Abstract
The many steps of gene expression, from the transcription of a gene to the production of its protein product, are well understood. Yet, transcriptional regulation has been the focal point for the study of gene expression during development. However, quantitative studies reveal that messenger RNA (mRNA) levels are not necessarily good predictors of the respective proteins’ levels in a cell. This discrepancy is, at least in part, the result of developmentally regulated, translational mechanisms that control the spatiotemporal regulation of gene expression. In this review, we focus on translational regulatory mechanisms mediating global transitions in gene expression: the shift from the maternal to the embryonic developmental program in the early embryo and the switch from the self-renewal of stem cells to differentiation in the adult.
The control of fate transitions is essential during development, the process by which an entire organism is built from a single totipotent cell, and for homeostasis in adults, in which stem cells sustain tissue integrity by contributing newly differentiated progeny cells. In both cases, changes in cellular fate rely on changes in gene expression, which instruct transitioning cells with new directives and provide the means to remodel cellular structure and to achieve a new identity. Not surprisingly, gene expression is regulated at practically every level during development and stem cell differentiation, from transcription and splicing of pre-messenger RNAs (mRNAs), through mRNA localization and decay, to translation and protein maturation, modification, and degradation.
From plants to mammals, accumulating evidence indicates that translational control plays critical roles during organismal development, tissue homeostasis, and tumorigenesis (Buszczak et al. 2014; Simsek and Barna 2017). Although global quantification of gene expression in different cell systems shows a good correlation between mRNA and protein levels, mRNA levels seem poor predictors for protein abundance during cell state transitions, particularly during early embryonic development (Peshkin et al. 2015; Liu et al. 2016). Yet, the study of spatiotemporal regulation of gene expression and its function in controlling fate transitions has been mostly limited to chromatin-based and transcriptional mechanisms. Among other reasons, this biased attention was facilitated by the advent of new technologies to systematically study changes in mRNA accumulation and chromatin modifications at the genome level. Indeed, until recently, the study of translational changes occurring in a given developmental transition was limited to small sets of individually tested candidate genes. With the advance of techniques able to tease apart translational changes from fluctuations in mRNA accumulation at the genome level and at nucleotide-resolution—such as ribosome profiling (Ingolia et al. 2009, 2018; Brar and Weissman 2015)—it is now possible to comprehensively visualize changes in translation during development. This has led to the realization of a much broader scale at which translation affects fate decisions and has considerably expanded our mechanistic understanding.
Historically, the most studied examples of translational control during development have been described at developmental periods during which transcriptional input is minimal, such as during oogenesis and early embryogenesis. In the absence of transcriptional activity, gene expression control is restricted to later regulatory steps, most notably translation, mRNA localization, and mRNA and protein decay. Indeed, a large amount of genetic, molecular, and biochemical evidence, mostly gathered from flies and frogs, has provided an impressive detailed picture of the multitude of posttranscriptional mechanisms driving development. More recently, genome-wide approaches revealed the widespread importance of certain regulatory mechanisms and allowed researchers to uncover new layers of regulation and to interrogate other important developmental and cell fate transitions, including stem cell differentiation and cancer.
Here, we review the knowledge gathered from studies conducted in animal models on how translational control plays essential roles during two critical developmental processes relying on cell fate transitions: embryonic development and adult stem cell differentiation. We argue that, as the processes of posttranscriptional regulation are closer to the final protein product than the process of mRNA transcription, this type of regulation may be preferred and advantageous during rapid cell fate transitions. Finally, we discuss how recent studies revealed new mechanisms controlling translation during development.
TRANSLATIONAL CONTROL DURING EMBRYO DEVELOPMENT
With little or no transcriptional input, the late stages of egg maturation and the first steps of embryo development rely exclusively on maternally provided gene product pools (mRNAs and proteins) to lay down the basic structures on which a new organism will be built. Far from being static, these pools rapidly evolve during the course of early development to both drive and adapt to transitioning states, creating a complex and synchronized ballet of dynamically changing mRNA and protein levels. At these developmental stages, sophisticated gene expression modules are in place to spatially and temporally regulate the expression of proteins required to control growth, establish the embryonic axes, and define new cellular fates (Fig. 1). The dependency on maternally provided pools ceases once the zygotic genome is activated and transcription is reestablished in a process known as the maternal-to-zygotic transition (MZT) (Fig. 1A) (Lee et al. 2014). Although the length of the transcriptional quiescence phase varies depending on the species, reflecting evolutionary and developmental strategies, its pervasiveness in animals and plants suggests a widespread requirement for posttranscriptional regulatory mechanisms during early embryo development (Baroux et al. 2008; Tadros and Lipshitz 2009).
Figure 1.
