Abstract
Objective
Prostate cancer (PCa) is one of the most prevalent types of cancer among men worldwide. The incidence of PCa is increasing in China. Therefore, there is an urgent need to identify novel diagnostic and prognostic markers for PCa to improve the treatment of the disease.
Methods
The Cancer Genome Atlas (TCGA) and GEO database were used to analyze the expression of miR-192, and the relationship between miR-192 and the clinical features of patients with PCa. Cell cycle and cell proliferation assay were used to detect the functional roles of miR-192 in PCa. Bioinformatic analysis for miR-192–5p was performed using gene ontology and KEGG analysis.
Results
By analyzing the dataset of TCGA, we found that miR-192 was overexpressed in PCa samples compared to normal tissues and was upregulated in high-grade PCa compared to low-grade PCa. We also observed that higher miR-192 expression was associated with a shorter biochemical recurrence-free survival time. Our results also demonstrated that miR-192 promoted PCa cell proliferation and cell cycle progression.
Conclusion
These results suggest that miR-192 may be considered for use as a potential diagnostic and therapeutic target of PCa.
Keywords: Prostate cancer, miR-192–5p, Proliferation, Cell cycle
Significance of the Study
Prostate cancer (PCa) is one of the most common cancers in men worldwide, for which there is an urgent need of good diagnostic and prognostic markers. We found that miR-192 is overexpressed in PCa tissue samples compared to normal tissues, and is higher in high-grade PCa compared to lower-grade cancer. We suggest that miR-192 may be considered for use as a potential diagnostic marker of PCa.
Introduction
MicroRNAs (miRNAs/miRs) are a kind of endogenous small noncoding RNA about 22 nucleotides in length. miRNAs had been reported to play an important role in various biological processes, including cell proliferation, apoptosis, migration, and autophagy [1, 2]. miRNAs usually act as gene expression posttranscriptional regulators by binding to the 3′ untranslated region (3′ UTR) of targets to inhibit their stability or translational efficiency [3, 4]. Recently, evidence indicates that miRNAs act as a bridge to mediate crosstalk among different types of RNAs in human cells, including long noncoding RNA, circular RNAs, mRNAs, and pseudogene. For example, Karreth et al. [5] reported that miR-653, miR-134, and miR543 mediated the effect of BRAFP1 on BRAF expression and MAPK activation. Li et al. [6] also found that miR-7, miR-17, and miR-214 could mediate the regulation of circ-ITCH in inhibiting esophageal squamous cell carcinoma development by suppressing the Wnt/β-catenin pathway. Therefore, exploring the functions of miRNAs could provide novel insights to identify the mechanisms underlying disease progression.
Prostate cancer (PCa) is one of the most prevalent type of cancer among men worldwide. Levels of prostate-specific antigen (PSA) is the most widely used biomarker for the prognosis of PCa. However, PSA testing has low specificity for PCa [7, 8, 9]. Therefore, there is an urgent need to identify novel diagnostic and prognostic markers for PCa to improve the treatment of the disease. Important roles for miRNAs have been implicated in PCa. A series of miRNAs, including miR-135a [10], miR-27a [11], miR-141 [12], and miR-424 [13] were found to be dysregulated in PCa. In our previous analysis, we observed that miR-192 was overexpressed in PCa. Several studies have demonstrated that miR-192 is involved in the progression of human cancer, including bladder cancer [14], acute lymphoblastic leukemia [15], and hepatocellular carcinoma [16]. However, the roles of miR-192 in tumor progression remain uncertain, and miR-192 could act as an oncogene. In hepatocellular carcinoma, miR-192 promotes the proliferation and metastasis of cancer cells by targeting SEMA3A [17]. miR-192 also promotes proliferation in esophageal squamous cell carcinoma by targeting Bim [18]. miR-192 could also serve as a tumor suppressor. For example, miR-192 suppressed the growth of bladder cancer cells by targeting cyclin D1, p21, p27, Bcl-2, Bax, and Mcl-1 [14]. However, the functional roles of miR-192 in PCa remains unclear.
