Abstract
Most of the eukaryotic genome is pervasively transcribed, yielding hundreds to thousands of long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs), some of which are well conserved during evolution. Functions have been described for a few lncRNAs and circRNAs but remain elusive for most. Both classes of RNAs play regulatory roles in translation by interacting with messenger RNAs (mRNAs), microRNAs (miRNAs), or mRNA-binding proteins (RBPs), thereby modulating translation in trans. Moreover, although initially defined as noncoding, a number of lncRNAs and circRNAs have recently been reported to contain functional open reading frames (ORFs). Here, we review current understanding of the roles played by lncRNAs and circRNAs in protein synthesis and discuss challenges and open questions in the field.
circRNAs: BIOGENESIS AND CHALLENGES IN DETECTION AND FUNCTIONAL STUDIES
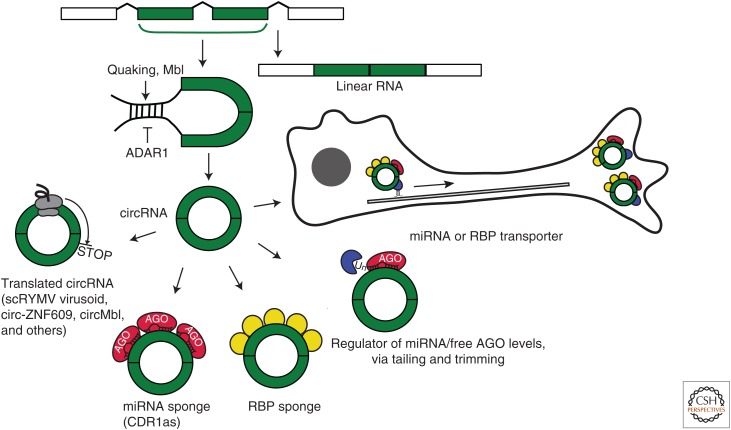
Hundreds to thousands of different circular RNAs (circRNAs) are expressed in eukaryotic cells (reviewed in Rybak-Wolf et al. 2015; Salzman 2016). circRNAs represent an important class of RNAs, created by head-to-tail splicing, or backsplicing, that link the 3′ end of an exon to the 5′ end of an upstream exon (Fig. 1) (Salzman et al. 2012, 2013; Jeck et al. 2013; Memczak et al. 2013). Among other forms of covalent RNA circles are certain RNA genomes (viroids, the hepatitis delta virus) and intron-derived RNAs (reviewed in Lasda and Parker 2014). In addition to canonical splicing factors and signals, backsplicing requires specific intronic sequences that mediate circularization, bringing the downstream donor and upstream acceptor sites into close proximity (Ashwal-Fluss et al. 2014; Zhang et al. 2014). Circularization can be promoted via complementary intronic sequences such as Alu repeats, or through the binding of introns by RNA-binding proteins (RBPs) (Dubin et al. 1995; Jeck et al. 2013; Ashwal-Fluss et al. 2014; Liang and Wilusz 2014; Zhang et al. 2014; Conn et al. 2015; Ivanov et al. 2015). For example, the splicing factor Muscleblind (Mbl) binds to introns flanking the second exon of its own transcript, facilitating circMbl formation (Ashwal-Fluss et al. 2014). It also has been proposed that the RBP Quaking may promote the biogenesis of multiple circRNAs via a similar mechanism by binding to specific sites in the flanking introns (Conn et al. 2015). ADAR1 negatively regulates circRNA biogenesis through A-to-I editing, thereby decreasing RNA base-pairing between the flanking introns (Ivanov et al. 2015).
Figure 1.
Scheme illustrating biogenesis and regulatory roles of circular RNAs (circRNAs) in translation. circRNAs form as a result of backsplicing, when flanking introns are brought into close proximity because of complementarity or through the binding of introns by specific RBPs. circRNAs play regulatory roles in translation, by acting as sponges and possibly transporting microRNAs (miRNAs) and RNA-binding proteins (RBPs), as well as regulating miRNA levels via tailing and trimming. Some circRNAs contain translatable open reading frames (ORFs). Regulatory proteins are shown, and examples are in parentheses. See text for more details.
circRNAs are mostly cytoplasmic (Salzman et al. 2012; Jeck et al. 2013; Memczak et al. 2013; Rybak-Wolf et al. 2015), raising a question as to the mechanism of their nucleocytoplasmic export. Messenger RNA (mRNA) export to the cytoplasm is mediated by the transport/export complex (TREX) that interacts with mRNA primarily via the exon junction complex (EJC), deposited on spliced mRNA (reviewed in Delaleau and Borden 2015). As circRNAs are spliced, their export to the cytoplasm might be mediated via similar mechanisms. As covalent circles, circRNAs are resistant to degradation by RNA exonucleases, and therefore have unusually long half-lives, exceeding 24–48 hours (Jeck et al. 2013; Memczak et al. 2013). That poses another question—how circRNAs are degraded—as it has been observed that during neuronal development, circRNAs appear to be cleared rapidly (Rybak-Wolf et al. 2015). Curiously, the circRNA CDR1as (cerebellar degeneration-related protein 1 antisense) has an almost perfectly complementary binding site for miR-671, leading to slicing of CDR1as (Hansen et al. 2011; Piwecka et al. 2017). Because of high complementarity of the target, miR-671 is destabilized via a mechanism that involves modification of microRNA (miRNA) ends, adding (“tailing”) and removing (“trimming”) nucleotides (reviewed in Duchaine and Fabian 2018). These data provide insight into the mechanisms of circRNA decay and release of the bound cargo. Based on the abundance of circRNAs in extracellular vesicles, Lasda and Parker (2014) suggested that formation of such vesicles might serve as an alternative mechanism for circRNA disposal.
