with heat maps and functional annotation clustering (using DAVID 6.7) showed 120 mRNAs that were upregulated (A) and 33. mRNAs downregulated (B) in MetS-MSCs.
Proteins
Of the 4,933 proteins identified in Lean- and MetS-MSCs, 36 (0.72%) were upregulated in MetS-MSCs compared to Lean-MSCs (Figure 2A), which were intra-cellular proteins with binding and catalytic activity, primarily involved in calcium signaling (Figure 2B). They showed diverse biological roles, with highest enrichment scores ascribed to regulation of protein transport and calcium ion binding, with the remaining categories, like ATP binding/protein kinase activity, cytoskeleton, and transmembrane proteins, evenly distributed (Figure 2C).
Figure 2.
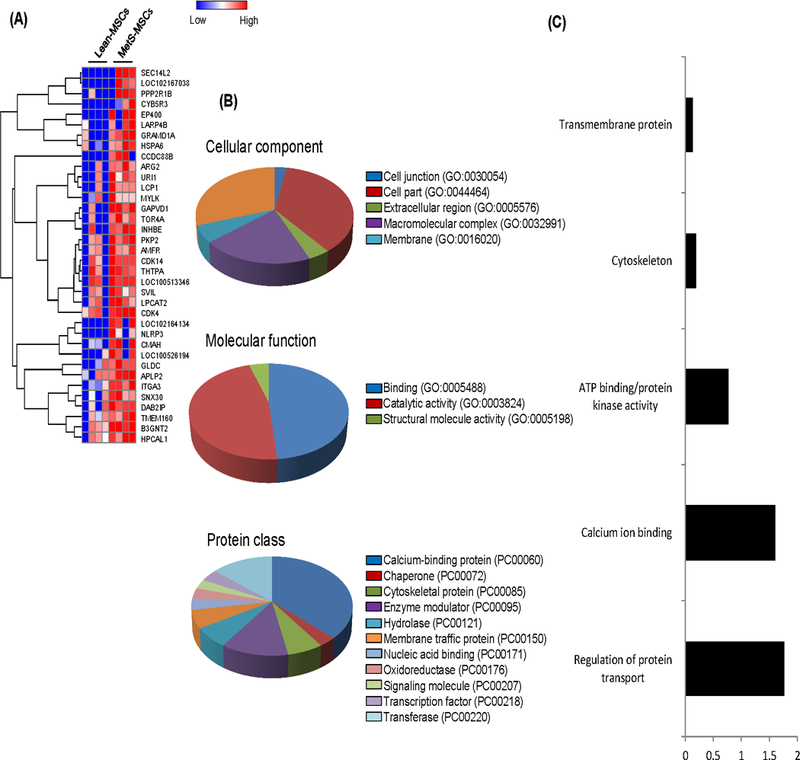
Evaluation of 36 proteins upregulated in MetS-MSCs compared to Lean MSCs using A. Heat map; B. Protein Analysis through Evolutionary Relationships (PANTHER) analysis of cellular components, molecular function, and protein class; C. Functional annotation clustering (DAVID 6.7).
Of 78 (1.58%) proteins downregulated in MetS-MSCs, most constituted intracellular and membrane proteins with binding, catalytic, ligase, and nucleic acid binding activities (Figure 3A–B). As per functional analysis, they primarily regulate mitochondrial function, cell development, apoptosis, blood vessel development, cation binding, as well as cytoplasmic vesicle and cell morphogenesis (Figure 3C).
Figure 3.
