Abstract
Metastasis is a complex process and a major contributor of death in cancer patients. Metastasis suppressor genes are identified by their ability to inhibit metastasis at a secondary site without affecting the growth of primary tumor. In this review, we have conducted a survey of the metastasis suppressor literature to identify common downstream pathways. The metastasis suppressor genes mechanistically target MAPK, G-protein-coupled receptor, cell adhesion, cytoskeletal, transcriptional regulatory, and metastasis susceptibility pathways. The majority of the metastasis suppressor genes are functionally multifactorial, inhibiting metastasis at multiple points in the cascade, and many operate in a context-dependent fashion. A greater understanding of common pathways/molecules targeted by metastasis suppressor could improve metastasis treatment strategies.
INTRODUCTION
Despite advances in diagnosis, surgery, radio- and chemotherapy, a strong need remains for improvements in metastasis-related cancer mortality.1,2 Most therapies address fundamental oncogenic events that are present in a metastasis, but an alternative is to address the metastatic process itself.
Clinical and translational advances pertaining to the metastatic process may be thwarted by our poor understanding of the molecular and biochemical mechanisms involved in the process of metastasis. Metastasis is a complex multistep sequential process involving invasion of cancer cells, intravasation, circulation, arrest in the capillary bed of secondary organ, extravasation, and colonization at the secondary site. Apart from being complex, metastasis is also highly inefficient. Experimental studies have shown that, among all the cancer cells injected into the circulation, very few (~ 0.01%) have the capacity to successfully metastasize to distant organ.3,4 This inefficiency in metastatic ability has been attributed to crucial rate-limiting steps of metastasis namely the host immune system in the circulation, extra-vasation, and post-extravasation survival in colonization.4–7Cells that metastasize to secondary sites may not necessarily proliferate immediately and could remain dormant for a long period, until the microenvironment at secondary site becomes congenial for its proliferation.8,9
Characterization of pathways/genes/proteins that specifically affect metastasis, metastasis suppressor or metastasis promoters, would provide insights into the molecular mechanisms of metastasis and could launch clinical- translational initiatives. This review will focus on the metastasis suppressor gene field, exploring the mechanistic pathways involved and suggested translational sequelae.
METASTASIS SUPPRESSORS AND THEIR FUNCTIONAL IDENTIFICATION
Like tumorigenesis, both stimulatory and inhibitory pathways regulate metastasis. Some inhibitory pathways are pertinent to both tumor growth and spread; the metastasis suppressor gene field has limited itself to those genes that are specific to the metastatic process. Metastasis suppressors are a class of genes which, when overexpressed (or re-expressed), cause inhibition of cancer metastasis to distant organs without affecting the size of the primary tumor. Because no in vitro assays exist for metastasis, credentialing of a new suppressor necessarily involves in vivo experiments. The gold standard is spontaneous metastasis assays, where a primary tumor forms and seeds out metastases; experimental (eg hematogenous) metastasis assays have been used in concert with separate primary tumor studies.
Metastasis suppressor genes have been identified in many ways, but often show reduced expression in highly metastatic cells compared to poorly or non-metastatic tumorigenic cells.10 The first metastasis suppressor gene, NM23 (NME), was identified by its overexpression in murine K-1735 melanoma lines with poor metastatic potential as compared with related lines of high metastatic capacity.11 Several techniques including microcell-mediated chromosome transfer, differential display, subtractive hybridization, microarrays, RNAseq and serial analysis of gene expression have been employed to identify genes or chromosomal regions which suppress metastasis.12 In addition to genes, multiple types of RNAs have been demonstrated to have metastasis suppressor functions.
Metastasis suppressors can act by inhibiting one or more steps in the metastatic cascade and regulate a wide range of biochemical signaling pathways.13 Most have been found to be active in several biochemical pathways, suggesting that exerting multiple control points on a genomically unstable cell population may be advantageous. Apart from inhibiting metastasis, metastasis suppressors may have a ‘normal’ function in the induction or maintenance of development and differentiation pathways.14–17 Examples include Raf kinase inhibitory protein (RKIP) (PEBP), which controls pulmonary airway tube formation in Drosophila, and brain, pancreas and retinal development in the mouse.18,19 NM23 controls the differentiation of presumptive adult tissue in the imaginal discs of Drosophila, and the knockout mouse has a mammary differentiation defect preventing nursing.20–22 The MKK4 (mitogen-activated protein kinase kinase 4) suppressor gene, as in mammalian cells, controls P38 and JNK-based responses to cell stress in Drosophila development, altering mitosis and motility.23 In contrast to their function in development and differentiation, where pathways are turned on and off in a controlled manner, these pathways operate in the instability of cancer cells as metastasis suppressors.
PATHWAYS TARGETED BY METASTASIS SUPPRESSOR GENES
We have categorized the metastasis suppressors genes into different pathways based on their predominant known mode of action. As will become apparent, few have only one biochemical function. An overview of metastasis suppressor pathways is listed in Table 1. A literature search reveals that the mitogen-activated protein kinase (MAPK) and cytoskeletal signaling pathways are the most common activities of metastasis suppressors: Whether that means that these pathways predominate biologically, or are just the best studied, remains an open question. Certainly, control of gene expression and other pathways may be as important, but less studied and more intricate.
Table 1.
A list of bona fide metastasis suppressor genes
| Pathways targeted | Genes | Tumor type/site | Mech. of action | Type of metastasis | |
|---|---|---|---|---|---|
| MAPK pathway | NM2337 | Breast | 1-Phosphorylation of KSR leading to reduced ERK1/2 activation | Reduces both experimental & spontaneous metastases | |
| Melanoma163–165 | |||||
| 2-LPA signaling54 | |||||
| Colon166,167 | |||||
| Oral36,168 | |||||
| RKIP60 | Prostate | Inhibits RAF mediated MAP/ERK kinase phosphorylation | Reduces spontaneous metastases | ||
| MKK466,70,71,72 | Prostate, ovarian cancer | MAP2K activates p38 and/or JNK | Reduces spontaneous metastases | ||
| MKK666 | Ovarian cancer | Activates MAPK p38 | Reduces experimental metastases | ||
| MKK772 | Prostate cancer | Activates MAPK JNK | Reduces spontaneous metastases | ||
| Adhesion proteins | KAI1169–171 | Prostate | Cell adhesion and Integrin signaling | Reduces spontaneous metastases | |
| CD4489,172,173,174,175 | Prostate | Affects cellular adhesion by hyaluronic acid and osteopontin signaling | Reduces spontaneous metastasis | ||
| E-cadherin176,177,178,179 | Colon, pancreatic and ovarian cancer | Ca2+ dependent cell adhesion | Reduces spontaneous metastases | ||
| Cytoskeletal signaling | Gelsolin106 | Melanoma | Actin-regulatory protein helping in actin polymerization | Reduces spontaneous metastasis | |
| Rho-Rac pathway: RhoGDI2109 | Bladder cancer | Rho-Rac signaling | Reduces experimental metastasis | ||
| DLC-1111 | Breast cancer | Rho GTPase activating protein which affects cytoskeletal remodeling | Reduces spontaneous metastasis | ||
| G-protein-coupled receptors pathway | KISS1114,115 | Melanoma | Cytoskeletal reorganization | Reduces both experimental and spontaneous metastases | |
| Breast116 | |||||
| Lungs | |||||
| Protein kinase A/C pathway: SSeCKS/Gravin/AKAP12120,121,180 | Prostate cancer | PKA-mediated activation of Src kinase | Reduces spontaneous metastasis | ||
| Apoptotic pathway | Caspase-8123,181,182 | Neuroblastoma | Increases integrin-mediated cell death | Reduces spontaneous metastasis | |
| GAS1124,183 | Melanoma | Induces apoptosis through caspases 3 and 9 | Reduces spontaneous metastasis | ||
