Diffuse large B-cell lymphoma (DLBCL) is the commonest lymphoma in both humans and dogs.1 Due to spontaneously high incidence, complex genetic interplay, aggressive clinical course, elevated frequency and the presence of an intact immune system, dogs with lymphoma are considered as an ideal comparative model for drug development for human lymphomas.1 The BCL2 inhibitor venetoclax and the BTK inhibitors ibrutinib and acalabrutinib are examples of drugs that benefited from such a model.2–4 Clinical heterogeneity of canine DLBCL (cDLBCL) is known and early insights into its biological and molecular mechanisms, revealing similarities with the human counterpart, have been reported, but a genomic characterization of this tumor has never been provided. Furthermore, a complete understanding of the molecular mechanisms driving the pathogenesis of cDLBCL is necessary to take more accurate advantage of this comparative model and, also, to improve the clinical outcome of the dogs. In this study, we report the first large integrated analysis with transcriptome sequencing, methylation genome-wide analysis, copy number variation analysis and clinical outcome in cDLBCL to identify the molecular mechanisms of this tumor comprehensively and to define its use in comparative medicine better.
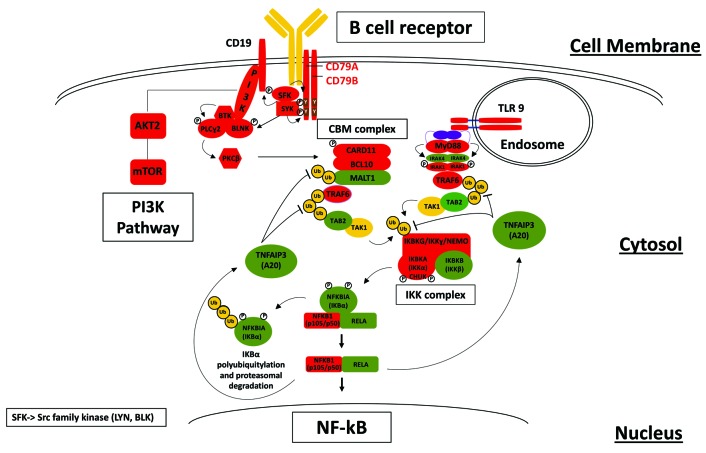
We first compared the transcriptome obtained via RNA-sequencing in 50 cDLBCL versus that from RNA-sequencing data of normal follicular B cells derived from lymph nodes of 11 healthy dogs (Online Supplementary Table S1; Online Supplementary Methods). Transcripts with levels higher in cDLBCL than in controls were significantly enriched in genes coding for proteins involved in the B-cell receptor (BCR), MYC signaling, PI3K/AKT/mTOR pathway, DNA replication, and cell cycle (Online Supplementary Figure S1). Genes coding for proteins involved in the nuclear factor-κB (NF-κB) pathway (CD79, CD19, SYK, LYN, CARD11, BCL10, BTK, TRAF6, MYD88, NFKB2, TLR7, TLR9) were also differentially expressed (Figure 1). The aberrant expression of these genes might contribute to enhance the proliferation of B cells and protect these cells from apoptosis by stimulating NF-κB activity. A similar mechanism has been described in human activated B-cell DLBCL, characterized by constitutive activation of NF-κB due to mutations in multiple genes belonging to this subgroup’s pathway.5 These data fit with the reported frequent inactivation of the NF-κB negative regulator TRAF3 in canine B-cell lymphomas6 and the anti-tumor activity of BTK inhibitors, specifically active in the human activated B-cell subtype of DLBCL and experimentally used in canine B-cell lymphomas.3,4
Figure 1.
Schematic representation of a potential nuclear factor-κB activation pathway obtained by RNA-sequencing analysis in canine diffuse large B-cell lymphomas. Red, upregulated genes. Green, downregulated genes. Yellow, genes that are neither upregulated or downregulated.
The protein-encoding gene LIN28B appeared as the most upregulated transcript in tumors. While LIN28B does not seem to be a relevant gene for human DLBCL, it is involved in the activation of the NF-κB pathway and MYC in different human and mouse tumor models and its overexpression causes murine peripheral T-cell lymphomas.7 We took advantage of the CLBL-1 cell line, the only in vitro model available of cDLBCL, which showed a profile similar to cDLBCL clinical specimens, with LIN28B as the most overexpressed gene compared to expression levels in normal lymph nodes (Online Supplementary Table S3). LIN28B silencing using locked nucleic acid-modified short interfering RNA led to a reduction in cell proliferation compared to that of the negative control (Online Supplementary Figure S2), opening new therapeutic perspectives, at least in cDLBCL.