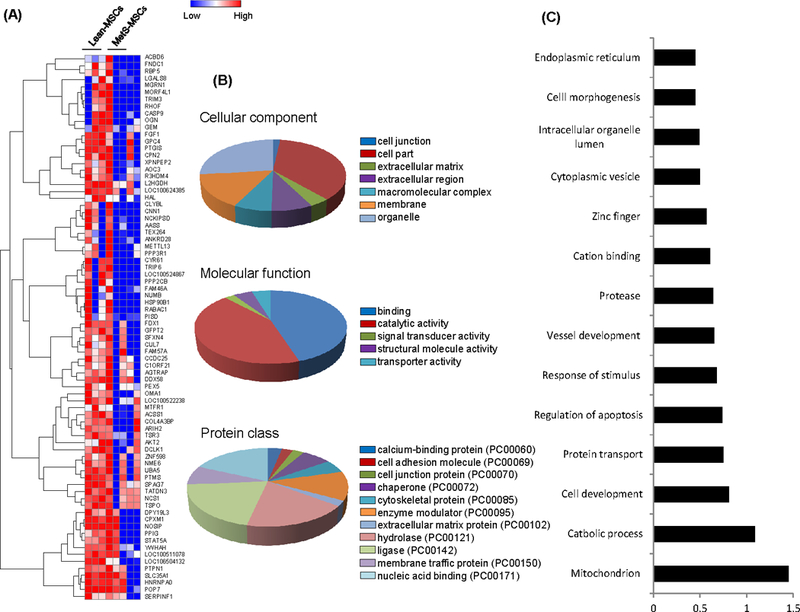
Seventy-eight proteins downregulated in MetS-MSCs compared to Lean-MSCs. A. Heat Map. B. Panther analysis of cellular component, molecular function, and protein class. C. Functional annotation clustering (DAVID 6.7).
MicroRNAs and integrated analysis
Analysis identified a total of 414 microRNAs, among which 12 (2.9%) were upregulated in MetS-MSCs (Figure 4A). Venn diagrams identified 11 mRNAs and 35 proteins downregulated in MetS-MSCs, which were targeted by microRNAs upregulated in MetS-MSCs (Figure 4B). Among them are proteins that participate in mitochondrial regulation (e.g.SLC25A36, PDHB, FDX1, SFXN4, MTFR1), Transcription (MITF, TLX1) Transmembrane proteins (LAPTM5), Apoptosis (CASP9, LGALS8), Angiogenesis (FGF1, PTGIS, and NOSIP) and Insulin signaling (GEM, XPNPEP2)
Figure 4.
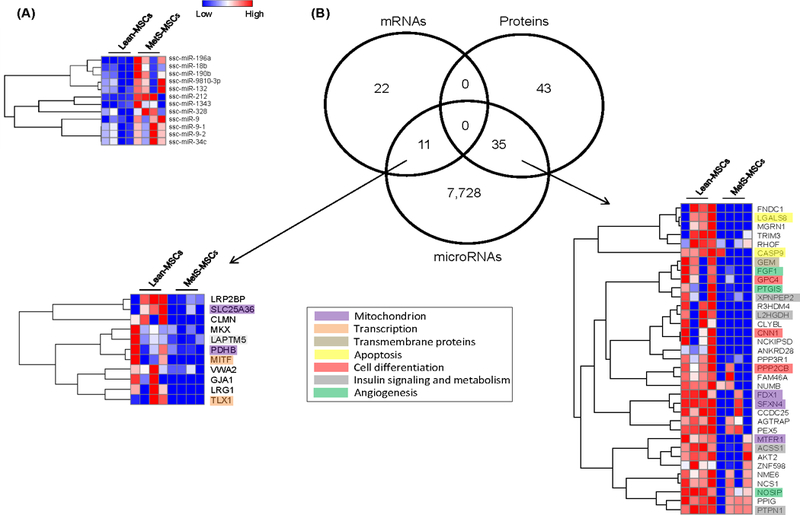
Twelve miRNA were upregulated in MetS-MSCs compared to Lean-MSCs. A. Heat map. B. Interactions among miRNA, mRNA and proteins in Lean and MetS-MSCs. Venn diagram showing distribution of upregulated miRNA, downregulated mRNA, and proteins in MSCs and their interactions.
We found 5 downregulated microRNAs in MetS-MSCs compared to Lean-MSCs (Figure 5A). Moreover, 42 mRNAs and 16 proteins upregulated in MetS-MSCs were targeted by microRNAs downregulated in MetS-MSCs (Figure 5B), and primarily involved in calcium signaling (TMEM63C, ADORA2A, CAMK2N2), Inflammation (CIITA, PCGF5A), and Senescence (SCMH1, EP400, CDK4).
