Follicular lymphoma (FL) is a common indolent lymphoma in which clinical and biological heterogeneity is increasingly recognized.1 Although median survival for newly diagnosed FL patients may be as high as 18 years,2 a significant subset of patients is at risk of adverse outcomes. In particular, patients experiencing disease progression within two years after rituximab-containing induction therapy have an increased risk of premature death, but can only be identified a posteriori.3–7 Recently, several molecular risk stratification methods have been reported, including the m7-Follicular Lymphoma International Prognostic Index (m7-FLIPI) clinicogenetic risk model,8 protein expression of FOXP1,9,10 and a predictor based on the expression of 23 genes (Huet et al., henceforth referred to as “Gene-Expression Profiling Score or GEP Score”).11 However, the uncertainty of whether these novel biomarkers identify similar biology is impeding the development of risk-adapted management strategies.
As the field would benefit from coherent risk models, we asked the question whether expression of FOXP1 and the GEP Score are inter-related. Here we demonstrate that the gene content from the GEP Score identifies patient groups with diverging outcomes in our own, independent dataset. We then show that expression of FOXP1 is strongly correlated with the GEP Score, demonstrating that they identify highly similar patient populations. Lastly, network analysis indicates gene expression interconnectivity between both biomarkers.
With regards to the methods, we had previously generated Illumina cDNA-mediated annealing, selection, extension and ligation (DASL) microarray expression profiles for 137 FL patients who had been treated with rituximab in combination with chemotherapy (cyclophosphamide, vincristine and prednisone).8 Of these, 113 patients had received rituximab maintenance by intention to treat.8 Microarray data have been deposited at the Gene Expression Omnibus (GEO) under accession number GSE119214. Gene names and coefficients from the GEP Score were taken from Huet et al.11 who had applied both Affymetrix and NanoString expression platforms to samples from FL patients treated with rituximab and a variety of chemotherapy regimens. In order to apply the GEP Score to our own data, we calculated risk scores (henceforth referred to as “modified Gene-Expression Profiling Score or mGEP Score”) from our DASL expression data using the weighted sum of quantile normalized gene expression levels for each patient. We used the coefficients described in Huet et al.11 for the Affymetrix platform. A binary score was defined by thresholding the scores at the value resulting in maximal value of the log-rank statistic. To account for multiple cut-point testing, P-values were adjusted using the method proposed by Miller and Siegmund,12 implemented in X-tile.13 To identify underlying subgroups in our data, we performed unsupervised, hierarchical clustering. The t-test was used to identify associations between gene expression scores and EZH2 mutation status, FOXP1 expression, and m7-FLIPI. Low and high expression of FOXP1 were defined as ≤10% and >10% of cells staining positive, as previously described.9 Differential gene expression analysis by FOXP1 expression was performed using limma.14 Gene set enrichment analysis was used to identify over-representation of relevant gene signatures.15 Gene co-expression similarities were determined and connections were calculated from standardized expressions scaled to zero mean and unit variance.