| Transcriptional complexes | BRMS1184,185 | Melanoma, ovarian cancer | Chromatin remodeling (polyubiquitinated p300, a histone acetyltransferase) and PI3K signaling | Reduces spontaneous metastasis | |
| MYC141 | Breast cancer | Transcriptional silencing of αv and β3 integrin subunits | Reduces spontaneous metastasis | ||
| Anti-metastatic miRNAs (Metastamirs) | miR-146a/b147,186 | Breast cancer | Reduces NFκB and EGFR pathway | Reduces experimental metastasis | |
| miR-206 | Breast cancer | Target transcription factor S0X4 and extracellular matrix component tenascin C | Reduces experimental metastasis | ||
| miR-335148 | |||||
| Long non-coding RNA (lncRNA) | LncRNA LET153,154 | Hepatocellular carcinoma and colon carcinoma | Binds to NF90 and causes its degradation | Reduces experimental metastasis | |
| NKILA (NFκB-activating lncRNA)155,156 | Breast cancer | Negative regulator of NFκB signaling | Reduces experimental metastasis | ||
| Metastasis susceptibility genes | Cadm1160 | Breast cancer | Sensitizes tumor cells to immune-surveillance mechanisms by CD8+ T cells | Reduces spontaneous metastasis | |
MAPK Pathway
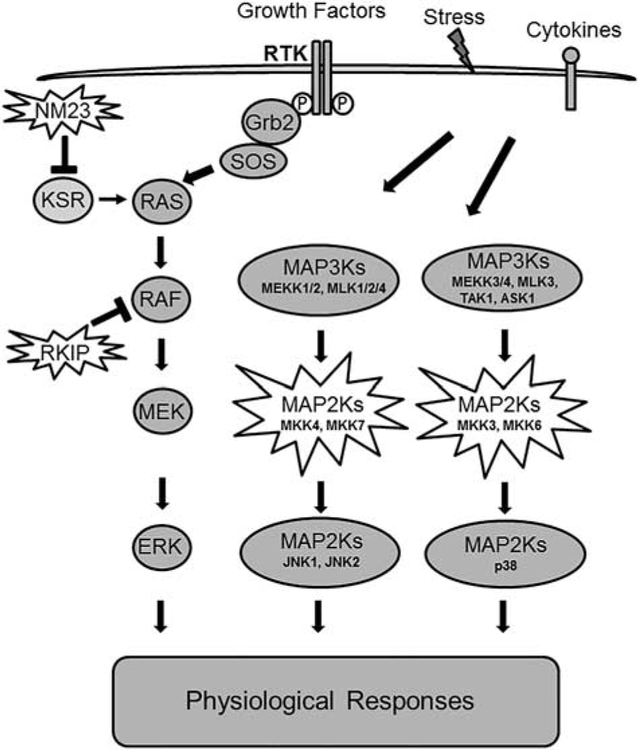
This signaling cascade starts with the binding of an extracellular mitogenic ligand to the membrane receptor (ie EGFR, PDGFR) leading to activation of Ras (a GTPase) which activates series of MAPKs via MAP3K, MAP2K, MAPK and finally activation of transcription factors (Figure 1). In simplistic terms, the ERK subpathway is thought to regulate mitosis, while the JNK and P38 subpathways regulate stress responses. The ERK–MAPK signaling plays a crucial role in cancer and metastasis. Studies on breast carcinomas, and head and neck squamous-cell carcinoma have reported a positive correlation between activated ERK expression, metastases and patient survival.24 Similarly, in studies using ERK inhibitors (PD 098059) or site-directed mutagenesis, ERK activation was functionally associated with in vivo metastasis,25,26 in vitromotility and invasion27,28 and the epithelial–mesenchymal transition (EMT).29 Metastasis suppressors with known ties to the ERK pathway are listed below.
Figure 1.
Mitogen-activated protein kinases (MAPK) pathway highlighting metastasis suppressor targeting sites.
NM23 (NME/NM23H1)
NM23 was the first metastasis suppressor identified, which showed reduced expression (by differential hybridization) in highly metastatic compared to poorly metastatic K-1735 murine melanoma cell lines. As a metastasis suppressor, its enforced expression suppressed metastasis without altering tumor growth in variety of cancer cell lines of different origins.30 Like its in vivo metastasis-inhibitory phenotype, overexpression of NM23 in several carcinoma cells (breast, melanoma, etc) reduced in vitro motility of cells to numerous chemoattractants and inhibited colony formation in soft agar, without affecting the cellular proliferation.31 In humans NM23/NDPK family consists of 10 members (NM23H1-H10) including NM23H1 and NM23H2 which share 88% homology.32 Like NM23H1, a variety of metastasis model systems have demonstrated NM23H2 as a metastasis suppressor gene and its expression levels inversely correlate with metastatic potential.33–36 NM23 overexpression in MDA-MB-435 breast carcinoma cells reduced ERK–MAPK activation levels compared to vector control.37,38
Biochemically, NM23 is known to have nucleoside diphosphate kinase (NDPK) activity,16,39–41 Histidine protein kinase activity (HPK)42,43 and a 5′–3′ exonuclease activity.44As an NDPK, it catalyzes the transphosphorylation of the γ-phosphate of a nucleoside triphosphate to a nucleoside diphosphate with a phosphorylated-histidine (H118) intermediate. As an HPK, the same phosphorylated-histidine 118 NM23 intermediate transfers a phosphate to a protein substrate, a histidine in all NME2 reported pathways but a histidine or serine in some NME1 pathways. Developmental studies in Drosophila demonstrated that the NDPK activity was necessary but not sufficient for normal differentiation, and roles for subcellular provision of nucleotides have been postulated.45 The motility-suppressive function of NM23 is well correlated with its HPK activity in site-directed mutagenesis experiments.46,47 The physiological substrates for HPK activity potentially leading to NM23-mediated metastasis suppression are incompletely known. Among several known HPK substrates of NM23, phosphorylation of kinase suppressor of ras (KSR) by NM23 is known to inhibit ERK–MAPK pathway (Figure 1).37,38,48 In this study, NM23phosphorylated KSR-serine (392) and altered its scaffold function, possibly by altering the docking of proteins and/or by altering KSR intracellular localization, leading to reduced ERK activation. Independent publications have reported a protein–protein interaction of NM23 and KSR affecting Caenorhabditis elegans development,49 and a functional NM23–KSR interaction regulating ERK activation was reported in HEK293 cancer cells.50 Similarly, tumor cells expressing an NM23 histidine-kinase-deficient mutant (P96S) showed high levels of ERK activation comparable to vector control.38 Therefore, it is very likely that differential expression of NM23 in metastatic vs non-metastatic tumor cells might impact ERK activation via KSR scaffold protein which could, at least in part, mediate NM23 actions.46,51
The mechanism of NM23 action leading to metastasis suppression is likely to be multifactorial. Apart from activities described above, NM23 binding proteins, which titer out ‘free NM23’ or reduce the titer of metastasis-promoting proteins (such as EBV), could lead to inhibition of its ability to suppress metastasis;52,53 altered gene expression of downstream targets (lysophosphatidic acid receptor LPA1/EDG2)54is potential candidates which has shown promising results for mediating NM23 activities, and recent observations on endocytic trafficking are likely contributory.20,21,45
Raf kinase inhibitory protein
RKIP binds Raf-1 and is an inhibitor of Raf -stimulated ERK– MAPK signaling (Figure 1);55 it is also a member of the phosphoethanolamine-binding protein (PEBP) family. RKIP inhibition is isoform specific and only works on Raf-1, and not on B-Raf.56 RKIP was identified as a metastasis suppressor in a screen comparing metastatic prostate cancer cell lines to non-metastatic cells.57 It has been proven as functional metastasis suppressor in multiple solid tumor types such as prostate and triple-negative breast cancers.57,58 Generally,RKIP expression is lost in poor-prognosis tumors.59 Studies using both in vitro and in vivo breast cancer models have demonstrated that expression of RKIP blocks multiple steps of the metastasis including angiogenesis, local invasion, intravasation, and colonization.57,60 Enforced expression of RKIP reduced the invasive potential of breast cancer cells without affecting their growth.57 Similarly, in a xenograft model its overexpression blocked intravasation, extravasation as well as colonization of tumor cells.
In addition to its RAF inhibitory action, RKIP has other biochemical activities which contribute to metastasis suppression. These include binding to and inhibition of signal transducer and activator of transcription 3 (STAT3),61binding to G-protein-coupled receptor kinase(GRK2),62MDA-9/Syntenin,63 centrosomes and kinetochores.64
These examples bring up a remaining question of the interaction between an antiproliferative pathway for ERK and a metastasis pathway independent of primary tumor growth. Multiple studies have indicated that tumor growth in a primary site is not necessarily the same as growth in a distant site,65 suggesting that context-dependent activation of a proliferative pathway may be key. Alternatively, ‘other’ functions of these suppressors may impact colonization by distinct pathways. Colonization is certainly more than just proliferation, involving stem cell function, viability pathways, stress responses, and immune evasion for example.
The stress activated protein kinase (SAPK) portion of the MAPK pathway terminates in P38 and JNK activation. The SAPK signaling pathways are generally activated by a variety of environmental stresses including pH changes, UV irradiation, hypoxia, cytokines or growth factor deprivation and treatment with chemotherapeutic agents.66 Intuitively, it is easy to envision a tumor cell arriving in a foreign tissue as being stressed, and lack of an adaptive response would be detrimental to colonization. P38 has also been functionally implicated in metastatic dormancy, which is a form of metastasis suppression.67–69 Other contributions of the SAPK, for instance in immune regulation, may also be functionally important.