Figure 5.
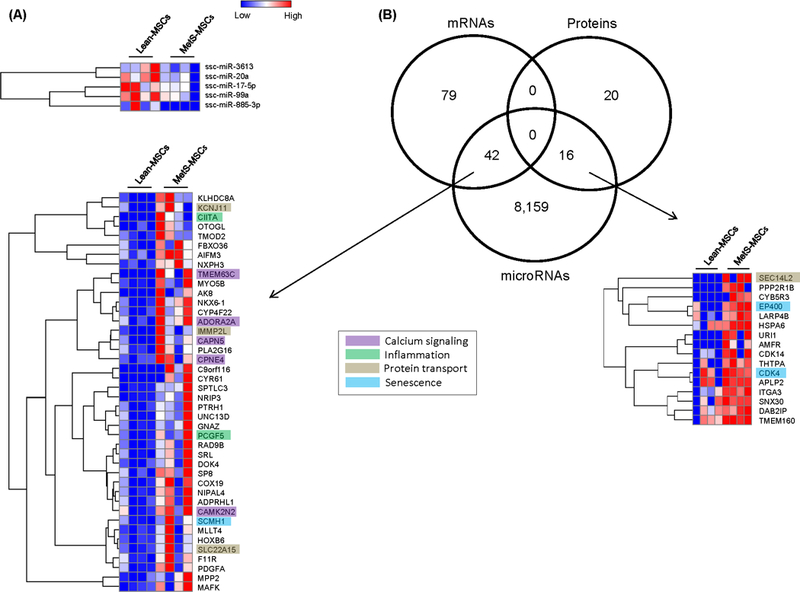
A. Five downregulated miRNA in MetS-MSCs compared to Lean-MSCs per Heat map. B. Interactions among miRNA, mRNA and proteins in Lean and MetS MSCs. Venn diagram showing distribution of downregulated miRNA, upregulated mRNA, and proteins in MSCs and their interactions and Heat maps.
MSC functional studies
NFkB was localized predominantly in the cytoplasm of Lean-MSCs, but in the nucleus of MetS-MSCs, suggesting NFkB nuclear translocation (Figure 6A), an index of inflammatory activation. SA-ß-GAL activity was higher and COX-IV activity was lower in MetS-MSCs compared to Lean-MSCs (Figure 6B–C), suggesting increased cellular senescence and decreased mitochondrial activity, respectively.
Figure 6.
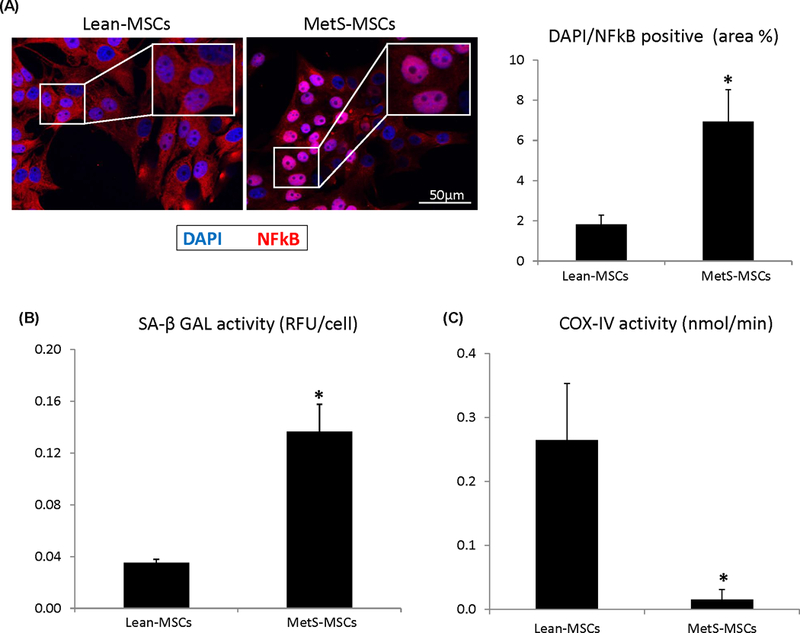
A. Nuclear factor kappa-B (NFkB) was localized predominantly in the nucleus of MetS-MSCs, associated with increased SA-β-Galactosidase (SA-β-GAL) and decreased cytochrome-c oxidase (COX)-IV activity. *p < 0.05 vs. Lean-MSCs.
