Abstract
Aberrant hypermethylation at CpG sites within the CDKN2A gene is associated with silencing and has been proposed as a target for reactivation using both DNA methylation and histone deacetylation inhibitors. This study investigates the role of selecting tumor samples with a silenced as compared to deleted CDKN2A locus when assessing the efficacy of DNA methyltransferase inhibitor, zebularine, combined with the HDAC inhibitor, depsipeptide. Non-small cell lung cancer cell lines with defined CDKN2A status were analyzed by MTS assay to determine the effect of zebularine or zebularine combined with depsipeptide on tumor cell growth. We observed that zebularine treatment resulted in inhibition of cell growth in 11 out of 12 lung cancer cell lines with silenced CDKN2A, but no cell growth inhibition was detected in the 7 lung cancer cell lines tested with deleted CDKN2A (p>0.001). In addition, we found that the combination of 30 μM zebularine and 6 or 7 nM depsipeptide resulted in a synergistic inhibition of cell growth in tumor cells with silenced CDKN2A (p<0.001, CI=0.70 and 0.57, respectively) but not in tumor cells with deleted CDKN2A. In conclusion, tumor cells with methylated CDKN2A are more sensitive to zebularine than cell lines with deleted CDKN2A and the combination of zebularine/depsipeptide results in a synergistic effect on cell growth inhibition that is also linked with the presence of silenced CDKN2A. Thus, combination of DNA methyltransferase and HDAC inhibitors may be a potential treatment for lung cancer patients, but careful selection of patients will be needed to optimize the benefit of this regimen.
Keywords: DNA methylation, HDAC inhibition, zebularine, depsipeptide, CDKN2A, lung cancer
Introduction
Aberrant hypermethylation at regulatory CpG sites in cancer genes is associated with both gene silencing and tumori-genesis (1,2). In addition, the observation that CDKN2A hypermethylation is mutually exclusive with somatic retinoblastoma (RB) or CDKN2A mutations in tumor samples suggests that hypermethylation is functionally comparable to somatic point mutations for cancer gene pathway inactivation (3). In contrast to acquired gene mutation, silenced genes may also serve as targets for reversible reactivation by DNA methylation inhibitors that efficiently re-induce mRNA and protein expression. For example, one of the most widely used drugs to inhibit DNA methylation and reactivate silenced genes is 5-aza-2’-deoxycytidine (DAC) (3,4). DAC is activated by deoxycytidine kinase (5) and is incorporated into newly synthesized DNA where it forms a covalent complex with DNA methyltransferase (DNMT) (6,7). Several studies have shown that incubation with DAC results in the reactivation of different bona-fide hypermethylated cancer genes including CDKN2A, MLH1, p15, p73, E-cadherin and VHL (3,8–12). While DAC has been used to treat a variety of leukemia, myelodysplastic diseases and solid tumors (13–16), it is rapidly inactivated in vivo by cytidine deaminase, decreasing its pharmacologic activity and its inhibitory effect (17). In addition, the use of DAC in patients has been complicated because of instability in solution and significant hematopoietic toxicity (18). Finally, the lack of a suitable biomarker to optimally pre-select patients for treatment may have contributed to the lack of efficacy of DNA demethylating agents in earlier clinical trials studying lung cancer (16).
In searching for a more stable and less toxic DNA methylation inhibitor, zebularine, a new synthetic analog of cytidine originally designed as a cytidine deaminase inhibitor because of a missing amino group at the C4 of the pyrimidine ring, was identified (19). Zebularine requires phosphorylation and conversion to its deoxynucleotide base before it is incorporated into DNA. Once incorporated it is paired with guanine forming a tight complex that can lead to inhibition of DNA methylation (20). Zebularine is stable in both acidic and neutral aqueous solutions and appears less cytotoxic in vitro than other DNA methylation inhibitors, thus allowing for continuous low-dose treatments (21,22). It has been shown that continuous treatment of T24 bladder cells with zebularine results in promoter demethylation of the CDKN2A gene and induction of its mRNA (23). Continuous zebularine treatment has also resulted in complete depletion of the DNMT1 enzyme required for maintaining methylation (21,24). In addition, oral or intra peritoneal administration of zebularine into nude mice with EJ6 xenograft tumors showed inhibition of tumor growth without significant animal toxicity (23), and appears to preferentially target cancer cells compared to normal cells with regard to growth inhibition, demethylation of the promoter region and DNMT1 depletion (24).
It has also been shown that DNA methylation inhibitors such as DAC can interact synergistically with histone deacetylase (HDAC) inhibitors to suppress cell growth (25,26). Histone deacetylation has been associated with both gene silencing and transcriptional repression, and HDAC inhibitors have been studied for their role in the reactivation of suppressor genes to inhibit tumor cell growth (1,27). The HDAC inhibitor, depsipeptide, is one of several HDAC inhibitors that has been shown to inhibit tumor cell growth by arresting cell cycle progression (28,29) and by inducing apoptotic cell death in many tumor types including lung cancer (30–32). For example, treatment of lung cancer cells with DAC followed by treatment with depsipeptide resulted in a significant enhancement of cytotoxicity and apoptosis over depsipeptide alone (30). In addition, enhanced CDKN2A/p16 protein expression was observed with this drug combination as compared to DAC treatment alone (33).
Since it is important to define the optimal subset of tumor samples for targeted therapies, in this report we investigated the effect of the DNA methylation inhibitor, zebularine, on the growth of lung and breast cancer cell lines with either homozygously deleted or with hypermethylated and silenced CDKN2A gene status. We observed that lung cancer cell lines with hypermethylated/silenced CDKN2A were more sensitive to zebularine treatment than cells with deleted CDKN2A suggesting that the ability to re-induce p16 protein expression correlates with cytotoxicity of the drug. We also observed that zebularine in combination with the HDAC inhibitor, depsipeptide, showed a synergistic effect on inhibition of tumor growth, which was also associated with reactivation of silenced CDKN2A. Moreover, we observed that the concentration of depsipeptide sufficient to enhance the zebularine effect was 1000-fold lower than the maximal tolerated dose administered to patients (34,35). These observations suggest that the use of DNA methylation inhibitors in combination with sub-toxic concentrations of HDAC inhibitors may prove to be beneficial for the subset of patients with silenced CDKN2A status.
