Abstract
B-cell acute lymphoblastic leukemia is the commonest childhood cancer. In infants, B-cell acute lymphoblastic leukemia remains fatal, especially in patients with t(4;11), present in ~80% of cases. The pathogenesis of t(4;11)/KMT2A-AFF1+ (MLL-AF4+) infant B-cell acute lymphoblastic leukemia remains difficult to model, and the pathogenic contribution in cancer of the reciprocal fusions resulting from derivative translocated-chromosomes remains obscure. Here, “multi-layered” genome-wide analyses and validation were performed on a total of 124 de novo cases of infant B-cell acute lymphoblastic leukemia uniformly diagnosed and treated according to the Interfant 99/06 protocol. These patients showed the most silent mutational landscape reported so far for any sequenced pediatric cancer. Recurrent mutations were exclusively found in K-RAS and N-RAS, were subclonal and were frequently lost at relapse, despite a larger number of non-recurrent/non-silent mutations. Unlike non-MLL-rearranged B-cell acute lymphoblastic leukemias, B-cell receptor repertoire analysis revealed minor, non-expanded B-cell clones in t(4;11)+ infant B-cell acute lymphoblastic leukemia, and RNA-sequencing showed transcriptomic similarities between t(4;11)+ infant B-cell acute lymphoblastic leukemias and the most immature human fetal liver hematopoietic stem and progenitor cells, confirming a “pre-VDJ” fetal cellular origin for both t(4;11) and RASmut. The reciprocal fusion AF4-MLL was expressed in only 45% (19/43) of the t(4;11)+ patients, and HOXA cluster genes are exclusively expressed in AF4-MLL-expressing patients. Importantly, AF4-MLL/HOXA-expressing patients had a significantly better 4-year event-free survival (62.4% vs. 11.7%, P=0.001), and overall survival (73.7 vs. 25.2%, P=0.016). AF4-MLL expression retained its prognostic significance when analyzed in a Cox model adjusting for risk stratification according to the Interfant-06 protocol based on age at diagnosis, white blood cell count and response to prednisone. This study has clinical implications for disease outcome and diagnostic risk-stratification of t(4;11)+ infant B-cell acute lymphoblastic leukemia.
Introduction
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is the most frequent cancer in children.1 Current 5-year survival rates in pediatric BCP-ALL approach 90%. However, BCP-ALL in infants (iBCP-ALL; <1 year of age) remains clinically challenging with an aggressive early clinical presentation in uniquely vulnerable hosts.2 Approximately 80% of iBCP-ALL are diagnosed with chromosomal rearrangements involving the mixed-lineage leukemia (KMTA2, also called MLL) gene, located on 11q23,3–5 which confers a dismal prognosis especially in patients carrying the t(4;11)/KMT2A-AFF1+ (MLL-AF4+).6–8
MLL is a H3K4 histone methyltransferase required for normal hematopoiesis and HOX gene expression.9,10 Leukemia transformation by MLL fusions requires the recruitment of the H3K79 histone methyltransferase Dot1L to the MLL transcriptional complex.11,12 Indeed, an H3K79 methylation profile defines both mouse and human t(4;11)/MLL-AF4+ BCP-ALL.13 Importantly, MLL rearrangements (MLLr) occur prenatally during embryonic/fetal hematopoiesis, and the concordance rate for iBCP-ALL in identical twins with a monochorionic placenta is close to 100%.14–17 This, coupled to the extremely short latency, suggests that MLL fusions might be sufficient for leukemogenesis.4 Accordingly, genome-wide studies using both single nucleotide polymorphism arrays and whole-genome sequencing revealed that MLLr iBCP-ALL has a very low frequency of somatic mutations with the predominant clone carrying ~1.3 non-silent mutations and one copy number alteration.18–20 Although these studies were performed at low coverage sequencing they reinforce the concept that MLLr iBCP-ALL requires few additional mutations to induce full transformation. In contrast, MLL-AF4-induced leukemogenesis has proven difficult to model.4,9 With the exception of a recent work by Lin et al.21,22 who fused human MLL to murine Af4, creating an artificial leukemogenic human-mouse chimeric fusion, current murine and humanized models of MLL-AF4+ BCP-ALL do not faithfully recapitulate the disease pathogenesis/phenotype, suggesting that MLL-AF4 per se is insufficient to initiate leukemogenesis.23–28
The few mutations and copy number alterations present in MLLr iBCP-ALL seem subclonal and not always retained at relapse.20 Intratumor heterogeneity drives clonal evolution in response to microenvironmental cues and cytotoxic treatment and therefore recurrent mutations at diagnosis and relapse may be found in minor but clinically relevant subclones.29 Here we aimed to address the clinical relevance of subclonal mutations and gene expression signatures in a large cohort of iBCP-ALL. To do this, we performed deeper exome sequencing along with whole-genome DNA- and RNA-sequencing on a large cohort of 50 MLLr and non-MLL iBCP-ALL patients uniformly treated and followed up according to an Interfant treatment protocol.30 Similarly to Anderson et al.,20 we report a silent mutational landscape in iBCP-ALL irrespective of the MLL rearrangement/status. However, strikingly, our genome-wide DNA and RNA analyses revealed new, clinically relevant information about disease outcome and cell-of-origin for t(4;11) and RAS mutations.
Methods
Patients
Bone marrow or peripheral blood samples from 124 infants (<12 months old) diagnosed with either pro-B or pre-B-cell ALL were used in this study. The discovery cohort of patients was composed of 42 de novo cases: 27 with the t(4;11) encoding for MLL-AF4, five with the t(9;11) encoding for KMT2A-MLLT3 (MLL-AF9) and ten without MLLr (non-MLL B-other BCP-ALL without numerical or structural chromosomal abnormalities reported at diagnosis). Additionally, for eight MLL-AF4+ iBCP-ALL patients matched diagnostic-relapse samples were available allowing for longitudinal studies. MLL rearrangements were confirmed by fluorescence in-situ hybridization.31,32 For validation, an additional cohort of patients, comprising 43 MLL-AF4+, 11 MLL-AF9+, and 28 non-MLL iBCP-ALL cases, was used. All patients were enrolled in the Interfant99 treatment study. Bone marrow samples were collected at Erasmus MC-Sophia Children’s Hospital (Rotterdam, the Netherlands), Armand Trousseau Hospital (Paris, France), and San Gerardo Pediatric Hospital (Monza, Italy). Complete remission bone marrow samples were available for all patients. The clinical and genetic features of the patients are presented in Online Supplementary Table S1. As a control for the RNA-sequencing studies, CD34+CD19+ healthy B-cell progenitors were purified by fluorescence-activated cell sorting (FACS) from 22-week old human fetal livers (FL) as previously described.32 FL hematopoietic stem and progenitor cells (HSPC) were processed and FACS-purified from second trimester human FL as previously described.33 Briefly, cells were processed and stained for flow cytometry with up to ten fluorophore-conjugated monoclonal antibodies [antibodies (clone): CD34PECy7 (8G12), CD45RA FITC (HI100), CD19APC (HIB19), CD123PE (9F5), CD90 PECy5 (5E10), CD38 Pacific blue (HIT2), lineage cocktail APC (CD2 (RPA-2.10)/CD3 (OKT3)/CD14 (61D3)/CD16(CB16)/CD19 (HIB19)/CD56 (TULY56)/CD235a (HIR2)]. FACS was performed using a BD FACSAria II (Becton Dickinson). Gates were set with unstained and fluorescence minus one controls, on viable cells. Data were analyzed using FlowJo software (Tree Star). Gating strategies are as described in the results section. The study was approved by the Barcelona Clinic Hospital (2013/8529) and Hammersmith and Queen Charlotte’s Hospital (04/Q0406/145) research ethics committees.
Statistical analysis
For quantitative variables, a one-tailed t-test was used to identify significant differences between groups. For qualitative variables, a Fisher exact test was used in order to identify significant differences between groups of patients. Software for analysis of mutations and gene expression have their own statistical models explained in detailed in the references. Where multiple tests were performed the significance is shown corrected for multiple testing. Mutation allele frequency evolution was plotted with the R package distribution Fishplot. Patterns Fisher exact test was used to assess the association between clinical characteristics and presence of RAS mutations or AF4-MLL expression. Event-free survival was defined as time from diagnosis to first event, i.e. resistance, relapse, death from any cause, or second malignant neoplasm. Observation periods were censored at the time of last contact when no events were reported. Event-free survival curves were estimated with the Kaplan-Meier method and standard errors (SE) were calculated according to Greenwood. Differences in event-free survival and overall survival between groups were compared with the log-rank test. Analysis of the prognostic relevance of AF4-MLL/HOXA expression in combination with risk stratification according to the Interfant-06 protocol (based on age at diagnosis, white blood cell count and response to prednisone) was performed with the Cox model and the Wald test. All tests were two-sided. Analyses were performed using SAS 9.2.
