Sepsis is a complex clinical condition that arises from multiple interacting tissues and unbalanced cell-specific host response mechanisms.1 Neutrophils are the most abundant circulating leukocytes and form the primary line of defense against pathogens.2 Upon activation and subsequent extravasation, neutrophils can engage pathogens by a broad set of actions, including phagocytosis, production of reactive oxygen species, release of antimicrobial peptides and by undergoing NETosis.3 Kinases play an integral role in regulating and enabling intracellular signaling cascades that control these, for neutrophil function, essential processes: for example, NADPH oxygenase assembly is regulated via Raf (RAF proto-oncogene serine/threonine-protein kinase), ERK (MAPK1/3) and MEK kinases3 and the balance of P38 MAPK and ERK upstream of GRK2 (G protein-coupled receptor kinase 2) as well as PAK and PI3K/AKT activity controls neutrophil migration.4,5 However, knowledge on kinome-wide regulation of kinases in primary neutrophils during sepsis and critical illness is limited. Using a kinome profiling approach we here uncover that, relative to patients with non-infectious critical illness, sepsis patients present a suppressed neutrophil kinase activity profile.
This study was incorporated in the MARS (Molecular Diagnosis and Risk Stratification of Sepsis) project (clinicaltrials.gov identifier NCT01905033).6 Consecutive patients aged >18 years admitted to the Intensive Care Unit of the Academic Medical Center (Amsterdam, the Netherlands) between April 2011 and May 2012 were included when they had at least two systemic inflammatory response syndrome (SIRS) criteria (body temperature ≤36°C or ≥38°C, tachycardia >90 beats/min, tachypnea >20 breaths/min or pCO2 <4.3 kPa, leukocyte count <4x109/L or >12x109/L) on the day of admission to the Intensive Care Unit, as described elsewhere.7 Patients with suspected or documented infection were classified as having sepsis, patients without evidence of infection as SIRS controls. Blood was obtained at 9 a.m. on the first morning after admission to the Intensive Care Unit. Age-matched healthy controls were included as controls. Neutrophils were isolated using Ficoll-Paque PLUS (GE Healthcare Life Science, Little Chalfont, UK) followed by erythrocyte lysis with ice-cold erythrocyte lysis buffer (Qiagen, Venlo, the Netherlands). Neutrophil purity was >90% (mean >96%), as determined by flow cytometry based on forward and side scatter, and CD14-PE and CD15-FITC expression on a FACS Calibur (BD Biosciences, San Jose, CA). Kinome profiling was performed using arrays (Pepscan, Lelystad, the Netherlands) containing triplicates of 976 kinase substrates (199 kinases; Online Supplementary Table S1), as described previously.8 Cytokines were measured in EDTA plasma by cytometric bead array (BD Biosciences). The study was approved by the Medical Ethics Committee of the Academic Medical Center, University of Amsterdam, and written informed consent was obtained from all patients (or legal representatives) and healthy subjects. The patients’ characteristics were compared using Student t-tests, or Fisher exact tests where appropriate. Kinome array results were quantified and subsequently analyzed in R 3.5.0 using limma.9 Interactions were analyzed using STRING-DB. Gene ontology term enrichment analysis was performed using clusterProfiler.10
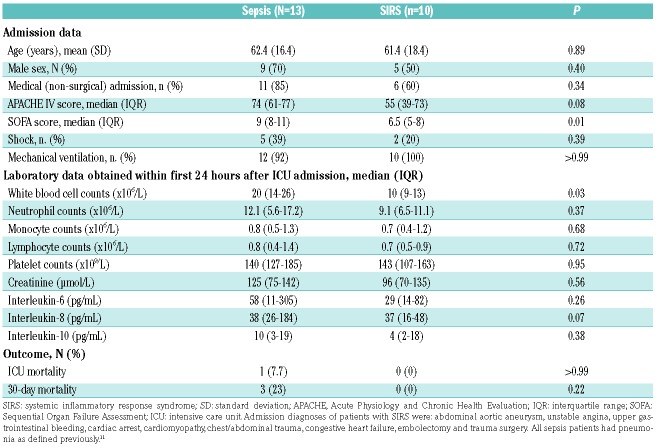
To elucidate the kinome state of blood neutrophils in critically ill patients we performed kinome profiling of neutrophils harvested from patients with sepsis (n=13) or SIRS (n=10) using kinomics array technology; the neutrophil kinome of healthy subjects [n=6, 33% male, mean (SD) age: 63 (14) years] was analyzed as a comparator. Pneumonia (defined as described previously11) was the primary source of infection in all sepsis patients. Sepsis patients presented with more severe disease with higher Sequential Organ Failure Assessment (SOFA) scores compared to those of SIRS patients; other clinical characteristics, including mortality in the Intensive Care Unit and at day 30, did not differ between the groups (Table 1).
Table 1.
Patients’ characteristics.