In the present study, we analyzed the pattern of expression of miR-192 in PCa samples by using public datasets. Furthermore, we performed bioinformatic analysis and experimental validation to reveal the potential mechanisms of miR-192 in PCa. The purpose of this study is to ascertain whether miR-192 could serve as a biomarker for PCa.
Materials and Methods
miRNA and mRNA Profile Data Collection
miRNA profile data were collected from the GEO database (www.ncbi.nlm.nih.gov/gds, GSE21036). A total of 28 normal samples, 99 primary PCa samples, and 14 metastasis PCa samples were included in GSE21036 [19]. GSE21036 included 58 Gleason 6 PCa samples, 39 Gleason 7 PCa samples, and 14 Gleason 8 and 9 PCa samples. The comparison of miRNA profiles between PCa samples and normal tissues were performed with the Limma package on the R platform using raw microarray data [20].
The expression data of miR-192 in PCa samples were downloaded from The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/tcga/) database. A total of 52 normal prostate samples and 495 PCa samples were included in TCGA datasets. The clinical features of the patients, including age at diagnosis, days to last follow-up, pathological tumor (T) stage and node (N) stage were retrospectively obtained from the patient records. The cut-off for significant differentially expressed miRNA was identified with t test methods by p < 0.05.
Cell Culture
We selected LNCaP, 22RV1, and PC-3 cells to explore the potential roles of miR-192. These cell lines are the most widely used cells in PCa research. LNCaP is an androgen-dependent PCa (ADPC) cell line, 22RV1 is weakly androgen dependent, and PC-3 is an androgen-independent PCa (AIPC) cell line without AR expression. LNCaP, 22RV1, and PC-3 cells were obtained from Shanghai Institutes for Biological Sciences Cell Resource Center. The three PCa cell lines were cultured in Ham's F12K media (Invitrogen, Beijing, China) supplemented with 10% fetal bovine serum.
Cell Transfection
Synthetic miR-192–5p mimic and its scrambled control miRNA (miR-NC) were purchased from Biotend (Shanghai, China). PCa cells were seeded into 6-well plates (Corning, New York, NY, USA) and then transfected using Lipofectamine 3000 reagent (Life, USA) and incubated overnight. Total RNAs were extracted 48 h after transfection.
RNA Extraction and Quantitative RT-PCR
Total RNA for RT-qPCR was extracted using miRNA Purification Kits (CWBIO, China). Reverse transcription was performed with an miRNA cDNA Synthesis Kit (CWBIO). miR-192–5p-specific reverse transcription primer was purchased from (HuaJin, China) SYBR Green Reagents (Bio-Rad, Hercules, CA, USA) was used to measure the microRNA expression. The expression level of miR-192–5p was normalized to U6. The PCR primers for mature miR-192–5p and U6 were designed and purchased from GenePharm. The relative miRNA expression fold change was determined as 2−ΔΔCt.
Cell Proliferation Assay
CCK-8 assays were performed to evaluate changes in cell viability. Cells were plated in 96-well plates in a final volume of 100 µL medium/well. Proliferation was detected at 0, 24, 48, 72, and 96 h. Cell viability was quantified by adding 10 µL of CCK-8 (Dojindo, Kumamoto, Japan) to each well. After incubation for 1.5 h at 37°C, the plates were monitored at specific time points using a PowerWave XS microplate reader (BioTek, Winooski, VT, USA), which measured absorbance at 450 nm. Absorbance at 630 nm was used as a reference.
Cell Cycle Assay
After the LNCaP, 22RV1, and PC-3 cells were transfected for 72 h, the cells were collected and incubated in 0.03% Triton X-100 and propidium iodide (50 ng/mL) for 15 min. Then the cells were detected by a fluorescence-activated cell sorting (FACS) flow cytometer (BD Biosciences, San Jose, CA, USA) and analyzed with ModFit software (Verity Software House, Topsham, ME, USA).