Initially, circRNAs were viewed as splicing artifacts or noise (Cocquerelle et al. 1993; Pasman et al. 1996). This opinion has recently changed in light of a number of findings. First, circRNAs have cell-type-specific expression patterns and are particularly abundant in the brain (Salzman et al. 2013; Rybak-Wolf et al. 2015; Szabo et al. 2015; You et al. 2015). Second, many circRNAs are regulated independently of their linear counterparts, and are well conserved (Jeck et al. 2013; Memczak et al. 2013; Ashwal-Fluss et al. 2014; Rybak-Wolf et al. 2015; You et al. 2015). Also, well-expressed circRNAs have enhanced conservation of nucleotides at third codon positions compared with upstream or downstream codons in bracketing coding sequences (Salzman et al. 2013; Rybak-Wolf et al. 2015; Szabo et al. 2015; You et al. 2015). Finally, biological functions have been reported for some, as discussed in the next section.
The number of circRNAs to which biological functions have been ascribed is limited. One of the reasons is the challenge of depleting circRNAs without affecting their linear counterparts. One method of doing so has been to design small interfering RNAs (siRNAs) that target the circRNA-specific splice junction. However, it is usually not possible to design more than a few siRNAs per junction, which makes it difficult to cope with off-target effects. Uncovering intronic elements that facilitate circRNA biogenesis (Jeck et al. 2013; Ashwal-Fluss et al. 2014; Ivanov et al. 2015) provided a new approach to knocking down circRNAs without affecting linear transcripts. Indeed, disrupting intronic complementary sequences with CRISPR/Cas strongly reduced the levels of circGCN1L1 (Zhang et al. 2016). However, for numerous circRNAs, there are no easily identifiable intronic complementary sequences.
The detection of circRNAs also poses several challenges. They can be identified in RNA sequencing data through reads that map to the backsplicing junction (Glazar et al. 2014; Jeck and Sharpless 2014; Gao et al. 2015). However, some of these reads might be the result of trans-splicing, making it important to validate the circularity of individual candidates by treatment with exonucleases to eliminate linear RNAs. This is also relevant when using vectors that overexpress circRNAs, as they also produce linear transcripts, sometimes as concatemers (Pamudurti et al. 2017), making it challenging to determine which form is responsible for a phenotype.
REGULATORY FUNCTIONS OF circRNAs IN TRANSLATION
The main regulatory function described for circRNAs so far is their action as sponges, which sequester miRNAs (Fig. 1) (Hansen et al. 2013; Memczak et al. 2013). miRNAs function as guides by pairing with partially complementary sites present in their target mRNAs, and subsequently recruiting a complex of proteins that cause mRNA deadenylation, decay, and translational repression (reviewed in Jonas and Izaurralde 2015; Duchaine and Fabian 2018). This means that RNAs with multiple miRNA-binding sites can compete for miRNA binding, leading to the stabilization and translational activation of miRNA-targeted mRNAs. circRNAs are particularly suited for this function because they are abundant and, as covalent circles without a poly(A) tail, they resist miRNA-mediated deadenylation and decay. Two recent studies (Hansen et al. 2013; Memczak et al. 2013) showed that the circRNA CDR1as contains 63 conserved binding sites for miR-7 and depletes miR-7 in neuronal tissues. Indeed, the expression of human CDR1as in zebrafish leads to defects in midbrain development that are reminiscent of an miR-7 knockdown, and a CDR1as knockout mouse showed impaired sensorimotor gating, a deficit associated with neuropsychiatric disorders (Piwecka et al. 2017). Moreover, biochemical miRNA-binding analyses in postmortem brains by “chimera analyses” (Grosswendt et al. 2014) showed that CDR1as is one of the most highly miRNA-bound transcripts in the human and mouse brain. circRNA deriving from the ZNF91 (zinc-finger 91) locus contains 24 miR-23 sites, making it the next best miRNA-sponge candidate (Guo et al. 2014). Another example of a circRNA hypothesized to function as an miRNA sponge is heart-related circRNA (HRCR) (Wang et al. 2016). This has been suggested to play a protective role in cardiac hypertrophy by sequestering miR-223 via its six binding sites. However, bioinformatic analyses suggest that most circRNAs do not function as miRNA sponges, as they do not harbor more AGO2/miRNA-binding sites than linear RNAs (Memczak et al. 2013; Guo et al. 2014; Rybak-Wolf et al. 2015; You et al. 2015). Such a function would also depend on the relative concentrations of the circRNAs and miRNAs in a specific subcellular compartment, but these numbers are difficult to estimate at present, especially in highly polarized cells such as neurons.
It is likely that circRNAs can also function as docking sites for RBPs to sequester them from their target RNAs or mediate their subcellular localization (Fig. 1). Curiously, circRNAs are particularly abundant in synaptosomes (Rybak-Wolf et al. 2015; You et al. 2015; Zappulo et al. 2017), suggesting that they may participate in local RNA regulation. For example, they might transport miRNAs and RBPs to synapses and release them in response to specific stimuli. One possible way of releasing bound factors from a circRNA is if they contain a perfectly complementary site to an miRNA, which can then be cleaved. The roles of circRNAs in the regulation of transcription and splicing are reviewed elsewhere (Ebbesen et al. 2016).
TRANSLATION OF circRNAs
The scanning model of initiation suggested that eukaryotic ribosomes require free 5′ mRNA ends to initiate translation, allowing them to thread onto mRNA like beads on a string (Kozak 1978, 1979; Konarska et al. 1981). This view was challenged by the discovery of internal ribosome entry site (IRES) elements that can directly recruit 40S ribosomal subunits (reviewed in Jackson 2005; Kwan and Thompson 2018). Indeed, an early study by Chen and Sarnow (1995) showed that eukaryotic small ribosomal subunits can initiate translation in vitro from covalent RNA circles containing the encephalomyocarditis virus (EMCV) IRES (Chen and Sarnow 1995). Consistent with this finding, IRES-bearing circRNAs generated from a reporter plasmid via backsplicing produced proteins (Wang and Wang 2015). Another study suggested that an artificial circRNA with an infinite open reading frame (ORF), that is, lacking a stop codon, can be translated even in the absence of a detectable IRES (Abe et al. 2015).