The integration of data on copy number variations and the transcriptome showed an association in 320 genes. A total of 239 genes were located in regions of gains and simultaneously overexpressed, while 81 genes were downregulated and mapped to regions of deletions (Online Supplementary Table S4). Gains of the entire chromosomes 13 and 31 and loss of chromosome 14 were the most frequent aberrations, occurring in >20% of the cases (Online Supplementary Figure S3). Survival analysis showed a trend to longer overall survival and event-free survival in dogs affected by this chromosome 13 aberration than in the remaining animals (P=0.134). A series of additional copy number variations, such as gains in chromosomes 4 and 27 and loss in chromosome 28, were observed in at least two dogs (Online Supplementary Figure S2).
A genome-wide next-generation sequencing-based methylation analysis, previously never performed in canine lymphomas, identified differentially methylated regions between cDLBCL and normal follicular B cells (Online Supplementary Table S4; Online Supplementary Figures S4 and S5). In accordance with RNA-sequencing data, genes that had a reduced degree of promoter methylation in cDLBCL were enriched for transcripts involved in cytokine/cytokine receptor interactions, BCR and JAK/STAT signaling. Genes participating in calcium signaling, cytoskeleton, Hedgehog/Wnt signaling and metabolism more commonly showed increased promoter methylation in neoplastic cells than in normal B cells and were also down-regulated (Online Supplementary Table S4). In line with the findings of our previous study,8 this observed increased promoter methylation supports an evaluation of the use of hypomethylating agents in the treatment of cDLBCL.
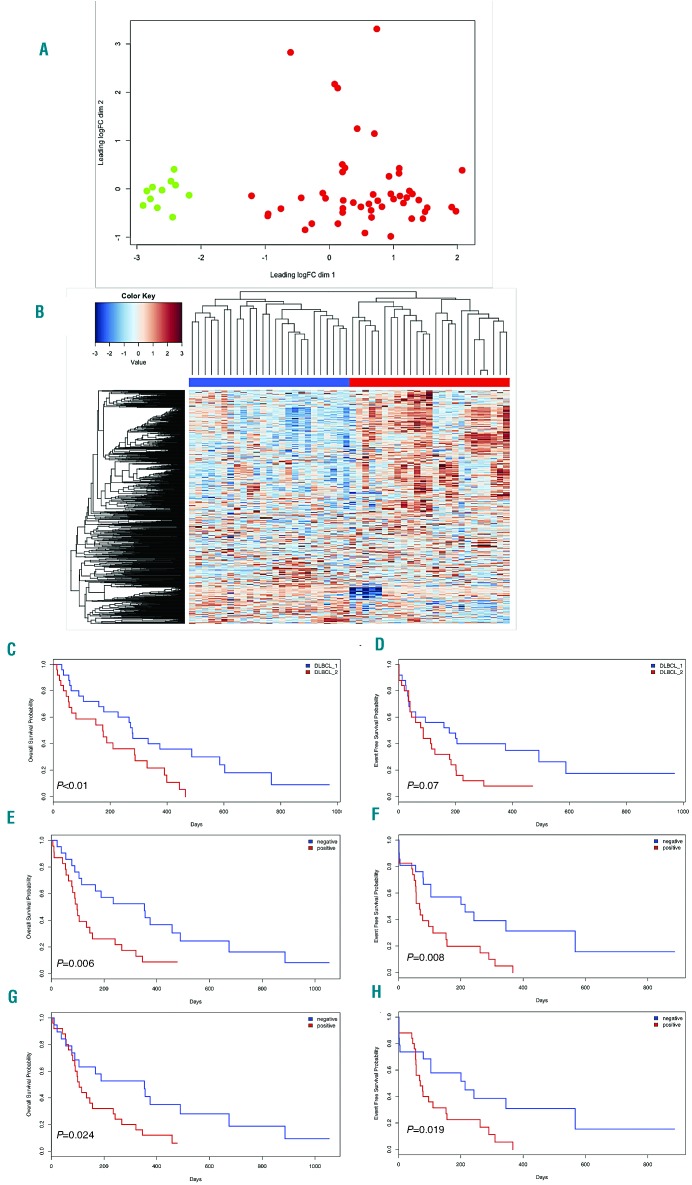
Unsupervised clustering of RNA-sequencing data separated cDLBCL from normal follicular B cells (Figure 2A) and identified two distinct subgroups within the tumors (Figure 2B). The two groups did not overlap with either the human germinal center B-cell or activated B-cell subtypes of DLBCL or the DLBCL consensus clusters,5 but they were associated with different overall survival (278 vs. 150 days, P<0.01) and event-free survival (179 vs. 113 days, P=ns) (Figure 2C,D). Tumors belonging to the group with worse prognosis were characterized by a higher expression of transcripts for proteins involved in apoptosis, JAK/STAT signaling, microenvironment, inflammatory response and the P53 pathway (Online Supplementary Table S5). In particular, dogs with the poorest outcome had a signature largely defined by markers of T-cell-mediated immune responses, with higher expression of transcripts involved in T-cell and macrophage regulation (CD163, CD96, PD-1, PDL-1, CTLA4, CD8a, CD4), as also shown by an enrichment of gene expression signatures of immune-inhibitory