Materials and methods
Cell culture and reagents.
Lung carcinoma cell lines were maintained in RPMI-1640 media supplemented with 10% FBS, 1% glutamine and 1% penicillin/streptomycin. MDAMB231 and MDA-MB435 breast cancer cells were maintained in DMEM media supplemented with 10% FBS, 1% gluta-mine and 1% penicillin/streptomycin. All lung cancer cell lines were obtained directly from a repository of lung cancer cell lines stored and developed at the Naval National Medical Center, NCI. All of these cells are available from ATCC. The two breast cancer cell lines were obtained from the Laboratory of Molecular Pharmacology, NCI. All cell lines were tested for mycoplasma and all cell lines were authenticated at the time of the experiments validating the status of p16 expression by immunoblot analysis and also by PCR analysis for confirmation of the homozygous deletion of the CDKN2A locus. In addition, the cells with silenced CDKN2A locus were further tested by DAC treatment followed by immunoblot analysis to show induction of the p16 protein. DAC (Sigma, St. Louis, MO) was dissolved in DMSO at a stock concentration of 20 mM and aliquots frozen at −20°C. Zebularine and depsipeptide were obtained from the NCI repository at the Developmental Therapeutics Program (DTP, NSC no. 309132) and were dissolved in water at a stock concentration of 200 and 10 μM, respectively.
Zebularine treatment, protein isolation and immunoblot analysis.
Zebularine was added 24 h after plating 2×106 cells into duplicate 100 cm2 dishes at a final concentration of 30 μM. Control and zebularine-treated cells were harvested after 5 days of drug treatment. A second set of the control and zebularine-treated cells was trypsinized and 2×106 cells were plated onto duplicate 100 cm2 dishes with zebularine added to the pretreated cells to maintain a final concentration of 30 μM. This process was repeated again on day 10. Cell pellets were harvested for protein lysate on day 0, 5, 10 and 15 of zebularine treatment.
Protein extracts were isolated as described previously (36). Briefly, cells were harvested in EBC lysis buffer supplemented with 0.5 mM DTT and a mix of protease inhibitors (Complete Mini, Roche cat no. 04693124001). Protein extract (30 μg) was resolved on a 4–20% SDS-PAGE, and immunoblotting was performed with p16 antibody (BD Biosciences, CA) at a concentration of 1.5 μg/ml overnight at 4°C as described previously (37,38). After washing with PBSTween the blot was incubated with goat anti-mouse secondary antibody (BioRad no. 172–1019, Hercules, CA) at room temperature for 1 h. Protein bands were detected using an ECL Western blotting detection kit (GE Healthcare, Piscataway, New Jersey).
Real-time RT-PCR analysis of CDKN2A expression.
For RNA isolation, cells were plated and treated with 15 or 30 μM zebularine as described above for extract preparation; and the cell pellet was used to isolate RNA using TRIzol reagent (Invitrogen, Carlsbad, CA). Real-time RT-PCR analyses were performed in triplicates with the iCycler iQ real-time PCR detection system (BioRad Laboratories) using the TaqMan EZ RT-PCR CORE Reagent kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s recommendation. For quantitation of gene expression, the endogenous reference 18s rRNA was used to normalize the difference between each sample. Specific hybridization probes for mouse 18s rRNA, and the CDKN2A gene labeled with 5-FAM-reporter dye were purchased from Applied Biosystems.
MTS cell proliferation assays.
Cells (1×104) were plated onto a 96-well microtiter plate and 18 h after plating, cells were treated with zebularine at a final concentration of 10, 30, 50, or 100 μM for 4 or 5 days, or cells were treated with depsipeptide at a final concentration of 1, 3, 5, 7, or 9 nM for 4 or 5 days. MTS assay was performed using the CellTiter 96 Aqueous One Solution kit as described by the manufacturer (Promega, Madison, WI). For DAC treatment, drug was added at a final concentration of 0.25, 0.5, 1, 2, or 3 μM 4 h after plating the cells and the media with fresh drug was changed every 24 h for a total of 72 h. The cells were then incubated without the drug for an additional 24 h before performing the MTS assay.
For combination studies with zebularine and depsipeptide, cells were pretreated with zebularine for 6 days before addition of depsipeptide. Cells (2×105) were plated in a 25 cm2 flask and zebularine was added to the cells 24 h after plating at a final concentration of 30 μM. After 4 days of incubation cells were re-plated into a 100 cm2 dish with fresh zebularine, incubated for an additional two days and then plated in triplicate into a 96-well dish at a density of 1×104 cells per well. Depsipeptide was added (4 h following cell plating) to either untreated control cells or to zebularine-pretreated cells maintaining 30 μM zebularine in the medium. The cells were incubated for an additional 4 or 5 days and assayed for cell growth and viability using the MTS assay.
For combination studies with DAC and 3 nM depsi-peptide, 1×104 cells were plated into a 96-well dish and both drugs were added 4 h later. Media with fresh depsipeptide or DAC were added every 24 h for a total of 72 h combined drug treatment. Cells were then incubated for an additional 24 h in drug-free medium before performing the MTS assay. For DAC and 50 nM depsipeptide combination treatment, 1×104 cells were plated into a 96-well dish and DAC was added 4 h later. Media with fresh DAC were added every 24 h for a total of 72 h. Cells were then incubated for additional 18 h in drug-free medium at which point depsipeptide was added at a final concentration of 50 nM and incubated for additional 6 h before performing the MTS assay.
Statistical, median effect and combination index analysis.
