SUMMARY
Heterochromatic repetitive satellite RNAs are extensively transcribed in a variety of human cancers, including BRCA1 mutant breast cancer. Aberrant expression of satellite RNAs in cultured cells induces the DNA damage response, activates cell cycle checkpoints, and causes defects in chromosome segregation. However, the mechanism by which satellite RNA expression leads to genomic instability is not well understood. Here we provide evidence that increased levels of satellite RNAs in mammary glands induce tumor formation in mice. Using mass spectrometry, we further show that genomic instability induced by satellite RNAs occurs through interactions with BRCA1-associated protein networks required for the stabilization of DNA replication forks. Additionally, de-stabilized replication forks likely promote the formation of RNA-DNA hybrids in cells expressing satellite RNAs. These studies lay the foundation for developing novel therapeutic strategies that block the effects of non-coding satellite RNAs in cancer cells.
In Brief
Heterochromatin-encoded satellite RNAs are often aberrantly expressed in human cancers. Zhu et al. show that these RNAs can promote breast cancer formation and that they bind BRCA1 and associated proteins that are important for replication fork stability and genomic instability.
Graphical Abstract
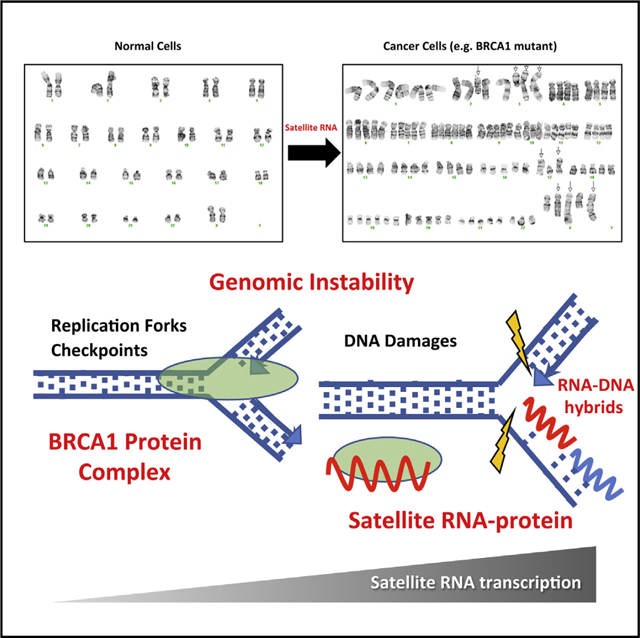
INTRODUCTION
Mutations in the tumor suppressor BRCA1 (breast cancer gene 1) represent the highest risk factor for developing hereditary forms of breast and ovarian cancer (King et al., 2003). In addition, data from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Network, 2012) reveal that BRCA1 is often mutated in sporadic forms of breast cancer as well (Nik-Zainal et al., 2016). Recent studies have demonstrated novel roles for BRCA1 in interstrand cross-link repair, stabilization of stalled DNA replication forks, regulation of homologous recombination, and RNA-DNA hybrid formation (R-loop) (Bunting et al., 2012; Hatchi et al., 2015; Schlacher et al., 2012; Willis et al., 2014). Our previous work indicates that one way in which BRCA1 functions as a tumor suppressor is by maintaining heterochromatin integrity and silencing the expression of non-coding pericentromeric satellite RNAs (Zhu et al., 2011).
Transcriptome and bioinformatic analyses have indicated that a large portion of the genome is transcribed into RNAs that do not encode proteins (i.e., non-coding RNAs) (Bertone et al., 2004; Birney et al., 2007). Mounting evidence suggests that these non-coding sequences play important roles in development and disease (Esteller, 2011). Studies during the past decade have identified a growing number of non-coding RNAs (microRNAs and long non-coding RNAs [lncRNAs] in particular) that drive tumorigenesis in humans (Morris and Mattick, 2014). Mouse models have been used to characterize the role of non-coding RNAs in cancer, revealing novel mechanisms of tumorigenesis. One major mechanism through which non-coding RNAs affect tumorigenesis is by binding to proteins required for maintaining genomic stability, such as chromatin modifiers and DNA damage response factors (Rinn and Chang, 2012). Satellite RNAs are a heterogeneous population of non-coding RNAs (20–1,500 nt in length) that are transcribed from satellite DNA, which includes large swaths of repetitive sequences at the centromere and telomeres of a variety of eukaryotic chromosomes (Chan and Wong, 2012; John and Miklos, 1979; Miklos and John, 1979; Probst and Almouzni, 2011). In a landmark study carried out in fission yeast, Volpe et al. (2002) demonstrated that the transcription of satellite RNAs is vital for the establishment and maintenance of pericentromeric heterochromatin. In the human genome, centromeric satellite DNAs are largely alpha satellite repeats that are 171 base pairs in length and arranged in tandem arrays that stretch for multiple megabases with minimal variations among chromosomal assigned arrays. There are also several satellite repeats in the pericentromeric heterochromatin, such as HSAT2 and HSAT3. In the mouse genome, repeat sequences are nearly, if not completely, identical between chromosomes, and only two sequence variants exist; namely, the major and minor satellite (Guenatri et al., 2004; Hayden and Willard, 2012). Another distinguishing feature of these satellite regions is that, under normal homeostatic conditions, they are rarely transcribed in adult mammalian organisms. Rather, expression of satellite repeats seems to be limited to embryonic tissues and embryonic stem cells (Probst and Almouzni, 2008). However, it has been shown that, in a variety of human cancers, including BRCA1 mutant breast cancer, these satellite DNAs are transcribed into non-coding RNAs (Ting et al., 2011; Zhu et al., 2011). In tumors derived from mouse models of pancreatic, colon, and lung cancers, satellite transcripts represent up to ~50% of total cellular RNA (Ting et al., 2011).
We have found that satellite RNAs are highly expressed in mouse and human breast tumors lacking BRCA1 (Zhu et al., 2011; Figure S1). Although the mechanism by which satellite DNAs are transcriptionally de-repressed is not well established, we have analyzed mammary epithelial cells and shown that BRCA1 monoubiquitinates histones associated with satellite DNA, preventing transcription of these regions (Zhu et al., 2011). Here we report that expression of satellite RNA transcripts is sufficient to activate DNA damage response pathways and to induce aneuploidy. Mice expressing satellite RNAs in the mammary glands are susceptible to tumor development. We further show that genomic instability induced by satellite RNAs occurs through interactions with BRCA1-associated protein networks that lead to destabilization of DNA replication forks, formation of RNA-DNA hybrids, and DNA damage.
RESULTS
Expression of Satellite RNA Directly Induces the DNA Damage Response and Chromosome Instability
To investigate whether it is the de novo synthesis of satellite RNA or the recruitment of satellite RNA to chromatin that is required to induce the DNA damage response, we injected in vitro-transcribed satellite RNA, which contains eight copies of the 171-base pair (bp) human satellite RNA (hSATa; see the sequence in the STAR Methods), into the nuclei of living primary human fibroblast cells and monitored H2AX phosphorylation (γH2AX), a marker for DNA damage. We chose to evaluate the effect of the hSATa sequence because our transcriptome data showed that the alpha satellite is the largest detectable expressed satellite and appears to be enriched in BRCA1-defcient tumors (Figure S1). All other sub-satellite RNA families have no consistent or preferential expression in BRCA1 mutant tumors (Figure S1). Elevated levels of γH2AX were observed 7 hr after the nuclear injection of satellite RNA compared with cells injected with a control non-polyadenylated red fluorescent protein (RFP) RNA (Figure 1A), indicating that the process of transcribing satellite RNA is not required to activate the DNA damage response; rather, the presence of elevated levels of satellite RNAs plays the critical role. We also transfected in vitro-transcribed satellite RNAs into primary human fibroblasts or human osteosarcoma U2OS cells and found that 12% of the transfected cells were positive for γH2AX 16 hr following transfection (Figure 1B). By comparison, less than 1% of cells transfected with a control polyadenylated GFP RNA, a satellite RNA from fission yeast (Grewal and Klar, 1997), or synthetic RNA consisting of a scrambled human satellite RNA sequence were γH2AX-positive (Figure 1C). Quantification by RNA sequencing (RNA-seq) analysis showed that the levels of satellite RNA transcripts represent approximately 10%–20% of the total cellular transcripts (Table S1), which is consistent with a previous report showing that pericentromeric satellite transcripts account for a mean of 12% (range, 1%–50%) of all cellular transcripts (Ting et al., 2011). In human breast tumors, as previously shown, in addition to overexpressed alpha satellite RNAs, the CFXr gamma-satellite RNAs, also induced the accumulation of γH2AX to the same degree under similar conditions (Zhu et al., 2011). Satellite RNA from pericentromeric major satellite DNAs in the mouse also induced DNA damage but was slightly less effective than RNA derived from human satellite DNAs. We further verified differences in the levels of γH2AX induction by western blotting of transfected cells (Figure 1D). In parallel, we found that the levels of another DNA damage marker, pS345 Chk1, also increased upon satellite RNA transfection (as quantified by ImageJ; Figure S2A). As a control, we found that another histone modification marker, histone H3 lysine 9 trimethylation (H3K9me3), was not affected by satellite RNA transfection (Figures 1C and 1D). To investigate whether the accumulation of γH2AX induced by satellite RNA requires localization of the satellite RNAs to the homologous DNA sequences, we performed co-staining of γH2AX with a centromere marker, anti-centromere antibody (ACA; Figure S2B), in human breast cancer cells (MCF7). These experiments showed that less than 6% of γH2AX induced by satellite RNAs localizes to the centromeric regions, indicating that the DNA damage responses induced by satellite RNAs occur beyond genomic regions with homologous DNA sequences. Moreover, we found that satellite RNAs also induced increased levels of replication protein A (RPA) but not 53BP1 staining (Figures S2D and S2E). Taken together, these data strongly suggest that high levels of satellite RNA directly induce the DNA damage response.
Figure 1. Satellite RNA Is Sufficient to Induce a DNA Damage Response.
