Abstract
Phosphorous is important for life but often limiting for plants. The symbiotic pathway of phosphate uptake via arbuscular mycorrhizal fungi (AMF) is evolutionarily ancient and today occurs in natural and agricultural ecosystems alike. Plants capable of this symbiosis can obtain up to all of the phosphate from symbiotic fungi, and this offers potential means to develop crops less dependent on unsustainable P fertilizers. Here, we review the mechanisms and insights gleaned from the fine-tuned signal exchanges that orchestrate the intimate mutualistic symbiosis between plants and AMF. As the currency of trade, nutrients have signaling functions beyond being the nutritional goal of mutualism. We propose that such signaling roles and metabolic reprogramming may represent commitments for a mutualistic symbiosis that act across the stages of symbiosis development.
Phosphorous is crucial for life by virtue of its unique chemistry. Phosphate is capable of chemical bonds that confer remarkable stability but at the same time facile manipulation (Westheimer 1987). Stable phosphodiester bonds enable the encoding of genetic information in nucleic acids. On the other hand, rapid and reversible transfer of phosphoryl groups onto proteins by kinases and phosphatases allow the regulation of cellular signaling processes and core metabolism (e.g., via ATP, GTP) (Westheimer 1987; Hunter 2012; Kamerlin et al. 2013). Phosphates also build phospholipid bilayers in membranes, enabling compartmentalization, electrophysiology, and signaling between organelles, cells, tissues, and organisms.
Plants acquire phosphate mainly in the form of inorganic orthophosphate (Pi) by the roots (Schachtman et al. 1998; Raghothama 1999; Marschner 2011). However, a substantial amount of phosphate in the soil remains inaccessible, precipitated with cations, or assimilated into organic compounds by microorganisms (Schachtman et al. 1998). Thus, a depletion zone of nutrients develops rapidly around the roots. To obtain the Pi necessary for growth, plants evolved intricate and coordinated responses to enhance Pi acquisition, while simultaneously conserving and remobilizing internal Pi stores (Raghothama 1999; Chiou and Lin 2011). Apart from direct uptake, plants also engage in an underground resource exchange, acquiring Pi via the indirect symbiotic pathway (Smith et al. 2003; Smith and Read 2008). Therefore, in the rhizosphere, the narrow region of soil surrounding the roots, plants, and microorganisms interact and reciprocally influence each other's fitness and biomass.
In this review, we explore the role of and our current understanding on symbiotic phosphate acquisition, from the underlying molecular mechanisms to the physiological, ecological, and agricultural implications; outline existing knowledge gaps and finally, propose that like other symbioses, metabolic reprogramming and dependency of both symbionts is crucial in the signaling and stability of a mutually beneficial symbiosis.
SYMBIOSIS AS THE NORM
In natural and agricultural ecosystems, plants exist in intimate associations with a plethora of microorganisms. Symbiosis, sensu de Bary (1879), is the rule (de Bary 1879; Oulhen et al. 2016). Bacteria, archaea, fungi, oomycetes, metazoans, and viruses reside in the rhizosphere (Vorholt 2012; Philippot et al. 2013), and the corresponding symbiotic outcomes exist along a spectrum—with respect to their effect on the host—from parasitism, to commensalism, to mutualism (Box 1 describes some of these associations). Adding to this is the growing nuance that symbiotic outcomes are context-dependent—the same host–symbiont combination can be mutually beneficial in one case, but parasitic in another (see Box 1 for details on how symbioses between the same agents shift across different conditions). For Arabidopsis thaliana, the fungus Colleotrichum toefieldiae has been shown be a beneficial endophyte only under nutrient-scarce (Pi-limiting) conditions (Hiruma et al. 2016).
Box 1.
The Symbiosis Spectrum
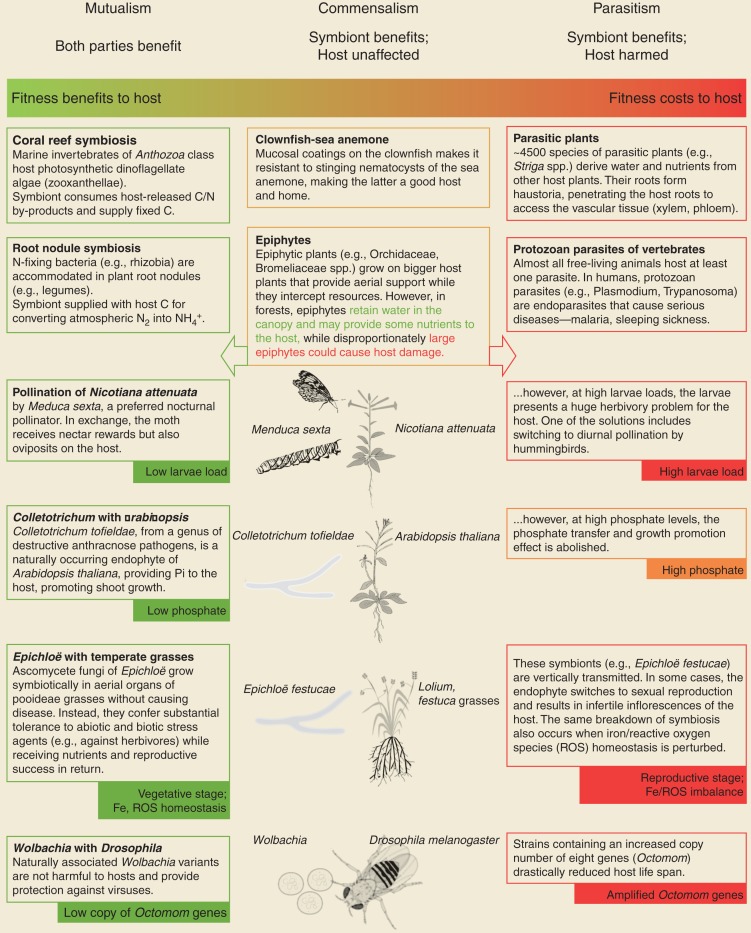
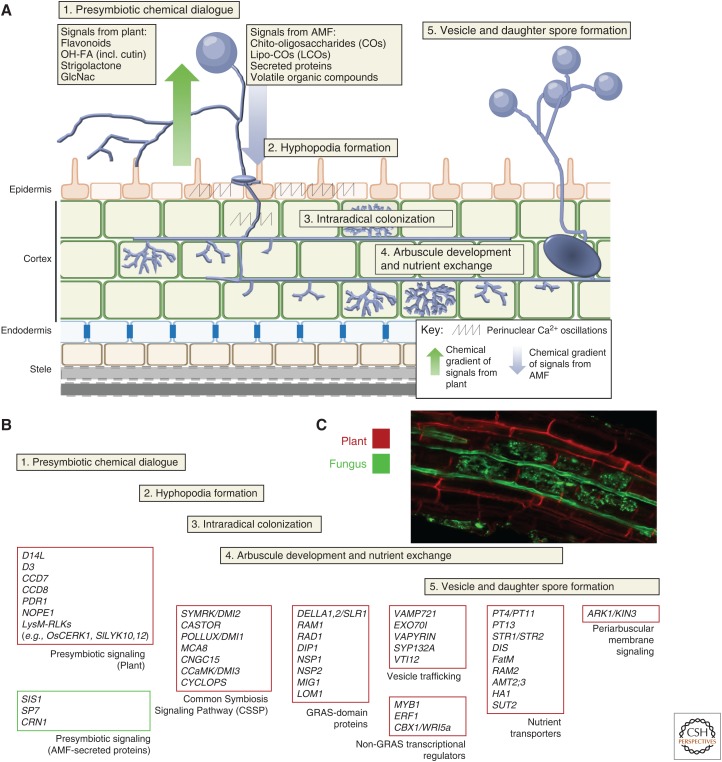
Among the multitude of associations plants engage in, the symbiotic relationship with arbuscular mycorrhizal fungi (AMF) is one of the most ubiquitous, ancient, and ecologically important (Remy et al. 1994; Redecker et al. 2000; Bidartondo et al. 2011; Strullu-Derrien et al. 2018). Around 71% of extant plants engage with and accommodate AMF in their roots (Wang and Qiu 2006; Brundrett and Tedersoo 2018). Recent phylogenomics approaches reveal that subsequent losses of the symbiosis in non-AM species appear to be associated with loss of symbiotic genes, although the drivers remain unknown (Favre et al. 2014; Delaux et al. 2015; Bravo et al. 2016). In the roots, highly branched tree-like structures eponymously known as arbuscules develop intracellularly for bidirectional nutrient exchange in the cortex (Fig. 1). This association involves extensive membrane biogenesis via dichotomous hyphal branching concomitant to invaginations of cortex cells; resulting in a symbiotic interface for signal and nutrient exchange without disrupting the host membrane integrity (Marx et al. 1982; Bonfante 2001). AMF are well known for their role in plant Pi acquisition, as the fungal hyphae are able to extend beyond the rhizosphere's depletion zone, mobilize and translocate nutrients, chiefly Pi, from regions inaccessible by roots in exchange for plant-fixed carbon in the form of sugars and fatty acids (FAs). Up to 22% of the photosynthetically fixed carbon is traded for symbiotic Pi, which could supply all (100%) of plant phosphate uptake (Jakobsen and Rosendahl 1990; Smith et al. 2003, 2011; Yang et al. 2012; Bravo et al. 2017; Jiang et al. 2017; Keymer et al. 2017; Luginbuehl and Oldroyd 2017; Roth and Paszkowski 2017).
Figure 1.
Mechanisms underlying establishment of arbuscular mycorrhizal symbiosis for nutrient transfer. (A) Signaling steps during arbuscular mycorrhizal (AM) symbiosis can be divided into five distinct stages beginning with precontact signaling mediated by diffusible molecules that reprogram both host and symbiont to commit to symbiosis, leading to contact and plant accommodation of arbuscular mycorrhizal fungi (AMF) intraradical hyphae inside the roots. At the cortex, precise regulation of cell biology leads to the development of arbuscules—the site of main nutrient exchange. A successful symbiosis enables the fungus to complete its life cycle by producing daughter spores that can then reinfect the same, as well as other hosts. In all cases, the symbiosis is asynchronous, so every stage is occurring in the host roots. (B) Molecular actors in the symbiosis. A list of key genes that have been identified to be required for the various stages of symbiosis signaling, divided according to their known roles (see MacLean et al. 2017 for an exhaustive list). (C) Fungal accommodation structures in roots stained with Alexa Fluor 488 wheat germ agglutinin showing intraradical spread of AMF and fully developed arbuscules (in green) in lateral roots of 5 weeks old (4 weeks postinoculation) rice (Oryza sativa). Plant cell walls are stained with propidium iodide (in red).
Other mycorrhizal associations (e.g., ectomycorrhizal, ericoid, arbutoid) exist but the arbuscular mycorrhizal symbiosis (also known as vesicular-arbuscular mycorrhiza [VAM] symbiosis) is unique as being a monophyletic fungal lineage that coevolved with land plants and maintained a wide host range (Peterson and Massicotte 2004; Smith and Read 2008; Martin et al. 2016). Structures strikingly similar to extant arbuscules have been identified in fossils dating as far back as the Rhynie Chert (Remy et al. 1994; Taylor et al. 1995; Redecker et al. 2000; Strullu-Derrien 2018; Strullu-Derrien et al. 2018). Although it is uncertain whether the symbiosis >400 million years ago (mya) was more parasitic or mutualistic, the ancient occurrence coupled with broad modern-day phylogenetic distribution of this symbiosis indicate its importance in the evolution of land plants (Humphreys et al. 2010; Field et al. 2015). Indeed, occurrence of AM symbiosis predates the evolution of true roots and has thus been attributed to the successful terrestrial colonization of plants from algal predecessors. Coevolution of plant (and roots) on Palaeozoic palaeosols alongside with AMF profoundly shaped terrestrial landscapes and, today, AM symbiosis remain an important aspect of global biogeochemical cycles (Retallack 1997; Raven and Edwards 2001; Brundrett 2002; Hetherington and Dolan 2018; Mills et al. 2018).