All experiments were carried out at least three times. For statistical analysis an average percentage of surviving cells after zebularine treatment was used from 11 cell lines with methylated CDKN2A gene (excluding the H1725 cells which were not responding to treatment) vs the 7 cell lines with a deletion in the CDKN2A gene. The statistical significance of the experimental results was determined by t-test and One-Way analysis of variance (Holm-Sidak method) using SigmaPlot software (San Jose, CA). Drug interactions were analyzed using CalcuSyn software (Biosoft, Cambridge, UK). Determination of synergy or antagonism was based on the multiple drug-effect equation of Chou and Talalay (39,40) and was quantified by the combination index (CI). CI<1 indicates synergy, CI=1 indicates additive effect and CI>1, antagonism.
Results
Inhibition of cell growth by zebularine is linked to re-induction of silenced CDKN2A.
Since zebularine has been previously shown to demethylate the CDKN2A gene in several distinct tumor cells (23,24), we investigated the effects of zebularine on cell growth, CDKN2A/p16 mRNA induction, and p16 protein expression in different lung cancer lines that have varying CDKN2A gene status. The H719 and H865 cells are hypermethylated in the 5’-promoter region resulting in silencing of the CDKN2A gene, while the H226 and H2228 cell lines have a deletion of the CDKN2A gene (41). Treatment of these 4 lung cancer cell lines with zebularine at concentrations ranging from 10 to 100 μM for 4 days resulted in a significant difference in the dose-response effect among the two groups of lung cancer cells (Fig. 1A). The H719 and H865 cells were sensitive to zebularine treatment, with an IC50 of 75 μM. In contrast, H226 and H2228 cells, with a deleted CDKN2A gene, were insensitive to zebularine treatment with an IC50 greater then 200 μM (Fig. 1A and data not shown). The H719 and H865 cells with a silenced CDKN2A gene showed a significantly greater cell killing following 4 days of treatment with 100 μM zebularine than the H226 and H2228 cells with deleted CDKN2A (65% killing vs 15%), p<0.001 (Fig. 1A). A statistically significant difference was also observed when the lung cancer cell lines were treated with 50 μM zebularine (p<0.001) as well as with 30 μM zebularine (p<0.001 and p<0.05) comparing H865 and H719 cells to H226 and H2228 cells, respectively (Fig. 1A).
Figure 1.

Tumor growth inhibition by zebularine and DAC in cells with silenced but not deleted CDKN2A. (A) Effect of zebularine on growth of lung cell lines H2228, H226 with deleted and H719 and H865 with silenced CDKN2A following a four-day treatment. (B) Effect of DAC on growth of lung cell lines H2228 and H226 with deleted and H719 and H865 with silenced CDKN2A. Lung cancer cells were treated with DAC (0.25–3.0 μM) for 72 h followed by 24 h incubation without drug. The data shown represent the mean ± SD of 3–5 experiments. (C) Effect of zebularine on growth of breast cancer cell lines MDA-MB231 with deleted and MDAMB435 with silenced CDKN2A following a four-day treatment.
We also tested if DAC, another DNA methylation inhibitor that is known to efficiently reactivate a silenced CDKN2A gene (3,33), had a similar effect as zebularine on tumor cell growth and CDKN2A status. Cells were exposed to concentrations of DAC ranging from 0.5 to 3 μM for 48 h and then harvested 96 h after the initial drug incubation. We again observed inhibition of cell growth in the H719 and H865 cells with hypermethylated CDKN2A gene, while no effect on cell growth was detected in either H226 or H2228 cells with a deleted CDKN2A gene (p<0.001, H719 and H865 vs H226 and H2228) (Fig. 1B). Thus, these results suggest that two distinct demethylating agents, zebularine and DAC, can inhibit lung tumor growth in cells with silenced but not deleted CDKN2A.
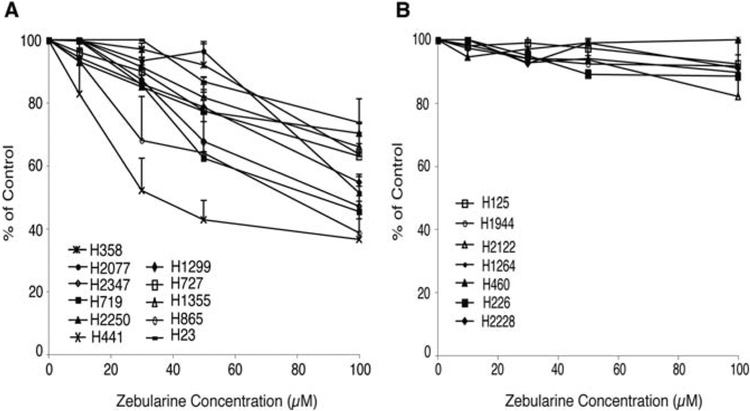
To confirm whether inhibition of cell growth by zebularine treatment correlated with the status of CDKN2A, 15 additional lung tumor cell lines were treated with zebularine. Ten of these cell lines (H23, H358, H441, H727, H1299, H1355, H1725, H2077, H2250, H2347) had a silenced CDKN2A locus, while 5 cell lines (H125, H460, H1264, H1944, H2122) had a deletion in CDKN2A gene (3,41). Of 12 cell lines with silenced CDKN2A locus tested, 11 were sensitive to zebularine treatment (Fig. 2A) (one cell line, H1725 was not sensitive to zebularine treatment and was not included in Fig. 2A). In contrast, growth inhibition was not detected in any of the 7 cell lines tested with deleted CDKN2A (Fig. 2B). Using a t-test analysis the difference between the tumor cells with silenced vs deleted CDKN2A were statistically significant with p<0.001 and p<0.05 at 100 and 50 μM of zebularine, respectively. To further test if the sensitivity to zebularine treatment depends on the status of the CDKN2A gene, we also examined the effect of zebularine on the breast cancer cell line, MDA-MB231 which has a deleted CDKN2A gene, and MDA-MB435 with low p16 expression that can be induced to higher levels by DAC (data not shown). The MDA-MB435 cells were sensitive to zebularine treatment with a calculated IC50 value of 75 μM, while the growth of the MDA-MB231 cells with deleted CDKN2A was unaffected by zebularine treatment (IC50 >100 μM) (Fig. 1C). These observations suggest that induction of p16 through CDKN2A demethylation may be linked with the ability to render the cells sensitive to zebularine treatment.