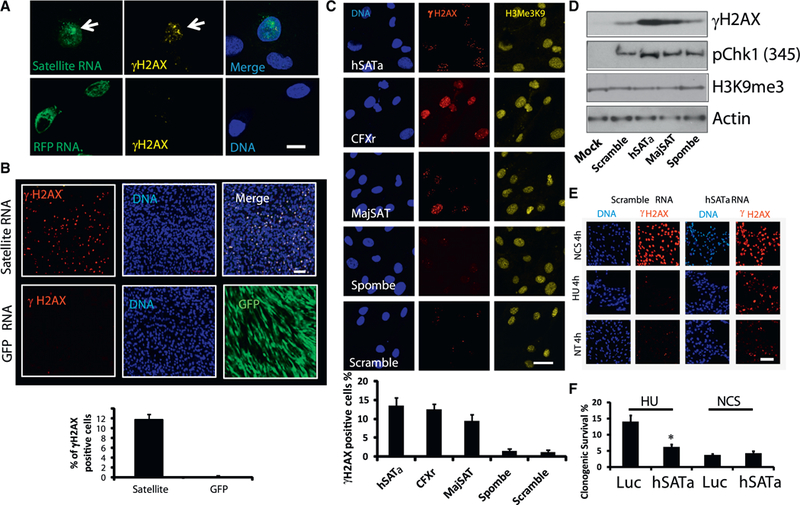
(A) Microinjection of in vitro-synthesized satellite RNA into primary human fibroblasts induces the accumulation of γH2AX. Seven hr after injection of fluorescein isothiocyanate (FITC)-labeled satellite RNA hSATa (top) or RFP RNA (bottom), the cells were fixed, immunostained, and imaged. Green, RNA; yellow, γH2AX; blue, DAPI. Scale bar, 10 γm.
(B) Transfection of in vitro-synthesized satellite RNA into primary human fibroblasts induces accumulation of γH2AX. Sixteen hr after transfection of satellite RNA hSATa (top) or GFP RNA (bottom), the cells were fixed, immunostained, and imaged. Green, GFP; red, γH2AX. Scale bar, 50 μm.
(C) The effect of satellite RNAs with different sequences on γH2AX accumulation. Transfected RNAs are indicated. Scale bar, 10 μm.
(D) Western blot analyses of protein lysates after RNA transfection.
(E and F) Satellite RNA-overexpressing cells are more sensitive to DNA replication stress (HU treatment) than to DNA DSBs (NCS treatment).
(E) Immunofluorescence staining of satellite RNA-transfected human osteosarcoma cells (U2OS). Red, γH2AX. Scale bar, 50 μm.
(F) Clonogenic survival assay of U2OS cells infected with lentiviruses expressing either satellite RNAs or a control RNA. Cells were treated with 3 mM HU or 1 μg/mL NCS for 24 hr and then grown into colonies for 8 to 10 days. Clonogenic survival is shown as the percentage of respective untreated cells. Values represent the mean of three independent experiments, and error bars represent the SEM. The asterisk indicates the significance level in two-tailed t tests (*p < 0.01).
See also Figures S1–S3 and Table S1.
We next asked whether the elevated levels of γH2AX induced by satellite RNAs resulted from cellular responses to DNA double-strand breaks (DSBs) or stalled DNA replication. We therefore transfected U2OS cells with either satellite or scrambled RNA and then treated them with neocarzinostatin (NCS), which induces DSBs, or hydroxyurea (HU), which causes DNA replication stress. Cells transfected with satellite or scrambled RNA showed similar levels of γH2AX-staining in response to 4 hr of NCS treatment (Figure 1E), suggesting that DSBs do not drive the DNA damage response caused by satellite RNA. In contrast, HU treatment resulted in higher levels of γH2AX in cells transfected with satellite RNA compared with cells transfected with control scrambled RNA. It thus appears that the DNA damage response induced by satellite RNAs is mediated by DNA replication stress. To examine the effect of drug treatment on the survival of satellite RNA-expressing cells, we performed clonogenic survival assays (Figure 1F). Following HU treatment, far fewer satellite RNA-expressing cells survived than those overexpressing a control luciferase (Luc) RNA (a 300-nt fragment of Luc RNA). Cellular proliferation assays further showed that satellite RNA-expressing cells were more sensitive to not only HU but also aphidicolin (another DNA replication stress reagent) (Figure S2C). These data indicate that cells overexpressing satellite RNA are more sensitive to DNA replication stress than to DNA DSBs.
To test whether the replication stress induced by satellite RNAs led to genome instability, we next analyzed the karyotype of primary human mammary epithelial cells (HMECs) infected with lentiviruses that expressed satellite RNA and a short hairpin RNA (shRNA) that downregulated the tumor suppressor p53 because cell death of satellite RNA-expressing cells was p53-dependent. Luc RNA control cells with p53 depletion demonstrated some abnormalities associated with chromosome numbers; however, cells expressing human or mouse satellite RNA (together with p53 depletion) exhibited many unique chromosomal aberrations and were karyotypically very abnormal (Figure S3A). We conclude that expression of satellite RNAs is sufficient to induce DNA damage and genomic instability.
Activation of Endogenous Satellite RNAs Induces the DNA Damage Response and Chromosome Instability
In previous experiments, we used either transfection or lentivirus vectors to express satellite RNAs. To activate the expression of endogenous satellite RNAs, we took advantage of CRISPR activation (CRISPRa) technology and induced transcription of satellite DNA repeats with guide RNAs that target the pericentromeric (major satellite; MajSAT) and centromeric (minor satellite; MinSAT) regions of the mouse genome (Konermann et al., 2015). This resulted in the elevated expression of both major and minor satellite RNAs, as measured by qRT-PCR experiments (Figure 2A). These cells also displayed mitotic defects (Figure 2B), which is reminiscent of reported mitotic errors because of satellite RNA derepression in vivo (Tasselli et al., 2016), formation of cells with multi- and micro-nuclei (Figure 2C), and γH2AX activation (Figure 2D). When we examined chromosomal integrity by G-banding karyotype analyses, we found that targeted expression of satellite DNAs led to chromosomal breakage (Figure 2E, red arrows), chromosomal radials (Figure 2E, blue box), and translocations (Figure S3B). Formation of chromosomal breaks and radials is a strong indicator of genomic instability. Collectively, these data show that the expression of endogenous satellite RNAs activates DNA damage response pathways and causes chromosome instability as found with transfected satellite RNAs.
Figure 2. CRISPR-Mediated Activation of Satellite RNA Expression Induces Chromosomal Instability.
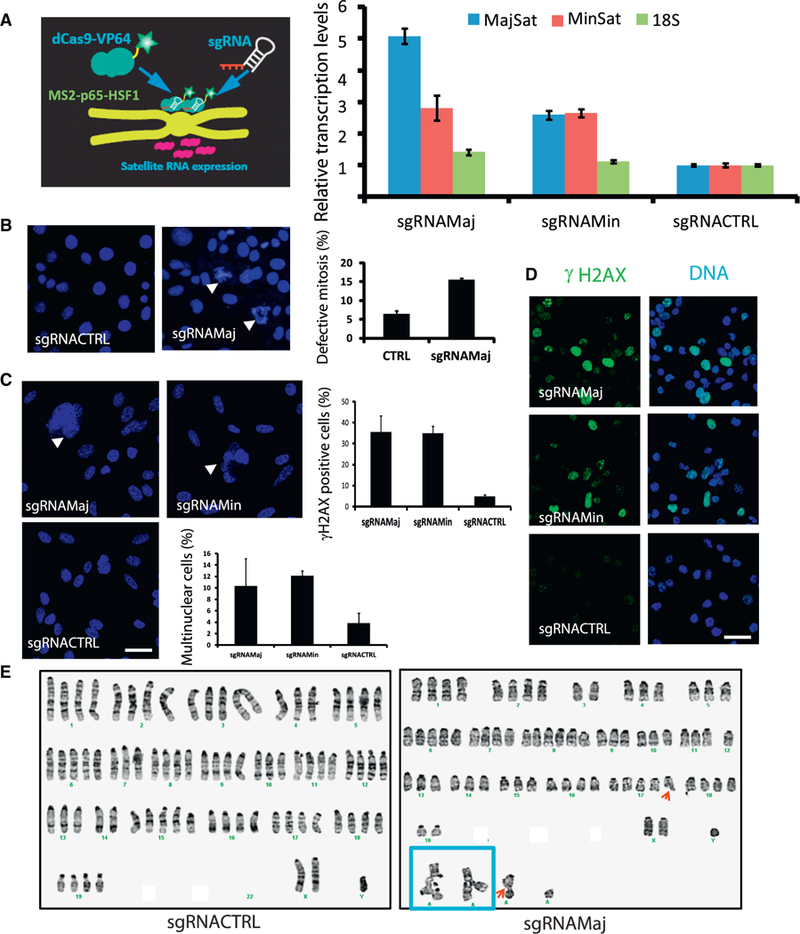
(A) Left: a diagram demonstrating that dCas9 mediates activation of endogenous satellite RNAs. Right: qRT-PCR experiments show that the expression of satellite RNAs is activated in cells expressing guide RNAs targeting mouse satellite RNAs.
(B–D) Activation of satellite RNAs induces(B) mitotic defects, (C) formation of multi-nucleated cells and micronuclei, and (D) γH2AX accumulation, as shown by representative fluorescence images. Bar graphs (left) represent the mean values from three randomly chosen fields of view; more than 50 cells were quantified per field of view. Data shown are representative of 3 independent experiments.
(A–D) Error bars represent the SEM. Mouse breast cancer cell line 67 NR was used in (A)–(D).
(E) chromosomal abnormalities as determined by karyotyping analysis in a mouse lung cancer cell line (Xia et al., 2014). Left: sgRNACTRL. Right: sgRNAMaj. Red arrows, chromosome breaks; blue box, chromosome radial formation.
Scale bars, 10 μm. See also Figure S3.
Induction of Breast Cancer by Satellite RNAs
To determine whether satellite RNAs induce tumor formation in mice, we injected lentiviruses expressing satellite RNAs (either human, hSATa, or mouse major; Zhu et al., 2011) and sh-p53 RNA into mouse mammary glands via ductal injection (Figure 3A). Using RNA-seq analyses, we determined that the levels of satellite RNA transcripts represent approximately 0.01%–0.02% of the total cellular transcripts, whether induced by lentiviral vector transduction or expressed in human BRCA1 mutant breast cancer (HCC1937) cells (Table S1). Five to eleven months following injection, over 60% of the mice (16 of 27) infected with satellite RNA lentiviruses developed a tumor mass at the injection site (Figures 3A and 3B). In contrast, ~7% (1 of 13) of the mice injected with luciferase RNA control (Luc) showed tumors, whereas 75% (3 of 4) of the mice injected with a vector containing oncogenic GTPase HRas (H-RAS) (G12V) developed tumors. Accordingly, mice infected with mouse or human satellite RNA had a decreased tumor-free time compared with controls (Figure 3B). Analysis of H&E-stained sections showed tumor cell masses (Figure 3C). These tumors arose from cells infected with the virus because all tumor cells expressed GFP. Based on transcriptome analysis of these tumors, we found that they were heterogeneous, belonging to basal-like or Her2 positive subtypes (Table S2). We confirmed this finding by immunohistochemistry, assessing levels of cytokeratin 8 and vimentin (Figure 3C). Upon analysis of RNA from these tissue samples by qRT-PCR, we found high levels of satellite RNA expression (Figure 3D). This is the first biological evidence showing that the expression of satellite RNA in the mouse mammary gland causes tumor formation.
