Abstract
Clavanin A (ClavA) is an antimicrobial peptide that enhances its antimicrobial activity in the presence of Zn(II) ions. The antimicrobial activity of ClavA has been shown to increase 16-fold in the presence of Zn(II) ions. In this study, we investigate potential sources of this enhancement, namely, the effect of Zn(II) binding on the helical conformation of ClavA and on the ClavA interaction with a model for gram-negative bacterial membranes. In addition, we investigate the effect of Zn(II) on the membrane mechanical properties. We employed all-atom equilibrium molecular dynamics (MD) simulations initiated from both fully helical and random coil structures of ClavA. We observe that Zn(II) can stabilize an existing helical conformation in the Zn(II)-binding region, but we do not observe induction of helical conformations in systems initiated in random coil configurations. Zn(II) binding to ClavA provides more favorable electrostatics for membrane association in the C-terminal region. This is evidenced by longer and stronger C-terminal-lipid interactions. Zn(II) is also capable of modulating the membrane properties in a manner which favors ClavA insertion and the potential for enhanced translocation into the cell. This work provides insights into the role of divalent metal cations in the antimicrobial activity of ClavA. This information can be useful for the development of synthetic antimicrobial peptides containing motifs that can bind metals (metalloAMPs) for therapeutic and medical purposes.
INTRODUCTION
Metal ions are abundant in all living organisms and their important role in various biological functions, such as biocatalysis through metalloproteins1–3 and immunity,4–6 are being widely and extensively studied.7 Iron and zinc ions, for example, can regulate expression of metal transporters in the bacterial pathogen Corynebacterium diphtheriae,8 while zinc and manganese ions are necessary to prevent growth of the fungus Aspergillus fumigatus during corneal infection.4 In the past few years, interest in the role of copper and zinc ions in the innate immune defense against bacterial pathogens has seen increased attention.5
Antimicrobial peptides (AMPs) are an important component of the innate immune system of many organisms, including humans.9–12 They are part of a larger class of peptides, called membrane-active peptides, as their mechanisms of action involves interaction with cell membranes.13 There are two common mechanisms of action that have been proposed: (1) AMPs cause membrane disruption either by creating pores or acting as a surfactant;13–15 and/or (2) AMPs penetrate the cell membrane, similar to cell-penetrating peptides, and act on intracellular targets, such as DNA, RNA or ribosomes.10,16–17
Numerous studies on a subclass of antimicrobial peptides containing motifs that can bind metals, hereinafter referred to as metalloAMPs, have shown that they exploit the naturally abundant metal ions in organisms for potentiation of their antimicrobial activity.10,16–20 These studies represent a growing body of evidence that highlights the crucial interactions of AMPs with metal ions, two important effectors of the immune system. Recently, the metalloAMP Clavanin A (ClavA, VFQFLGKIIHHVGNFVHGFSHVF-NH2), initially isolated from the tunicate Styela clava, was shown to have a 16-fold increase in antimicrobial activity in the presence of Zn(II) ions.10 ClavA is a cationic and amphipathic, 23-amino acid peptide that is active against a broad spectrum of pathogens in vitro, including methicillin-resistant Staphylococcus aureus, with a maximal activity at pH 5.5.21 More significantly, ClavA has exhibited efficacy against an in vivo S. aureus wound model, with low to no cytotoxicity in mammalian cells, suggesting that ClavA can be used for novel peptide-based therapeutic strategies in treating wound and sepsis infections.22 The HXXXH motif located from positions 17 to 21 in ClavA, where X is any canonical amino acid, is known to form a putative binding site for transition metal ions, including Zn(II).23–24 To our knowledge, there are no experimental or computational reports that investigate Zn(II) binding on the structural properties of ClavA. Here, we investigate the hypothesis that Zn(II) can act as a “staple” that can induce and stabilize helical conformations at the Zn(II)-binding region of the peptide in an attempt to elucidate a structure-function relationship for Zn(II) bound ClavA.
All-atom classical molecular dynamics (MD) simulations have been widely used to investigate how AMPs behave, both in solution and in the presence of model membranes.25–29 Due to limitations in computational power, some MD simulations fail to reach relevant time scales or sample the relevant configurational space and therefore are limited in their ability to provide complete and accurate information on the mechanism of action of AMPs.30 However, computational studies are useful in providing various insights that can guide experimental design efforts. Also, MD simulations are useful in providing mechanistic insights by calculating energy landscapes for conformational transitions, and the ability to observe high energy transition states that are difficult to observe experimentally.31–32
In this work, we investigate the influence of Zn(II) on the conformational properties of ClavA and its interaction with a general model for a gram-negative model membrane. To our knowledge, this is the first time that classical MD simulations have been used to investigate metalloAMPs in the presence of a two-component model membrane. We show that Zn(II) can stabilize helical conformations in the Zn(II)-binding region. We also find that Zn(II) provides more favorable electrostatics for the C-terminal region of ClavA to interact with the membrane. Furthermore, we observe a critical threshold for the molar ratio of Zn(II) to lipids that will induce increased lateral deformability for the E. coli membrane model. With these insights, we present a model of how Zn(II) affects the structure of ClavA and its overall interaction with the membrane.
METHODS
Simulation Models.
The simulations of ClavA were initiated from helical and extended conformations (Figure 1). The helical structure was determined by solution state NMR PDBID:6C4133, in which an α-helical segment was observed from residues Phe2 to Val22. This structure was obtained in 35% 2,2,2-trifluoroethanol (TFE)-d3, a solvent which provides a low dielectric constant similar to a membrane, 10% D2O, and 55% buffer at pH 4.3. The extended structure was generated using the PDB utilities server (https://spin.niddk.nih.gov/bax/). Chemical modifications to the peptide were performed using CHARMM-GUI34–36 which included C-terminal amidation and protonation of histidines (His) in systems modeling a pH 5 environment (Chart 1).
Figure 1.
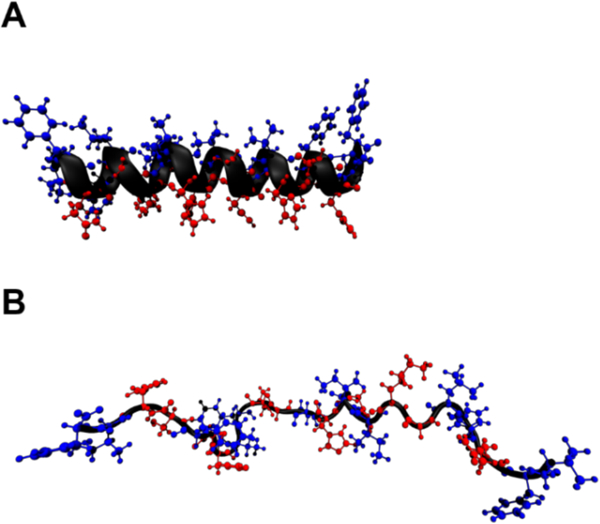
Two simulation models of ClavA used as initial structures in the simulations: (A) helical and (B) random coil. Residues are shown using ball-and-stick model, with hydrophobic residues in blue and hydrophilic residues in red.
Chart 1.
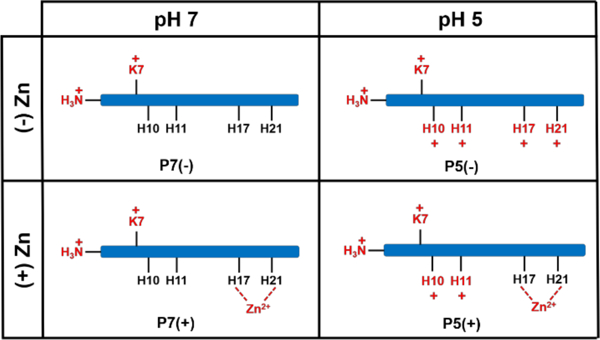
Illustration of the peptide models used in the simulations and their shorthand names in the paper. There is one Lys and four His residues that can titrate depending on the system pH. His residues are protonated at pH 5, except when these residues are bound to Zn(II).
The peptide models were solvated with the TIP3P water model and 150 mM NaCl. Both the ordered and disordered peptide models were also simulated in the presence of a gram-negative bacterial membrane model.37–38 The membrane model consisted of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (POPG) lipids. The membrane contained 120 lipids (60 in each leaflet) with a ratio of 80:20 POPE:POPG. The peptide models were initially placed approximately 3 nm above the membrane. The peptide-membrane system was generated the using CHARMM-GUI server 34–36.