Discussion
Our study demonstrates that metabolic syndrome alters the content of genes, proteins, and microRNAs of swine adipose tissue-derived MSCs, and our three-pronged analysis shed light on these effects. MetS upregulated in MSCs genes involved in regulation of inflammation, cellular activation, and adhesion, but downregulated transcription, transmembrane, and extracellular regulation. MetS also upregulated in MetS-MSCs proteins with intra-cellular binding and catalytic activity, and microRNAs that target genes and proteins implicated in multiple cellular processes, including mitochondrial function, transcription, apoptosis, differentiation, insulin signaling, and angiogenesis. Functionally, we found that MetS-MSCs have increased inflammation and senescence, but impaired mitochondrial respiratory function. Therefore, MetS induces upregulation of genes, and to a lower extent proteins and microRNAs, that overall regulate cellular activation and adaptive responses. These observations suggest that MetS modulates MSC content to adapt to the harsh milieu that it introduces, but might in turn interfere with their functionality.
Cardiovascular risk factors frequently coexist with CKD, and may impact endogenous reparative mechanisms, including cellular repair systems. Alterations in MSC might have functional consequences for both tissue healing and for the choice of cell-based treatment for subjects with CKD and MetS. While stem cell therapy is emerging as a promising therapy for patients with kidney injury, autologous MSCs isolated from a MetS milieu might have aberrant protein and gene expression, associated with impaired proliferative and immunomodulatory potential.
This study shows that of all identified genes, 5% were upregulated in MetS-MSCs compared to Lean-MSCs, and were chiefly involved in regulation of inflammation, cell activation, and adhesion. Only 0.3% of the genes was downregulated in MetS-MSCs, and was involved in cellular processes like transcription, transmembrane, and extracellular regulation. We also found that the 0.7% of total proteins that were upregulated in MetS-MSCs had intra-cellular binding and catalytic activity, whereas many of the 1.6% downregulated proteins were enzyme regulators and cytoskeletal proteins. Of microRNA, ~3% were upregulated in MetS-MSCs, with target genes and proteins implicated in multiple cellular processes, including mitochondrial function, transcription, apoptosis, differentiation, insulin signaling, and angiogenesis. A fraction (0.01%) of all microRNAs was downregulated, with targets implicated in calcium signaling, inflammation, and cellular senescence. Therefore, MetS induces primarily upregulation of genes, and to a lesser extent of proteins and microRNAs, that regulate cellular activation and adaptive responses. Modest modulation is also exerted by downregulation of genes, proteins and microRNA. Interestingly, almost half of the changes in gene and protein expression in MetS-MSCs can be accounted for by orchestrated changes in the expression of microRNA that target them, underscoring the significance of these particular pathways. These observations suggest that MetS regulates MSC content to adapt to the harsh milieu that it introduces.
The cellular processes affected by MetS appear to be fundamental for cellular function, such as those modulated by mitochondria. Several mitochondrial genes were downregulated in MetS-MSC, including SLCA2536 (Solute Carrier Family-25 Member-36), PDHB (Pyruvate dehydrogenase E1-Beta Subunit), FDX1 (Ferredoxin), SFXN4 (Sideroflexin) and MTFR1 (Mitochondrial Fission Regulator-1). SLC25A36 is involved in mitochondrial cytosolic and matrix function, specifically import/export of pyrimidine nucleotides into and from mitochondria, and helps in transport steps essential for mitochondrial DNA, RNA synthesis, and breakdown (4, 37). We also observed downregulation of genes coding for proteins such as MTFR1, which regulates mitochondrial fission and apoptosis. Mtfr1 is a direct target of miR-324–5p, which attenuates mitochondrial fission (38). Marycz et al (39) have shown previously that stem cells in equine MetS have increased number of impaired mitochondria due to ruptured membranes, disarrayed cristae, and vacuole formation. They also showed that toxic compounds accumulating in the mitochondria under oxidative stress lead to alterations in morphology and may be responsible for senescent phenotype and decreased proliferation potential (34). This suggests that MetS may affect cellular function by impacting on mitochondrial function.