Figure 3. Ductal Injection of Lentiviruses Overexpressing Satellite RNA Induces Tumor Formation in the Mammary Gland.
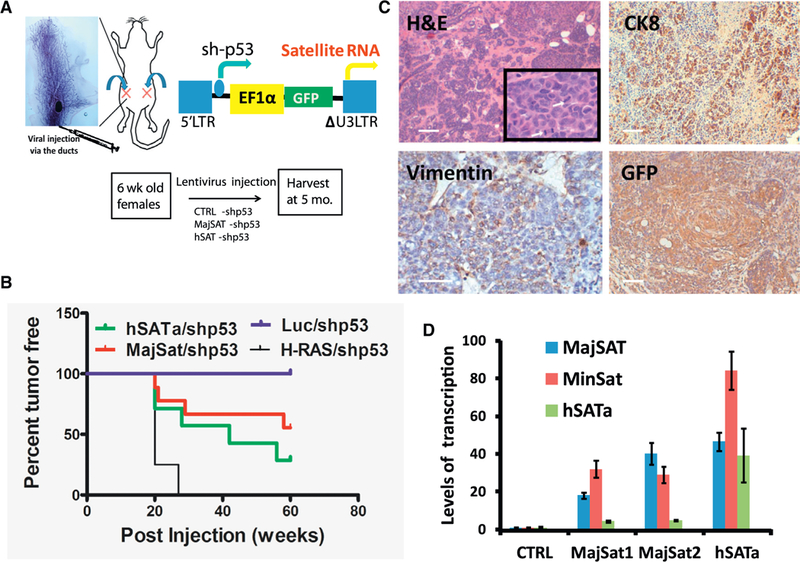
We injected 1 × 106 virus particles in 10 μL of PBS and 0.2% of trypan blue with a Hamilton syringe into the duct of the number 4 mammary gland in a 6-to8-week-old female mouse according to the protocol described previously (Krause et al., 2013).
(A) Top: the ductal injection of lentiviruses into the number 4 mammary glands. Bottom left: the lentiviral vector for overexpressing satellite RNA. Bottom right: representative image of mammary glands or tumors harvested from mice 5 months after the injection.
(B) Kaplan-Meier survival curves in mice injected with lentivirus-overexpressing satellite RNAs or control RNA using a log rank (Mantel-Cox) test. p < 0.0001.
(C) H&E (top) and IHC staining (bottom) of tumor sections. Tumors resulted from the infection of satellite RNA-expressing lentiviruses.
(D) qRT-PCR analysis of tumors from mice injected with satellite RNA-expressing lentiviruses. Error bars represent SEM.
Scale bars, 100 μm. See also Table S2.
Satellite RNAs Bind to a Network of Proteins Associated with BRCA1
We reasoned that the DNA damage response may require recruitment of satellite RNAs and associated proteins to chromatin. Therefore, we set out to identify proteins that bind hSATa. We used a proteomics approach to identify proteins associated with satellite RNA transcripts (Zhou et al., 2002). We constructed a lentivector with doxycycline (Dox)-inducible expression of satellite RNA (eight copies of the 171-bp human satellite RNA as in Figure 1) tagged with the MS2 coat protein binding site (Figure 4A). This vector also expresses the MS2-GFP fusion protein. Because the U2OS cells showed DNA damage response upon satellite RNA expression, we infected U2OS cells and sorted for MS2-GFP-positive cells to ensure maximum satellite RNA expression in all cells. To isolate satellite RNA-binding proteins, we treated sorted U2OS cells with Dox for 24 hr (the time required to reach maximal satellite RNA expression; Figure S4A) before harvesting the cell lysates. To enrich for proteins bound to satellite RNA, we performed affinity purification using beads bound to a GFP-specific single-chain antibody (Chromotek, GFP-Trap). As a negative control, we used satellite RNA from fission yeast and an mRNA encoding the mCherry fluorescent protein (Figure S4B).
Figure 4. Mass Spectrometry Experiments Identified Proteins that Bind Satellite RNAs.
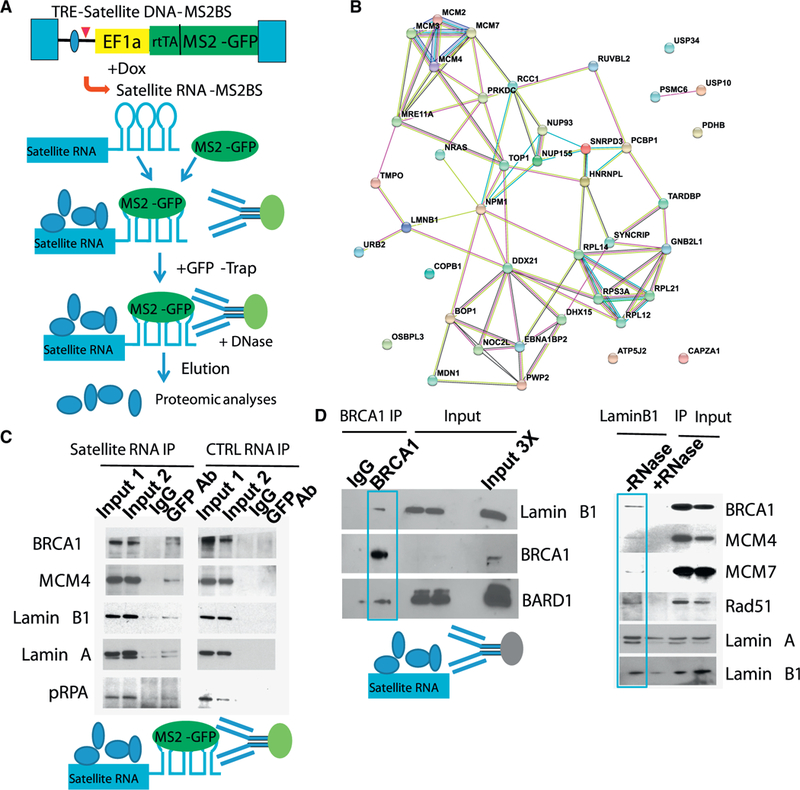
(A) A diagram of the workflow.
(B) The protein-protein interaction network pulled down using satellite RNA is positively correlated with BRCA1 function (visualized with StringDB).
(C) RNA IP followed by western blotting confirmed the binding of satellite RNA to the proteins, as indicated, but not a control RNA.
(D) Co-IP followed by western blotting revealed (left) interaction between BRCA1 and Lamin B1, as shown by BRCA1 IP. BARD1 is shown as a positive control for BRCA1 interaction. Right: interactions between Lamin B1 and the indicated factors. GFP-Trap was used to pull down Lamin B1 in a LaminB1-GFP knockin U2OS cell line (the controls are IgG proteins from rabbits, mice, or rats instead of alpaca, from which the GFP antibody is derived).
Our mass spectrometry (MS) analyses revealed a number of proteins that specifically bound to satellite RNA (Table S3). Interestingly, the majority of these proteins belong to a network that is associated with the BRCA1 protein (Figure 4B). Using RNA-immunoprecipitation (IP) experiments, we verified that satellite RNA bound MCM4, Lamin B1, and Lamin A (Figure 4C). Although not identified in the initial MS experiment, BRCA1 was also associated with satellite RNA, as determined by western blotting of the bound proteins (Figure 4C). This unexpected finding led us to determine whether BRCA1 binds other satellite RNA-binding proteins. Indeed, we found that BRCA1 associated with Lamin B1 in an RNA-dependent fashion (Figure 4D) because the binding could be abolished by treatment with RNase. Thus, we have identified a BRCA1-associated protein network that interacts with satellite RNAs.
Satellite RNA Induces the DNA Damage Response and Chromosome Instability by Interacting with the BRCA1-Associated DNA Replication Fork Protection Network
We next sought to determine which satellite RNA-binding proteins are required for satellite RNA expression-mediated genomic instability. Because Lamin B1 is important for chromatin stability (Butin-Israeli et al., 2015), we asked whether Lamin B1 is involved in the DNA damage response downstream of satellite RNA expression. We used shRNA to knock down Lamin B1 in U2OS cells (Figure 5A), and when these cells were transfected with satellite RNA, the levels of γH2AX were significantly reduced compared with controls. Our data indicate that satellite RNA-binding proteins such as Lamin B1 are required, at least in part, for the satellite RNA-elicited DNA damage response.
Figure 5. Satellite RNA Overexpression-Induced Genomic Instability Is Mediated through Protein-Binding Partners.
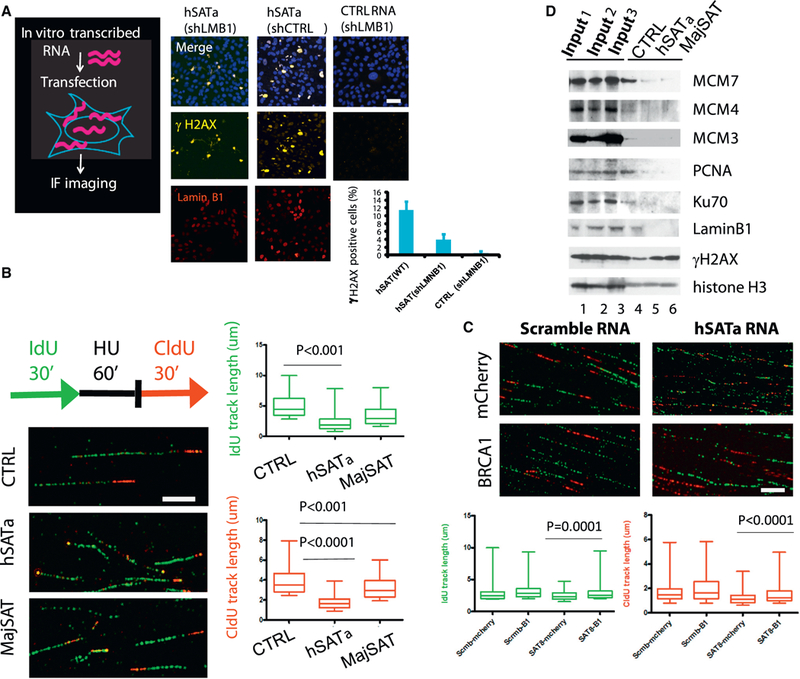
(A) Lamin B1 is required for the satellite RNA-mediated DNA damage response. U2OS cells were stably infected with lentiviruses expressing shRNA targeting Lamin B1 (shLMB1) or non-targeting shRNA (shCTRL) prior to transfection of satellite RNA or a control scrambled RNA. Sixteen hr after transfection, cells were fixed, immunostained, and imaged. Yellow, γH2AX; red, Lamin B1. Scale bar, 50 μm.