Figure 2.
Tumor cell growth inhibition by zebularine in a panel of lung cell lines with silenced but not deleted CDKN2A. (A) A panel of eleven cell lines with silenced CDKN2A that respond to four days of zebularine treatment. (B) A panel of seven cell lines with deleted CDKN2A that do not respond to four days of zebularine treatment.
Synergistic effect of zebularine and depsipeptide on the inhibition of cell growth is linked to re-induction of silenced CDKN2A gene.
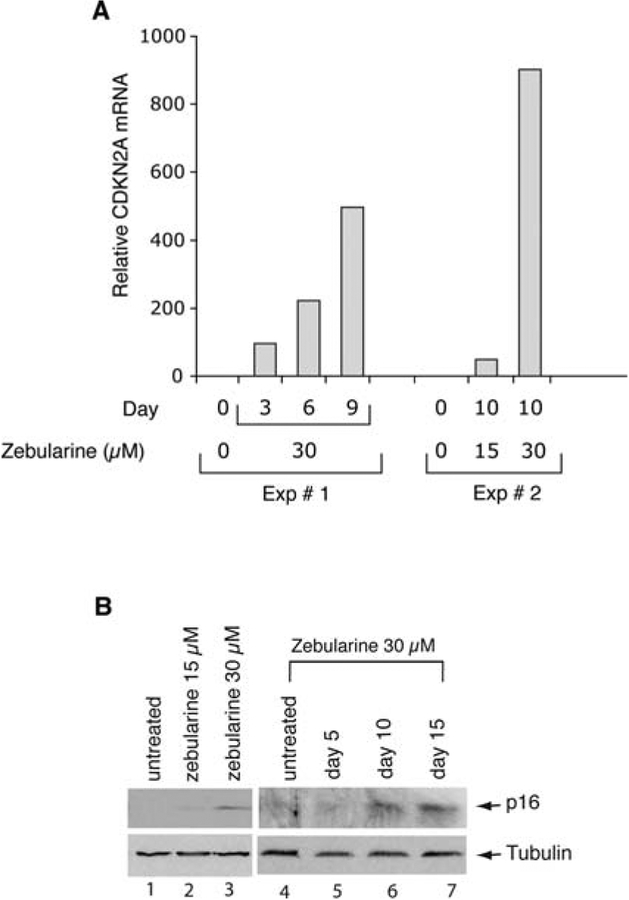
It has been shown that DNA methylation inhibitors such as DAC can interact synergistically with histone deacetylase (HDAC) inhibitors such as depsipeptide to suppress growth of lung cancer cells (25,26). To study the effect of zebularine in combination with depsipeptide, we chose 30 μM zebularine since that concentration can be safely achieved in murine plasma following a daily dose of 5 mg/ml in drinking water (unpublished data). We have also shown that 30 μM zebularine alone exhibits minimal cell growth inhibition in vitro (Fig. 1A). To test if 30 μM zebularine treatment is sufficient to re-induce CDKN2A gene expression, we isolated RNA from H719 cells 3, 6, 9 and 10 days after treatment. We found that 30 μM zebularine treatment was sufficient to induce both CDKN2A mRNA (Fig. 3A) and p16 protein levels (Fig. 3B, lanes 3, 6 and 7) in H719 cells, however, this was the minimal effective concentration as we detected only negligible CDKN2A mRNA levels and p16 protein when 15 μM zebularine was used (Fig. 3A and B). To determine the effect of the length of treatment on the induction of p16 protein expression, we treated H719 cells with 30 μM zebularine for 5, 10 and 15 days. Immunoblot analysis detected p16 protein levels in the H719 cells after 10 days of zebularine treatment that remained constant through day 15 of treatment (Fig. 3B, lanes 6 and 7), while exposure for only 5 days did not result in detectable expression of p16 protein in these cells (Fig. 3B, lane 5). These data suggest that a continuous prolonged exposure to zebularine is required for re-induction of a silenced CDKN2A gene.
Figure 3.
Zebularine treatment can reactivate silenced p16 mRNA and protein levels. (A) Real-time RT-PCR analysis of CDKN2A mRNA isolated from zebularine-treated H719 cells. (B) Immunoblot analysis of p16 expression in zebularine treated H719 cells; (15–30 μM) for 10 days (lane 2 and 3) and 30 μM for 5–15 days (lanes 5–7). p16 protein and tubulin are indicated by arrows.