An additional important cellular category affected by MetS is transcription, reflected in suppression of MITF and TLX1 genes, which affect specific differentiation pathways. Transcription activity is an indicator of stem cell pluripotency, which also contributes to their plasticity and lineage specification, and is driven by the actively transcribed portion of the genome. TLX1 (HOX11) regulates differentiation of hematopoietic stem/progenitor cells and is critical for the development of splenic extramedullary hematopoiesis (40). Transmembrane proteins have several important functions, including serving as cell surface markers, responsiveness to growth factors, reuse of developmental signaling cascade, interaction with molecules of extracellular matrix, and protection against cellular stress (41). LAPTM5 is responsible for encoding a receptor associated with lysosomes, GEM may play a role in signal transduction, and XPNPEP2 is associated with protein metabolism. Therefore, downregulation of these genes in MetS-MSC might have far reaching ramification for their functionality.
Cell differentiation was targeted by upregulated microRNAs with the associated suppressed genes CNN1 (CCN family member-1) and PPP2CB. CNN1 participates in cell migration, adhesion, proliferation, differentiation, survival and apoptosis (42), as well as in vessel development (43, 44). Alteration of apoptosis in MetS-MSC was suggested by downregulation of LGALS8 and CASP-9; Caspases are a family of cysteine proteases that are activated during apoptosis, and CASP-9 suppression may be linked to MSC senescence. We also observed suppression of genes involved in insulin signaling, including XPNPEP2, L2HGDH, GEM, ACSS1, and, PTPN1, extending our previous observations (17). Blunted angiogenic capability was suggested by downregulation of FGF1, PTGIS, and NOSIP. NOSIP expression interferes with endogenous nitric oxide that is critical to cell cycle-related actions such as apoptosis and proliferation (45), while FGF1 promotes angiogenic proliferation (46).
The expression of genes and proteins is often regulated by microRNAs. We therefore performed an additional integrated analysis to explore the impact of downregulated microRNA in concert with their target mRNA and proteins in the same cells. Major categories impacted by downregulated microRNA were calcium signaling, inflammation, protein transport, and cellular senescence. Genes like TMEM63C, ADORA2A, CAPN5, CPNE4, and CAMK2N2 are involved in calcium signaling, which is important for promoting stem cell development and differentiation (47). The effect of MetS on inflammation was denoted by upregulation of CIITA, a MHC-II trans-activator, and PCGF5. Therefore, MetS might interfere with the immunomodulatory and anti-inflammatory properties of MSC (48).
KCNJ11, IMMP2L, SLC22A15 and SEC14L2 that were upregulated in MetS are involved in protein transport. The KCNJ11 gene encodes for proteins in ATP-sensitive potassium channels, including glucose-stimulated insulin secretion from beta cells, and mutation in this gene may cause neonatal diabetes (49). IMMP2L (subunit of mitochondrial inner membrane peptidase) is involved in encoding a protein that directs proteins to mitochondria, and its loss results in overproduction of mitochondrial reactive oxygen species and early aging (50). We have also shown before that MetS-MSC have increased propensity for senescence (4). Genes involved in senescence that were found in this study to be upregulated in MetS-MSC included SCMH1, EP400, and CDK4. Studies have shown that CDK (Cyclin dependent kinase) in a complex with corresponding cyclins have a role in cell differentiation and senescence. Hence, their upregulation might lead MSC towards early senescence.