(B) The DNA fiber assay was used to analyze the stability of DNA replication forks. Replication forks were labeled in U2OS cells infected with lentiviruses overexpressing hSATa or MajSAT or a control Luc RNA, respectively, with IdU and CldU before (green) and after (red) HU stalling (3 mM, 1 hr), as indicated in the scheme. DNA fibers were prepared on glass slides, and tracks of labeled DNA were detected by immunofluorescent staining. Replication track lengths (n > 100) were measured and are shown as boxplots. Green tracks of control (CTRL): 4.950 μm ± 0.07980, n = 523; hSATa: 2.365 μm ± 0.08680, n = 297; MajSAT: 3.475 μm ± 0.07494, n = 523. Red tracks of CTRL: 3.902 μm ± 0.06607, n = 393; hSATa 1.799 mm ± 0.07122, n = 103; MajSAT: 3.278 μm ± 0.05516, n = 393. The p values from unpaired two-tailed t test are shown. Scale bar, 5 μm.
(C) Overexpression of BRCA1 (B1) by transient transfection rescued DNA replication fork defects in satellite RNA-overexpressing cells. The DNA fiber assay was performed as in (B), but a plasmid overexpressing human BRCA1 protein or an mCherry protein as CTRL was transfected into U2OS cells 8hr before satellite RNA or a scrambled RNA transfection prior to HU treatment and DNA fiber preparation. Replication track lengths (n > 100)were measured and are shown as boxplots. The p values from two-tailed t test are shown. Scale bar, 10 μm.
(D) iPOND assay followed by western blotting showed that satellite RNA overexpression significantly reduced the amount of proteins bound to DNA replication forks. Fork-associated proteins were purified from CTRL or hSATa- or MajSAT-overexpressing U2OS cells by iPOND after stalling with HUfor1 hr. Western blots from EdU pull-down were probed with the indicated antibodies. 1% of each cell lysate was loaded as input (1, 2, and 3 in lanes 1, 2, and 3, respectively) of each captured sample (CTRL, hSATa, and MajSAT in lanes 4, 5, and 6, respectively).
See also Figure S5
To investigate whether the binding of satellite RNA to MCM proteins (Figure 4D), which are the essential components of the DNA replication machinery, has any functional consequence, we performed DNA fiber assays. In these assays, the nascent DNA is labeled by incorporation of 5-iododeoxyuridine (IdU) and 5-chlorodeoxyuridine (CldU), which are added to the medium before and after stalling, respectively. Thus, the progression of individual replication forks can be visualized by immunostaining of single-molecule DNA fibers. We found that U2OS cells transfected with hSATa RNA had shorter DNA fork track lengths before stalling than the luciferase RNA control (green IdU tracks in Figure 5B). This difference indicates nascent strand degradation during the stalling period (Schlacher et al., 2011). Control cells restarted fork progression following HU removal, producing red CldU tracks adjacent to the green tracks (Figure 5B). hSATa RNA- and MajSAT RNA-overexpressing cells, however, showed significantly shorter CldU tracks. We next showed that defects in DNA replication and replication fork restart in satellite RNA-expressing cells could be rescued by ectopic expression of wild-type BRCA1 (Figures S5A and 5C; compare the green tracks of SAT8-mcherry, 2.554 μm ± 0.06611, n = 138 with SAT8-B1, 3.262 μm ± 0.1088, n = 155; unpaired t test, p < 0.0001; compare the red tracks of SAT8-mcherry, 1.259 μm ± 0.04152, n = 178 with SAT8-B1, 1.531 μm ± 0.04949, n = 238, unpaired t test, p < 0.0001). These data suggest that DNA replication in satellite RNA-expressing cells was either delayed and/or that the forks progressed more slowly after restart. Importantly, overexpression of BRCA1 could overcome these DNA replication defects.
To identify protein complexes at the DNA replication fork that are affected by satellite RNA expression in the presence of HU, we used a method for isolating proteins on nascent DNA (isolating proteins on nascent DNA [iPOND]) (Sirbu et al., 2011), in which nascent DNA is labeled by incorporation of 5-ethynyl- 2′-deoxyuridine (EdU), followed by a biotinylation reaction and streptavidin bead pull-down of associated complexes. Using this assay, we found that satellite RNA expression significantly increased the accumulation of γH2AX in the vicinity of replication forks in HU-treated cells (Figure 5D, compare lanes 5 and 6 with the control in lane 4). Concomitantly, we observed a stark reduction of replicating DNA binding in the levels of MCM3, MCM4, MCM7, and proliferating cell nuclear antigen (PCNA) required for DNA replication in hSATa- and MajSAT-expressing cells. Similarly, the levels of Ku70 and Lamin B1 binding to replicating DNA were also reduced in satellite RNA-expressing cells, although the total protein level of Lamin B1 was not affected (data not shown). We also found a significant reduction of proteins that are associated with the replication forks in satellite RNA expressed in human BRCA1−/− breast cancer cells (HCC1937) compared with cells re-expressing BRCA1 (Zhu et al., 2011; Figure S5B). Altogether, these data show that satellite RNA expression impairs the function of BRCA1-associated proteins that protect DNA replication forks and prevent DNA damage. This defect may directly lead to chromosome instability and the emergence of chromosomes with deleterious structural changes observed in BRCA1-deficient cells.
Satellite RNA Expression-Induced DNA Replication Defects Require RNA-DNA Hybrid Formation
To further understand the mechanism by which satellite RNA mediated DNA replication defects, we examined the presence of RNA-DNA hybrids in satellite RNA-expressing cells. We found increased RNA-DNA hybrid formation in U2OS cells upon transfection with hSATa RNA compared with scrambled RNA following staining with an antibody that recognizes RNA-DNA hybrids (S9.6) (Figure 6A). When we overexpressed RNase H1, which degrades RNA in RNA-DNA hybrids, in such cells, we found that the amount of S9.6 antibody staining as well as γH2AX staining was greatly reduced (Figures 6B and 6C and S6A and S6B). To examine the effect of RNase H1 expression on DNA replication defects mediated by satellite RNA, we performed Click-IT EdU experiments to quantify the amount of replicating DNA (Figure 6D). In the presence of hSATa, the DNA content of the cells, as measured by DAPI staining in flow cytometry experiments, was not significantly altered (Figure S6B). However, the amount of EdU-labeled replicating DNA was significantly reduced (by 50%; Figure 6D, boxed S phase area), suggesting increased stalling of DNA replication forks. The expression of RNase H1 increased the percentage of replicating DNA. This result demonstrates that formation of RNA-DNA hybrids significantly contributes to the satellite RNA-mediated DNA damage response.
Figure 6. Satellite RNA-Mediated RNA-DNA Hybrid Formation and γH2AX Accumulation Are Sensitive to RNase H Treatment.
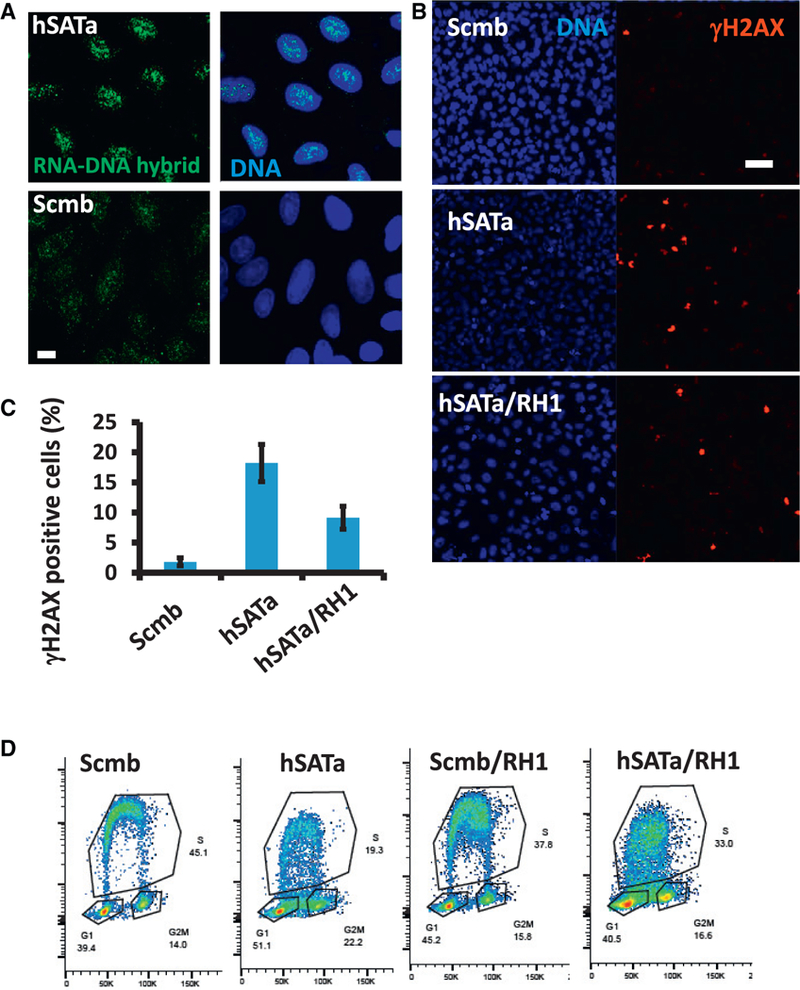
U2OS cells were infected with Dox-inducible RNase H1-expressing lentiviruses prior to RNA transfection with either satellite RNA or a scrambled RNA.
(A) Increased levels of RNA-DNA hybrids in hSATa-transfected cells as shown by S9.6 antibody staining and imaged by confocal microscopy. Scale bar, 10 μm.
(B and C) Cells were fixed and immunostained with γH2AX antibody (B). The number of γH2AX staining foci were quantified with Imaris software and are plotted in (C). Scale bar, 50 μm.
(D) Cell cycle analysis of satellite RNA-transfected U2OS cells as stained by Click-IT chemistry and DAPI in flow cytometry. The boxed S phase areas indicate differences compared with hSATa RNA transfection.
See also Figure S6.