Mitogen-activated protein kinase kinase 4
MKK4 is a member of the MAP kinase kinase family which directly phosphorylates and activates JNK or P38 in the SAPK. Human prostate cancer primary tumors with higher Gleason grade, and metastatic ovarian cancer patients, showed reduced expression of MKK4. MKK4 acts as a functional metastasis suppressor wherein its ectopic expression in highly metastatic cells suppressed metastasis significantly (80–90%) in both ovarian and prostate cancer cells.70,71 Interestingly, it was observed that for suppressing metastasis in ovarian cancer models, MKK4 signaled through the p38 arm,66 whereas in prostate cancer models, MKK4 signaled through the JNK arm.72
Mitogen-activated protein kinase kinase 6/7
Mitogen-activated protein kinase kinase 6/7 (MKK 6 and 7) play a central role in the SAPK signaling pathway. MKKs 6 and 7 occupy slightly different positions in the MAPK cascade: In response to cellular stress generally MAPKKK is activated which can, in turn, activate MKK4, MKK7, or MKK3/MKK6. Activation of MKK3, MKK6, and/or MKK4 (but not of MKK7) can lead to phosphorylation of all the four members of the mammalian p38 MAPK family members (p38α, p38β, p38γ, and p38δ).73 Activation of p38 in metastatic cells is required for its survival in dormant state at the secondary site.74,75 It is speculated that activation of p38-induced dormancy resulted in an unfolded protein response, which is an adaptive pathway that might help the survival of dormant tumor cells at the secondary site.67 However, depending on the model system and the isoform involved, MKK data show inconsistencies in literature. In an ovarian cancer model, MKK6 overexpression suppressed metastatic colonization whereas MKK7 had no effect. However, in prostate cancer ectopic expression of the MKK7 (JNK-specific kinase) suppresses formation of overt metastases, whereas the p38-specific kinase MKK6 had no effect.66,72,76 The inconsistency of MKK4, 6, 7 effects in the various models may indicate the importance of the pathway, but redundant mechanisms for its traversal. Alternatively, do the MKK proteins serve other, non-MAPK functions? It is noteworthy, however, that the SAPK pathway has been widely reported as an oncogene,77,78 and that its function may be context dependent.
Adhesion Proteins
Adhesion proteins modulate cell:cell and cell:extracellular matrix interactions, with resultant effects on proliferation, survival, motility, immune function, and other aspects of metastasis. Multiple adhesion molecules have been credentialed as metastasis suppressors.
KAI1
KAI1/CD82 is a member of tetraspanin family, which is characterized by its four transmembrane and two extracellular domains. It was first identified in T-cell activation study;79however, its role as a metastasis suppressor was highlighted later in a somatic cell hybridization of highly metastatic and non-metastatic rat prostate cancer cells.80 Decreased expression of KAI1 has been widely correlated with an aggressive cancer phenotype in several cancer types, including pancreatic, hepatocellular, bladder, breast, and non-small cell lung cancers.81–83 Similarly, in breast cancer cells its over-expression led to poor colonization in soft agar, reduced invasion, and significantly suppressed both spontaneous and experimental metastasis.84 KAI functions as an adaptor protein that forms oligomeric complexes with its binding partners, including integrins, EGFR, GTPases, and other cell adhesion molecules. Since the MAPK pathway lies downstream of some of these targets, it may be functionally involved. KAI1-mediated metastasis suppression has also been linked to abrogation of EGFR and integrin signaling, possibly by increasing their endocytosis.85 This hypothesis is also supported by the presence of internalization consensus sequence in KAI1, which is unique in the tetraspanin family.86
CD44
CD44 is a transmembrane glycoprotein involved in cell–cell and cell–matrix interactions and acts as a receptor for the extracellular matrix components hyaluronic acid87 and osteopontin.88 CD44, like a metastasis suppressor gene, has downregulated mRNA and protein expression in highly metastatic prostate cancer.89 Similarly, overexpression of CD44 (Mr 85 000, standard form) in highly metastatic AT3.1 rat prostatic cells suppressed their metastatic ability to the lungs without affecting their in vivo growth rate or tumorigenicity.89 CD44 exists in multiple isoforms, some of which have been demonstrated to increase metastasis.90 Switching of CD44 isoforms through cancer progression has been reported.91 The range of CD44 isoforms and their contributions to metastasis remain incompletely understood. The role(s) of CD44 in metastasis suppression are likely multifactorial and can involve immune function,92 interactions with receptor tyrosine kinase signaling,93 cytoskeletal changes,94,95 particularly in response to hyaluronic acid, stem cell function, TGF-β function,96 etc. The CD44–hyaluronic acid pathway is the subject of multiple translational initiatives.
E-Cadherin
Cadherins are transmembrane glycoproteins that form Ca+2 dependent homotypic complexes and mediate cell–cell interactions mainly in epithelial cells. There are 20 members of cadherin family, which show tissue-specific expression patterns.97 Loss of E-cadherin in a variety of tumor cell types is associated with the EMT and correlates with increased invasion and metastasis.98 Re-expression of E-cadherin in highly invasive epithelial tumors suppressed the invasive potential of these cells.99 Similarly, high E-cadherin expression in primary tumor inhibits shedding of tumor cells from the primary tumor,100,101 highlighting a metastasis suppressor role. E-cadherin modulation of metastasis has been reported,98,102,103 with the caveat that tumor suppressive roles have also been reported;104 metastasis modulation may involve adhesive or other signaling events.
Cytoskeletal Signaling
Cellular migration and other cellular deformability events in response to microenvironmental cues are guided by a dynamic cytoskeleton. Multiple proteins in traditional motility/invasion signaling pathways have been credentialed as metastasis suppressors, affecting the mechanics of ratcheting a cell forward. Less clear in the literature is the contribution of cytoskeletal pathways to growth in a distant site, metastatic colonization. Cytoskeletal pathways may affect viability (such as the anoikis process), immune intravasation, mechanical signaling from environmental stiffness, and other aspects which need further study. These end points may be as useful as motility in translating this field toward clinical application. Multiple molecules acting at different level of cytoskeletal signaling have been credentialed as metastasis suppressors.
Gelsolin
Gelsolin is an actin-binding protein that regulates cell motility by severing actin. Gelsolin also regulates cell morphology, proliferation, and apoptosis. Studies on Gelsolin expression have highlighted its reduced expression in several cancer types, including breast, colon, ovary, and prostate cancer. Overexpression and knockdown studies on Gelsolin have highlighted its role in inhibiting the EMT in breast cancer,105and as a suppressor of metastasis in B16 melanoma cells.106 Gelsolin functions as a switch that controls E- and N-cadherin conversion, that is, knockdown of gelsolin causes a reduction in E-cadherin and enhanced expression of N-cadherin via a transcription factor, Snail.
RhoGDI
The Rho/Rac proteins constitute a subgroup of the Ras superfamily of small GTP hydrolases which were originally implicated in the control of cytoskeletal events. However, it is also known to regulate diverse cellular functions, including cell polarity, vesicular trafficking, cell cycle, transcription, and proliferation.107 Upon activation, it switches from an inactive GDP-bound form to an active GTP-bound form which can interact with a plethora of different effectors mediating its different cellular functions.107 Rho GTPases are known to contribute to most steps of cancer initiation and progression, including proliferation potential, survival and evasion from apoptosis, angiogenesis, tissue invasion, and the establishment of metastases. RhoGDI stabilizes the GDP-bound form of Rho and sequesters it in an inactive, non-membrane localized, cytoplasmic compartment, inactivating Rho–Rac signaling.108RhoGDI2 expression was diminished as a function of primary tumor stage and grade in human bladder and in several other cancers.109 Enforced expression of RhoGDI2, into cells having reduced expression with metastatic ability, suppressed experimental lung metastasis without affecting in vitro growth, colony formation, and in vivo tumorigenic potential.109 RhoGDI2 also affects gene expression by an incompletely characterized mechanism, which may also contribute to metastasis suppression.110
DLC-1
When overexpressed in sublines of the MDA-MB-435 breast carcinoma, DLC-1 resulted in reduced migration, invasion, and significantly suppressed pulmonary metastases.111 DLC-1 is a GTPase activating proteins (GAP) important for switching the active GTP-bound state to the inactive GDP-bound state of Rho proteins.112 A caveat is that, DLC-1 is also known to have potential tumor suppressor activity in hepatocellular carcinoma.113
G-Protein-Coupled Receptors
G-protein-coupled receptors (GPCRs) constitute a large class of druggable signaling proteins with a plethora of downstream activities. GPCR family members contains a typical seven transmembrane α-helical region. They detect many extra-cellular signals and transduce them to heterotrimeric G proteins, which further carry these signals to intracellular effectors (cAMP, PKA, PI3K, etc), and thereby play an important role in various cellular responses.