Several recent studies have reported that endogenously produced circRNAs can be translated in vivo (AbouHaidar et al. 2014; Legnini et al. 2017; Pamudurti et al. 2017; Yang et al. 2017). AbouHaidar et al. (2014) detected the translation product of a circular satellite RNA virusoid associated with rice yellow mottle virus (scRYMV) in a wheat germ in vitro translation system. The protein corresponds to an unusual ORF within scRYMV, combining initiation and termination codons in a UGAUGA sequence.
Another example of circRNA translation is provided by a circular form of the ZNF609 transcript (circ-ZNF609) that regulates myoblast proliferation (Legnini et al. 2017). As circ-ZNF609 contains an ORF that overlaps with the main ORF of linear ZNF609, the investigators tested whether the circular form could be translated. They tagged the endogenous ZNF609 locus with a Flag tag in a way such that the Flag peptide would be produced only from the circular form of the transcript. Mass spectrometry analysis of anti-Flag immunoprecipitates identified a ZNF609-specific peptide, supporting the idea of circ-ZNF609 translation. The mechanism by which this occurs is unclear, but it is ∼100-fold less efficient than the translation of its linear counterpart and is dependent on splicing. Importantly, the investigators observed a strong increase in the translation of circ-Flag-ZNF609 on heat-shock stress. Stress leads to eIF4G proteolysis, separating its cap-binding function from its helicase and ribosome-binding activity (reviewed in Holcik and Sonenberg 2005). As a result, cap-dependent translation is inhibited, and the translation of IRES-containing mRNAs is promoted. This result led the investigators to speculate that the translation of circ-ZNF609 might be low by default but activated in response to specific stimuli that inhibit cap-dependent translation.
The study of Pamudurti et al. (2017) searched for ribosome-associated circRNAs (ribo-circRNAs) in the heads of Drosophila using ribosome profiling (Riboseq), a technique that can yield a genome-wide snapshot of ribosome footprints on mRNAs (Ingolia et al. 2009, 2018). As circRNAs contain a unique head-to-tail splice junction that is not present in their linear counterparts, they can be distinguished from linear transcripts in RNAseq and Riboseq data. To distinguish reads reflecting bona fide translation from background signals, the authors examined the pattern of Riboseq reads across the circRNA-specific splice junction. A known hallmark of translation is the three-nucleotide (nt) phasing of Riboseq reads, which reflects the codon-by-codon movement of translating ribosomes (Calviello et al. 2016). In addition, accumulation of reads around start and stop codons is often observed for translated ORFs. As start codons and most of the ORF are usually shared between the linear and circular transcripts, Pamudurti et al. (2017) concentrated on the stop codons. Indeed, an enrichment of Riboseq reads around the putative stop codon of the most abundant ribo-circRNA, circMbl, supported the translation of this circRNA. The mbl locus produces several circRNAs (Ashwal-Fluss et al. 2014). The authors used targeted mass spectrometry to search for peptides that could be produced from these circRNAs. An analysis of MBL immunoprecipitates identified a peptide that could only be produced by circMbl3, but not by linear Mbl. In findings similar to those of Legnini et al. (2017), Pamudurti et al. (2017) suggested that translation of circMbl is modulated in response to specific stimuli that interfere with cap-dependent translation.
A study by Yang et al. (2017) suggested that the translation of circRNAs might be driven by m6A RNA modifications (m6A effects on protein synthesis are reviewed in Peer et al. 2018). The authors generated split GFP reporters that would restore the GFP ORF on circularization. Surprisingly, GFP protein was detected for all of the transfected constructs, independently of whether they contained an IRES element or not. Yang et al. (2017) hypothesized that this is caused by the m6A motif RRACH (R = G or A; H = A, C, or U) (Csepany et al. 1990; Harper et al. 1990), which had by chance been included in all the plasmids. Mutations of the corresponding motifs reduced levels of GFP. To search for endogenous circRNAs that were potentially translated in human 293 cells, the authors employed three different approaches: (1) identification of m6A-modified circRNAs; (2) searching for circRNAs associated with polysomes; and (3) looking for peptides matching ORF-containing circRNAs. Each approach resulted in dozens to hundreds of potential candidates, although it is difficult to estimate the degree of overlap between the approaches, due to differences in the sensitivity of the methods used and the usage of different cell lines for different types of analyses. The underlying mechanisms of translation remain mysterious. Based on small hairpin RNA (shRNA) depletion and coimmunoprecipitation experiments, Yang et al. (2017) suggested that the m6A-binding protein YTH3 recruits eIF4G2/DAP5 to promote cap-independent initiation. This is plausible given a study from Liberman et al. (2015), which showed that eIF4G2, instead of canonical eIF4G, takes part in initiation at a subset of cellular IRESes.
Interestingly, an earlier study of Meyer et al. (2015) used in vitro translation systems to show that m6A in 5′UTRs (untranslated regions) can indeed initiate translation independently of the cap, via the recruitment of eIF3. However, this mechanism did not involve internal ribosome entry and required free 5′ ends. Meyer et al. (2015) showed this in several types of experiments. First, by incubating m6A-containing mRNAs bearing two start codons using 40S subunits, eIF1, -1A, -2 and -3, and Met-tRNAiMet, they showed that 48S complexes assemble almost exclusively at the first AUG. This outcome can be easily explained if the preinitiation complex is recruited to the 5′ end and initiates at the first suitable start codon, but it is unlikely to result from internal-entry initiation. Second, when translated in HeLa lysates, an m6A-modified reporter mRNA with two in-frame start codons produced almost exclusively the longer protein product, which relied on initiation at the first AUG. Finally, the insertion of a stable hairpin at the extreme 5′ end to block 5′ end-dependent initiation inhibited the translation of the m6A reporter. Interestingly, a similar mode of initiation—independent of the cap, but dependent on the 5′-end—was described for an mRNA reporter carrying an eIF4G-binding domain from the EMCV IRES (Terenin et al. 2013), indicating that simply binding to the 40S subunit is not sufficient for internal initiation—an IRES is also providing for a mechanism for internal entry. These data suggested that m6A alone is rather unlikely to drive translation from circRNAs.