Energy minimization and equilibration steps were performed according to the CHARMM-GUI protocol, employing a steepest-descent minimization for 500 steps and a canonical ensemble equilibration for 25 ps with a timestep of 1 fs for the helical ClavA in solution simulations. For the ClavA in the presence of a membrane model, three successive canonical ensemble equilibrations for 25 ps (1 fs timestep) followed by three successive isothermal-isobaric ensemble equilibrations for 50 ps (2 fs timestep) were employed. Semiisotropic pressure coupling was used to generate a zero surface tension ensemble, using the Berendsen barostat. During the equilibration steps, positional restraints on protein heavy atoms and lipid phosphorus atoms were employed. Dihedral restraints were placed on the lipid glycerol and tail moieties, around the unsaturated bond. The force constants for these restraints were gradually decreased after each equilibration step.
For the helical ClavA in solution system, three independent MD simulations were performed at four different temperatures (300 K, 320 K, 340 K, and 360 K), resulting in twelve independent simulations. Another set of 12 simulations were run for the helical ClavA with Zn(II) bound to His17 and His21. Zn(II) was restrained by applying a distance restraint of 2.095 Å to each δ-nitrogen atom of the His residues, with a force constant of 20,710.8 kJ mol−1 nm−2. These values were obtained by averaging the experimentally obtained coordination length and force constant39 of Zn(II) to the nitrogen atoms of imidazole, an analog of Histidine. The simulations were run for 100 ns each using a timestep of 2 fs. Temperature was maintained using a Nose-Hoover thermostat40–41 with a coupling time constant of 1 ps. Pressure was maintained at 1 bar by implementing an isotropic pressure coupling scheme using the Parrinello-Rahman barostat with a coupling constant of 5 ps.42 Van der Waals interactions were cut off at 1.2 nm, with the interactions being modified using the force-switch method between 1.0 and 1.2 nm. Long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) method, with a real-space cutoff of 1.2 nm. The first 30 ns of the simulations were considered as further equilibration, leaving the remaining 70 ns for analysis (Table 1).
Table 1.
Summary of simulation details.
| System Description |
Initial Conformation |
# of Simulations |
Simulation Length |
Equilibration Time |
Analysis Time | Total Sampling |
Temperature | pH | Zn(II) bound to ClavA ? |
|---|---|---|---|---|---|---|---|---|---|
| Helical Stability Studies |
Helical ClavA in solution | 24 | 100 ns | 30 ns | 70 ns | 1.68 μs | 300 K 320 K 340 K 360 K |
7 | Yes/N o |
| P7(−) P5(−) |
Random Coil ClavA in presence of model membrane | 12 | 500 ns | 100 ns | 400 ns | 4.8 μs | 300 K | 7 5 |
No |
| P7(+) P5(+) |
7 5 |
Yes | |||||||
| hP7(−) hP7(+) |
Helical ClavA in presence of model membrane | 6 | 500 ns | 100 ns | 400 ns | 2.4 μs | 300 K | 7 | Yes/No |
| Zn(II) Effect on Membran e Propertie s |
Model membrane with Zn(II) ions (no ClavA) |
6 | 500 ns | 100 ns | 400 ns | 2.4 μs | 300 K | 7 | n/a |
For the system containing the ClavA in the presence of a membrane, isothermal-isobaric ensemble simulations were run at 300 K for 500 ns. For each of the conditions shown in Chart 1, triplicate simulations were conducted from a random coil configuration. The pH 7 states (P7(+), P7(−)) were also simulated in triplicate starting from a helical conformation. . The temperature was maintained using the Nose-Hoover thermostat40–41 with a coupling time constant of 1 ps, while a semiisotropic pressure coupling scheme was implemented using the Parrinello-Rahman barostat42 with a coupling constant of 5 ps. The lipid bilayer compressibility was set to 4.5 × 10−5 bar−1, and the reference pressures in the bilayer plane (XY) and bilayer normal (Z) were set to 1 bar. Treatment of non-bonded interactions were consistent with the ClavA in solution systems, described above. The first 100 ns of the simulation was considered as further equilibration, leaving the remaining 400 ns for analysis.
Another set of simulations to investigate the effect of ClavA and Zn(II) on membrane properties were also conducted. A total of six simulations with varying Zn(II):lipid ratios, specifically 0:120, 1:120, 1:60, 1:30, 1:15, and 1:7.5, were conducted. The ions were placed approximately 3 nm above the upper leaflet of the membrane. To keep the ions interacting with only the upper leaflet, a flat bottom restraint was applied 6 nm above the membrane. The systems were solvated in 150 mM NaCl and TIP3P water model. The production runs were performed in an isothermal-isobaric ensemble at 300 K and 1 bar for 500 ns. The temperature and pressure coupling parameters, and the non-bonded parameters used in the extended peptide-membrane simulations were also used for the membrane only simulations. The first 100 ns of the simulations were considered as further equilibration, leaving the remaining 400 ns for analysis (Table 1).
All simulations were performed using the GROMACS 201643 program and the CHARMM3644-45 force field. As a control simulation, the ability of the force field to maintain the helical conformation in TFE was evaluated in a 100 ns MD simulation. A superposition of the MD equilibrated and NMR structures (PDBID:6C41) [root-mean-squared deviation (RMSD) = 2.9 Å] showed that ClavA in TFE retained its overall secondary structure and helicity during the simulation (Figure S1). The trajectories were visually analyzed using VMD46, and further quantitative analyses were performed using a combination of GROMACS tools, MDTraj47 and inhouse Python scripts.
Membrane Properties Analysis.
To evaluate the effect of ClavA and Zn(II) on the structural and mechanical properties of membranes, the membrane thickness, area per lipid and area compressibility, were measured as a function of Zn(II) concentration. The membrane thickness was defined by the Z-coordinate difference between the center of mass of the upper and lower leaflet phosphates. The area per lipid was calculated by dividing the box area (in X-Y plane) by the number of lipids per leaflet (60). The area compressibility modulus, KA, of the membrane was calculated from48 where N is the number of lipids in each leaflet and A and A0 are the instantaneous and equilibrium area per lipid, respectively, kB is Boltzmann’s constant and T is the system temperature. All membrane quantities were averaged over the final 400 ns of the simulations (Table 1).
Statistical Analyses.
All reported error estimates are standard errors. For all analyses that required determination of significant difference, the standard Welch’s t-test was used for unequal variances. Survival probability analysis was also used to determine significant differences in association duration times. To summarize, this analysis plots the probability of a region to stay bound before unbinding over some time interval. Log rank test was used to compare significant differences between survival probability plots.
RESULTS
Zn(II) effects on ClavA α-helical stability in Solution
We examined the stability of an α-helical segment located at the Zn(II)-binding region (His17 to His21) by running 100 ns simulations initiated from a helical configuration of ClavA in 150 mM NaCl solution at four different temperatures: 300 K, 320 K, 340 K, and 360 K. For Zn(II)-bound ClavA systems, Zn(II) was bound by applying distance restraints to the δ-nitrogens of His17 and His21 (see Materials and Methods). We observed that, on average, there are four ligands coordinated to Zn(II) ions throughout the simulations at all temperatures (Figure S10). In addition to the coordinating His δ-nitrogens atoms, two water molecule oxygen atoms are found in the first coordination sphere of Zn(II). This is consistent with other Zn(II)-binding proteins that bind Zn(II) in a tetrahedral fashion.49–51 During the simulations a decrease in the average helicity of ClavA without Zn(II) as temperature is increased is observed, as expected since high thermal energy can denature the peptide secondary structure. Interestingly, we did not observe a consistent decrease in helicity in the ClavA-Zn(II) complex as a function of increasing temperature (Figure 2A). At 340 K and above the average helicity of ClavA-Zn(II) remained at about 40%, and did not further decrease in the temperature range we examined. In contrast ClavA without Zn(II) displayed continuously decreasing helicity as temperature increased. Moreover, at temperatures 320 K and 360 K, our simulations showed significantly greater helicity when Zn(II) was bound as compared to that of the Zn(II)-free peptide. The difference in helicity between the two systems could possibly be due to the Zn(II)-binding region being restricted to remain in an α-helical conformation. Looking at the snapshots at the end of the simulations of a representative trial at 360 K (Figure 2B), we observed that the Zn(II)-binding region remains α-helical. To quantify our observations, we used the DSSP analysis tool to calculate the most probable secondary structures of each residue, at 1 ns intervals (Figure 2C). In the same representative trial, we observed that the α-helical conformation of the Zn(II)-binding region is stable for the duration of the 100 ns simulations. The helical stabilization in Zn(II) bound systems was concluded from three independent trials at each temperature. The secondary structure evolution for all simulations is presented in Figures S2–S9. We observed stable helical conformations in regions other than the Zn(II)-binding region, but the helical regions were not consistent across all trials. On longer timescales, we expect that these helical conformations outside the Zn(II)-binding region will eventually destabilize, whereas the region with the metal ion bound to it will remain helical.
Figure 2.