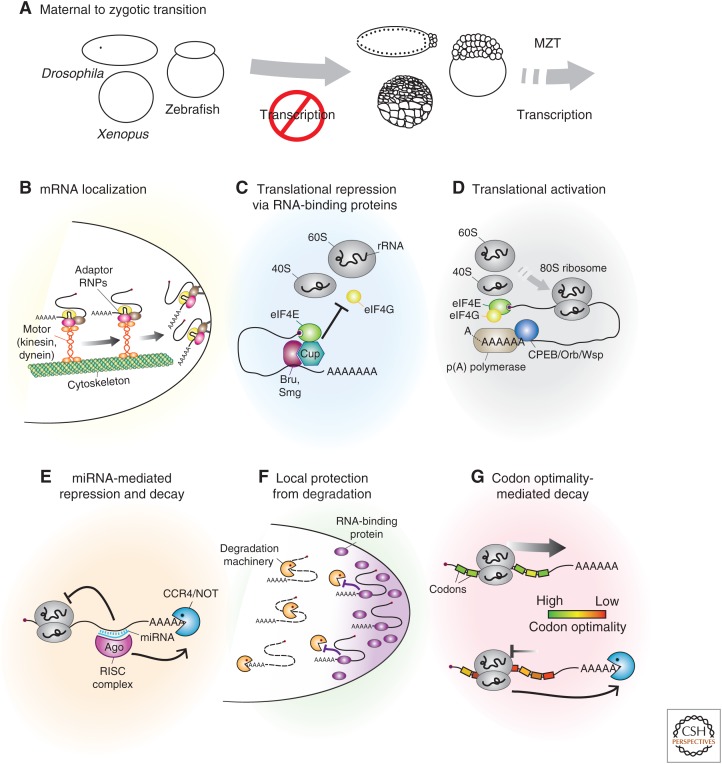
Mechanisms of translational control during embryo development. Examples of modules regulating protein synthesis during the first steps of embryo development, a period during which the zygotic genome is mostly transcriptionally quiescent. (A) Maternal-to-zygotic transition (MZT). (Left) Fertilized Drosophila (top), Xenopus (bottom), and zebrafish (right) embryos are transcriptionally silent and contain maternally synthesized gene products that sustain development and lay down the basic structures and embryonic axes. (Right) During the MZT, maternal products are largely degraded, and zygotic transcription ensues. This allows asynchrony in cell divisions and morphogenetic cell movements of gastrulation needed for body plan building. (B) Messenger RNA (mRNA) localization is mediated by RNA-binding proteins that recognize secondary structures present in the 3′ untranslated region (UTR) of target mRNAs. Assembled adaptor ribonucleoprotein complexes (RNPs) are recruited to molecular motors and delivered to specific intracellular domains. (C) Translational repression can be actively established on targets by RNA-binding proteins—such as Bruno (Bru) and Smaug (Smg)—that recognize sequences within the transcript 3′UTRs and recruit other factors to block translation initiation. (D) Translational activation is associated with the recruitment and activity of poly(A) [p(A)] polymerases and interactions between the poly(A)-binding protein and translation eukaryotic initiation factor (eIF)4G and eIF4E on target mRNA transcripts. (E) Base complementarity-targeted microRNA regulation is mediated by the RNA-induced silencing complex (RISC) and induces large-scale translational repression and CCR4/NOT-dependent target mRNA decay during the MZT. (F) Local protection of evenly distributed mRNAs against bulk degradation, which is mediated by localized RNA-binding proteins recognizing specific motifs in the 3′UTR of target mRNAs leads to spatially regulated expression at the subcellular level. (G) Slower ribosome translocation rates over transcripts containing suboptimal codons trigger mRNA deadenylation and decay during embryo development. rRNA, ribosomal RNA; miRNA, microRNA.
mRNA Localization and the Spatial Control of Translation
The most compelling and well-characterized examples of spatially regulated expression of proteins at the subcellular level were delineated by work focusing on embryonic axis specification in Drosophila. In this system, the anteroposterior and dorsoventral embryonic axes are predetermined during oogenesis by the targeted intracellular localization of mRNAs encoding key factors that are at the top of cascades establishing embryonic protein gradients. At first, an active and polarized microtubule-dependent mechanism (minus-end-directed), driven by dynein (Bullock and Ish-Horowicz 2001; Dienstbier et al. 2009), and dependent on Bicaudal-D and on the RNA-binding protein Egalitarian (Egl), delivers master-regulator mRNAs—namely, oskar (osk), bicoid (bcd), and gurken (grk)—from transcribing nurse cells to the transcriptionally quiescent oocyte. Once inside the oocyte, osk mRNA-containing particles are brought to the posterior side of the Drosophila oocyte through a kinesin-dependent mechanism (Brendza et al. 2000; Zimyanin et al. 2008; Lehmann 2016; Gaspar et al. 2017). Concurrently, bcd and grk mRNAs are actively delivered to the anterior and anterodorsal cortexes, respectively, through the action of dynein motors (plus-end directed) (Davidson et al. 2016; Trovisco et al. 2016). Localization specificity is mostly provided by sequences and secondary structures present in the 3′ untranslated region (UTR) of target mRNAs, which are in turn recognized by double-stranded RNA-binding proteins like Egl and Staufen (Stau) (Fig. 1B) (St Johnston et al. 1991, 1992; Micklem et al. 2000; Jambor et al. 2011, 2014; Le Bouteiller et al. 2013). mRNA–protein interactions required for transport are normally established in the cytoplasm, and form ribonucleoprotein complexes (RNPs) that may contain multiple mRNA copies per particle and are charged into the microtubule network (Jambor et al. 2011; Little et al. 2015; Trcek et al. 2015; Gaspar et al. 2017). Nevertheless, meaningful protein–mRNA interactions can equally well be established inside the transcribing nuclei (Chin and Lecuyer 2017). In the best-studied example, osk mRNA splicing and the interaction of osk mRNA with the exon junction complex (EJC) are critical for its final posterior oocyte localization (Hachet and Ephrussi 2004; Ghosh et al. 2012; Marchand et al. 2012). This indicates that at least in some cases, target mRNA recognition is initiated before nuclear export (Chin and Lecuyer 2017).
Regardless of the mechanism required for RNP assembly and recruitment to molecular motors, the general role for localized mRNA in determining embryo axial polarity seems to be widespread in animals. In fact, the importance of localized mRNA in defining cellular polarity goes far beyond the above-described example, as it has been implicated in the polarization of migrating cells, in the establishment of apicobasal structure, and in asymmetric cell divisions (reviewed in Medioni et al. 2012; Chin and Lecuyer 2017; Moor et al. 2017).