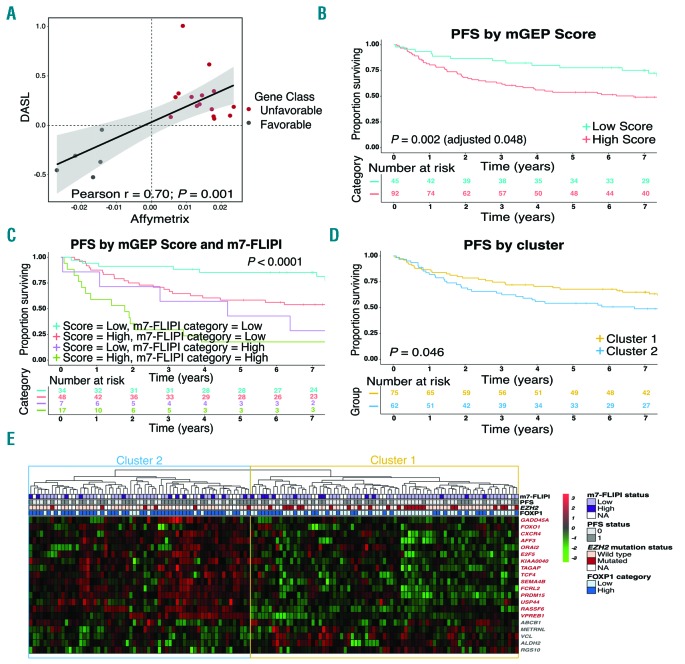
Twenty genes from the GEP Score (87%) were found to be present in our dataset. All downstream analyses were performed using these 20 genes. We correlated the expression of each of these 20 genes with progresion-free survival (PFS) using univariate Cox regression analysis and showed that the coefficients from Cox regression were significantly correlated with the coefficients reported by Huet et al.11 (Pearson r = 0.7; P<0.001) (Figure 1A). All genes that had previously been found to portend poor-risk were also associated with poor PFS in our data, and vice versa. Having information on only 20 genes out of the 23, we calculated an mGEP Score, that was found to be significantly associated (as a continuous variable) with PFS in univariate Cox regression analysis (P=0.002). Using the maximally selected log-rank statistic, we defined an optimal cut-off to dichotomize the distribution of the mGEP Score. We defined a cut-off of 1.14, and using this we identified 92 (67%) cases with a high-risk score who had inferior PFS compared to 45 (33%) cases with low-risk score (5-year PFS 54% vs. 78%; P=0.002 and adjusted P=0.048) (Figure 1B). The corresponding 5-year overall survival (OS) rates were 72% and 87% (P=0.025) (Online Supplementary Figure S1A). We then assessed the combined effect of the mGEP Score and the m7-FLIPI and found that the mGEP Score added prognostic value to both m7-FLIPI risk groups (Figure 1C and Online Supplementary Figure S1B).
Figure 1.
Coefficients, scores and clusters generated based on the modified Gene-Expression Profiling (mGEP) Score reveal distinct patient outcomes. (A) Plot showing the coefficients from Cox regression analysis from our cohort (cDNA-mediated annealing, selection, extension and ligation, DASL) and coefficients from Huet et al.11 (Affymetrix). (B) Progression-free survival (PFS) by mGEP Score in our cohort. Five-year PFS was 54% versus 78% in patients with high and low scores, respectively. (C) PFS by mGEP Score and m7-Follicular Lymphoma International Prognostic Index (m7-FLIPI) category in our cohort. (D) PFS by cluster in our cohort. Five-year PFS was 54% versus 68% in patients with Cluster 2 and Cluster 1, respectively. (E) Heatmap showing the genes along rows and patients along columns. Three genes, SHISA8, EML6 and DCAF12, from the GEP score were not included in the mGEP Score calculations. Gene names in red are associated with poor outcome in the mGEP Score. Cluster 1 and Cluster 2 were characterized by high expression of genes associated with favorable and poor outcome, respectively.
To be confident that the genes from the mGEP Score identify distinct biological subgroups, we performed unbiased hierarchical clustering. We identified two main clusters, Cluster 1 and Cluster 2. Patients from Cluster 2 experienced worse PFS compared to patients in Cluster 1 (P=0.046; 5-year PFS 54% vs. 68%) (Figure 1D). Cluster 1 and Cluster 2 were characterized by high expression of genes associated with favorable and poor outcome, respectively (Figure 1E). The 5-year OS was 72% for patients in Cluster 2 vs. 81% in Cluster 1 (P=0.13) (Online Supplementary Figure S1C). The association between PFS and clusters remained significant after adjusting for FLIPI and rituximab maintenance [Hazard Ratio (HR) 1.86; P=0.01]. The clusters were not significantly associated with individual FLIPI factors, with the exception that age was slightly lower for patients belonging to Cluster 2 (Online Supplementary Table S1). Collectively, our results strongly suggest that the findings from Huet et al.11 hold true in our independent dataset, lending support to the robustness of their predictor.
