Abstract
Cells under stress must adjust their physiology, metabolism, and architecture to adapt to the new conditions. Most importantly, they must down-regulate general gene expression, but at the same time induce synthesis of stress-protective factors, such as molecular chaperones. Here, we investigate how the process of phase separation is used by cells to ensure adaptation to stress. We summarize recent findings and propose that the solubility of important translation factors is specifically affected by changes in physical–chemical parameters such temperature or pH and modulated by intrinsically disordered prion-like domains. These stress-triggered changes in protein solubility induce phase separation into condensates that regulate the activity of the translation factors and promote cellular fitness. Prion-like domains play important roles in this process as environmentally regulated stress sensors and modifier sequences that determine protein solubility and phase behavior. We propose that protein phase separation is an evolutionary conserved feature of proteins that cells harness to regulate adaptive stress responses and ensure survival in extreme environmental conditions.
Organisms exposed to stressful conditions, such as temperature variations, changes in nutrient availability, injuries or infections, must sense these changes and mount stress-specific defense mechanisms. But how do organisms adapt to changes in their environment and protect their molecular constituents from damage?
One cellular component that is very sensitive to changing physical–chemical conditions are proteins. Proteins constitute more than one-half of the dry mass of a cell and they exert numerous functions in metabolism, signaling, and cell architecture. Proteins are synthesized by the ribosome as linear polymers from activated amino acids. While being synthesized, the nascent polypeptide emerges from the ribosomal exit tunnel and begins its complex folding process, often with the assistance of molecular chaperones (also see Deuerling et al. 2018). The folding pathway of a given polypeptide is determined by numerous precise noncovalent interactions between specific sets of amino acids in its primary sequence (Daggett and Fersht 2003; Balchin et al. 2016). As a result, every polypeptide sequence adopts a unique structure and a unique shape. Once folding is completed, the folded polypeptide has acquired its biologically active form. Maintenance of the so-called native state is essential for protein functionality and loss of the native state usually results in a loss of activity.
Under conditions of stress, many proteins are no longer able to attain their native structure, a process known as protein misfolding. When proteins unfold, they gain conformational flexibility and expose hydrophobic surfaces and unstructured regions, which render them insoluble and prone to undergo intermolecular interactions. Consequently, they may aggregate and form insoluble precipitates (also see Hartl 2018). Unlike protein assembly or oligomerization, which are specific associations of polypeptide chains, aggregation describes the nonspecific association of misfolded polypeptides.
Because protein aggregation is a collective physical process, it can be described as a phase transition. A phase transition is a conversion of one state of matter into another. In the case of protein aggregation, it is the transition from a soluble to an assembled state with solid-like properties. This solid-like state may be a crystal, as in the case of amyloid fibrils, or a gel or glass, as in the case of amorphous aggregates. The most important difference between these solid-like states is the degree of molecular order. Amyloid fibrils are highly ordered because their polypeptide chains are arranged into intermolecular β-sheets that are oriented perpendicular to a fibril axis; amorphous aggregates are more structurally disordered, and their formation may or may not involve intermolecular β-sheets.
Protein aggregation is a form of protein damage and it is generally considered to be detrimental. Indeed, protein aggregation often coincides with loss of protein activity and protein aggregates are frequently associated with age-related diseases. One example of such a disease is amyotrophic lateral sclerosis (ALS). In ALS, specific proteins misfold and aggregate with increasing age in specific cells and tissues; in the case of ALS, aggregates accumulate in motor neurons (Taylor et al. 2016; Alberti et al. 2017). Such pathological or aberrant phase transitions are often irreversible.
Given the limited stability of proteins and the complexity of protein folding within cells, it seems surprising that cells are usually free of aggregates. Extensive research over the past decades has uncovered a comprehensive protein homeostasis network of molecular chaperones and protein degradation machines that surveys the folding state of the proteome (also see Hegde and Zavodszky 2018). These machines maintain the folding state by assisting in de novo protein folding, as well as by inhibiting nonproductive interactions among proteins and their subsequent aggregation (Balchin et al. 2016). Some of these machines can even revert the detrimental effects of protein aggregation (Sanchez and Lindquist 1990; Glover and Lindquist 1998; Motohashi et al. 1999; Shorter 2011; Rampelt et al. 2012; also see Shorter and Southworth 2018). Because molecular chaperones increase the folding efficiency and play essential roles in proteome homeostasis, synthesis of molecular chaperones is one of the most important cellular defense mechanisms against protein misfolding and aggregation.
But are all assemblies of high-molecular weight aggregates? Increasing evidence suggests that many assemblies within cells are not protein aggregates in the classical sense but are rather disordered multimeric states that serve physiological functions. This has been most clearly shown for intracellular compartments that lack surrounding membranes, such as the centrosomes (Woodruff et al. 2017) or the nucleolus (Feric et al. 2016). Such membrane-less compartments form by the process of liquid–liquid phase separation, in which an initially homogeneous solution of proteins demixes into a protein-rich phase that stably coexists with a dilute protein phase (Banani et al. 2017; Shin and Brangwynne 2017). The dense and the dilute phase continuously exchange molecules while maintaining the steep concentration gradient across the phase boundary. The dense phase often matures into a more solid-like state with the material properties of gels or glasses (Patel et al. 2015; Alberti and Hyman 2016; Alberti 2017). Assemblies that form by phase separation inside of cells were termed biomolecular condensates (Banani et al. 2017; Shin and Brangwynne 2017) and, unlike protein aggregates, appear to be tightly linked to the regulation and organization of biological processes.
Protein phase transitions, and phase separation in particular, are exquisitely sensitive to changing physical–chemical conditions. Thus, in principle, they could be used to detect and respond to changes in the environment. In this review, we discuss emerging evidence suggesting that regulated protein phase separation and condensate formation is a widely used and underappreciated cellular mechanism to adapt to fluctuating and stressful environments. We pay special attention to prion-like low-sequence complexity regions (LCRs) that are frequently found in phase-separating proteins. These domains act as protein-specific modifier sequences with chaperone-like functions that cooperate with other domains in the same protein to regulate reversible condensate formation in response to stress. Regulation of the protein phase behavior by LCRs may be as important for cell survival as the well-characterized process of regulating protein misfolding by stress-induced molecular chaperones.