Very important for the successful establishment of a protein gradient, localized mRNAs have to be translationally silenced during transport. Frequently, translational silencing is actively established on targets by RNA-binding proteins that recognize sequences within the transcript UTRs. In contrast to mechanisms involved in recruitment of mRNAs to molecular motors, which seem to rely on secondary-structure RNA motifs, translational repression is regulated by sequence-specific RNA-binding proteins that directly interact with motifs in the 3′UTR of the target RNA (Fig. 1C). For instance, specific motifs in the osk 3′UTR (known as Bruno response elements [BREs]) are recognized by the RNA-binding protein Bruno (Bru) (Kim-Ha et al. 1995; Webster et al. 1997; Snee et al. 2008), which in turn is thought to recruit Cup. The Cup protein binds to the cap-binding protein eukaryotic initiation factor (eIF)4E and antagonizes eIF4E binding to eIF4G, thereby blocking osk translation (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; Zappavigna et al. 2004; Chekulaeva et al. 2006). The fact that Cup can interact with eIF4E in an RNA-independent manner further shows the key role of sequence-specific RNA-binding proteins such as Bru in specifying the set of translationally regulated mRNAs (Nakamura et al. 2004). Similarly, Bru binds to the grk 3′UTR and, together with another RNA-binding protein (Squid), is thought to repress grk translation during transport (Norvell et al. 1999; Saunders and Cohen 1999; Filardo and Ephrussi 2003; Caceres and Nilson 2009; Weil et al. 2012). In addition to repressive mechanisms relying on protein–protein interactions, it is nonetheless likely that translational repression during transport may also be accomplished by physical exclusion of the translation initiation apparatus, either as a result of the fact that target mRNAs are compacted into large RNPs (Chekulaeva et al. 2006) or because of the preferential enrichment of translational activators only at the final destination (Davidson et al. 2016).
Much less understood are the mechanisms by which localized mRNAs become translationally active. Generally, poly(A) polymerases are recruited to target mRNA transcripts during translational activation or derepression (Fig. 1D) (Castagnetti and Ephrussi 2003; Norvell et al. 2015). For instance, osk and grk translation is activated during oogenesis and requires the Oo18 RNA-binding protein (Orb, the homolog of the Xenopus cytoplasmic polyadenylation element-binding protein [CPEB]), a protein that is enriched inside the oocyte and that can directly associate with the poly(A) polymerases (Chang et al. 1999, 2001; Castagnetti and Ephrussi 2003; Weil et al. 2012; Norvell et al. 2015). Although grk and osk activation differ in many other mechanistic aspects, such as the fact that Orb directly recognizes sequences in the osk 3′UTR while the physical link between Orb and grk has not been reported, the poly(A) tails of both grk and osk mRNAs increase in length during activation. Indeed, translational activation and poly(A) polymerase activity are often correlated, although translation of maternally deposited mRNA per se does not necessarily require an increase in poly(A) tail length during early embryogenesis (Eichhorn et al. 2016). Such is the case of bcd mRNA, which is silenced during oogenesis and activated during embryogenesis. Global analysis of mRNA poly(A) traits in Drosophila oocytes and early embryos revealed that bcd mRNA translational activation is not associated with an increase in poly(A) tail length, suggesting that its temporal regulation is defined by the release of a repressive state rather than the promotion of translational activation and poly(A) polymerase activity (Eichhorn et al. 2016).
During oogenesis, regulated translation of localized osk mRNA at the posterior cortex of the forming egg nucleates a specialized cytoplasm—known as germ plasm—and is required for the entrapment of a set of mRNAs passively moving in the cytoplasmic stream created during oogenesis (Lehmann 2016). Among these is nos, a master determinant of posterior fate, as well as many other mRNAs required for the formation of germ cells at the embryonic posterior cortex, including pgc and gcl (Wang and Lehmann 1991; Gavis and Lehmann 1992, 1994; Little et al. 2015; Trcek et al. 2015). Posterior localization is determined by sequences and secondary structures present in their 3′UTRs, even though mechanistically the movement of mRNAs does not include their recruitment to molecular motors or the assembly of large RNPs (Crucs et al. 2000; Forrest et al. 2004; Rangan et al. 2009; Little et al. 2015; Trcek et al. 2015). Regardless of such differences, translation of germ plasm-enriched mRNAs is also tightly regulated, with most of them being repressed during oogenesis and individually activated during embryogenesis (Rangan et al. 2009). For instance, nos mRNA translation is repressed during oogenesis by Glorund and Smaug (Smg), two RNA-binding proteins that interact with the nos 3′UTR (Smibert et al. 1999; Kalifa et al. 2006; Tamayo et al. 2017). Similarly to Bru, Smg interacts with Cup, which then binds to eIF4E and inhibits nos translation initiation by precluding the eIF4E–eIF4G interaction and formation of the preinitiation complex at the 5′ end of nos mRNA (Nelson et al. 2004; Andrews et al. 2011).
Transcript-specific translational regulation may be achieved via association of mRNAs within large, membraneless RNP granules. For example, at the Drosophila oocyte anterior cortex, translationally active grk is preferentially distributed at the edges of RNP granules, although translationally repressed bcd is found both in the center and at the edge of the granule (Weil et al. 2012). This correlation is not observed for mRNAs localizing to the germ granules at the posterior pole. Here, multiple copies of a single RNA organize into homotypic RNA clusters, which occupy fixed locations with respect to the protein germ granule. This spatial organization, however, does not reflect translational activity (Trcek et al. 2015). In line with the importance of such posttranscriptional mechanisms for embryonic development, 71% of the 2314 maternally deposited mRNAs tested in Drosophila embryos were shown to be subcellularly localized (Lecuyer et al. 2007). Beyond embryos, accumulating evidence indicates that mRNA localization is more prevalent than expected and may regulate other aspects of cellular biology, including cell signaling, neuronal axon growth, and dendritic plasticity (reviewed in Chin and Lecuyer 2017).
Translational Efficiency and mRNA Decay
Thousands of maternally transcribed mRNAs are deposited into the transcriptionally quiescent oocyte during oogenesis and the vast majority of these mRNAs are found evenly distributed in the egg cytoplasm. To drive the first steps of embryonic development, their translation is temporally—and in some cases even spatially—regulated. For instance, prominent developmental transitions such as oocyte maturation and egg activation, as well as the activation of the zygotic genome during the MZT, are characterized by large-scale changes in gene expression (Fig. 1A) (reviewed in Laver et al. 2015; Yartseva and Giraldez 2015). Mechanistically, these changes are mostly achieved by the bulk regulation of translation efficiency and mRNA decay, although global changes coexist with the more targeted regulatory mechanisms described in the previous section.