DNA, RNA and B-cell receptor (VDJ) repertoire genome-wide analyses and data analysis
Preparation and analysis of all DNA and RNA genome-wide high-throughput sequencing is detailed in the Online Supplementary Methods, Online Supplementary Figure S1 and Online Supplementary Table S2.
Results
At diagnosis infant B-cell precursor acute lymphoblastic leukemia shows a silent mutational landscape irrespective of MLL gene status
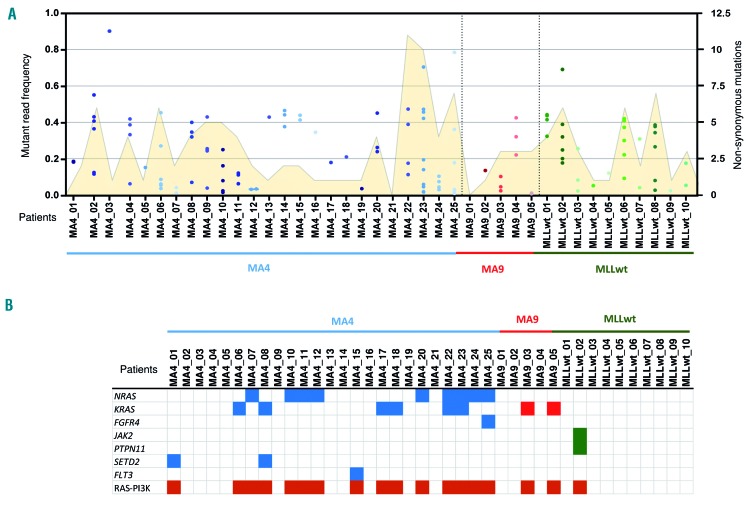
Whole-exome sequencing and whole-genome sequencing analyses showed a silent mutational landscape in the three iBCP-ALL subtypes studied here: MLL-AF4+, MLL-AF9+ and non-MLL (n=42 patients, Online Supplementary Table S1). Our study revealed an average of one genomic rearrangement and 2.5 non-silent single nucleotide variants, a 2-fold higher number than that reported by Andersson et al.,20 likely reflecting the 3-fold larger sequencing coverage (Figure 1A, Online Supplementary Figure S1 and Online Supplementary Table S3). All mutations found at diagnosis were validated using orthogonal methods. This mutational frequency is the lowest described for any other pediatric tumor type according to recent reports34 (Online Supplementary Figure S2). Intriguingly, one third of the mutations validated showed a mutant allele frequency (MAF) <20% indicating that iBCP-ALL contains genetically different intratumoral subclones despite its genomic stability, likely explaining the higher mutational load than that reported by Andersson et al.20 (Figure 1A and Online Supplementary Table S3). Despite the paucity of mutations, ~80% of the validated protein-coding mutations (90/116) are predicted to produce deleterious effects on the protein (Online Supplementary Figure S3A) which might support a strong selective pressure in iBCP-ALL. To gain insights into the molecular mechanisms underlying the accumulation of mutations, we analyzed the enrichment of specific mutational signatures as described by Alexandrov et al.35 In the MLL-AF4+ iBCP-ALL subgroup we identified a significant enrichment of signature 1 characterized by the accumulation of C>T/G>A transitions, linked to a spontaneous deamination of 5-methylcytosine (Online Supplementary Figure S3B,C).35 This mutational signature has also been described in other pediatric tumors, suggesting that iBCP-ALL is not subjected to a specific mutational signature.
Figure 1.
Somatic mutations detected by whole-exome sequencing in the discovery cohort of infant B-cell precursor acute lymphoblastic leukemia. (A) Total number of mutations identified in each individual patient. The total number of non-synonymous mutations (yellow area, right Y axis) and mutant allele frequency (MAF) for each mutation (individual dots, left Y axis) are represented. (B) Oncodrive software identified the PI3K-RAS pathway as the only recurrently mutated pathway in infant B-cell precursor acute lymphoblastic leukemia. The distribution of mutations in genes of the PI3K-RAS pathway is shown for all patients within the three iBCP-ALL subgroups: [total 42 patients: 27 t(4;11)+, 5 t(9;11)+ and 10 MLLwt].
We also determined the molecular breakpoint of all MLLr at the base-pair level. In t(4;11)/MLL-AF4+ iBCP-ALL, the AF4 breakpoints were almost invariably localized within intron 3 whereas MLL breakpoints were found between introns 9 and 11 (Online Supplementary Table S4).3 We found whole-genome sequencing reads compatible with an AF4-MLL reciprocal rearrangement in all samples (Online Supplementary Figure S4). AF4-MLL genomic breakpoints were validated by polymerase chain reaction capillary sequencing and they were located nearby MLL-AF4 breakpoints, confirming a reciprocal chromosomal translocation.
RAS-PI3K is the only recurrently mutated pathway in infant B-cell precursor acute lymphoblastic leukemia with NRAS mutations being significantly more frequent in t(4;11)+ patients
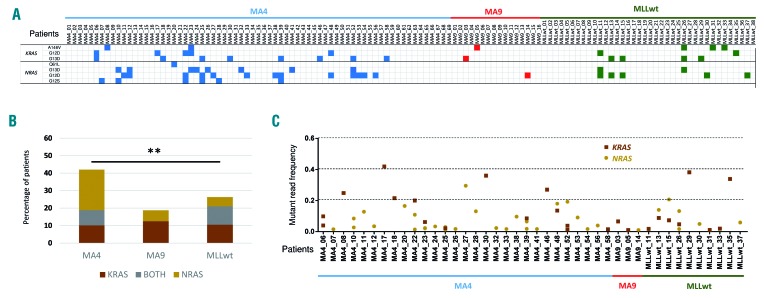
Despite the low number of mutations found per sample, 38% of the sequenced iBCP-ALL patients displayed activating/gain-of-function mutations in either KRAS or NRAS. Additional mutations in other genes members of the RAS-PI3K pathway such as FGFR4, JAK2, PTPN11, SETD2, or FLT3 were also identified (Figure 1B). To further validate the unique recurrence of KRAS and NRAS mutations, we performed targeted sequencing of these mutations in a large, additional validation cohort of infant patients (n=82) and confirmed that 34% of the iBCP-ALL cases carry mutations in either KRAS or NRAS36 (Figure 2A). Interestingly, the overall frequency of RAS mutations differed slightly between the different cytogenetic subgroups of iBCP-ALL, with the MLL-AF4+ subgroup showing the highest frequency (42%) and the MLL-AF9+ subgroup the lowest (19%). This difference was basically attributed to the frequency of NRAS mutations, which was 6-fold more common in the MLL-AF4+ subgroup (32% vs. 6%, Fisher exact test P=0.01) (Figure 2A,B).
Figure 2.
Frequent somatic mutations in RAS genes in both the discovery and validation cohorts. (A) Specific KRAS and NRAS mutations recurrently found in each patient by high coverage targeted sequencing. (B) Proportions of patients with mutations in KRAS (brown), NRAS (yellow) or both (gray) within the three infant B-cell precursor acute lymphoblastic leukemia subgroups. (C) Mutant allele frequency of KRAS (brown squares) or NRAS (yellow circles) mutations in each individual patient. Discovery cohort, n=42; validation cohort, n=82.
Surprisingly, we observed that many iBCP-ALL patients had mutations in both KRAS and NRAS, or more than one (different) mutation in the same gene (Figure 2A,C). To further analyze the biological contribution of KRAS and NRAS mutations, we calculated the MAF of individual mutations and observed that the majority of patients who had a single RAS mutation (either KRAS or NRAS) had MAF scores between 0.20 and 0.45, suggesting that the mutation is present in a major leukemic subclone (P=0.0025). By contrast, those patients harboring two or more RAS mutations displayed MAF scores between 1% and 20%, compatible with these RAS mutations being in distinct and smaller leukemic subclones. We then analyzed the impact of RAS mutations on disease outcome and found no clinical correlation of RAS mutations with either clinical outcome (overall survival, event-free survival, central nervous system infiltration) or diagnostic parameters (gender, age, percentage of blasts and white blood cells) (Online Supplementary Figure S5).