PROTEIN-INTRINSIC FACTORS THAT REGULATE PROTEIN SOLUBILITY AND PHASE BEHAVIOR
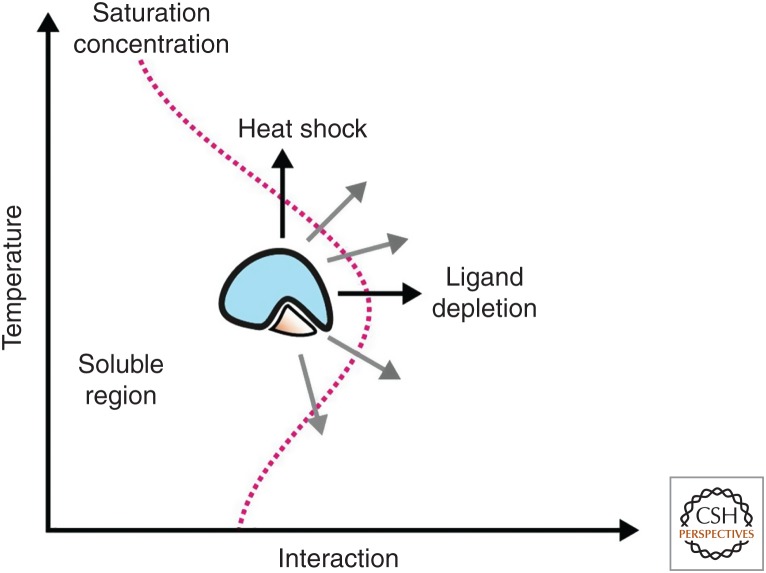
The solubility of proteins is an important requirement for their function. It is thus surprising that many proteins are barely soluble in the intracellular environment even under physiological conditions (Ciryam et al. 2013, 2017; also see Sormanni and Vendruscolo 2018). Proteins with low solubility in the cytosol have been termed supersaturated proteins. Many supersaturated proteins have been linked to protein aggregation diseases, suggesting that their protein structures are metastable and prone to misfolding and aggregation. In agreement with this, studies have shown that a large fraction of cellular proteins is on the verge of misfolding (Foit et al. 2009; Niwa et al. 2012). This phenomenon is also referred to as marginal protein stability. This means that only little energy is required to unfold a large portion of the proteome (Fig. 1). In agreement with this, even mild physiological fluctuations, such as progression through the cell cycle, have strong effects on the thermal stability of the proteome (Becher et al. 2018). Moreover, many proteins aggregate when, for example, yeast cells are exposed to physiological stress conditions, such as glucose depletion or mild heat stress (Narayanaswamy et al. 2009; Olzscha et al. 2011; Picotti et al. 2013; O'Connell et al. 2014; Wallace et al. 2015). Likewise, the abundant presence of misfolded polypeptides with weak folding mutations disrupts the balance of the protein-folding quality control and can establish aberrant physical and genetic interactions that interfere with the folding of marginally stable proteins (Gidalevitz et al. 2006, 2009). Remarkably, directed evolution experiments have shown that more stable proteins can readily be engineered from marginally stable proteins (Foit et al. 2009; Sachsenhauser and Bardwell 2018). This shows that most cellular proteins have not been optimized for maximum but optimum stability and suggests that the stability of the proteome is subject to functional constraints. But what could be these constraints that render a large fraction of proteins marginally stable?
Figure 1.
Schematic showing the solubility as a function of temperature and self-interaction strength for a ligand-binding protein with a deep level of supersaturation. The cellular concentration of many proteins is tuned to be close to the saturation concentration. Small impacts, such as a change in physical–chemical conditions, lead to stronger interactions among the proteins and therefore induce a phase transition.
One hypothesis suggests that marginal stability is the result of neutral evolution and thus a constraint of the evolutionarily accessible protein sequence-space that is independent of functional selection (Williams et al. 2007). Another hypothesis suggests that marginal stability is a product of protein-specific functional constraints because protein structure and function are intimately related. For example, steroid receptors (SRs) must be in an “open” conformation to be able to bind activating ligands in the cytoplasm (Echeverria and Picard 2010; Levin and Hammes 2016; also see Biebl and Buchner 2018). On ligand binding, SRs undergo conformational changes into an active form and then translocate into the nucleus. However, the ability to bind a ligand and undergo conformational changes also renders SRs prone to misfold and aggregate (Kirschke et al. 2014; Lorenz et al. 2014). Thus, the marginal stability of SRs appears to be a trade-off between functionality and stability.
Another example is the RNA-binding protein fused in sarcoma (FUS). FUS is a protein with a deep level of supersaturation (Ciryam et al. 2017) and its solubility depends on the interaction with RNA (Monahan et al. 2017; Maharana et al. 2018). Changes in the level of FUS supersaturation through changes in local RNA concentration are harnessed by cells to promote the formation of physiological membrane-less organelles by condensation (Altmeyer et al. 2015; Patel et al. 2015; Monahan et al. 2017; Maharana et al. 2018).
Ligand-binding proteins, such as SRs or FUS, are often less stable in the absence of their ligands (Luque et al. 2002). This also means that under conditions of limited ligand availability, these proteins become conformationally more heterogeneous and have an increased tendency to misfold and aggregate. As discussed in the next section, such conditions could be induced by sudden changes in the environment, in which the intracellular concentration of metabolites changes drastically.
EXTRINSIC FACTORS REGULATING PROTEIN SOLUBILITY AND PHASE BEHAVIOR
The physical–chemical conditions inside cells shape and put constraints on protein evolution. For a given cytoplasmic protein, these are largely determined by the physical and chemical properties of the cytosol.
The cytosol contains many macromolecules of different sizes and shapes. A considerable fraction of these macromolecules are proteins and nucleic acids. These molecules expose large surfaces that are covered with positive and negative charges. Consequently, many binding events in the cytosol—functional and nonfunctional—are mediated by electrostatic interactions. Studies have shown that the presence of highly charged macromolecules such as RNAs can have important effects on protein structure and function. For example, RNA can promote the folding of proteins (Choi et al. 2008; Docter et al. 2016; Horowitz and Bardwell 2016). The large volume fraction of proteins and nucleic acids also turns the cytosol into a highly crowded environment (Ellis 2001; Soranno et al. 2014; Delarue et al. 2018). This has additional functional consequences, because macromolecular crowding indirectly affects protein folding and protein–protein interactions.
The cytosol also contains a high concentration of small molecules and ions. This is important, because protein solubility is strongly dependent on the ionic strength and the pH of the cytosol. To maintain protein solubility, cells invest large amounts of energy keeping the salt concentration and the pH in a narrow range. In yeast, for example, the pH is regulated through ATP-driven pumps that continuously remove protons from the cytosol to the outside (Orij et al. 2011).
Cells also regulate their water content. A decrease in water content reduces the hydration shell that surrounds proteins and concomitantly favors interactions among macromolecules. This can cause widespread assembly of proteins into higher-order structures. Important differences between organisms exist. For example, plants, fungi, and roundworms are frequently exposed to water-limiting conditions and have evolved mechanisms to cope with severe water loss (Erkut et al. 2011, 2013, 2016; Tapia and Koshland 2014; Boothby et al. 2017). Often, they produce small osmolytes, such as trehalose or glycerol, which can increase protein stability. Moreover, many organisms have one or more vacuoles that serve as an osmoregulatory organelle.
The physical parameter with the strongest effect on protein solubility is temperature. Many organisms including bacteria, fungi, or plants cannot regulate temperature and therefore must withstand large temperature-induced fluctuations in protein solubility. As we will discuss below, these organisms have evolved mechanisms to limit the detrimental effects of temperature changes by regulating protein solubility. More generally, this suggests that the solubility and stability of a given proteome is adapted to the specific habitat of an organism.