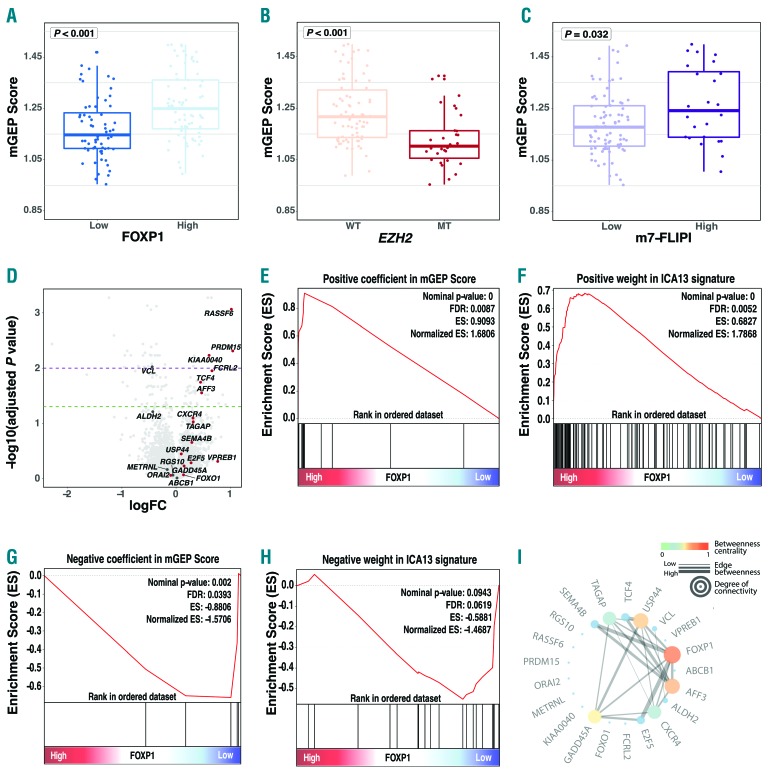
Previously, a gene expression signature (ICA13), that is highly correlated with the GEP Score, was found to be expressed at high levels in centroblasts residing in the dark zone of the germinal center.11 In parallel, we had found that a dark zone-related gene set was enriched in FL biopsies with high FOXP1 expression.9 These observations prompted us to test whether FOXP1 expression is associated with expression of either the ICA13 signature and/or the mGEP Score. We found that the mean mGEP Score was significantly higher in cases with high expression of FOXP1 (P<0.001) (Figure 2A). The mGEP Score as a categorical variable was also significantly associated with FOXP1 expression using the χ2 test (P=0.002). Interestingly, the mean mGEP Score was significantly lower in cases with EZH2 mutation (P<0.001) (Figure 2B) and it was also lower in cases with low m7-FLIPI risk score (P=0.032) (Figure 2C). The latter observations are consistent with EZH2 mutation status having a strong negative coefficient (associated with favorable risk) in the m7-FLIPI risk model. Differential gene expression analysis between FOXP1-high and low expressors identified that 7 out of the 20 genes from the mGEP Score were differentially expressed in this comparison (Figure 2D).
Figure 2.
Convergence of outcome prediction tools in follicular lymphoma (FL). (A, B and C) The mean modified Gene-Expression Profiling (mGEP) Score was significantly higher in cases who had high FOXP1 expression, were EZH2 mutation wild-type and were classified as high-risk m7-Follicular Lymphoma International Prognostic Index (m7-FLIPI) category. (D) Volcano plot, with genes from the mGEP score highlighted by labeling. Positive log fold-change (logFC) indicates higher expression in cases with high FOXP1 expression, and vice versa. The horizontal, dashed, green line indicates -log10(adjusted P=0.05) and the dashed, purple line indicates -log10(adjusted P=0.01). Adjusted P-values were calculated using the Benjamini and Hochberg method. (E and F) Gene set enrichment analysis (GSEA) shows that genes with positive coefficients from the mGEP Score and genes with a positive weight in the ICA13 signature were significantly enriched in cases with high FOXP1 expression. (G and H) GSEA shows that genes with negative coefficients from the mGEP score and genes with a negative weight in the ICA13 signature were significantly enriched in cases with low FOXP1 expression. (I) Gene connections based on related expression.