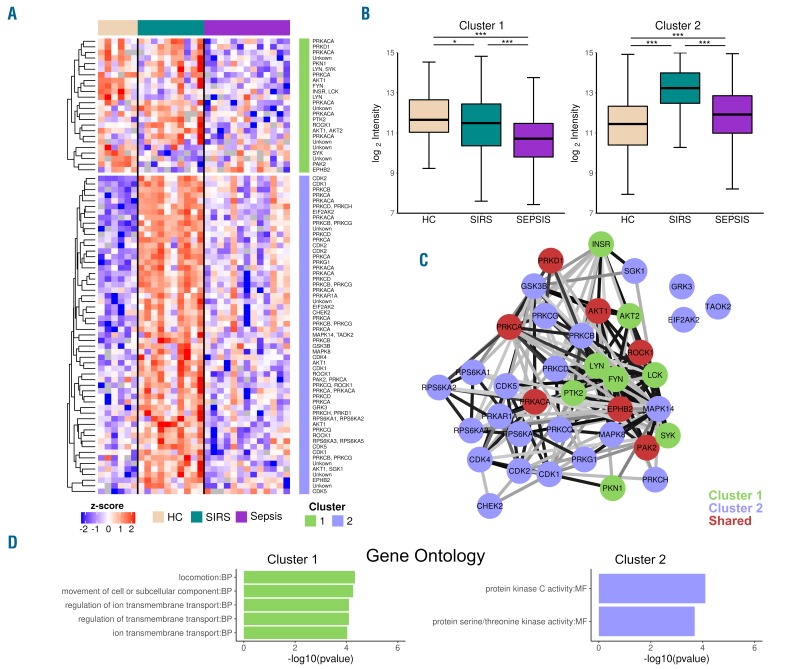
We detected 81 differentially phosphorylated substrates; of these, ten lacked an upstream kinase annotation, while 16 were associated with two upstream kinases. In total 38 kinases displayed differential activity (Figure 1A). For most kinases, multiple substrates were differentially phosphorylated [e.g., 5 substrates for AKT1 (RAC-alpha serine/threonine-protein kinase) and 10 substrates for PRKCA (protein kinase C alpha type)], providing more weight to the interpretation of kinase activities. Two patterns could be observed: a cluster (1: 24 substrates, 15 kinases) with decreased kinase activity, and a larger cluster (2: 57 substrates, 30 kinases) with increased kinase activity in SIRS and sepsis patients compared to that in healthy subjects (Figure 1A,B). In both clusters sepsis kinase activities were greatly reduced compared to those of SIRS neutrophils. Most kinases were unique to one cluster, although substrates for AKT1, PAK2 (serine/threonine-protein kinase PAK 2), ROCK1 (rho-associated protein kinase 1), EPHB2 (ephrin type-B receptor 2), PRKCA (protein kinase C alpha type), PRKD1 (serine/threonine-protein kinase D1) and PRKACA (cAMP-dependent protein kinase catalytic subunit alpha) were present in both clusters (Figure 1C). Thus, overall kinase activities were lower in sepsis patients than in SIRS patients; in cluster 1 kinase activities in neutrophils from sepsis patients were even lower than those in neutrophils from healthy controls. Collectively these findings suggest that neutrophils from sepsis patients have relatively impaired kinase activity, especially when compared to those from SIRS patients. To the best of our knowledge, this has not been shown previously, and literature on kinase activities of primary cells from critically ill patients is very limited. Of note, however, reduced neutrophil effector functions have been described in critically ill sepsis patients.1,12 We did not observe altered activities for kinases generally assessed in targeted studies; for example, of kinase substrates relevant for NFκB activation contained within the array (i.e. for MAP3K8, EIF2AK2, RPS6KA1, MAP3K14, MAP3K14, PRKCZ, CHUK, GSK3B, IKBKB, PRKACA, PRKCZ) of which none was significantly altered (Online Supplementary Table S1). These data are in agreement with a recent study from our group showing that the intracellular levels of phosphorylated NFκB are not altered in neutrophils of patients with sepsis or SIRS, relative to those of healthy controls.13 Our study is limited in that we did not determine the proportions of immature neutrophils in patients; differences in these proportions between sepsis and SIRS patients may influence the results obtained. In addition, the sample size is too small to determine whether sepsis and/or SIRS are associated with specific kinase signatures, which potentially could provide additional information on pathogenesis and possible therapeutic targets.
Figure 1.