DEALING WITH CHANGING PHYSICAL—CHEMICAL CONDITIONS INSIDE CELLS
Many enzymes show high catalytic activity at neutral cytosolic pH, at which cell growth is optimal. Keeping the pH neutral requires constant expenditure of energy to remove protons from the cytosol (Dechant et al. 2010; Orij et al. 2011). However, stress conditions, such as a depletion of nutrients can cause a rapid drop in cellular energy levels and a concomitant cytosolic acidification for example in yeast. This has direct effects on the activity of enzymes, but it also drastically reduces protein solubility. Indeed, protein solubility is usually lowest when the solution pH is close to a protein's isoelectric point and many cytosolic proteins have an isoelectric point in the mildly acidic range. As a result, these proteins assemble into higher-order structures as soon as the cytosol acidifies (Munder et al. 2016).
There are two ways in which an organism can deal with such a situation: (1) It can keep the internal physical–chemical conditions constant at all costs. This appears to be the strategy used by many motile organisms with a nervous system. To maintain homeostasis, these organisms use a large amount of energy and avoid stressful conditions. (2) The organism can mitigate the effects of stressful environments by directly regulating protein solubility and phase behavior. This seems to be the method of choice for many plants, fungi, or bacteria. These organisms have in common that they are often sessile and cannot escape from stressful conditions.
Which proteins become insoluble under stressful conditions? A few proteome-wide studies have investigated the proteins that change their solubility on stress. In energy-depleted yeast cells, many metabolic enzymes and proteins involved in translation regulation assemble into higher-order structures (Narayanaswamy et al. 2009; O'Connell et al. 2014). In heat-shocked human and bacterial cells, the most unstable proteins are those that bind cofactors and nucleic acids (Leuenberger et al. 2017). In yeast, the proteins that are most sensitive to heat are RNA-binding proteins (Wallace et al. 2015). It is unclear why these specific proteins are so sensitive to changing conditions. One possibility is that these proteins are vulnerable because they have metastable folds and have been optimized for conditions occurring in unstressed, growing cells. Another intriguing possibility is that these exceptional protein properties have been harnessed and shaped by evolution to turn these proteins into environmental stress sensors that mount adaptive responses. Indeed, accumulating evidence suggests a functional link between the stress response and condition-responsive intrinsically disordered LCRs that may intimately be involved in regulating protein solubility and phase behavior.
REGULATION OF PHASE SEPARATION BY INTRINSICALLY DISORDERED LCRs
Intrinsically disordered proteins (IDPs) or regions (IDRs) are frequently found in eukaryotic proteomes. IDPs/IDRs do not fold into a single native structure that corresponds to the lowest energy state. Rather, they sample an ensemble of conformational states with similar intrinsic energies that is defined by their specific protein sequence. Most IDPs/IDRs are readily identified because of their low-sequence complexity. This means that their primary sequences consist only of a subset of all possible amino acids. Frequently, they also contain repetitive motifs. Accordingly, these regions are called LCRs.
What are the functions of IDRs? One abundant class of IDRs serves as linker sequences that connect folded domains (Oldfield and Dunker 2014). Another class of IDRs are display sites. These regions contain amino acids that can be modified by posttranslational modifications, such as phosphorylation. In recent years, evidence for a class of IDRs that regulates protein phase behavior has been emerging. One example comes from investigations of the helicase Ddx4, which is a constituent of membrane-less germ granules. The IDR/LCR of Ddx4 self-interacts to promote phase separation into protein-rich liquid-like droplets (Nott et al. 2015). The diverse functions of IDRs are not mutually exclusive, and IDRs that promote phase separation may also function as display sites and/or linker sequences.
Phase separation requires the formation of many reversible interactions between phase-separating proteins. The number and strength of these interactions sets the concentration threshold of phase separation (called the saturation concentration), as well as the material properties of the assembled condensates. Proteins that are able to phase separate often have multiple motifs or domains that are able to undergo weak and transient interactions (Li et al. 2012; Brangwynne et al. 2015; Ruff et al. 2018). For example, the LCR of Ddx4 is characterized by blocks of alternating charges and a high number of glycine-arginine and phenylalanine-glycine motifs (Nott et al. 2015). These sequence features promote many weak binding events mediated through charge–charge, pi–pi, or cation–pi interactions. Such multivalent interactions are the main driver of protein phase separation in cells.
Phase separation not only requires many weak and transient interactions, but the polymer chain length and its flexibility are also important. LCRs are intrinsically disordered and explore more conformations than folded proteins, a feature that has been shown to promote phase separation. At low-protein concentrations, LCRs can undergo intrachain interactions. Above the saturation concentration, interchain interactions become increasingly dominant leading to phase separation of the LCR-containing protein. For example, the LCR of LAF-1, a helicase similar to Ddx4, undergoes large conformational fluctuations that promote self-interaction and phase separation at protein concentrations as low as 1.5 µm (Wei et al. 2017). More generally, the specific sequence of an LCR determines its conformational ensemble and thus its geometry. LCRs with a high net charge usually adopt extended chain geometries, whereas LCRs enriched for polar and depleted of charged residues often adopt collapsed ensemble geometries (Holehouse and Pappu 2018). Regulation of the ensemble geometries through posttranslational modifications, protonation, or changes in the ionic strength of a solution therefore provide a direct means of condition-specific control of protein function by phase separation (Ruff et al. 2018). Such behavior is in stark contrast to the phase behavior of globular proteins. Globular proteins can also undergo condition-specific phase separation, but this usually requires protein concentrations orders of magnitude higher than for LCR-containing proteins.
In summary, intrinsically disordered LCRs are being recognized as key elements involved in the formation of cellular structures by phase separation. One family of LCRs that we have not yet discussed is prion-like LCRs. This class of LCRs has received particular attention in recent years; prion-like LCRs are frequently found in proteins that have a high tendency to form protein aggregates and the aggregation of these proteins is associated with human diseases. Importantly, our understanding of the functional role of prion-like LCRs has been subject to various modifications over the years owing to conceptual advances. For this reason, we will first give a historical account of the discoveries that have led to the current view of prion-like LCRs.
PRION-LIKE LCRs: THEIR DISCOVERY IN ASSOCIATION WITH YEAST PRIONS
As the name suggests, prion-like LCRs have first been described in association with self-propagating forms of proteins, called prions. Prion proteins in budding yeast, such as the translation termination factor Sup35, can switch between a normal and a heritable prion state that is transmitted from generation to generation (Tuite and Serio 2010; Newby and Lindquist 2013; Harvey et al. 2018). These prion states were proposed to generate heritable phenotypic variation with adaptive value in fluctuating environments (True and Lindquist 2000; True et al. 2004). However, this hypothesis has been contested and others have proposed that yeast prions are diseases (Nakayashiki et al. 2005; Wickner et al. 2011).
The structural basis for the heritability of prions is the conversion of the prion protein into an amyloid-like state. Importantly, the information for prion formation is contained in specific LCRs within prion proteins (Edskes et al. 1999; Li and Lindquist 2000; Santoso et al. 2000; Osherovich and Weissman 2001). When these regions are removed, prion proteins lose the ability to function as prions; conversely, when transferred onto other proteins, those proteins gain the ability to function as prions. These observations led to the hypothesis that prion proteins contain a specific domain that holds all the information for prion behavior: the prion domain (PrD).