Yet another mechanism, involving the recruitment of the YHT-domain containing family protein 1 (YTHDF1), has been proposed to explain the involvement of 3′UTR-located m6A in translation (Wang et al. 2015). The authors detected a number of proteins among interactors of YTHDF1, including eIF3 subunits, YBX1, IGF2BP1, G3BP1, and PCBP2, and suggested that they might function collectively to affect the translation of methylated mRNA. So, there is no consensus on the matter whether m6A can drive internal initiation and by which mechanisms.
To sum up, several recent studies have provided evidence that some circRNAs can be translated. The evidence includes the presence of ORFs, association with polysomes, enrichment of Riboseq reads around putative stop codons, and identification of peptides corresponding to circRNA sequences based on mass spectrometry. Translated circRNAs possess special properties such as a high stability and independence from the 5′ cap and poly(A) tail that are required for the translation of most eukaryotic mRNAs. Thus, circRNAs could have adapted in ways that lead to translation in special cases where these properties are advantageous. Indeed, reports indicate that the translation of circRNAs is inefficient under normal conditions and is preferentially activated when cap-dependent translation is inhibited (Legnini et al. 2017; Pamudurti et al. 2017). A number of mechanistic questions remain to be resolved through future studies. What drives initiation on circRNAs, and which translation factors are required? If a ribosome terminates translation on a circRNA, does it reinitiate on the same message? Do circRNAs contain ORFs lacking an in-frame termination codon, producing long repetitive proteins as a result of multiple rounds of translation? How is the translation of circRNAs regulated? The functions of the proteins produced from circRNAs remain unclear as well. One point to note is that many circRNAs share the 5′ part of their potential ORFs with their linear counterparts, which would give them the potential to encode proteins containing only the amino-terminal portion of the protein and to function as dominant negatives.
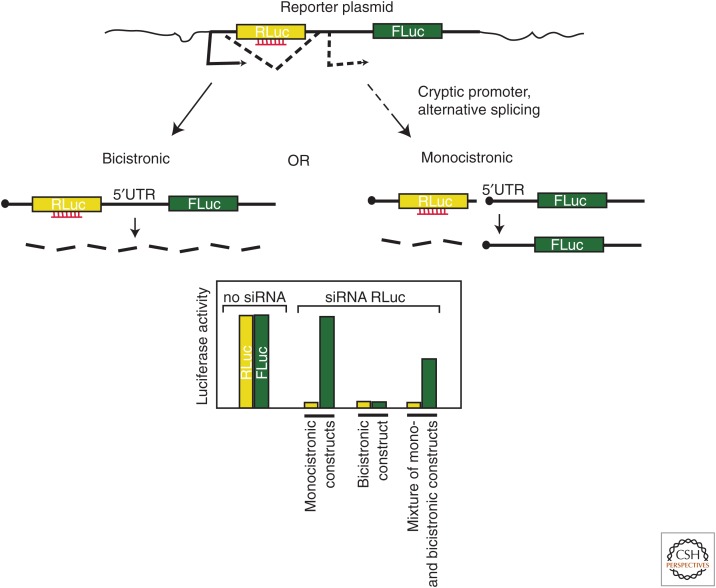
Rigorous technical procedures will be required to dissect the mechanisms underlying circRNA translation. For example, Pamudurti et al. (2017) observed that circRNA plasmid reporter constructs tend to generate not only circRNAs, but also linear concatemers that encode the same peptides. This makes it important to test the true circularity of the RNA products by using RNase R treatment. It is also essential to include a test for internal initiation, such as a bicistronic assay and/or in vitro circularized RNA, instead of relying exclusively on the cap-independency test (reviewed in Terenin et al. 2017). One must take into account the fact that bicistronic assays are prone to artifacts that result from cryptic promoters or splicing events that produce monocistronic capped mRNAs. Lloyd and colleagues (Van Eden et al. 2004) proposed a stringent test to assess the integrity of bicistronic constructs (Fig. 2): RNAi against the first cistron is performed on cells transfected with either the bicistronic construct or two corresponding monocistronic constructs. In the first case, the production of proteins from both cistrons is expected to be equally reduced. In the second case, the production of the protein produced from the first cistron will be reduced, but the second will remain unaffected. If the result for a bicistronic construct lies somewhere in between (i.e., the product of the second cistron is reduced, but less efficiently than that of the first), this probably means that some amount of monocistronic, possibly capped, mRNA has been produced from the second cistron. Given the fact that tested circRNAs and the corresponding bicistronic reporters have generally produced low amounts of protein, it is particularly important to control for the integrity of the mRNAs that are analyzed. For the same reason, assessing the ratios of two cistrons through qPCR does not suffice: even if capped monocistronic mRNA represents such a small percentage of bicistronic mRNA that it cannot be detected by qPCR (∼1%–2%), it could still be responsible for most of the protein product.
Figure 2.
Small interfering RNA (siRNA) test for the integrity of bicistronic reporters. (Top) Transcription gives rise to a bicistronic mRNA (left) or to two monocistronic mRNAs (right), with the RNAi fates depicted. (Bottom) Predicted results of luciferase assays. See text for details.