DISCUSSION
We have previously shown that non-coding satellite RNAs are aberrantly overexpressed in BRCA1 mutant breast tumors (Zhu et al., 2011) as a result of the failure to form condensed pericentromeric heterochromatin in the absence of BRCA1. Here we demonstrate that overexpression of satellite RNAs is oncogenic in the mouse mammary gland. Satellite RNAs co-operate with loss of other tumor suppressors, such as p53, in promoting tumor formation. We also show that satellite RNA expression induces the DNA damage response and chromosome instability by sequestering factors in the BRCA1-associated protein network, causing destabilization of DNA replication forks, leading to formation of R-loops and DNA damage.
Satellite RNA Silencing in Mammals
Maintaining heterochromatin integrity promotes genomic stability and the reliable propagation of established gene expression programs. In yeast and plants, the transcription of repetitive RNAs is tightly associated with the RNA interference pathway, which is essential for establishing heterochromatin (Reyes-Turcu and Grewal, 2012). Although the mechanisms by which heterochromatin is formed and maintained are well documented in other eukaryotes, they are unresolved in mammalian systems. Our current data strongly suggest that overexpression of non-coding satellite RNAs promotes tumorigenesis in mice. We demonstrate that accumulation of heterochromatic repetitive RNAs adversely affects cell function in mammals.
Satellite RNAs and Cancer
lncRNAs are emerging as important players in human cancer formation, progression, and metastasis (Quinn and Chang, 2016). Dysregulation of lncRNAs (i.e., upregulation of oncogenic lncRNAs or downregulation of tumor-suppressive lncRNAs) causes malignancies in a variety of tissues. In this report, we have demonstrated that overexpression of satellite RNAs promote tumorigenesis in the breast. Several molecular mechanisms have been proposed for lncRNA-mediated genomic instability and tumorigenesis. Some lncRNAs function as molecular decoys, binding to proteins and disrupting their normal interactions and function. lncRNAs can sequester microRNAs and mRNAs, referred to as competitive endogenous RNAs (ceRNAs), to regulate gene expression. We and others have observed that satellite RNAs are overexpressed (Leonova et al., 2013; Ting et al., 2011; Zhu et al., 2011) in a variety of human solid tumors. In some cases, the amount of satellite RNA can represent up to 50% of total cellular transcripts (Ting et al., 2011). The amount of satellite RNA in tumor cells is in stark contrast to the low levels of most lncRNAs in normal cells. This represents a significant barrier to understanding the function of these RNAs and identification of physiologically relevant RNA-protein interactions. We took advantage of the high levels of satellite RNAs in tumors to identify proteins that are bound by satellite RNAs. These efforts revealed molecular mechanisms by which satellite RNAs induce genomic instability and tumorigenesis.
Satellite RNAs and DNA Replication
Complete and accurate replication of the genome is a complex task, and failure to achieve faithful replication causes genomic instability (Aguilera and Gómez-González, 2008). A number of proteins, including MCM2, MCM3, MCM4, MCM6, and MCM7 and PCNA are critical for the initiation and progression of DNA replication. Emerging evidence suggests that BRCA1 plays a role in stabilizing and repairing stalled replication forks (Joukov et al., 2014; Pathania et al., 2011, 2014; Schlacheret al., 2012); i.e., in suppressing DNA replication stress. Using a MS approach, we identified a number of proteins that are bound by the satellite RNAs (Figure 4) and may participate in this process. Unexpectedly, we found that BRCA1 forms a complex with MCM proteins and Lamins in an RNA-dependent manner. It has been shown that Lamin B1 is important for the stability of DNA replication forks (Butin-Israeli et al., 2015). Indeed, our iPOND experiments revealed that satellite RNA overexpression leads to lower levels of MCM and other proteins at nascent DNA replication forks (Figure 5). Consequently, both DNA replication and replication restart following stalling are impaired. These deficits can be rescued by overexpression of BRCA1. Two mechanisms may be involved; BRCA1 can protect heterochromatin through histone mono-ubiquitination (Zhu et al., 2011) but can also neutralize deleterious effects of satellite RNA overexpression by monitoring the stability of the replication forks, acting in parallel on chromosome ends and at replication forks. This new pathway could be a second line of defense for accidental and/or transient expression of satellite RNA in normal cells. It also explains why cells overexpressing satellite RNA are more sensitive to DNA replication stress reagents and how satellite RNA overexpression leads to chromosome breaks and radials, similar to those seen in BRCA1-deficient Fanconi anemia cells (Schlacher et al., 2012).
A large body of evidence supporting the role of BRCA1 in maintaining genomic stability has emerged to demonstrate BRCA1 as part of the DNA replication fork protection complexes (Schlacher et al., 2012) as well as its association with genomic foci exhibiting RNA-DNA hybrid structures (Hatchi et al., 2015). Because of the detrimental consequences of satellite RNA on DNA replication fork stability, we investigated the possibility of RNA-DNA hybrid formation induced by satellite RNA expression. Indeed, we found evidence of accumulation of such hybrids in satellite RNA-expressing cells, which is in agreement with earlier reports (Zeller et al., 2016; Bersani et al., 2015). Furthermore, RNase H treatment partially diminished the effect of satellite RNA expression on DNA damage and replication (Figure 6). Additionally, BRCA1 expression could rescue the DNA replication defect resulting from satellite RNA expression (Figure 5). One open question remains: what is the sequence specificity for formation of such RNA-DNA hybrids? Because both human hSAT and mouse MajSAT essentially induce tumor formation and DNA replication defects, it is tentative to postulate that mismatch base-pairing of satellite RNAs to single-stranded DNA happens at stalled DNA replication forks, promoting further destabilization of the replication forks.
Taken together, we propose a model that supports the notion that satellite RNA expression-mediated genomic instability occurs through promoting the formation of both RNA-protein sequestration and RNA-DNA hybrids at replication forks (Figure S6D). Loss of BRCA1 leads to overexpression of satellite RNAs, sequestering BRCA1-associated protein networks that function to protect DNA replication forks, leading to genomic instability and tumorigenesis. Our work provides compelling evidence that aberrantly expressed repetitive non-coding RNAs can induce oncogenesis.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-g-H2AX | EMD Millipore | 05–636, RRID: AB_309864 |
| Rabbit polyclonal anti-phospho-Chk1 (Ser345) | Cell Signaling | 2348, RRID: AB_331212 |
| Rabbit polyclonal anti-phospho-RPA32 (Ser4/Ser8) | Bethyl | A300–245A, RRID: AB_210547 |
| Mouse monoclonal anti-BrdU | BD | BDB555627 |
| Rat monoclonal anti-BrdU | Thermo Fisher | MA1–82718, RRID: AB_927213 |
| Mouse monoclonal S9.6 antibody | Kerafast | ENH001, RRID: AB_2687463 |
| rabbit-anti-trimethyl-Histone H3 K9 | EMD Millipore | 07–442, RRID: AB_306848 |
| rabbit anti-GFP | Thermo Fisher | A-11122, RRID: AB_221569 |
| rabbit anti-cytokeratin 5 | Abcam | Ab_53121, RRID: AB_869889 |
| rabbit anti-cytokeratin 8 | Abcam | Ab_59400 |
| rabbit anti-vimentin | Abcam | Ab_92547 |
| rabbit anti-estrogen receptor | Santa Cruz | Sc_542 |
| Mouse anti-b-Actin | Sigma-Aldrich | A2228, RRID: AB_476697 |
| Rabbit anti-MCM3 | Cell Signaling | 4003 |
| Rabbit anti-MCM4 | Cell Signaling | 12973 |
| Rabbit anti-MCM7 | Cell Signaling | 4018 |
| Rabbit anti-PCNA | Abcam | ab2426, RRID:AB_303062 |
| Rabbit anti-Rad51 | EMD Millipore | PC-130 |
| Rabbit anti-BRCA1 | Santa Cruz | Sc-642 |
| Rabbit anti-BARD1 | Santa Cruz | Sc-11438 |
| Rabbit anti-histone H3 | Cell Signaling | 9715, RRID:AB_331563 |
| Rabbit anti-lamin A | Sigma-Aldrich | L1293, RRID:AB_532254 |
| Rabbit anti-Ku70 | Abcam | Ab83501, RRID: |
| Rabbit anti-lamin B1 | Abcam | ab16048, RRID:AB_443298 |
| Chemicals, Peptides, and
Recombinant Proteins | ||
| IdU | ACROS Organics | AC122350010 |
| CIdU | Sigma-Aldrich | C6891–100MG |
| DAPI | Thermo Fisher | D1306 |
| Doxycycline | Sigma Aldrich | D9891 |
| Crystal Violet | Sigma Aldrich | C3886 |
| Hydroxyurea | Sigma Aldrich | H8627 |
| Aphidicolin | Sigma Aldrich | A0781 |
| neocarzinostatin | Sigma Aldrich | N9162 |
| Zeocin | GIBCO | R25005 |
| Blasticidin S | Thermo Fisher | ant-bl-1 |
| puromycin | Thermo Fisher | A1113803 |
| Paraformaldehyde | Electron Microscope Sciences | 15714 |
| RiboLock RNase Inhibitor | Thermo Fisher | FEREO0382 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | A25742 |
| TRIzol Reagent RNA extraction | Thermo Fisher | 15596026 |
| RNase H | New England Biolabs | M0297S |
| Complete Protease Inhibitor Cocktail Tablet | Sigma | 11697498001 |
| ProLong Gold Anti-fade Mountant | Thermo Fisher | P36930 |
| Critical Commercial
Assays | ||
| GFP-Trap_A Kit | ChromoTek | Gta_20 |
| mMESSAGE mMACHINE T7 Kit | Thermo Fisher | AM1344 |
| mMESSAGE mMACHINE SP6 Kit | Thermo Fisher | AM1340 |
| qScript cDNA Supermix | Quanta BioScience | 95048–025 |
| Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Thermo Fisher | C10424 |
| TruSeq Stranded Total RNA Library Prep Kit | Illumina | 20020597 |
| MTT Assay Kit (Cell proliferation) | Abcam | ab211091 |
| Pierce Silver Stain Kit | Thermo Fisher | 24612 |
| Trans | Mirus | Mir2225 |
| Deposited Data | ||
| Sequencing data of RNA-seq experiments | This study | GEO: GSE88961 |
| Experimental Models: Cell
Lines | ||
| Human: HEK293T cells | ATCC | CRL-1273 |
| Human: U-2 OS cells | ATCC | HTB-96 |
| Human: HCC1937 cells | Laboratory of Genetics, Salk | |
| Human: MCF7 cells | ATCC | HTB-22 |
| Mouse: M3L2 | Laboratory of Genetics, Salk | |
| Mouse: 67 NR | Laboratory of Jing Yang, UCSD | |
| Experimental Models:
Organisms/Strains | ||
| B6D2 female mice | Jackson Laboratory | |
| Oligonucleotides | ||
| Primers for qRT-PCR | This study | Table S4 |
| Recombinant DNA | ||
| pBK-SAT8 | This study | |
| pBK-Maj6 | This study | |
| pBK-Scrm | This study | |
| pBK-CFXr | This study | |
| pBK-Spom | This study | |
| Lenti MS2-P65-HSF1_Hygro | Addgene | 61426 |
| Lenti dCAS-VP64_Blast | Addgene | 61425 |
| lenti sgRNA(MS2)_zeo | Addgene | 61427 |
| LV-SAM-sgMaj | This study | |
| LV-SAM-sgMin | This study | |
| LV-SAM-sgCTRL | This study | |
| pRAR-MS2-SAT8 | This study | |
| pRAR-MS2-mcherry | This study | |
| pRAR-shLMB1 | This study | |
| pRAR-shCTRL | This study | |
| pRAR-MS2-Spom | This study | |
| pRAR-RH1 | This study | |
| 2si-THM-hSAT | This study | |
| 2si-THM-Maj | This study | |
| 2si-THM-LucN | This study | |
| Software and
Algorithms | ||
| ImageJ | NA | https://imagej.nih.gov/ij/download.html |
| Imaris | Bitplane | |
| Bowtie2 | (Langmead and Salzberg, 2012) | |
| AIMS package | Paquet and Hallett, 2014 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| RawXtract (version 1.9.9) | (McDonald et al., 2004 | Bioconductor (Huber et al., 2015) |
| ProLuCID algorithm | Eng et al., 1994 | |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tony Hunter (hunter@salk.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ductal injection of lentiviruses
Wild-type B6D2 female mice of 6–8 weeks old were purchased from the Jackson Laboratory. All mice were maintained under pathogen-free conditions at the Salk Institute for Biological Studies, and all procedures performed in this study were approved by the Institutional Animal Care and Use Committee. Mice were injected with lentiviruses under anesthesia. Lentivirus constructs that overexpress satellite RNAs or the control RNA were described in Zhu et al. (Zhu et al., 2011). MMTV-Cre transgenic female mice were injected with a lentivirus that expresses oncogenic H-RAS (G12V) and shRNA targeting tumor suppressor p53 (Friedmann-Morvinski et al., 2012) as a positive control for the ductal injection experiments. We injected 1×106 virus particles in 10 μL of PBS, 0.2% of Trypan Blue with a Hamilton syringe into the duct of the number 4 mammary gland according to the protocol described (Krause et al., 2013).