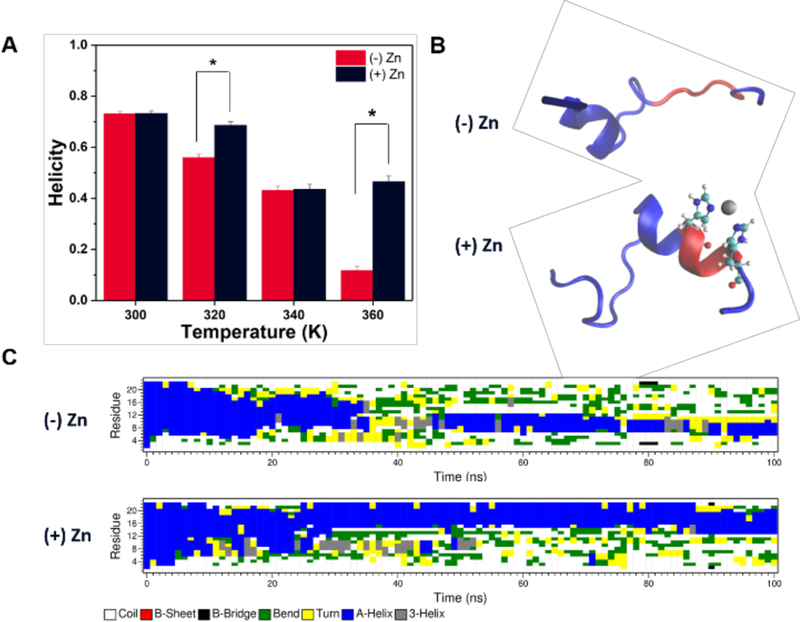
(A) Average helicity of ClavA in the absence or presence of Zn(II) at four different temperatures. Bars represent the average of three trials ± the standard error. Welch’s t-test: *p < 0.05 (B) Final snapshots of representative trials at 360 K. The secondary structure of ClavA is shown, with Zn(II)-binding region represented in red, Zn(II) ion represented in gray, and His17 and H21 represented as a ball-and-stick model. (C) Secondary structure evolution (DSSP analysis) of the same representative trials at 360 K throughout 100 ns simulations.
Zn(II) effects on ClavA a-helical stability in presence of a membrane
To characterize which regions of ClavA have stable helical conformation in the presence of a membrane, simulations were initiated with a helical ClavA placed 3 nm above the membrane. We observed that residues near the C-terminal region, particularly residues His17 to Phe23 lose helicity as early as 80 ns (Figure 3A). In one of the trials (Figure S11), the same region was observed to lose helicity transiently for about 100 ns. It should be noted that helicity in the N-terminal region is not very stable in the presence of a membrane. The general observation in all our helical peptide-membrane simulations is that helicity in the central region, Ile8 to Val16, is more stable than the terminal regions. When Zn(II) was bound to His17 and His21 of ClavA, the helicity of the C-terminal region increased, with it remaining helical for the full 500 ns in all three simulations (Figure 3B, S12). This implies that Zn(II) provided helical stability to the C-terminal region even in the presence of a membrane, as was similarly observed in solution. We also ran triplicate simulations of ClavA in the presence of a membrane, but with 32 free (unrestrained) Zn(II) ions in solution. The results were similar to the ClavA-membrane system without Zn(II), where the terminal regions were not as stable as the central region (Figure 3C). No sustained spontaneous formation of Zn(II)-histidine coordination was observed in the free Zn(II) simulations.
Figure 3.
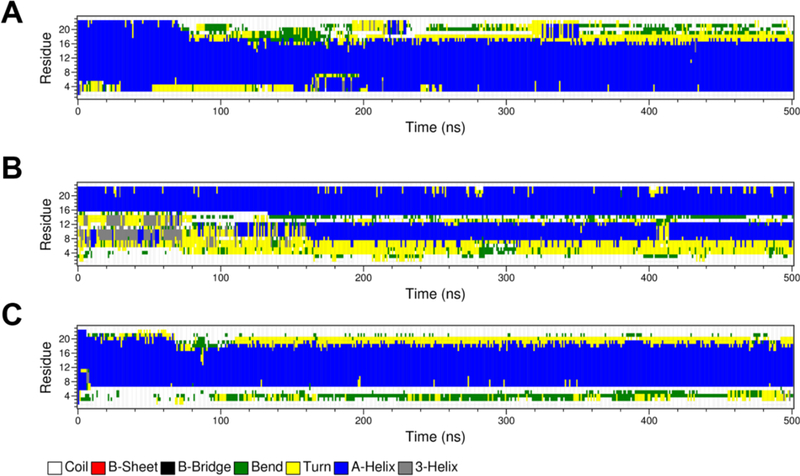
Secondary structure evolution of different peptide models in the presence of a gramnegative bacterial model membrane: (A) ClavA only, (B) ClavA with Zn(II) restrained to His17 and His21, and (C) ClavA with unrestrained 32 Zn(II) ions. The secondary structure evolution plots of the other replicates are presented in the Supporting Information (Figures S11-S13).
We next examined whether Zn(II) influences formation of α-helical structure in ClavA in the presence of a membrane by starting from a random coil conformation of the peptide. The peptide was placed in the presence of the gram-negative bacterial membrane model, composed of 4:1 ratio of POPE and POPG lipids (see Methods). Antimicrobial peptides have been observed, both experimentally and computationally, to form ordered secondary structures in the presence of a membrane due to favorable electrostatic interactions with the lipid headgroups and hydrophobic interactions with the lipid tails.52-54
We did not observe any formation of α-helical structure that can be attributed to binding Zn(II) to ClavA (Figure 4). In the representative trial of ClavA at pH 7 (Figure 4A), P7(−), we observed formation of a-helices and β-sheets outside the Zn(II)-binding region for about 100 ns, but these conformations are short-lived and not stable throughout the simulation. On the other hand, in one trial of the Zn(II)-bound ClavA (Figure S14), P7(+), we observed formation of a stable α-helix outside the Zn(II)-binding region, specifically from residues Lys7 to Val12. To determine whether Zn(II) could possibly have long-range effect of inducing helix formation, we extended the simulation for an additional 500 ns and removed the Zn(II). The helix remained stable even without the Zn(II), thus, we do not attribute the helical formation to Zn(II) binding.
Figure 4.

Secondary structure evolution of representative 500 ns simulation trials initiated from a disordered peptide in the presence of a membrane at neutral pH (A), and at acidic pH (B). In both pH conditions simulations were conducted with and without Zn(II) bound to the peptide.
Because the activity of ClavA is greater at pH 5.5 than at pH 7,21 we proceeded to investigate helical formation under acidic conditions (P5(−) and P5(+) in Chart 1). The simulations of the P5 systems showed no formation of helical structures (Figure 4B). This observation indicates that having an additional four (from P7(−) to P5(−)) or two (from P7(+) to P5(+)) positive charges decreases the probability of forming ordered secondary structures, due to the increased electrostatic repulsion between the protonated residues. However, we do not eliminate the possibility that pKa shifts occur as the peptide inserts and interacts with lower dielectric regions of the membrane.
ClavA Membrane Binding Analysis
Throughout 500 ns membrane binding simulations (Figures 5 and S15–S26), we observed that Phe residues of ClavA insert the deepest into the membrane. Phe residues reach a depth of 1 nm below the phosphate groups. In a previous study, mutation of the Phe residues to other hydrophobic residues, such as isoleucine or tryptophan, did not show any significant differences with the wild type at low pH.55 Though this indicates that Phe residues are not crucial at low pH, our simulations present a striking observation that phenylalanines (Phe) can easily overcome the energetic barrier of crossing the headgroup region of the membrane, and its hydrophobicity provides stabilization for these residues to insert deeper into the hydrophobic region of the membrane.
Figure 5.
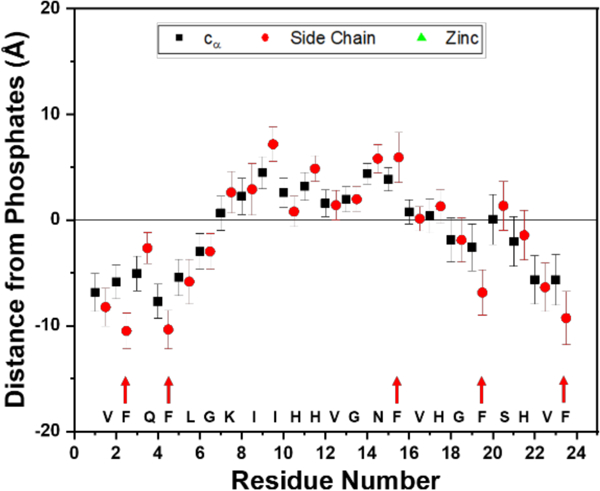
Insertion depths of the residues with respect to the center of mass of the upper leaflet phosphates. Phenylalanine residues insert the deepest, indicated by red arrows. Data is from the last 50 ns of one trial of the system without Zn(II). Data points represent the mean ± standard deviation of the mean.
