Abstract
Background
The aim of this study was to evaluate the frequency of toxigenic C. difficile and C. perfringens infections at health care facility-onset (HCFO) and community-onset (CO), in two health care centers (HCC) in Bogotá, Colombia. A total of 220 stool samples from patients presenting diarrhea acquired at HCFO or CO were analyzed by several PCR tests.
Results
We found that 65.5% (n = 144) of the population had C. difficile infection, followed by toxigenic C. difficile with 57.3% (n = 126), and finally toxigenic C. perfringens with a frequency of 32.7% (n = 72).
Conclusions
This study is the first molecular detection and characterization of C. difficile and C. perfringens in HCFO and CO in Latin America and demonstrates a relevant frequency of these two species, including coinfection and strikingly diverse toxigenic profiles, especially in the CO.
Keywords: Clostridium perfringens, Clostridium difficile, Diarrhea, Community-onset, Health care facility onset
Introduction
Clostridium difficile is one of the most studied clostridial species, as it leads to developing diarrhea associated with the use of antibiotics at the hospital level [1]. The main virulence factors of C. difficile are Toxin A (TcdA) and Toxin B (TcdB), belonging to the large family of Clostridial toxins with glucosyltransferase activity [2]. These toxins are encoded by genes located in a region of the chromosome of approximately 20 Kb, which constitutes the pathogenicity locus (PaLoc). Some strains of C. difficile can produce a third toxin called binary, which is encoded by a chromosomal region called CdtLoc, located downstream of PaLoc, which contains the cdtA and cdtB genes, coding for its two components, in addition to a regulator for these genes (cdtR) [3].
On the other hand, diarrhea can also be caused by C. perfringens, a species that is widely distributed in various hosts and environments, and that has been related to histotoxic and intestinal infections in animals and humans. C. perfringens has been identified in humans as the main etiologic agent of gas gangrene, also being able to cause other complications such as diarrheal disease associated with food poisoning, necrotizing enteritis, and other nonspecific gastrointestinal manifestations [4]. Historically, it has been considered that C. perfringens produces the following four main toxins: alpha (CPA), beta (CPB), epsilon (ETX), and iota (ITX). However, recently some authors have added two more toxins to its toxin repertoire: enterotoxin (CPE), and necrotic enteritis B-like (NetB) toxin; all these toxins can be differentially produced [5] and determine the clinical spectrum of infection by this species [6]. CPA is recognized as the main virulence factor in humans, causing hemolytic and dermonecrotic effects, characteristic of clostridial myonecrosis, which can be lethal [5]. Different gene regions coding for the above toxins have been previously implemented for C. perfringens molecular detection. The cpa gene encoding for CPA, has been described as the best molecular target, which is located in a stable region of the genome present in the seven identified toxinotypes (A–G), according to the most recent reclassification of the species [6].
The importance of these two pathogens in terms of public health mediated by the broad toxigenic arsenal, leads to the need to evaluate the coinfection frequency (defined as a positive result simultaneously for the two species). In this context, this study aimed at determining the frequency of C. perfringens and C. difficile (any type or toxigenic specifically) present in health care facility-onset (HCFO) or community-onset (CO) diarrhea, at two health care centers in Bogotá, Colombia.
Methods
Study population
A total of 220 stool samples from patients with diarrhea [7], were collected during the period from September 2015 to April 2017 at two health care centers (HCC), located in the city of Bogotá, Colombia [the Hospital Universitario Mayor—Méderi (HCC-1) and the Fundación Clínica Shaio (HCC-2)]. Participant selection (HCFO and CO groups) was carried out following the guidelines of the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America [7].
Molecular detection and toxinotyping of C. difficile and C. perfringens
The clostridial species were identified using several conventional polymerase chain reaction (PCR) tests. Two sets of consensus primers targeting constitutive genes coding for the 16S ribosomal subunit (rRNA-16S) and for the glutamate dehydrogenase enzyme (GDH) as reported elsewhere [8, 9] were initially used for C. difficile detection (toxigenic or non-toxigenic). Subsequently, the toxigenic profiles of the C. difficile positive samples were determined using six independent amplification tests, four of them directed to the PaLoc regions that code for the main toxins of C. difficile [10, 11], and the other two to the CdtLoc, where the coding regions for the binary toxin are located [12]. A positive result for any of these genes led to the assignment in the ‘tox_C. difficile’ category. In the case of C. perfringens, the detection was carried out by conventional PCR directed to the cpa gene, as reported elsewhere [13], considered as an indicator of the presence of ‘tox_C. perfringens’.