Gene Ontology and KEGG Pathway Analysis
The Molecule Annotation System (MAS, version 3.0), provided by CapitalBio Corporation (Beijing, China; bioinfo.capitalbio.com/mas3/) was used to determine the biological roles of differentially expressed mRNAs. Gene functions were classified into three subgroups: biological process, cellular component, and molecular function. The enriched gene ontology (GO) terms were presented by enrichment scores. KEGG pathway analysis was carried out to determine the involvement of differentially expressed mRNAs in different biological pathways. The recommended hypergeometric p value used as the cut-off was p < 0.05.
Statistical Analysis
The numerical data are presented as the mean ± standard deviation (SD) of at least three determinations. Statistical comparisons between groups of normalized data were performed using the t test or Mann-Whitney U test according to the test condition. Statistical comparisons among multiple groups of normalized data were performed using one-way analysis of variance followed by a Dunnett's post hoc test. p < 0.05 was considered statistically significant with a 95% confidence level.
Results
miR-192–5p Is Upregulated in PCa
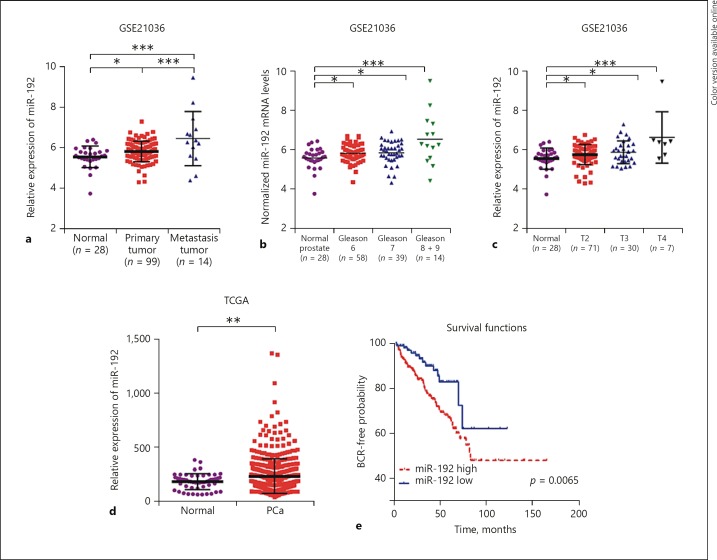
By analyzing the GSE21036 dataset, we found that miR-192-5p was significantly upregulated in PCa tissues compared to nontumor tissues (Fig. 1a–c) and was overexpressed in metastatic samples compared to primary tumor tissues (p < 0.001; Fig. 1a). Furthermore, we evaluated the expression levels of miR-192 with clinical varies. As shown in Figure 1c, we found miR-192 to be overexpressed in patients with T2 (p < 0.05), T3 (p < 0.05), and T4 (p < 0.001) PCa compared to healthy controls (Fig. 1c). Subsequently, the patients in GSE21036 were categorized based on Gleason grades; the results demonstrated that miR-192 expression levels in Gleason grade 6 (p < 0.05), 7 (p < 0.05), 8 and 9 (p < 0.001) PCa samples were higher than those in matched normal tissues (Fig. 1b).
Fig. 1.
miR-192–5p is upregulated in PCa. In the GSE21036 dataset, miR-192–5p was significantly upregulated in metastatic samples compared to primary tumor tissues (a), was upregulated in Gleason grades 6 (p < 0.05), 7 (p < 0.05), 8 and 9 (p < 0.001) PCa compared to normal tissues (b), and was overexpressed in patients with T2 (p < 0.05), T3 (p < 0.05), and T4 (p < 0.001) PCa compared to normal controls (c). In the TCGA dataset, miR-192 was significantly upregulated in PCa samples compared to matched normal samples (d, p < 0.01) and the BCR-free survival rates were higher in patients with low miR-192 (e). * p < 0.05; ** p < 0.01; *** p < 0.001.