molecules (Online Supplementary Figure S6), and genes associated with the ATP-binding cassette transporter. The clustering into two distinct DLBCL subgroups was also supported by methylation data and differences in copy number variations (Online Supplementary Figure S7). Cases with inferior outcome had a higher degree of promoter methylation (P<0.005), and a higher number of aberrations (P<0.01) compared to the remaining cases, but no equivalent to the human 9p24 amplification associated with expression of PD-L1, which could be mainly supported by the observed activation of the JAK/STAT pathway.
Figure 2.
Unsupervised clustering of canine diffuse large B-cell lymphoma RNA-sequencing data and prognostic impact of PD-L1 and PD-1 expression. (A) Multidimensional scaling plot of all the samples demonstrated a clear separation between normal B cells (green dots) and canine diffuse large B-cell lymphomas (cDLBCL) (red dots). (B) Two cDLBCL subgroups (blue and red) were defined by hierarchical clustering. (C,D) Kaplan-Meier plots of overall survival (C) and event-free survival (D) of cDLBCL grouped on the basis of their RNA-sequencing profiles. (E-H) Prognostic impact of PD-L1 tumor expression and PD-1 residual lymphocyte expression on overall survival and event-free survival in cDLBCL: Kaplan-Meier plots of overall survival (E,G) and event-free survival (F,H) based on immunohistochemical scores of PDL-1 and PD-1, respectively.
We first validated the pattern of expression of PDL-1, PD-1, CTLA-4 and CD5 by immunohistochemistry (Online Supplementary Figure S8). All 25 samples from the group with a poor outcome were strongly positive for PDL-1 and CTLA-4 (tumor proportion score: >50%), while cases in the other cluster were completely negative (n=5, 20%) or weakly positive (tumor proportion score: 1%-40%; n=20, 80%). The same cases also presented a higher number of PD-1+ and CD5+ cells (higher or lower than 20 per high power field). We then analyzed an independent series of 44 cDLBCL, all treated with CHOP (cyclophosphamide, doxorubicin, vincristine and prednisone) plus vaccine.9 Staining for PDL-1 and PD-1 was positive in 21 (48%) and 25 cases (57%), respectively, and conferred inferior outcome (Figure 2E,F). CTLA-4 and CD5 were expressed in 18 (41%) and 22 cases (50%), respectively, but were not associated with clinical outcome. In a multivariate analysis including bone marrow infiltration and substage, PD-L1 and PD-1 maintained their prognostic impact on overall survival, but not on event-free survival (Online Supplementary Table S6). The results obtained represent a solid foundation on which to evaluate novel regimens based on immune-checkpoint inhibitors in canine lymphomas before moving to the human setting. A combination of immune-checkpoint modulators with active vaccines9 would also be worth exploring.