To determine if alterations in the protein cargo of MetS-MSCs affects the function of the cells, we compared NFkB activation, senescence, and mitochondrial energy production between Lean- and MetS- MSCs. We found that NFkB was localized predominantly in the nucleus of MetS-MSCs, suggesting activation of pro-inflammatory transcription. This was associated with increased SA-β-Galactosidase and decreased cytochrome-c oxidase-IV activity, suggesting that MetS-MSCs also have increased senescence, but impaired mitochondrial respiratory function. Hence, these changes observed in transcriptome and proteome in MetS-MSCs translates into aberrant functionality.
We acknowledge several limitations to this study, including a modest number of samples and inability to identify MSC post-translational/epigenetic changes. MSC at the 3rd passage possess a phenotype comparable to their parent cells (51), and we have previously validated trends observed in RNAseq using PCR and Western Blots (17, 51–55). Future studies will also need to define the functional limitations that MetS impose on MSC, and the mechanisms by which it modifies genetic and protein cellular information. Despite these caveats, this study has a number of strengths, and used cutting-edge techniques to highlight the comprehensive regulation of the mRNA, miRNA, and proteomic cargo of adipose tissue-derived MSCs.
In summary, we found that MetS modifies the cargo of porcine MSC, which might in turn modulate several important cellular functions and cell fate. Genetic message and proteins related to mitochondrial function, transcription, apoptosis, differentiation, insulin signaling, and angiogenesis were all downregulated in Mets-MSCs, whereas inflammatory pathways were activated. Therefore, MetS might decrease MSC survival and sternness. Further studies are needed to determine whether these changes adversely impact MSC reparative function.
Acknowledgements
This study was partly supported by the NIH grant numbers: DK120292, DK104273, HL123160, DK102325, and DK106427.
Footnotes
Conflict of interests
The authors declare no conflict of interest.
References
- 1.Morbidity and Mortality in Patients With Chronic Kidney Disease. American Journal of Kidney Diseases. 2012;59(1):e59–e68. [Google Scholar]
- 2.Ojo A Addressing the global burden of chronic kidney disease through clinical and translational research. Trans Am Clin Climatol Assoc. 2014;125:229–43; discussion 43–6. [PMC free article] [PubMed] [Google Scholar]
- 3.Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6(2):230–47. [PubMed] [Google Scholar]
- 4.Zhu XY, Ma S, Eirin A, Woollard JR, Hickson LJ, Sun D, et al. Functional Plasticity of Adipose-Derived Stromal Cells During Development of Obesity. Stem Cells Transl Med. 2016;5(7):893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.in’t Anker PS, Scherjon SA, Kleijburg-van der Keur C, de Groot-Swings GMJS, Claas FHJ, Fibbe WE, et al. Isolation of mesenchymal stem cells of fetal or maternal origin from human placenta. Stem Cells. 2004;22(7):1338–45. [DOI] [PubMed] [Google Scholar]
- 6.Papazova DA, Oosterhuis NR, Gremmels H, van Koppen A, Joles JA, Verhaar MC. Cell-based therapies for experimental chronic kidney disease: a systematic review and meta-analysis. Disease Models & Mechanisms. 2015;8(3):281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.In ‘t Anker PS, Scherjon SA, Kleijburg-van der Keur C, de Groot-Swings GM, Claas FH, Fibbe WE, et al. Isolation of mesenchymal stem cells of fetal or maternal origin from human placenta. Stem Cells. 2004;22(7):1338–45. [DOI] [PubMed] [Google Scholar]
- 8.Eirin A, Zhu X-Y, Ebrahimi B, Krier JD, Riester SM, van Wijnen AJ, et al. Intra-renal delivery of mesenchymal stem cells and endothelial progenitor cells attenuates hypertensive cardiomyopathy in experimental renovascular hypertension. Cell transplantation. 2015;24(10):2041–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119(24):5621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saad A, Dietz AB, Herrmann SMS, Hickson LJ, Glockner JF, McKusick MA, et al. Autologous Mesenchymal Stem Cells Increase Cortical Perfusion in Renovascular Disease. J Am Soc Nephrol. 