In order to further validate our findings, we analyzed an independent dataset – TCGA. The results demonstrated that miR-192 was significantly upregulated in PCa samples compared to matched normal samples (Fig. 1d, p < 0.01). In order to detect whether miR-192 was specifically upregulated in PCa, we evaluated the expression pattern of miR-192 in several other types of human cancer, including kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), and liver hepatocellular carcinoma (LIHC) by analyzing TCGA datasets. Interestingly, we observed that in miR-192 expression was not altered in KIRC compared to normal samples (online suppl. Fig. 1A; for all online suppl. material, see www.karger.com/doi/10.1159/000496206), and was downregulated in KIRP (online suppl. Fig. 1B) and LIHC (online suppl. Fig. 1C).
The Kaplan-Meier method was also performed to further investigate the prognostic value of miR-192 in PCa. As demonstrated in Figure 1e, compared with patients with high miR-192 expression, the biochemical recurrence (BCR)-free survival rates were higher in patients with low miR-192 expression in this dataset. These results suggested the important roles of miR-192 in PCa.
Overexpression of miR-192–5p Promoted the Proliferation of PCa Cells
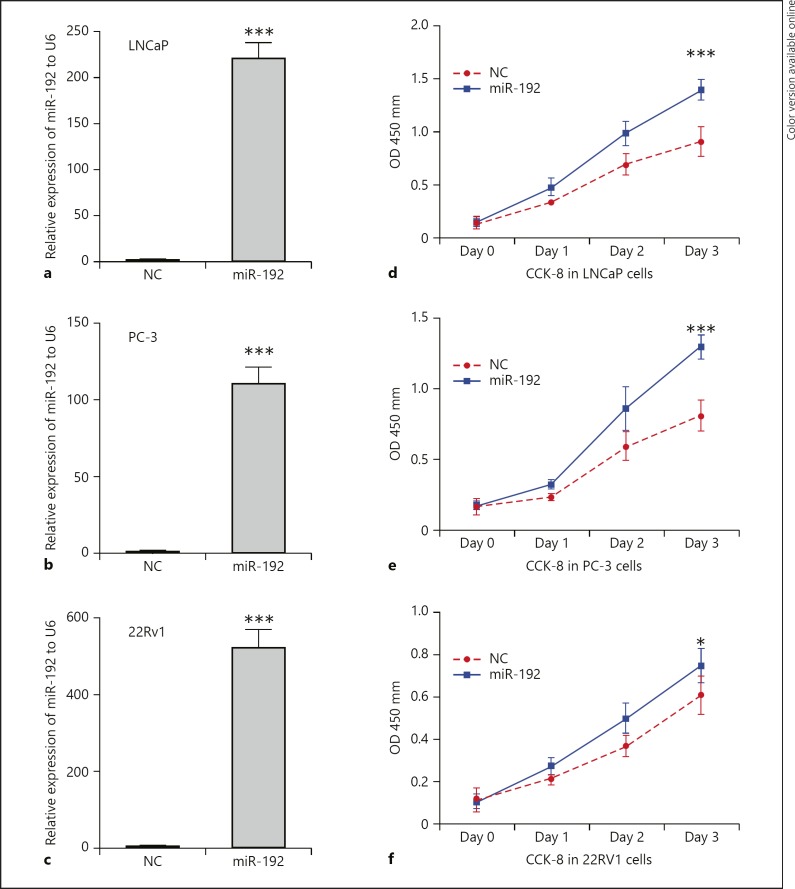
To explore the potential effect of miR-192–5p on the proliferation of PCa cells, we performed the CCK-8 assay by transfecting LNCaP, PC-3, and 22RV1 with NC or miR-192–5p mimics. As shown in Figure 2, we found that miR-192–5p significantly promoted the growth rate of LNCaP (p < 0.001), PC-3 (p < 0.001), and 22RV1(p < 0.05) cells. The transfection efficiency is shown in Figure 2a–c.
Fig. 2.