KISS1
KISS1 was identified as a metastasis suppressor gene by subtractive hybridization on metastatic melanoma cells (C8161) with and without human chromosome 6.114,115 Overexpression of KISS1 inhibited metastases formed by breast and melanoma cells, without altering the size of primary tumors.116 The mechanism of action of KISS1 took an unexpected turn when it was reported to encode a secreted neuropeptide (designated as Metastin and Kisspeptin) for an orphan G-protein-coupled receptor (hGPR54).117,118 Mela-noma cells overexpressing hGPCR54 developed reduced metastases upon treatment with Metastin, while in in vitro assays it inhibited chemotaxis and invasion. These ligands (Metastin, Kisspeptin) stimulate PIP2 hydrolysis, Ca2+ mobilization, arachidonic acid release, ERK1/2 and p38 MAP kinase phosphorylation and stress fiber formation, leading to reduced cellular proliferation.118 In another study, constitutive upregulation of KISS1 in HT10810 cells resulted in decreased nuclear factor-κB (NFκB) activation, which in turn led to reduced MMP9 transcription and resultant changes in invasion.
Src-suppressed C kinase substrate
Src-suppressed C kinase substrate (SSeCKS) is a scaffolding protein for many important signaling intermediates. It has metastasis suppressive activity (complicated by reports of tumor suppressive activity). In studies of motility, loss of SSeCKs expression increased chemotactic velocity and linear directionality, correlating with replacement of leading edge lamellipodia with fascin-enriched filopodia-like extensions, the formation of longitudinal F-actin stress fibers reaching to filopodial tips, relative enrichments at the leading edge of phosphatidylinositol (3,4,5)P3 (PIP3), Akt, PKC-zeta, Cdc42-GTP and active Src (SrcpoY416), and a loss of Rac1.119 SSeCKs is also thought to be a scaffold for protein kinase A, with resultant effects on angiogenesis120 and a scaffold protein for protein kinase C, with downstream effects on the ERK pathway.121,122
Apoptotic Pathway
Apoptosis is defined as a programmed cell death mediated by intrinsic or extrinsic pathways. The intrinsic pathway is activated predominantly by intracellular stimuli, including DNA damage, oxidative stress and growth factor deprivation, and involves formation of apoptosome (comprised of procaspase-9, Apaf-1, and cytochrome c). The extrinsic pathway of apoptosis is initiated by the binding of death ligands (Fas ligands) to death receptors which leads to formation of death-inducing signaling complex (DISC). Following this, DISC can either directly induce cell death by activating effector caspases (caspase-3, −6, and −7) or cleave the Bcl-2 family member Bid, to activate the mitochondria-mediated intrinsic apoptotic pathway.
Caspase-8
Caspase-8 is a member of the cysteine–aspartic acid protease (caspase) family that is an effector of apoptosis. In neuroblastoma, greater metastases were observed in caspase-8-deficient tumors, whereas tumors expressing caspase-8 have less frequent metastases, highlighting caspase-8 role as a metastasis suppressor.123 Interestingly, caspase-8-deficient cells showed no difference in tumor formation and growth in vivo. It was observed that caspase-8 expression was generally associated with increased apoptosis in highly invasive tumor cells,123 suggesting a specific linkage of apoptotic and invasion pathways.
Growth Arrest Specific 1
Growth Arrest Specific 1 (GAS1) was isolated as a metastasis suppressor gene by a genome-wide RNAi screening in mouse melanoma cells.124 Overexpression of GAS1 reduced spontaneous metastasis in melanoma cells by promoting apoptosis in the disseminated cancer cells at the secondary site.124 Interestingly, expression of GAS1 could predict metastasis recurrence in stage II and III colorectal carcinoma.125 Recent data on GAS1 mechanism of action highlight its independent roles in suppressing aerobic glycolysis and AMPK/mTOR/p70S6K signaling.126
Transcriptional Complexes
Given the many intricate functions involved in the metastatic process, it could be expected that suppressors could exert multiple points of control, either through altered protein function or altered expression of proteins. Several examples of metastasis suppressors that act as transcriptional regulators have been reported.
Breast cancer metastasis suppressor 1
Breast cancer metastasis suppressor 1 (BRMS1) was metastasis suppressive in MDA-MB-231 and MDA-MB-435 breast cancer cells127 without affecting the cancer cells growth in vitro or in vivo. Subsequently, BRMS1 metastasis suppression was also demonstrated in ovarian cancer, melanoma, non-small cell lung cancer, and bladder cancer.128 BRMS1affects several key events of metastasis, including inhibition of migration, invasion, and initiation of growth by single cells, and promotion of anoikis.129,130 BRMS1 overexpression alsorestore homotypic and heterotypic gap junction’s intercellular communication.131 A role for BRMS1 in transcriptional regulation was proposed by its identification in the mSin3 histone deacetylase complex, with a functional readout of transcriptional repression.132 In support of this hypothesis, the nuclear localization signal within BRMS1 was found to be necessary for metastasis suppression.133 BRMS1 also poly ubiquitinated p300, a histone acetyltransferase, and this activity was essential to its metastasis suppressor function.134
MYC
MYC is a pleiotropic transcription factor that participates in the regulation of a wide variety of genes known to be involved in cell cycle, growth, apoptosis, cell adhesion, and genomic stability.135–137 MYC overexpression is widely known to transform cells and these cells can form tumors in experimental animals.138,139 Its overexpression has been reported in several cancer types.140 A complete paradigm shift occurred when it was reported that overexpression of MYC stimulated proliferation of breast cancer cells (both in vitro and in vivo), but suppressed motility and invasiveness in culture.141 In xenografted tumors, MYC overexpression inhibited metastases to liver and lungs. Both the above phenotypes have been attributed to MYC-mediated repression of αvβ3 integrin transcription.141 In an independent study, genetic or pharmacological inhibition of MYC synergized to increase TGF-β induced metastasis.142 A role for MYC control of protein translation is also implicated in its suppressive role.143 MYC expression has been reported as a favorable prognostic factor.144 MYC metastasis suppression is likely context dependent.145,146 This type of data provides a strong cautionary tale for the advancement of translational projects without metastasis data.142
Anti-Metastatic miRNAs (Metastamirs)
miRNAs which are associated with the regulation of metastasis (pro or anti) are commonly referred to as Metastamirs/metastasis-associated miRNAs. Metastamirs are generally identified using in vitro screens for several of the metastatic cascade steps including EMT, adhesion, migration, invasion, apoptosis, and/or angiogenesis. Metastasis-suppressing Metastamirs include:
miR-146a/b
miR-146 was identified as a molecule responsible for the BRMS1 mediated metastasis suppression. Its expression is altered by BRMS1 and both are associated with decreased signaling through the NFκB pathway. Transduction of miR-146a/b into MDA-MB-231 cells downregulated expression of EGFR and inhibited invasion, migration, and significantly suppressed experimental lung metastasis.147
miR-335 and -206
In a search for microRNAs whose expression was gradually lost with increasing metastatic potential in human breast cancer cells, these miRNAs were identified. Re-expression of these microRNAs in malignant cells suppressed lung and bone metastasis without affecting the size of primary tumor. miR-335-mediated suppression of metastasis targeted the progenitor cell transcription factor SOX4 and extracellular matrix component tenascin C.148
The molecular identification of Metastamirs is in a nascent stage. With other miRNA projects, metastamirs have potential therapeutic implications through direct or indirect delivery.