Cell culture and virus infection
Cells, 293T, U2-OS, MCF7, 67 NR, M3L2 were cultured in DMEM medium with 10% FBS and glutamax. HCC 1937 cells were cultured in DMEM/F12 with 20% FBS, glutamax, non-essential amino acids. Cultured cells were infected with lentivirus by a 12-h incubation with an MOI of 5 and harvested at the time indicated in corresponding figure legends. All the cells were cultured at 37°C and 5% CO2. Cells purchased commercially are authenticated by STR DNA typing. Cells from other sources were not authenticated. All cell lines are negative for mycoplasma contamination. Please also see detailed information of each cell line in Key Resources Table.
METHOD DETAILS
RNA in vitro synthesis, microinjection, and transfection
In vitro RNA synthesis was performed using T7 or SP6 mMESSAGE mMACHINE Kits (Thermo Fisher Scientific) and following manufacturer’s instruction. 8 copies of the hSATa were cloned into pBluescript vector: hSATa. Similar vectors were generated for 6 copies of the MajSAT: MajSAT, a 1.4 kb of the gamma satellite RNACFXr (Zhu et al., 2011), a control RNA of the S. pombe satellite RNA, a scramble RNA with 1.4 kb in size, RFP and GFP RNA. Purified RNA (1 μg) was injected into the nucleus of culture cells with a microinjector (Eppendorf Transjector) or transfected into cultured cells using the TransIT®-mRNA Transfection Kits (Mirus).
hSATa
CTGTCTAGTTTTCATATGAAGATATCCCGTGTCCAACGAAATCCTCAAAGGTATCAAAATATCCACTTGCAGATTCTACAAAAAGAGT GCTTCAAAACTGCTCTGTCAAAAGGAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGCTGCTAGA TCTGTCTAGTTTTCATATGAAGATATCCCGTGTCCAACGAAATCCTCAAAGGTATCAAAATATCCACTTGCAGATTCTACAAAAAGAG TGCTTCAAAACTGCTCTGTCAAAAGGAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGCTGCTAG ATCTGTCTAGTTTTCATATGAAGGTATTCCCGTATCCATCGAAATCGCTAGAGCTATCCAAATATCCTCTTGCAGATTCTACAAAAAG ATGTTTCCAAACTGCTGTATCAATAGACAGGTTGTACTCTGTTAGTTGAGGACACACATCAAAAAGAAGTTTCTGAGAATGCTGCTAG ATCTGTCTAGTTTTCATATGAAGATATCCCGTGTCCAACGAAATCCTCAAAGGTATCAAAATATCCACTTGCAGATTCTACAAAAAGA GTGCTTCAAAACTGCTCTGTCAAAAGGAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGCTGCTA GATCTGTCTAGTTTTCATATGAAGATATCCCGTGTCCAACGAAATCCTCAAAGGTATCAAAATATCCACTTGCAGATTCTACAAAAAG AGTGCTTCAAAACTGCTCTGTCAAAAGGAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGCTGCT AGATCTGTCTAGTTTTCATATGAAGATATCCCGTGTCCAACGAAATCCTCAAAGGTATCAAAATATCCACTTGCAGATTCTACAAAAA GAGTGCTTCAAAACTGCTCTGTCAAAAGGAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGCTGC TAGATCTGTCTAGTTTTCATATGAAGATATCCCGTTTCCAAAGAAATCCTCAAATGTATCCAAATATCTACTTCCAGATTCTACAAAAA GACTGTTTCAAAAC GGCTCTGTCAAAAGTAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGCTGC TAGATCTGTCTAGTTTTCATATGAAGATATCCCGTGTCCAACGAAATCCTCAAAGGTATCAAAATATCCACTTGCAGATTCTACAAAA AGAGTGCTTCAAAACTGCTCTGTCAAAAGGAAGGTTCAACTCTGTTACTTGAGTACACACATCACAAGGAAGTTTCTGAGAATGC
MajSAT
AAAAAGGTGGAAAATTTAGAAATTTCCACAATAGGACGTGGAATATGGCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCAC TTGACGACTTGAAAAATGACAAAATCCCTGAAAAACGTGAAAAATGAGAAATGCACACTGTAGGAGCTGGAATATGGCGAGAAAAC TGAAAATCACGGAAAATGAGAAATACACACTTTAGGACGTGAATCTAGCTATGGCGAGGATAACTGAAAAAGGTGGAAAATTTAGAA ATGTCCACTCTAGGACGTGGAAAATGGCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACA AAATCACTAAAAAATGTGAAAAATGAGAAATGCACACTGAAGGACCTGGAATATGGCGAGAAAACTGAAATTCACGGAAAATGAGA AATACACACTTTAGGACGTGAATCTAGAACTAGTGGATCTCTGCTATGGCGAGGATAACTGAAAAAGGTGGAAAATTTAGAAATGTC CACTCTAGGACGTGGAATATGGCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACGAAATC ACTAAAAAACGTGAAAAATGAGAAATGCACAATGAAAAACCTGGAATATGGCAAGAAAACTGAAAATCACGGAAAATGAGAAATACA CACTTTAGGACGTGAATCTAGCTATGGCGAGGATAACTGAAAAAGGTGGAAAATTTAGAAATTTCCACAATAGGACGTGGAATATG GCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACAAAATCCCTGAAAAACGTGAAAAATGA GAAATGCACACTGTAGGAGCTGGAATATGGCGAGAAAACTGAAAATCACGGAAAATGAGAAATACACACTTTAGGACGTGAATCTA GCTATGGCGAGGATAACTGAAAAAGGTGGAAAATTTAGAAATGTCCACTCTAGGACGTGGAAAATGGCAAGAAAACTGAAAATCAT GGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACAAAATCACTAAAAAATGTGAAAAATGAGAAATGCACACTGAAGGACC TGGAATATGGCGAGAAAACTGAAATTCACGGAAAATGAGAAATACACACTTTAGGACGTGAATCTAGAACTAGTGGATCTCTGCTAT GGCGAGGATAACTGAAAAAGGTGGAAAATTTAGAAATGTCCACTCTAGGACGTGGAATATGGCAAGAAAACTGAAAATCATGGAAA ATGAGAAACATCCACTTGACGACTTGAAAAATGACGAAATCACTAAAAAACGTGAAAAATGAGAAATGCACAATGAAAAACCTGGA ATATGGCAAGAAAACTGAAAATCACGGAAAATGAGAAATA
Scramble
GTTTGGCGCGTGTAACACGTTTCTGATTACTATTAGTTCATTTATACATTCAAACATAATCTCGTAAAGGAGGTCGTCAACCATAAAT GTTTATGCCTGAACTTCTTTTAATGAATTATACGGCAGATACCCTTATCTTTTGTTGGTCTCATCAGTAAAAAACTGCCCAAGTAAAA GTTCGGTTGAACACAGACTCGTTTATAGATATGCGCCATAGGCCCAATCATGTCGAATACTCACCTTGTAAGTGACGGAAACCACT AAGGTTATTATCGTGATACCCTATCGACTTATAGGTAGTTCTCCATTAGTCGAATTCACTTGAGGGTCTCTGTAGGATACCTGAACT CGTCAATGTGAGTGACAATTGAGAGAATCGTTCCTATTAACTTATTTCAAGGTGCTACATTATAATAAAACTTCTCCAGACCTGGCAG GTTAGTAAAGACTTAGTAAAGTGTTAATCTCAAACACCTCAAGGAGGTCATCATCTTAATAATGTAGGTCTTATCGTTGTACTTAAGA AGTCTCTATTAGCCGGAATGAAAAGTCGTACCGTCGATAGATAAAGTAAATAAGTATACCATCTGCAGTTCGCAGCCTTAACTGAAT AACCAGCAAACACAACACCAGACTGGCCAGGCCGGTCGACCATAGTTACAGACTACATTAACTTAACGTGATGGCCATAGCTACT TATCCAACAGAAGGTCTTTGTAATAAATTATGAACCAACTAACGATCTTAAGTTATCAGCCTGCGACCCTTCTACAAGTTTCTTGTCAC CTTTTCCTCCGCCTAGATAATCCTATACTTGTCGAACTGAAGGATACTTCACGCACATTTCTTTACACGAGTAGTGATGAATGTATAG TATCGTCCTGACCTAGTCGATCTTGTTTTTTTAGCATAAAATAATGAGCAACCGAAGATTCTACATGAACGTAGCGCTGGCAGGACC CCCCGCCAGATTACACAAATGGTAGCGGTGATTGCTCAAGATATCAAACCACATGGTCCACATAACACCAAATAACAAGCAAAAGA TTGTCACTTAAATGAGTGGTTATTCGAACTACAAATGTCAATAAAAACGAGTTCACTCTTGCAGTTACTGAAAAGAATAGGGTAAAGA ACAGTGGAACCGAAAATAAGGGCCTGACTATTAAGGTTTAGTAATAAACAGCAACAACGTCTTTTGCTCACTAGTTATTCACGAGTCT AACAATGTCTCTGGTAACACACAAGAGTGAACTAAACGATAAGGAAATCGACACTGAAAAATCCAGATTTCGACACTCACTTAA ACCTATTATGCAATCTCAAAAGACTACTCAATTGCAATTGATCAGTAAGTATTCAATAAAACAATATCCTGTACTAAATTTCCACGTATTACCG
Immunohistochemical analysis
Immunohistochemical staining was performed as described (Zhu et al., 2011). Primary antibodies included: mouse –anti-phospho-H2AX (1:1,000, Millipore), rabbit-anti-trimethyl-Histone H3 Lys9 (1:100, Millipore), rat anti-BrdU (1:250, Thermo Fisher), mouse anti-BrdU (1:250, BD), rabbit anti-GFP (1:500, Invitrogen), rabbit anti-cytokeratin 5 (1:150, Abcam), rabbit anti-cytokeratin 8 (1:150, Abcam), rabbit anti-estrogen receptor (1:75, Santa Cruz), rabbit anti-vimentin (1:500, Abcam), human anti-centromere antigen (1:500, a gift from Drs. Karen Oegema and Arshad Desai), Mouse anti-S9.6 (1:500, Kerafast).