Mechanisms Underlying Establishment of Arbuscular Mycorrhizal Symbiosis
AM colonization occurs asynchronously across the root system, but follows a set of distinct stages as illustrated in Figure 2, and detailed in recent reviews by (Luginbuehl and Oldroyd 2017; MacLean et al. 2017; Choi et al. 2018; Pimprikar and Gutjahr 2018). Presymbiotic molecular dialog involves the reciprocal perception of diffusible molecules: root-released flavonoids, hydroxy-FAs, strigolactones (SLs), and N-acetylglucosamine (GlcNAc) derivatives are perceived by AMF (Harrison and Dixon 1993; Larose et al. 2002; Akiyama et al. 2005; Besserer et al. 2006; Steinkellner et al. 2007; Kretzschmar et al. 2012; Wang et al. 2012; Nadal et al. 2017), whereas a cocktail of fungal molecules, including chito-oligosaccharides (COs), lipochito-oligosaccharides (LCOs), proteins, and possibly volatile organic compounds, are perceived by plants (Kloppholz et al. 2011; Maillet et al. 2011; Genre et al. 2013; Sun et al. 2015a,b; Tsuzuki et al. 2016; Carotenuto et al. 2017). These signals trigger perinuclear Ca2+ oscillations, cytoskeletal rearrangements, as well as symbiosis gene expression that involve and also transcend the common symbiosis signaling pathway (CSSP) (Kistner et al. 2005; Gutjahr et al. 2008; Parniske 2008; Gutjahr and Parniske 2013; Camps et al. 2015). The CSSP, as its name suggests, represents a conserved signaling pathway for intracellular symbiont accommodation, from which arose the nodulation symbiosis in the eurosids ∼100 mya (Parniske 2008). Meanwhile, host perception triggers hyphal growth and branching in AMF. Hyphopodia, a swollen, branched fungal structure forms upon contact at the root surface. The penetration hypha invades the rhizodermal cell layer, whereby the plant directs the route of growth for the fungi, eventually forming arbuscules in the cortex (Genre et al. 2005, 2008; Gutjahr and Parniske 2013). Most of the symbiotic nutrient transfer occurs at these highly ramified yet ephemeral structures. Concurrently, lipid storage vesicles and spores emerge as the AMF completes its life cycle and reinfects the roots (Alexander et al. 1988; Kobae and Hata 2010; Kobae and Fujiwara 2014).
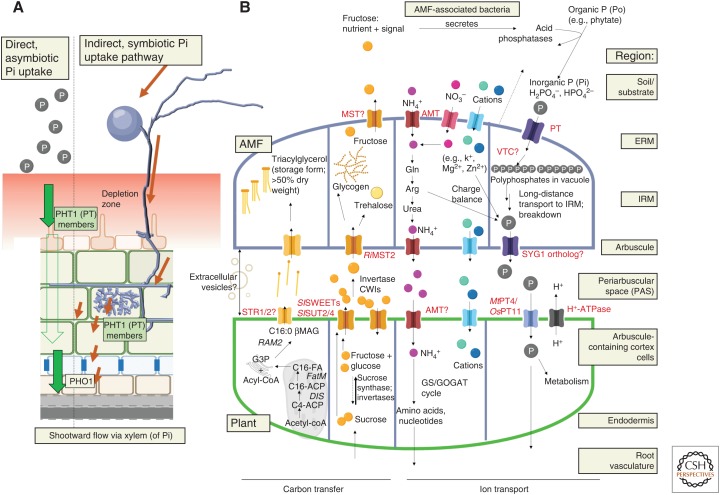
Figure 2.
Mechanisms of phosphate acquisition in plants. All arbuscular mycorrhizal (AM) host plants are capable of direct and indirect uptake of inorganic orthophosphate (Pi). (A) Direct uptake occurs via the asymbiotic route where plant phosphate transporters (PTs) are directly involved in the uptake of Pi from the soil into the root and eventually translocate the net flux toward the shoot. However, limited accessibility of Pi rapidly builds a depletion zone of Pi (red gradient). Plant PTs (also known as PHT1 in the literature) belong to the phosphate: H+-symporter (PHS) family in the major facilitator superfamily (MFS) and comprise 12 transmembrane domains (TMDs) with a conserved GGDYPLSATIxSE signature in TMD4. (B) Indirect, symbiotic pathway. The long hyphae of arbuscular mycorrhizal fungi (AMF) enable them to extend outside of the depletion zone for Pi uptake via fungal PTs, as well as via another symbiotic relationship between AMF hyphae and its associated microbiome that secretes acid phosphatases to solubilize organic phosphate (P) into Pi. Pi taken up by AMF is assimilated as polyphosphates into the vacuole and moved over long distances into the intraradical mycelium. At the arbuscule, polyphosphates are broken down into Pi for release into periarbuscular space, where plant PTs (including but not limited to the OsPT11/MtPT4 type) expressed in the arbuscule and localized to the plant periarbuscular membrane (PAM) takes up Pi with the help of H+-ATPases. Apart from Pi, the plant receives other nutrients as well, including N (as NH4+) and metal ions. Bidirectional exchange is a hallmark and also a functional necessity in AM symbiosis. In return, plants provided reduced, organic carbon in the form of sugars and more importantly fatty acids (FAs) to the AMF. AMFs are FA auxotrophs as they lack genes for long-chain FA biosynthesis. De novo FA synthesis is regulated in arbuscule-containing cortex cells via WRI5b/ERF1/WRI5a/CBX1 (transcription factors), to produce FA in a pathway involving DIS (FA synthase), FatM (acyl transferase), and RAM2 (GPAT6). PAM-localized lipid transporter candidates STR/STR2 then shuttles the FA into the periarbuscular space for the AMF. Circles denote nutrients and the known/proposed/unknown transporters are indicated in red.
MECHANISMS OF PHOSPHATE UPTAKE IN PLANTS
Two pathways for uptake, direct and indirect, exist for plants capable of entering AM symbiosis (Smith et al. 2003, 2011). In the direct uptake pathway, Pi is acquired by low and high H+/Pi symporters primarily on the epidermis (Mudge et al. 2002; Misson et al. 2004; Shin et al. 2004). These PHOSPHATE TRANSPORTER1 (PHT1; abbreviated and used henceforth as PT) family transporters are either constitutively expressed or inducible upon Pi starvation, and have overlapping and spatiotemporally complementary functions in Pi uptake (Nussaume et al. 2011; Gu et al. 2016). In the roots, Pi is assimilated and loaded into the xylem via PHO1 transporters toward the shoot (Hamburger et al. 2002; Stefanovic et al. 2011; Arpat et al. 2012), whereas organic forms (ATP, hexose phosphates) move in the phloem (Rausch and Bucher 2002). Figure 2 illustrates the physiological route of direct and indirect Pi uptake, with greater detail on the latter.
Indirect Phosphate Pathway—Uptake by Fungus
In the indirect mycorrhizal pathway, P is acquired by H+/Pi or Na+/Pi symporters of the AMF extraradical hyphae (Harrison and Buuren 1995; Maldonado-Mendoza et al. 2001; Tisserant et al. 2012; Kikuchi et al. 2014; Xie et al. 2016). The Pi is imported presumably via the vacuolar transporter chaperone complex (Hothorn et al. 2009) into the tonoplast and translocated as polyphosphates to the intraradical mycelium (IRM) probably driven by mobile tubules in vacuole and/or aquaporin-mediated water flow driving bulk flow (Cox et al. 1980; Solaiman et al. 1999; Rasmussen et al. 2000; Viereck et al. 2004; Kuga et al. 2008; Tani et al. 2009; Hijikata et al. 2010; Kikuchi et al. 2014; Ezawa and Saito 2018).
Organic P sources (Po, e.g., phytate) are abundant in nature and can also be mobilized (Feng et al. 2002; Shibata and Yano 2003), possibly via acid phosphatases secreted either by AMF (Sato et al. 2015), by colonized plant roots (Ezawa et al. 2005), or by AMF-associated bacteria (Kim et al. 1997; Toro et al. 1997; Zhang et al. 2016, 2018; Battini et al. 2017). AMF-associated bacteria have long been hypothesized to solubilize Po to enhance phosphate uptake by AMF. It was recently demonstrated that C delivered from the plant to fungus is then delivered as fructose to the bacteria, which acts as a nutrient and signal for the latter to secrete acid phosphatases to solubilize Po for uptake by AMF (Fig. 2). Thus, nutrient exchanges also exist between AMF and its microbiome (Zhang et al. 2018). Similarly, AMF-associated bacteria was also shown to be crucial determinants of growth-promoting and suppressive soils (Svenningsen et al. 2018). These recent insights highlight the tripartite association and multitude of interactions that, through AMF symbiosis, determine soil health and productivity.
Breakdown of polyphosphates in the IRM might involve acid and alkaline phosphatases, and this huge negative charge generated by Pi could explain the simultaneous uptake of cations observed (Ezawa et al. 2001; Aono et al. 2004; Kojima and Saito 2004; Kikuchi et al. 2014). More unclear is how Pi from AMF is released at the arbuscular interface, as suggested by presence of phosphatase activities and evidenced by polyphosphate accumulation in mtpt4 mutants where symbiotic transfer is disrupted (Gianinazzi et al. 1979; Capaccio and Callow 1982; van Aarle et al. 2001, 2002; Javot et al. 2007a).
Taken together, significant knowledge gaps exist regarding the mediators and mechanisms underlying fungal phosphate metabolism. The notorious genetic intractability of AMF, compounded by the presence of hundreds to thousands of coenocytic nuclei per individual, has severely hampered functional characterization of genes involved in symbiosis (Sanders and Croll 2010; but see Helber and Requena 2007). Nonetheless, functional characterization is possible, as exemplified by the charcterization of Gigaspora margarita phosphate transceptor GigmPT (Xie et al. 2016). Recent efforts in obtaining AMF genomes and transcriptomes of various strains, species, and life stages have greatly facilitated comparative functional analyses (Tisserant et al. 2012, 2013; Lin et al. 2014; Handa et al. 2015; Salvioli et al. 2016; Tang et al. 2016; Kamel et al. 2017; Chen et al. 2018; Kobayashi et al. 2018; Mathieu et al. 2018; Sun et al. 2018a; Zeng et al. 2018). For instance, a list of candidate genes has since been proposed (Ezawa and Saito 2018), including yeast SYG1 orthologs in AMF as putative Pi exporters at the arbuscule (Azevedo and Saiardi 2017). With techniques such as host-induced gene silencing and the careful validation of symbiont-specific gene silencing (and not on hosts), we are witnessing an increasing understanding of AMF genes during symbiosis (Helber et al. 2011; Kloppholz et al. 2011; Tsuzuki et al. 2016; Xie et al. 2016; Sun et al. 2018b; Voß et al. 2018).
Plant Uptake of Symbiotic Pi Requires Specialized Phosphate Transporters
Plant-encoded H+/Pi symporters are specifically expressed in arbuscule-containing cortex cells and are localized to the plant periarbuscular membrane (PAM) for Pi uptake (Javot et al. 2007a; Yang et al. 2012; Watts-Williams et al. 2015). Arbuscules are believed to be the main site of nutrient transfer, where plants obtain primarily P/N (among other nutrients), while providing carbohydrates and FAs to the fungus. Since the first identification of AM-specific PTs in plants (Rausch et al. 2001; Harrison et al. 2002; Paszkowski et al. 2002), PTs that are specifically expressed or are involved in Pi uptake during symbiosis have been described in an expanded list of angiosperm species, summarized in Table 1 (Gümil et al. 2005; Nagy et al. 2005; Poulsen et al. 2005; Maeda et al. 2006a; Wegmüller et al. 2008; Loth-Pereda et al. 2011; Xie et al. 2013; Walder et al. 2015; Liu et al. 2016a; Sawers et al. 2017). Not all of the identified transporters have been characterized for expression and localization activity, and species-specific differences exist (e.g., expression in other tissues, with roles not limited to symbiotic uptake) as discussed below (see also Table 1). In all of these species, there appears to be at least one phylogenetically conserved PT4/PT11 transporter. These form a clade closely related to Physcomitrella patens and Selaginella moellendorffii PTs, suggesting an ancient origin and functional conservation (Yang et al. 2012); although a more recent analysis suggests the relationship to be nonorthologous in the nonvascular plants (Delaux et al. 2015). From a phylogenetic perspective, it will be exciting to investigate the existence of functional conservation in symbiotic pteridophytes and bryophytes to better understand how the symbiotic transporters were recruited.
Table 1.