At pH 7, binding of Zn(II) will effectively increase the net charge of the peptide from +2 to +4, which we predict will lead to stronger electrostatic interactions and preferential interaction of the C-terminal (Zn(II)-binding) region. It has been postulated that the mechanism through which ClavA kills is distinctly different at pH 7 and at pH 5.56 The membrane is not efficiently permeabilized at low pH, suggesting a killing mechanism where there is an internal cell target.10,56 While at a physiological pH, the mechanism involves nonspecific membrane permeabilization, possibly via a carpet model, indicating that peptide-membrane interactions are more critical at pH 7.56–57 Thus, we examined the influence of Zn(II) on the membrane interaction of ClavA in both neutral and acidic pH conditions. The insertion depth profiles of the α-carbon, side chains, and Zn(II) were analyzed (Figures S15–S26). At both pHs, we observed that throughout the simulation, the C-terminal region of ClavA interacts more consistently and has longer association times to the membrane than the N-terminal region, when Zn(II) was bound. In the systems where ClavA has the least positive charge (P7(−)), the peptide approaches the membrane in a fashion where the N-terminal region initially interacts with the membrane and the C-terminal region floats in solution (Figure 6A). With Zn(II) bound at pH 7, visual inspection of the trajectories show that the N-terminal region still associates with the membrane, but the C-terminal region has increased interactions with the membrane (Figure 6B). The snapshots for the initial approach in all trials of all systems are shown in Figures S27–S30.
Figure 6.

Snapshots of the peptide, in the (A) absence or (B) presence of Zn(II), when the N-terminal or C-terminal approaches the model membrane. The snapshots are taken from the longest binding event of one representative trajectory.
Binding of ClavA segments was further quantified by analyzing the number of lipid contacts made by the C-terminal and N-terminal regions for the first 100 ns during the longest binding event of each terminal region. This approach does not inform on how long the regions remain bound, but it provides information on the frequency and amount of contacts. We designated the first 8 residues (VFQFLGKI) as the N-terminal region and the last 8 residues (VHGFSHVF) as the C-terminal region. Each region contains two Phe residues and can carry up to a +2 charge, depending on the pH. A residue contact is considered to be formed when the side chain center of mass (COM) is located within 4 Å of the COM of an upper leaflet phosphate or when the side chain COM is between the phosphates of both leaflets. To evaluate whether there is a preferential interaction of one of the regions, differences in the number of N-terminal contacts and C-terminal contacts were calculated between different simulation conditions (Figure 7). At pH 7, binding of Zn(II) via H17 and H21 significantly increased the number of C-terminal contacts, accompanied by a decrease in the N-terminal contacts, indicating that Zn(II) biases the C-terminal region toward the membrane and keeps it from unbinding. Visual observations from the trajectories indicate that the decrease in interaction of the N-terminal region does not completely prevent this region from associating with the membrane. The total number of contacts between the two systems at pH 7 are not significantly different. This implies that the peptide maintains its affinity with the membrane during the initial approach by decreasing the N-terminal-membrane interactions while Zn(II) increases the C-terminal-membrane interactions. Interestingly, binding Zn(II) at pH 5 did not display any significant difference in the individual N-terminal and C-terminal contacts but the total number of contacts significantly decreased. This further supports that the observed increased contacts at pH 7 is due to the stronger electrostatic interactions, attributed to the presence of bound Zn(II). At pH 5, the C-terminal region has a net charge of +2, regardless of the presence of Zn(II) (Chart 1). Another interesting observation is that when the overall charge of the peptide is increased either by lowering the pH or binding Zn(II), we consistently observe a significant increase in the C-terminal contacts and decrease in the N-terminal contacts.
Figure 7.

Membrane contact analysis. There are significantly greater number of C-terminal region contacts than the number of N-terminal region contacts when Zn(II) is present at pH 7. Bars represent the average of three trials ± the standard error of the mean. Welch’s t-test: *p < 0.05.
We also employed survival analysis to further confirm the preferential interaction of the C-terminal region. In this analysis, each binding incident was treated as an event, and the temporal length of each binding event was measured. The probability of the bound region to unbind was defined as the survival probability. This was plotted against the temporal length of the binding events. As a complement to the first approach, survival analysis does not give complete information on how frequent the contacts are, but rather analyzes the length of each binding event before the region unbinds.
At pH 7 when Zn(II) is present, the probability of having a long binding event (> 50 ns) for the C-terminal region was significantly larger than for the N-terminal region (Figure 8A, right). This was not observed for the systems at pH 5 (Figure S31). In comparison, the binding times of the two terminal regions are not significantly different when Zn(II) is absent (Figure 8A, left). In examining the C-terminal regions, we observed a significant difference between the binding times when Zn(II) is absent and when Zn(II) is present (Figure 8B).
Figure 8.

Survival probability analysis of: (A) the binding times between the N-terminal region and C-terminal region for ClavA (left) (χ2 < 3.84) and ClavA-Zn(II) (right) (χ2 > 3.84), and (B) the binding times of the C-terminal region when Zn(II) is present and when Zn(II) is absent (χ2 > 3.84). Log-rank test: χ2 > 3.84 indicates significant difference between the survival probabilities of the two samples.
We went on to examine the preference for ClavA to interact with the different lipids in our model membrane. Increased positive charge of the peptide should lead to an increased preference of some residues to interact with anionic POPG lipids. We calculated the average ratio of POPE-residue interactions to POPG-residue interactions. One residue-lipid interaction (whether POPE or POPG) is defined as when the region of interest is located within 4 Å of the upper leaflet phosphates, along the bilayer normal. This criterion was set because the tails are chemically identical between POPE and POPG and therefore there should not be any specificity of interactions with the tails. The bulk ratio of POPE:POPG lipids in the bilayer is 4:1, therefore any ratio below that will indicate a preference for POPG while any ratio above that will indicate a preference for POPE. In our simulations, there are two scenarios where the charge of the peptide is increased: (1) decreasing the pH and (2) binding of Zn(II) (Chart 1). Upon binding Zn(II) at pH 7, there is a drastic shift from POPE preference to POPG preference from Val16 to His21 (Figure 9A), where Zn(II) is locally bound, and this is attributed to the increase in positive charge brought by the Zn(II). On the other hand, this was not observed at pH 5. Instead, all C-terminal residues except His17 retained their preference to POPE (Figure 9B). At pH 5 without Zn(II), His17 and His21 are both positively charged, and binding Zn(II) will not increase the positive charge because these His residues will be deprotonated to accommodate Zn(II) binding. The lipid preferences of each residue and Zn(II) in the four different systems are shown in Figure S32.
Figure 9.

Comparison between different systems: (a) P7(−) vs. P7(+), (b) P5(−) vs. P5(+), and (c) P7(+) vs. P5(+) showing the ratio of POPE interactions to POPG interactions of each residue normalized against the bulk ratio of POPE:POPG (4:1) in the model membrane. Bars represent the average of three trials ± the standard error of the mean. The amino acid sequence of Clavanin A is written below the x-axis, with blue and red letters indicating nonpolar and polar residues, respectively.
Moreover, we observed that at pH 5, most residues in the N-terminal region (Gln3 to Ile8) have preference for POPG, peaking at Gly6 (Figure 9B). This is because His10 and His11 become protonated at pH 5, effectively increasing POPG interactions towards the N-terminal region. This is also observed at pH 5 with Zn(II), but the preference is lower in magnitude and it peaks at Ile8. We also compared whether Zn(II) the preference of ClavA to interact with POPE or with POPG at the two pH values. Zn(II) was observed to have more interactions with POPG at pH 7, while there are more favorable interactions with POPE at pH 5 (Figure 9C). This does not indicate that Zn(II) changed its preferential interaction from POPG to POPE as pH is decreased. Our hypothesis is that the two additional protonated His residues at positions 10 and 11 elicit more electrostatic interactions with POPG at pH 5 than with Zn(II). Zn(II) will still want to interact with POPG, but it will have to compete with the protonated residues in terms of binding to POPG, and there will be less available POPG for Zn(II) to interact with. To support the hypothesis, we calculated the probability density of POPG lipids around each residue at both pH levels averaged over the last 400 ns of the simulation and all trials. This analysis shows the probability of having one POPG lipid within 4 Å of each residue. To calculate for the probability for each residue, the total number of POPG lipids within 4 Å was determined and divided by 400 because the total analysis time is 400 ns. A probability of 1 means that, on average, one POPG is around the residue every nanosecond, and a probability of 12 means that all POPGs in the leaflet are found around a given residue every nanosecond. The time course of number of POPG lipids around each residue during the full simulation are shown in Figure S33. At pH 7, large densities of POPG were present around residues Gly18 to Phe23 which spans the C-terminal region of the peptide (Figure 10). Moreover, there is a large density of POPG around Zn(II) at pH 7. At pH 5, the POPG densities in the C-terminal region decreased drastically, while some residues in the N-terminal region (Vall, Gln3, Gly6, and Lys7) and on the central region (Val12 to Asn14) increased in POPG density. The implication is that the protonated His residues at positions 10 and 11 significantly contributed in the increase of anionic POPG lipid interactions towards the N-terminal region. The individual density heat maps of POPG lipids around each residue are shown in Figures S34 and S35.