The transcriptome data had potential therapeutic implications. First, cDLBCL showed gene expression signatures suggestive of MYC activation and we therefore assessed the BET inhibitor birabresib (MK-8628/OTX015)10 and the BRD4 degrader MZ1 (alongside its inactive epimer cis MZ1)11 for their ability to block the proliferation of CLBL-1 cells.12 Both drugs inhibited proliferation, although MZ1 was more potent (Online Supplementary Figure S9A), in accordance with data obtained in human DLBCL models.13 Interestingly, MZ1 significantly downregulated not only MYC, but also LIN28B (Online Supplementary Figure S9B). Second, also in agreement with a transcriptome study based on a small number of canine B-cell lymphomas,14 genes belonging to the PI3K/AKT/mTOR pathway and/or to gene expression signatures obtained in human DLBCL cell lines exposed to BCR inhibitors15 were enriched for transcripts expressed in cDLBCL and in the CLBL-1 cell line (Online Supplementary Figure S10). We therefore tested a dual PI3K/mTOR inhibitor bimiralisib (PQR309) and PI3Kδ inhibitor idelalisib. Both drugs were active in the CLBL-1 cell line with half maximal inhibitory concentration (IC50) values of 150 nM and 80 nM, respectively (Online Supplementary Figure S9C). A xenograft model with CLBL-1 implanted in NOD-SCID mice showed that both bimiralisib and MZ1, given at the dose of 100 mg/kg, significantly affected tumor growth in comparison with that in the control group, with reductions in the tumor volume on day 8 of 43% and 33%, respectively, compared to the control tumor volume (P<0.05) (Online Supplementary Figure S9D).
In conclusion, we identified deregulated pathways and individual transcripts providing therapeutic targets, including an immune-related signature affecting the outcome of a subgroup of cDLBCL. Our study provides a series of novel findings that allow a better understanding of cDLBCL as a comparative model for human DLBCL but will also lead to improvements in the management of dogs in veterinary clinical practice.
Supplementary Material
Footnotes
Funding: SIR (Scientific Independence of Young Researchers) 2014, Ministero dell’Istruzione, dell’Universita’ e della Ricerca (n. RBSI14EDX9) (to LA) and the Gelu Foundation (to FB).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Aresu L. Canine Lymphoma, More than a morphological diagnosis: what we have learned about diffuse large B-cell lymphoma. Front Vet Sci. 2016;3:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202–208. [DOI] [PubMed] [Google Scholar]
- 3.Harrington BK, Gardner HL, Izumi R, et al. Preclinical evaluation of the novel BTK inhibitor acalabrutinib in canine models of B-cell non-Hodgkin lymphoma. PLoS One. 2016;11(7):e0159607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honigberg LA, Smith AM, Sirisawad M, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107(29):13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasqualucci L, Dalla-Favera R. Genetics of diffuse large B-cell lymphoma. Blood. 2018;131(21):2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushell KR, Kim Y, Chan FC, et al. Genetic inactivation of TRAF3 in canine and human B-cell lymphoma. Blood. 2015;125(6):999–1005. [DOI] [PubMed] [Google Scholar]
- 7.Beachy SH, Onozawa M, Chung YJ, et al. Enforced expression of Lin28b leads to impaired T-cell development, release of inflammatory cytokines, and peripheral T-cell lymphoma. Blood. 2012;120(5):1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferraresso S, Arico A, Sanavia T, et al. DNA methylation profiling reveals common signatures of tumorigenesis and defines epigenetic prognostic subtypes of canine diffuse large B-cell lymphoma. Sci Rep. 2017;7(1):11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marconato L, Frayssinet P, Rouquet N, et al. Randomized, placebo-controlled, double-blinded chemoimmunotherapy clinical trial in a pet dog model of diffuse large B-cell lymphoma. Clin Cancer Res. 2014;20(3):668–677. [DOI] [PubMed] [Google Scholar]
- 10.Boi M, Gaudio E, Bonetti P, et al. The BET Bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res. 2015; 21(7):1628–1638. [DOI] [PubMed] [Google Scholar]
- 11.Zengerle M, Chan KH, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10(8):1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutgen BC, Willenbrock S, Reimann-Berg N, et al. Authentication of primordial characteristics of the CLBL-1 cell line prove the integrity of a canine B-cell lymphoma in a murine in vivo model. PLoS One. 2012;7(6):e40078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarantelli C, Ekeh H, Moscatello C, et al. Abstract A179: The BRD4 degrader MZ1 exhibits potent antitumoral activity in diffuse large B cell lymphoma of the activated B cell-like type. Mol Cancer Ther. 2018;17(1 Supplement):A179–A179. [Google Scholar]
- 14.Mooney M, Bond J, Monks N, et al. Comparative RNA-Seq and microarray analysis of gene expression changes in B-cell lymphomas of Canis familiaris. PLoS One. 2013;8(4):e61088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarantelli C, Gaudio E, Arribas AJ, et al. PQR309 Is a novel dual PI3K/mTOR inhibitor with preclinical antitumor activity in lymphomas as a single agent and in combination therapy. Clin Cancer Res. 2018;24(1):120–129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.