During the stages involving oocyte maturation and egg activation in which meiotic divisions proceed toward completion, regulation of gene expression is mostly achieved at the translational level, with minimal fluctuation at the mRNA accumulation level (Su et al. 2007; Tadros et al. 2007; Thomsen et al. 2010; Chen et al. 2011; Kronja et al. 2014; Eichhorn et al. 2016). In Drosophila, ∼1350 out of ∼5800 mRNAs assessed by ribosome profiling were shown to be under translational control during egg activation, with 70% of those being translationally activated and 30% repressed (Kronja et al. 2014). Interestingly, most of the observed changes in translational efficiency depend on the activity of the serine/threonine kinase PAN GU (Tadros et al. 2007; Kronja et al. 2014), although the details of how this is achieved and which PAN GU substrates are involved in translational regulation are not yet fully understood. Given that translational changes during egg activation are mainly determined by changes in mRNA poly(A) tail length, which are dictated by the opposing activities of deadenylases and the poly(A) polymerase Wispy (Benoit et al. 2008; Cui et al. 2013; Subtelny et al. 2014), it is possible that PAN GU acts directly upstream of such modifying enzymes (Eichhorn et al. 2016).
In contrast to the prevalence of translational regulation during oocyte maturation and egg activation, MZT is largely accompanied by widespread changes in mRNA levels (affecting 30%–40% of maternal mRNAs) that are mainly driven by mRNA decay mechanisms (reviewed in Laver et al. 2015; Yartseva and Giraldez 2015). Maternal mRNA clearance starts before zygotic transcriptional activation but is intensified on its onset. Accumulating evidence indicates that mRNA destabilization is often preceded by translational inhibition, and that both types of regulation are mediated by the same machineries involving the deadenylation of target mRNAs (Fig. 1E). For instance, the RNA-binding protein Smaug mediates most of the maternal mRNA translational repression and decay before zygotic activation in Drosophila (Nelson et al. 2004; Tadros et al. 2007; Benoit et al. 2009; Chen et al. 2014). Indeed, PAN GU activates Smaug mRNA translation during egg activation (Tadros et al. 2007), with Smaug protein accumulating during early embryogenesis but decreasing soon after the MZT (Benoit et al. 2009). Smaug binds to secondary structure RNA motifs (Smaug recognition elements [SREs]) (Dahanukar et al. 1999; Smibert et al. 1999) within target mRNAs, and mediates either decay by recruiting the CCR4/NOT deadenylase complex or translational repression by interacting with Cup (Nelson et al. 2004; Laver et al. 2015; Eichhorn et al. 2016). However, it is still unclear what determines the recruitment of either downstream effector.
On genome activation, zygotically transcribed microRNAs play a crucial role in translational control and maternal mRNA clearance in many animal species (Fig. 1F). This is notably the case for miR-430 in zebrafish, miR-309 in Drosophila, and miR-427 in Xenopus, which are responsible for destabilization of hundreds of transcripts during early embryogenesis (Giraldez et al. 2006; Bushati et al. 2008; Lund et al. 2009). Mechanistically, microRNA specificity is determined by base complementarity with target mRNAs (Duchaine and Fabian 2018), but the mode of action can vary between translational repression and target mRNA cleavage and decay. Interestingly, careful analysis of miR-403-mediated regulation in zebrafish using genome-wide approaches revealed that targeting initially leads to translation initiation, which is then followed by mRNA deadenylation and decay (Bazzini et al. 2012).
In parallel to the regulation mediated by RNA-binding proteins and microRNAs, recent work shed light on a new mechanism by which the process of translation elongation influences mRNA stability (see Dever et al. 2018) and dictates mRNA decay during the MZT in Drosophila, zebrafish, Xenopus, and mouse (Presnyak et al. 2015; Bazzini et al. 2016; Mishima and Tomari 2016). Genome-wide approaches indicate that mRNAs encoding open reading frames (ORFs) enriched with uncommon, suboptimal codons decay faster than those enriched with optimal codons. “Codon optimality” is mostly defined by transfer RNA (tRNA) availability, and slower ribosome translocation rates over transcripts containing suboptimal codons trigger mRNA deadenylation by the CCR4/NOT complex, promoting decay (Fig. 1G). Interestingly, deadenylation activity is increased when suboptimal codons are placed closer to the stop codon and in mRNAs containing shorter 3′UTRs (Mishima and Tomari 2016), suggesting that physical proximity between the ribosome and the poly(A) tail may be required for the decay. The molecular mechanism by which ribosome elongation speed is interpreted and how this connects to the decay machinery remain to be determined. Nevertheless, analysis of mRNA decay kinetics during zebrafish embryo development indicates that the magnitude of codon composition-mediated mRNA decay regulation is comparable to that observed for microRNA regulation (Bazzini et al. 2016). In addition, a third mechanism, which can promote the degradation of transcripts enriched for the RNA modification N6-methyladenosine (m6A) (Wang et al. 2014; Peer et al. 2018), complements microRNA- and codon composition-mediated decay during the MZT in zebrafish embryos (Zhao et al. 2017).
Finally, translational regulation of evenly distributed mRNAs is not solely regulated in time but can also be differentially regulated in space. A notable example is provided by the uniformly distributed hunchback mRNA (hb) in Drosophila (Lehmann and Nusslein-Volhard 1987; Tautz 1988; Irish et al. 1989). Like thousands of other mRNAs, hb is maternally provided and only translated during early embryogenesis. hb translation is inhibited at the posterior and forms a protein gradient that is required during the MZT for regulating the expression of genes defining the anterior–posterior embryonic axis. hb translation repression is mediated by the sequence-specific binding of Pumilio (Pum) and its partners Nanos and Brain tumor (Brat) to the 3′UTR of hb mRNA (Irish et al. 1989; Wharton and Struhl 1991; Murata and Wharton 1995; Chagnovich and Lehmann 2001; Sonoda and Wharton 2001). Mechanistically, Nanos modulates Pum target specificity and binding to the Nanos response element (NRE) found at the 3′UTRs (Murata and Wharton 1995; Weidmann et al. 2016). In the case of hb mRNA, Nanos also recruits Brat, forming the NRE complex (Sonoda and Wharton 2001). Brat then interacts with the eIF4E-homologous protein (4EHP), which binds to the mRNA 5′ cap structure and negatively regulates translation by blocking eIF4E binding and formation of the preinitiation complex at the cap (Cho et al. 2006; Arvola et al. 2017). Spatially, this regulation is defined by Nanos protein distribution because of the posterior localization and local translation of nanos mRNA (Gavis and Lehmann 1992). Therefore, the spatial regulation of evenly distributed mRNAs may ultimately depend on an initial subcellular enrichment of translational repressors.