ANNOTATION OF lncRNAs
Long noncoding RNAs (lncRNAs) are broadly defined as noncoding RNAs longer than 200 nt, to distinguish them from transfer RNAs (tRNAs), miRNAs, Piwi-interacting (piRNAs), and other classes of noncoding RNAs (reviewed in Perry and Ulitsky 2016). lncRNAs are classified based on their location relative to protein-coding genes: most are encoded by intergenic regions (termed lincRNAs for long intergenic, or intervening, RNAs), but some overlap with protein-coding genes. Although thousands of loci in vertebrates produce lncRNAs, biological functions have been ascribed to only a small fraction of them. lncRNAs exhibit lower levels of synthesis, processing, and stability than mRNAs (Mukherjee et al. 2017). They also exhibit a lower level of conservation than protein-coding genes. For example, of ∼1000 lncRNAs that are moderately to highly expressed in humans, only ∼300 are conserved outside mammals in other vertebrates (Hezroni et al. 2015). Despite the popular belief that lncRNAs are primarily nuclear, recent studies showed that they exhibit a variety of localization patterns, from nuclear to almost exclusively cytoplasmic localization (Derrien et al. 2012; Djebali et al. 2012; Cabili et al. 2015; Mukherjee et al. 2017). Thus, analysis of 31 lncRNAs in three cell lines with single-molecule RNA-FISH showed that ∼55% of them were mostly nuclear, ∼40% exhibited both nuclear and cytoplasmic localization, and ∼5% were mostly cytoplasmic. Consistently, analysis of RNAseq data showed that the nuclear/cytoplasmic ratio is moderately higher for lncRNAs than for mRNAs (Derrien et al. 2012; Djebali et al. 2012; Cabili et al. 2015; Mukherjee et al. 2017). A number of lncRNAs have been implicated in the regulation of cell proliferation, apoptosis, response to stress, and other processes. Based on the mechanisms by which they function, lncRNAs can be loosely divided into three groups (reviewed in Perry and Ulitsky 2016). The first group includes lncRNAs for which only the fact of transcription itself is important, for example, through modification of chromatin in the locus. The second group comprises cis-acting lncRNAs that accomplish their functions by attracting trans-acting factors to the chromatin locus, thereby modulating transcription. The third group, trans-acting lncRNAs, function independently of their transcription sites by recruiting other RNAs or RBPs. Some of these lncRNAs play a role in the regulation of mRNA translation and stability. We discuss this group in more detail below.
REGULATORY ROLES OF lncRNAs IN TRANSLATION
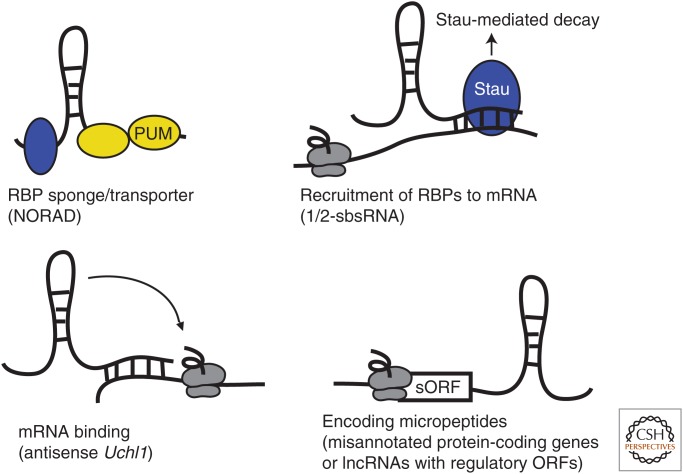
Some lncRNAs sequester and modulate the activity of RBPs and thus affect translation and stability of the mRNAs that these proteins target (Fig. 3). For example, recent studies (Lee et al. 2016; Tichon et al. 2016) showed that the lncRNA NORAD (noncoding RNA activated by DNA damage) harbors 17 binding sites for the mammalian PUF family proteins Pumilio1 and Pumilio2. PUF family proteins are conserved from yeast to animals and plants, in which they deadenylate and repress the translation of their target mRNAs (reviewed in Miller and Olivas 2011). Because Pumilio targets are enriched in genes involved in chromosome segregation, the sequestration of Pumilio by NORAD plays an important role in maintaining genomic stability. The importance of a precise regulation of Pumilio levels can be gleaned from observations that Pumilio1 haploinsufficiency leads to neurodegeneration in mice (Gennarino et al. 2015).
Figure 3.
Scheme illustrating regulatory roles of long noncoding RNAs (lncRNAs) in translation. lncRNAs act in sequestering messenger RNA (mRNA)-binding proteins (RBPs), facilitating recruitment of RBPs to mRNAs, or interacting with mRNAs directly. A number of annotated lncRNAs encode micropeptides and thus represent bona fide protein-coding genes. See text for more details. sORF, Short ORF.
Carrieri et al. (2012) have described a mechanism by which antisense lncRNA can activate translation. Antisense Uchl1 is a neuron-specific lncRNA transcribed from the opposite strand of the protein-coding gene Uchl1. UCHL1 protein is a deubiquitinating enzyme whose loss leads to ataxia and axonal degeneration (Saigoh et al. 1999). Antisense Uchl1 regulates protein synthesis through the combined activities of two domains: the 5′ antisense region provides specificity through its complementarity to the sense mRNA, and a SINEB2 repeat confers translational activation via an unknown mechanism. The localization of antisense Uchl1 to the cytoplasm is controlled by signaling pathways such as mammalian target of rapamycin (mTOR) (Carrieri et al. 2012). The authors suggest that antisense Uchl1 belongs to a new functional class of antisense lncRNAs that act via similar mechanisms involving SINEB2 repeats (Fig. 3).
lncRNAs have also been reported to function as trans-acting factors in Staufen-mediated mRNA decay (SMD) (Gong and Maquat 2011). SMD degrades translationally active mRNAs bound by the dsRNA-binding protein Staufen. Staufen-binding sites can be formed through imperfect base-pairing between an Alu element in the 3′UTR of the target mRNA and an Alu element in a lncRNA. For example, the Alu elements within SERPINE1 and FLJ21870 3′UTRs have the potential to base-pair with the lncRNA AF087999 (designated as 1/2-sbsRNA). This finding shows another functional strategy employed by lncRNAs in modulating mRNA metabolism by helping to recruit specific RBPs (Fig. 3).