PrDs have an unusual amino acid composition of mostly polar amino acids, such as glutamine, asparagine, serine, glycine, proline, and tyrosine. Subsequent biophysical studies showed that this amino acid composition determines the self-interacting intrinsically disordered nature of PrDs and promotes nucleation into a rare self-templating prion form (Mukhopadhyay et al. 2007). In agreement with this, experiments have shown that overexpression of either prion proteins or PrDs alone can induce a prion state (Alberti et al. 2009; Chakrabortee et al. 2016).
These powerful experiments revealed the following hallmarks of prions: (1) The information for prion behavior is contained in disordered LCRs with specific amino acid composition, (2) self-interaction at high protein concentrations promotes conformational conversion into a prion state, and (3) the specificity of prion-mediated aggregation is conferred by self-templating amyloid structures. These discoveries have prompted a search for proteins with similar sequences. Proteome-wide screens revealed dozens of prion-like proteins in yeast (Alberti et al. 2009; Chakrabortee et al. 2016) and humans (King et al. 2012). Many of these prion-like proteins are RNA-binding proteins. Remarkably, a large number of the identified human prion candidates are also associated with age-related protein misfolding diseases (Cushman et al. 2010; King et al. 2012). For example, mutations in the RNA-binding protein TDP-43 cause ALS and frontotemporal dementia (FTD). In patients carrying these mutations, TDP-43 adopts a characteristic amyloid-like state in neurons (Taylor et al. 2016). Many of these mutations map to prion-like LCRs and increase the propensity of the protein to aggregate. This corroborates that many of these domains may not have evolved to promote prion formation but rather exert other functions.
PRION-LIKE LCRs REGULATE THE FORMATION OF MEMBRANE-LESS COMPARTMENTS
One important step forward in understanding the functional role of prion-like LCRs was the discovery that many prion-like proteins localize to ribonucleoprotein particle (RNP) granules, in particular to stress-inducible granules or stress granules (SGs). SGs consist of many proteins and RNAs and they form when cells are exposed to heat, oxidative stress, or nutrient depletion. The role of prion-like LCRs in the formation of SGs was first investigated in genetic studies (Gilks et al. 2004; Decker et al. 2007). When LCRs were genetically removed from various prion-like proteins, SG formation was impaired. Conversely, the LCRs aggregated in the absence of stress when overexpressed, suggesting that prion-like aggregation of proteins underlies the formation of SGs.
Support for this idea came from in vitro experiments with prion-like RNA-binding proteins such as FUS (Han et al. 2012; Kato et al. 2012; Kato and McKnight 2018). The prion-like LCR of FUS was shown to form hydrogels at high-protein concentrations. Electron microscopy of the hydrogels revealed the presence of amyloid-like fibers. The fibers were suggested to form scaffold-like structures that stabilize membrane-less compartments in cells. This collective set of experiments led to the proposal that SGs and other membrane-less compartments have properties of hydrogels that form by protein polymerization into amyloid-like structures.
The hypothesis of β-sheet-driven polymerization of LCRs was very appealing because it provided a solution to one of the key problems in the field: How do cells ensure the specificity of compartment formation in the complex intracellular environment? However, the dynamic nature of membrane-less compartments suggested that there must be other ways of assembly. For example, fluorescence recovery after photobleaching (FRAP) experiments of RNA-binding proteins such as FUS showed that the exchange rates of these proteins within SGs are on the order of seconds (Molliex et al. 2015; Patel et al. 2015). Such dynamic behavior is difficult to reconcile with the idea that these SGs are hydrogels permeated by a network of amyloid-like fibrils.
LCRs AS REGULATORS OF PROTEIN PHASE BEHAVIOR
The fluid-like nature of RNP granules in living cells suggests another assembly mechanism, namely, that they may form by liquid–liquid phase separation. This idea is increasingly supported by evidence from in vitro experiments. Prion-like proteins, such as FUS and hnRNPA1 phase separate in vitro into liquid droplets (Molliex et al. 2015; Patel et al. 2015). Importantly, phase separation of these proteins occurs already at physiological concentrations. Recent studies revealed that the LCR of FUS and FUS-like proteins, synergizes with other regions in the protein to drive phase separation (Qamar et al. 2018; Wang et al. 2018). Phase separation is driven by a molecular grammar that involves segregated cohesive motifs known as stickers (Wang et al. 2018). In FUS, these are the tyrosine residues in the prion-like LCR and arginine residues in the mostly disordered RNA-binding domain (RBD). Productive interactions among tyrosine and arginine residues are regulated by complementary electrostatic interactions, which are provided by negatively charged amino acids in the prion-like LCR. These negatively charged residues also reduce self-interaction among prion-like LCRs and promote productive interactions among the LCRs and RBDs. Accordingly, homotypic LCR–LCR interactions are disfavored at low physiological protein concentrations and instead interactions among prion-like LCRs and other parts of the protein are the preferred type of interaction. These findings suggest that LCRs are protein-specific modulators of intra- and intermolecular interactions.
But why do proteins such as FUS need LCRs to regulate their phase behavior? Currently we can only speculate about the reasons. The carboxyl terminus of FUS contains an arginine-rich RNA-binding region. The interaction of FUS and RNA regulates the formation of FUS-containing membrane-less organelles in cells (Maharana et al. 2018). One possibility is that the amino-terminal LCR of FUS competes with RNA for binding to the arginine-rich RBD. In this scenario, the LCRs would interact with the RNA-free carboxyl terminus of FUS. On RNA binding, the LCR is displaced from the carboxyl terminus and becomes free to undergo interactions with itself or other proteins. Indeed, there is increasing evidence that phase separation of FUS is regulated by RNA. For example, low concentrations of RNA promote phase separation, whereas high concentrations prevent phase separation (Banerjee et al. 2017; Maharana et al. 2018). This suggests that the phase behavior of FUS is dependent on the local RNA concentration and that LCRs have been added to the protein to regulate RNA-dependent phase behavior.
As explained above, FUS can also form gels and fibrils. Is there a function associated with these material states? One possibility is that compartments adopt a more solid-like state with time and that gels and fibrils have important unknown functions in cells. Another explanation is provided by studies showing that gelation and fiber formation is promoted by disease-associated mutations (Molliex et al. 2015; Patel et al. 2015). Thus, gels and fibrils could also be pathological states that cells must prevent. This view has been summarized in the continuum model: LCRs can adopt a range of material states from liquids to gels to fibers (Alberti and Hyman 2016). These different states can have physiological or pathological roles depending on the type of protein and the condition.
In summary, the following picture emerges: Prion-like LCRs act as modifier sequences that regulate the phase behavior of proteins. In FUS, this involves heterotypic interactions between the amino-terminal LCR and the carboxy-terminal RBD. These interactions seem necessary for adapting the phase behavior of the protein to the local concentration of ligands, such as RNA.
LCRs AND THE REGULATION OF PROTEIN PHASE BEHAVIOR IN STRESSED CELLS
How general is this responsiveness of prion-like proteins to changing conditions such as local RNA concentrations? Does this sensitivity to changing conditions also apply to prion proteins in yeast? As described above, yeast cells experience massive fluctuations in physical–chemical conditions, such as changes in pH or temperature that strongly affect the solubility of proteins. How do yeast cells cope with such sudden changes in the solubility of the proteome?