Long Non-Coding RNA (lncRNA)
LncRNAs are long transcribed RNA molecules (greater than 200 nucleotides) and they do not have protein-coding capacity. They are transcribed by RNA pol II, are generally capped, polyadenylated, and frequently spliced.149 LncRNAs are known to function by their interactions with proteins, RNAs, and DNAs.150 They can also function as a transcriptional guide (recruitment of chromatin-modifying enzymes),151 a decoy (miR sponges)149 and as a scaffold (stabilization of ribonucleoprotein complexes).152
LncRNA LET
Its ectopic expression in hepatocellular carcinoma cells (SMMC-7721 and HCCLM3) and colon carcinoma (SW480) cells resulted in inhibition of lung colonization after tail vein injection.153 LncRNA LET has been proposed to work by binding to NF90 (a double-stranded RNA binding protein), resulting in ubiquitination and degradation of NF90.154
NFκB-activating lncRNA
NFκB-activating lncRNA (NKILA) is an inhibitor of metastasis in both in vitro and in vivo model systems and is reported to work by a protein–RNA interaction. It was identified as an lncRNA induced by NFκB-signaling activating inflammatory cytokines (TNFα and IL-1β) in breast cancer cells and acts as a negative regulator of NFκB signaling.155,156 NKILA shows low expression in breast carcinomas (without distant or regional lymph node metastasis) and its expression was further reduced in metastasis and predicted a poor clinical outcome.155 Ectopic expression of NKILA in MDA-MB-231 cells reduces invasion and in xenografts model inhibited metastasis to the lungs, liver, and lymph nodes and prolonged survival.155
Metastasis Susceptibility Genes
These refer to group of genes that are associated with inherited susceptibility for the development of metastatic lesions.157,158 Metastasis susceptibility genes display a germ-line polymorphism which modifies tumor cell metastatic capability, indicating that heritable genetic variability can predetermine a tumor cell’s propensity to metastasize. These genes were identified using a complex genetics screen that exploits the differential heritable metastatic susceptibility observed among different strains of inbred mice.159
Cadm1
Among several, Cadm1 is a validated member of the metastatic susceptibility genes group, overexpression of which specifically suppress metastasis without any significant difference in primary tumor growth. The molecular mechanism of Cadm1 involves sensitization of tumor cells to immune-surveillance mechanisms by CD8+ T cells.160
Conclusions
We have attempted to survey the metastasis suppressor literature for evidence of their efficacy and underlying mechanisms. The very incomplete nature of the data available may stem from the difficulty of performing metastasis experiments, with no single in vitro assay. Based on the available literature, it can be inferred that majority of the metastasis suppressor effects are multifactorial and some may show their effect in context-dependent fashions. It is likely that, with continued gene expression and mutation profiling, additional candidate metastasis suppressors will be identified and validated.
Clinical application of metastasis suppressors is in nascent stage. It is currently proposed as a prognostic marker which can predict better or worse outcome based on recurrence.161,162 However, it is anticipated that, with the discovery of detailed mechanisms of action, new metastasis suppressor genes, translational approaches to enhance their activity, and better clinical trials design to target metastasis, the field can be of direct therapeutic importance.
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer 2005;5:591–602. [DOI] [PubMed] [Google Scholar]
- 2.Brabletz T, Lyden D, Steeg PS, et al. Roadblocks to translational advances on metastasis research. Nat Med 2013;19:1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss L Metastatic inefficiency. Adv Cancer Res 1990;54:159–211. [DOI] [PubMed] [Google Scholar]
- 4.Fidler IJ. Metastasis: quantitative analysis of distribution and fate of tumor emboli labeled with 125 I-5-iodo-2’-deoxyuridine. J Natl Cancer Inst 1970;45:773–782. [PubMed] [Google Scholar]
- 5.Hanna N Role of natural killer cells in control of cancer metastasis. Cancer Metastasis Rev 1982;1:45–64. [DOI] [PubMed] [Google Scholar]
- 6.Key ME. Macrophages in cancer metastases and their relevance to metastatic growth. Cancer Metastasis Rev 1983;2:75–88. [DOI] [PubMed] [Google Scholar]
- 7.Chambers AF, MacDonald IC, Schmidt EE, et al. Steps in tumor metastasis: new concepts from intravital videomicroscopy. Cancer Metastasis Rev 1995;14 279–301. [DOI] [PubMed] [Google Scholar]
- 8.Luzzi KJ, MacDonald IC, Schmidt EE, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol 1998;153 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naumov GN, MacDonald IC, Weinmeister PM, et al. Persistence of solitary mammary carcinoma cells in a secondary site: a possible contributor to dormancy. Cancer Res 2002;62:2162–2168. [PubMed] [Google Scholar]
- 10.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer 2003;355–63. [DOI] [PubMed] [Google Scholar]
- 11.Steeg PS, Bevilacqua G, Kopper L, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988;80200–204. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida BA, Sokoloff MM, Welch DR, et al. Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst 2000;921717–1730. [DOI] [PubMed] [Google Scholar]
- 13.Robinson VL, Kauffman EC, Sokoloff MH, et al. The basic biology of metastasis. Cancer Treat Res 2004;118:1–21. [DOI] [PubMed] [Google Scholar]
- 14.Ma X, Chen Y, Zhang S, et al. Rho1-Wnd signaling regulates loss-of-cell polarity-induced cell invasion in Drosophila. Oncogene 2016;35: 846–855. [DOI] [PubMed] [Google Scholar]
- 15.Biggs J, Tripoulas N, Hersperger E, et al. Analysis of the lethal interaction between the prune and Killer of prune mutations of Drosophila. Genes Dev 1988;2:1333–1343. [DOI] [PubMed] [Google Scholar]
- 16.Biggs J, Hersperger E, Steeg PS, et al. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell 1990;63:933–940. [DOI] [PubMed] [Google Scholar]
- 17.Risinger JI, Custer M, Feigenbaum L, et al. Normal viability of Kai1/Cd82 deficient mice. Mol Carcinog 2014;53:610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theroux S, Pereira M, Casten KS, et al. Raf kinase inhibitory protein knockout mice: expression in the brain and olfaction deficit. Brain Res Bull 2007;71:559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardo FN, Altirriba J, Pradas-Juni M, et al. The role of Raf-1 kinase inhibitor protein in the regulation of pancreatic beta cell proliferation in mice. Diabetologia 2012;55:3331–3340. [DOI] [PubMed] [Google Scholar]
- 20.Dammai V, Adryan B, Lavenburg KR, et al. Drosophila awd, the homolog of human nm23, regulates FGF receptor levels and functions synergistically with shi/dynamin during tracheal development. Genes Dev 2003;17:2812–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woolworth JA, Nallamothu G, Hsu T. The Drosophila metastasis suppressor gene Nm23 homolog, awd, regulates epithelial integrity during oogenesis. Mol Cell Biol 2009;29:4679–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ignesti M, Barraco M, Nallamothu G, et al. Notch signaling during development requires the function of awd, the Drosophila homolog of human metastasis suppressor gene Nm23. BMC Biol 2014;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geuking P, Narasimamurthy R, Lemaitre B, et al. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PLoS ONE 2009;4:e7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartholomeusz C, Gonzalez-Angulo AM, Liu P, et al. High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients. Oncologist 2012;17:766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb CP, Van Aelst L, Wigler MH, et al. Signaling pathways in Ras-mediated tumorigenicity and metastasis. Proc Natl Acad Sci USA 1998;95:8773–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward Y, Wang W, Woodhouse E, et al. Signal pathways which promote invasion and metastasis: critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol Cell Biol 2001;21:5958–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spector M, Nguyen VA, Sheng X, et al. Activation of mitogen-activated protein kinases is required for alpha1-adrenergic agonist-induced cell scattering in transfected HepG2 cells. Exp Cell Res 2000;258:109–120. [DOI] [PubMed] [Google Scholar]
- 28.Simon C, Hicks MJ, Nemechek AJ, et al. PD 098059, an inhibitor of ERK1 activation, attenuates the in vivo invasiveness of head and neck squamous cell carcinoma. Br J Cancer 1999;80:1412–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellenrieder V, Hendler SF, Boeck W, et al. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res 2001;61:4222–4228. [PubMed] [Google Scholar]
- 30.Salerno M, Ouatas T, Palmieri D, et al. Inhibition of signal transduction by the nm23 metastasis suppressor: possible mechanisms. Clin Exp Metastasis 2003;20:3–10. [DOI] [PubMed] [Google Scholar]