TRANSLATION OF lncRNAs
By definition, noncoding RNAs lack conserved protein-coding reading frames. Yet, a number of recent studies have reported the translation of RNAs formerly annotated as noncoding (Guttman et al. 2013; Slavoff et al. 2013; Ingolia et al. 2014; Prabakaran et al. 2014; Mackowiak et al. 2015; D’Lima et al. 2017; for reviews, see Andrews and Rothnagel 2014; Saghatelian and Couso 2015). Several approaches have been used to find RNAs with coding potential among sequences previously assumed to be noncoding: (1) a computational search for translatable ORFs and evolutionary conservation analysis; (2) ribosome profiling that uses filters to detect known hallmarks of translation; and (3) mass spectrometry to identify peptides encoded by ORFs that are found exclusively in RNAs that have been annotated as noncoding.
sORFs IN ANNOTATED NONCODING TRANSCRIPTS
An ORF is defined as a continuous stretch of in-frame sense codons that begins with a start codon and ends with a stop codon. However, not all ORFs are translated and defining the parameters that distinguish those that are translated from those that are not has been challenging. Historically, protein-coding genes were discovered using strategies that assumed that true ORFs would be relatively long, usually >100 codons (Claverie 1997). Such strategies were based on the assumptions that (1) shorter ORFs can occur through random processes, and (2) peptides with fewer than 100 amino acids are unlikely to form structures stable enough to perform biological functions. A more recent algorithm, PhyloCSF, is based on conservation and broadens the size range used to predict coding ORFs (Lin et al. 2011; Bazzini et al. 2014). This fits with an increasing body of evidence demonstrating that short ORFs (sORFs) can be translated to produce proteins with important biological functions (reviewed in Andrews and Rothnagel 2014; Saghatelian and Couso 2015).
Such findings have motivated recent studies to search for sORFs among RNAs that have long been annotated as noncoding. An analysis of intergenic regions has identified ∼41,000 sORFs in mice (Frith et al. 2006) and between 7159 (Hanada et al. 2007) and 33,809 (Lease and Walker 2006) in Arabidopsis. Taking into account additional parameters, such as evolutionary conservation and a sequence’s location in transcribed regions, led to predictions that ∼5% of these sORFs might be translated (Hanada et al. 2007). Recent work (Mackowiak et al. 2015) offered an integrated computational pipeline for the identification of conserved sORFs in human, mouse, zebrafish, Drosophila, and Caenorhabditis elegans. By filtering potential amino acid sequences for conservation, the authors identified ∼2000 sORFs in mRNA 3′ and 5′UTRs and in RNAs that had been annotated as noncoding. Cross-checking this prediction with experimental ribosome profiling data has offered evidence in support of translation for 110 sORFs, and peptidomic evidence has been found for 74 sORFs, 36 of which were mapped to sequences annotated as lncRNAs. Interestingly, the peptides they encode are enriched in disordered regions and short linear interaction motifs that match motifs from the ELM (eukaryotic linear motifs) database (Dinkel et al. 2016).
EVIDENCE FROM RIBOSOMAL PROFILING
The computational prediction of ORFs is a powerful technique that is most efficient for conserved ORFs under selective pressure to preserve a specific amino acid sequence that results in a functional protein. However, this approach fails to detect species-specific and very short ORFs. Translation could play a regulatory role by modulating RNA stability, for example, in which case no selective pressure on the protein sequence is expected. Therefore, ribosome profiling, a technique that identifies ribosome-protected RNA fragments (Ingolia et al. 2009), has been used to map ORFs that are translated within RNAs that have been annotated as noncoding. The first studies showed that ribosome occupancy per se is not sufficient to define translated RNAs. For nearly half of the lncRNAs, the density of ribosome footprints was similar to known protein-coding transcripts (Ingolia et al. 2011). To distinguish between ribosome profiling reads that reflect bona fide translation from the background signal, a number of additional criteria were suggested. One approach was based on the fact that ribosomes generally dissociate from mRNA at the end of the coding sequence after translation. Guttman et al. (2013) introduced a ribosome release score (RRS), defined as the ratio between the number of reads within the putative ORF and those within the 3′UTR. For lncRNAs, this produced a median RRS ∼100 times lower than for known protein-coding RNAs, suggesting that lncRNAs are not translated. An important parameter in separating the true ribosome footprints and background reads resulting from secondary structures and nonribosomal RNPs is the observation that ribosome-protected RNA fragments have a characteristic length of 28–30 nt (Ingolia et al. 2014). To account for this, Ingolia et al. (2014) introduced a fragment length organization similarity score (FLOSS), which reflects how well the lengths of fragments that are detected agree with true 80S ribosome footprints. Several studies further pointed to the 3-nt periodicity of ribosome profiling reads, reflecting the codon-by-codon movement of ribosomes along mRNA during translation (Michel et al. 2012; Bazzini et al. 2014; Duncan and Mata 2014). This property led Calviello et al. (2016) to develop RiboTaper, a computational tool intended to identify translated ORFs with a high degree of confidence. RiboTaper drew on HEK293 cell ribosome profiling data to identify ORFs in 504 noncoding genes, including pseudogenes, antisense, and lncRNAs. Interestingly, these ORFs were generally nonconserved, suggesting that their translation might be playing a regulatory role rather than resulting in the production of stable functional proteins.