List of symbiotic phosphate transporters in plants identified to date
| Species | Gene | Mutant phenotype | P transport activity demonstrated? | Transcript/promoter or protein localization or expression pattern | References |
|---|---|---|---|---|---|
| Oryza sativa | OsPT11 | Reduced % root length colonization (RLC) and % arbuscules (A) | Yes; 33P uptake in yeast pho84 | Arbuscules (promoter:uidA reporter for GUS staining; GUS) | Paszkowski et al. 2002; Kobae and Hata 2010; Yang et al. 2012 |
| Stunted arbuscules | PT11-GFP to PAM | ||||
| Abolished symbiotic Pi transport | |||||
| OsPT13 | Same as pt11 mutants, but functional symbiotic Pi uptake | Does not complement | Arbuscules (GUS) | ||
| Zea mays | ZmPT6 | Phenocopies ospt11 in sterile culture | Complements pho84 growth | Arbuscules (in situ hybridization; ISH) | Glassop et al. 2005; Willmann et al. 2013; Liu et al. 2016a; Sawers et al. 2017; Liu et al. 2018 |
| Transcomplementation with chive nurse plants increases %RLC but not %A; %RLC WT-like in soil, reduced %A | |||||
| ZmPT9 | p35S:PT9 lines in Lotus hairy roots reduced %RLC | Complements pho84 growth | Colonized regions (pZmPT9-GFP; Lotus hairy roots) | ||
| ZmPT2,4, 7, 11 | n.d. | Induced by low Pi or AMF | |||
| Brachypodium distachyon | BdPT3, 13 | All n.d. | All n.d. | Both root-specific and AM-specific | Hong et al. 2012 |
| BdPT7, 12 | Both AMF-induced; also expressed in shoots | ||||
| Sorghum bicolour | SbPT10, 11 | n.d. (SbPT10: Putative OsPT11 function) | n.d. | Both highly induced | Walder et al. 2015; Watts-Williams et al. 2018 |
| SbPT8, 9, 11 | n.d. (SbPT9: putative OsPT13 function) | n.d. | Both have modest AMF-induction | ||
| Medicago truncatula | MtPT4 | Phenocopies ospt11 | Yes; P uptake in pam2 and NS219 mutants | Arbuscules (GUS, hairy roots) | Harrison et al. 2002; Javot et al. 2007; Breuillin-Sessoms et al. 2015; Floss et al. 2017 |
| Transcomplementation with leek nurse plants increases %RLC but not stunted arbuscules | Immunolocalization onto PAM; surrounding branches | ||||
| Phenotype rescued with N-starvation; dependent on AMT2;3 | |||||
| Lotus japonicus | LjPT3 | RNAi (hairy roots): reduced %RLC and %A; reduced growth and Pi uptake; co-inoculation of AMF and rhizobia gave necrotic nodules | Yes; 32P uptake in pam2 | Arbuscules (ISH) | Maeda et al. 2006 |
| LjPT4 | RNAi (hairy roots): reduced colonization, stunted arbuscule morphology, reduced root P content | n.d. | Arbuscules and root tips (GUS, hairy roots) | Volpe et al. 2016 | |
| Astralagus sinicus | AsPT4 | Phenocopy ospt11 | Does not complement MB192 | Both: Arbuscules (GUS, hairy roots, and ISH) | Xie et al. 2013 |
| Required for symbiotic Pi uptake (total shoot P levels, not 32/33P) | Complements; transport Pi | ||||
| AsPT1 | Phenocopy ospt13; As-PT1OE increases colonization in hairy roots | ||||
| Glycine max | GmPT10, 11 | n.d. | Yes; 32P uptake in pam2 | Branch domains of arbuscules (prom:GFP, hairy roots) | Tamura et al. 2012; Inoue et al. 2014 |
| GmPT7 | n.d. | n.d. | Mature noncollapsing arbuscules, root tips, LR primordia, vein endings of senescent leaves (GUS, hairy roots) | ||
| Solanum tuberosum | StPT3 | n.d. | Yes, 32P uptake in pam2 | All: Arbuscules (GUS, hairy roots); PT3 is also expressed in asymbiosis and shoots | Rausch et al. 2001; Karandashov et al. 2004; Nagy et al. 2005 |
| StPT4 | n.d. | Pam2 data unclear; n.d. for PT5 | |||
| StPT5 | n.d. | PT4 and PT5 AMF-induced and root-specific | |||
| Solanum lycopersicum | LePT3 | n.d. | n.d. | All are AMF-induced | Nagy et al. 2005; Balestrini et al. 2007; Xu et al. 2007 |
| LePT4 | Dispensable for Pi uptake and symbiosis | pam2 data unclear | PT4: Arbuscules (GUS, hairy roots) | ||
| LePT5 | n.d. | n.d. | LePT1 to 5 all detected in LCM of arbuscules | ||
| Petunia hybrida | PhPT3, 4, 5 | All n.d. | n.d. | AMF induced; PT4 might be AM-specific | Wegmüller et al. 2008 |
| Solanum melongena | PT4, 5 | All n.d. | n.d. | Root-specific, AMF-specific | Chen et al. 2007 |
| Capsicum frutescens | PT3 | Induced by AMF, also expressed in shoots | |||
| Nicotiana tabacum | PT2 | Root-specific, induced by AMF and Pi starvation | |||
| Populus trichocarpa | PtPT10 | All n.d. | No growth complementation of pho84 for PT10; others n.d. | Root-specific, AM-specific | Loth-Pereda et al. 2011 |
| PtPT9, 12 | Induced by AM and ECM; also in senescing autumn leaves | ||||
| Vitis vinifera | VvPT1, VvPT2 | n.d. | n.d. | Both are root-specific (not in leaves at least), AM-specific | Valat et al. 2018 |
Phosphate transporters (PTs) highlighted in yellow are specifically induced during arbuscular mycorrhizal (AM) symbiosis and phylogenetically cluster with the OsPT11/MtPT4 clade; while other PTs may also be involved in Pi uptake at the arbuscule during symbiosis but have low levels of expression during asymbiosis; or expression in other tissues. Where subsequent functional characterization was conducted, the phenotypes and observations are listed in the respective columns; for mutant phenotypes, complementation of P uptake/transport in yeast mutants; as well as expression/localization pattern of the transcript/protein where investigated. Nonstably transformed plants are indicated “hairy roots” in parentheses to indicate Agrobacterium rhizogenes–mediated transformation of root organ culture.
AMF, Arbuscular mycorrhizal fungi; ECM, ectomycorrhizal; LCM, laser capture microdissection; n.d., not determined; WT, wild type.
Inside plant cells, excess Pi is stored in vacuoles while a net flux toward the shoot occur via PHO1-mediated xylem loading. The remobilization of vacuolar Pi stores, best understood in non-AM hosts Arabidopsis, is important for Pi homeostasis, and recent work have identified vacuolar Pi transporters (VPTs; also PHT5 family members) that possess both SYG1/PHO81/XPR1 (SPX) and major facilitator family (MFS) domains, to primarily mediate vacuolar influx (Liu et al. 2016b; Xu et al. 2019). Although the role of SPX-MFS3 in mediating vacuolar Pi efflux in rice is contested (Wang et al. 2015; Xu et al. 2019), a recent study revealed that intriguingly, an ancestral plasma membrane-localized glycerol-3-phosphate transporter (GlpT; at least in Escherichia coli) is instead directed to the plant tonoplast and mediates vacuolar Pi efflux in rice, Marchantia polymorpha and P. patens (Xu et al. 2019). These studies have since advanced our understanding on how Pi shuttles in and out of the vacuolar stores. However, how Pi is specifically partitioned and transported from the ports of entry (epidermis or cortex during asymbiosis and symbiosis, respectively) into the endodermis remains to be elucidated, although most models suggest nutrients to move via apoplastic, symplastic, or a trans-cellular pathway coupling both mechanisms for inward radial movement of nutrients.
Energetics of Nutrient Transfer
Nutrients exchanged at the symbiotic interface, regardless of their direction, identity, precursor, and subsequent assimilation pathways, traverse the same cell membranes. Because the membrane potential is negative inside relative to the outside, both parties require cation/proton channels to power the transport of neutral and negatively charged molecules across membranes. Accordingly, the periarbuscular space has been described as an acidic compartment, and H+-ATPases were proposed to provide the proton gradient and electrochemical energy (Gianinazzi-Pearson et al. 2000; Guttenberger 2000; Ferrol et al. 2002; Krajinski et al. 2002; Ramos et al. 2009). Indeed, H+-ATPases have since been demonstrated to be necessary for transport of phosphate, arbuscule development, and fungal colonization in both Medicago and rice (Krajinski et al. 2014; Wang et al. 2014).
Recent modeling and biophysical descriptions of nutrient exchange at the arbuscule suggest that the main classes of nutrient traded to our knowledge is relatively comprehensive, although the underlying mechanistic understanding is unclear (Dreyer et al. 2018). Using either minimal transporter networks for P/C exchange (Schott et al. 2016), or without transporter identity but simply first principles of thermodynamics and H+-ATPase-powered transport as the sole assumptions (Dreyer et al. 2018), P (or P/N) trade for C at the interface was shown to be sufficient as the general minimal model to which more nutrients can be added. These models reveal that AMF is likely to unload Pi via hitherto elusive H+/Pi symporters because it is energetically favorable and ensures increased reward in the form of increased C flux (Dreyer et al. 2018). Whereas for the plant, although electroneutral transport of C (as hexoses or FAs) is not the most energetically favorable, it enables additional P uptake as long as C is not limiting. Importantly, these models show that even when viewed from a competitive perspective, the relative ease of access to P or C stores by each party (i.e., self-interest), is sufficient to drive a robust mutualistic exchange.
Multiple Evolutionary Trajectories of Plant Phosphate Transporters
With their key roles in symbiosis, plant symbiotic PTs have been studied in considerable detail relative to other transporters. Table 1 summarizes the various symbiotic phosphate transporters identified across a wide range of species to date. Apart from the conserved MtPT4/OsPT11 transporters, individual mycorrhizal plant families also appear to have evolved phylogenetically divergent PTs that may serve redundant or nonredundant roles in symbiosis. In rice, OsPT11 and OsPT13 are specifically expressed in arbuscules and serve nonredundant roles in the regulation of symbiosis (Yang et al. 2012). Whereas OsPT11 belongs to the phylogenetically conserved AM-specific PT, OsPT13 represents a phylogenetically distant innovation in the monocotyledons (Yang and Paszkowski 2011; Yang et al. 2012). OsPT11 alone is necessary and sufficient for symbiotic Pi uptake. OsPT13, in contrast, does not contribute to symbiotic Pi transport, although it is expressed at the arbuscule, and also failed to display Pi transport activity in yeast pam2 mutants (Yang et al. 2012). Interestingly, ospt13 mutants, despite having reduced colonization and stunted arbuscule development, have wild-type levels of OsPT11 transcripts and symbiotic Pi uptake (Yang et al. 2012). Therefore, whether OsPT13 functions as a nontransporting sensor to monitor Pi levels at the periarbuscular interface—that is, as a transceptor—remains to be shown. Transceptor activity has been identified for yeast Pho84, which transports Pi and activates protein kinase A signaling in A. thaliana nitrate transporter NRT1.1; and more recently has also been suggested for GigmPT (Bun-Ya et al. 1991; Giots et al. 2003; Walch-Liu and Forde 2008; Xie et al. 2016).
Neofunctionalization of symbiotic phosphate transporters for asymbiotic roles is also observed in certain plant families or species, and may have thus conferred additional tools for the plant P transport and allocation over evolutionary time. Gene duplication in the Solanaceae gave rise to two orthologs of OsPT11 in tomato and potato (SlPT4/LePT4 and SlPT5/LePT5) (Nagy et al. 2005; Poulsen et al. 2005). LePT4 is exclusively expressed during symbiosis, but it is dispensable for symbiotic P uptake; whereas LePT5 is also expressed during asymbiosis and is induced >10-fold during fruit ripening (Xu et al. 2007; Chen et al. 2014). These examples suggest that gene duplications can give rise to new regulation and functions in P partitioning. Similarly, OsPT11/MtPT4 orthologs in maize (ZmPT6) and Brachypodium (BdPT7) accumulate in both mycorrhizal roots and also noncolonized roots and leaves of Pi-starved plants (Nagy et al. 2006; Hong et al. 2012). This raises a question regarding the ancestral state and subsequent evolution of phosphate transporters in the plant kingdom—were the roles of the ancestral phosphate transporter(s) symbiotic, asymbiotic, or both?
Spatial Physiology of Symbiotic Pi Exchange Remains Unclear
One neglected aspect of symbiotic and asymbiotic Pi uptake is the relative contributions of various root types to the overall uptake and assimilation of Pi. Changes in root architecture is one of the common physiological responses investigated in asymbiotic nutrient signaling pathways. In non-AM A. thaliana, primary root growth is arrested and lateral root density (LRD) increases (Péret et al. 2014) during Pi starvation. In monocots where genetic architecture of root development is different, changes to the primary embryonic roots of seedling roots are subtle, whereas LRD decreased in maize but increased in rice (Péret et al. 2014; Hochholdinger et al. 2018). More importantly, lateral rooting correlates positively with Pi acquisition under low P conditions (Zhu and Lynch 2004; Gamuyao et al. 2012; Vejchasarn et al. 2016). Where and how phosphate transporters are expressed in different root types, and contribute to Pi uptake, remain to be fully characterized.
AM symbiosis, meanwhile, increases LRD in the leguminous and monocot hosts examined thus far (Oláh et al. 2005; Gutjahr et al. 2009; Maillet et al. 2011; Mukherjee and Ané 2011; Chiu et al. 2018). With AMF preferentially colonizing LRs in both monocots and dicots, this effectively increases the symbiotic interface for nutrient exchange (Gutjahr et al. 2009). Colonization by AMF elicited a fundamental reprogramming of the rice crown root transcriptome, whereby the profiles of secondary cell wall metabolism, hormone, and transport-related genes indicated a switch in functional relationships of root types upon entering symbiosis (Gutjahr et al. 2015). To this end, better physiological characterization of contributions of the various root types to symbiotic Pi uptake will help define an “ideal” root architecture for crop-breeding programs.