Evidence of clone selection and genomic instability at relapse
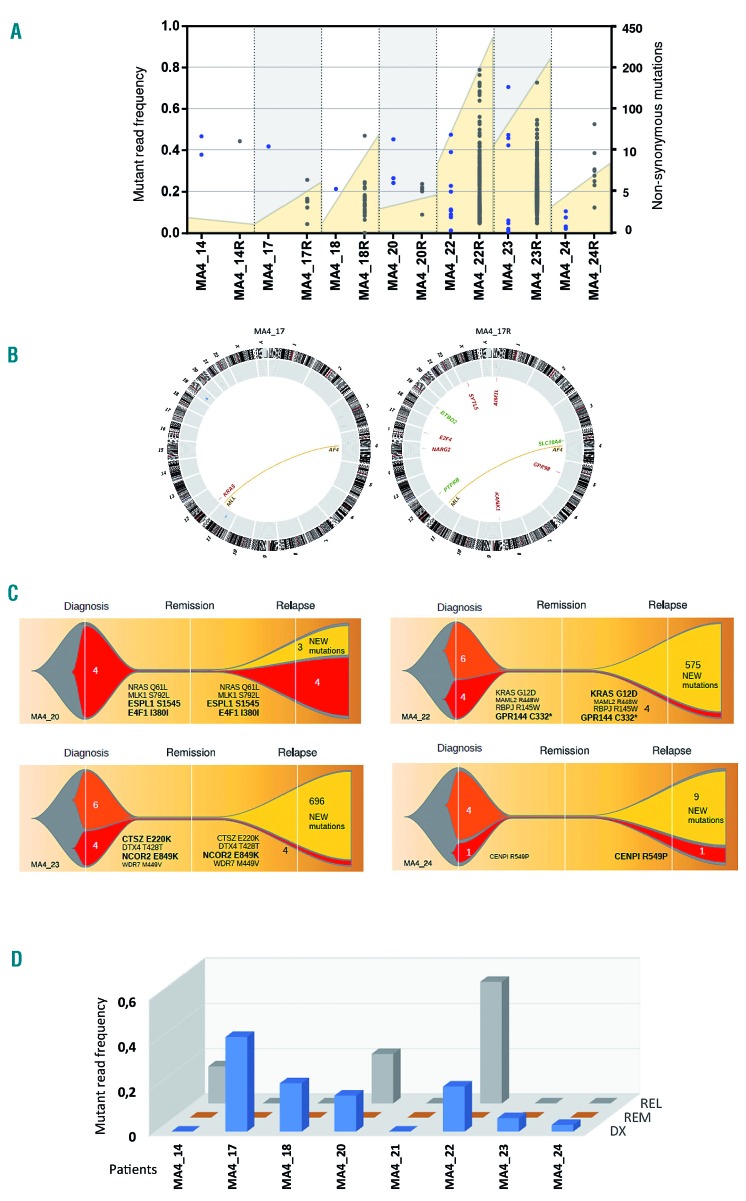
Paired diagnostic-relapse samples were available for eight MLL-AF4+ iBCP-ALL patients, permitting longitudinal studies. Whole-exome sequencing revealed an 8-fold increase in the number of somatic non-synonymous mutations at relapse (19.5 mutations/patient, range:1-434, paired t-test P=0.03) (Figure 3A,B and Online Supplementary Table S3). We performed orthogonal validation for 160 random mutations, and 90% and 75% of mutations with MAF >15% and <15%, respectively, were confirmed (data not shown). Similarly to diagnosis, the majority of the somatic mutations found at relapse had MAF commonly <30%, suggesting the existence of multiple leukemic sub-clones (Figure 3A). Importantly, none of the new de novo somatic mutations found at relapse was found in more than one patient, likely reflecting an intrinsic genomic instability of leukemic clones surviving induction/consolidation chemotherapy. This is further reflected by a significant enrichment of signature 6 associated with defective DNA mismatch repair, including a higher number of small indels, observed in MLL-AF4+ patients at relapse (Online Supplementary Figure S6A).
Figure 3.
Clonal evolution and genomic instability at relapse. (A) Total number of mutations identified for each patient in paired diagnostic-relapse samples. Total number of non-synonymous mutations (yellow area, right Y axis) and mutant allele frequency (MAF) for each mutation (individual dots, left Y axis) are represented for paired diagnostic and relapsed (R) samples. (B) Circos plot representation of the total number of mutations identified at diagnosis and relapse for a representative patient (MA4_17). Genomic rearrangements are represented by lines connecting both breakpoints. Copy number alterations (blue=gains, red=losses) are represented by the outer gray circle. Somatic mutations (both single nucleotide variants and indels) are depicted in the center of the circle and the affected gene is indicated. (C) Graphic representation of clonal evolution in paired diagnostic (DX)-relapsed (RL) samples. The number of unique somatic mutations called at diagnosis (orange), relapse (yellow) or shared between DX and REL (red) are indicated. Bigger gene names indicate higher MAF for the mutations shared at DX and REL. (D) Dynamics of RAS-mutated clones identified as MAF in matched DX-Remission-REL trios (n=8).
To delineate the evolutionary clonal structure from diagnosis to relapse, we performed high-coverage targeted sequencing on the identified mutations in paired diagnostic-remission-relapse samples.37 Importantly, the main leukemic clone at relapse was always present at diagnosis although in some cases with a very low MAF, suggesting a chemotherapy-induced clonal pressure selecting for resistant/adapted leukemic subclones (Figure 3C). Interestingly, we found a correlation between the number of mutations and time to relapse in MLL-AF4+ patients, with a trend towards a higher mutational load in patients with late relapses (Online Supplementary Figure S6B). We next analyzed the clonal evolution of RAS-mutated leukemic clones at relapse. We found that the contribution of the RAS mutations varied among patients: one-third of the iBCP-ALL patients had RAS-mutated clones at relapse (MA4_20 and MA4_22 increased the size of the RAS-mutated initial clone and in MA4_14 a de novo RAS mutation emerged), whereas it was lost in two-thirds of the patients (MA4_17, MA4_18, MA4_23, MA4_24) (Figure 3D). This indicates that infants with MLL-AF4+ BCP-ALL relapse irrespective of the status of KRAS and NRAS. Thus, subclones carrying KRAS mutations do not exert an advantage over non-mutated clones, despite representing a recurrent genetic insult at diagnosis. Hence, this would argue against a leukemia-initiating role for RAS mutations.38 Alternatively, RAS mutations might indeed be leukemogenic drivers, but the treatment-induced genetic instability observed at relapse may compensate de novo RAS mutations, acting as new leukemia drivers cooperating with MLL-AF4 during relapse.