During stress, many yeast proteins form amorphous aggregates attributable to unfolding and subsequent aggregation, but others coprecipitate with RNA. The latter assemblies have been termed SGs and they are analogous to stress-inducible assemblies in mammalian cells. Initially, both types of assemblies, amorphous aggregates and SGs, were regarded as distinct entities, yet more recent studies showed that at least in yeast cells, heat-induced SGs and protein aggregates are found within one and the same assembly (Cherkasov et al. 2013; Kroschwald et al. 2015). The role of SG proteins and their assembly with RNA remains enigmatic, but the current view is that SGs bind and stabilize messenger RNAs (mRNAs) that have been released by the ribosome and protect them against degradation (Anderson and Kedersha 2008; Buchan et al. 2008).
One yeast protein whose role in mRNA metabolism and SG formation has been investigated in depth is the poly(A)-binding protein Pab1. Pab1 binds the poly(A) tail of many mRNAs and is involved in mRNA biogenesis, stability, and translation (Otero et al. 1999). Pab1 is also a prion-like, LCR-containing RNA-binding protein (Alberti et al. 2009). Recently, Pab1 was shown to phase separate and form gel condensates in response to physiological heat shock and pH stress (Riback et al. 2017). Phase separation of Pab1 is dependent on its proline-rich prion-like LCR. Importantly, this LCR is not required for phase separation. Instead it modulates the stress-induced demixing process that is otherwise driven by the folded RNA recognition motif (RRM) domains. Mutations that affect the temperature sensitivity of Pab1 also altered the cellular fitness during growth at high temperature and during energy depletion. Importantly, cellular stress tolerance correlates with the ability of Pab1 to phase separate at the physiological stress temperature. This indicates that the sensitivity of Pab1 to form condensates helps yeast cells adapt to stress-associated pH and temperature changes. Why Pab1 phase separation is beneficial during stress is still unclear. One possibility is that Pab1 represses translation of stress-protective mRNAs during growth and that these mRNAs are released from Pab1 for translation when it forms condensates.
Another SG protein that is highly sensitive to stress is the poly(U)-binding protein Pub1. Like Pab1, Pub1 is a prion-like protein (Alberti et al. 2009). Physiological stresses, such as changes in temperature and/or pH affect the solubility of Pub1 and lead to the formation of Pub1 condensates (Kroschwald et al. 2018). Importantly, different stresses, as well as stress intensities induce condensates with different properties. Pub1 condensates induced by acidic pH behaved as reversible gels that readily dissolve when the pH is neutralized; heat-induced condensates instead are more solid-like and their dissolution requires chaperone-catalyzed protein disaggregation. As for Pab1, condensate formation of Pub1 is driven by folded RRM domains and modulated by its prion-like proline-rich LCR. Removal of this LCR destabilizes Pub1 and prevents dissolution of Pub1 assemblies in the stress recovery phase. This suggests that the solubility of Pub1 is maintained at a critical level to allow autonomous stress-sensing of Pub1 via phase separation. Whether condensate formation is coupled to a release of stress-protective mRNA for translation of stress genes, such as molecular chaperones, remains to be determined.
Together, these findings suggest that prion-like proteins may act as stress sensor proteins that directly respond to changes in environmental conditions through protein phase separation. Most, if not all, prion-like LCRs are linked to folded domains (Alberti et al. 2009), suggesting that prion-like proteins carry evolutionarily tuned LCRs that modulate the phase boundary of the protein's folded domains. We speculate that coevolution of prion-like LCRs and the folded domains set the sensitivity of these proteins to stress and the threshold at which these proteins enter a condensed state. At the same time, these LCRs modify the material properties of the condensates such that the proteins contained in the condensed phase can be retrieved when the stress subsides. Modulation of the phase behavior of folded domains by IDRs may be a more general principle with important biological significance (Franzmann and Alberti 2018; Mittal et al. 2018). The stimulus-dependent formation of condensates from proteins with folded domains could also act as a signal for a protein-folding problem that could be harnessed by cells to orchestrate the stress response.
This view is very different from the historical view of prion-like LCRs as autonomous drivers of protein aggregation and suggests that regulation of protein phase behavior may be the ancestral function of prion-like LCRs. This idea was tested by analyzing the phase behavior of the canonical prion protein Sup35 (Franzmann et al. 2018). On stress, Sup35 forms protective gels via pH-regulated liquid phase separation followed by gelation. Importantly, removal of the PrD destabilizes the protein and leads to the formation of irreversible aggregates from which the functional protein can no longer be retrieved. In agreement with this, phase separation promoted yeast cell survival by rescuing the essential Sup35 translation factor from stress-induced damage. Thus, prion-like and PrDs represent conserved environmental stress sensors that facilitate rapid adaptation in unstable environments by modifying protein phase behavior.
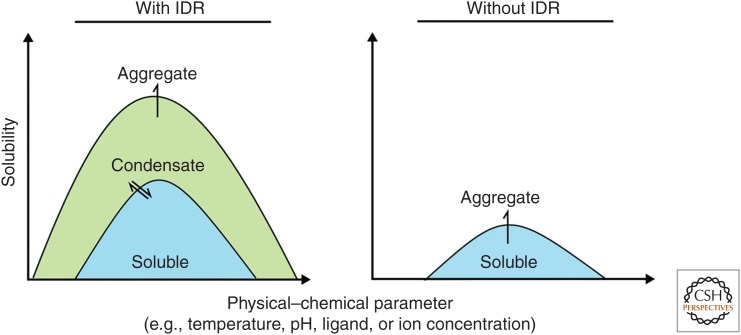
Based on these findings, we speculate that most prion-like LCRs are protein-specific modifiers of protein phase behavior (Fig. 2). This includes Pab1, Pub1, Sup35, and potentially many human RNA-binding proteins such as FUS. These proteins have complicated domain arrangements and folds that render them prone to misfolding and aggregation in response to stress. LCRs with chaperone-like functions extend the soluble phase space of these proteins, reducing the likelihood of them undergoing aberrant interactions and modulating condensate formation.
Figure 2.
Schematic solubility profile of a protein with (left) and without (right) an intrinsically disordered region (IDR). Compared with the situation without IDR, a protein shows greater solubility when the IDR is present. Importantly, the solubility increases with the ability of the protein to explore a greater phase space, including the reversible transition to form a condensate. By this the effective aggregation concentration is increased and the protein persists in a recoverable state at a broader range of physical–chemical conditions. In the absence of the IDR, the range of physical–chemical conditions that a protein can explore is much smaller and consequently the overall saturation concentration is small.
Does this mean that prion states are aberrant states of these proteins? Indeed, prion induction in yeast often requires overexpression. In agreement with this, in vitro experiments have shown that the soluble and amyloid/prion states are separated by a large kinetic energy barrier. For example, aggregation of the PrD of Sup35 requires high protein concentrations and excessive shaking (Glover et al. 1997; Serio et al. 2000). In fact, this appears to hold true for most identified yeast prion proteins, including the 25 that were originally identified using a prion detection algorithm (Alberti et al. 2009). Increasing evidence now indicates that prion proteins can also form condensates under more physiological conditions. This suggests that the prion algorithm not only identifies proteins that have a high propensity to adopt amyloid-like states but also proteins that form biomolecular condensates via phase separation.