Plant Carbon Transfer to AMF in Exchange for Pi
Reciprocal nutrient exchange is a hallmark of this mutualism—in exchange for Pi, plants deliver C to AMF. Most plants are facultative symbionts—they allow AMF colonization and benefit from symbiotic Pi uptake, but can grow axenically without them. On the other hand, AMF, being FA auxotrophs, absolutely rely on plant-fixed carbon to complete their life cycle (Bago and Bécard 2002). Radiolabeling experiments demonstrated that fixed C is transferred to the fungus as hexoses and lipids via the IRM (Shachar-Hill et al. 1995; Solaiman and Saito 1997; Bago et al. 2000, 2002, 2003). Rhizophagus irregularis high-affinity monosaccharide transporter MST2, for example, is able to transport glucose, mannose, fructose, and is expressed in IRM—suggesting that apart from arbuscules, IRM are possible sites of symbiotic nutrient exchange (Helber et al. 2011; Ait Lahmidi et al. 2016; Roth and Paszkowski 2017). Noteworthy also are Medicago truncatula della mutants, where extensive IRM growth suggest hexose transfer but the severe lack of arbuscules and lipid transfer result in the failure of AMF to complete its life cycle (Floss et al. 2013; Yu et al. 2014).
Meanwhile, FAs have emerged as the central form of C required by AMF to complete their life cycle. Substantial amounts of C are stored as triacylglycerols in all fungal structures (Bago et al. 2002), and the central role of lipids for AMF metabolism contrasted starkly against the lack of type I fatty acyl synthase subunits for palmitic acid synthesis from AMF genomes and transcriptomes (Tisserant et al. 2013; Wewer et al. 2014; Tang et al. 2016). Lipid transfer from plants to AMF was conclusively demonstrated via two independent approaches. First, using a synthetic method, Umbellularia californica fatty acyl-ACP thioesterase (UcFatB) introduced into Medicago roots resulted in the production of 12:0FA (lauric acid), which does not occur naturally in Medicago. Detection of lauric acid in R. irregularis spores demonstrated the existence of lipid transfer (Jiang et al. 2017; Luginbuehl et al. 2017). In addition, isotopolog profiling of FA compositions with 13C-labeled glucose together with fungal-specific 16:1ω5 FA enabled the monitoring of FA transfer. The close mirroring of 16:0 and 16:1 FA, and the impaired 16:1FA accumulation in Lotus japonicus FA biosynthesis mutants demonstrated their roles in lipid transfer (Keymer et al. 2017; Brands et al. 2018). STUNTED ARBUSCULES1 (STR1) and STR2, both half-size ABC transporters of the G family, are therefore widely hypothesized to transport lipids as 16:0βMAG across the plant PAM (Bravo et al. 2017; Keymer et al. 2017; MacLean et al. 2017), but their direct transport activity remains to be demonstrated, and elucidating transport activity of ABC transporters is nontrivial (Lefevre et al. 2015). Plant carbon nourishment of AMF is reviewed extensively in Luginbuehl and Oldroyd (2017), MacLean et al. (2017), Roth and Paszkowski (2017), and Keymer and Gutjahr (2018).
REGULATION OF ARBUSCULE DEVELOPMENT—THE OLD AND NEW
Transcriptional Regulation of Arbuscule Development
Arbuscules are often considered the “heart of the symbiosis,” for the symbiosis culminates in these highly branched structures where nutrient exchange occurs. Years of work by various groups have revealed how the plant coordinates and orchestrates the development of the arbuscule, revealing the transcriptional and traffic control required for a functional nutrient exchange interface to be established. In the meantime, new frontiers of signaling have also emerged.
Trafficking of Plant Symbiotic Transporters
During arbuscule development, root cortical cells undergo significant transcriptional reprogramming, creating a PAM that carries a distinct composition of proteins otherwise contiguous with the plasma membrane. These include the aforementioned symbiosis-specific phosphate, ammonium, sugar, ABCG transporters (e.g., STRs), and H+-ATPases. How the arbuscule achieves such precise spatiotemporal localization of proteins is not completely understood. For example, MtPT4/OsPT11/GmPT10/GmPT11 specifically localizes to the branch domains of periarbuscular membrane (Kobae and Hata 2010; Tamura et al. 2012). Genetic analyses predominantly in legumes reveal the involvement of polarized exocytosis involving an EXO70I subunit of the EXOCYST complex, v-SNARE: VESICLE ASSOCIATED MEMBRANE PROTEIN (VAMP721d,e) and a splice variant of t-SNARE (SYNTAXIN OF PLANTS, SYP132), which are all symbiosis-specific members of big gene families, as well as symbiosis-induced VAPYRIN (Feddermann et al. 2010; Pumplin et al. 2010; Murray et al. 2011; Ivanov et al. 2012; Zhang et al. 2015; Huisman et al. 2016; Pan et al. 2016). Systematic promoter analysis in Medicago revealed that precise localization is achieved by the timing of gene expression and transient reorientation of the secretory pathway (Pumplin et al. 2012; reviewed in Harrison and Ivanov 2017). Demonstrating the conservation in other AM host species, and identifying the temporal regulators of expression and secretory activity will generate better understanding and potentially useful biological applications.
Recently, small anionic lipids—phosphoinositides and phosphatidic acid—have also been shown to possess distinct subcellular distribution during arbuscule development (Ivanov and Harrison 2018). The specific distribution of these anionic lipids known to influence cellular processes ranging from membrane trafficking, signaling, exocytosis, and endocytosis raises questions on their roles in regulating arbuscule development.
Network of GRAS-Domain Proteins May Integrate Host Nutrient and Growth Status
A suite of transcriptional regulators, especially GRAS-domain proteins, have emerged as key players that regulate arbuscule development, possibly through their spatiotemporal expression and combinatorial control of symbiotic gene expression in successive “transcriptional waves” (Gutjahr and Parniske 2013; Luginbuehl and Oldroyd 2017; MacLean et al. 2017; Choi et al. 2018; and detailed in Pimprikar and Gutjahr 2018). Perception of AMF activates the CSSP via CALCIUM/CALMODULIN DEPENDENT KINASE (CCaMK) and via CYCLOPS, a transcription factor (TF) to activate symbiotic gene expression, enabling intraradical accommodation of AMF. DELLA proteins (SLENDER1 in rice), members of the GRAS-domain family, and known transcriptional suppressors of gibberellic acid signaling, form a complex with CCaMK and CYCLOPS essential for arbuscule development (Jin et al. 2016; Pimprikar et al. 2016). This includes REQUIRED FOR ARBUSCULAR MYCORRHIZA1 (RAM1), a GRAS-domain TF that activates EXO70I, FatM, RAM2, STR1 expression, suggesting a key role of this transcriptional module in orchestrating lipid biosynthesis and membrane biogenesis. Another GRAS-domain protein, REQUIRED FOR ARBUSCULE DEVELOPMENT1 (LjRAD1), interacts with RAM1 and DELLAs in vivo, and is required for arbuscule development at the fine branch stage (Park et al. 2015; Rich et al. 2015, 2017; Xue et al. 2015; Pimprikar et al. 2016). Joining the concert of GRAS-domain proteins regulating arbuscule development is MYCORRHIZA-INDUCED GRAS (MtMIG1), which is also required for arbuscule branching and together with DELLA proteins activate radial cell expansion of the cortex (Heck et al. 2016). Taken together, the pervasive role of GRAS domain TFs (Fig. 1), especially in the case of DELLA proteins, could provide a means for plants to integrate growth, developmental, and nutritional signals via phytohormones with arbuscule development.
Transcriptional Module Regulating Bidirectional Nutrient Exchange
Recently, two independent groups working on different leguminous species identified and functionally characterized WRINKLED1-like AP2 TFs that work downstream of RAM1. CTTC MOTIF-BINDING TRANSCRIPTION FACTOR1 (LjCBX1) and Medicago WRINKLED5a (MtWRI5a) (Jiang et al. 2018; Xue et al. 2018) are TFs that bind to the AW-box motif and regulate the expression of fatty-acid biosynthesis genes, phosphate transporters (MtPT4/LjPT4), as well as H+-ATPases, suggesting the existence of a transcriptional network directly regulating bidirectional nutrient exchange (Jiang et al. 2018; Xue et al. 2018). With the plethora of actors identified in the plant host, we look forward to the identification of the fungal counterparts.
Receptor-Like Kinase Signaling at the Periarbuscular Membrane
In addition to the increasingly complex transcriptional network and nutrient exchange during arbuscule development, recent work characterizing the proteome and transcriptome of mycorrhizal roots and arbusculated cells, respectively, demonstrated for the first time existence of signaling cascades mediated by receptor-like kinases (RLKs) at the PAM (Roth et al. 2018). In rice, ARBUSCULAR RECEPTOR-LIKE KINASE 1 (ARK1), a family of serine/threonine RLK present in AM host species, is a PAM-localized RLK required for sustaining fungal fitness at the latter stages of symbiosis. OsARK1 is orthologous to M. truncatula KINASE3 (MtKIN3) (Bravo et al. 2016; Roth et al. 2018). Although arbuscule development remained normal, vesicle development of AMF was impaired and symbiosis collapsed over prolonged time periods. The presence of a common mycorrhizal network supported by wild-type plants, however, rescued the ark1 mutant phenotype. Following arbuscule senescence, vesicles often emerge as lipid storage bodies and could be crucial for subsequent rounds of infection affected in ark1 mutants (Roth et al. 2018). Whereas the formation of vesicles and daughter spores in AMF reflects fungal fitness, the dearth of knowledge on the mechanisms specifying and underlying these processes invite further studies.
INSIGHT: NUTRIENTS ARE ALSO SIGNALS FOR SYMBIOSIS
Beyond transcriptional and trafficking changes orchestrated in the developing arbuscule, a key insight into regulation of arbuscule development is the regulatory role played by nutrients (Yang and Paszkowski 2011). Functional characterization of symbiotic transporters have revealed the regulatory role of nutrients in arbuscule development. Consistently, mutants of symbiotic PTs (MtPT4, OsPT11, OsPT13), genes involved in lipid biosynthesis/delivery (OsSTR1,2, MtSTR1,2, LjFatM, DIS, RAM2), as well as H+-ATPases powering nutrient transfer all result in disrupted nutrient transfer, reduced colonization, stunted arbuscule development, altered arbuscule morphology, and in the case of mtpt4/ospt11 mutants, increased arbuscule turnover (Javot et al. 2007a; Yang et al. 2012; Xie et al. 2013; Krajinski et al. 2014; Wang et al. 2014; Bravo et al. 2016, 2017; Keymer et al. 2017; Brands et al. 2018). Radiotracer experiments showed that like mtpt4/ospt11, symbiotic Pi uptake is also abolished in mtha1 mutants (Krajinski et al. 2014). Beyond P, uptake of other nutrients is also affected in mtha1 mutants (Hubberten et al. 2015). The Rhizobia-Medicago nodule symbiosis, however, appears unaffected (Krajinski et al. 2014; Wang et al. 2014). Thus, proton gradients established by MtHA1/OsHA1 appear crucial for AM symbiosis development and exchange of neutral and negatively charged nutrients. It will be interesting to investigate whether symbiotic Pi transfer is abolished in lipid transfer mutants, and whether the lack of transfer is a result of plant or fungal signaling.
Interestingly, in the case of Medicago pt4, the mutant phenotype is rescued by nitrogen deprivation, in a manner dependent on an ammonium transporter, MtAMT2-3 (Javot et al. 2011; Breuillin-Sessoms et al. 2015). As MtAMT2-3 is unable to complement a yeast ammonium transport mutant, it has since been suggested to have a signaling role in arbuscule maintenance—possibly in a similar fashion to OsPT13 (Yang et al. 2012; Breuillin-Sessoms et al. 2015). Screening for the suppression of the mtpt4 phenotype revealed the role of MYB1, a MYB-like TF that regulates arbuscule degeneration. The requirement of a MYB1-DELLA-NSP1 complex to degenerate arbuscules lends further support to the theme that GRAS-domain protein complexes have temporal and combinatorial control of arbuscule development (Floss et al. 2017).
Collectively, it appears that nutrient delivery is a signaling requirement. Nonproductive colonization can be detected and terminated by the plant host, and possibly by the AMF symbiont as well (e.g., undernourishment without FAs). How each party measures the delivery of P/FAs and senses the lack of symbiotic nutrient exchange to terminate arbuscule development is currently unclear. Nonetheless, it is also plausible that H+/Pi/C fluxes may have a self-organizing feature capable of driving and sustaining transcriptional activities to allow full arbuscule development without requiring a “sensor.”
Why Are Arbuscules Transient?
Given the multitude of transport and regulatory activity occurring during symbiosis, the transient nature of arbuscules may appear enigmatic. Arbuscules have short life spans of ∼4–15 days before they mature and collapse, leaving the cortical cells competent for reinfection (Toth and Miller 1984; Alexander et al. 1988; Kobae and Hata 2010; Kobae and Fujiwara 2014). As such, in the same plant (and likewise fungus), symbiosis development is highly asynchronous, with new infection units co-occurring alongside fully developed arbuscules and senescing ones.
Here, we propose that rather than being a futile cycle of infection and iterative membrane biogenesis, the transitory nature of an arbuscule may be crucial to its symbiotic nature, from enabling bidirectional uptake in the short term to allowing evolutionary stability in the long run. A substantial amount of membrane phospholipids is generated per arbuscule, which in itself provides an increased surface area for symbiotic nutrient and signal exchange. Phospholipids constitute two of the key currencies of the trade—phosphates and fatty acids. We speculate that the sustained development of a PAM requires commitment by both parties to direct P and C to the interface, which allows the monitoring and policing of symbiotic nutrient exchange, and provides, with the ensuing senescence, a possible second pathway of symbiotic nutrient exchange. This policing could ensure that nonproductive arbuscules (and nonproductive AMF species) become terminated instead of maintained.