Kinome profiles of neutrophils from critically ill patients reveal sepsis kinome anergy. (A,B) Heatmap of 81 differentially phosphorylated substrates between healthy controls (HC, lobster pink), critically ill patients with SIRS (turquoise) or sepsis (purple), annotated for upstream kinases (A) and clustering of K-means discriminates between decreased (cluster 1) and increased (cluster 2) kinase activity (B). The clustering is depicted as a side annotation (cluster 1, green; cluster 2, lilac) in the heatmap and as box and whisker plots (based on log2 intensities). Box and whisker plots depict the median (horizontal lines in the boxes), lower and upper quantiles (boxes), and the smallest and largest observations (whiskers). ***P<0.001, *P<0.05 as determined by the Kruskal-Wallis test with a pairwise Wilcoxon rank sum post-hoc test. (C) STRING protein association network of differentially active kinases: colors indicate kinase substrate cluster membership (cluster 1: green; cluster 2: lilac; kinases with substrates in both clusters: red). Edge opacity is based on the STRING analysis combined score, ranging from 0.5 (light gray) to 1 (black). (D) Top most enriched gene ontology terms per cluster (cluster 1, green; cluster 2, lilac). MF: molecular function, BP: biological process.
To visualize the interaction network of the differentially active kinases we applied STRING analysis (Figure 1C). The members of both clusters displayed a high connectivity, indicating that related processes are regulated in these clusters. As expected, close proximity was observed within cluster 1 between the highly related SRC kinase family members: LYN (tyrosine-protein kinase Lyn), LCK (tyrosine-protein kinase Lck) and FYN (tyrosine-protein kinase Fyn); when applying gene ontology term analyses (Figure 1D), this cluster was enriched for terms of transmembrane transport and locomotion. Others have shown a role for SRC family kinases in neutrophil integrin-dependent responses, such as migration.14 In addition, GRK3 (ADRBK2, beta-adrenergic receptor kinase 2) activity was enhanced in SIRS patients. GRK3 acts as a negative regulator of CXCR4 (C-X-C chemokine receptor type 4) and decreased GRK3 expression was found to enhance T-cell migration.15 For cluster 2, the molecular function term protein kinase C activity was enriched.
In conclusion, by using an unbiased approach subjecting neutrophils from critically ill patients to kinome analysis we highlight that isolated primary neutrophils of sepsis patients have impaired kinase activity when compared to those of critically ill patients without infection. Thus, our findings provide a novel kinome-based perspective to the suppression of neutrophil effector functions in sepsis.
Supplementary Material
Footnotes
Funding: this work was supported by the Center for Translational Molecular Medicine (http://www.ctmm.nl), project MARS (grant 04I-201).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017;17(7):407–420. [DOI] [PubMed] [Google Scholar]
- 2.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33(5):657–670. [DOI] [PubMed] [Google Scholar]
- 3.Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol. 2014;15(7):602–611. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Ma B, Malik AB, et al. Bidirectional regulation of neutrophil migration by mitogen-activated protein kinases. Nat Immunol. 2012;13(5):457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuhler GM, Knol GJ, Drayer AL, Vellenga E. Impaired interleukin-8-and GROalpha-induced phosphorylation of extracellular signal-regulated kinase result in decreased migration of neutrophils from patients with myelodysplasia. J Leukoc Biol. 2005;77(2):257–266. [DOI] [PubMed] [Google Scholar]
- 6.van Vught LA, Klein Klouwenberg PMC, Spitoni C, et al. Incidence, risk factors, and attributable mortality of secondary infections in the intensive care unit after admission for sepsis. JAMA. 2016; 315(14):1469–1479. [DOI] [PubMed] [Google Scholar]
- 7.van Vught LA, Wiewel MA, Hoogendijk AJ, et al. Reduced responsiveness of blood leukocytes to lipopolysaccharide does not predict nosocomial infections in critically ill patients. Shock. 2015;44(2):110–114. [DOI] [PubMed] [Google Scholar]
- 8.Alves MM, Fuhler GM, Queiroz KCS, et al. PAK2 is an effector of TSC1/2 signaling independent of mTOR and a potential therapeutic target for tuberous sclerosis complex. Sci Rep. 2015;5:14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10(2):946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. Omics. 2012; 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein Klouwenberg PMC, Ong DSY, Bos LDJ, et al. Interobserver agreement of Centers for Disease Control and Prevention criteria for classifying infections in critically ill patients. Crit Care Med. 2013; 41(10):2373–2378. [DOI] [PubMed] [Google Scholar]
- 12.Kovach MA, Standiford TJ. The function of neutrophils in sepsis. Curr Opin Infect Dis. 2012;25(3):321–327. [DOI] [PubMed] [Google Scholar]
- 13.Hoogendijk AJ, Garcia-Laorden MI, van Vught LA, et al. Sepsis patients display a reduced capacity to activate nuclear factor-κB in multiple cell types. Crit Care Med. 2017;45(5):e524–e531. [DOI] [PubMed] [Google Scholar]
- 14.Futosi K, Mócsai A. Tyrosine kinase signaling pathways in neutrophils. Immunol Rev. 2016;273(1):121–139. [DOI] [PubMed] [Google Scholar]
- 15.Balabanian K, Levoye A, Klemm L, et al. Leukocyte analysis from WHIM syndrome patients reveals a pivotal role for GRK3 in CXCR4 signaling. J Clin Invest. 2008;118(3):1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.