LCRs AND MOLECULAR CHAPERONES: TWO WAYS OF DEALING WITH PROTEIN FOLDING
In a living cell, nonproductive protein folding is in many, if not most, cases prevented by the action of a highly conserved set of proteins termed molecular chaperones. In this section, we discuss the importance of chaperones and how they could interact and/or synergize with intrinsically disordered LCRs.
Protein folding is a highly cooperative process in which a protein can populate multiple intermediate states on the pathway to its native fold. This is often depicted as an energy landscape in which the native state has the lowest energy. Folding intermediates are local energy minima on the way to the native state that are transiently populated. Proteins can be kinetically trapped in these intermediate states. Hence, the overall folding rate is often determined by the number and energy state of these intermediates in a protein's folding pathway. In addition to on-pathway states, there are off-pathway states. This is because the folding polypeptide exposes hydrophobic surfaces, which are typically buried in the native state. To prevent protein misfolding, folding should be fast and aberrant intermolecular interactions with other proteins should be avoided. Moreover, any condition that destabilizes the native state or shifts the balance away from on-pathway to off-pathway folding should be prevented at all costs.
To ensure efficient folding in the complex environment of the cell, chaperones receive the nascent protein chain emerging from the ribosomal exit tunnel and guide it along a productive folding pathway to the native state. However, the final native state rarely corresponds to a single rigid conformation; it is often characterized by various dynamic protein structures that constantly interconvert. Because of these dynamics and the rather large conformational changes, a given protein must be under constant surveillance by an integrated network of chaperones and protein degradation machineries to maintain protein homeostasis, even after a protein has adopted its native state.
Molecular chaperones adopt different functional roles in the cell and, accordingly, there are different classes (Kim et al. 2013). However, all chaperones have in common that they bind to misfolded proteins and by doing so inhibit aberrant interactions and the formation of off-pathway conformations. In contrast to protein degradation pathways, chaperones act to recover and refold proteins that cannot attain or have lost their native state. Chaperones can also modify the material properties of assemblies. Small heat shock proteins (sHsps), for example, bind to and sequester misfolded proteins to form sHsps–client complexes from which misfolded client proteins are more easily recoverable (Lee and Vierling 2000; Mogk et al. 2003a,b; Cashikar et al. 2005; Haslbeck et al. 2005).
Which of these chaperone functions can also be executed by LCRs and which are unique to LCRs? There are many commonalities, but also some differences. The most obvious difference is that LCRs act in cis and not in trans. This means that LCRs coevolve with the rest of a given protein and thus are more protein-specific in their mode of action. In contrast, chaperones must maintain their ability to interact with a broad range of client proteins. Analogous to the role of chaperones in protein folding, it is conceivable that LCRs shape the energy landscape of proteins and alter intermediate folding states during de novo folding, but also when the native state is lost secondarily. Moreover, when the native state is lost, LCRs could inhibit intramolecular and intermolecular interactions to increase the overall solubility of the nonnative state and, analogous to the mode of action of sHsps, LCRs could weaken the interactions among polypeptides to form soft material assemblies from which proteins are more easily recoverable.
One important observation is that many chaperones contain LCRs themselves. One example is the yeast small heat shock protein Hsp42. The LCR of Hsp42 is prion-like, binds misfolded proteins, and modifies the properties of forming assemblies that sequester these misfolded proteins (Haslbeck et al. 2004a; Ungelenk et al. 2016; Grousl et al. 2018). Likewise, the disordered regions of many other small heat shock proteins, including Hsp26, were shown to be essential for the interaction with misfolded clients (Stromer et al. 2003, 2004; Haslbeck et al. 2004b; Basha et al. 2006; Haslbeck and Vierling 2015; also see Janowska et al. 2018). Interestingly, the activation of the chaperone function of Hsp26 (Franzmann et al. 2008), as well as the activation of the redox-regulated chaperone Hsp33 from Escherichia coli (Reichmann et al. 2012), coincide with a substantial increase in structural disorder within the chaperones.
Why do some proteins rely on chaperones and others on LCRs? We propose that proteins with complex folds/folding pathways use LCRs to ensure productive folding and recoverability after stress-induced condensation. These proteins may have specific folding demands that cannot be met by general chaperones. In the future, it will be important to mechanistically understand these protein-specific folding problems and how they are modulated by LCRs.
CONCLUDING REMARKS
We have made a lot of progress in understanding how organisms adapt to stress. Stress conditions generate specific protein-folding problems and the cellular response to these problems must be well calibrated to ensure organism survival. Here, we have laid out the evidence for a new stress-coping mechanism. This mechanism is based on intrinsically disordered LCRs that detect changes in the environment, alter the folding pathways of proteins, and orchestrate stress-specific cellular responses. We predict that environmentally responsive LCRs have a similar functional significance in proteostasis as the well-characterized process of chaperone-mediated folding. Many questions remain to be addressed in the future. Most urgently, the molecular details of how LCRs function are largely unknown. How do LCRs modify protein phase behavior and the physical properties of condensates? How do LCR-containing proteins sense changes in the environment? Do chaperones and LCRs synergize to regulate the formation of stress-induced condensates? Could LCRs also act as chaperone-binding regions? Do organisms that live in highly fluctuating environments have more LCRs? These questions and many others urgently await answers to improve our understanding of a newly emerging mechanism with fundamental importance in biology.