2017;28(9):2777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iimori S, Naito S, Noda Y, Sato H, Nomura N, Sohara E, et al. Prognosis of chronic kidney disease with normal-range proteinuria: The CKD-ROUTE study. PLoS ONE. 2018;13(1):e0190493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bundy JD, Chen J, Yang W, Budoff M, Go AS, Grunwald JE, et al. Risk factors for progression of coronary artery calcification in patients with chronic kidney disease: The CRIC study. Atherosclerosis. 2018;271:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins AJ, Foley RN, Chavers B, Gilbertson D, Herzog C, Johansen K, et al. ‘United States Renal Data System 2011 Annual Data Report: Atlas of chronic kidney disease & end-stage renal disease in the United States. Am J Kidney Dis. 2012;59(1 Suppl 1):A7, e1–420. [DOI] [PubMed] [Google Scholar]
- 14.Cameron AJ, Shaw JE, Zimmet PZ. The metabolic syndrome: prevalence in worldwide populations. Endocrinol Metab Clin North Am. 2004;33(2):351–75, table of contents. [DOI] [PubMed] [Google Scholar]
- 15.Pawar AS, Zhu XY, Eirin A, Tang H, Jordan KL, Woollard JR, et al. Adipose tissue remodeling in a novel domestic porcine model of diet-induced obesity. Obesity (Silver Spring). 2015;23(2):399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. Obesity-induced mitochondrial dysfunction in porcine adipose tissue-derived mesenchymal stem cells. J Cell Physiol. 2018;233(8):5926–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley SM, Zhu XY, Eirin A, Tang H, Lerman A, van Wijnen AJ, et al. Metabolic syndrome alters expression of insulin signaling-related genes in swine mesenchymal stem cells. Gene. 2018;644:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng Y, Eirin A, Zhu XY, O’Brien DR, Lerman A, van Wijnen AJ, et al. The metabolic syndrome modifies the mRNA expression profile of extracellular vesicles derived from porcine mesenchymal stem cells. Diabetol Metab Syndr. 2018;10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. The metabolic syndrome alters the miRNA signature of porcine adipose tissue-derived mesenchymal stem cells. Cytometry A. 2018;93(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galili O, Versari D, Sattler KJ, Olson ML, Mannheim D, McConnell JP, et al. Early experimental obesity is associated with coronary endothelial dysfunction and oxidative stress. Am J Physiol Heart Circ Physiol. 2007;292(2):H904–11. [DOI] [PubMed] [Google Scholar]
- 21.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. [DOI] [PubMed] [Google Scholar]
- 22.Ebrahimi B, Eirin A, Li Z, Zhu X-Y, Zhang X, Lerman A, et al. Mesenchymal Stem Cells Improve Medullary Inflammation and Fibrosis after Revascularization of Swine Atherosclerotic Renal Artery Stenosis. PLoS ONE. 2013;8(7):e67474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O’Brien D, et al. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;551(1):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu XY, Urbieta-Caceres V, Krier JD, Textor SC, Lerman A, Lerman LO. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem Cells. 2013;31(1):117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 28.Sun Z, Evans J, Bhagwate A, Middha S, Bockol M, Yan H, et al. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC Genomics. 2014;15:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan MC, Johnson KL, Zenka RM, Charlesworth MC, Madden BJ, Mahoney DW, et al. Subfractionation, characterization, and in-depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int. 2014;85(5):1225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogan MC, Bakeberg JL, Gainullin VG, Irazabal MV, Harmon AJ, Lieske JC, et al. Identification of Biomarkers for PKD1 Using Urinary Exosomes. J Am Soc Nephrol. 2015;26(7):1661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KI, van de Wiel MA. Effects of dependence in high-dimensional multiple testing problems. BMC Bioinformatics. 2008;9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33(Database issue):D284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem. 2014;115(10):1816–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng Y, Eirin A, Zhu XY, Tang H, Hickson LJ, Lerman A, et al. Micro-RNAS Regulate Metabolic Syndrome-induced Senescence in Porcine Adipose Tissue-derived Mesenchymal Stem Cells through the P16/MAPK Pathway. Cell Transplant. 