Statistical analyses
Descriptive analyses were carried out to determine the frequencies in terms of percentages with respect to the total population, for each event of interest. χ2 tests were performed to identify potential associations between the variables analyzed. A binomial logistic regression analysis was used to estimate the association between infection by C. perfringens, C. difficile or tox_C. difficile (taken as dependent variables) and the different factors evaluated (hospital center and hospital stay) taken as independent variables within the analysis. Additionally, the strength of association between the existing coinfections (C. perfringens and C. difficile, C. perfringens and tox_C. difficile) was calculated using odds ratios (ORs) with their corresponding 95% confidence intervals (CIs). The adjustment of the ORs (AdOR) was carried out from hospital (HCC-1 and HCC-2) and place of stay (HCFO and CO), as confounding variables. All analyses were performed using STATA14® (StataCorp LLC, College Station, TX, USA). The level of significance was established at p < 0.05.
Results
Frequency of C. difficile/C. perfringens infection and/or coinfection
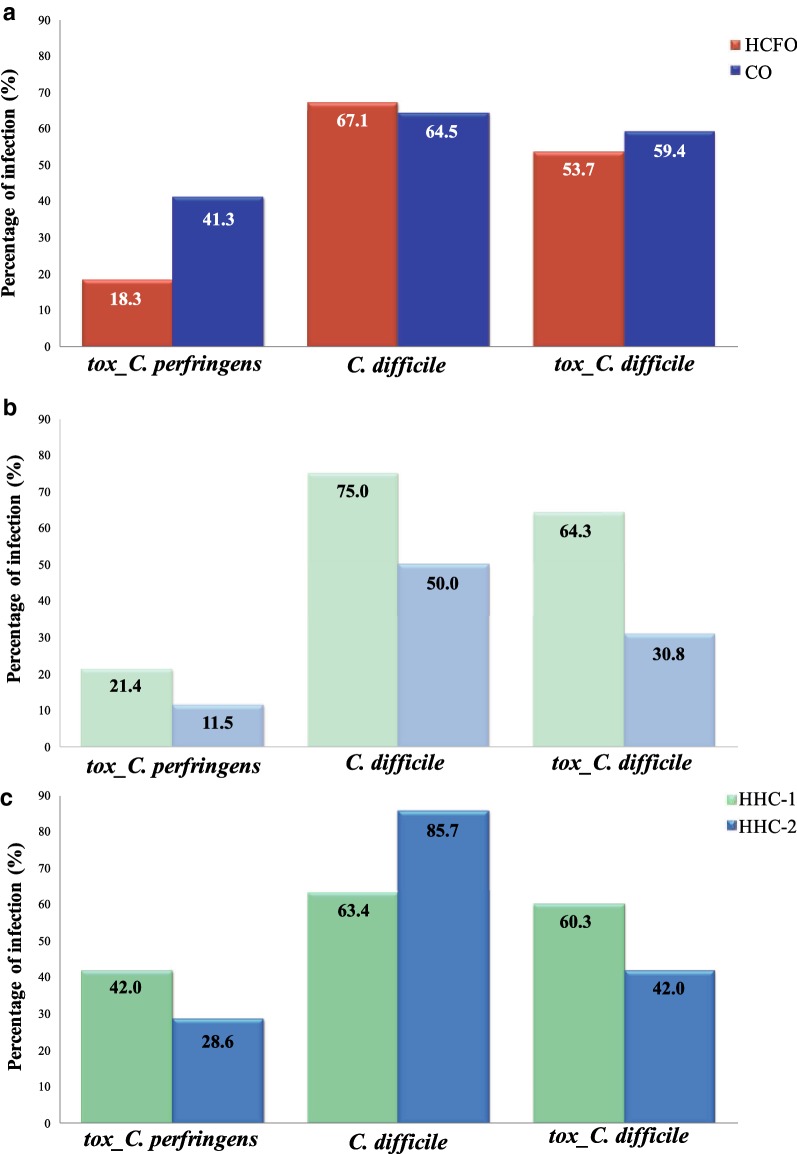
A total of 85.0% (n = 187) of stool samples from patients with diarrhea collected for this study came from HCC-1, and the remaining 15.0% (n = 33) were from HCC-2. Of the total samples collected in HCC-1, the majority were obtained from CO patients (70.0%, n = 131), while in the case of HCC-2, the majority came from HCFO patients (78.8%, n = 26). Regarding the distribution of the evaluated species, the results showed that 65.5% (n = 144) of the population had C. difficile infection, followed by tox_C. difficile with 57.3% (n = 126), and finally tox_C. perfringens with a frequency of 32.7% (n = 72). When evaluating the distribution of the frequency of the species according to the stay (HCFO and CO), the frequency of C. difficile was higher in patients coming from HCFO compared to those of CO (67.1% and 64.5%, respectively; p = 0.697). In contrast, the frequency of infection for C. perfringens was higher in CO patients compared with those of HCFO (18.3% and 41.3%, respectively, p = 0.004); this same distribution was observed for tox_C. difficile (53.7% and 59.4%, respectively, p = 0.032) (Fig. 1a).