TRANSLATIONAL CONTROL DURING STEM CELL MAINTENANCE AND DIFFERENTIATION
Throughout development and adult life, precursor cells transit between quiescent, self-renewing, and differentiating states in a spatiotemporally coordinated fashion to promote embryonic development, tissue formation, and later to sustain organismal homeostasis. Although particularities exist in different stem cell lineages and developmental stages, transitions are fundamentally driven and accompanied by changes in gene expression, including translational control (Buszczak et al. 2014). For instance, many stem cell lineages present lower global protein synthesis rates in comparison to their immediate differentiating daughters. This was observed both in vitro (Sampath et al. 2008; Ingolia et al. 2011) and in stem cell systems in vivo, the latter of which includes Drosophila adult germline stem cells (Sanchez et al. 2016), mouse adult hematopoietic stem cells (Signer et al. 2014, 2016), mouse adult neural stem cells (NSCs) (Llorens-Bobadilla et al. 2015), mouse adult skeletal muscle stem cells (or satellite cells) (Zismanov et al. 2016), and mouse adult hair follicle stem cells (Fig. 2) (Blanco et al. 2016). In the studied cases, changes in protein synthesis rate during self-renewal and differentiation were mediated by regulatory mechanisms rather than being a consequence of changes in cell size, proliferation rate, or cell-cycle status (Sampath et al. 2008; Buszczak et al., 2014; Signer et al. 2014; Blanco et al. 2016; Sanchez et al. 2016).
Figure 2.
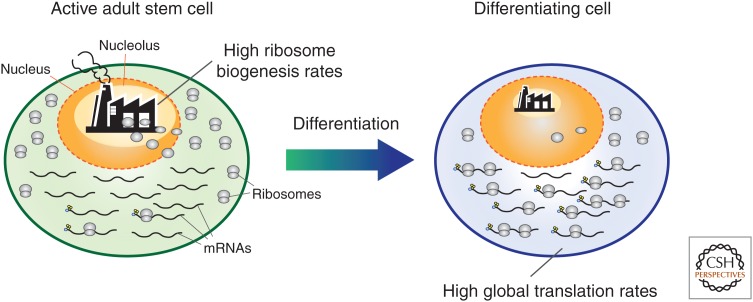
Translational control during stem cell maintenance and differentiation. Actively dividing stem cells (left) present lower global protein synthesis rates (fewer ribosomes attached to messenger RNAs [mRNAs]) in comparison to their immediate differentiating daughters (right). Yet, ribosomal RNA synthesis and ribosome biogenesis (shown by a black factory) are up-regulated compared with their differentiated progeny. This has been observed in several stem cells, such as Drosophila adult female germline and mouse embryonic stem cells, indicating that ribosome biogenesis and protein synthesis rates may be uncoupled in such stem cell systems.
Genetic experiments using mutations affecting translational control indicate that changes in protein synthesis rates are required and sufficient to modulate self-renewal and differentiation capacities in many stem cell systems. For instance, mutations inducing a global reduction in protein synthesis favor the accumulation of undifferentiated cells at the expense of differentiation. This is the case for Nsun2-deficient mouse hair follicle stem cells and male germline stem cells (Blanco et al. 2011; Hussain et al. 2013), for knockdowns of mechanistic target of rapamycin (mTOR) or transformation/transcription domain-associated protein (TRRAP)—both belonging to the phosphatidylinositol 3-kinase-related kinase (PIKK) family—in Drosophila female germline stem cells (Sanchez et al. 2016), and for mTOR-inhibited cultured mouse embryonic stem cells (Bulut-Karslioglu et al. 2016). Similarly, phosphorylation of eIF2α decreases protein synthesis rates and promotes mouse skeletal muscle stem cell self-renewal rather than differentiation (Zismanov et al. 2016). Muscle stem cells unable to phosphorylate eIF2α exit the quiescent state and differentiate. Furthermore, pharmacological treatment inhibiting eIF2α dephosphorylation promotes the regenerative capacity of muscle stem cells (Zismanov et al. 2016). Consistent with this, a genetically or chemically induced increase in protein synthesis, for instance by ectopically activating the mTOR pathway in stem cells, induces premature differentiation and stem cell depletion in many of the above-mentioned systems (Yilmaz et al. 2006; Zhang et al. 2006; Castilho et al. 2009; Sun et al. 2010; Bonaguidi et al. 2011; Deng et al. 2015). The central role for protein synthesis in this process is exemplified by the fact that a hypomorphic mutation affecting the ribosomal protein Rpl24 is sufficient to counterbalance the activation of the mTOR pathway in mouse hematopoietic stem cells, restoring normal self-renewal and differentiation functions (Signer et al. 2014). Yet, strong translation abrogation by means of affecting ribosome biogenesis can lead to stem cell loss as well as anomalous differentiation (Fichelson et al. 2009; LaFever et al. 2010; Le Bouteiller et al. 2013; Zhang et al. 2014; Sanchez et al. 2016). This shows how fine-tuning mechanisms, rather than drastic changes in protein synthesis, balance self-renewal and differentiation functions, possibly in a lineage-specific manner (Buszczak et al. 2014).