Figure 10.

Probability density of POPG lipids around each residue of Clavanin A. Residue number 24 represents Zn(II). A high probability of POPG lipids surrounding Gly18 to Phe23 and Zn(II) at pH 7 is observed. The probability decreased at pH 5 with an increase in the probability around the N-terminal residues.
Effect of Zn(II) on membrane properties
Zn(II) will also interact with the membrane, by coordinating with the negatively charged headgroup phosphates when the peptide is bound to the lipid. Thus, we were interested in how Zn(II) can modify different properties of the membrane, including membrane thickness, area per lipid, and area compressibility modulus, when the Zn(II) ions are only interacting with one leaflet of the bilayer (see Methods). At Zn(II):lipid ratios of 1:120 to 1:30, there were no significant changes in any of the membrane properties (Figure 11). Starting at a ratio of 1:15, the membrane increased its thickness and the area per lipid decreased. This indicates that the membrane started to lengthen along the membrane normal and compress laterally. Meanwhile, the compressibility modulus decreased dramatically at a ratio of 1:15 and above, indicating that the membrane can more easily undergo in-plane deformations because of the increased fluctuations in area per lipid. To validate whether the observed trend is dependent on the timescale of analysis, we have plotted the different Ka values calculated using different analysis windows (Figure S36). The absolute values of Ka show some variation, but a significant change in the properties is always observed at Zn(II):lipid ratios of 1:15 and higher. We also conducted three simulations each for the system with no Zn(II) and system with 1:7.5 Zn(II):lipid ratio, where the lateral pressure is set to different values to produce a non-zero surface tension on the lipid bilayer. This is an alternative method of calculating the area compressibility modulus by fitting the changes in area per lipid (A) against the surface tension (γ), according to the following equation: The simulations were run for 100 ns and only the last 60 ns was considered for data analysis (Figure S37A). We have obtained Ka values (151.92 mN/m for no Zn(II) and 137.51 mN/m for 1:7.5 Zn(II):lipid) that are different from our previous methodology, but we still observe a significant decrease in area compressibility modulus at the highest number of Zn(II) ions interacting with the membrane (Figure S37B).
Figure 11.

(left) Effect of Zn(II) concentration on membrane thickness, area per lipid, and area compressibility modulus of the gram-negative bacterial membrane model. (right) Effect of peptide interactions in different system conditions: pH 7 or pH 5, Zn(II) absent, Zn(II) restrained to His17 and His21, or 32 Zn(II) ions unrestrained. Significant changes in the membrane properties were observed at Zn(II) concentrations at or above 1:15 Zn(II):lipid ratio.
We also examined how the membrane properties were affected by ClavA (with or without Zn(II)). Interestingly, we observe an opposite effect in membrane thickness and area per lipid for peptide-membrane simulations in both Zn(II)-absent and Zn(II)-restrained systems. Interaction of ClavA with the membrane creates additional surface area in the bilayer plane leading to increased area per lipid and membrane thinning (Figure 11). The compressibility modulus of the membranes in ClavA systems without Zn(II) decreased significantly, similar to the Zn(II)-membrane systems at ratios of 1:15 and 1:7.5. Binding Zn(II) to ClavA neutralizes this effect, restoring the compressibility modulus to the value observed in membrane only systems. An additional system was considered with ClavA and 32 free Zn(II) ions (1:7.5 Zn(II):lipid ratio). We ran three trials of this system, and the effects on the membrane were similar to the Zn(II)-membrane system with 1:7.5 Zn(II):lipid ratio.
DISCUSSION
There are three main effects of Zn(II) that were observed from this simulation study: (i) Zn(II) stabilizes helical conformations in the C-terminal region by coordinating to His residues in the HXXXH motif and act as a structural staple, (ii) Zn(II) influences the behavior of the C-terminal interactions with gram-negative bacterial membrane model, and (iii) both peptide and Zn(II) causes changes to structural and mechanical properties of the bilayer causing a lower energy cost for in-plane stretching deformations. There is a wide variety of studies done on peptide stapling, including those that use all-hydrocarbon molecules as staples.58–59 These staples decrease the conformational space available for the peptide backbone, restrict the peptide to its helical conformation, and in effect, stabilize the helicity of the peptides. There should be a balance between hydrophilic and hydrophobic residues for an AMP to penetrate the cell membrane, as electrostatic interactions are crucial in approaching the polar headgroups while hydrophobic effects are important when interacting and crossing the hydrophobic core of the bilayer. Cationic and amphipathic peptides, such as ClavA, are advantageous for cell entry as they have an amphipathic character. However, this amphipathic character is most pronounced when the peptide is in an ordered secondary structure, such as an α-helix. ClavA is a short, disordered peptide in solution, and it can be inferred that a helical conformation will be marginally stable (or unstable) due to the loss of configurational entropy upon folding. In this respect, Zn(II) can act to reduce the configurational space in the unfolded state and hence reduce the entropy loss upon folding. We believe this effect will stabilize an α-helical segment in the C-terminal region in the presence of the membrane. The stabilization of the helix facilitates membrane penetration by the hydrophobic effect when ordered waters are released into the bulk when the peptide hydrophobic face buries into the bilayer.
When ClavA is inserted into the bilayer, the hydrophobic environment of the membrane was found to stabilize the overall a-helical structure of ClavA. The backbones atoms of Val1 and Phe23 of ClavA interacted through weak hydrogen bonds with the water molecules present at the surface of the membrane. (Figure S38). Superposition of ClavA and ClavA-Zn(II) showed that the two structures were quite similar with an overall RMSD of 3.05 Å. However, His17 in the Zn(II)-free form was flipped away from the position it adopts in the Zn(II) bound system, while His21 maintained the same orientation in both forms.
The importance of the C-terminal region of ClavA is also highlighted by the observation that binding Zn(II) resulted in longer and more favorable interaction of the C-terminal region with the lipids, as compared to that of the N-terminal region. Moreover, the C-terminal region exhibited a shift in preference to POPG interactions when it is Zn(II)-bound at pH 7. On the other hand, at pH 5, the Zn(II)-bound C-terminal region retained its interaction to POPE because two protonated His residues, His10 and His11, compete with Zn(II) in interacting with POPG. We hypothesize that this will consequently pull POPG towards the N-terminal region and away from the C-terminal region, leaving Zn(II) to interact with POPE. All these observations show the importance of the charge state of both the peptide and the membrane.
The initial contact of metalloAMP to the membrane is due to the electrostatic interactions of the anionic lipids in the model membrane and two components of the metalloAMP: (i) positively charged residues, such as histidines (at pH 5) and lysines (at pH 7 and pH 5), or (ii) the metal cation bound to the peptide. In this case, we propose the metal cation shifting the equilibrium toward the folded state which then in turn favors interactions with the membrane due to the amphipathicity of the helical peptide. However, we hypothesize that the metal ion should not bind too tightly to the metalloAMP, as the metal ion will not be able to efficiently translocate across the membrane due to highly unfavorable interactions with the lipid tails and favorable interactions with the lipid headgroups. Enhancement in the binding of ClavA was also observed in the simulations through the survival analysis which showed longer binding times for the Zn(II) bound-C terminal region as compared to ClavA without Zn(II).
In studying the peptide-membrane interactions of cationic metalloAMPs using classical MD simulations, it is informative to examine the local environment of the metalloAMP as it interacts with the model membrane. We see that the Phe residues can readily cross the polar headgroups region of the bilayer, and insert deeply into the hydrophobic region of the bilayer. A previous study on ClavA highlighted the importance of Phe residues by providing balanced hydrophobicity, membrane affinity, and conformational flexibility of ClavA.55 T ogether with our findings, this gives us insight on the crucial role of Phe residues in the antimicrobial activity of ClavA and why these residues are highly conserved in clavanins (Figure S39). A potential future direction would be to investigate ClavA mutants involving Phe mutations to other hydrophobic residues, such as isoleucine or tryptophan, and evaluate the effect on membrane binding and insertion. Second, we would like to note that we have not observed the ClavA models to be completely buried in the membrane. Charged residues are important for membrane binding but it is believed that most charged residues will lose their charge as they enter the low dielectric environment of the membrane interior.60–61 However, this feature cannot be captured in classical MD simulations due to the fixed charge model of the force field. Fluctuating charge models, such as polarizable force fields for proteins and lipids or constant pH MD simulations,62–65 can in principle be employed to overcome this limitation.