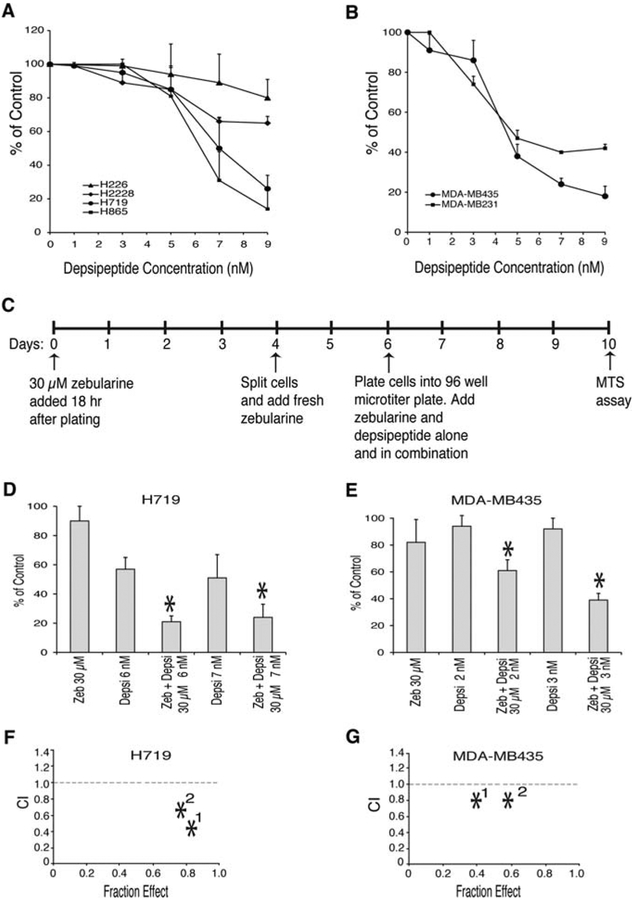
To determine whether depsipeptide could enhance the ability of 30 μM zebularine treatment to inhibit cell growth in tumor cells with a silenced CDKN2A gene, we first used a dose titration to establish the concentration of depsipeptide that would not inhibit cell growth after 4 days of treatment. Using the MTS assay, we observed a dose-dependent cell killing with depsipeptide in the hypermethylated tumor lines, H719 and H865 with a IC50 of 7.5 nM and 6.5 nM respectively and in the CDKN2A deleted tumor cell lines H226 and H2228 with with IC50 > 12 nM (Fig. 4A and data not shown). The H719 and H865 cells showed a steep decrease of cell growth within a narrow range of 5 to 7 nM (Fig. 4A). In addition, we observed a dose-dependent effect of depsi-peptide on growth inhibition of two breast cancer cells (MDA-MB435 and MDA-MB231) with IC50 values = 4.5 and 5.0 nM, respectively (Fig. 4B).
Figure 4.
Enhanced growth inhibition by combined zebularine and depsipeptide treatment in lung and breast cancer cells with hypermethylated CDKN2A. (A) and (B) Effect of depsipeptide on growth of lung and breast cancer cells. (A) Lung cancer (H226, H2228, H865 and H719) and (B) breast cancer (MDAMB231, MDA-MB435) cell lines were treated with the indicated concentrations of depsipeptide as described in Materials and methods. Inhibition of cell growth measured by MTS assay was determined as compared to untreated cells. Data shown represent the mean ± SD of 3–5 experiments. (C) Schematic representation of continuous zebularine and depsipeptide treatment. (D) H719 and (E) MDA-MB435 cells were treated with zebularine (Zeb) alone, depsipeptide (Depsi) alone, or a combination at the concentrations indicated as described in Materials and methods. Inhibition of cell growth is presented as comparison to the untreated controls. (F) CalcuSyn analysis of drug combination in H719 cells showing synergistic drug interaction where no. 1 corresponds to 30 μM zeb and 6 nM depsipeptide and no. 2 corresponds to 30 μM zebularine and 7 nM depsipeptide (G) CalcuSyn analysis of drug combination treatment in MDA-MB435 cells showing additive effect where no. 1 corresponds to 30 μM zebularine and 2 nM depsipeptide and no. 2 corresponds to 30 μM zebularine and 3 nM depsipeptide.
To investigate the effect of 30 μM zebularine in combination with depsipeptide we examined whether 30 μM zebularine combined with depsipeptide at 3, 5, 6 and 7 nM would synergistically or additively inhibit cell growth in lung cancer cells. Since a prolonged zebularine exposure was required to reactivate CDKN2A expression (Fig. 3), we pretreated cells with 30 μM zebularine for 6 days before depsipeptide was added for an additional 4 days of treatment (Fig. 4C). Control cells were treated with either zebularine alone for 10 days, depsipeptide alone for 4 days, or left untreated. We found that in H719 cells with silenced CDKN2A gene, the combination of either 6 or 7 nM depsipeptide with 30 μM zebularine resulted in a synergistic inhibition of cell growth (Fig. 4D) while concentrations less or equal to 5 nM depsipeptide combined with 30 μM zebularine showed a negligible effect on cell growth inhibition (data not shown). For the combination of either 6 and 7 nM depsipeptide with 30 μM zebularine treatment the percent of tumor cell growth adjusted to untreated control cells was 21±5 and 24±9, respectively, which is significantly different from the percent of tumor cell growth for zebularine alone (90±10) and either 6 and 7 nM depsipeptide treatment alone (57±10 and 51±16) with a p<0.001. CalcuSyn analysis confirmed a synergistic effect on the inhibition of growth of lung cancer H719 cells treated with a combination of 30 μM zebularine and either 6 or 7 nM depsipeptide, with CI of 0.70 and 0.57, respectively (Fig. 4F).
To investigate the effect of 30 μM zebularine in combination with depsipeptide on the growth of MDA-MB435 cells, we used 2 and 3 nM depsipeptide to test whether concentrations which are not cytotoxic to these cells are sufficient to inhibit cell growth (Fig. 4E). While the percent cell growth inhibition for 30 μM zebularine alone or 3 nM depsipeptide alone was 82±17 and 92±8, respectively, the combination of 30 μM zebularine with 3 nM depsipeptide was 38.5±5, p<0.001 (Fig. 4E). Using 2 nM depsipeptide alone, and zebularine combined with 2 nM depsipeptide, the percent growth inhibition of the combination therapy was reduced as compared to inhibition with the 3 nM depsipeptide combination, but still statistically significant, p<0.05 (Fig. 4E). Using CalcuSyn software (BioSoft) we found that combination of zebularine with either 2 or 3 nM of depsipeptide yielded an additive growth inhibitory effect with CI of 0.93 and 0.92, respectively (Fig. 4G).
Since zebularine alone did not affect the growth of H226, H1264 and MDA-MB231 cells with a deleted CDKN2A gene, we asked whether the combination of zebularine with depsipeptide would have any growth inhibitory effect on these cells. We treated the H226, H1264 and MDA-MB231 cells with 30 μM zebularine in combination with a range of 3 to 14 nM depsipeptide. While depsipeptide enhanced the effect of 30 μM zebularine in H719 and MDA-MB435 cells, we did not detect an increase in growth inhibition of H226, H1264 and MDA-MB231 cells, as compared to either drug treatment alone (Fig. 5A–C). Since the IC50 for H226 cells treated with depsipeptide is higher than for H719 cells (14 vs 7.5 nM), we also investigated the effect of zebularine on H226 and H1264 cells combined with 14 nM depsipeptide and again did not observe any enhancement of growth inhibition as compared to either drug alone (Fig. 5A and C).