HOXA cluster genes are only expressed in t(4;11)+ patients expressing the reciprocal fusion AF4-MLL which determines clinical outcome
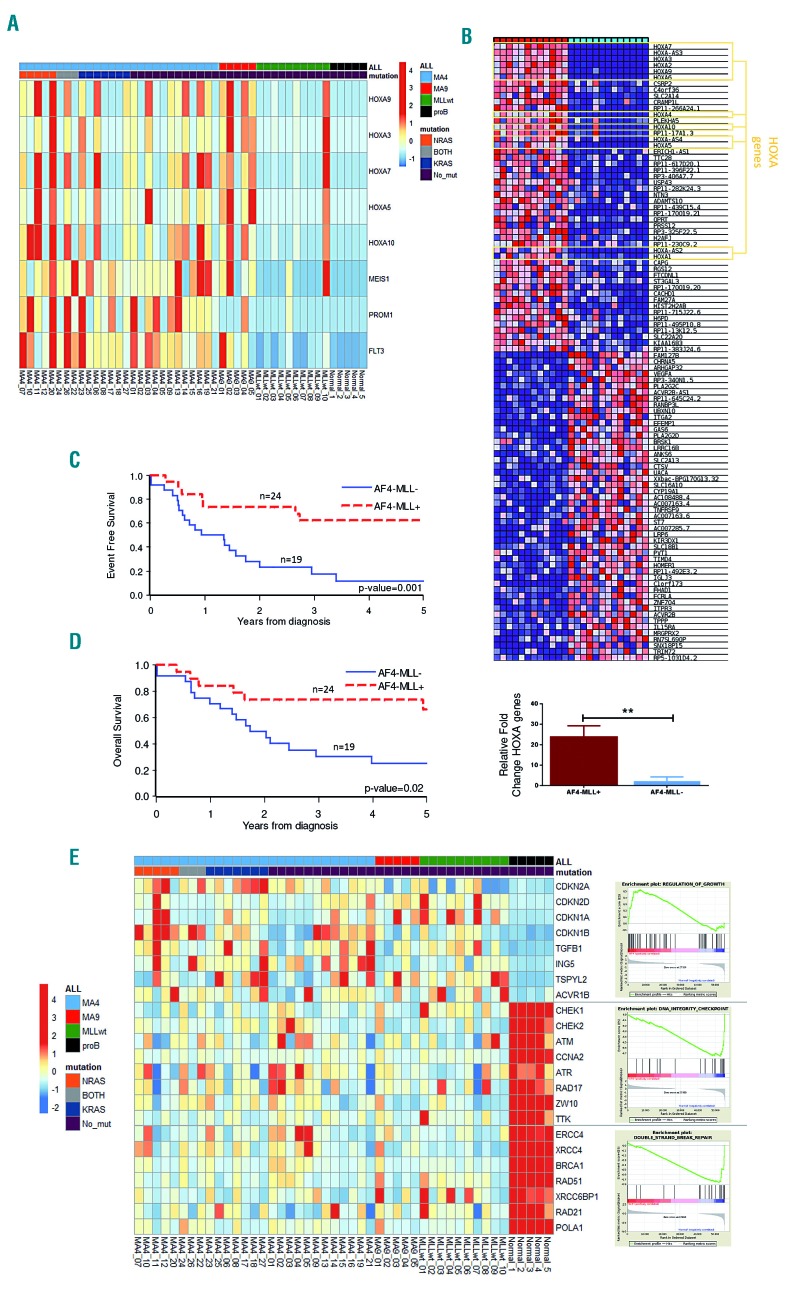
To gain insights into the mechanisms underlying leukemogenesis in these mutationally silent MLLr and MLL germline iBCP-ALL patients, we performed RNA-sequencing in the discovery cohort of patients (n=42) using FL-derived CD34+CD19+ healthy B-cell progenitors as controls, as these cells most likely represent the healthy counterparts of the leukemic blast stalled at the pro/pre-B-cell differentiation stage. We first surveyed the expression of the genes previously reported to be specific to either MLLr iBCP-ALL or specifically to MLL-AF4+ iBCP-ALL39. RNA-sequencing profiling confirmed that these genes segregate patients according to the molecular subtype, MLL-AF4+, MLL-AF9+ and MLL germline (Online Supplementary Figure S7). We also observed, at diagnosis, a strong upregulation of the MLL target genes FLT3,40 MEIS1, PROM1 and HOXA genes in many of our MLLr iBCP-ALL samples but not in MLL germline samples (t-test, P<0.05) (Figure 4A), thus validating our RNA-sequencing approach.
Figure 4.
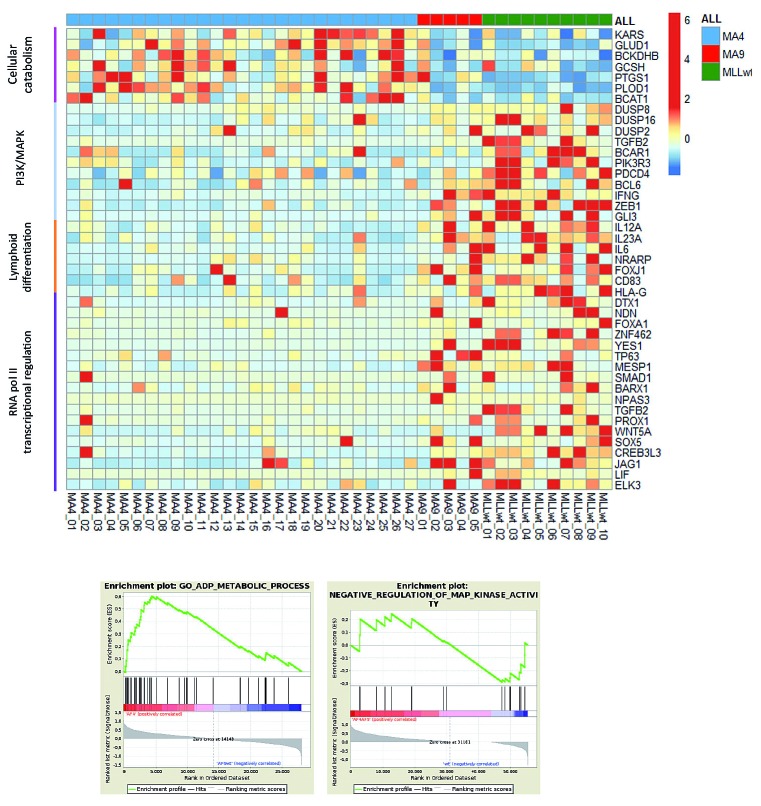
Transcriptional signature of infant B-cell precursor acute lymphoblastic leukemia samples. (A) Heatmap representing FLT3, PROM1, MEIS1 and HOXA gene expression according to the infant B-cell precursor acute lymphoblastic leukemia (iBCP-ALL) cytogenetic group and RAS mutations. (B) Top panel: heatmap showing HOXA cluster gene expression according to the expression of the reciprocal fusion AF4-MLL. Bottom panel: quantitative polymerase chain reaction validating high expression of HOXA cluster genes in t(4;11) iBCP-ALL patients expressing AF4-MLL. (C,D) Four-year event-free survival (C) and overall survival (D) Kaplan-Meier curves for t(4;11) iBCP-ALL patients according to AF4-MLL expression, n=43 t(4;11)+ patients. (E) Heatmap representation of selected genes for the signaling pathways most significantly deregulated. Right panels represent positive pathway enrichment called by gene set enrichment analysis software. Total 42 patients: 27 t(4;11)+, 5 t(9;11)+ and 10 MLLwt.
Strikingly, the reciprocal AF4-MLL fusion gene was discernibly expressed in 19/43 (45%) of the t(4;11)+ iBCP-ALL samples, and its expression was always maintained at relapse (data not shown). We then compared the genes differentially expressed between AF4-MLL-expressing and non-expressing t(4;11)+ patients and found a striking positive correlation between the expression of the HOXA gene cluster and overexpression of the reciprocal AF4-MLL fusion (t-test, P=0.002) (Figure 4B). These AF4-MLL/HOXA-expressing patients (n=19) had a significantly better prognosis than those lacking AF4-MLL/HOXA expression (n=24). Four-year event-free and overall survival rates were 62.4% (SE, 11.3%) versus 11.7% (SE, 10.2%) (P=0.001) (Figure 4C), and 73.7% (SE, 10.1%) versus 25.2% (SE, 10.3%) (P=0.016) (Figure 4D), respectively. When “AF4-MLL expression” was analyzed in a Cox model adjusting for risk stratification (medium risk or high risk according to the Interfant-06 protocol based on age at diagnosis, white blood cell count and response to prednisone), it retained its prognostic significance with a hazard ratio for patients lacking AF4-MLL expression of 3.42 [95% confidence interval (95% CI): 1.35-8.63; P=0.01) compared to those expressing AF4-MLL/HOXA, while risk group was not significant (HR for high risk vs. medium risk, 1.34; 95% CI: 0.59-3.03; P=0.49). This is the first study showing that AF4-MLL overexpression correlates very well with transcriptional deregulation of the HOXA gene cluster in iBCP-ALL and that the co-expression of AF4-MLL and HOXA gene cluster identifies a subgroup of t(4;11)+ iBCP-ALL with a very more favorable clinical outcome.