Meanwhile, ultrastructural analyses of the periarbuscular space reveal the presence of interkingdom trafficking of extracellular vesicles (EVs) between the plant and AMF (Ivanov et al. 2019; Roth et al. 2019), which invites speculation on the cargo constituents in these EVs. In Prochlorococcus, EVs carry organic carbon and nucleic acids with importance in nutrient fluxes in marine ecosystems (Biller et al. 2014). In Paracoccus, EVs carry hydrophobic N-acyl-homoserine lactones for quorum sensing (Toyofuku et al. 2017). In a plethora of human organs, EVs are capable of regulating local and global metabolism and associated cellular processes (Iraci et al. 2016, 2017). In AMF symbiosis, could these EVs be involved in signaling and potential nutrient exchange as well?
AGRICULTURAL SIGNIFICANCE OF SYMBIOTIC PHOSPHATE UPTAKE
Understanding molecular and physiological processes of symbiotic P uptake have translational importance to agriculture where P is often limiting crop growth and supplemented by fertilizers. In all plant species tested, the direct and indirect P uptake pathways appear to be distinct, nonadditive physiological alternatives (Smith 2003; Bucher 2007). Using radioisotopes of 32P/33P accessible by AMF hyphae but not plant roots allows the relative contributions of direct and indirect P to be determined. Early radiotracer experiments showed that the indirect pathway can dominate total P uptake, even when the plants (e.g., cucumber, wheat, barley, and tomato) do not display increased biomass or total P increase over the nonmycorrhizal plants (Pearson and Jakobsen 1993; Morel and Plenchette 1994; Ravnskov and Jakobsen 1995; Schweiger and Jakobsen 1999; Zhu et al. 2003; Smith et al. 2004; Yang et al. 2012). In species such as tomato and flax, the mycorrhizal pathway contributed 100% of Pi acquired by the plant (Smith et al. 2004). Such physiological experiments also demonstrate that the quantities of Pi delivered by AMF correlate strongly with the extraradical hyphae abundance, rather than percentage of intraradical structures (Schweiger and Jakobsen 1999; Jakobsen et al. 2001; Yao et al. 2001; Schnepf et al. 2008). Interestingly, a similar correlation between mycorrhizal Pi delivery and extraradical hyphae abundance was also observed in a diversity panel of 30 maize lines (Sawers et al. 2017), and it will be interesting to investigate whether a recent diversity analysis of 14 Sorghum accessions produces the same observations (Watts-Williams et al. 2018).
Importantly, these physiological observations imply that in agricultural and natural ecosystems, symbiotic P uptake might be the predominant uptake pathway for AM host species and is therefore crucial for crop nutrition. For example, in rice, AMF contributes up to 70% of the P acquired by the plant (Yang et al. 2012). Second, the nonadditive interaction may explain the lack of simple additive growth/Pi acquisition benefits with AMF observed in physiological experiments. Third, experiments such as those in Sawers et al. (2017), Watts-Williams et al. (2018), and Zhang et al. (2018) suggest that plant genetics, fungal genotypes, and associated bacteria together determine mycorrhizal benefit derived from symbiosis, forming the basis for complex crop-breeding programs in the future (see Toju et al. 2018). Crop-domestication and -breeding programs to date might have selected for AM independence under high Pi, well-fertilized conditions (Martín-Robles et al. 2018). Thus, beyond Pi-directed growth benefits, the additional benefits such as drought, salinity (abiotic stress) tolerance, and resistance to other biotic agents conferred by AMF (Newsham et al. 1995; Herre et al. 2007) should also be considered in modeling and implementing mycorrhizal enrichment as well as plant-breeding programs. Finally, it will be interesting to investigate whether derepressing the direct uptake pathway during symbiosis, if possible, could significantly increase total P uptake or the replacement of Pi fertilizers, and whether it reciprocally affects stages or the extent of AM symbiosis.
Looking forward, significant benefits of symbiotic Pi delivery can be harnessed, considering that modern industrial agricultural practices including the intensive use of fertilizers, fungicides, long fallow periods, and tilling have diminished the contribution of symbiotic Pi to crop nutrition (Sawers et al. 2008, 2018; Pérez-Jaramillo et al. 2016). The nonrenewable and politically tenuous nature of mined rock phosphates as well as pollutive impact of intense fertilizer application in industrial agriculture have led to the growing awareness and drive for more sustainable alternative mechanisms to improve crop P acquisition and P use efficiency (Cordell et al. 2009; Carpenter and Bennett 2011; Elser and Bennett 2011; Steffen et al. 2015).
ECOLOGICAL SIGNIFICANCE OF AM SYMBIOSIS
Empirically, AMF have variable effects on their hosts ranging from growth promotion to growth depression; and conversely AMF proliferation and sporulation varies with host species. Although surprising in deviating from the definition of a pure mutualistic symbiosis, it is less surprising considering that symbiotic trade necessitates costs and benefits, and the fitness benefits need not necessarily manifest as growth increases.
Plants and AMF May Select for More Cooperative Partners
In a mycorrhizal network, one plant can be colonized by multiple AMF species at once, and each AMF species can infect multiple hosts of the same or different plant species (Giovannetti et al. 2004). This has lent the symbiosis to many imaginative metaphors (e.g., “wood-wide web”; underground economy, etc.). Indeed, trade of commodities (nutrients) between multiple actors with different comparative advantages (in harnessing phosphate versus reduced carbon) invites analyses via the biological market theory and other models that allow hypotheses on resource exchange in different contexts to be formulated and tested (Noë and Hammerstein 1995; Kiers et al. 2003; Werner et al. 2014; Walder and van der Heijden 2015; Noë and Kiers 2018). Given the benefits of “cheating” in mutualism, does the plant and fungus select for a more cooperative partner (West et al. 2007; Kiers and Denison 2008)?
Several experiments suggest that a discrimination mechanism for reciprocal exchange exists—plants provided more C to fungal species that delivered more Pi (Bever et al. 2009; Kiers et al. 2011) and, likewise, AMF species provided more N/Pi to unshaded plants that likely delivered more C (Fellbaum et al. 2014). Collectively, these mechanisms might drive the amplification of size inequality in a network (Merrild et al. 2013; Weremijewicz and Janos 2013; Weremijewicz et al. 2016). However, in contrast to these observations, flax derived more nutrients than sorghum despite contributing less into the mycorrhizal network (Walder et al. 2012). Nevertheless, direct comparisons may be difficult, as the relative costs of C for a C4 plant (sorghum) may be different from that of a C3 plant (flax).
Cheating Partners May Be an Evolutionarily Stable Outcome
Overall, it appears that preferential resource allocation is able to drive evolution of mutualism even when the costs are immense, whereas coexistence of an exploitative partner is possible as a negative feedback in the long run in a mixed colonization situation (Foster and Kokko 2006; Steidinger and Bever 2016). These conclusions may help explain the diversity of plant-AMF interaction outcomes, as well as the evolutionary emergence of mycoheterotrophs that epitomize exploitative plant partners. Ultimately, symbiotic nutrient exchange occurs between a kingdom of plants and a subdivision of Glomeromycotina fungi, notwithstanding other symbioses. The diversity in trade terms is therefore not unexpected, and remains to be characterized across many other combinations and hierarchies with modeling tools.
Less Well-Understood Traits of AMF: Ecological and Agricultural Applications
Moreover, there are several aspects regarding the lifestyle and genetics of AMF that we are only beginning to understand. If and how diversity may lead to functional differences in plant symbiotic uptake is still unknown. Although ecologically successful, AMF only comprise ∼300 species in contrast to their broad host range (∼200,000 plant species). Its mycelium contains hundreds to thousands of nuclei within one continuous cytoplasm without undergoing formal sexual reproduction but capable of anastomosis. This has led to the long-standing enigma of how AMF of limited morphological diversity, are capable of colonizing roots of disparate genetic diversity over 400 million years (Judson and Normark 1996; Sanders 1999; Corradi and Brachmann 2017). Large-scale sequencing approaches have since revealed some insight into how diversity can be generated in spite of an apparent asexual lifestyle. First, AMF possess massive intraspecific genotypic variation, with the common laboratory strain R. irregularis alone possessing a pangenome of 150,000 genes and likely, as a result, considerable phenotypic variation even in a morphologically defined species (Mathieu et al. 2018). In addition, single-nucleus sequencing of AMF revealed that genetic diversity can be generated via internuclear recombination in a dikaryotic stage (two distinct mating loci) (Chen et al. 2018). Although rare in laboratory-cultured strains, how widespread are these processes in natural ecosystems? How functionally important is recombination toward symbiosis and long-term ecological fitness? Can an optimized, highly “cooperative” strain be developed to boost plant symbiotic P uptake?
Symbiont in a Symbiont: A Symbiotic Matryoshka?
In another symbiosis almost as old as AM symbiosis (Naumann et al. 2010; Mondo et al. 2012; Pawlowska et al. 2018), most AMF themselves harbor bacterial endosymbionts in their cytoplasm that are obligate biotrophs, although their function in symbiotic Pi uptake or in forming a tripartite mutualism remains to be fully demonstrated (Bianciotto et al. 2003, 2004; Naumann et al. 2010). Sequenced genomes of Candidatus Glomeribacter gigasporum (CaGg) or the more widespread Mollicutes-related endobacteria (MRE) reveal their lack of crucial metabolic pathways and consequent dependency on the fungal host for essential nutrients and energy (Ghignone et al. 2012; Naito et al. 2015; Torres-Cortés et al. 2015). Intriguingly, MRE form a sister lineage to the Mycoplasma groups, which are biotrophic extracellular parasites of animals (Torres-Cortés et al. 2015). Although the endobacteria are unculturable without a host, they are dispensable for AMF survival insofar that the few species of AMF cured of bacteria examined to date can still establish root symbiosis (Lumini et al. 2007; Bonfante 2014; Bonfante and Desirò 2017). The existence of endobacteria dependent on AMF, which themselves are dependent on plants, raises exciting questions on the dynamics and function of the interphylum interactions and nutrient transfer in the plant-fungi-bacteria holobiont. As the fungal–bacterium association is vertically transmitted, pan-global, evolutionarily stable, and ancient, it is likely that they provide fitness benefits to the fungal host (Bianciotto et al. 2004; Mondo et al. 2012; Bonfante and Desirò 2017; Pawlowska et al. 2018). Possible roles of endobacteria have been proposed to include nutrient transfer to and activating metabolic reprograming of AMF to increase success of colonizing host plants (Lumini et al. 2007; Salvioli et al. 2016; Vannini et al. 2016; Venice et al. 2017; Dearth et al. 2018), as well as transkingdom gene transfer for putative effector-like proteins with an extended phenotype on the plant (Torres-Cortés et al. 2015).
PERSPECTIVES AND CONCLUSION
Perspective: The Paradox of Phosphate Starvation during AM Symbiosis
Phosphate not only regulates local arbuscule development; it also has well-known systemic regulatory roles in symbiosis signaling. AM colonization is repressed under high Pi supply (Mosse 1973; Branscheid et al. 2010; Balzergue et al. 2011, 2013; Kobae et al. 2016) and infection attempts in maize roots were found to be inversely proportional to shoot Pi status (Braunberger et al. 1991). This regulation is systemic, as split-root experiments demonstrated that high Pi on one side suppressed AM symbiosis globally (Branscheid et al. 2010; Breuillin et al. 2010; Balzergue et al. 2011). The root-to-shoot signal remains elusive. Overexpression of miR399, members of which are well-described systemic Pi-starvation signals, failed to restore AM colonization under high Pi levels (Branscheid et al. 2010). In addition, high phosphate suppresses SL biosynthesis, which attenuates the level of plant-to-fungus signal during presymbiotic signaling. Nevertheless, the exogenous application of a synthetic SL, GR24, failed to restore AM colonization at high Pi levels (Breuillin et al. 2010; Balzergue et al. 2011), indicating that reduced SLs in the rhizosphere is insufficient to explain the suppressive effect. However, perinuclear Ca2+-oscillations were still generated in response to AMF hyphopodia under high Pi levels (Balzergue et al. 2013). This suggests that the Ca2+-oscillation machinery is not affected, but does not rule out Pi-suppression of presymbiotic signals leading to hyphopodia formation, or of the transcriptional activation to accommodate AMF downstream of Ca2+ oscillations. Finally, we also cannot rule out intrinsic inhibitory responses of AMF under high Pi. How AMF sense and respond to high Pi and subsequent root colonization dynamics is also relatively unclear. Evidence so far suggest that high Pi treatment may decrease expression of secreted AMF proteins, including STRIGOLACTONE-INDUCED SECRETED PROTEIN1 (SIS1) that positively regulates AMF colonization; as well as cell-cycle regulatory genes, DNA replication, and mitosis-related genes in the IRM but not ERM extraradical mycelium (Kikuchi et al. 2014; Tsuzuki et al. 2016; Sugimura and Saito 2017). It is, however, a challenge to uncouple intrinsic AMF responses from plant/exudate-mediated responses in a plant-AMF coculture system.