PROTEOMIC EVIDENCE AND THE FUNCTIONALITY OF MICROPEPTIDES
Proof that a given ORF is indeed translated is the detection of the corresponding peptides through mass spectrometry. Banfai et al. (2012) analyzed two data sets produced by the Encyclopedia of DNA elements (ENCODE) project involving tandem mass spectrometry (MS/MS) and RNAseq data for the cell lines K562 and GM12878. The authors detected matching peptides for 69 lncRNAs, amounting to ∼8% of annotated lncRNAs, concluding that most of the cases represented protein-coding genes that had been incorrectly annotated as noncoding. Slavoff et al. (2013) performed peptidomic analysis to detect products of sORFs in human cells. A small fraction of the peptides that were detected matched annotated lincRNAs (8 of 1866). A study by Prabakaran et al. (2014) compared transcriptomic and proteomic data from mouse cortical neurons. This analysis identified 250 unique peptides that mapped to regions that had been annotated as noncoding, including UTRs of known protein-coding genes, pseudogenes, introns, antisense RNAs, and intergenic regions. Of 25 peptides that mapped to intergenic regions, three were also found in the ribosomal profiling data of Ingolia et al. (2011). It should be noted that proteomic approaches are generally less sensitive than transcriptomic because of the PCR-based amplification step in RNAseq protocols. Indeed, even for known protein-coding transcripts, proteomic evidence is normally obtained for only 25%–30% of the transcripts typically detected by RNAseq-based methods. Extrapolating from this observation, the real number of proteins that originate from annotated noncoding RNAs can be expected to be 3–4 times higher than currently confirmed by proteomics studies.
Importantly, some proteins that originated from intergenic regions and RNAs annotated as noncoding were reported to have biological functions. Hanada et al. (2013) overexpressed 473 coding intergenic sORFs in Arabidopsis, of which 49 produced visible phenotypes, suggesting that the peptides played some role in plant development. A clear functional example comes from the peptide produced by four tandem sORFs within the Drosophila tarsal-less (tal) lncRNA (Galindo et al. 2007; Kondo et al. 2007, 2010). Mutations that disrupt the translation of these sORFs lead to an embryonic lethal phenotype similar to mutations in the transcription factor Ovo/Svb (Kondo et al. 2007, 2010). Tal peptides trigger the amino-terminal truncation of the Ovo protein, which converts it from a repressor to an activator. Interestingly, homologs of tal have been found in insect species but not in vertebrates (Li et al. 2002). These functional data have led to a recent reclassification of tal as a polycistronic mRNA.
The study of Magny et al. (2013) described two peptides, sarcolamban A and B, that are encoded by the putative Drosophila noncoding RNA pncr003:2L. These peptides are conserved from flies to humans and function as regulators of calcium transport and cardiac muscle contraction. Myoregulin (MLN) and dwarf open reading frame (DWORF) are recently discovered micropeptides encoded by annotated lncRNAs that have turned out to be important for muscle function (Anderson et al. 2015; Nelson et al. 2016). Muscle contraction and relaxation depend on the release of Ca2+ from the sarcoplasmic reticulum (SR) and its reuptake by the membrane pump SERCA. MLN functions as a SERCA inhibitor, together with homologous proteins phospholamban (PLN) and sarcolipin (SLN). DWORF, on the other hand, enhances SERCA activity by displacing SERCA inhibitors. Mice lacking MLN show improved performance in exercise, and the deletion of DWORF leads to delays in Ca2+ clearance and relaxation.
D’Lima et al. (2017) used proteomics approaches to identify a small protein they called NoBody, for nonannotated P-body dissociating polypeptide. NoBody is encoded by LINC01420/LOC550643 RNA and is specific to mammals. The protein localizes to P bodies and interacts with components of the decapping machinery. Its overexpression resulted in the dissolution of P bodies, suggesting that NoBody has a function in the regulation of mRNA metabolism.
Curiously, mitochondrial 12S rRNA was also reported to contain a functional sORF encoding a 16-amino-acid peptide MOTS-c (mitochondrial ORF of the 12S rRNA-c) (Lee et al. 2015). MOTS-c inhibits the folate cycle primarily in skeletal muscles, thereby regulating insulin sensitivity and metabolic homeostasis.
To summarize, annotated lncRNAs can be divided into three groups based on their status with regard to translation: (1) actual lncRNAs that do not encode proteins; (2) lncRNAs with conserved sORFs that are protein-coding genes misannotated as lncRNAs because the proteins they encode are shorter than the limit of 100 amino acids used as a benchmark; and (3) lncRNAs with nonconserved sORFs. The question of functions is most interesting and controversial for the last group. A minor but important proportion of such nonconserved sORFs encode functional micropeptides, but the lack of conservation suggests that for most of them their translation is likely to play a regulatory rather than coding role, for example by modulating RNA stability and/or localization. Smith et al. (2014) showed that translation of sORFs from annotated lncRNAs in yeast targets them for nonsense-mediated RNA decay (reviewed in Karousis and Mühlemann 2018). Such sORFs could also represent intermediate steps in de novo protein evolution (Carvunis et al. 2012; Ruiz-Orera et al. 2014), and it is certainly possible that some lncRNAs with nonconserved sORFs play the dual role of encoding functional peptides and regulating the fates of other RNAs.
CONCLUSIONS AND FUTURE PROSPECTS
In conclusion, there is solid evidence that some lncRNAs and circRNAs play regulatory roles in translation. Such functions involve interactions with trans-acting translational regulators, such as RBPs and miRNAs. These interactions can have diverse consequences for the sequestration, storage, or intracellular localization of trans-acting factors. Moreover, lncRNAs can directly interact with mRNAs and regulate their fates by recruiting specific RBPs. More regulatory mechanisms undoubtedly remain to be discovered.
Besides playing regulatory roles in translation, some circRNAs are translated in vivo, but future work is needed to test whether the products of this translation are functionally important. As circRNAs lack a cap and poly(A) tail, they may have adapted to undergo translation in special situations when cap-dependent translation is inhibited. In the future, it will be important to intensify efforts toward understanding the mechanisms that govern circRNA translation. The translation of sORFs has also been reported for a number of RNAs annotated as lncRNAs. Some represent missannotations of protein-coding genes that undergo corrections when corresponding peptides are found but the generally low conservation of lncRNA sORFs makes it likely that their translation plays more of a regulatory role. Further work is needed to understand the significance of this process. Clearly, the future will see the identification of novel interaction partners for circRNAs and lncRNAs and increasing clarity in defining their functions.