Fig. 1.
Frequency of infection of the Clostridial species evaluated. a In the Global population; b in HCFO and c in CO
Statistical associations
In addition, the distribution of infections according to the stay (HCFO and CO) and the hospital of origin of the patients (HCC-1 and HCC-2) was determined. At the HCFO level, infections were higher for the three species in the patients from the HCC-1 healthcare center compared to HCC-2, the distribution observed for C. difficile being statistically significant (p = 0.0250) (Fig. 1b). For CO patients, C. difficile infections were higher in HCC-2 compared to HCC-1 (p = 0.2285), in contrast to C. perfringens whose infections were higher than those observed in HFCO. In community patients from HCC-1, the frequency of occurrence was higher than that observed for HCC-2 (p = 0.2810). A similar pattern was found for the distribution of tox_C. difficile between hospital centers which was statistically significant (p = 0.0001) (Fig. 1c).
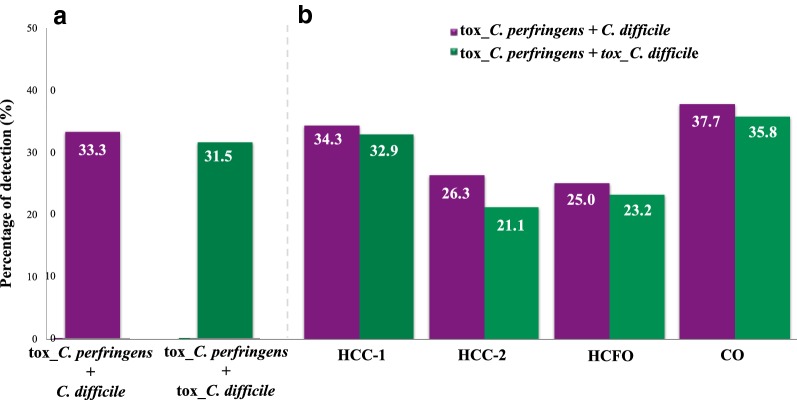
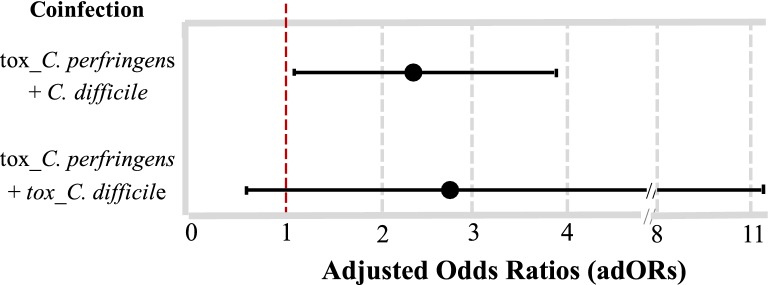
The evaluation of the coinfection frequencies between the two species evaluated revealed a global percentage of 33.3% for the case of tox_C. perfringens + C. difficile and 31.5% for tox_C. perfringens + tox_C. difficile (Fig. 2a). The analysis by HCC and by population, showed that the coinfection frequencies ranged between 21.1% for tox_C. perfringens + tox_C. difficile in HCC-2, and up to 37.7% for tox_C. perfringens + C. difficile in CO (Fig. 2b). The results of the OR between the clostridial infections with HCC and stay, showed a positive association for community patients and tox_C. perfringens infections (AdOR: 2.69 CI 95% 1.35–5.35). In contrast, a minor association was observed between HCC-2 and tox_C. difficile infection (AdOR: 0.14 CI 95% 0.04–0.46) (Table 1). In the same context, AdORs were calculated by the association between the types of coinfections present in the populations evaluated; only positive association was observed for the combination of C. perfringens and C. difficile (AdOR: 2.05 CI 95% 1.07–3.93) (Fig. 3).