To further test whether defined signatures are associated with FOXP1 expression, we performed gene set enrichment analysis.15 This analysis revealed that the genes with positive coefficients in the mGEP Score, as well as the genes with positive weight in the ICA13 signature (i.e. the genes associated with poor PFS) were enriched in the FOXP1-high phenotype [false discovery rate (FDR) 0.009 and 0.005, respectively] (Figure 2E and F). Conversely, genes with negative coefficients in the mGEP Score (i.e. the genes associated with favorable PFS) were enriched in the FOXP1-low phenotype (FDR 0.039) (Figure 2G); and genes with negative weight in the ICA13 signature had a negative enrichment score in the FOXP1-low phenotype (FDR 0.062) (Figure 2H). As FOXP1 and other transcription factors represented in the mGEP Score likely operate in tightly regulated networks, we computed a weighted network based on gene expression correlation (Figure 2I). This analysis revealed a co-expression network (among SEMA4B, TAGAP, TCF4, USP44, VCL, AFF3, ALDH2, CXCR4, E2F5, GADD45A and FOXP1), with FOXP1 emerging as a central gene with one of the highest amounts of interconnectivity (Figure 2I). Taken together, these findings illustrate that high expression of FOXP1 and high-risk assignment by the mGEP Score identify similar disease biology.
In summary, we were able to not only independently validate the prognostic relevance of the mGEP Score, but we also observed a striking association between high expression of FOXP1 protein by immunohistochemistry and high expression of poor-risk gene signatures identified by Huet et al.11 Although recent studies used a variety of approaches to define prognostic models, we demonstrate biomarker convergence on a common phenotype: FOXP1 expression, EZH2 wild-type status and expression of dark zone-related genes define an FL phenotype with adverse outcome following front-line treatment with rituximab and chemotherapy. Despite the associations demonstrated herein, it is evident that the various biomarkers are not perfectly concordant, as evidenced, for example, by the additional prognostic information that is gained when combining the m7-FLIPI and the mGEP Score. Nonetheless, our observations confirm that the clinical validity of biomarkers such as the GEP Score is now established. Future studies will need to test whether they have predictive value and whether alternative treatment options may be better suited to offer high-risk patients improved outlooks.
Supplementary Material
Footnotes
Funding: this work was supported by a Terry Fox Research Institute New Investigator Award (Grant #1079), a Leukemia & Lymphoma Society of Canada Operating Grant (Grant #513421) and the Princess Margaret Cancer Foundation.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Lackraj T, Goswami R, Kridel R. Pathogenesis of follicular lymphoma. Best Pract Res Clin Haematol. 2018;31(1):2–14. [DOI] [PubMed] [Google Scholar]
- 2.Tan D, Horning SJ, Hoppe RT, et al. Improvements in observed and relative survival in follicular grade 1–2 lymphoma during 4 decades: the Stanford University experience. Blood. 2013;122(6):981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mozessohn L, Cheung MC, Crump M, et al. Chemoimmunotherapy resistant follicular lymphoma: predictors of resistance, association with transformation and prognosis. Leuk Lymphoma. 2014;55(11):2502–2507. [DOI] [PubMed] [Google Scholar]
- 4.Casulo C, Byrtek M, Dawson KL, et al. Early Relapse of Follicular Lymphoma After Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone Defines Patients at High Risk for Death: An Analysis From the National LymphoCare Study. J Clin Oncol. 2015;33(23):2516–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurer MJ, Bachy E, Ghesquières H, et al. Early event status informs subsequent outcome in newly diagnosed follicular lymphoma. Am J Hematol. 2016;91(11):1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurinovic V, Kridel R, Staiger AM, et al. Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood. 2016;128(8):1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kridel R, Chan FC, Mottok A, et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016;13(12):e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastore A, Jurinovic V, Kridel R, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015;2045(15):1–12. [DOI] [PubMed] [Google Scholar]
- 9.Mottok A, Jurinovic V, Farinha P, et al. FOXP1 expression is a prognostic biomarker in follicular lymphoma treated with rituximab and chemotherapy. Blood. 2018;131(2):226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musilova K, Devan J, Cerna K, et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood. 2018;132(22):2389–2400. [DOI] [PubMed] [Google Scholar]
- 11.Huet S, Tesson B, Jais J, et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: a retrospective training and validation analysis in three international cohorts. Lancet Oncol. 2018;19(4):549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller R, Siegmund D. Maximally Selected Chi Square Statistics. Biometrics. 1982;38(4):1011. [Google Scholar]
- 13.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. [DOI] [PubMed] [Google Scholar]
- 14.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.