In addition, recent work on biotic interactions in the non-AM host Arabidopsis illustrates that the continuity from mutualism to parasitism along the symbiosis spectrum (Box 1) can shift with phosphate availability and phosphate starvation signaling. For one, C. toefieldiae acts as a Pi-delivering endophyte without causing disease only under Pi limitation. When Pi is abundant, plant defense genes are induced and growth-promoting benefits of the endophyte are abolished (Hiruma et al. 2016). Whereas in mutants of phr1, phl1 where transcriptional phosphate starvation responses (PSRs) are lost, defense genes were up-regulated and the root microbiota composition was atypical to that of wild-type in a synthetic community setup. This suggests that PSRs, under homeostatic conditions, have regulatory roles on the defense response and correspondingly shape the root microbiome (Castrillo et al. 2017). Finally, in the AM-host maize, high Pi activated defense responses, shifted fungal community, and reduced AMF colonization especially in lateral roots (Yu et al. 2018).
Therefore, during symbiosis, AMF experience a “Pi regulation conundrum” (Fig. 3). PSR promotes AMF colonization, but symbiotic Pi exchange and subsequent mobilization to the shoot could relieve the PSR and activate defense gene expression to terminate symbiosis. Indeed, symbiotic Pi uptake mediated by MtPT4/OsPT11 is required for down-regulating the direct Pi uptake arm of PSR (Harrison et al. 2002; Paszkowski et al. 2002; Javot et al. 2007a; Yang et al. 2012; Watts-Williams et al. 2015). However, AMF colonization steadily increases in wild-type plants, suggesting that symbiosis signaling also down-regulates certain aspects of the PSR to favor fungal accommodation over exclusionary defense. The exact molecular mechanisms mediating the differential regulation of PSR during symbiosis, however, is unclear.
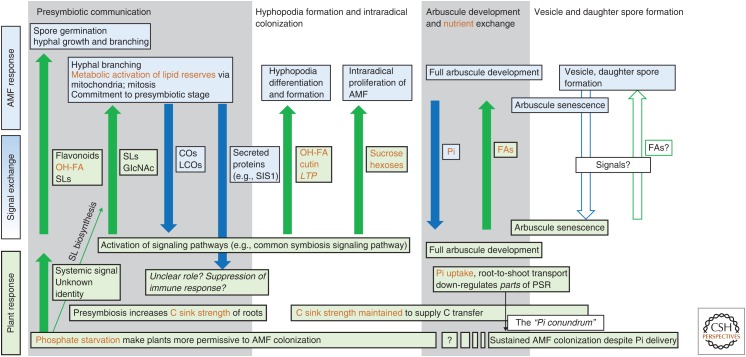
Figure 3.
Nutrients play important signaling roles at various stages of symbiosis. The first stages of symbiosis signaling result in substantial reprogramming of both plant and arbuscular mycorrhizal fungi (AMF) metabolism. Crucially, the reprogramming commits the plant to supply AMF with carbon via hydroxyl fatty acids (OH-FA), cutin, and lipid transfer proteins (LTPs) at the presymbiotic and contact stages and via sugars in the intraradical stage. At the heart of the symbiosis, a fully functional bidirectional exchange of Pi and fatty acids is required for signaling as much as nutritional benefit. Together, the plant and fungus elicit, via and as a result of signal exchanges, metabolic changes in the other party. Several gaps in understanding remain—including the nature of signals exchanged in the last stage of symbiosis where arbuscules degenerate, while vesicles and daughter spores form. Importantly, how AMF manages to deliver Pi without down-regulating the entirety of PSR to continue to allow sustained symbiosis development at new, secondary infection sites remains unclear.
Conclusion: Metabolic Reprogramming and Nutrient Dependency Is a Hallmark of Symbiosis
Symbioses enabled the first eukaryotes to gain complex metabolic capabilities with the establishment of mitochondria and plastids (Mereschkowsky 1905; Margulis 1991); and continued to provide evolutionary innovations to enable functional and ecological diversity to occupy new niches. The symbiosis between plants and AMF is one of the many symbioses, but it is remarkable for its widespread occurrence, evolutionary success, and our level of mechanistic understanding. Metabolic interdependence has been invoked to explain symbiotic dependence, in particular between sap-feeding insects and obligate symbionts, where the latter provide amino acids, vitamins, and cofactor synthesis in exchange for host-derived sugars (Zientz et al. 2004; Wu et al. 2006; Luan et al. 2015; Wilson and Duncan 2015). Evolutionarily younger, artificial symbioses similarly reveal the gradual evolution of auxotrophy and nutritional interdependence for mutualism to develop. This includes the Chlamydomonas reinhardtii—Saccharomyces cerevisiae exchange of carbon and nitrogen (Hom and Murray 2014); Lobomonas rostrate—Mesorhizobium loti exchange of carbon for Vitamin B12 (Helliwell et al. 2018); as well as Synechocystis PCC6803—Paramecium bursaria exchange of carbon for nitrogen (Sørensen et al. 2016), to name a few.
Nutrient interdependency occurs in the AM symbiosis too. Plants, when colonized, can become predominantly reliant on AMF for phosphate nutrition, whereas the fungal symbiont, probably as a result of the readily available C transfer from plants, lost the ability to synthesize fatty acids over evolutionary time. Moreover, on top of long-term evolutionary dynamics, the signaling processes required for successful symbiosis establishment appear to involve nutrient dependency at all stages (Fig. 3). Presymbiotic signaling itself induces transcriptional and metabolic reprogramming in hosts, increasing C flux and C sink strength. On the fungal end, perception of SL, uptake of plant-released fatty acids (e.g., of hydroxyfatty acids, or of cutin), and monosaccharides drive metabolism, although potentially maintaining an altered plant metabolic status for its growth inside the root (Nagahashi et al. 2010; Helber et al. 2011; Wang et al. 2012). Especially at the stage of arbuscule development, it is now clear that sustained nutrient transfer is not just the ultimate goal, but a necessary signal. It will be exciting to unravel exactly how nutrient signals recur and mediate the various stages of symbiosis and vice versa. For one, emerging frontiers such as EVs at the biogenesis and collapse of the arbuscule offer further, yet uncharacterized opportunities for ways to mediate nutrient and signal exchanges between organisms. Understanding how plants, AMF, as well as the AMF-associated microbiome engage in the symbiotic nutrient trade strategies maintained over millions of years will, importantly, help provide solutions for the phosphate challenge modern agriculture faces.
ACKNOWLEDGMENTS
C.H.C. is supported by a Gates Cambridge PhD Scholarship. Research in the laboratory of U.P. is supported by the BBSRC Grant No. BB/N008723/1.
Footnotes
Editor: Pamela C. Ronald
Additional Perspectives on Engineering Plants for Agriculture available at www.cshperspectives.org
REFERENCES
- Ait Lahmidi N, Courty PE, Brulé D, Chatagnier O, Arnould C, Doidy J, Berta G, Lingua G, Wipf D, Bonneau L. 2016. Sugar exchanges in arbuscular mycorrhiza: RiMST5 and RiMST6, two novel Rhizophagus irregularis monosaccharide transporters, are involved in both sugar uptake from the soil and from the plant partner. Plant Physiol Biochem 107: 354–363. 10.1016/j.plaphy.2016.06.023 [DOI] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Alexander T, Meier R, Toth R, Weber HC. 1988. Dynamics of arbuscule development and degeneration in mycorrhizas of Triticum aestivum L. and Avena sativa L. with reference to Zea mays L. New Phytol 110: 363–370. 10.1111/j.1469-8137.1988.tb00273.x [DOI] [Google Scholar]
- Aono T, Maldonado-Mendoza IE, Dewbre GR, Harrison MJ, Saito M. 2004. Expression of alkaline phosphatase genes in arbuscular mycorrhizas. New Phytol 162: 525–534. 10.1111/j.1469-8137.2004.01041.x [DOI] [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y. 2012. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J 71: 479–491. 10.1111/j.1365-313X.2012.05004.x [DOI] [PubMed] [Google Scholar]
- Azevedo C, Saiardi A. 2017. Eukaryotic phosphate homeostasis: The inositol pyrophosphate perspective. Trends Biochem Sci 42: 219–231. 10.1016/j.tibs.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Bago B, Bécard G. 2002. Bases of the obligate biotrophy of arbuscular mycorrhizal fungi. In Mycorrhizal technology in agriculture: From genes to bioproducts (ed. Gianinazzi S, Schüepp H, Barea JM, Haselwandter K), pp. 33–48. Birkhäuser, Basel, Switzerland. [Google Scholar]
- Bago B, Pfeffer PE, Shachar-Hill Y. 2000. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol 124: 949–958. 10.1104/pp.124.3.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Zipfel W, Williams RM, Jun J, Arreola R, Lammers PJ, Pfeffer PE, Shachar-Hill Y. 2002. Translocation and utilization of fungal storage lipid in the arbuscular mycorrhizal symbiosis. Plant Physiol 128: 108–124. 10.1104/pp.010466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Pfeffer PE, Abubaker J, Jun J, Allen JW, Brouillette J, Douds DD, Lammers PJ, Shachar-Hill Y. 2003. Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol 131: 1496–1507. 10.1104/pp.102.007765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R, Gómez-Ariza J, Lanfranco L, Bonfante P. 2007. Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol Plant-Microbe Interact 20: 1055–1062. [DOI] [PubMed] [Google Scholar]
- Balzergue C, Puech-Pagès V, Bécard G, Rochange SF. 2011. The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J Exp Bot 62: 1049–1060. 10.1093/jxb/erq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzergue C, Chabaud M, Barker DG, Bécard G, Rochange SF. 2013. High phosphate reduces host ability to develop arbuscular mycorrhizal symbiosis without affecting root calcium spiking responses to the fungus. Front Plant Sci 4: 426 10.3389/fpls.2013.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battini F, Grønlund M, Agnolucci M, Giovannetti M, Jakobsen I. 2017. Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci Rep 7: 4686 10.1038/s41598-017-04959-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. 2006. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226 10.1371/journal.pbio.0040226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. 2009. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol Lett 12: 13–21. 10.1111/j.1461-0248.2008.01254.x [DOI] [PubMed] [Google Scholar]
- Bianciotto V, Lumini E, Bonfante P, Vandamme P. 2003. “Candidatus Glomeribacter gigasporarum” gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int J Syst Evol Microbiol 53: 121–124. 10.1099/ijs.0.02382-0 [DOI] [PubMed] [Google Scholar]
- Bianciotto V, Genre A, Jargeat P, Lumini E, Bécard G, Bonfante P. 2004. Vertical transmission of endobacteria in the arbuscular mycorrhizal fungus Gigaspora margarita through generation of vegetative spores. Appl Environ Microbiol 70: 3600–3608. 10.1128/AEM.70.6.3600-3608.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, Duckett JG. 2011. The dawn of symbiosis between plants and fungi. Biol Lett 7: 574–577. 10.1098/rsbl.2010.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. 2014. Bacterial vesicles in marine ecosystems. Science 343: 183–186. 10.1126/science.1243457 [DOI] [PubMed] [Google Scholar]
- Bonfante P. 2001. At the interface between mycorrhizal fungi and plants: The structural organization of cell wall, plasma membrane and cytoskeleton. In The mycota: Fungal associations (ed. Hock B), pp. 45–61. Springer-Verlag, Berlin. [Google Scholar]
- Bonfante P. 2014. The endless tale of endobacteria: A conversation with Paola Bonfante. Trends Plant Sci 19: 744–746. 10.1016/j.tplants.2014.08.008 [DOI] [PubMed] [Google Scholar]
- Bonfante P, Desirò A. 2017. Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in Mucoromycota. ISME J 11: 1727–1735. 10.1038/ismej.2017.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands M, Wewer V, Keymer A, Gutjahr C, Dörmann P. 2018. The Lotus japonicus acyl–acyl carrier protein thioesterase FatM is required for mycorrhiza formation and lipid accumulation of Rhizophagus irregularis. Plant J 95: 219–232. 10.1111/tpj.13943 [DOI] [PubMed] [Google Scholar]
- Branscheid A, Sieh D, Pant BD, May P, Devers EA, Elkrog A, Schauser L, Scheible WR, Krajinski F. 2010. Expression pattern suggests a role of miR399 in the regulation of the cellular response to local Pi increase during arbuscular mycorrhizal symbiosis. Mol Plant Microbe Interact 23: 915–926. 10.1094/MPMI-23-7-0915 [DOI] [PubMed] [Google Scholar]
- Braunberger PG, Miller MH, Peterson RL. 1991. Effect of phosphorus nutrition on morphological characteristics of vesicular-arbuscular mycorrhizal colonization of maize. New Phytol 119: 107–113. 10.1111/j.1469-8137.1991.tb01013.x [DOI] [PubMed] [Google Scholar]
- Bravo A, York T, Pumplin N, Mueller LA, Harrison MJ. 2016. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat Plants 2: 15208 10.1038/nplants.2015.208 [DOI] [PubMed] [Google Scholar]
- Bravo A, Brands M, Wewer V, Dörmann P, Harrison MJ. 2017. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol 214: 1631–1645. 10.1111/nph.14533 [DOI] [PubMed] [Google Scholar]
- Breuillin F, Schramm J, Hajirezaei M, Ahkami A, Favre P, Druege U, Hause B, Bucher M, Kretzschmar T, Bossolini E, et al. 2010. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J 64: 1002–1017. 10.1111/j.1365-313X.2010.04385.x [DOI] [PubMed] [Google Scholar]