- 31.Kantor JD, McCormick B, Steeg PS, et al. Inhibition of cell motility after nm23 transfection of human and murine tumor cells. Cancer Res 1993;53:1971–1973. [PubMed] [Google Scholar]
- 32.Stahl JA, Leone A, Rosengard AM, et al. Identification of a second human nm23 gene, nm23-H2. Cancer Res 1991;51:445–449. [PubMed] [Google Scholar]
- 33.Bevilacqua G NM23 gene expression and human breast cancer metastases. Pathol Biol (Paris) 1990;38:774–775. [PubMed] [Google Scholar]
- 34.Nakayama T, Ohtsuru A, Nakao K, et al. Expression in human hepatocellular carcinoma of nucleoside diphosphate kinase, a homologue of the nm23 gene product. J Natl Cancer Inst 1992;84: 1349–1354. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi A, Urano T, Goi T, et al. Expression of human nm23-H1 and nm23-H2 proteins in hepatocellular carcinoma. Cancer 1994;73: 2280–2284. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki H, Fukuda M, Ishijima Y, et al. Overexpression of nm23-H2/NDP kinase B in a human oral squamous cell carcinoma cell line results in reduced metastasis, differentiated phenotype in the metastatic site, and growth factor-independent proliferative activity in culture. Clin Cancer Res 1999;5:4301–4307. [PubMed] [Google Scholar]
- 37.Hartsough MT, Morrison DK, Salerno M, et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem 2002;277: 32389–32399. [DOI] [PubMed] [Google Scholar]
- 38.Salerno M, Palmieri D, Bouadis A, et al. Nm23-H1 metastasis suppressor expression level influences the binding properties, stability, and function of the kinase suppressor of Ras1 (KSR1) Erk scaffold in breast carcinoma cells. Mol Cell Biol 2005;25:1379–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilles AM, Presecan E, Vonica A, et al. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J Biol Chem 1991;266:8784–8789. [PubMed] [Google Scholar]
- 40.Munoz-Dorado J, Inouye S, Inouye M. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J Biol Chem 1990;265:2707–2712. [PubMed] [Google Scholar]
- 41.Wallet V, Mutzel R, Troll H, et al. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. J Natl Cancer Inst 1990;82:1199–1202. [DOI] [PubMed] [Google Scholar]
- 42.Wagner PD, Vu ND. Phosphorylation of ATP-citrate lyase by nucleo-side diphosphate kinase. J Biol Chem 1995;270:21758–21764. [DOI] [PubMed] [Google Scholar]
- 43.Crovello CS, Furie BC, Furie B. Histidine phosphorylation of P-selectin upon stimulation of human platelets: a novel pathway for activation-dependent signal transduction. Cell 1995;82:279–286. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q, McCorkle JR, Novak M, et al. Metastasis suppressor function of NM23-H1 requires its 3’−5’ exonuclease activity. Int J Cancer 2011;128:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boissan M, Montagnac G, Shen Q, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 2014;344: 1510–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freije JM, Blay P, MacDonald NJ, et al. Site-directed mutation of Nm23-H1. Mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J Biol Chem 1997;272:5525–5532. [DOI] [PubMed] [Google Scholar]
- 47.Wagner PD, Steeg PS, Vu ND. Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci USA 1997;94:9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison DK. KSR: a MAPK scaffold of the Ras pathway? J Cell Sci 2001;114:1609–1612. [DOI] [PubMed] [Google Scholar]
- 49.Masoudi N, Fancsalszky L, Pourkarimi E, et al. The NM23-H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans. Development 2013;140:3486–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tso PH, Wang Y, Yung LY, et al. RGS19 inhibits Ras signaling through Nm23H1/2-mediated phosphorylation of the kinase suppressor of Ras. Cell Signal 2013;25:1064–1074. [DOI] [PubMed] [Google Scholar]
- 51.MacDonald NJ, Freije JM, Stracke ML, et al. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem 1996;271:25107–25116. [DOI] [PubMed] [Google Scholar]
- 52.Marino N, Marshall JC, Steeg PS. Protein-protein interactions: a mechanism regulating the anti-metastatic properties of Nm23-H1. Naunyn Schmiedebergs Arch Pharmacol 2011;384:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian C, Cotter MA 2nd, Robertson ES. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med 2001;7:350–355. [DOI] [PubMed] [Google Scholar]
- 54.Horak CE, Mendoza A, Vega-Valle E, et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res 2007;67:11751–11759. [DOI] [PubMed] [Google Scholar]
- 55.Yeung K, Seitz T, Li S, et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999;401:173–177. [DOI] [PubMed] [Google Scholar]
- 56.Trakul N, Menard RE, Schade GR, et al. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem 2005;280: 24931–24940. [DOI] [PubMed] [Google Scholar]
- 57.Dangi-Garimella S, Yun J, Eves EM, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 2009;28:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yesilkanal AE, Rosner MR. Raf kinase inhibitory protein (RKIP) as a metastasis suppressor: regulation of signaling networks in cancer. Crit Rev Oncog 2014;19:447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinho O, Pinto F, Granja S, et al. RKIP inhibition in cervical cancer is associated with higher tumor aggressive behavior and resistance to cisplatin therapy. PLoS ONE 2013;8:e59104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Z, Smith PC, Zhang L, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst 2003;95:878–889. [DOI] [PubMed] [Google Scholar]
- 61.Yousuf S, Duan M, Moen EL, et al. Raf kinase inhibitor protein (RKIP) blocks signal transducer and activator of transcription 3 (STAT3) activation in breast and prostate cancer. PLoS ONE 2014;9:e92478. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Deiss K, Kisker C, Lohse MJ, et al. Raf kinase inhibitor protein (RKIP) dimer formation controls its target switch from Raf1 to G protein-coupled receptor kinase (GRK) 2. J Biol Chem 2012;287:23407–23417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das SK, Bhutia SK, Sokhi UK, et al. Raf kinase inhibitor RKIP inhibits MDA-9/syntenin-mediated metastasis in melanoma. Cancer Res 2012;72:6217–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eves EM, Shapiro P, Naik K, et al. Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol Cell 2006;23:561–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer 2009;9:302–312. [DOI] [PubMed] [Google Scholar]
- 66.Hickson JA, Huo D, Vander Griend DJ, et al. The p38 kinases MKK4 and MKK6 suppress metastatic colonization in human ovarian carcinoma. Cancer Res 2006;66:2264–2270. [DOI] [PubMed] [Google Scholar]
- 67.Ranganathan AC, Zhang L, Adam AP, et al. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res 2006;66:1702–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 2014;14: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diehl NL, Enslen H, Fortner KA, et al. Activation of the p38 mitogen-activated protein kinase pathway arrests cell cycle progression and differentiation of immature thymocytes in vivo. J Exp Med 2000;191: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamada SD, Hickson JA, Hrobowski Y, et al. Mitogen-activated protein kinase kinase 4 (MKK4) acts as a metastasis suppressor gene in human ovarian carcinoma. Cancer Res 2002;62:6717–6723. [PubMed] [Google Scholar]
- 71.Yoshida BA, Dubauskas Z, Chekmareva MA, et al. Mitogen-activated protein kinase kinase 4/stress-activated protein/Erk kinase 1 (MKK4/SEK1), a prostate cancer metastasis suppressor gene encoded by human chromosome 17. Cancer Res 1999;59:5483–5487. [PubMed] [Google Scholar]
- 72.Vander Griend DJ, Kocherginsky M, Hickson JA, et al. Suppression of metastatic colonization by the context-dependent activation of the c-Jun NH2-terminal kinase kinases JNKK1/MKK4 and MKK7. Cancer Res 2005;65:10984–10991. [DOI] [PubMed] [Google Scholar]
- 73.Remy G, Risco AM, Inesta-Vaquera FA, et al. Differential activation of p38MAPK isoforms by MKK6 and MKK3. Cell Signal 2010;22:660–667. [DOI] [PubMed] [Google Scholar]
- 74.Adam AP, George A, Schewe D, et al. Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer Res 2009;69:5664–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aguirre-Ghiso JA, Estrada Y, Liu D, et al. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38 (SAPK). Cancer Res 2003;63:1684–1695. [PubMed] [Google Scholar]
- 76.Hubner A, Mulholland DJ, Standen CL, et al. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc Natl Acad Sci USA 2012;109:12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Pan Y, Dai JL. Evidence of MKK4 pro-oncogenic activity in breast and pancreatic tumors. Oncogene 2004;23:5978–5985. [DOI] [PubMed] [Google Scholar]
- 78.Khatlani TS, Wislez M, Sun M, et al. c-Jun N-terminal kinase is activated in non-small-cell lung cancer and promotes neoplastic transformation in human bronchial epithelial cells. Oncogene 2007;26:2658–2666. [DOI] [PubMed] [Google Scholar]