In previous studies, the interaction of Zn(II) with the phospholipid 1-palmitoyl-2-oleoyl-sn-3-glycero-phosphocholine (POPC) was shown to create Zn(II)-bridges between adjacent lipid molecules that stabilize the gel phase of the bilayer.66 This bridge causes the POPC membrane to be less laterally compressible.66 Interestingly, our simulations show an opposite behavior. The area compressibility modulus of the membrane model decreases starting at a Zn(II):lipid ratio of 1:15, which indicates that the membranes can be deformed easier. This could be due to the differences in the membrane composition, as the larger size of choline headgroups relative to the glycerol and ethanolamine could create a different mechanical response. On the other hand, our results agree with an experimental study that shows an increase in membrane thickness in 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC).67 It has been observed that both Ca(II) and Zn(II) binding results in membrane thickening, but a critical concentration of Zn(II) swells the DPPC membrane to a maximum thickness, with no further thickening at concentrations higher than the critical concentration.67 Higher concentration of Ca(II) with respect to DPPC, on the other hand, results in bilayer thinning.67 Another interesting observation is the ability of ClavA to decrease the area compressibility modulus of the membrane model comparable what is observed at high Zn(II) concentrations (1:15 Zn(II):lipid ratio), but with decreased membrane thickness and increased area per lipid. This could be a contributing factor to the membrane penetration of the peptide as it attacks its target. Oddly, binding Zn(II) inhibits this effect and this opens up a further area of investigation as to how this happens. A possible implication of this phenomenon is that once Zn(II) has served its role of pulling the peptide closer to the membrane, it will unbind from the peptide and interact with the anionic lipids, and this will prevent the membrane from transitioning into a less compressible state.
In this study we have employed equilibrium constant temperature simulations, performed in replicates to statistically analyze different effects on the peptide conformation and membrane interactions. Due to the timescale limitations in standard MD, we were unable to observe Zn(II) induced spontaneous helical formation in the C-terminal region. In MD simulations, significant formation of secondary structures is well-observed at a timescale of microseconds to milliseconds, which is beyond the timescales simulated in the current study.30,68 There are various simulation approaches which can be employed to access longer time scale phenomena from shorter simulations lengths. These approaches, known collectively as enhanced sampling methods, accelerate the sampling through temperature-based methods (e.g. replica exchange), by applying biases along specific variables of the system (e.g. umbrella sampling, metadynamics) or by stitching together independent short simulations which sample different regions of phase space (e.g. weighted ensemble, Markov state models).69–73 In future studies we aim to further examine ClavA folding through an enhanced sampling approach to assess thermodynamics quantities which can only be accurately calculated from converged equilibrium probabilities. Another limitation in the current study is we were unable to observe spontaneous formation of Zn(II)-histidine complexes when we expose ClavA to unrestrained Zn(II) ions. CHARMM36 is a standard unpolarized force field that model charges as being fixed. Therefore, our current system cannot accurately depict the polarization effects of the metal center Zn(II) to its potential histidine ligands.74 To explore spontaneous binding of Zn(II) to ClavA, force fields such as the Extended Zinc AMBER Force Field (EZAFF) or polarizable force fields, such as AMOEBA, are likely to provide increased accuracy in modeling the ClavA-Zn(II) complex.74–77
CONCLUSIONS
We present a domino-effect model on how Zn(II) facilitates ClavA membrane interactions that could potentially contribute in the enhancement of antimicrobial activity (Scheme 1). First, Zn(II) binding to ClavA increases the net positive charge in the C-terminal region of the peptide, driving the peptide closer to the membrane, increasing the effective concentration and possibly causing peptide localization on the cell membrane, as observed experimentally.10 An alternative possibility is that Zn(II) will first bind to the membrane via electrostatic interactions with the anionic lipids, which could then enhance the affinity of the C-terminal region of ClavA to the membrane. However, the peptide interactions with a Zn(II) bound membrane was not explored in the current study. Once ClavA is in contact with the membrane, the Phe residues start to penetrate the membrane. ClavA will then have an increased probability of forming an α-helical conformation as the peptide positions its polar residues along the polar headgroups and its nonpolar residues along the nonpolar lipid tails. Zn(II) can then act to restrict conformational sampling and favor folding as well as act as a peptide staple, stabilizing the helical conformation once formed. Once stable, Zn(II) can unbind from the peptide, interact with the membrane, and cause an easier lateral deformation of the membrane. This will increase the probability of having lipids spaced out away from each other, such that ClavA can insert itself into the bilayer and transition to a transmembrane state. We do not suggest that all AMPs can take advantage of the lateral membrane-deforming effect brought by Zn(II) binding to the membrane. For example, the antimicrobial activity of magainin 2, clavanin C, and the anionic peptide maximin H5, do not get potentiated in the presence of Zn(II).10,78 In contrast, the AMP kappacin and an anionic hexapeptide from human amniotic fluid potentiate their activities in the presence of Zn(II).79–80 Interestingly, all AMPs we identified which have enhanced antimicrobial activity in the presence of Zn(II) contain the HXXXH motif, except for the ovine pulmonary surfactant-associated anionic peptides. (Figure S40).
Scheme 1.
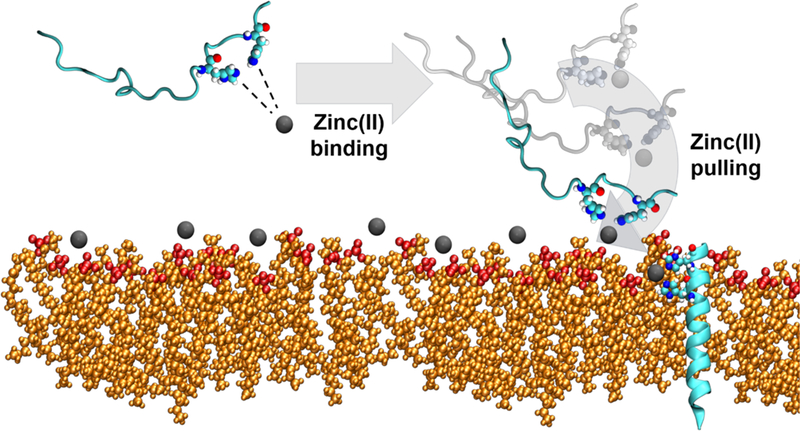
Proposed domino-effect model for the role of Zn(II) in the helical stability, enhanced membrane interaction of C-terminal region, and the role of Zn(II) in greater bilayer deformability.
We would like to emphasize that our studies have only examined single peptides and that larger amounts of peptides and Zn(II), likely lead to oligomerization. Alternatively, it has been suggested that transmembrane organization does not happen for ClavA,81 and we will explore these phenomena in future studies. Also, even though the simulation models present that the same Zn(II) is bound to ClavA throughout the simulation, it is important to take note that Zn(II) binding is dynamic. The binding dynamics is an advantage for this mechanism because Zn(II) will be able to easily alternate roles of stabilizing helical conformations, driving ClavA to the membrane, and modifying membrane properties. This study provides new data and also opens new questions to the metalloAMP field to further understand the role of metal ions in metalloAMPs and how we can utilize this knowledge in developing novel AMP-based therapeutics.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the University of Connecticut for granting access to the Storrs HPC cluster.
Funding Sources
This work was supported by the grants from National Science Foundation (Grant MCB1715494 to AMA-B and Grant CHE-1664926 to RP) and the National Institutes of Health (Grant R35GM119762 to ERM). Computational resources have been provided by the NSF XSEDE program (grant TG-MCB170151).
ABBREVIATIONS
- ClavA
Clavanin A
- AMP
antimicrobial peptides
- metalloAMP
antimicrobial peptide with metal ion/s bound to it via coordination
- TFE
2,2,2-trifluoroethanol
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol)
- MD
molecular dynamics
- PDB
protein data bank
- CHARMM
Chemistry at Harvard Macromolecular Mechanics
- CHARMM-GUI
Chemistry at Harvard Macromolecular Mechanics – Graphical User Interface
- GROMACS
Groningen Machine for Chemical Simulations software
- VMD
visual molecular dynamics software
- TIP3P
three-site transferable intermolecular potential
- PME
particle mesh Ewald
- DSSP
Define Secondary Structure of Proteins algorithm
- COM
center of mass
- DNA
deoxyribonucleic acid
- NMR
nuclear magnetic resonance
- ATCUN
amino-terminal copper and nickel binding motif.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
The following files are available free of charge.