CRISPR SAM experiments
Activation of endogenous satellite RNAs by CRISPR SAM system was performed as described (Konermann et al., 2015) in a mouse lung cancer cell line M3L2 (Xia et al., 2014) or mouse breast cancer cell line 67NR (gift from Dr. Jing Yang). The sequences for the guide RNAs targeting the mouse satellites are: Minor: aaaaacacattcgttggaaa (GenBank id Z22159);Major: aaatgagaaatgcacactgt; gaaatgcacactgtaggagc (Hörz and Altenburger, 1981); control: acgtggagctgggggaag
Western blotting analysis
Western blotting was performed using 3%–8% Tris-Acetate or 4%–12% Bis-Tris gradient precast Novex gels (Invitrogen) and XCell Blot Modules (Invitrogen) according to manufacturer’s protocol. anti-pS345 Chk1 (1:200, Cell Signaling), anti-actin (1:1,000, Santa Cruz), anti-MCM3, MCM4, MCM7 (1:200, Cell Signaling), PCNA (1:100, Abcam), anti-Rad51 (1:500, Millipore), Lamin A (1:200, Sigma), anti-Lamin B1 (1:200, Abcam), anti-pRPA32 (S4/S8) (1:200, Bethyl), anti-Ku70 (1:500, Abcam), anti-BRCA1 (1:200, Santa Cruz), anti-BARD1 (1:250, Santa Cruz) and anti-histone H3 (1:500, Cell Signaling).
Clonogenic survival assay
U2-OS cells (500–1000) were seeded in 6-cm culture dishes, allowed to adhere for 16 hr, and treated for 24 hr or as indicated. Drugs were removed by washing with PBS and cells grown in media for 8–10 days to form colonies. Cells were stained with 0.5% crystal violet in 20% methanol. Dishes were scanned, and colonies (> 50 cells) counted using ImageJ software. Clonogenic survival is given as percentage of respective untreated controls.
Immunofluorescent staining and imaging
Immunofluorescent staining was performed as described in Pao et al. (2014) (#3996). Slides were mounted using Anti-fade Prolong Gold (Life Technology), and images acquired on LSM 710 or LSM 780 or LSM 880, laser scanning confocal (Zeiss) at the Waitt Advanced Biophotonics Core. Image quantification analyses were performed using Imaris (BitPlane) or ImageJ. ACA antibody (1:1000) is a gift from Drs. Arshad Desai and Karen Oegema.
Cell cycle analysis and flow cytometry
Cells were washed with cold PBS and collected for fixation with 70% ice-cold ethanol overnight. Fixed cells were re-suspended in freshly made 0.5% Triton X-100 with 100 μg/ml RNase A and incubated for 30 min at room temperature. The cells were washed and resuspended in 1 μg/ml DAPI (Thermo Fisher) in PBS before analysis by flow cytometry at the Salk Flow Cytometry Core. Or cells were processed according to the manufacturers’ instruction Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit.
RNA-seq analysis
Total RNA was isolated using Trizol reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. Libraries were prepared using TruSeq Stranded Total RNA Library Prep Kit with Rib-Zero (Illumina) and sequenced with paired-end 150 chemistry on a Next Seq 500 at the Next Generation Sequencing Core Facility in the Salk Institute for Biological Studies. Raw reads were filtered and trimmed with BBDuk v38.82 (https://sourceforge.net/projects/bbmap/) and then aligned to GRCm38/mm10 using STAR v2.5.0c (Dobin et al., 2013). Transcripts per million were quantified by RSEM v1.2.25 (Li and Dewey, 2011) using Gencode M9 annotations (Mudge and Harrow, 2015). Gene-expression profiles for each sample were subjected to classification of the breast cancer intrinsic subtypes (Parker et al., 2009) using the AIMS package (Paquet and Hallett, 2014) in Bioconductor (Huber et al., 2015).
RNA-seq satellite RNA analysis
RNA sequence datasets were mapped against the human reference assembly (GRCh38; GCA_000001405.15), including alternative reference assemblies and a database of repeat sequences missing from the human assembly, as previously described (Miga et al., 2015). Any remaining unmapped reads were further studied using previously published databases to screen for alpha satellite (Hayden et al., 2013) and/or human satellite 2,3 sequences (Altemose et al., 2014). In both cases, specific satellite families and/or chromosome assignment were determined using exact matches with a k-mer strategy (where k = 24 base pair windows) with previously characterized subfamily sequences.
DNA fiber assay
Cells were labeled in media containing 10 μg/ml of IdU orCldU (Sigma) as indicated in figure legends and harvested by trypsinization. Lysis, DNA fiber spreading on glass slides, and fixation were done as described previously (Jackson and Pombo, 1998). After denaturation with 2.5 M HCl for 30 min, slides were blocked 3% BSA in PBS containing 0.1% Tween 20. Immunofluorescent staining was done with sequential incubation with the BrdU antibody B44 (Becton Dickinson; mouse monoclonal, recognizes IdU) at 4C over-night and secondary antibody anti-mouse Alexa488 at room temperature for 2h, followed by BrdU antibody BU1/75 (Thermo; rat monoclonal, recognizes CldU) at 1:200 dilution at 4C, overnight and secondary anti-rat Alexa594 (The Jackson Immunology) were used at 1:200. Slides were mounted using Anti-fade Prolong Gold (Life Technology), and images acquired on LSM 710 laser scanning confocal microscope or the Elyra Structure Illumination Superresolution Microscope (Zeiss). For the analysis of replication track lengths, more than 100 individual tracks from multiple fields were measured. A MATLAB routine was used to automatically segment the images using an [1] MCT algorithm (Maximum Correlation Thresholding). The segmented regions were then visually inspected for accuracy before measurements were taken. The segmented regions were further filtered based on a circularity-metric to exclude small non-linear segments. The major axis lengths of all remaining segmented regions were measured. Boxplots of the track length were generated with Prism software.
iPOND assay
EdU-labeling and pull-down were done as previously described (Leung et al., 2013; Sirbu et al., 2011) with minor modifications. Briefly, 5 × 107 of U2OS cells were labeled with 10 μM EdU (Invitrogen), washed and harvested for nuclei enrichment. The harvested nuclei were biotinylated with 4 μM biotin-azide in 1 mL Click-it reaction buffer (Invitrogen) according to the manufacturer’s instructions. After washing in PBS, the nuclei were sonicated and chromatin sheared by 4 rounds of pulses of 15 s with an ultrasonic processor (Active Motif). Insoluble material was removed by centrifugation at 21000 g, and biotin complexes purified for 1 hr at 4°C on 40 μl streptavidine agarose (Invitrogen). Beads were washed three times and complexes eluted by boiling in LDS sample buffer.
Quantitative RT-PCR
Reverse transcription was carried out using Quanta BioScience kit. The quantitation of PCR products was analyzed with SYBR Green using ABI PRISM 7900 Sequence Detection system software (Applied Biosystems). Please also see detailed reagent information in Key Resources Table
Tissue preparation, histology, and karyotyping
Animals were anesthetized with ketamine/xylazine and perfused transcardially with 0.9% saline followed by 4% paraformaldehyde in 0.1M phosphate buffer before tissues and or tumors were dissected out. Histological analyses were performed at the Mores Cancer Center, University of California, San Diego. Karyotyping experiments were performed at the Cytogenetics Research Services Laboratory, Oregon Health and Science University.
Mass spectrometry analysis
The protein lysates were prepared using the GFP-Trap kit (Chromotek) according to the manufacturer’s instruction with minor modifications. Briefly, the cells were lysed with RIPA lysis buffer (10 mM Tris pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% Deoxycholate, 0.02% Thimerosal, 2.5 mM MgCl2, RNase-free DNaseI (Thermo Fisher Scientific), proteinase inhibitor cocktails (Sigma) and either RNase (Roche) or RNase inhibitors (Thermo Fisher Scientific)), and centrifuged at 20,000 g for 10 min at 4°C. The supernatants were incubated with agarose beads coupled with anti-GFP VHH for 2 h at 4°C. The beads were washed three times with wash buffer (10 mM Tris pH 7.5,1 M NaCl, 0.5 mM EDTA, 0.02% Thimerosal), and then the proteins were eluted with Urea elution buffer (8 M Urea, 20 mM Tris pH7.5,100mM NaCl) and analyzed by silver staining (Bio-Rad).The proteins were trypsinized and applied to the Mass-Spec and analyzed at the Scripps Research Institute. Samples were reduced and alkylated with 10 mM TCEP and 55 mM iodoacetamide respectively and then diluted to 2 M urea with 100 mM Tris pH 8.5 prior to digestion with trypsin [Promega] overnight at 37°C.