We next explored new molecular pathways involved in the pathogenesis of iBCP-ALL, by performing an unbiased transcriptional analysis of the RNA-sequencing data from the iBCP-ALL patients. We found deregulated expression of a total of 3,905 genes, of which 2,575 (66%) were upregulated and 1,330 (34%) downregulated as compared with those of healthy FL-derived B-cell progenitors, illustrating the global transcriptional activation nature of MLL fusions (Online Supplementary Figure S8).25,41 Furthermore, a significant upregulation of genes involved in the control of cell growth, including the CDK inhibitors P21, P16, P19, P27 and components of the transforming growth factor-β pathway such as TGFB1, SMAD and ACVR1B, was observed in iBCP-ALL (Figure 4D and Online Supplementary Figure S9). By contrast, iBCP-ALL showed a robust downregulation of genes involved in DNA integrity checkpoints such as CHEK1, CHEK2, ATM, ATR and RAD17, and in double-strand break repair genes including ERCC4, BRCA1, POLA1 and RAD51 (Figure 4E and Online Supplementary Figure S9). These transcriptional changes were validated by quantitative reverse transcriptase polymerase chain reaction in ten patients per group (Online Supplementary Figure S10). Deregulation of DNA integrity checkpoints and double-strand break repair genes may well contribute to the genomic instability observed at relapse, and might explain the enrichment in C>T/G>A transitions, associated with the spontaneous deamination of 5-methylcytosine (Online Supplementary Figures S3 and S6).
By using FL-derived normal B-cell progenitors as controls, differences between leukemic blasts and their normal counterparts could be identified but this does not allow the definition of transcriptomic differences within the iBCP-ALL cytogenetic groups. We, therefore, analyzed the RNA-sequencing data comparing the genes differentially expressed in MLL-AF4+ versus MLL-AF9+ and MLL-wildtype iBCP-ALL patients, without considering normal B-cell progenitors as controls. A gene ontology analysis (gene set enrichment analysis, GSEA) performed with the genes differentially expressed revealed that MLL-AF4+ patients show, as compared to both MLL-AF9+ and MLL-wildtype patients, a significant upregulation of genes associated with cellular catabolism, coupled to a significant downregulation of negative regulators of the PI3-MAPK pathway, as well as of genes involved in lymphoid differentiation and RNApol II transcriptional regulation (Figure 5). This suggests, respectively, a metabolic change in MLL-AF4+ cells towards rapid energy generation while reinforcing the basal hyperactivation of the PI3-MAPK pathway by RAS mutations (Figures 1 and 2), a poorly differentiated cellular origin of t(4;11), and an impairment of the normal function of AF4, a key component of the RNApol II transcription complex.
Figure 5.
Specific transcriptional differences between MLL-AF4+ and MLL-AF9+ or MLLwt infant B-cell precursor acute lymphoblastic leukemia patients. Here, FL-derived CD34+CD19+ progenitors were not included as normalizers in the analysis in order to avoid potential bias. Gene set enrichment analysis (GSEA) was performed with the genes differentially expressed between MLL-AF4+ patients and MLL-AF9+ or MLLwt patients. MLL-AF4+ infant B-cell precursor acute lymphoblastic leukemia (iBCP-ALL) patients showed a significant overexpression of genes associated with cellular catabolism, coupled to a significant downregulation of negative regulators of the PI3-MAPK pathway, as well as of genes involved in lymphoid differentiation and RNApol II transcriptional regulation as compared to both MLL-AF9+ and MLLwt iBCP-ALL patients. The bottom panels represent positive pathway enrichment called by GSEA software. Total 42 patients: 27 t(4;11)+, 5 t(9;11)+ and 10 MLLwt.
Deep-sequencing analysis of B-cell receptor repertoires suggests a hematopoietic stem cell/early pre-VDJ progenitor as the cell-of-origin for t(4;11) and RAS mutations
We next analyzed BCR repertoires to gain insights into the immunoglobulin heavy chain (IgH) rearrangement clonal composition of paired diagnostic-relapse samples from t(4;11)+ iBCP-ALL (4 pairs). BCR are generated through DNA recombination during B-cell differentiation and represent unique markers for each B-cell clone. Because the BCR sequence provides a molecular tag for each B-cell clone, high-throughput sequencing of BCR provides a detailed analysis of B-cell population dynamics and clone tracking.42,43 BCR sequencing was therefore performed to address whether t(4;11)+ iBCP-ALL cells expressed fully rearranged BCR from which increased levels of B-cell clonal expansion may be observed and to determine whether there are detectable levels of B-cell clonal persistence over time indicative of B-cell clonal survival. BCR sequencing was performed on t(4;11)/MLL-AF4+ iBCP-ALL peripheral blood samples (blasts >98%) using a polymerase chain reaction-based method37 with additional incorporation of unique molecular barcodes, allowing for accurate quantitation of relative B-cell clone frequency. After BCR sequence filtering, each sample yielded between 1,583-46,863 BCR (1,213-38,426 unique BCR) (Online Supplementary Table S5).
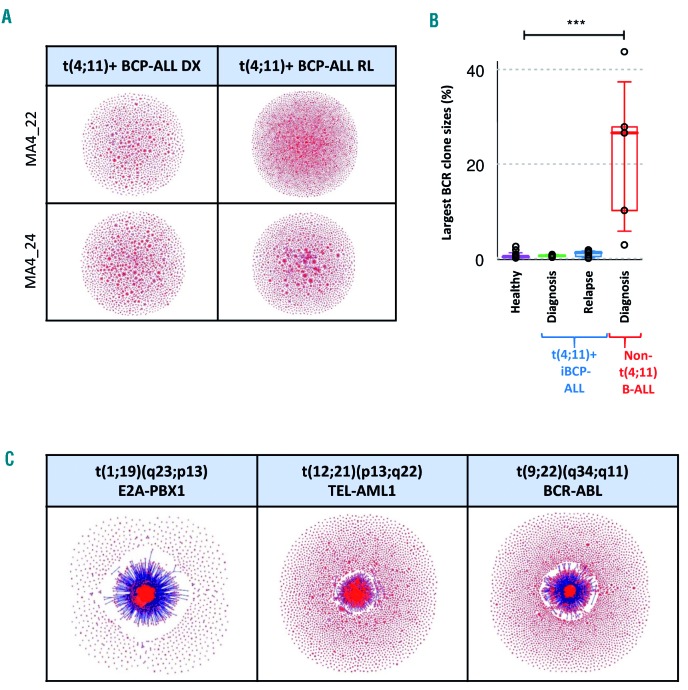
We first delineated the relative clonality in these patients, and found that the BCR repertoires from t(4;11)+ patients did not exhibit significantly expanded VDJ-rearranged B-cell clones (Figure 6A) either at diagnosis or relapse compared to healthy peripheral blood samples (Figure 6B). This is in contrast to non-MLL BCP-ALL patients (n=5) including three patients with t(1;19)/TCF3-PBX1 (EF2-PBX1), one with t(12;21)/ETV6-RUNX1 (TEL-AML1) and one with t(9;22)/BCR-ABL1, which were all found to be significantly clonal, with large B-cell clones comprising ~3-40% of total BCR (Figure 6B,C).37 Given the persistence of both t(4;11) and RAS mutations in MLL-AF4+ iBCP-ALL, the lack of B-cell clonal expansion or persistence supports the model that t(4;11)/MLL-AF4+ iBCP-ALL malignant cells are developmentally stalled at the pro-B stage, and that the cellular origin of such genomic drivers has to be pre-VDJ stem/progenitor cells.
Figure 6.
Analysis of B-cell receptor repertoires suggest a hematopoietic stem cell/early pre-VDJ progenitor as the cell-of-origin for t(4;11)/MLL-AF4+ infant B-cell precursor acute lymphoblastic leukemia. (A) Cloud-plots of B-cell receptor (BCR) repertoires from two representative t(4;11)+ infant B-cell precursor acute lymphoblastic leukemia (iBCP-ALL) patients depicting the existence of many minor non-expanded B-cell clones either at diagnosis or relapse. Each vertex represents a unique BCR sequence, and the relative vertex size is proportional to the number of identical reads. (B) Largest BCR clone size in t(4;11)+ iBCP-ALL, healthy individuals and non-t(4;11)+ pediatric BCP-ALL. (C) Cloud-plots of BCR repertoires of representative t(1;19)/E2A-PBX1+, t(12;21)/TEL-AML1+ and t(9;22)/BCR-ABL+ patients showing high clonality of B-cell clones. The samples from the iBCP-ALL patients who were BCR-sequenced were four MLL-AF4+ diagnostic-relapse pairs, three E2A-PBX1+ samples, one TEL-AML1+ sample and one BCR-ABL+ sample.