Figure 5.

Lack of enhanced growth inhibition by combined zebularine and depsipeptide treatment in lung and breast cancer cells with deleted CDKN2A gene. (A) H226 and (B) H1264 lung cancer cell lines and (C) MDA-MB231 breast cancer cell line were treated with zebularine (Zeb) alone, depsipeptide (Depsi) alone, or a combination at the indicated concentrations. The amount of cell growth was measured by MTS assay and the growth inhibition of treated cells is presented as a percentage of surviving cells as compared to untreated controls.
To compare the effect of zebularine with another demethylating agent, DAC, we tested 3 nM depsipeptide in combination with either 0.5 or 1 μM DAC and measured growth inhibition of tumor cells by MTS assay after 72 h of drug treatment. To confirm the previously published synergy of 50 nM depsipeptide combined with 1 μM DAC (3,42), we also treated H719 cells with 1 μM DAC for 72 h followed by a 6 h incubation with depsipeptide. We observed a significant enhancement of growth inhibition by DAC in combination with either 3 or 50 nM depsipeptide as compared to either drug alone, (p<0.001) (Fig. 6A). We also observed that the lower 0.5 μM DAC concentration in combination with 3 nM depsipeptide was sufficient to detect enhancement of growth inhibition as compared to either drug alone (Fig. 6B) (p<0.001). CalcuSyn analysis of H719 cells treated with 0.5 or 1 μM DAC in combination with 3 nM depsipeptide indicated a strong synergistic growth inhibitory effect with CIs of 0.31 and 0.22, respectively (Fig. 6C).
Figure 6.
Enhanced growth inhibition with combined DAC and depsipeptide in cells with hypermethylated but not deleted CDKN2A. (A) DAC (1 μM) combined with 50 or 3 nM depsipeptide (Depsi) enhanced inhibition of cell growth in hypermethylated H719 cells. (B) DAC (0.5 μM) combined with 3 nM depsipeptide enhanced inhibition of cell growth in H719 cells. (C) CalcuSyn analysis of drug combination in H719 cells showing synergistic drug interaction where no. 1 corresponds to 0.5 μM DAC and 3 nM depsipeptide and no. 2 corresponds to 1 μM DAC and 3 nM depsipeptide. (D) Lack of enhanced growth inhibition in H226 cells with deleted CDKN2A with indicated concentrations of DAC and depsipeptide in combination. (E) Lack of enhanced growth inhibition with 7, 14 or 50 nM depsipeptide combined with 1 μM DAC in CDKN2A deleted H460 cells.
Since treatment of the H226 cells (deleted CDKN2A), with combined depsipeptide and zebularine did not show enhanced tumor growth inhibition (Fig. 5A), we investigated whether these cells would also show resistance to the combination of depsipeptide and DAC. Treatment with 0.5 or 1 μM DAC in combination with 3 nM depsipeptide or treatment with 1 μM DAC in combination with 50 nM depsipeptide did not show an enhanced effect on cell growth inhibition (Fig. 6D). It was previously shown, however, that treatment with 0.1 μM DAC for 3 days followed by a 6-h incubation with 50 nM depsipeptide of lung cancer cell line H460, with a deleted CDKN2A, resulted in synergistic inhibition of cell growth as compared to treatment with either drug alone (43). Since we observed that cells with deleted CDKN2A do not respond to DAC/depsipeptide treatment (Fig. 5), we treated the H460 cells with 1 μM DAC for 3 days and 50 nM depsipeptide for 6 h as previously described; and we did not observe any inhibition of cell growth (Fig. 6E). In addition, we treated the H460 cells similarly to the H226 cells (as described in Materials and methods) and as expected, no inhibition of cell growth was observed (Fig. 6E).
Discussion
Zebularine, an inhibitor of DNA methyltransferase, has demonstrated activity in vitro and in vivo in several distinct cancer cell types (24,44,45). Although inhibition of cell growth in vitro and tumor growth in mouse xenograft models following zebularine treatment was accompanied by re-induction of CDKN2A gene expression (21,23), this agent is predicted to effect global genome hypomethylation (47). To address if CDKN2A re-expression is an important biomarker for response to zebularine, we set out to determine whether tumor cells with a silenced CDKN2A gene would respond preferentially to zebularine treatment as compared to tumor cells lacking the CDKN2A gene due to homozygous gene deletion. We demonstrated that all 7 non-small cell lung cancer (NSCLC) tumor lines with a deleted CDK2A gene showed no or minimal inhibition of cell growth upon zebularine treatment, while 11 out of 12 NSCLC tumor lines (91.6%) with silenced CDKN2A gene were sensitive to zebularine treatment. These results suggests that re-induction of CDKN2A gene by demethylating agents may be an important bio-marker to predict response of cells to zebularine treatment. CDKN2A, however, is not the only gene that can be regulated by zebularine treatment. For example, microarray analysis identified approximately 20 genes that can be differentially regulated in bladder, colon and pancreatic cancer cell lines(24). Thus, it remains to be established whether other gene products may cooperate with the CDNK2A/p16 to exert cytotoxic effect on lung cancer cells following zebularine treatment.