The mechanisms by which the protein synthesis rate is limited in wild-type stem cells are still poorly understood, but current evidence suggests that it may be achieved by different means depending on the specific stem cell lineage. For instance, translation inhibition in mouse adult hematopoietic stem cells and in mouse embryonic stem cells is at least partially mediated by the regulation of the eukaryotic initiation factor 4E-binding proteins 4E-BP1 and 4E-BP2 (Signer et al. 2016; Tahmasebi et al. 2016). In these systems, 4E-BP proteins are preferentially hypophosphorylated; this favors 4E-BP binding to the eIF4E, leading to inhibition of translation initiation. As a consequence, the translation of certain subsets of target mRNAs becomes more strongly inhibited (Roux and Topisirovic 2012; Morita et al. 2013; Proud 2018). In mouse skeletal muscle stem cells, on the other hand, high levels of eIF2α phosphorylation, another fulcrum of translation initiation regulation (Hinnebusch and Lorsch 2012; Merrick and Pavitt 2018; Wek 2018), are responsible for limiting protein synthesis (Zismanov et al. 2016). In contrast to these systems, mouse NSCs seem to use ribosome biogenesis regulation rather than translation initiation to modulate protein synthesis; here, stem cells express lower levels of ribosomal subunits in comparison to their immediate daughters (Llorens-Bobadilla et al. 2015). This type of regulation may not be generally used in other stem cell systems, however. For instance, ribosomal RNA synthesis and ribosome biogenesis are up-regulated in Drosophila adult germline stem cells compared with their differentiated progeny; yet, stem cells show lower protein synthesis rates (Fig. 2) (Neumuller et al. 2008; Zhang et al. 2014; Sanchez et al. 2016; reviewed in Brombin et al. 2015). It remains to be determined how ribosome biogenesis and protein synthesis rates are uncoupled in some stem cell systems. Recurrent observations in several independent systems suggest that active, yet unknown processes limit protein synthesis specifically in stem cells (such as the above-mentioned regulation of translation initiation) and/or conversely boost it in differentiating progeny. Given that differentiation relies on higher protein synthesis rates, it has been proposed that a surfeit of ribosome biogenesis in stem cells may prime daughter cells with enough ribosomes required for efficient differentiation (Ingolia et al. 2011; Sanchez et al. 2016).
In parallel to the global regulation of the translation machinery, specific translational regulators drive fate transitions during adult stem cell self-renewal and differentiation, with many examples being described in the Drosophila female germline stem cell system. In this system, a series of RNA-binding proteins initially identified through genetic screens delineates a regulatory cascade that is both required and sufficient for self-renewal and differentiation (reviewed in Slaidina and Lehmann 2014). These include conserved translational repressors, such as Nanos and Pum (Forbes and Lehmann 1998; Tsuda et al. 2003; Miller and Olivas 2011), as well as the DExH-box RNA helicase Benign gonial cell neoplasm (Bgcn) (Bailey et al. 2017), the TRIM-NHL tumor suppressor proteins Mei-P26 and Brat (Neumuller et al. 2008; Harris et al. 2011; Insco et al. 2012), and the cytoplasmic RNA-binding Fox 1 (Rbfox1) (Carreira-Rosario et al. 2016). It is important to note that, in addition to its function as translational repressor, Mei-P26 also regulates translation by directly interacting with Ago1 and inhibiting the microRNA pathway (Neumuller et al. 2008).
By regulating each other’s expression through mutual translational repression, the translational repressor factors establish a self-limiting regulatory network that promotes precise fate transitions during female germline stem cell differentiation. Similar to the mechanisms described in embryos, translational silencing during germline stem cell self-renewal and differentiation is generally mediated by repressor complexes that bind to sequences within the target mRNA 3′UTRs. In germline stem cells, for instance, translational repressors Nanos and Pum cooperate to promote self-renewal by inhibiting the translation of differentiation factors including Brat and Mei-P26, likely through the recruitment of the CCR4/NOT decay machinery to target mRNAs (Harris et al. 2011; Joly et al. 2013). In differentiating daughter cells, the expression of the differentiation factor Bag-of-marbles (Bam) leads to the assembly of a complex containing the sex determination factor Sex-lethal (Sxl) and translational repressors Bgcn and Mei-P26 (Li et al. 2009, 2013; Chau et al. 2012). Sxl and Mei-P26 are able to interact with the Nanos mRNA 3′UTR and repress its translation in differentiating daughter cells (Chau et al. 2012; Li et al. 2012, 2013), although the mechanism by which this repression is mediated remains to be determined. In the absence of Nanos, Pum is free to interact with Brat, and the Pum–Brat complex inhibits the translation of other self-renewal factors in differentiating cells, including transcription factors Myc and Mad (Harris et al. 2011). Interestingly, in more advanced differentiation stages, cytoplasmic Rbfox1 is expressed and binds to a sequence-specific motif in Pum mRNA 3′UTR, thereby repressing Pum translation and ensuring the progression to terminally differentiated states (Tastan et al. 2010; Carreira-Rosario et al. 2016).
Finally, it has recently been proposed that changes in tRNA concentrations may account for some of the gene expression differences observed in proliferating and differentiating cells (Gingold et al. 2014). Given that codon optimality is likely to be determined by tRNA availability (Presnyak et al. 2015; Bazzini et al. 2016), it is to be expected that in addition to a direct effect on translation (Gingold et al. 2014), the observed variation in tRNA abundance in these two states may also induce changes in mRNA stability like those observed during embryogenesis (Bazzini et al. 2016). Although it remains to be determined whether tRNA repertoires vary during stem cell fate transitions, it has recently been shown that changes in the expression of specific tRNAs modulate protein expression and metastatic activity during cancer progression through both translational control and mRNA decay (Goodarzi et al. 2016).
CONCLUDING REMARKS
Regulation at the posttranscriptional level is increasingly recognized as an important enabler of cell fate transitions in a variety of developmental and stem cell systems. At a global level, regulation of translational efficiency and RNA or protein turnover may be used to facilitate transitions between cell states by relinquishing entire gene programs to make way for synthesis of a new program. It may not be surprising that these posttranscriptional mechanisms are found so prominently at transitions that require gross turnover, like the transition from the maternally provided gene products of the egg to new gene synthesis by the embryo or during the transition from long-term, self-renewing stem cells to differentiating progeny. While broad, these transitions do not affect all mRNAs and our understanding of the mechanisms of regulation and target selection is still limited. Although this review has focused on 3′UTR-mediated regulation during the early stages of embryogenesis and global regulation at the stem cell–differentiation transition, additional mechanisms mediate translational efficiency via 5′ upstream sequences. These include control via RNA secondary structures, internal ribosome entry sites, upstream ORFs, and specialized ribosomes (Hinnebusch 2005; Chew et al. 2016; Johnstone et al. 2016; Shi et al. 2017; Leppek et al. 2018).