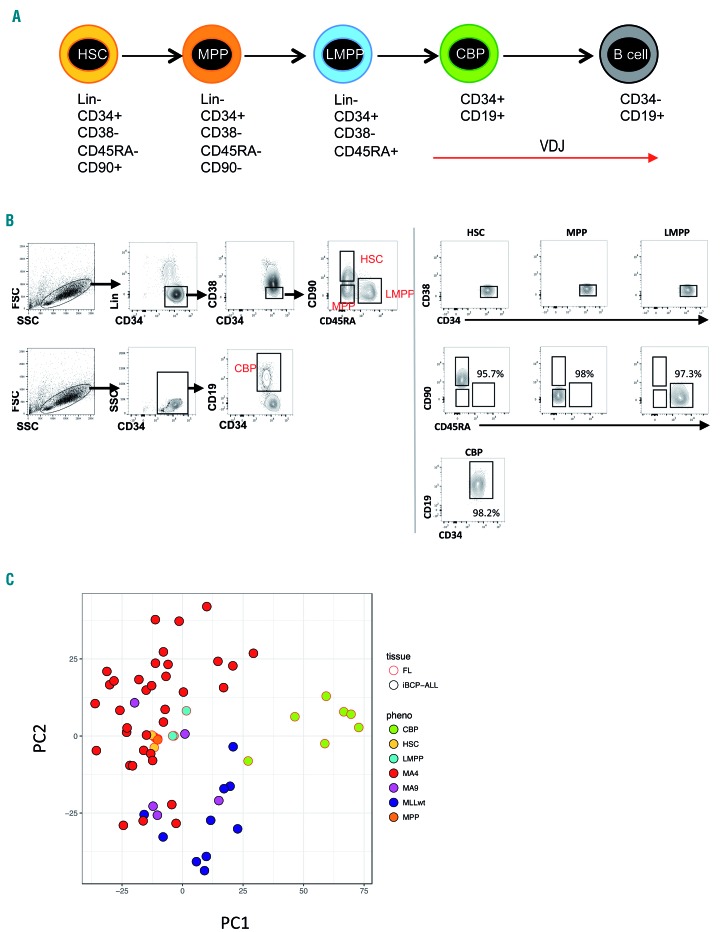
Finally, in order to understand whether the fetal cell-of-origin in iBCP-ALL lies upstream of committed B progenitors, we compared the transcriptome of iBCP-ALL blasts (n=42) with that of highly purified human FL HSPC populations (3-7 for each population) (Figure 7A,B and Online Supplementary Table S6) by RNA-sequencing. In keeping with the results of the BCR analysis, our principal component analysis revealed a gene expression signature for primitive Lin−CD34+CD38−CD19− FL HSPC (hematopoietic stem cells, multipotent progenitors, and lymphoid-primed multipotent progenitors, which lie upstream of B progenitors) very similar to t(4;11)+ iBCP-ALL, while FL-committed B progenitors clustered as a transcriptionally different entity (Figure 7C).
Figure 7.
Comparison of the transcriptome of human fetal CD34+ hematopoietic stem and progenitor cell populations to infant B-cell precursor acute lymphoblastic leukemia. (A) Schematic representation of B-cell development in human fetal liver (FL) showing immunophenotypic definitions for hematopoietic stem cells (HSC), multipotent progenitors (MPP), lymphoid-primed multipotent progenitors (LMPP), committed B progenitors (CBP) and B cells. The onset and expected patterns of IgH rearrangements59,60 are depicted as red arrows. (B) Sorting strategy for FL hematopoietic stem and progenitor (HSPC) populations by fluorescence-activated cell sorting. The sorting gates for each population are shown in representative flow plots on the left. The purity of the sorted populations is depicted on the right demonstrating >95% purity. (Lin, Lineage cocktail). (C) Principal component analysis of gene expression of infant B-cell precursor acute lymphoblastic leukemia (iBCP-ALL) samples (n=42) and FL HSPC populations (n=3-7) using the top 1,000 variably expressed genes, as determined by RNA-sequencing. FL HSPC as in (A); MAF4, MLL-AF4+ iBCP-ALL; MA9, MLL-AF9+ iBCP-ALL; MLLwt, MLL wildtype iBCP-ALL.
Discussion
We set out to perform multi-layered sequencing on a large cohort iBCP-ALL patients, all enrolled in the international, collaborative Interfant treatment protocol. The fact that all patients were identically treated provides legitimacy and confidence in potential correlations of clinical value. Our study revealed an average of 2.5 non-silent single nucleotide variants, a 2-fold higher number than that reported by Andersson et al.,20 likely reflecting the 3-fold larger sequencing coverage. This silent mutational landscape, even in non-MLL iBCP-ALL, likely reflects the very young age of these patients, reinforcing the notion that infant cancer is a developmental disease with not enough time to develop somatic mutations. We also found the only recurrent, but subclonal, mutations occur in the KRAS and NRAS genes (gain-of-function mutations), although the frequency of subclonal NRAS mutations is significantly higher in t(4;11)+ patients. In line with our previous work we found no recurrent mutations in the FLT3 gene.40
Analysis of clonal evolution of RAS-mutated clones from diagnosis to relapse revealed that one-third of the patients still carry RAS mutations at relapse, whereas the other two-thirds of patients who relapse have lost the diagnostic RAS mutation. This is in accordance with recently published data by Trentin et al.,36 and suggests that the therapy is able to eliminate the RAS-mutated clone in some patients, while in other patients the RAS mutation seems to confer chemoresistance, allowing these clones to evade treatment.44 Intriguingly, ~25% of the patients carry more than one RAS-mutated clone at diagnosis, indicating a selection bias towards mutations in the RAS genes, or activated RAS pathways during leukemic transformation. From this perspective, the occurrence of patients carrying multiple distinct clones with activated RAS pathways may point to convergent evolution of clones capable of controlling the proliferation rate. However, arguing against this is the substantial representation of patients not carrying RAS mutations at all. Hence, the role of RAS mutations in t(4;11)+ iBCP-ALL remains obscure, and the available data suggest that RAS pathway mutations are unlikely leukemia-initiating lesions. Indeed, Tamai et al.45 showed that leukemogenesis of transgenic mice expressing human MLL-AF4 could be significantly accelerated by KRAS mutations. However, although activated KRAS did cooperate with MLL-AF4 in human cord blood-derived CD34+ HSPC to promote extramedullary infiltration and central nervous system infiltration it failed to initiate leukemia in engrafted mice.27 Importantly, we report a lack of correlation between RAS status and parameters associated with diagnosis or disease outcome such as overall survival, event-free survival, central nervous system infiltration, gender, percentage of blasts and white blood cells and age, further supporting the concept that RAS mutations are not leukemia-initiating/propagating lesions.
Clearly, this brings us back to the central question of whether or not MLL-AF4 by itself is sufficient to initiate BCP-ALL in humans. The silent mutational landscape observed in this study and by others20 certainly votes in favor of MLL-AF4+ iBCP-ALL being initiated by a single “big-bang” transformation hit, probably in a short-lived but highly proliferative prenatal B-cell progenitor.4 This hypothesis is supported by recent work by Lin et al., who indeed demonstrated that enforced expression of a fusion transcript consisting of human MLL and murine Af4 in cord blood-derived CD34+ HPSC is sufficient to induce pro-B ALL in xenografted immunodeficient mice.21,22 Yet, similar results using a human MLL-AF4 transcript remain to be established.