Overexpression of miR-192–5p promotes the proliferation of PCa cells. a–c The transfection efficiency. Overexpression of miR-192–5p significantly promotes the growth rate of LNCaP (d, p < 0.001), PC-3 (e, p < 0.001), and 22RV1 (f, p < 0.05) cells. Data are presented as the mean ± SD. * p < 0.05; *** p < 0.001.
Overexpression of miR-192–5p Promotes Cell Cycle Progression in vitro
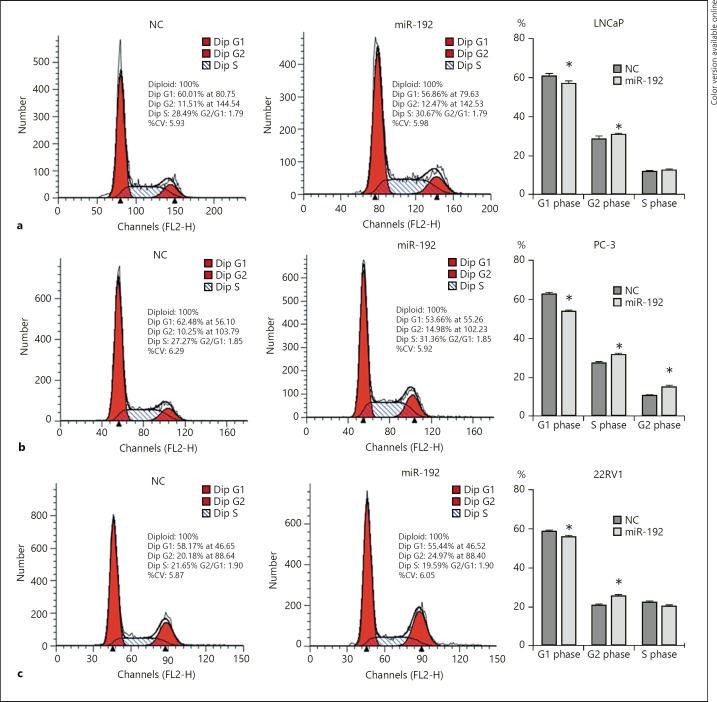
We then assessed the function of miR-192–5p on cell cycle profiles of LNCaP, PC-3, and 22RV1 cells. Flow cytometric analysis revealed that overexpression of miR-192–3p in LNCaP (Fig. 3a), PC-3 (Fig. 3b), and 22RV1 (Fig. 3c) cells resulted in a significant increase in the proportion of cells in the S phase and a decrease in the proportion of cells in the G1 phase. Taken together, these results suggested miR-192–5p inhibits proliferation of PCa cells by inducing G1‑phase cell cycle arrest.
Fig. 3.
Overexpression of miR-192–5p promotes cell cycle progression in vitro. Overexpression of miR-192–5p significantly induces an increase in the proportion of cells in the S phase and a decrease in the proportion of cells in the G1 phase in LNCaP (a), PC-3 (b), and 22RV1 (c) cells. Data are presented as the mean ± SD (n = 3). * p < 0.05.