- 79.Gaugitsch HW, Hofer E, Huber NE, et al. A new superfamily of lymphoid and melanoma cell proteins with extensive homology to Schistosoma mansoni antigen Sm23. Eur J Immunol 1991;21:377–383. [DOI] [PubMed] [Google Scholar]
- 80.Ichikawa T, Ichikawa Y, Isaacs JT. Genetic factors and suppression of metastatic ability of prostatic cancer. Cancer Res 1991;51:3788–3792. [PubMed] [Google Scholar]
- 81.Yang X, Welch DR, Phillips KK, et al. KAI1, a putative marker for metastatic potential in human breast cancer. Cancer Lett 1997;119: 149–155. [DOI] [PubMed] [Google Scholar]
- 82.Guo X, Friess H, Graber HU, et al. KAI1 expression is up-regulated in early pancreatic cancer and decreased in the presence of metastases. Cancer Res 1996;56:4876–4880. [PubMed] [Google Scholar]
- 83.Guo XZ, Friess H, Di Mola FF, et al. KAI1, a new metastasis suppressor gene, is reduced in metastatic hepatocellular carcinoma. Hepatology 1998;28:1481–1488. [DOI] [PubMed] [Google Scholar]
- 84.Yang X, Wei LL, Tang C, et al. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res 2001;61:5284–5288. [PubMed] [Google Scholar]
- 85.Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol 2000;10:1009–1012. [DOI] [PubMed] [Google Scholar]
- 86.Bienstock RJ, Barrett JC. KAI1, a prostate metastasis suppressor: prediction of solvated structure and interactions with binding partners; integrins, cadherins, and cell-surface receptor proteins. Mol Carcinog 2001;32:139–153. [DOI] [PubMed] [Google Scholar]
- 87.Underhill C CD44: the hyaluronan receptor. J Cell Sci 1992;103(Pt 2): 293–298. [DOI] [PubMed] [Google Scholar]
- 88.Weber GF, Ashkar S, Glimcher MJ, et al. Receptor-ligand interaction between CD44 and osteopontin (Eta-1). Science 1996;271:509–512. [DOI] [PubMed] [Google Scholar]
- 89.Gao AC, Lou W, Dong JT, et al. CD44 is a metastasis suppressor gene for prostatic cancer located on human chromosome 11p13. Cancer Res 1997;57:846–849. [PubMed] [Google Scholar]
- 90.Gunthert U, Hofmann M, Rudy W, et al. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 199165: 13–24. [DOI] [PubMed] [Google Scholar]
- 91.Brown RL, Reinke LM, Damerow MS, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelialmesenchymal transition and breast cancer progression. J Clin Invest 2011;121:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arch R, Wirth K, Hofmann M, et al. Participation in normal immune responses of a metastasis-inducing splice variant of CD44. Science 1992;257:682–685. [DOI] [PubMed] [Google Scholar]
- 93.van der Voort R, Taher TE, Wielenga VJ, et al. Heparan sulfate-modified CD44 promotes hepatocyte growth factor/scatter factor-induced signal transduction through the receptor tyrosine kinase c-Met. J Biol Chem 1999;274:6499–6506. [DOI] [PubMed] [Google Scholar]
- 94.Bourguignon LY. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin Cancer Biol 2008;18:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Murai T, Miyazaki Y, Nishinakamura H, et al. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem 2004;279:4541–4550. [DOI] [PubMed] [Google Scholar]
- 96.Bourguignon LY, Singleton PA, Zhu H, et al. Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor beta receptor I in metastatic breast tumor cells. J Biol Chem 2002;277:39703–39712. [DOI] [PubMed] [Google Scholar]
- 97.Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: diversity in form and function. J Cell Sci 2001;114:629–641. [DOI] [PubMed] [Google Scholar]
- 98.Onder TT, Gupta PB, Mani SA, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 2008;68:3645–3654. [DOI] [PubMed] [Google Scholar]
- 99.Vleminckx K, Vakaet L Jr., Mareel M, et al. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 1991;66:107–119. [DOI] [PubMed] [Google Scholar]
- 100.Frixen UH, Behrens J, Sachs M, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol 1991;113:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perl AK, Wilgenbus P, Dahl U, et al. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature 1998;392:190–193. [DOI] [PubMed] [Google Scholar]
- 102.Sawada K, Mitra AK, Radjabi AR, et al. Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res 2008;68:2329–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mbalaviele G, Dunstan CR, Sasaki A, et al. E-cadherin expression in human breast cancer cells suppresses the development of osteolytic bone metastases in an experimental metastasis model. Cancer Res 1996;56:4063–4070. [PubMed] [Google Scholar]
- 104.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci 1999;24:73–76. [DOI] [PubMed] [Google Scholar]
- 105.Tanaka H, Shirkoohi R, Nakagawa K, et al. siRNA gelsolin knockdown induces epithelial-mesenchymal transition with a cadherin switch in human mammary epithelial cells. Int J Cancer 2006;118:1680–1691. [DOI] [PubMed] [Google Scholar]
- 106.Fujita H, Okada F, Hamada J, et al. Gelsolin functions as a metastasis suppressor in B16-BL6 mouse melanoma cells and requirement of the carboxyl-terminus for its effect. Int J Cancer 2001;93:773–780. [DOI] [PubMed] [Google Scholar]
- 107.Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal 2010;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ueda T, Kikuchi A, Ohga N, et al. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem 1990;265: 9373–9380. [PubMed] [Google Scholar]
- 109.Gildea JJ, Seraj MJ, Oxford G, et al. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res 2002;62: 6418–6423. [PubMed] [Google Scholar]
- 110.Titus B, Frierson HF Jr., Conaway M, et al. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res 2005;65: 7320–7327. [DOI] [PubMed] [Google Scholar]
- 111.Goodison S, Yuan J, Sloan D, et al. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res 2005;65:6042–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Popescu NC, Goodison S. Deleted in liver cancer-1 (DLC1): an emerging metastasis suppressor gene. Mol Diagn Ther 2014;18: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liao YC, Lo SH. Deleted in liver cancer-1 (DLC-1): a tumor suppressor not just for liver. Int J Biochem Cell Biol 2008;40:843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee JH, Miele ME, Hicks DJ, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst 1996;88: 1731–1737. [DOI] [PubMed] [Google Scholar]
- 115.Lee JH, Welch DR. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer 1997;71:1035–1044. [DOI] [PubMed] [Google Scholar]
- 116.Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res 1997;57:2384–2387. [PubMed] [Google Scholar]
- 117.Ohtaki T, Shintani Y, Honda S, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature 2001;411:613–617. [DOI] [PubMed] [Google Scholar]
- 118.Kotani M, Detheux M, Vandenbogaerde A, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem 2001;276: 34631–34636. [DOI] [PubMed] [Google Scholar]
- 119.Ko HK, Guo LW, Su B, et al. Suppression of chemotaxis by SSeCKS via scaffolding of phosphoinositol phosphates and the recruitment of the Cdc42 GEF, Frabin, to the leading edge. PLoS ONE 2014;9: e111534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gelman IH. Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Rev 2012;31:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Su B, Bu Y, Engelberg D, et al. SSeCKS/Gravin/AKAP12 inhibits cancer cell invasiveness and chemotaxis by suppressing a protein kinase CRaf/MEK/ERK pathway. J Biol Chem 2010;285:4578–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guo LW, Gao L, Rothschild J, et al. Control of protein kinase C activity, phorbol ester-induced cytoskeletal remodeling, and cell survival signals by the scaffolding protein SSeCKS/GRAVIN/AKAP12. J Biol Chem 2011;286:38356–38366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stupack DG, Teitz T, Potter MD, et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature 2006;439:95–99. [DOI] [PubMed] [Google Scholar]
- 124.Gobeil S, Zhu X, Doillon CJ, et al. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev 2008;22:2932–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiang Z, Xu Y, Cai S. Down-regulated GAS1 expression correlates with recurrence in stage II and III colorectal cancer. Hum Pathol 2011;42: 361–368. [DOI] [PubMed] [Google Scholar]
- 126.Li Q, Qin Y, Wei P, et al. Gas1 inhibits metastatic and metabolic phenotypes in colorectal carcinoma. Mol Cancer Res 2016;14:830–840. [DOI] [PubMed] [Google Scholar]
- 127.Seraj MJ, Samant RS, Verderame MF, et al. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res 2000;60:2764–2769. [PubMed] [Google Scholar]
- 128.Hurst DR, Welch DR. Metastasis suppressor genes at the interface between the environment and tumor cell growth. Int Rev Cell Mol Biol 2011;286:107–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Phadke PA, Vaidya KS, Nash KT, et al. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol 2008;172: 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hedley BD, Vaidya KS, Phadke P, et al. BRMS1 suppresses breast cancer metastasis in multiple experimental models of metastasis by reducing solitary cell survival and inhibiting growth initiation. Clin Exp Metastasis 2008;25:727–740. [DOI] [PubMed] [Google Scholar]