ACKNOWLEDGMENTS
We thank Jordina Guillen-Boixet, Christine Desroche, Anne Eßlinger, and Edgar Boczek for discussions and comments on the manuscript. We gratefully acknowledge funding from the Max Planck Society. This work is further supported by the MaxSynBio consortium, jointly funded by the Federal Ministry of Education and Research of Germany the German Federal Ministry of Research and Education (BMBF 031A359A to T.M.F.). and the Max Planck Society. We further acknowledge funding by the Human Frontiers Program for Grant RGP0034/2017 to S.A. and the Volkswagen “Life?” initiative for a grant to S.A.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alberti S. 2017. The wisdom of crowds: Regulating cell function through condensed states of living matter. J Cell Sci 130: 2789–2796. 10.1242/jcs.200295 [DOI] [PubMed] [Google Scholar]
- Alberti S, Hyman AA. 2016. Are aberrant phase transitions a driver of cellular aging? Bioessays 38: 959–968. 10.1002/bies.201600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158. 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Mateju D, Mediani L, Carra S. 2017. Granulostasis: Protein quality control of RNP granules. Front Mol Neurosci 10: 84 10.3389/fnmol.2017.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask M-BD, Streicher W, Jungmichel S, Nielsen ML, et al. 2015. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6: 8088 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. 2016. In vivo aspects of protein folding and quality control. Science 353: aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PR, Milin AN, Moosa MM, Onuchic PL, Deniz AA. 2017. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew Chem Int Ed Engl 56: 11354–11359. 10.1002/anie.201703191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Friedrich KL, Vierling E. 2006. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem 281: 39943–39952. 10.1074/jbc.M607677200 [DOI] [PubMed] [Google Scholar]
- Becher I, Andrés-Pons A, Romanov N, Stein F, Schramm M, Baudin F, Helm D, Kurzawa N, Mateus A, Mackmull MT, et al. 2018. Pervasive protein thermal stability variation during the cell cycle. Cell 173: 1495–1507.e18. 10.1016/j.cell.2018.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Biebl MM, Buchner J. 2018. Structure, function, and regulation of Hsp90 machinery. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a034017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B. 2017. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65: 975–984.e5. 10.1016/j.molcel.2017.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, Pappu RV. 2015. Polymer physics of intracellular phase transitions. Nat Phys 11: 899–904. 10.1038/nphys3532 [DOI] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183: 441–455. 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL. 2005. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem 280: 23869–23875. 10.1074/jbc.M502854200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Byers JS, Jones S, Garcia DM, Bhullar B, Chang A, She R, Lee L, Fremin B, Lindquist S, et al. 2016. Intrinsically disordered proteins drive emergence and inheritance of biological traits. Cell 167: 369–381.e12. 10.1016/j.cell.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. 2013. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23: 2452–2462. 10.1016/j.cub.2013.09.058 [DOI] [PubMed] [Google Scholar]
- Choi SI, Han KS, Kim CW, Ryu KS, Kim BH, Kim KH, Kim SI, Kang TH, Shin HC, Lim KH, et al. 2008. Protein solubility and folding enhancement by interaction with RNA. PLoS ONE 3: e2677 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. 2013. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep 5: 781–790. 10.1016/j.celrep.2013.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Lambert-Smith IA, Bean DM, Freer R, Cid F, Tartaglia GG, Saunders DN, Wilson MR, Oliver SG, Morimoto RI, et al. 2017. Spinal motor neuron protein supersaturation patterns are associated with inclusion body formation in ALS. Proc Natl Acad Sci 114: E3935–E3943. 10.1073/pnas.1613854114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. 2010. Prion-like disorders: Blurring the divide between transmissibility and infectivity. J Cell Sci 123: 1191–1201. 10.1242/jcs.051672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett V, Fersht AR. 2003. Is there a unifying mechanism for protein folding? Trends Biochem Sci 28: 18–25. 10.1016/S0968-0004(02)00012-9 [DOI] [PubMed] [Google Scholar]
- Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M. 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J 29: 2515–2526. 10.1038/emboj.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G, et al. 2018. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174: 338–349.e20. 10.1016/j.cell.2018.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Deuerling E, Gamerdinger M, Kreft SG. 2018. Chaperone interactions at the ribosome. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docter BE, Horowitz S, Gray MJ, Jakob U, Bardwell JCA. 2016. Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res 44: 4835–4845. 10.1093/nar/gkw291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria PC, Picard D. 2010. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochim Biophys Acta 1803: 641–649. 10.1016/j.bbamcr.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Edskes HK, Gray VT, Wickner RB. 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci 96: 1498–1503. 10.1073/pnas.96.4.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. 2001. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr Opin Struct Biol 11: 114–119. 10.1016/S0959-440X(00)00172-X [DOI] [PubMed] [Google Scholar]
- Erkut C, Penkov S, Khesbak H, Vorkel D, Verbavatz JM, Fahmy K, Kurzchalia TV. 2011. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21: 1331–1336. 10.1016/j.cub.2011.06.064 [DOI] [PubMed] [Google Scholar]
- Erkut C, Vasilj A, Boland S, Habermann B, Shevchenko A, Kurzchalia TV. 2013. Molecular strategies of the Caenorhabditis elegans dauer larva to survive extreme desiccation. PLoS ONE 8: e82473 10.1371/journal.pone.0082473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkut C, Gade VR, Laxman S, Kurzchalia TV. 2016. The glyoxylate shunt is essential for desiccation tolerance in C. elegans and budding yeast. eLife 5: e13614 10.7554/eLife.13614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JCA. 2009. Optimizing protein stability in vivo. Mol Cell 36: 861–871. 10.1016/j.molcel.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann T, Alberti S. 2018. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J Biol Chem 10.1074/jbc.TM118.001190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Menhorn P, Walter S, Buchner J. 2008. Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol Cell 29: 207–216. 10.1016/j.molcel.2007.11.025 [DOI] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. 2018. Phase separation of a yeast prion protein promotes cellular fitness. Science 359: eaao5654 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474. 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. 2009. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet 5: e1000399 10.1371/journal.pgen.1000399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398. 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89: 811–819. 10.1016/S0092-8674(00)80264-0 [DOI] [PubMed] [Google Scholar]
- Grousl T, Ungelenk S, Miller S, Ho CT, Khokhrina M, Mayer MP, Bukau B, Mogk A. 2018. A prion-like domain in Hsp42 drives chaperone-facilitated aggregation of misfolded proteins. J Cell Biol 217: 1269–1285. 10.1083/jcb.201708116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. 2012. Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell 149: 768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- *.Hartl U. 2018. Protein folding and misfolding in the cytoplasm. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey ZH, Chen Y, Jarosz DF. 2018. Protein-based inheritance: Epigenetics beyond the chromosome. Mol Cell 69: 195–202. 10.1016/j.molcel.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Vierling E. 2015. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J Mol Biol 427: 1537–1548. 10.1016/j.jmb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J. 2004a. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23: 638–649. 10.1038/sj.emboj.7600080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Ignatiou A, Saibil H, Helmich S, Frenzl E, Stromer T, Buchner J. 2004b. A domain in the N-terminal part of Hsp26 is essential for chaperone function and oligomerization. J Mol Biol 343: 445–455. 10.1016/j.jmb.2004.08.048 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. 2005. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem 280: 23861–23868. 10.1074/jbc.M502697200 [DOI] [PubMed] [Google Scholar]
- *.Hegde RS, Zavodszky E. 2018. Spatial quality control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse AS, Pappu RV. 2018. Collapse transitions of proteins and the interplay among backbone, sidechain, and solvent interactions. Annu Rev Biophys 10.1146/annurev-biophys-070317-032838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Bardwell JCA. 2016. RNAs as chaperones. RNA Biol 13: 1228–1231. 10.1080/15476286.2016.1247147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Janowska MK, Baughman HER, Woods CN, Klevitt RE. 2018. Mechanisms of small heat shock proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a034025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL. 2018. A solid-state conceptualization of information transfer from gene to message to protein. Annu Rev Biochem 87: 351–390. 10.1146/annurev-biochem-061516-044700 [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82: 323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. 2012. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462: 61–80. 10.1016/j.brainres.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. 2014. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157: 1685–1697. 10.1016/j.cell.2014.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4: e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S. 2018. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep 23: 3327–3339. 10.1016/j.celrep.2018.05.041 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E. 2000. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198. 10.1104/pp.122.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger P, Ganscha S, Kahraman A, Cappelletti V, Boersema PJ, von Mering C, Claassen M, Picotti P. 2017. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 355: eaai7825 10.1126/science.aai7825 [DOI] [PubMed] [Google Scholar]