Bioinformatic Analysis for miR-192–5p in PCa
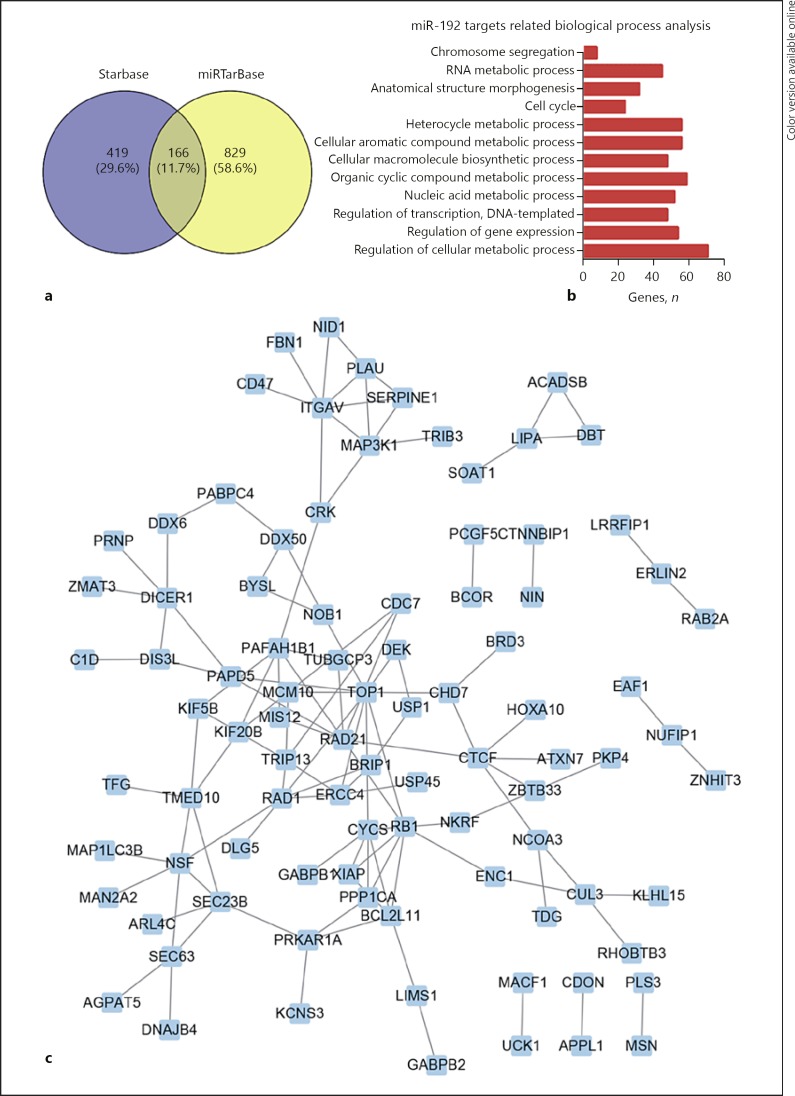
We performed bioinformatics analysis for miR-192–5p to explore its potential mechanisms in PCa by using its targets. The Starbase [21, 22] and miRTarBase datasets were used to identify validated targets of miR-192–5p (Fig. 4a). A total of 166 targets of miR-192–5p was used to perform the GO categories analysis. Our data revealed that miR-192–5p affects the regulation of the cellular metabolic process, regulation of gene expression, DNA-templated regulation of transcription, nucleic acid metabolic process, organic cyclic compound metabolic process, cellular macromolecule biosynthetic process, cellular aromatic compound metabolic process, heterocycle metabolic process, cell cycle, anatomical structure morphogenesis, RNA metabolic process, and chromosome segregation (Fig. 4b). Moreover, we constructed a protein-protein interaction network based on the miR-192 target protein (Fig. 4c).
Fig. 4.
Bioinformatic analysis of miR-192–5p in PCa. a The targets of miR-192–5p which were validated by the Starbase and miRTarBase datasets. b GO analysis of 166 targets of miR-192–5p. c The protein-protein interaction network involved in miR-192 targets.
Discussion
PCa is a leading cause of cancer-related deaths in men worldwide. The Gleason grade, PSA, and clinical stage are the most widely used pretreatment prognostic parameters for PCa patients. However, the prognosis of PCa patients is limited due to the limitation of these prognostic biomarkers [8, 23, 24]. Emerging studies have started to identify novel biomarkers for PCa and have found that noncoding RNAs are associated with PCa progression. Recent reports showed the prognostic value of noncoding RNAs in the diagnosis of PCa diagnosis. For example, PCA3 showed better performance than PSA in urinary detection of PCa [25, 26, 27]. In the present study, we found that miR-192 is significantly upregulated in PCa and associated with a shorter BCR-free survival time. We also analyzed the expression pattern of miR-192 in several types of human cancers, including KIRC, KIRP, and LIHC. Interestingly, we observed that miR-192 expression was not altered in KIRC compared to normal samples, and was downregulated in KIRP and LIHC. These results suggest that miR-192 is specifically upregulated in PCa.