The protein digests were pressure-loaded onto 250 micron i.d. fused silica capillary [Polymicro Technologies] columns with a Kasil frit packed with 3 cm of 5 micron Partisphere strong cation exchange (SCX) resin [Whatman] and 3 cm of 5 micron C18 resin [Phenomenex]. After desalting, each bi-phasic column was connected to a 100 micron i.d. fused silica capillary [Polymicro Technologies] analytical column with a 5 micron pulled-tip, packed with 10 cm of 5 micron C18 resin [Phenomenex].
Each MudPIT column was placed in line with an 1200 quaternary HPLC pump [Agilent Technologies] and the eluted peptides were electrosprayed directly into an LTQ Orbitrap XL mass spectrometer [Thermo Scientific]. The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A), 80% acetonitrile/0.1% formic acid (buffer B) and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). An eleven-step MudPIT was run with salt pulses of 0%, 10%, 20%, 30%, 40%, 50%, 60%, 70% and 100% buffer C and 90% buffer C/10% buffer B (twice). The 120 minute elution gradient had the following profile: 10% buffer B beginning at 15 minutes to 40% buffer Bat 105 minutes, continuing to 110 minutes. A cycle consisted of one full scan mass spectrum (400–1600 m/z) at 60 K resolution followed by 5 data-dependent collision induced dissociation (CID) MS/MS spectra. Charge state exclusion was enabled with +1 and unassigned charge states rejected for fragmentation. Dynamic exclusion was enabled with a repeat duration of 30 s, a count of 500 and an exclusion duration of 180 s. Early expiration was enabled with a count of 3 and a signal-to-noise ratio of 3. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system [Thermo Scientific].
MS/MS spectra were extracted using RawXtract (version 1.9.9) (McDonald et al., 2004). MS/MS spectra were searched with the ProLuCID algorithm (Eng et al., 1994) against a human Uniprot database supplemented with the MS2-GFP sequence that was concatenated to a decoy database in which the sequence for each entry in the original database was reversed (Peng et al., 2003). The ProLuCID search was performed using no enzyme specificity and static modification of cysteine due to carboxyamidomethylation (57.02146). The data were searched using a precursor mass tolerance of 50 ppm and a fragment ion mass tolerance of 600 ppm. The ProLuCID search results were assembled and filtered using the DTASelect (version 2.0) algorithm (Tabb et al., 2002). A minimum of one peptide was required for each protein identification, and peptides were required to be fully tryptic. All peptide-spectra matches had less than 10 ppm mass error. The protein false positive rate was below one percent for all experiments. Identified proteins were both analyzed for functional enrichment and visualized using String DB (Szklarczyk et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
In studies in which statistical analyses were performed, a 2-tailed Student’s t test was used to generate p values. p values less than or equal to 0.05 were considered significant. Data shown are from one representative result of multiple experimental replications.
For animal studies, animals were randomly chosen and concealed allocation and outcome assessment was blinded. All data are expected to have normal distribution. Statistical analysis and graphical presentation was performed using Prism 6.0 (GraphPad). Log-rank (Mantel-Cox) tests were used to plot Kaplan–Meier survival curves in mice injected with lentivirus overexpressing satellite RNAs or control RNA. “n” represents number of animals in each group and the values are included in the main and supplemental figures.
Fluorescence microscopic image quantification: Image quantification analyses were performed using Imaris (BitPlane) or ImageJ. For the analysis of replication track lengths, more than 100 individual tracks from multiple fields were measured. A MATLAB routine was used to automatically segment the images using an [1] MCT algorithm (Maximum Correlation Thresholding). The segmented regions were then visually inspected for accuracy before measurements were taken. The segmented regions were further filtered based on a circularity-metric to exclude small non-linear segments. The major axis lengths of all remaining segmented regions were measured.
For analysis of cell survival experiments, data represent mean ± SEM from two biologically independent experiments using technical triplicates per data point.
Supplementary Material
Highlights.
Satellite RNAs induce DNA damage
Elevated satellite transcripts induce tumor formation
Satellite RNAs bind to the BRCA1 protein complex
Increased levels of satellite RNAs destabilize DNA replication forks
ACKNOWLEDGMENTS
We thank Dr. Ruben Alvarez Rodriguez for providing a Dox-inducible shRNA construct against Lamin B1 and Nina Tonnu for assistance with mouse work and Dr. Tushar Menon, Greg Park, and Benjamin Lewis for assistance with generating the CRISPR constructs. We are thankful to Dr. Stefan Aigner from the Gene Yeo lab for the gift of the MS2-tag plasmid and Drs. Karen Oegema and Arshad Desai for the gift of the ACA antibody. We also thank Dr. David O’Keefe for editorial work in preparing the manuscript. Q.Z. was supported by the California Breast Cancer Research Program, the Ruth L. Kirschstein National Research Service Award, and an NIH CCSG Pilot Grant (2 P30 CA014195). I.M.V. is an American Cancer Society Professor of Molecular Biology and holds the Irwin and Joan Jacobs Chair in Exemplary Life Science. This work was also supported in part by a pilot grant from the NIH Cancer Center Core (P30 CA014195–38), the H. N. and Frances C. Berger Foundation, and Leona M. and Harry B. Helmsley Charitable Trust grant 2012-PG-MED002. T. Hunter is supported by NIH CA080100 and CA082683, is a Frank and Else Schilling American Cancer Society Professor, and holds the Renato Dulbecco Chair in Cancer Research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and four tables and can be found with this article online at https://doi.org/10.1016/j.molcel.2018.04.023.
DATA AND SOFTWARE AVAILABILITY
All software used in this study is listed in the Key Resources Table.
RNA-seq data accession
The accession number for the RNA-seq data reported in this paper is GEO: GSE88961. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE88961.
Original imaging data have been deposited to Mendeley Data and are available at: https://doi.org/10.17632/8xvd7h85tm.1.
REFERENCES
- Aguilera A, and Gómez-González B (2008). Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217. [DOI] [PubMed] [Google Scholar]
- Altemose N, Miga KH, Maggioni M, and Willard HF (2014). Genomic characterization of large heterochromatic gaps in the human genome assembly. PLoS Comput. Biol. 10, e1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersani F, Lee E, Kharchenko PV, Xu AW, Liu M, Xega K, MacKenzie OC, Brannigan BW, Wittner BS, Jung H, et al. (2015). Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 112, 15148–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. (2004). Global identification of human transcribed sequences with genome tiling arrays. Science 306,2242–2246. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ; ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Kozak ML, Kim JM, Wong N, López-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. (2012). BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell 46, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Jain N, Otte GL, Neems D, Wiesmüller L, Berger SL, and Goldman RD (2015). Role of lamin b1 in chromatin instability. Mol. Cell. Biol. 35, 884–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, and Wong LH (2012). Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 40, 11178–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, and Yates JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Esteller M (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, and Verma IM (2012). Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338, 1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SIS, and Klar AJS (1997). A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146, 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, and Almouzni G (2004). Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. (2015). BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE, and Willard HF (2012). Composition and organization of active centromere sequences in complex genomes. BMC Genomics 13, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, and Willard HF (2013). Sequences associated with centromere competency in the human genome. Mol. Cell. Biol. 33, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W, and Altenburger W (1981). Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 9, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, and Pombo A (1998). Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140,1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, and Miklos GL (1979). Functional aspects of satellite DNA and heterochromatin. Int. Rev. Cytol. 58, 1–114. [DOI] [PubMed] [Google Scholar]
- Joukov V, Walter JC, and De Nicolo A (2014). The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol. Cell 55, 578–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Marks JH, and Mandell JB; New York Breast Cancer Study Group (2003). Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Brock A, and Ingber DE (2013). Intraductal injection for localized drug delivery to the mouse mammary gland. J. Vis. Exp. 80, 50692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, Sen GC, Komarova EA, and Gudkov AV (2013). p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl. Acad. Sci. USA 110, E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KH, Abou El Hassan M, and Bremner R (2013). A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques 55, 204–206. [DOI] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, and Yates JR 3rd (2004). MS1, MS2, and SQT-three unifed, compact, and easily parsed fle formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168. [DOI] [PubMed] [Google Scholar]
- Miga KH, Eisenhart C, and Kent WJ (2015). Utilizing mapping targets of sequences underrepresented in the reference assembly to reduce false positive alignments. Nucleic Acids Res. 43, e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos GL, and John B (1979). Heterochromatin and satellite DNA in man: properties and prospects. Am. J. Hum. Genet. 31, 264–280. [PMC free article] [PubMed] [Google Scholar]
- Morris KV, and Mattick JS (2014). The rise of regulatory RNA. Nat. Rev. Genet. 15, 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, and Harrow J (2015). Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome 26, 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. (2016). Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54, advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet ER, and Hallett MT (2014). Absolute assignment of breast cancer intrinsic molecular subtype. J. Natl. Cancer Inst. 107, 357. [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao GM, Zhu Q, Perez-Garcia CG, Chou SJ, Suh H, Gage FH, O’Leary DD, and Verma IM (2014). Role of BRCA1 in brain development. Proc Natl Acad Sci USA 111, E1240–E1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, Feunteun J, and Livingston DM (2011). BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell 44, 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, Su Y, Ting DT, Polyak K, Richardson AL, et al. (2014). BRCA1 haploin-sufficiency for replication stress suppression in primary cells. Nat. Commun. 5, 5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, and Gygi SP (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50. [DOI] [PubMed] [Google Scholar]
- Probst AV, and Almouzni G (2008). Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation 76,15–23. [DOI] [PubMed] [Google Scholar]
- Probst AV, and Almouzni G (2011). Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 27, 177–185. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, and Chang HY (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, and Grewal SI (2012). Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr. Opin. Genet. Dev. 22, 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, and Chang HY (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, and Jasin M (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, and Jasin M (2012). A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA½. Cancer Cell 22, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, and Cortez D (2011). Analysisofprotein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, and Yates JR 3rd (2002). DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, and Chua KF (2016). SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 23, 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, and Martienssen RA (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, Deng C, and Scully R (2014). BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature 510, 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Liu YL, Xie Y, Zhu W, Guerra F, Shen S, Yeddula N, Fischer W, Low W, Zhou X, et al. (2014). A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin. Sci. Transl. Med. 6, 263ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V,Tijsterman M, and Gasser SM (2016). Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48, 1385–1395. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, and Reed R (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, and Verma IM (2011). BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