Secondary structure evolution, insertion depth profiles, additional experimental details for all simulation trials, and sequence alignment of Clavanins and other antimicrobial peptides (PDF)
The authors declare no competing financial interests.
REFERENCES
- 1.Poole K, At the nexus of antibiotics and metals: The impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol 2017, 25 (10), 820–832. [DOI] [PubMed] [Google Scholar]
- 2.On advances and challenges in biocatalysis. Nature Catalysis 2018, 1 (9), 635–636. [Google Scholar]
- 3.Ball ZT, Designing enzyme-like catalysts: A rhodium(ii) metallopeptide case study. Acc. Chem. Res 2013, 46 (2), 560–570. [DOI] [PubMed] [Google Scholar]
- 4.Clark HL; Jhingran A; Sun Y; Vareechon C; de Jesus Carrion S; Skaar EP; Chazin WJ; Calera JA; Hohl TM; Pearlman E, Zinc and manganese chelation by neutrophil S100A8/A9 (Calprotectin) limits extracellular Aspergillus fumigatus hyphal growth and corneal infection. J Immunol 2016, 196 (1), 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djoko KY; Ong CL; Walker MJ; McEwan AG, The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 2015, 290 (31), 18954–18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer LD; Skaar EP, Transition metals and virulence in bacteria. Annu Rev Genet 2016, 50, 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conklin SE; Bridgman EC; Su Q; Riggs-Gelasco P; Haas KL; Franz KJ, Specific histidine residues confer Histatin peptides with copper-dependent activity against Candida albicans. Biochemistry 2017, 56 (32), 4244–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng ED; Oram DM; Battistel MD; Lyman LR; Freedberg DI; Schmitt MP, Iron and zinc regulate expression of a putative ABC metal transporter in Corynebacterium diphtheriae. JBacteriol 2018, 200 (10), e00051–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fjell CD; Hiss JA; Hancock RE; Schneider G, Designing antimicrobial peptides: Form follows function. Nat Rev Drug Discov 2011, 11 (1), 37–51. [DOI] [PubMed] [Google Scholar]
- 10.Juliano SA; Pierce S; deMayo JA; Balunas MJ; Angeles-Boza AM, Exploration of the innate immune system of Styela clava: Zn2+ binding enhances the antimicrobial activity of the tunicate peptide Clavanin A. Biochemistry 2017, 56 (10), 1403–1414. [DOI] [PubMed] [Google Scholar]
- 11.Paulmann M; Arnold T; Linke D; Ozdirekcan S; Kopp A; Gutsmann T; Kalbacher H; Wanke I; Schuenemann VJ; Habeck M et al. , Structure-activity analysis of the dermcidin-derived peptide DCD-1l, an anionic antimicrobial peptide present in human sweat. J Biol Chem 2012, 287 (11), 8434–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sass V; Schneider T; Wilmes M; Korner C; Tossi A; Novikova N; Shamova O; Sahl HG, Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun 2010, 78 (6), 2793–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henriques ST; Melo MN; Castanho MA, Cell-penetrating peptides and antimicrobial peptides: How different are they? Biochem J 2006, 399 (1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balandin SV; Ovchinnikova TV, Antimicrobial peptides of invertebrates. Part 2. Biological functions and mechanisms of action. Russian Journal of Bioorganic Chemistry 2016, 42 (4), 343–360. [Google Scholar]
- 15.Shai Y, Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by a-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochimica et Biophysica Acta 1999, 1462, 55–70. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y; Cowan JA, Targeted cleavage of HIV Rev response element RNA by metallopeptide complexes. J. Am. Chem. Soc 2006, 128 (2), 410–411. [DOI] [PubMed] [Google Scholar]
- 17.Libardo MDJ; Bahar AA; Ma B; Fu R; McCormick LE; Zhao J; McCallum SA; Nussinov R; Ren D; Angeles-Boza AM et al. , Nuclease activity gives an edge to host-defense peptide Piscidin 3 over Piscidin 1, rendering it more effective against persisters and biofilms. FEBS J 2017, 284 (21), 3662–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunden LS; Nolan EM, Bioinorganic explorations of Zn(ii) sequestration by human S100 host-defense proteins. Biochemistry 2018, 57 (11), 1673–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Łoboda D; Koziowski H; Rowińska-Żyrek M, Antimicrobial peptide–metal ion interactions – a potential way of activity enhancement. New Journal of Chemistry 2018, 42 (10), 7560–7568. [Google Scholar]
- 20.Libardo MDJ; Gorbatyuk VY; Angeles-Boza AM, Central role of the copper-binding motif in the complex mechanism of action of Ixosin: Enhancing oxidative damage and promoting synergy with Ixosin B. ACS Infectious Diseases 2016, 2 (1), 71–81. [DOI] [PubMed] [Google Scholar]
- 21.Lee IH; Cho Y; Lehrer RI, Effects of pH and salinity on the antimicrobial properties of Clavanins. Infect Immun 1997, 65 (7), 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva ON; Fensterseifer IC; Rodrigues EA; Holanda HH; Novaes NR; Cunha JP; Rezende TM; Magalhaes KG; Moreno SE; Jeronimo MS et al. , Clavanin A improves outcome of complications from different bacterial infections. Antimicrob Agents Chemother 2015, 59 (3), 1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzsimons MP; Barton JK, Design of a synthetic nuclease: DNA hydrolysis by a zinc-binding peptide tethered to a rhodium intercalator. J. Am. Chem. Soc 1997, 119 (14), 3379–3380. [Google Scholar]
- 24.Zaykov AN; Popp BV; Ball ZT, Helix induction by dirhodium: Access to biocompatible metallopeptides with defined secondary structure. Chemistry 2010, 16 (22), 6651–6659. [DOI] [PubMed] [Google Scholar]
- 25.Perrin BS Jr.; Fu R; Cotten ML; Pastor RW, Simulations of membrane-disrupting peptides II: AMP Piscidin 1 favors surface defects over pores. Biophys J 2016, 111 (6), 1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D; Forsman J; Woodward CE, Molecular simulations of melittin-induced membrane pores. J Phys Chem B 2017, 121 (44), 10209–10214. [DOI] [PubMed] [Google Scholar]
- 27.Li J; Liu S; Lakshminarayanan R; Bai Y; Pervushin K; Verma C; Beuerman RW, Molecular simulations suggest how a branched antimicrobial peptide perturbs a bacterial membrane and enhances permeability. Biochim Biophys Acta 2013, 1828 (3), 1112–1121. [DOI] [PubMed] [Google Scholar]
- 28.Khatami MH; Bromberek M; Saika-Voivod I; Booth V, Molecular dynamics simulations of histidine-containing cod antimicrobial peptide paralogs in self-assembled bilayers. Biochim Biophys Acta 2014, 1838 (11), 2778–2787. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y; Schlamadinger DE; Kim JE; McCammon JA, Comparative molecular dynamics simulations of the antimicrobial peptide CM15 in model lipid bilayers. Biochim Biophys Acta 2012, 1818 (5), 1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y; Zhao T; Wei D; Strandberg E; Ulrich AS; Ulmschneider JP, How reliable are molecular dynamics simulations of membrane active antimicrobial peptides? Biochim Biophys Acta 2014, 1838 (9), 2280–2288. [DOI] [PubMed] [Google Scholar]
- 31.Nangia S; May ER, Influence of membrane composition on the binding and folding of a membrane lytic peptide from the non-enveloped flock house virus. Biochim Biophys Acta Biomembr 2017, 1859 (7), 1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nangia S; Pattis JG; May ER, Folding a viral peptide in different membrane environments: Pathway and sampling analyses. J Biol Phys 2018, 44 (2), 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva ON; Alves ES; de la Fuente-Nunez C; Ribeiro SM; Mandal SM; Gaspar D; Veiga AS; Castanho MA; Andrade CA; Nascimento JM et al. , Structural studies of a lipid-binding peptide from tunicate hemocytes with anti-biofilm activity. Sci Rep 2016, 6, 27128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu EL; Cheng X; Jo S; Rui H; Song KC; Davila-Contreras EM; Qi Y; Lee J; Monje-Galvan V; Venable RM et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem 2014, 35 (27), 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jo S; Lim JB; Klauda JB; Im W, CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys J 2009, 97 (1), 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jo S; Kim T; Im W, Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2007, 2 (9), e880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epand RF; Savage PB; Epand RM, Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (ceragenins). Biochim Biophys Acta 2007, 1768 (10), 2500–2509. [DOI] [PubMed] [Google Scholar]
- 38.Pandit KR; Klauda JB, Membrane models of E.coli containing cyclic moieties in the aliphatic lipid chain. Biochim Biophys Acta 2012, 1818 (5), 1205–1210. [DOI] [PubMed] [Google Scholar]