In recent years, several studies have demonstrated that miR-192 is involved in the progression of human cancer. miR-192 could act as either an oncogene or a tumor suppressor in human cancers. For example, miR-192 promotes the proliferation of cancer cell in hepatocellular carcinoma [17] and esophageal squamous cell carcinoma [18] by inhibiting their targets. However, miR-192 inhibits cell proliferation of bladder cancer cells via targeting cyclin D1, p21, p27, Bcl-2, Bax, and Mcl-1 [14]. The functional roles of miR-192 in PCa remains unclear. In this study, we performed a gain of function assay to evaluate the roles of miR-192 in PCa. We found that overexpression of miR-192 significantly promotes cell proliferation by reducing G1‑phase cell cycle arrest. Our results strongly suggest that miR-192 may act as an oncogene in PCa.
Although several studies have shown that miR-192 could target YY1, CCNT2, and GATA6 in human cancers, the mechanism of miR-192 in regulating PCa progression remains unclear [28, 29]. In the present study, we performed bioinformatics analysis for miR-192–5p to explore its potential mechanisms in PCa by using its targets. Our data revealed that miR-192–5p affects several biological processes, including regulation of the cellular metabolic process, regulation of transcription, and cell cycling.
Our study suggests that miR-192–5p could serve as a novel biomarker for PCa. However, several limitations of this study should be noted. First, we did not know whether the increase of miR-192 in public datasets is reflected in PCa cells or in other cell types, including fibroblasts, vascular endothelial cells, and so on. In future studies, we should evaluate the expression pattern of miR-192 in PCa cells, fibroblasts, and vascular endothelial cells. We suggest that this will help determine whether miR-192 could serve as a specific target for PCa.
Conclusion
In this study, we found that miR-192 is overexpressed in PCa samples compared to normal samples. The analysis using the dataset of TCGA revealed that miR-192 is upregulated in high-grade PCa compared to low-grade PCa. We also observed that higher miR-192 expression is associated with a shorter BCR-free survival time. These results demonstrated that miR-192 may promote proliferation of PCa cells and cell cycle progression. The results suggest that miR-192 may be considered as a potential diagnostic marker and therapeutic target of PCa.
Statement of Ethics
No human participants and/or animals were included in the present study.
Disclosure Statement
The authors declare no conflicts of interest.
Supplementary Material
Supplementary data
References
- 1.Chiaretti S, Gianfelici V, Ceglie G, Foà R. Genomic characterization of acute leukemias. Med Princ Pract. 2014;23((6)):487–506. doi: 10.1159/000362793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Ahmad A, Kong D, Bao B, Sarkar FH. Targeting MicroRNAs for personalized cancer therapy. Med Princ Pract. 2013;22((5)):415–7. doi: 10.1159/000353562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002 Apr;30((4)):363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 4.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005 Dec;123((6)):1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 5.Tang J, Ning R, Zeng B, Li Y. Molecular evolution of PTEN pseudogenes in mammals. PLoS One. 2016 Dec;11((12)):e0167851. doi: 10.1371/journal.pone.0167851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017 Mar;8((2)):8. doi: 10.1002/wrna.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovac E, Elshafei A, Tay KJ, Mendez MH, Polascik T, Jones JS. 5-Year biochemical Progression-Free survival following salvage Whole-Gland prostate cryoablation: defining success with nadir PSA. J Endourol. 2016 doi: 10.1089/end.2015.0719. [DOI] [PubMed] [Google Scholar]
- 8.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012 Mar;4((127)):127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jalloh M, Cooperberg MR. Implementation of PSA-based active surveillance in prostate cancer. Biomarkers Med. 2014;8((5)):747–53. doi: 10.2217/bmm.14.5. [DOI] [PubMed] [Google Scholar]