Fig. 2.
Co-infection frequency between the two evaluated species. a In the global population and b by Health care center (HCC) and population. HCFO Health Care Facility-Onset, CO Community Onset)
Table 1.
Logistic regression modeling showing the relationship between a positive result for infection and the hospital and acquisition of infection
| C. perfringens infection | C. difficile infection | Toxigenic C. difficile infection | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AdOR | [95% CI] | p | AdOR | [95% CI] | p | AdOR | [95% CI] | p | |
| Health care center | |||||||||
| HCC-1 | Reference | Reference | Reference | ||||||
| HCC-2 | 0.50 | [0.17–1.45] | 0.206 | 0.59 | [0.26–1.34] | 0.208 | 0.14 | [0.04–0.46] | 0.001 |
| Service | |||||||||
| HCFO | Reference | Reference | Reference | ||||||
| CO | 2.69 | [1.35–5.35] | 0.005 | 0.76 | [0.40–1.44] | 0.413 | 1.98 | [0.66–5.94] | 0.221 |
Adjusted OR by population and health-care center
Italic values indicate significance of p value
Fig. 3.
Diagram of strength of association between Clostridial species evaluated. Odds ratios (OR), and their corresponding 95% confidence intervals, which indicate the strength of association between both species according with the type of infection
Discussion
The present study identified the frequency of C. difficile (general and tox_ C. difficile), but also tox_C. perfringens in two HCCs of Bogotá, Colombia through molecular detection [5]. It is important to note that previous reports indicate frequent development of diarrhea in individuals in whom the microbiota has been altered by the effect of antimicrobial agents, favoring the proliferation of pathogens such as those belonging to the genus Clostridium [14]. This could explain the presence of tox_C. perfringens in HCFO diarrhea. However, the results reported in this study indicate greater frequency of infection in patients with CO diarrhea (Fig. 1), which could be related either to the established association of infection by this species with diarrheal disease caused by food poisoning [4], or with the presence of some other factor that could be involved with the development of dysbiosis and acquiring the role of a pathogen [14]. Although, due to the lack of clinical and sociodemographic information of the individuals included in this study, it is not possible to establish an association of causality between the presence of C. perfringens or C. difficile in patients with diarrhea in Colombia. We found interesting associations between C. perfringens and C. difficile (AdOR: 2.05 CI 95% 1.07–3.93) (Fig. 3). This suggests that coinfection plays a relevant role in the population. Future studies should consider both species in terms of clinical and sociodemographic data which might provide novel insights regarding the impact of both species in a particular population.
One limitation of our study was the inability to recover the isolates and conduct molecular characterization using genome sequencing or Multilocus Sequence Typing. Therefore, we encourage the scientific community to develop novel studies in the region aimed at unraveling the molecular features of these two species. The findings herein identified represent a baseline about the high coexistence of these two Clostridial species towards depicting the epidemiological panorama in the country and Latin-America.
Acknowledgements
We thank Clara Lina Salazar from Universidad de Antioquia for providing the reference strain of C. perfringens. We also thank Analisa Avila, ELS, of Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Authors’ contributions
AJF, MM, DIRC and JDR: led the project and contributed to definition of experimental approach. MC, SCS, MM and JDR: analyzed the results and conducted the statistical analyzes. CB, DP, JMP, DFJ and MAP: led the clinical component, from the inclusion of samples to analysis of the results at the epidemiological level. MAP and JDR: revised the final version. All authors read and approved the final manuscript.