Although the intricacies of transcriptional regulation have long been recognized to modulate gene expression, posttranscriptional mechanisms were thought of as “housekeeping” and permissive. Recent discoveries and technologies challenge this perception and promise opportunities for continued discoveries of fundamental significance. As developmental biology moves toward single-cell level analysis, one of the challenges is the development of technologies to globally assess changes in translational control in individual cells, thereby complementing the recently established techniques (such as single-cell RNA-sequencing) that measure mRNA accumulation levels at single-cell resolution.
ACKNOWLEDGMENTS
We apologize to our colleagues whose work was not cited due to space limitations. F.K.T. is a Wellcome Trust and Royal Society Sir Henry Dale Fellow (206257/Z/17/Z). R.L. is supported by National Institutes of Health (NIH) R37HD41900 and is a Howard Hughes Medical Institute (HHMI) Investigator.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference in also in this collection.
- Andrews S, Snowflack DR, Clark IE, Gavis ER. 2011. Multiple mechanisms collaborate to repress nanos translation in the Drosophila ovary and embryo. RNA 17: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvola RM, Weidmann CA, Tanaka Hall TM, Goldstrohm AC. 2017. Combinatorial control of messenger RNAs by Pumilio, Nanos and brain tumor proteins. RNA Biol 14: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, Fuller MT. 2017. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6: e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Autran D, Gillmor CS, Grimanelli D, Grossniklaus U. 2008. The maternal to zygotic transition in animals and plants. Cold Spring Harb Symp Quant Biol 73: 89–100. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35: 2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. 2009. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 136: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. 2011. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 7: e1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. 2016. Stem cell function and stress response are controlled by protein synthesis. Nature 534: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. 2011. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145: 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Weissman JS. 2015. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM. 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289: 2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombin A, Joly JS, Jamen F. 2015. New tricks for an old dog: Ribosome biogenesis contributes to stem cell homeostasis. Curr Opin Genet Dev 34: 61–70. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Ish-Horowicz D. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414: 611–616. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Biechele S, Jin H, Macrae TA, Hejna M, Gertsenstein M, Song JS, Ramalho-Santos M. 2016. Inhibition of mTOR induces a paused pluripotent state. Nature 540:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. 2008. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol 18: 501–506. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Signer RA, Morrison SJ. 2014. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159: 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L, Nilson LA. 2009. Translational repression of gurken mRNA in the Drosophila oocyte requires the hnRNP Squid in the nurse cells. Dev Biol 326: 327–334. [DOI] [PubMed] [Google Scholar]
- Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, Buszczak M. 2016. Repression of Pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev Cell 36: 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A. 2003. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development 130: 835–843. [DOI] [PubMed] [Google Scholar]
- Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. 2009. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 5: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnovich D, Lehmann R. 2001. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci 98: 11359–11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Tan L, Schedl P. 1999. The Drosophila CPEB homolog, Orb, is required for oskar protein expression in oocytes. Dev Biol 215: 91–106. [DOI] [PubMed] [Google Scholar]
- Chang JS, Tan L, Wolf MR, Schedl P. 2001. Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 128: 3169–3177. [DOI] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. 2012. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci 109: 9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533. [DOI] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. 2011. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 25: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Dumelie JG, Li X, Cheng MH, Yang Z, Laver JD, Siddiqui NU, Westwood JT, Morris Q, Lipshitz HD, et al. 2014. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol 15: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF. 2016. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun 7: 11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A, Lecuyer E. 2017. RNA localization: Making its way to the center stage. Biochim Biophys Acta 1861: 2956–2970. [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. 2006. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol 16: 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crucs S, Chatterjee S, Gavis ER. 2000. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol Cell 5: 457–467. [DOI] [PubMed] [Google Scholar]
- Cui J, Sartain CV, Pleiss JA, Wolfner MF. 2013. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev Biol 383: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. 1999. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell 4: 209–218. [DOI] [PubMed] [Google Scholar]
- Davidson A, Parton RM, Rabouille C, Weil TT, Davis I. 2016. Localized translation of gurken/TGF-α mRNA during axis specification is controlled by access to Orb/CPEB on processing bodies. Cell Rep 14: 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Lei X, Zhang X, Zhang H, Liu S, Chen Q, Hu H, Wang X, Ning L, Cao Y, et al. 2015. mTOR signaling promotes stem cell activation via counterbalancing BMP-mediated suppression during hair regeneration. J Mol Cell Biol 7: 62–72. [DOI] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstbier M, Boehl F, Li X, Bullock SL. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 23: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Duchaine TF, Fabian MR. 2018. Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. 2016. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 5: e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR. 2009. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol 11: 685–693. [DOI] [PubMed] [Google Scholar]
- Filardo P, Ephrussi A. 2003. Bruno regulates gurken during Drosophila oogenesis. Mech Dev 120: 289–297. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125: 679–690. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Clark IE, Jain RA, Gavis ER. 2004. Temporal complexity within a translational control element in the nanos mRNA. Development 131: 5849–5857. [DOI] [PubMed] [Google Scholar]
- Gaspar I, Sysoev V, Komissarov A, Ephrussi A. 2017. An RNA-binding atypical tropomyosin recruits kinesin-1 dynamically to oskar mRNPs. EMBO J 36: 319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R. 1992. Localization of nanos RNA controls embryonic polarity. Cell 71: 301–313. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R. 1994. Translational regulation of nanos by RNA localization. Nature 369: 315–318. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gaspar I, Ephrussi A. 2012. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol 19: 441–449. [DOI] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. 2016. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165: 1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963. [DOI] [PubMed] [Google Scholar]
- Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. 2011. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell 20: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb Perspect Biol 4: a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M. 2013. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 33: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, Tam CH, Fuller MT. 2012. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell 11: 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. 1989. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338: 646–648. [DOI] [PubMed] [Google Scholar]
- Jambor H, Brunel C, Ephrussi A. 2011. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17: 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Mueller S, Bullock SL, Ephrussi A. 2014. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA 20: 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. 2016. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35: 706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly W, Chartier A, Rojas-Rios P, Busseau I, Simonelig M. 2013. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep 1: 411–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. 2006. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell 10: 291–301. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. [DOI] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr-Weaver TL. 2014. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep 7: 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L, Feoktistov A, Hsu HJ, Drummond-Barbosa D. 2010. Specific roles of target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development 137: 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver JD, Marsolais AJ, Smibert CA, Lipshitz HD. 2015. Regulation and function of maternal gene products during the maternal-to-zygotic transition in Drosophila. Curr Top Dev Biol 113: 43–84. [DOI] [PubMed] [Google Scholar]