Although MLL-AF4 by itself may be sufficient to induce BCP-ALL without significant contributions from cooperative genetic lesions, the contribution of the MLL-AF4 and RAS mutations to leukemogenesis should take into account the nature of both the fetal target cell for transformation and the leukemia-initiating cell, according to the increasingly accepted stochastic stem cell model of BALL.46,47 Here, we employed high-throughput BCR-sequencing of the IgH locus to delineate the dynamics of clonality of B-cell populations in paired diagnosis-relapse samples of t(4;11)/MLL-AF4+ iBCP-ALL. While pediatric patients with E2A-PBX1+, TEL-AML1+ and BCR-ABL1+ BALL all had significantly clonal disease, with a major VDJ rearranged B-cell IgH clone accounting for up to 40% of all BCR, infants with MLL-AF4+ BCP-ALL exhibited a BCR repertoire composed of thousands of minor, non-expanded VDJ rearranged IgH B-cell clones. Because MLL fusions are clonal and RAS mutations are found in clones of relative big size, this suggests that MLL fusions with or without RAS mutations are likely to originate in primitive fetal progenitors that have a germline or an incompletely rearranged (DJ) IgH locus.48 Indeed, an unsupervised comparison of the transcriptome of FL HSPC populations and iBCP-ALL blasts suggests that while the gene expression of primitive FL HSPC (Lin−CD38−CD34+CD19− populations) is similar to that of iBCP-ALL, FL B progenitors (CD34+CD19+) are transcriptionally distinct. Our data elegantly reinforces previous fluorescence in-situ hybridization findings suggesting that a primitive “pre-VDJ” stem/progenitor cell (perhaps CD34+CD19−) may represent the cell in which both t(4;11) and RAS mutations arise.14,31,49
Cooperative leukemogenic events in iBCP-ALL may need to be sought beyond genetic insults; for instance, epigenetic and transcriptomic deregulation. MLL-AF4 might only induce BCP-ALL in cells that meet certain epigenetic and transcriptomic make-up criteria, either influenced by microenvironmental cues, or characteristic of the cell-of-origin.31 Indeed, lesions such as RAS mutations may contribute to disease pathogenesis only against certain intrinsic epigenetic or transcriptomic backgrounds present in the cell in which the MLL translocations occurred50,51. This is supported by the limited impact of RAS mutations in transcriptomic signatures associated with leukemia origin, development and pathogenesis, although this is likely due to the subclonal nature of RAS mutations.38 However, in line with the reported contribution of RAS mutations to extramedullary infiltration of MLLr BCP-ALL blasts,27 RAS-mutated patients displayed a transcriptomic signature associated with migration.
The functional and molecular contribution of the reciprocal fusion genes resulting from the derivative translocated chromosomes remains obscure in cancer. The AF4-MLL genomic fusion was previously detected in 80-85% of t(4;11)+ patients.5,52 Our “multi-layered omics” approach allowed for the exact characterization of the t(4;11) molecular DNA/RNA break points and the identification of those patients expressing the reciprocal AF4-MLL fusion. We now report that the AF4-MLL reciprocal fusion is expressed in only 50% of t(4;11)+ iBCP-ALL patients. Strikingly, there was a previously unrecognized and very significant positive correlation between the upregulation of the HOXA gene cluster and the expression of AF4-MLL. Of note, a recent study showed that approximately half of t(4;11)+ patients do not have an activated HOXA signature.44,53,54 Furthermore, in the recent MLL-Af4-induced B-ALL xenograft model MLL-Af4 failed to bind to HOXA genes and therefore HOXA gene expression was not upregulated.21 This is experimentally supported by chromatin immunoprecipitation-sequencing analysis performed in human embryonic stem cells transduced with MLL-AF4, AF4-MLL or both showing a significant enrichment of H3K79 methylated regions specifically associated with HOX-A cluster genes in double fusion-expressing hematopoietic derivatives, establishing a functional and molecular cooperation between MLL-AF4 and AF4-MLL fusions during human hematopoietic development (data not shown). Strikingly, AF4-MLL-expressing patients had a 5-fold longer event-free survival and a 3-fold longer overall survival compared to t(4;11)+ iBCP-ALL patients lacking AF4-MLL expression, which is in line with previous reports suggesting that high HOXA gene expression is associated with improved survival and lower risk of relapse.22,39 Because the expression of AF4-MLL is not analyzed in routine molecular diagnosis, our “multi-layered omics” approach was critical to unraveling the association between AF4-MLL and HOXA expression, thus identifying a novel subgroup of t(4;11)+ iBCP-ALL with better clinical outcome. It is very important for routine diagnostic and clinical practice that when the expression of AF4-MLL was evaluated in a Cox model adjusting for risk stratification (medium risk or high risk according to the Interfant-06 protocol), it retained its prognostic significance.
Mechanistically, AF4-MLL contains the SET domain disrupted from its “specification domain”, the N-terminal portion of MLL, which binds to MEN1 and LEDGF thus shaping the gene-targeting module of the MLL gene. When AF4-MLL is expressed, the N-terminal portion is substituted by the AF4 N-terminus (AF4N) which is the crucial domain for binding to and strongly activating RNA polymerase II (RNAP II) for transcriptional elongation. Thus, expression of A4M-MLL may induce robust RNAP II-dependent gene transcription by overwriting the elongation control process in a dominant fashion.55–58 We hypothesize that a likely function of AF4-MLL could be to prepare the ground for MLL-AF4 or other transcription factors to skew normal and leukemic hematopoietic cell fate decisions. This also explains why MLL-AF4, but not AF4-MLL, seems to be necessary in 100% of patients.
Despite being a developmental cancer, iBCP-ALL patients did not show reactivation of pluripotent or embryonic-like gene expression signatures as revealed by RNA-sequencing. Additional research is required to decipher the nature of the insults initiating MLLr iBCP-ALL, as so far we can only speculate on the data currently available. Whole-genome pyrosequencing will likely provide unique insights into the DNA methylome landscape of this mutationally silent iBCP-ALL. This study has clinical implications in the diagnostic risk-stratification of t(4;11)+ iBCP-ALL.
Supplementary Material
Acknowledgments
We would like to thank the Santander Supercomputing Service for IT support. The human fetal material was provided by the Joint MRC/Wellcome Trust (grant# MR/R006237/1) Human Developmental Biology Resource (http://hdbr.org). This work was supported by the European Research Council (CoG-2014-646903 to PM; and StG-2014-637904 to IV), the Spanish Ministry of Economy and Competitiveness (SAF-SAF2013-43065 to PM and SAF2016-76758-R to IV), the Asociación Española Contra el Cáncer (AECC-CI-2015), FERO Foundation, and the ISCIII (PI14-01191) to CB. PM/IV also acknowledge financial support from The Obra Social La Caixa-Fundaciò Josep Carreras, The Inocente Inocente Foundation, Fundación Ramón Areces and The Generalitat de Catalunya (SGR330). IR was supported by a Programme Grant from Bloodwise (LLR 13001) and by the Oxford NIHR Biomedical Centre based at Oxford University Hospitals NHS Trust and University of Oxford. PM is an investigator of the Spanish Cell Therapy cooperative network (TERCEL). AR was supported by a Clinician Scientist Fellowship from Bloodwise (14041). This work was motivated by our patients and it honors the vital example given to us by the family of AMC.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/6/1176
References
- 1.Pui C-H, Evans WE. A 50-year journey to cure childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50(3):185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Mullighan CG, Evans WE, Relling MV. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120(6):1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer C, Burmeister T, Gröger D, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32(2):273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanjuan-Pla A, Bueno C, Prieto C, et al. Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia. Blood. 2015;126(25):2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2011;152(2):141–154. [DOI] [PubMed] [Google Scholar]
- 6.Ribeiro RC, Pui CH. Prognostic factors in childhood acute lymphoblastic leukemia. Hematol Pathol. 1993;7(3):121–142. [PubMed] [Google Scholar]
- 7.Pieters R, Schrappe M, De Lorenzo P, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370(9583):240–250. [DOI] [PubMed] [Google Scholar]
- 8.Biondi A, Cimino G, Pieters R, Pui C-H. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96(1):24–33. [PubMed] [Google Scholar]
- 9.Milne TA. Mouse models of MLL leukemia: recapitulating the human disease. Blood. 2017;129(16):2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10(5): 1119–1128. [DOI] [PubMed] [Google Scholar]
- 11.Chen CW, Armstrong SA. Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond. Exp Hematol. 2015;43(8):673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean CM, Karemaker ID, van Leeuwen F. The emerging roles of DOT1L in leukemia and normal development. Leukemia. 2014;28(11):2131–2138. [DOI] [PubMed] [Google Scholar]