2018;27(10):1495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Li ZL, Crane JA, Jordan KL, Pawar AS, Textor SC, et al. Valsartan regulates myocardial autophagy and mitochondrial turnover in experimental hypertension. Hypertension. 2014;64(1):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Noia MA, Todisco S, Cirigliano A, Rinaldi T, Agrimi G, Iacobazzi V, et al. The human SLC25A33 and SLC25A36 genes of solute carrier family 25 encode two mitochondrial pyrimidine nucleotide transporters. J Biol Chem. 2014;289(48):33137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Zhang DL, Long B, An T, Zhang J, Zhou LY, et al. NFAT4-dependent miR-324–5p regulates mitochondrial morphology and cardiomyocyte cell death by targeting Mtfr1. Cell Death Dis. 2015;6:e2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marycz K, Kornicka K, Szlapka-Kosarzewska J, Weiss C. Excessive Endoplasmic Reticulum Stress Correlates with Impaired Mitochondrial Dynamics, Mitophagy and Apoptosis, in Liver and Adipose Tissue, but Not in Muscles in EMS Horses. Int J Mol Sci. 2018;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda A, Tezuka T, Ueno Y, Hosoda S, Amemiya Y, Notsu C, et al. Niche-induced extramedullary hematopoiesis in the spleen is regulated by the transcription factor Tlx1. Scientific Reports. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurer MH. Proteomic definitions of mesenchymal stem cells. Stem Cells Int. 2011;2011:704256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dotterweich J, Ebert R, Kraus S, Tower RJ, Jakob F, Schutze N. Mesenchymal stem cell contact promotes CCN1 splicing and transcription in myeloma cells. Cell Commun Signal. 2014;12:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien TP, Lau LF. Expression of the growth factor-inducible immediate early gene cyr61 correlates with chondrogenesis during mouse embryonic development. Cell Growth Differ. 1992;3(9):645–54. [PubMed] [Google Scholar]
- 44.Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol. 2002;22(24):8709–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schleicher M, Brundin F, Gross S, Muller-Esterl W, Oess S. Cell cycle-regulated inactivation of endothelial NO synthase through NOSIP-dependent targeting to the cytoskeleton. Mol Cell Biol. 2005;25(18):8251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoseini SJ, Ghazavi H, Forouzanfar F, Mashkani B, Ghorbani A, Mahdipour E, et al. Fibroblast Growth Factor 1-Transfected Adipose-Derived Mesenchymal Stem Cells Promote Angiogenic Proliferation. DNA Cell Biol. 2017;36(5):401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tonelli FM, Santos AK, Gomes DA, da Silva SL, Gomes KN, Ladeira LO, et al. Stem cells and calcium signaling. Adv Exp Med Biol. 2012;740:891–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–49. [DOI] [PubMed] [Google Scholar]
- 50.George SK, Jiao Y, Bishop CE, Lu B. Mitochondrial peptidase IMMP2L mutation causes early onset of age-associated disorders and impairs adult stem cell self-renewal. Aging Cell. 2011;10(4):584–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. The metabolic syndrome alters the miRNA signature of porcine adipose tissue-derived mesenchymal stem cells. Cytometry Part A. 2018;93a(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace JA, Kagele DA, Eiring AM, Kim CN, Hu R, Runtsch MC, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129(23):3074–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. Obesity-induced mitochondrial dysfunction in porcine adipose tissue-derived mesenchymal stem cells. Journal of Cellular Physiology. 2018;233(8):5926–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eirin A, Zhu XY, Jonnada S, Lerman A, van Wijnen AJ, Lerman LO. Mesenchymal Stem Cell-Derived Extracellular Vesicles Improve the Renal Microvasculature in Metabolic Renovascular Disease in Swine. Cell Transplantation. 2018;27(7):1080–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng Y, Eirin A, Zhu XY, O’Brien DR, Lerman A, van Wijnen AJ, et al. The metabolic syndrome modifies the mRNA expression profile of extracellular vesicles derived from porcine mesenchymal stem cells. Diabetology & Metabolic Syndrome. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]