- Le Bouteiller M, Souilhol C, Beck-Cormier S, Stedman A, Burlen-Defranoux O, Vandormael-Pournin S, Bernex F, Cumano A, Cohen-Tannoudji M. 2013. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J Exp Med 210: 2351–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. 2016. Germ plasm biogenesis—An Oskar-centric perspective. Curr Top Dev Biol 116: 679–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. 1987. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol 119: 402–417. [DOI] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M. 2018. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19: 158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. 2009. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci 106: 9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M. 2012. Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development 139: 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M. 2013. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS ONE 8: e58301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Sinsimer KS, Lee JJ, Wieschaus EF, Gavis ER. 2015. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat Cell Biol 17: 558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell 165: 535–550. [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. 2015. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17: 329–340. [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. 2009. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15: 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Gaspar I, Ephrussi A. 2012. An intracellular transmission control protocol: Assembly and transport of ribonucleoprotein complexes. Curr Opin Cell Biol 24: 202–210. [DOI] [PubMed] [Google Scholar]
- Medioni C, Mowry K, Besse F. 2012. Principles and roles of mRNA localization in animal development. Development 139: 3263–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR, Adams J, Grunert S, St Johnston D. 2000. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J 19: 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Olivas WM. 2011. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA 2: 471–492. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Tomari Y. 2016. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61: 874–885. [DOI] [PubMed] [Google Scholar]
- Moor AE, Golan M, Massasa EE, Lemze D, Weizman T, Shenhav R, Baydatch S, Mizrahi O, Winkler R, Golani O, et al. 2017. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 357: 1299–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18: 698–711. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 6: 69–78. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. 2004. Drosophila cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 23: 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schupbach T. 1999. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev 13: 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvell A, Wong J, Randolph K, Thompson L. 2015. Wispy and Orb cooperate in the cytoplasmic polyadenylation of localized gurken mRNA. Dev Dyn 244: 1276–1285. [DOI] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Wühr M, Pearl E, Haas W, Freeman RM Jr, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. 2015. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell 35: 383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG, Lehmann R. 2009. Temporal and spatial control of germ-plasm RNAs. Curr Biol 19: 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Topisirovic I. 2012. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 4: a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. 2008. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2: 448–460. [DOI] [PubMed] [Google Scholar]
- Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R. 2016. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18: 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C, Cohen RS. 1999. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell 3: 43–54. [DOI] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, Barna M. 2017. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83.e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Qi L, Zhao Z, Thompson D, Sigova AA, Fan ZP, DeMartino GN, Young RA, Sonenberg N, Morrison SJ. 2016. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev 30: 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Barna M. 2017. An emerging role for the ribosome as a nexus for post-translational modifications. Curr Opin Cell Biol 45: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaidina M, Lehmann R. 2014. Translational control in germline stem cell development. J Cell Biol 207: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Lie YS, Shillinglaw W, Henzel WJ, Macdonald PM. 1999. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA 5: 1535–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee M, Benz D, Jen J, Macdonald PM. 2008. Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol 5: 1–9. [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. 2001. Drosophila brain tumor is a translational repressor. Genes Dev 15: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. 1991. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66: 51–63. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG, Jantsch M. 1992. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci 89: 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. 2007. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol 302: 104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Quan Z, Zhang B, Wu T, Xi R. 2010. TSC1/2 tumour suppressor complex maintains Drosophila germline stem cells by preventing differentiation. Development 137: 2461–2469. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: A play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos-Pournatzis T, Matta-Camacho E, Tsukumo Y, Yanagiya A, Li W, Atlasi Y, Caron M, et al. 2016. Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc Natl Acad Sci 113: 12360–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo JV, Teramoto T, Chatterjee S, Hall TMT, Gavis ER. 2017. The Drosophila hnRNP F/H homolog Glorund uses two distinct RNA-binding modes to diversify target recognition. Cell Rep 19: 150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastan OY, Maines JZ, Li Y, McKearin DM, Buszczak M. 2010. Drosophila ataxin 2-binding protein 1 marks an intermediate step in the molecular differentiation of female germline cysts. Development 137: 3167–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. 1988. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature 332: 281–284. [DOI] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. 2010. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol 11: R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. 2015. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun 6: 7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovisco V, Belaya K, Nashchekin D, Irion U, Sirinakis G, Butler R, Lee JJ, Gavis ER, St Johnston D. 2016. bicoid mRNA localises to the Drosophila oocyte anterior by random Dynein-mediated transport and anchoring. eLife 5: e17537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. 2003. Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241. [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R. 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66: 637–647. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. 1997. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev 11: 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Qiu C, Arvola RM, Lou TF, Killingsworth J, Campbell ZT, Tanaka Hall TM, Goldstrohm AC. 2016. Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. eLife 5: e17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al. 2012. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 14: 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. 1991. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67: 955–967. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. 2003. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol 163: 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yartseva V, Giraldez AJ. 2015. The maternal-to-zygotic transition during vertebrate development: A model for reprogramming. Curr Top Dev Biol 113: 191–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441: 475–482. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC. 2004. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Natl Acad Sci 101: 14800–14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441: 518–522. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shalaby NA, Buszczak M. 2014. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. 2017. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542: 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, Crist C. 2016. Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18: 79–90. [DOI] [PubMed] [Google Scholar]