Prior studies using zebularine at 100 and 500 μM concentration showed consistent re-induction of CDKN2A mRNA in vitro (21), however, subsequent animal studies demonstrated that 30 μM was the limit for achievable concentrations of zebularine in mouse plasma (46 and unpulished observation). In this study we show that 30 μM zebularine was sufficient to re-induce both silenced CDKN2A mRNA and the encoded p16 protein. However, since 30 μM of zebularine was only minimally cytotoxic to cells in vitro, we combined zebularine with the HDAC inhibitor, depsipeptide, and tested whether this combination could enhance inhibition of tumor cell growth as compared to either drug alone. In this study we demonstrated enhanced inhibition of tumor cell growth with combined zebularine and depsipeptide treatment in both lung cancer and breast cancer cell lines with hypermethylated and silenced CDKN2A. In addition, we demonstrated that combination treatment enhanced the inhibition of tumor cell growth in cells with silenced CDKN2A (H719 and MDMB435), while combining zebularine or DAC with depsipeptide had no effect on the inhibition of cell growth in cell lines (H226, H1264 and MD231) with a deleted CDKN2A gene.
One exception to this model was the previous report that H460 lung cancer cells, that carried a CDKN2A gene deletion, showed inhibition of cell growth with the combination of DAC and depsipeptide treatment (43). In our experiments, however, treatment of these cells with combined DAC and depsipeptide or with zebularine alone did not inhibit H460 cell growth. In addition, a 10-fold higher concentration of DAC (1 μM) that was previously reported did not result in cell growth inhibition consistent with our data on other tumor cell lines with deleted CDKN2A.
Zebularine has been tested for safety and pharmacokinetics in mice, rats and primates (44). Although repetitive administration at high dose in primates resulted in toxicity in an NCI, DTP study (unpublished observation), recent animal studies suggest that zebularine works best after prolonged administration at lower doses, capable of reaching a minimal plasma concentration after continuous oral administration in drinking water (47). Depsipeptide, however, has been extensively tested in patients at micromolar concentrations and we estimated that the dose of 17.8 mg/m2 of depsipeptide administered to patients (34,35) is equal to 2.1 μM concentration of which is approximately 1000-fold higher than the nanomolar concentration of depsipeptide required in our study to achieve a synergistic effect on cell growth inhibition when combined with either zebularine or DAC. Our data suggest that combination of DNA methyltransferase and HDAC inhibitors may be a potential treatment for lung cancer patients with silenced CDKN2A status, but selection of patients may be needed to optimize the benefit of this regimen. For example, a recent phase II trial in lung cancer using 5-azacitidine and an experimental HDAC-inhibitor, entinostat (SNDX-275), observed a low response rate (<10%) in non-small cell lung cancer, however, two patients experienced a drastic tumor reduction, including one complete remission (Juergens et al, ASCO, Abst. 8055, 2009). Our data strongly support the importance of prospective pre-selection of patients in future clinical trials and suggests CDKN2A status as a key biomarker for demethylation/HDAC inhibition studies.
Acknowledgments
We thank Carmen Allegra for critical reading of this manuscript and CTEP, DCTD, NCI for providing depsipeptide and zebularine. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute and by Department of Medicine and Department of Anatomy and Cell Biology of University of Florida, Gainesville, Florida.
References
- 1.Baylin SB: DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2 (Suppl. 1): S4–S11, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA and Baylin SB: The epigenomics of cancer. Cell 128: 683–692, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otterson GA, Khleif SN, Chen W, Coxon AB and Kaye FJ: CDKN2 gene silencing in lung cancer by DNA hypermethylation and kinetics of p16INK4 protein induction by 5-aza 2’deoxycytidine. Oncogene 11: 1211–1216, 1995. [PubMed] [Google Scholar]
- 4.Yoo CB and Jones PA: Epigenetic therapy of cancer: Past, present and future. Nat Rev Drug Discov 5: 37–50, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Momparler RL and Derse D: Kinetics of phosphorylation of 5-aza-2’-deoxycytidine by deoxycytidine kinase. Biochem Pharmacol 28: 1443–1444, 1979. [DOI] [PubMed] [Google Scholar]
- 6.Juttermann R, Li E and Jaenisch R: Toxicity of 5-aza-2’-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA 91: 11797–11801, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santi DV, Norment A and Garrett CE: Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA 81: 6993–6997, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graff JR, Gabrielson E, Fujii H, Baylin SB and Herman JG: Methylation patterns of the E-cadherin 5’ CPG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem 275: 2727–2732, 2000. [DOI] [PubMed] [Google Scholar]
- 9.Herman JG, Umar A, Polyak K, et al. : Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95: 6870–6875, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano S, Miller CW, Gombart AF, et al. : Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood 94: 1113–1120, 1999. [PubMed] [Google Scholar]
- 11.Wong IH, Ng MH, Huang DP and Lee JC: Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: Potential prognostic implications. Blood 95: 1942–1949, 2000. [PubMed] [Google Scholar]
- 12.Herman JG, Latif F, Weng Y, et al. : Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA 91: 9700–9704, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Issa JP, Garcia-Manero G, Giles FJ, et al. : Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103: 1635–1640, 2004. [DOI] [PubMed] [Google Scholar]
- 14.Momparler RL and Ayoub J: Potential of 5-aza-2’-deoxycytidine (decitabine) a potent inhibitor of DNA methylation for therapy of advanced non-small cell lung cancer. Lung Cancer 34 (Suppl. 4): S111–S115, 2001. [DOI] [PubMed] [Google Scholar]
- 15.Momparler RL, Rivard GE and Gyger M: Clinical trial on 5-aza-2’-deoxycytidine in patients with acute leukemia. Pharmacol Ther 30: 277–286, 1985. [DOI] [PubMed] [Google Scholar]