- 131.Kapoor P, Saunders MM, Li Z, et al. Breast cancer metastatic potential: correlation with increased heterotypic gap junctional intercellular communication between breast cancer cells and osteoblastic cells. Int J Cancer 2004;111:693–697. [DOI] [PubMed] [Google Scholar]
- 132.Meehan WJ, Samant RS, Hopper JE, et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem 2004;279:1562–1569. [DOI] [PubMed] [Google Scholar]
- 133.Hurst DR, Xie Y, Thomas JW, et al. The C-terminal putative nuclear localization sequence of breast cancer metastasis suppressor 1, BRMS1, is necessary for metastasis suppression. PLoS ONE 2013;8:e55966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, Mayo MW, Nagji AS, et al. BRMS1 suppresses lung cancer metastases through an E3 ligase function on histone acetyltransferase p300. Cancer Res 2013;73:1308–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grandori C, Cowley SM, James LP, et al. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000;16:653–699. [DOI] [PubMed] [Google Scholar]
- 136.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 1999;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oster SK, Ho CS, Soucie EL, et al. The myc oncogene: MarvelouslY Complex. Adv Cancer Res 2002;84:81–154. [DOI] [PubMed] [Google Scholar]
- 138.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 2005;6:635–645. [DOI] [PubMed] [Google Scholar]
- 139.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976–990. [DOI] [PubMed] [Google Scholar]
- 140.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene 1999;18:3004–3016. [DOI] [PubMed] [Google Scholar]
- 141.Liu H, Radisky DC, Yang D, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of alphav and beta3 integrin subunits. Nat Cell Biol 2012;14:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cichon MA, Moruzzi ME, Shqau TA, et al. MYC is a crucial mediator of TGFbeta-induced invasion in basal breast cancer. Cancer Res 2016;76: 3520–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Elkon R, Loayza-Puch F, Korkmaz G, et al. Myc coordinates transcription and translation to enhance transformation and suppress invasiveness. EMBO Rep 2015;16:1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee KS, Kwak Y, Nam KH, et al. Favorable prognosis in colorectal cancer patients with co-expression of c-MYC and ss-catenin. BMC Cancer 2016;16:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Andrews FH, Singh AR, Joshi S, et al. Dual-activity PI3K-BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis. Proc Natl Acad Sci USA 2017;114:E1072–E1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin X, Sun R, Zhao X, et al. C-myc overexpression drives melanoma metastasis by promoting vasculogenic mimicry via c-myc/snail/Bax signaling. J Mol Med (Berl) 2017;95:53–67. [DOI] [PubMed] [Google Scholar]
- 147.Hurst DR, Edmonds MD, Scott GK, et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 2009;69:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012;9703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013;491083–1096. [DOI] [PubMed] [Google Scholar]
- 154.Langley RR, Fidler IJ. The seed and soil hypothesis revisited—the role of tumor-stroma interactions in metastasis to different organs. Int J Cancer 2011;128:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370–381. [DOI] [PubMed] [Google Scholar]
- 156.Huber MA, Azoitei N, Baumann B, et al. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest 2004;114:569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hunter KW, Broman KW, Voyer TL, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res 2001;61: 8866–8872. [PubMed] [Google Scholar]
- 158.Park YG, Zhao X, Lesueur F, et al. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet 2005;37: 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bai L, Yang HH, Hu Y, et al. An integrated genome-wide systems genetics screen for breast cancer metastasis susceptibility genes. PLoS Genet 2016;12e1005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Faraji F, Pang Y, Walker RC, et al. Cadm1 is a metastasis susceptibility gene that suppresses metastasis by modifying tumor interaction with the cell-mediated immunity. PLoS Genet 2012;8:e1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fu Z, Kitagawa Y, Shen R, et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate 2006;66:248–256. [DOI] [PubMed] [Google Scholar]
- 162.Hennessy C, Henry JA, May FE, et al. Expression of the antimetastatic gene nm23 in human breast cancer: an association with good prognosis. J Natl Cancer Inst 1991;83:281–285. [DOI] [PubMed] [Google Scholar]
- 163.Miele ME, De La Rosa A, Lee JH, et al. Suppression of human melanoma metastasis following introduction of chromosome 6 is independent of NME1 (Nm23). Clin Exp Metastasis 1997;15: 259–265. [DOI] [PubMed] [Google Scholar]
- 164.Leone A, Flatow U, King CR, et al. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 1991;65:25–35. [DOI] [PubMed] [Google Scholar]
- 165.Baba H, Urano T, Okada K, et al. Two isotypes of murine nm23/nucleoside diphosphate kinase, nm23-M1 and nm23-M2, are involved in metastatic suppression of a murine melanoma line. Cancer Res 1995;55:1977–1981. [PubMed] [Google Scholar]
- 166.de la Rosa A, Williams RL, Steeg PS. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays 1995;17:53–62. [DOI] [PubMed] [Google Scholar]
- 167.Hsu S, Huang F, Wang L, et al. The role of nm23 in transforming growth factor beta 1-mediated adherence and growth arrest. Cell Growth Differ 1994;5:909–917. [PubMed] [Google Scholar]
- 168.Otsuki K, Alcalde RE, Matsumura T, et al. Immunohistochemical analysis of nucleoside diphosphate kinases in oral squamous cell carcinomas. Oncology 1997;54:63–68. [DOI] [PubMed] [Google Scholar]
- 169.Dong JT, Suzuki H, Pin SS, et al. Down-regulation of the KAI1 metastasis suppressor gene during the progression of human prostatic cancer infrequently involves gene mutation or allelic loss. Cancer Res 1996;56:4387–4390. [PubMed] [Google Scholar]
- 170.Kawana Y, Komiya A, Ueda T, et al. Location of KAI1 on the short arm of human chromosome 11 and frequency of allelic loss in advanced human prostate cancer. Prostate 1997;32:205–213. [DOI] [PubMed] [Google Scholar]
- 171.Dong JT, Lamb PW, Rinker-Schaeffer CW, et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995;268:884–886. [DOI] [PubMed] [Google Scholar]
- 172.Lou W, Krill D, Dhir R, et al. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res 1999;59: 2329–2331. [PubMed] [Google Scholar]
- 173.Noordzij MA, van Steenbrugge GJ, Schroder FH, et al. Decreased expression of CD44 in metastatic prostate cancer. Int J Cancer 1999;84:478–483. [DOI] [PubMed] [Google Scholar]
- 174.Nagabhushan M, Pretlow TG, Guo YJ, et al. Altered expression of CD44 in human prostate cancer during progression. Am J Clin Pathol 1996;106:647–651. [DOI] [PubMed] [Google Scholar]
- 175.De Marzo AM, Bradshaw C, Sauvageot J, et al. CD44 and CD44v6 downregulation in clinical prostatic carcinoma: relation to Gleason grade and cytoarchitecture. Prostate 1998;34:162–168. [DOI] [PubMed] [Google Scholar]
- 176.Kashima T, Nakamura K, Kawaguchi J, et al. Overexpression of cadherins suppresses pulmonary metastasis of osteosarcoma in vivo. Int J Cancer 2003;104:147–154. [DOI] [PubMed] [Google Scholar]
- 177.Suyama K, Shapiro I, Guttman M, et al. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2002;2:301–314. [DOI] [PubMed] [Google Scholar]
- 178.Qi J, Chen N, Wang J, et al. Transendothelial migration of melanoma cells involves N-cadherin-mediated adhesion and activation of the beta-catenin signaling pathway. Mol Biol Cell 2005;16:4386–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res 2001;61:3819–3825. [PubMed] [Google Scholar]
- 180.Su B, Zheng Q, Vaughan MM, et al. SSeCKS metastasis-suppressing activity in MatLyLu prostate cancer cells correlates with vascular endothelial growth factor inhibition. Cancer Res 2006;66:5599–5607. [DOI] [PubMed] [Google Scholar]
- 181.Stupack DG, Puente XS, Boutsaboualoy S, et al. Apoptosis of adherent cells by recruitment of caspase-8 to unligated integrins. J Cell Biol 2001;155:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhao H, Ross FP, Teitelbaum SL. Unoccupied alpha(v)beta3 integrin regulates osteoclast apoptosis by transmitting a positive death signal. Mol Endocrinol 2005;19:771–780. [DOI] [PubMed] [Google Scholar]
- 183.Del Sal G, Ruaro ME, Philipson L, et al. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell 1992;70: 595–607. [DOI] [PubMed] [Google Scholar]
- 184.Shevde LA, Samant RS, Goldberg SF, et al. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res 2002;273:229–239. [DOI] [PubMed] [Google Scholar]
- 185.Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer 2006;16:522–531. [DOI] [PubMed] [Google Scholar]
- 186.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res 2009;69: 7495–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]