Funding
The Departamento Administrativo de Ciencia, Tecnología e Innovación (Colciencias) financed the PhD program to MM within the framework of the National Program for Promoting Research Training (sponsorship call 617). This work was funded By DIRECCIÓN DE INVESTIGACIÓN E INNOVACIÓN from Universidad del Rosario.
Availability of data and materials
All the data are within the manuscript.
Ethics approval and consent to participate
An initial study aimed to detect C. difficile infections in fecal samples from patients with diarrhea was approved by the Universidad del Rosario’s Research Ethics Committee (Approval Act No. 290, July 27, 2015). In addition, an addendum was approved, which authorized the additional use of samples for research purposes aimed at the description and characterization of any microorganisms present in the human gastrointestinal tract (Approval Act No. 312 April 28, 2016). All patients (adult) signed the written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alex J. Forero and Marina Muñoz contributed equally as first author
Contributor Information
Alex J. Forero, Email: ajfode@gmail.com
Marina Muñoz, Email: claudia.munoz@urosario.edu.co.
Milena Camargo, Email: sandramilena.camargopinzon@gmail.com.
Sara C. Soto-De León, Email: soto.sarac@gmail.com
Dora I. Ríos-Chaparro, Email: dora.rios@urosario.edu.co
Claudia Birchenall, Email: cibirchenall@yahoo.com.
Darío Pinilla, Email: dario.pinilla@mederi.com.co.
Juan M. Pardo, Email: juan.pardo@urosario.edu.co
Diego F. Josa, Email: diego.josa@shaio.org
Manuel A. Patarroyo, Email: mapatarr.fidic@gmail.com
Juan D. Ramírez, Phone: (+57-1) 297 0200, Email: juand.ramirez@urosario.edu.co
References
- 1.Dubberke ER, Olsen MA. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis. 2012;55(Suppl 2):S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rineh A, Kelso MJ, Vatansever F, Tegos GP, Hamblin MR. Clostridium difficile infection: molecular pathogenesis and novel therapeutics. Exp Rev Anti-Infect Ther. 2014;12(1):131–150. doi: 10.1586/14787210.2014.866515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18(2):247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, Adams V, Moore RJ, Rood JI, McClane BA. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Fut Microbiol. 2014;9(3):361–377. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarro MA, McClane BA, Uzal FA. Mechanisms of action and cell death associated with Clostridium perfringens toxins. Toxins. 2018;10(5):212. doi: 10.3390/toxins10050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rood JI, Adams V, Lacey J, Lyras D, McClane BA, Melville SB, Moore RJ, Popoff MR, Sarker MR, Songer JG, et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe. 2018;53:5–10. doi: 10.1016/j.anaerobe.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH, Society for Healthcare Epidemiology of A, Infectious Diseases Society of A Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31(5):431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 8.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, Group ES Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377(9759):63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 9.Du P, Cao B, Wang J, Li W, Jia H, Zhang W, Lu J, Li Z, Yu H, Chen C, et al. Sequence variation in tcdA and tcdB of Clostridium difficile: ST37 with truncated tcdA is a potential epidemic strain in China. J Clin Microbiol. 2014;52(9):3264–3270. doi: 10.1128/JCM.03487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48(3):770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson S, Jensen JN, Olsen KE. Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J Clin Microbiol. 2011;49(12):4299–4300. doi: 10.1128/JCM.05161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186(2):307–312. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- 13.Roussan DA, Shaheen IA, Totanji WS, Khawaldeh GY, Al Rifai RH. Simultaneous detection of Clostridium perfringens and Clostridium colinum by duplex-polymerase chain reaction. Poult Sci. 2012;91(12):3080–3085. doi: 10.3382/ps.2012-02368. [DOI] [PubMed] [Google Scholar]
- 14.Leslie JL, Young VB. The rest of the story: the microbiome and gastrointestinal infections. Curr Opin Microbiol. 2015;23:121–125. doi: 10.1016/j.mib.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data are within the manuscript.