- 10.Wan X, Pu H, Huang W, Yang S, Zhang Y, Kong Z, et al. Androgen-induced miR-135a acts as a tumor suppressor through downregulating RBAK and MMP11, and mediates resistance to androgen deprivation therapy. Oncotarget. 2016 Aug;7((32)):51284–300. doi: 10.18632/oncotarget.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo W, Zhang J, Li X, Meng D, Gao Y, Yang S, et al. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8((2)):e56592. doi: 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao J, Gong AY, Eischeid AN, Chen D, Deng C, Young CY, et al. miR-141 modulates androgen receptor transcriptional activity in human prostate cancer cells through targeting the small heterodimer partner protein. Prostate. 2012 Oct;72((14)):1514–22. doi: 10.1002/pros.22501. [DOI] [PubMed] [Google Scholar]
- 13.Oneyama C, Kito Y, Asai R, Ikeda J, Yoshida T, Okuzaki D, et al. MiR-424/503-mediated Rictor upregulation promotes tumor progression. PLoS One. 2013 Nov;8((11)):e80300. doi: 10.1371/journal.pone.0080300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y, Lu J, Wen J, Shen Y, Wen X. Regulation of growth of human bladder cancer by miR-192. Tumour Biol. 2015 May;36((5)):3791–7. doi: 10.1007/s13277-014-3020-8. [DOI] [PubMed] [Google Scholar]
- 15.Schotte D, Akbari Moqadam F, Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R, et al. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011 Sep;25((9)):1389–99. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- 16.Lian J, Jing Y, Dong Q, Huan L, Chen D, Bao C, et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016 Jan;7((3)):2672–83. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan-Chun L, Hong-Mei Y, Zhi-Hong C, Qing H, Yan-Hong Z, Ji-Fang W. MicroRNA-192-5p promote the proliferation and metastasis of hepatocellular carcinoma cell by targeting SEMA3A. Appl Immunohistochem Mol Morphol. 2017 Apr;25((4)):251–60. doi: 10.1097/PAI.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Li F, Niu R, Zhang H, Cui A, An W, et al. Mir-192 suppresses apoptosis and promotes proliferation in esophageal aquamous cell caicinoma by targeting Bim. Int J Clin Exp Pathol. 2015 Jul;8((7)):8048–56. [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010 Jul;18((1)):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008 Jul;24((13)):1556–8. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014 Jan;42((Database issue)):D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011 Jan;39((Database issue suppl_1):D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venderbos LD, Roobol MJ. PSA-based prostate cancer screening: the role of active surveillance and informed and shared decision making. Asian J Androl. 2011 Mar;13((2)):219–24. doi: 10.1038/aja.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy MJ. Tumor markers in clinical practice: a review focusing on common solid cancers. Med Princ Pract. 2013;22((1)):4–11. doi: 10.1159/000338393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hessels D, Klein GJ, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, Kiemeney LA, Witjes JA, Schalken JA. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8-15–15-16. doi: 10.1016/s0302-2838(03)00201-x. [DOI] [PubMed] [Google Scholar]
- 26.Deras IL, Aubin SM, Blase A, Day JR, Koo S, Partin AW, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008 Apr;179((4)):1587–92. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 27.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002 May;62((9)):2695–8. [PubMed] [Google Scholar]
- 28.Ke S, Li RC, Lu J, Meng FK, Feng YK, Fang MH. MicroRNA-192 regulates cell proliferation and cell cycle transition in acute myeloid leukemia via interaction with CCNT2. Int J Hematol. 2017 Aug;106((2)):258–65. doi: 10.1007/s12185-017-2232-2. [DOI] [PubMed] [Google Scholar]
- 29.Moore R, Ooi HK, Kang T, Bleris L, Ma L. MiR-192-Mediated positive feedback loop controls the robustness of Stress-Induced p53 oscillations in breast cancer cells. PLOS Comput Biol. 2015 Dec;11((12)):e1004653. doi: 10.1371/journal.pcbi.1004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data