- Levin ER, Hammes SR. 2016. Nuclear receptors outside the nucleus: Extranuclear signalling by steroid receptors. Nat Rev Mol Cell Biol 17: 783–797. 10.1038/nrm.2016.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lindquist S. 2000. Creating a protein-based element of inheritance. Science 287: 661–664. 10.1126/science.287.5453.661 [DOI] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz OR, Freiburger L, Rutz DA, Krause M, Zierer BK, Alvira S, Cuéllar J, Valpuesta JM, Madl T, Sattler M, et al. 2014. Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol Cell 53: 941–953. 10.1016/j.molcel.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Luque I, Leavitt SA, Freire E. 2002. The linkage between protein folding and functional cooperativity: Two sides of the same coin? Annu Rev Biophys Biomol Struct 31: 235–256. 10.1146/annurev.biophys.31.082901.134215 [DOI] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén-Boixet J, Franzmann T, et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360: 918–921. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A, Holehouse AS, Cohan MC, Pappu RV. 2018. Sequence-to-conformation relationships of disordered regions tethered to folded domains of proteins. J Mol Biol 430: 2403–2421. 10.1016/j.jmb.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. 2003a. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 50: 585–595. 10.1046/j.1365-2958.2003.03710.x [DOI] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Friedrich KL, Schönfeld HJ, Vierling E, Bukau B. 2003b. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem 278: 31033–31042. 10.1074/jbc.M303587200 [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O'Meally R, Dignon GL, Conicella AE, Zheng W, et al. 2017. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J 36: 2951–2967. 10.15252/embj.201696394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M. 1999. Heat-inactivated proteins are rescued by the DnaK·J-GrpE set and ClpB chaperones. Proc Natl Acad Sci 96: 7184–7189. 10.1073/pnas.96.13.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. 2007. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci 104: 2649–2654. 10.1073/pnas.0611503104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder MC, Midtvedt D, Franzmann T, Nüske E, Otto O, Herbig M, Ulbricht E, Müller P, Taubenberger A, Maharana S, et al. 2016. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5: e09347 10.7554/eLife.09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci 102: 10575–10580. 10.1073/pnas.0504882102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci 106: 10147–10152. 10.1073/pnas.0812771106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby GA, Lindquist S. 2013. Blessings in disguise: Biological benefits of prion-like mechanisms. Trends Cell Biol 23: 251–259. 10.1016/j.tcb.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Niwa T, Kanamori T, Ueda T, Taguchi H. 2012. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc Natl Acad Sci 109: 8937–8942. 10.1073/pnas.1201380109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JD, Tsechansky M, Royall A, Boutz DR, Ellington AD, Marcotte EM. 2014. A proteomic survey of widespread protein aggregation in yeast. Mol Biosyst 10: 851–861. 10.1039/c3mb70508k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Dunker AK. 2014. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 83: 553–584. 10.1146/annurev-biochem-072711-164947 [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144: 67–78. 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- Orij R, Brul S, Smits GJ. 2011. Intracellular pH is a tightly controlled signal in yeast. Biochim Biophys Acta 1810: 933–944. 10.1016/j.bbagen.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. 2001. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 106: 183–194. 10.1016/S0092-8674(01)00440-8 [DOI] [PubMed] [Google Scholar]
- Otero LJ, Ashe MP, Sachs AB. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J 18: 3153–3163. 10.1093/emboj/18.11.3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Picotti P, Clément-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Röst H, Sun Z, Rinner O, Reiter L, et al. 2013. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 494: 266–270. 10.1038/nature11835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Ströhl F, et al. 2018. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173: 720–734.e15. 10.1016/j.cell.2018.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. 2012. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J 31: 4221–4235. 10.1038/emboj.2012.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. 2012. Order out of disorder: Working cycle of an intrinsically unfolded chaperone. Cell 148: 947–957. 10.1016/j.cell.2012.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. 2017. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168: 1028–1040.e19. 10.1016/j.cell.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff KM, Roberts S, Chilkoti A, Pappu RV. 2018. Advances in understanding stimulus responsive phase behavior of intrinsically disordered protein polymers. J Mol Biol 430: 4619–4635. 10.1016/j.jmb.2018.06.031 [DOI] [PubMed] [Google Scholar]
- Sachsenhauser V, Bardwell JC. 2018. Directed evolution to improve protein folding in vivo. Curr Opin Struct Biol 48: 117–123. 10.1016/j.sbi.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist SL. 1990. HSP104 required for induced thermotolerance. Science 248: 1112–1115. 10.1126/science.2188365 [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. 2000. Molecular basis of a yeast prion species barrier. Cell 100: 277–288. 10.1016/S0092-8674(00)81565-2 [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. 2000. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289: 1317–1321. 10.1126/science.289.5483.1317 [DOI] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shorter J. 2011. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 6: e26319 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shorter J, Southworth DR. 2018. Spiraling in control: Structures and mechanisms of the Hsp104 disaggregase. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a034033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranno A, Koenig I, Borgia MB, Hofmann H, Zosel F, Nettels D, Schuler B. 2014. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc Natl Acad Sci 111: 4874–4879. 10.1073/pnas.1322611111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sormanni P, Vendruscolo M. 2018. Protein solubility predictions using the CamSol method in the study of protein homeostasis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromer T, Ehrnsperger M, Gaestel M, Buchner J. 2003. Analysis of the interaction of small heat shock proteins with unfolding proteins. J Biol Chem 278: 18015–18021. 10.1074/jbc.M301640200 [DOI] [PubMed] [Google Scholar]
- Stromer T, Fischer E, Richter K, Haslbeck M, Buchner J. 2004. Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation: The N-terminal domain is important for oligomer assembly and the binding of unfolding proteins. J Biol Chem 279: 11222–11228. 10.1074/jbc.M310149200 [DOI] [PubMed] [Google Scholar]
- Tapia H, Koshland DE. 2014. Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol 24: 2758–2766. 10.1016/j.cub.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr, Cleveland DW. 2016. Decoding ALS: From genes to mechanism. Nature 539: 197–206. 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. 10.1038/35035005 [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431: 184–187. 10.1038/nature02885 [DOI] [PubMed] [Google Scholar]
- Tuite MF, Serio TR. 2010. The prion hypothesis: From biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol 11: 823–833. 10.1038/nrm3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungelenk S, Moayed F, Ho C-T, Grousl T, Scharf A, Mashaghi A, Tans S, Mayer MP, Mogk A, Bukau B. 2016. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat Commun 7: 13673 10.1038/ncomms13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace EWJ, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. 2018. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174: 688–699.e16. 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MT, Elbaum-Garfinkle S, Holehouse AS, Chen CCH, Feric M, Arnold CB, Priestley RD, Pappu RV, Brangwynne CP. 2017. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat Chem 9: 1118–1125. 10.1038/nchem.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A. 2011. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion 5: 258–262. 10.4161/pri.17748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PD, Pollock DD, Goldstein RA. 2007. Functionality and the evolution of marginal stability in proteins: Inferences from lattice simulations. Evol Bioinform Online 2: 91–101. [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. 2017. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169: 1066–1077.e10. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]