- 39.Carballo R; Castiñeiras A; Covelo B; Garcĺ’a-Martĺ’nez E; Niclós J; Vzquez-López EM, Solid state coordination chemistry of mononuclear mixed-ligand complexes of Ni(ii), Cu(ii) and Zn(ii) with a-hydroxycarboxylic acids and imidazole. Polyhedron 2004, 23 (9), 1505–1518. [Google Scholar]
- 40.Hoover WG, Canonical dynamics: Equilibrium phase-space distributions. Physical Review A 1985, 31 (3), 1695–1697. [DOI] [PubMed] [Google Scholar]
- 41.Nosé S, A unified formulation of the constant temperature molecular dynamics methods. The Journal of Chemical Physics 1984, 81 (1), 511–519. [Google Scholar]
- 42.Parrinello M; Rahman A, Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics 1981, 52 (12), 7182–7190. [Google Scholar]
- 43.Abraham MJ; Murtola T; Schulz R; Pall S; Smith JC; Hess B; Lindahl E, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar]
- 44.Klauda JB; Venable RM; Freites JA; O’connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD Jr.; Pastor RW, Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 2010, 114 (23), 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Best RB; Zhu X; Shim J; Lopes PE; Mittal J; Feig M; Mackerell AD Jr., Optimization of the additive charmm all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput 2012, 8 (9), 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Humphrey W; Dalke A; Schulten K, VMD - Visual Molecular Dynamics. J. Molec. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 47.McGibbon RT; Beauchamp KA; Harrigan MP; Klein C; Swails JM; Hernandez CX; Schwantes CR; Wang LP; Lane TJ; Pande VS, Mdtraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys J 2015, 109 (8), 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marrink SJ; de Vries AH; Mark AE, Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B 2004, 108 (2), 750–760. [Google Scholar]
- 49.Wong AH; Zhou D; Rini JM, The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J Biol Chem 2012, 287 (44), 36804–36813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen TT; Chang SC; Evnouchidou I; York IA; Zikos C; Rock KL; Goldberg AL; Stratikos E; Stern LJ, Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol 2011, 18 (5), 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin H; Andersen GR; Yatime L, Crystal structure of human S100A8 in complex with zinc and calcium. BMC Struct Biol 2016, 16 (1), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nymeyer H; Woolf TB; Garcia AE, Folding is not required for bilayer insertion: Replica exchange simulations of an alpha-helical peptide with an explicit lipid bilayer. Proteins 2005, 59 (4), 783–790. [DOI] [PubMed] [Google Scholar]
- 53.Gong Z; Ikonomova SP; Karlsson AJ, Secondary structure of cell-penetrating peptides during interaction with fungal cells. Protein Sci 2018, 27 (3), 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W; Smith DK; Moulding K; Chen HM, The dependence of membrane permeability by the antibacterial peptide Cecropin B and its analogs, CB-1 and CB-3, on liposmes of different composition. J. Biol. Chem. 1998, 273 (42), 27438–27448. [DOI] [PubMed] [Google Scholar]
- 55.van Kan EJM; Demel RA; van der Bent A; de Kruijff B, The role of the abundant phenylalanines in the mode of action of the antimicrobial peptide clavanin. Biochimica et Biophysica Acta (BBA) - Biomembranes 2003, 1615 (1–2), 84–92. [DOI] [PubMed] [Google Scholar]
- 56.van Kan EJM; Demel RA; Breukink E; van der Bent A; de Kruijff B, Clavanin permeabilizes target membranes via two distinctly different pH-dependent mechanisms. Biochemistry 2002, 41 (24), 7529–7539. [DOI] [PubMed] [Google Scholar]
- 57.van kan EJM; van der Bent A; Demel RA; de Kruijff B, Membrane activity of the peptide antibiotic clavanin and the importance of its glycine residues. Biochemistry 2001, 40 (21), 6398–6405. [DOI] [PubMed] [Google Scholar]
- 58.Schafmeister CE; Po J; Verdine GL, An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J. Am. Chem. Soc 2000, 122 (24), 5891–5892. [Google Scholar]
- 59.Verdine GL; Hilinski GJ, Stapled peptides for intracellular drug targets. Methods Enzymol 2012, 503, 3–33. [DOI] [PubMed] [Google Scholar]
- 60.Panahi A; Brooks CL 3rd, Membrane environment modulates the pKa values of transmembrane helices. J Phys Chem B 2015, 119 (13), 4601–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gleason NJ; Vostrikov VV; Greathouse DV; Koeppe RE, 2nd, Buried lysine, but not arginine, titrates and alters transmembrane helix tilt. Proc Natl Acad Sci 2013, 110 (5), 1692–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khandogin J; Brooks CL 3rd, Constant pH molecular dynamics with proton tautomerism. Biophys J 2005, 89 (1), 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H; Chowdhary J; Huang L; He X; MacKerell AD Jr.; Roux B, Drude polarizable force field for molecular dynamics simulations of saturated and unsaturated zwitterionic lipids. J Chem Theory Comput 2017, 13 (9), 4535–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J; Lopes PE; Roux B; MacKerell AD Jr., Recent advances in polarizable force fields for macromolecules: Microsecond simulations of proteins using the classical drude oscillator model. J Phys Chem Lett 2014, 5 (18), 3144–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopes PE; Huang J; Shim J; Luo Y; Li H; Roux B; Mackerell AD Jr., Force field for peptides and proteins based on the classical drude oscillator. J Chem Theory Comput 2013. 9 (12), 5430–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Binder H; Arnold K; Ulrich AS; Zschornig O, Interaction of Zn2+ with phospholipid membranes. Biophys. Chem. 2001, 90, 57–74. [DOI] [PubMed] [Google Scholar]
- 67.Kucerka N; Dushanov E; Kholmurodov KT; Katsaras J; Uhrikova D, Calcium and zinc differentially affect the structure of lipid membranes. Langmuir 2017, 33 (12), 3134–3141. [DOI] [PubMed] [Google Scholar]
- 68.Cino EA; Choy WY; Karttunen M, Comparison of secondary structure formation using 10 different force fields in microsecond molecular dynamics simulations. J Chem Theory Comput 2012, 8 (8), 2725–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Husic BE; Pande VS, Markov state models: From an art to a science. J Am Chem Soc 2018, 140 (7), 2386–2396. [DOI] [PubMed] [Google Scholar]
- 70.Zuckerman DM; Chong LT, Weighted ensemble simulation: Review of methodology, applications, and software. Annu Rev Biophys 2017, 46, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barducci A; Bussi G; Parrinello M, Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Phys Rev Lett 2008, 100 (2), 020603. [DOI] [PubMed] [Google Scholar]
- 72.Sugita Y; Okamoto Y, Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett 1999, 314, 141–151. [Google Scholar]
- 73.Torrie GM; Valleau JP, Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comput Phys 1977, 23, 187–199. [Google Scholar]
- 74.Li P; Merz KM Jr., Metal ion modeling using classical mechanics. Chem Rev 2017, 117 (3), 1564–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu Z; Li P; Merz KM Jr., Extended Zinc Amber Force Field (EZAFF). J Chem Theory Comput 2018, 14 (1), 242–254. [DOI] [PubMed] [Google Scholar]
- 76.Wu R; Lu Z; Cao Z; Zhang Y, A transferable nonbonded pairwise force field to model zinc interactions in metalloproteins. J Chem Theory Comput 2011, 7, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J; Yang W; Piquemal JP; Ren P, Modeling structural coordination and ligand binding in zinc proteins with a polarizable potential. J Chem Theory Comput 2012, 8 (4), 1314–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lai R; Liu H; Lee WH; Zhang Y, An anionic antimicrobial peptide from toad Bombina maxima. Biochem Biophys Res Commun 2002, 295, 796–799. [DOI] [PubMed] [Google Scholar]
- 79.Schlievert P; Johnson W; Galask RP, Isolation of a low molecular weight antibacterial system from human amniotic fluid. Infect Immun 1976, 14 (5), 1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dashper SG; O’Brien-Simpson NM; Cross KJ; Paolini RA; Hoffmann B; Catmull DV; Malkoski M; Reynolds EC, Divalent metal cations increase the activity of the antimicrobial peptide Kappacin. Antimicrob Agents Chemother 2005, 49 (6), 2322–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van kan EJM; Ganchev DN; Snel MME; Chupin V; van der Bent A; De Kruijff B, The peptide antibiotic Clavanin A interacts strongly and specifically with lipid bilayers. Biochemistry 2003, 42 (38), 11366–11372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