ACKNOWLEDGMENTS
We are grateful to the BIMSB faculty, Sergey Dmitriev (MSU), and Russ Hodge (MDC) for comments on the manuscript. We apologize to authors whose work could not be cited in this review because of size limitations.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al. 2015. Rolling circle translation of circular RNA in living human cells. Sci Rep 5: 16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. 2014. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci 111: 14542–14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. 2015. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SJ, Rothnagel JA. 2014. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 15: 193–204. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56: 55–66. [DOI] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22: 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. 2015. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, Ohler U. 2016. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 13: 165–170. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. 2012. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–457. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. [DOI] [PubMed] [Google Scholar]
- Claverie JM. 1997. Computational methods for the identification of genes in vertebrate genomic sequences. Hum Mol Genet 6: 1735–1744. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. 1993. Mis-splicing yields circular RNA molecules. FASEB J 7: 155–160. [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134. [DOI] [PubMed] [Google Scholar]
- Csepany T, Lin A, Baldick CJ Jr, Beemon K. 1990. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem 265: 20117–20122. [PubMed] [Google Scholar]
- Delaleau M, Borden KL. 2015. Multiple export mechanisms for mRNAs. Cells 4: 452–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Van Roey K, Michael S, Kumar M, Uyar B, Altenberg B, Milchevskaya V, Schneider M, Kuhn H, Behrendt A, et al. 2016. ELM 2016—Data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res 44: D294–D300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke-Andersen J, Saghatelian A, Slavoff SA. 2017. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 13: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H. 1995. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167: 245–248. [DOI] [PubMed] [Google Scholar]
- *.Duchaine TF, Fabian MR. 2018. Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CD, Mata J. 2014. The translational landscape of fission-yeast meiosis and sporulation. Nat Struct Mol Biol 21: 641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen KK, Kjems J, Hansen TB. 2016. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 1859: 163–168. [DOI] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM. 2006. The abundance of short proteins in the mammalian proteome. PLoS Genet 2: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zhao F. 2015. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim JY, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, et al. 2015. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell 160: 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, Rajewsky N. 2014. circBase: A database for circular RNAs. RNA 20: 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. 2007. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17: 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. 2013. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci 110: 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. 2011. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30: 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL. 1990. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res 18: 5735–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. 2015. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 11: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. 2015. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10: 170–177. [DOI] [PubMed] [Google Scholar]
- Jackson RJ. 2005. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans 33: 1231–1241. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. [DOI] [PubMed] [Google Scholar]
- *.Karousis ED, Mühlemann O. 2018. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M, Filipowicz W, Domdey H, Gross HJ. 1981. Binding of ribosomes to linear and circular forms of the 5′-terminal leader fragment of tobacco-mosaic-virus RNA. Eur J Biochem 114: 221–227. [DOI] [PubMed] [Google Scholar]
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. 2007. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9: 660–665. [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1978. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell 15: 1109–1123. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1979. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature 280: 82–85. [DOI] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R. 2014. Circular RNAs: Diversity of form and function. RNA 20: 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease KA, Walker JC. 2006. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, et al. 2015. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 21: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. 2016. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. 2017. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66: 22–37.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dai Q, Li L, Nair M, Mackay DR, Dai X. 2002. Ovol2, a mammalian homolog of Drosophila ovo: Gene structure, chromosomal mapping, and aberrant expression in blind-sterile mice. Genomics 80: 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. 2014. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A, Sonenberg N. 2015. DAP5 associates with eIF2β and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res 43: 3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak SD, Zauber H, Bielow C, Thiel D, Kutz K, Calviello L, Mastrobuoni G, Rajewsky N, Kempa S, Selbach M, et al. 2015. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol 16: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. 2013. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. 2012. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res 22: 2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Olivas WM. 2011. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA 2: 471–492. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Calviello L, Hirsekorn A, de Pretis S, Pelizzola M, Ohler U. 2017. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol 24: 86–96. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, et al. 2016. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351: 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. 2017. Translation of CircRNAs. Mol Cell 66: 9–21.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman Z, Been MD, Garcia-Blanco MA. 1996. Exon circularization in mammalian nuclear extracts. RNA 2: 603–610. [PMC free article] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RB, Ulitsky I. 2016. The functions of long noncoding RNAs in development and stem cells. Development 143: 3882–3894. [DOI] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357: eaam8526. [DOI] [PubMed] [Google Scholar]
- Prabakaran S, Hemberg M, Chauhan R, Winter D, Tweedie-Cullen RY, Dittrich C, Hong E, Gunawardena J, Steen H, Kreiman G, et al. 2014. Quantitative profiling of peptides from RNAs classified as noncoding. Nat Commun 5: 5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. 2014. Long non-coding RNAs as a source of new peptides. eLife 3: e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58: 870–885. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Couso JP. 2015. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol 11: 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, et al. 1999. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet 23: 47–51. [DOI] [PubMed] [Google Scholar]
- Salzman J. 2016. Circular RNA expression: Its potential regulation and function. Trends Genet 32: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7: e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9: e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. 2013. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 9: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, Coller J, Baker KE. 2014. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep 7: 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. 2015. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Andreev DE, Dmitriev SE, Shatsky IN. 2013. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res 41: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Smirnova VV, Andreev DE, Dmitriev SE, Shatsky IN. 2017. A researcher’s guide to the galaxy of IRESs. Cell Mol Life Sci 74: 1431–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I. 2016. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun 7: 12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. 2004. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 10: 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z. 2015. Efficient backsplicing produces translatable circular mRNAs. RNA 21: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. 2016. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37: 2602–2611. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. 2017. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18: 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulo A, van den Bruck D, Ciolli Mattioli C, Franke V, Imami K, McShane E, Moreno-Estelles M, Calviello L, Filipchyk A, Peguero-Sanchez E, et al. 2017. RNA localization is a key determinant of neurite-enriched proteome. Nat Commun 8: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. 2014. Complementary sequence-mediated exon circularization. Cell 159: 134–147. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. 2016. The biogenesis of nascent circular RNAs. Cell Rep 15: 611–624. [DOI] [PubMed] [Google Scholar]