- 16.Schrump DS, Fischette MR, Nguyen DM, et al. : Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res 12: 5777–5785, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Laliberte J, Marquez VE and Momparler RL: Potent inhibitors for the deamination of cytosine arabinoside and 5-aza-2’-deoxycytidine by human cytidine deaminase. Cancer Chemother Pharmacol 30: 7–11, 1992. [DOI] [PubMed] [Google Scholar]
- 18.Stresemann C and Lyko F: Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer 123: 8–13, 2008. [DOI] [PubMed] [Google Scholar]
- 19.Kim CH, Marquez VE, Mao DT, Haines DR and McCormack JJ: Synthesis of pyrimidin-2-one nucleosides as acid-stable inhibitors of cytidine deaminase. J Med Chem 29: 1374–1380, 1986. [DOI] [PubMed] [Google Scholar]
- 20.Hurd PJ, Whitmarsh AJ, Baldwin GS, et al. : Mechanism-based inhibition of C5-cytosine DNA methyltransferases by 2-h pyrimidinone. J Mol Biol 286: 389–401, 1999. [DOI] [PubMed] [Google Scholar]
- 21.Cheng JC, Weisenberger DJ, Gonzales FA, et al. : Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol 24: 1270–1278, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquez VE, Kelley JA, Agbaria R, et al. : Zebularine: A unique molecule for an epigenetically based strategy in cancer chemotherapy. Ann NY Acad Sci 1058: 246–254, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Cheng JC, Matsen CB, Gonzales FA, et al. : Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst 95: 399–409, 2003. [DOI] [PubMed] [Google Scholar]
- 24.Cheng JC, Yoo CB, Weisenberger DJ, et al. : Preferential response of cancer cells to zebularine. Cancer Cell 6: 151–158, 2004. [DOI] [PubMed] [Google Scholar]
- 25.Cameron EE, Bachman KE, Myohanen S, Herman JG and Baylin SB: Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Primeau M, Gagnon J and Momparler RL: Synergistic antineoplastic action of DNA methylation inhibitor 5-aza-2’-deoxycytidine and histone deacetylase inhibitor depsipeptide on human breast carcinoma cells. Int J Cancer 103: 177–184, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Wu LP, Wang X, Li L, et al. : Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CPG and H3K9 methylation on the promoter. Mol Cell Biol 28: 3219–3235, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandor V, Senderowicz A, Mertins S, et al. : P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer 83: 817–825, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates SE and Piekarz RL: Histone deacetylase inhibitors in combinations: Will the preclinical promises be kept? Cancer J 13: 80–83, 2007. [DOI] [PubMed] [Google Scholar]
- 30.Zhu WG, Lakshmanan RR, Beal MD and Otterson GA: DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res 61: 1327–1333, 2001. [PubMed] [Google Scholar]
- 31.Yu XD, Wang SY, Chen GA, et al. : Apoptosis induced by depsipeptide FK228 coincides with inhibition of survival signaling in lung cancer cells. Cancer J 13: 105–113, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Tan J, Zhuang L, Jiang X, Liu ET and Yu Q: Inhibitors of histone deacetylases target the RB-E2F1 pathway for apoptosis induction through activation of proapoptotic protein bim. Proc Natl Acad Sci USA 102: 16090–16095, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu WG, Dai Z, Ding H, et al. : Increased expression of unmethylated CDKN2D by 5-aza-2’-deoxycytidine in human lung cancer cells. Oncogene 20: 7787–7796, 2001. [DOI] [PubMed] [Google Scholar]
- 34.Klimek VM, Fircanis S, Maslak P, et al. : Tolerability, pharmacodynamics, and pharmacokinetics studies of depsipeptide (romidepsin) in patients with acute myelogenous leukemia or advanced myelodysplastic syndromes. Clin Cancer Res 14: 826–832, 2008. [DOI] [PubMed] [Google Scholar]
- 35.Sandor V, Bakke S, Robey RW, et al. : Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res 8: 718–728, 2002. [PubMed] [Google Scholar]
- 36.Rahman L, Voeller D, Rahman M, et al. : Thymidylate synthase as an oncogene: A novel role for an essential DNA synthesis enzyme. Cancer Cell 5: 341–351, 2004. [DOI] [PubMed] [Google Scholar]
- 37.Alexandrova N, Niklinski J, Bliskovsky V, et al. : The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol 15: 5188–5195, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niklinski J, Claassen G, Meyers C, et al. : Disruption of Myctubulin interaction by hyperphosphorylation of c-Myc during mitosis or by constitutive hyperphosphorylation of mutant c-Myc in Burkitt’s lymphoma. Mol Cell Biol 20: 5276–5284, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou TC and Talalay P: Analysis of combined drug effects - a new look at a very old problem. Trends Pharmacol Sci 4: 450–454, 1983. [Google Scholar]
- 40.Chou TC and Talaly P: A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J Biol Chem 252: 6438–6442, 1977. [PubMed] [Google Scholar]
- 41.Otterson GA, Kratzke RA, Coxon A, Kim YW and Kaye FJ: Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wild-type RB. Oncogene 9: 3375–3378, 1994. [PubMed] [Google Scholar]
- 42.Yu X, Guo ZS, Marcu MG, et al. : Modulation of p53, Erbb1, Erbb2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst 94: 504–513, 2002. [DOI] [PubMed] [Google Scholar]
- 43.Steiner FA, Hong JA, Fischette MR, et al. : Sequential 5-aza 2’-deoxycytidine/depsipeptide FK228 treatment induces tissue factor pathway inhibitor 2 (TFPI-2) expression in cancer cells. Oncogene 24: 2386–2397, 2005. [DOI] [PubMed] [Google Scholar]
- 44.Herranz M, Martin-Caballero J, Fraga MF, et al. : The novel DNA methylation inhibitor zebularine is effective against the development of murine T-cell lymphoma. Blood 107: 1174–1177, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Scott SA, Lakshimikuttysamma A, Sheridan DP, Sanche SE, Geyer CR and DeCoteau JF: Zebularine inhibits human acute myeloid leukemia cell growth in vitro in association with p15INK4B demethylation and reexpression. Exp Hematol 35: 263–273, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Holleran JL, Parise RA, Joseph E, et al. : Plasma pharmacokinetics, oral bioavailability, and interspecies scaling of the DNA methyltransferase inhibitor, zebularine. Clin Cancer Res 11: 3862–3868, 2005. [DOI] [PubMed] [Google Scholar]
- 47.Yoo CB, Chuang JC, Byun HM, et al. : Long-term epigenetic therapy with oral zebularine has minimal side-effects and prevents intestinal tumors in mice. Cancer Prev Res 1: 233–240, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]