- 13.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer. 2009;14(5):355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102(7):2321–2333. [DOI] [PubMed] [Google Scholar]
- 15.Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363(6427):358–360. [DOI] [PubMed] [Google Scholar]
- 16.Gale KB, Ford AM, Repp R, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci U S A. 1997;94(25):13950–13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bueno C, Montes R, Catalina P, Rodríguez R, Menendez P. Insights into the cellular origin and etiology of the infant pro-B acute lymphoblastic leukemia with MLL-AF4 rearrangement. Leukemia. 2011;25(3):400–410. [DOI] [PubMed] [Google Scholar]
- 18.Bardini M, Galbiati M, Lettieri A, et al. Implementation of array based whole-genome high-resolution technologies confirms the absence of secondary copy-number alterations in MLL-AF4-positive infant ALL patients. Leukemia. 2011;25(1):175–178. [DOI] [PubMed] [Google Scholar]
- 19.Dobbins SE, Sherborne AL, Ma YP, et al. The silent mutational landscape of infant MLL-AF4 pro-B acute lymphoblastic leukemia. Genes Chromosom Cancer. 2013;52(10): 954–960. [DOI] [PubMed] [Google Scholar]
- 20.Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(4):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, Luo RT, Ptasinska A, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30(5):737–749. [DOI] [PubMed] [Google Scholar]
- 22.Lin S, Luo RT, Shrestha M, Thirman MJ, Mulloy JC. The full transforming capacity of MLL-Af4 is interlinked with lymphoid lineage commitment. Blood. 2017;130(7):903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto H. Successful sustained engraftment after reduced-intensity umbilical cord blood transplantation for adult patients with severe aplastic anemia. Blood. 2011;116(26): 6123–6132. [DOI] [PubMed] [Google Scholar]
- 24.Montes R, Ayllón V, Prieto C, et al. Ligand-independent FLT3 activation does not cooperate with MLL-AF4 to immortalize/transform cord blood CD34+ cells. Leukemia. 2014;28(3):666–674. [DOI] [PubMed] [Google Scholar]
- 25.Bueno C, Montes R, Melen GJ, et al. A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res. 2012;22(6):986–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bueno C, Ayllón V, Montes R, et al. FLT3 activation cooperates with MLL-AF4 fusion protein to abrogate the hematopoietic specification of human ESCs. Blood. 2013;121(19):3867–3878. [DOI] [PubMed] [Google Scholar]
- 27.Prieto C, Stam RWRW, Agraz-Doblas A, et al. Activated KRAS cooperates with MLL-AF4 to promote extramedullary engraftment and migration of cord blood CD34+ HSPC but is insufficient to initiate leukemia. Cancer Res. 2016;76(8):2478–2489. [DOI] [PubMed] [Google Scholar]
- 28.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. [DOI] [PubMed] [Google Scholar]
- 29.Varela I, Menendez P, Sanjuan-Pla A. Intratumoral heterogeneity and clonal evolution in blood malignancies and solid tumors. Oncotarget. 2017;8(39):66742–66746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Driessen E, Lorenzo PMC, et al. Outcome of congenital acute lymphoblastic leukemia treated on the Interfant-99 protocol. Blood. 2009;114(18):3764–3768. [DOI] [PubMed] [Google Scholar]
- 31.Menendez P, Catalina P, Rodríguez R, et al. Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J Exp Med. 2009;206(13):3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muñoz-López A, Romero-Moya D, Prieto C, et al. Development refractoriness of MLL-rearranged human B cell acute leukemias to reprogramming into pluripotency. Stem Cell Reports. 2016;7(4):602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy A, Cowan G, Mead AJ, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci U S A. 2012;109(43):17579–17584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gröbner SN, Worst BC, Weischenfeldt J, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–327. [DOI] [PubMed] [Google Scholar]
- 35.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trentin L, Bresolin S, Giarin E, et al. Deciphering KRAS and NRAS mutated clone dynamics in MLL-AF4 paediatric leukaemia by ultra deep sequencing analysis. Sci Rep. 2016;6:34449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashford-Rogers RJM, Nicolaou KA, Bartram J, et al. Eye on the B-ALL: B-cell receptor repertoires reveal persistence of numerous B-lymphoblastic leukemia sub-clones from diagnosis to relapse. Leukemia. 2016;30(12):2312–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prelle C, Bursen A, Dingermann T, Marschalek R. Secondary mutations in t(4;11) leukemia patients. Leukemia. 2013;27(6):1425–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stam RW, Schneider P, Hagelstein JAP, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood. 2010;115(14):2835–2844. [DOI] [PubMed] [Google Scholar]
- 40.Chillón MC, Gómez-Casares MT, López-Jorge CE, et al. Prognostic significance of FLT3 mutational status and expression levels in MLL-AF4 and MLL-germline acute lymphoblastic leukemia. Leukemia. 2012;26(11): 2360–2366. [DOI] [PubMed] [Google Scholar]
- 41.Boelens JJ, Aldenhoven M, Purtill D, et al. Outcomes of transplantation using various hematopoietic cell sources in children with Hurler syndrome after myeloablative conditioning Key Points. Blood. 2013;121(10): 3981–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bashford-Rogers RJM, Palser AL, Huntly BJ, et al. Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Res. 2013;23(11):1874–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruggemann M, Schrauder A, Raff T, et al. Standardized MRD quantification in European all trials: Proceedings of the Second International Symposium on MRD assessment in Kiel, Germany, 18-20 September 2008. Leukemia. 2010;24(3):521–535. [DOI] [PubMed] [Google Scholar]
- 44.Driessen EMC, van Roon EHJ, Spijkers-Hagelstein JAP, et al. Frequencies and prognostic impact of RAS mutations in MLL-rearranged acute lymphoblastic leukemia in infants. Haematologica. 2013;98(6):937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamai H, Miyake K, Takatori M, et al. Activated K-Ras protein accelerates human MLL/AF4-induced leukemo-lymphomogenicity in a transgenic mouse model. Leukemia. 2011;25(5):888–891. [DOI] [PubMed] [Google Scholar]
- 46.Elder A, Bomken S, Wilson I, et al. Abundant and equipotent founder cells establish and maintain acute lymphoblastic leukaemia. Leukemia. 2017;31(12):2577–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prieto C, López-Millán B, Roca-Ho H, et al. NG2 antigen is involved in leukemia inva-siveness and central nervous system infiltration in MLL-rearranged infant B-ALL. Leukemia. 2018;32(3):633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jansen MWJC, Corral L, van der Velden VHJ, et al. Immunobiological diversity in infant acute lymphoblastic leukemia is related to the occurrence and type of MLL gene rearrangement. Leukemia. 2007;21(4):633–641. [DOI] [PubMed] [Google Scholar]
- 49.Hotfilder M, Röttgers S, Rosemann A, et al. Leukemic stem cells in childhood high-risk ALL/t(9;22) and t(4;11) are present in primitive lymphoid-restricted CD34+CD19-cells. Cancer Res. 2005;65(4):1442–1449. [DOI] [PubMed] [Google Scholar]
- 50.Bergmann AK, Castellano G, Alten J, et al. DNA methylation profiling of pediatric B-cell lymphoblastic leukemia with KMT2A rearrangement identifies hypomethylation at enhancer sites. Pediatr Blood Cancer. 2017;64(3):e26251. [DOI] [PubMed] [Google Scholar]
- 51.Malouf C, Ottersbach K. The fetal liver lymphoid-primed multipotent progenitor provides the prerequisites for the initiation of t(4;11) MLL-AF4 infant leukemia. Haematologica. 2018. June 14 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kowarz E, Burmeister T, Lo Nigro L, et al. Complex MLL rearrangements in t(4;11) leukemia patients with absent AF4MLL fusion allele. Leukemia. 2007;21(6):1232–1238. [DOI] [PubMed] [Google Scholar]
- 53.Trentin L, Giordan M, Dingermann T, Basso G, Te Kronnie G, Marschalek R. Two inde pendent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol. 2009;83(5):406–419. [DOI] [PubMed] [Google Scholar]
- 54.Kühn A, Löscher D, Marschalek R. The IRX1/HOXA connection: insights into a novel t(4;11)- specific cancer mechanism. Oncotarget. 2016;7(23):35341–35352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson AC, Ballabio E, Geng H, et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013;3(1):116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benedikt A, Baltruschat S, Scholz B, et al. The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia. 2011;25(1): 135–144. [DOI] [PubMed] [Google Scholar]
- 57.Scholz B, Kowarz E, Rössler T, Ahmad K, Steinhilber D, Marschalek R. AF4 and AF4N protein complexes: recruitment of P-TEFb kinase, their interactome and potential functions. Am J Blood Res. 2015;5(1):10–24. [PMC free article] [PubMed] [Google Scholar]
- 58.Mück F, Bracharz S, Marschalek R. DDX6 transfers P-TEFb kinase to the AF4/AF4N (AFF1) super elongation complex. Am J Blood Res. 2016;6(3):28–45. [PMC free article] [PubMed] [Google Scholar]
- 59.van Zelm MC, van der Burg M, de Ridder D, et al. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175(9): 5912–5922. [DOI] [PubMed] [Google Scholar]
- 60.Böiers C, Richardson SE, Laycock E, et al. A human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6-RUNX1. Dev Cell. 2018;44(3):362–377.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.