- Breuillin-Sessoms F, Floss DS, Gomez SK, Pumplin N, Ding Y, Levesque-Tremblay V, Noar RD, Daniels DA, Bravo A, Eaglesham JB, et al. 2015. Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 Family Protein AMT2;3. Plant Cell 27: 1352–1366. 10.1105/tpc.114.131144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC. 2002. Coevolution of roots and mycorrhizas of land plants. New Phytol 154: 275–304. 10.1046/j.1469-8137.2002.00397.x [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Tedersoo L. 2018. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol 220: 1108–1115. 10.1111/nph.14976 [DOI] [PubMed] [Google Scholar]
- Bucher M. 2007. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173: 11–26. 10.1111/j.1469-8137.2006.01935.x [DOI] [PubMed] [Google Scholar]
- Bun-Ya M, Nishimura M, Harashima S, Oshima Y. 1991. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11: 3229–3238. 10.1128/MCB.11.6.3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C, Jardinaud MF, Rengel D, Carrère S, Hervé C, Debellé F, Gamas P, Bensmihen S, Gough C. 2015. Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol 208: 224–240. 10.1111/nph.13427 [DOI] [PubMed] [Google Scholar]
- Capaccio LCM, Callow JA. 1982. The enzymes of polyphosphate metabolism in vesicular-arbuscular mycorrhizas. New Phytol 91: 81–91. 10.1111/j.1469-8137.1982.tb03294.x [DOI] [Google Scholar]
- Carotenuto G, Chabaud M, Miyata K, Capozzi M, Takeda N, Kaku H, Shibuya N, Nakagawa T, Barker DG, Genre A. 2017. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol 214: 1440–1446. 10.1111/nph.14539 [DOI] [PubMed] [Google Scholar]
- Carpenter SR, Bennett EM. 2011. Reconsideration of the planetary boundary for phosphorus. Environ Res Lett 6: 014009 10.1088/1748-9326/6/1/014009 [DOI] [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, et al. 2017. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543: 513–518. 10.1038/nature21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Hu J, Sun S, Xu G. 2007. Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173: 817–831. [DOI] [PubMed] [Google Scholar]
- Chen A, Chen X, Wang H, Liao D, Gu M, Qu H, Sun S, Xu G. 2014. Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol 14: 61 10.1186/1471-2229-14-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EC, Mathieu S, Hoffrichter A, Sedzielewska-Toro K, Peart M, Pelin A, Ndikumana S, Ropars J, Dreissig S, Fuchs J, et al. 2018. Single nucleus sequencing reveals evidence of inter-nucleus recombination in arbuscular mycorrhizal fungi. eLife 7: e39813 10.7554/eLife.39813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206. 10.1146/annurev-arplant-042110-103849 [DOI] [PubMed] [Google Scholar]
- Chiu CH, Choi J, Paszkowski U. 2018. Independent signalling cues underpin arbuscular mycorrhizal symbiosis and large lateral root induction in rice. New Phytol 217: 552–557. 10.1111/nph.14936 [DOI] [PubMed] [Google Scholar]
- Choi J, Summers W, Paszkowski U. 2018. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annu Rev Phytopathol 56: 135–160. 10.1146/annurev-phyto-080516-035521 [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. 2009. The story of phosphorus: Global food security and food for thought. Glob Environ Change 19: 292–305. 10.1016/j.gloenvcha.2008.10.009 [DOI] [Google Scholar]
- Corradi N, Brachmann A. 2017. Fungal mating in the most widespread plant symbionts? Trends Plant Sci 22: 175–183. 10.1016/j.tplants.2016.10.010 [DOI] [PubMed] [Google Scholar]
- Cox G, Moran KJ, Sanders F, Nockolds C, Tinker PB. 1980. Translocation and transfer of nutrients in vesicular-arbuscular mycorrhizas. III: Polyphosphate granules and phosphorous translocation. New Phytol 84: 649–659. wiley.com/10.1111/j.1469-8137.1980.tb04778.x [Google Scholar]
- Dearth SP, Castro HF, Venice F, Tague ED, Novero M, Bonfante P, Campagna SR. 2018. Metabolome changes are induced in the arbuscular mycorrhizal fungus Gigaspora margarita by germination and by its bacterial endosymbiont. Mycorrhiza 28: 421–433. 10.1007/s00572-018-0838-8 [DOI] [PubMed] [Google Scholar]
- de Bary AH. 1879. De la symbiose. Rev Int Sci 3: 301–309. [Google Scholar]
- Delaux PM, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, Sekimoto H, Nishiyama T, Melkonian M, Pokorny L, et al. 2015. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci 112: 13390–13395. 10.1073/pnas.1515426112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer I, Spitz O, Kanonenberg K, Montag K, Handrich M, Ahmad S, Schott-Verdugo S, Navarro-Retamal C, Rubio-Meléndez ME, Gomez-Porras JL, et al. 2018. Nutrient exchange in arbuscular mycorrhizal symbiosis from a thermodynamic point of view. New Phytol 10.1111/nph.15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser J, Bennett E. 2011. A broken biogeochemical cycle. Nature 478: 29–31. 10.1038/478029a [DOI] [PubMed] [Google Scholar]
- Ezawa T, Saito K. 2018. How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol 220: 1116–1121. 10.1111/nph.15187 [DOI] [PubMed] [Google Scholar]
- Ezawa T, Smith SE, Smith FA. 2001. Differentiation of polyphosphate metabolism between the extra- and intraradical hyphae of arbuscular mycorrhizal fungi. New Phytol 149: 555–563. [DOI] [PubMed] [Google Scholar]
- Ezawa T, Hayatsu M, Saito M. 2005. A new hypothesis on the strategy for acquisition of phosphorus in arbuscular mycorrhiza: Up-regulation of secreted acid phosphatase gene in the host plant. Mol Plant Microbe Interact 18: 1046–1053. 10.1094/MPMI-18-1046 [DOI] [PubMed] [Google Scholar]
- Favre P, Bapaume L, Bossolini E, Delorenzi M, Falquet L, Reinhardt D. 2014. A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol 14: 333 10.1186/s12870-014-0333-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddermann N, Duvvuru Muni RR, Zeier T, Stuurman J, Ercolin F, Schorderet M, Reinhardt D. 2010. The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J 64: 470–481. 10.1111/j.1365-313X.2010.04341.x [DOI] [PubMed] [Google Scholar]
- Fellbaum CR, Mensah JA, Cloos AJ, Strahan GE, Pfeffer PE, Kiers ET, Bücking H. 2014. Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytol 203: 646–656. 10.1111/nph.12827 [DOI] [PubMed] [Google Scholar]
- Feng G, Su Y, Li X, Wang H, Zhang F, Tang C, Rengel Z. 2002. Histochemical visualisation of phosphatase released by arbuscular mycorrhizal fungi in soil. J Plant Nutr 25: 1 10.1081/PLN-120003932 [DOI] [Google Scholar]
- Ferrol N, Pozo MJ, Antelo M, Azcón-Aguilar C. 2002. Arbuscular mycorrhizal symbiosis regulates plasma membrane H+-ATPase gene expression in tomato plants. J Exp Bot 53: 1683–1687. 10.1093/jxb/erf014 [DOI] [PubMed] [Google Scholar]
- Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. 2015. Symbiotic options for the conquest of land. Trends Ecol Evol 30: 477–486. 10.1016/j.tree.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Floss DS, Levy JG, Lévesque-Tremblay V, Pumplin N, Harrison MJ. 2013. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 110: E5025–E5034. 10.1073/pnas.1308973110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DS, Gomez SK, Park HJ, MacLean AM, Müller LM, Bhattarai KK, Lévesque-Tremblay V, Maldonado-Mendoza IE, Harrison MJ. 2017. A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr Biol 27: 1206–1212. 10.1016/j.cub.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Foster KR, Kokko H. 2006. Cheating can stabilize cooperation in mutualisms. Proc R Soc B Biol Sci 273: 2233–2239. 10.1098/rspb.2006.3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. 2012. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539. 10.1038/nature11346 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. 2005. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499. 10.1105/tpc.105.035410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. 2008. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420. 10.1105/tpc.108.059014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, et al. 2013. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 190–202. 10.1111/nph.12146 [DOI] [PubMed] [Google Scholar]
- Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L, Cruveiller S, Bianciotto V, Piffanelli P, Lanfranco L, et al. 2012. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J 6: 136–145. 10.1038/ismej.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi S, Gianinazzi-Pearson V, Dexheimer J. 1979. Enzymatic studies on the metabolism of vesicular-arbuscular mycorrhiza. III: Ultrastructural localisation of acid and alkaline phosphatase in onion roots infected by Glomus mossae (Nicol. & Gerd.). New Phytol 82: 127–132. 10.1111/j.1469-8137.1979.tb07566.x. [DOI] [Google Scholar]
- Gianinazzi-Pearson V, Arnould C, Oufattole M, Arango M, Gianinazzi S. 2000. Differential activation of H+-ATPase genes by an arbuscular mycorrhizal fungus in root cells of transgenic tobacco. Planta 211: 609–613. 10.1007/s004250000323 [DOI] [PubMed] [Google Scholar]
- Giots F, Donaton MCV, Thevelein JM. 2003. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 47: 1163–1181. 10.1046/j.1365-2958.2003.03365.x [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Sbrana C, Avio L, Strani P. 2004. Patterns of below-ground plant interconnections established by means of arbuscular mycorrhizal networks. New Phytol 164: 175–181. 10.1111/j.1469-8137.2004.01145.x [DOI] [PubMed] [Google Scholar]
- Glassop D, Smith SE, Smith FW. 2005. Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222: 688–698. [DOI] [PubMed] [Google Scholar]
- Gu M, Chen A, Sun S, Xu G. 2016. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol Plant 9: 396–416. 10.1016/j.molp.2015.12.012 [DOI] [PubMed] [Google Scholar]
- Gümil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Ne Docquier M, Descombes P, et al. 2005. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci 102: 8066–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M. 2013. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29: 593–617. 10.1146/annurev-cellbio-101512-122413 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U. 2008. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005. 10.1105/tpc.108.062414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Casieri L, Paszkowski U. 2009. Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol 182: 829–837. 10.1111/j.1469-8137.2009.02839.x [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Sawers RJH, Marti G, Andrés-Hernández L, Yang SY, Casieri L, Angliker H, Oakeley EJ, Wolfender JL, Abreu-Goodger C, et al. 2015. Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc Natl Acad Sci 112: 6754–6759. 10.1073/pnas.1504142112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenberger M. 2000. Arbuscules of vesicular-arbuscular mycorrhizal fungi inhabit an acidic compartment within plant roots. Planta 211: 299–304. 10.1007/s004250000324 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. 2002. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902. 10.1105/tpc.000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Nishide H, Takeda N, Suzuki Y, Kawaguchi M, Saito K. 2015. RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol 56: 1490–1511. 10.1093/pcp/pcv071 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dixon RA. 1993. Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant Microbe Interact 6: 643–654. 10.1094/MPMI-6-643 [DOI] [Google Scholar]
- Harrison MJ, Ivanov S. 2017. Exocytosis for endosymbiosis: Membrane trafficking pathways for development of symbiotic membrane compartments. Curr Opin Plant Biol 38: 101–108. 10.1016/j.pbi.2017.04.019 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, van Buuren ML. 1995. A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378: 626–629. 10.1038/378626a0 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. 2002. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429. 10.1105/tpc.004861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck C, Kuhn H, Heidt S, Walter S, Rieger N, Requena N. 2016. Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr Biol 26: 2770–2778. 10.1016/j.cub.2016.07.059 [DOI] [PubMed] [Google Scholar]
- Helber N, Requena N. 2007. Expression of the fluorescence markers DsRed and GFP fused to a nuclear localization signal in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 177: 537–548. [DOI] [PubMed] [Google Scholar]
- Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. 2011. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23: 3812–3823. 10.1105/tpc.111.089813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Pandhal J, Cooper MB, Longworth J, Kudahl UJ, Russo DA, Tomsett E V, Bunbury F, Salmon DL, Smirnoff N, et al. 2018. Quantitative proteomics of a B12-dependent alga grown in coculture with bacteria reveals metabolic tradeoffs required for mutualism. New Phytol 217: 599–612. 10.1111/nph.14832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herre EA, Mejía LC, Kyllo DA, Rojas E, Maynard Z, Butler A, Van Bael SA. 2007. Ecological implications of anti-pathogen effects of tropical fungal endophytes and mycorrhizae. Ecology 88: 550–558. 10.1890/05-1606 [DOI] [PubMed] [Google Scholar]
- Hetherington AJ, Dolan L. 2018. Stepwise and independent origins of roots among land plants. Nature 561: 235–238. 10.1038/s41586-018-0445-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata N, Murase M, Tani C, Ohtomo R, Osaki M, Ezawa T. 2010. Polyphosphate has a central role in the rapid and massive accumulation of phosphorus in extraradical mycelium of an arbuscular mycorrhizal fungus. New Phytol 186: 285–289. 10.1111/j.1469-8137.2009.03168.x [DOI] [PubMed] [Google Scholar]
- Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O'Connell RJ, et al. 2016. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165: 464–474. 10.1016/j.cell.2016.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Yu P, Marcon C. 2018. Genetic control of root system development in maize. Trends Plant Sci 23: 79–88. 10.1016/j.tplants.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Hom EFY, Murray AW. 2014. Niche engineering demonstrates a latent capacity for fungal–algal mutualism. Science 345: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JJ, Park YS, Bravo A, Bhattarai KK, Daniels DA, Harrison MJ. 2012. Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta 236: 851–865. 10.1007/s00425-012-1677-z [DOI] [PubMed] [Google Scholar]
- Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, et al. 2009. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324: 513–516. 10.1126/science.1168120 [DOI] [PubMed] [Google Scholar]
- Hubberten HM, Sieh D, Zöller D, Hoefgen R, Krajinski F. 2015. Medicago truncatula Mtha1-2 mutants loose metabolic responses to mycorrhizal colonization. Plant Signal Behav 10: e989025 10.4161/15592324.2014.989025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman R, Hontelez J, Mysore KS, Wen J, Bisseling T, Limpens E. 2016. A symbiosis-dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host-microbe interface in symbiosis. New Phytol 211: 1338–1351. 10.1111/nph.13973 [DOI] [PubMed] [Google Scholar]
- Humphreys CP, Franks PJ, Rees M, Bidartondo MI, Leake JR, Beerling DJ. 2010. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun 1: 103 10.1038/ncomms1105 [DOI] [PubMed] [Google Scholar]
- Hunter T. 2012. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci 367: 2513–2516. 10.1098/rstb.2012.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Kobae Y, Omoto E, Tanaka A, Banba M, Takai S, Tamura Y, Hirose A, Komatsu K, Otagaki S, et al. 2014. The soybean mycorrhiza-inducible phosphate transporter gene, GmPT7, also shows localized expression at the tips of vein endings of senescent leaves. Plant Cell Physiol 55: 2102–2111. [DOI] [PubMed] [Google Scholar]
- Iraci N, Leonardi T, Gessler F, Vega B, Pluchino S. 2016. Focus on extracellular vesicles: Physiological role and signalling properties of extracellular membrane vesicles. Int J Mol Sci 17: 171 10.3390/ijms17020171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraci N, Gaude E, Leonardi T, Costa ASH, Cossetti C, Peruzzotti-Jametti L, Bernstock JD, Saini HK, Gelati M, Vescovi AL, et al. 2017. Extracellular vesicles are independent metabolic units with asparaginase activity. Nat Chem Biol 13: 951–955. 10.1038/nchembio.2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Harrison MJ. 2018. Accumulation of phosphoinositides in distinct regions of the periarbuscular membrane. New Phytol 221: 2213–2227. 10.1111/nph.15553 [DOI] [PubMed] [Google Scholar]
- Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, Bisseling T. 2012. Rhizobium–legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc Natl Acad Sci 109: 8316–8321. 10.1073/pnas.1200407109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Austin J, Berg RH, Harrison MJ. 2019. Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat Plants 5: 194–203. [DOI] [PubMed] [Google Scholar]
- Jakobsen I, Rosendahl L. 1990. Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plants. New Phytol 115: 77–83. 10.1111/j.1469-8137.1990.tb00924.x [DOI] [Google Scholar]
- Jakobsen I, Gazey C, Abbott LK. 2001. Phosphate transport by communities of arbuscular mycorrhizal fungi in intact soil cores. New Phytol 149: 95–103. 10.1046/j.1469-8137.2001.00006.x [DOI] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. 2007a. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 104: 1720–1725. 10.1073/pnas.0608136104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Pumplin N, Harrison MJ. 2007b. Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ 30: 310–322. 10.1111/j.1365-3040.2006.01617.x [DOI] [PubMed] [Google Scholar]
- Javot H, Varma Penmetsa R, Breuillin F, Bhattarai KK, Noar RD, Karen Gomez S, Zhang Q, Cook DR, Harrison MJ. 2011. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J 68: 954–965. 10.1111/j.1365-313X.2011.04746.x [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D, et al. 2017. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175. 10.1126/science.aam9970 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie Q, Wang W, Yang J, Zhang X, Yu N, Zhou Y, Wang E. 2018. Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol Plant 11: 1344–1359. 10.1016/j.molp.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Jin Y, Liu H, Luo D, Yu N, Dong W, Wang C, Zhang X, Dai H, Yang J, Wang E. 2016. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat Commun 7: 12433 10.1038/ncomms12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson OP, Normark BB. 1996. Ancient asexual scandals. Trends Ecol Evol 11: 41–46. 10.1016/0169-5347(96)81040-8 [DOI] [PubMed] [Google Scholar]
- Kamel L, Tang N, Malbreil M, San Clemente H, Le Marquer M, Roux C, Frei dit Frey N. 2017. The comparison of expressed candidate secreted proteins from two arbuscular mycorrhizal fungi unravels common and specific molecular tools to invade different host plants. Front Plant Sci 8: 124 10.3389/fpls.2017.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerlin SCL, Sharma PK, Prasad RB, Warshel A. 2013. Why nature really chose phosphate. Q Rev Biophys 46: 1–132. 10.1017/S0033583512000157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandashov V, Nagy R, Wegmüller S, Amrhein N, Bucher M. 2004. Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 101: 6285–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer A, Gutjahr C. 2018. Cross-kingdom lipid transfer in arbuscular mycorrhiza symbiosis and beyond. Curr Opin Plant Biol 44: 137–144. 10.1016/j.pbi.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Keymer A, Pimprikar P, Wewer V, Huber C, Brands M, Bucerius SL, Delaux PM, Klingl V, Röpenack-Lahaye EV, Wang TL, et al. 2017. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6: e29107 10.7554/eLife.29107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Denison RF. 2008. Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Annu Rev Ecol Evol Syst 39: 215–236. 10.1146/annurev.ecolsys.39.110707.173423 [DOI] [Google Scholar]
- Kiers ET, Rousseau RA, West SA, Denison RF. 2003. Host sanctions and the legume–rhizobium mutualism. Nature 425: 78–81. 10.1038/nature01931 [DOI] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, et al. 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882. 10.1126/science.1208473 [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Hijikata N, Yokoyama K, Ohtomo R, Handa Y, Kawaguchi M, Saito K, Ezawa T. 2014. Polyphosphate accumulation is driven by transcriptome alterations that lead to near-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus. New Phytol 204: 638–649. 10.1111/nph.12937 [DOI] [PubMed] [Google Scholar]
- Kim KY, Jordan D, McDonald GA. 1997. Effect of phosphate-solubilizing bacteria and vesicular-arbuscular mycorrhizae on tomato growth and soil microbial activity. Biol Fertil Soils 26: 79–87. 10.1007/s003740050347 [DOI] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al. 2005. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229. 10.1105/tpc.105.032714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppholz S, Kuhn H, Requena N. 2011. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21: 1204–1209. 10.1016/j.cub.2011.06.044 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Fujiwara T. 2014. Earliest colonization events of Rhizophagus irregularis in rice roots occur preferentially in previously uncolonized cells. Plant Cell Physiol 55: 1497–1510. 10.1093/pcp/pcu081 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Hata S. 2010. Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol 51: 341–353. 10.1093/pcp/pcq013 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Ohmori Y, Saito C, Yano K, Ohtomo R, Fujiwara T. 2016. Phosphate treatment strongly inhibits new arbuscule development but not the maintenance of arbuscule in mycorrhizal rice roots. Plant Physiol 171: 566–579. 10.1104/pp.16.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Maeda T, Yamaguchi K, Kameoka H, Tanaka S, Ezawa T, Shigenobu S, Kawaguchi M. 2018. The genome of Rhizophagus clarus HR1 reveals a common genetic basis for auxotrophy among arbuscular mycorrhizal fungi. BMC Genomics 19: 465 10.1186/s12864-018-4853-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Saito M. 2004. Possible involvement of hyphal phosphatase in phosphate efflux from intraradical hyphae isolated from mycorrhizal roots colonized by Gigaspora margarita. Mycol Res 108: 610–615. 10.1017/S0953756204009980 [DOI] [PubMed] [Google Scholar]
- Krajinski F, Hause B, Gianinazzi-Pearson V, Franken P. 2002. Mtha1, a plasma membrane H+-ATPase gene from Medicago truncatula, shows arbuscule-specific induced expression in mycorrhizal tissue. Plant Biol 4: 754–761. 10.1055/s-2002-37407 [DOI] [Google Scholar]
- Krajinski F, Courty PE, Sieh D, Franken P, Zhang H, Bucher M, Gerlach N, Kryvoruchko I, Zoeller D, Udvardi M, et al. 2014. The H+-ATPase HA1 of Medicago truncatula is essential for phosphate transport and plant growth during arbuscular mycorrhizal symbiosis. Plant Cell 26: 1808–1817. 10.1105/tpc.113.120436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. 2012. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344. 10.1038/nature10873 [DOI] [PubMed] [Google Scholar]
- Kuga Y, Saito K, Nayuki K, Peterson RL, Saito M. 2008. Ultrastructure of rapidly frozen and freeze-substituted germ tubes of an arbuscular mycorrhizal fungus and localization of polyphosphate. New Phytol 178: 189–200. 10.1111/j.1469-8137.2007.02345.x [DOI] [PubMed] [Google Scholar]
- Larose G, Chênevert R, Moutoglis P, Gagné S, Piché Y, Vierheilig H. 2002. Flavonoid levels in roots of Medicago sativa are modulated by the developmental stage of the symbiosis and the root colonizing arbuscular mycorrhizal fungus. J Plant Physiol 159: 1329–1339. 10.1078/0176-1617-00896 [DOI] [Google Scholar]
- Lefevre F, Baijot A, Boutry M. 2015. Plant ABC transporters: Time for biochemistry? Biochem Soc Trans 43: 931–936. 10.1042/BST20150108 [DOI] [PubMed] [Google Scholar]
- Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DGO, Mu D, Pang E, Cao H, Cha H, Lin T, et al. 2014. Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet 10: e1004078 10.1371/journal.pgen.1004078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xu Y, Jiang H, Jiang C, Du Y, Gong C, Wang W, Zhu S, Han G, Cheng B, et al. 2016a. Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int J Mol Sci 17: 930 10.3390/ijms17060930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Huang T-K, Yang S-Y, Hong Y-T, Huang S-M, Wang F-N, Chiang S-F, Tsai S-Y, Lu W-C, Chiou T-J. 2016b. Identification of plant vacuolar transporters mediating phosphate storage. Nat Commun 7: 11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xu Y, Han G, Wang W, Li X, Cheng B. 2018. Identification and functional characterization of a maize phosphate transporter induced by mycorrhiza formation. Plant Cell Physiol 59: 1683–1694. [DOI] [PubMed] [Google Scholar]
- Loth-Pereda V, Orsini E, Courty PE, Lota F, Kohler A, Diss L, Blaudez D, Chalot M, Nehls U, Bucher M, et al. 2011. Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol 156: 2141–2154. 10.1104/pp.111.180646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan JB, Chen W, Hasegawa DK, Simmons AM, Wintermantel WM, Ling KS, Fei Z, Liu SS, Douglas AE. 2015. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol 7: 2635–47. 10.1093/gbe/evv170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuehl LH, Oldroyd GED. 2017. Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr Biol 27: R952–R963. 10.1016/j.cub.2017.06.042 [DOI] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan G V, Breakspear A, Oldroyd GED, Eastmond PJ. 2017. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178. 10.1126/science.aan0081 [DOI] [PubMed] [Google Scholar]
- Lumini E, Bianciotto V, Jargeat P, Novero M, Salvioli A, Faccio A, Bécard G, Bonfante P. 2007. Presymbiotic growth and sporal morphology are affected in the arbuscular mycorrhizal fungus Gigaspora margarita cured of its endobacteria. Cell Microbiol 9: 1716–1729. 10.1111/j.1462-5822.2007.00907.x [DOI] [PubMed] [Google Scholar]
- MacLean AM, Bravo A, Harrison MJ. 2017. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 29: 2319–2335. 10.1105/tpc.17.00555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda D, Ashida K, Iguchi K, Chechetka SA, Hijikata A, Okusako Y, Deguchi Y, Izui K, Hata S. 2006. Knockdown of an arbuscular mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol 47: 807–817. 10.1093/pcp/pcj069 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63. 10.1038/nature09622 [DOI] [PubMed] [Google Scholar]
- Maldonado-Mendoza IE, Dewbre GR, Harrison MJ. 2001. A phosphate transporter gene from the extra-radical mycelium of an arbuscular mycorrhizal fungus Glomus intraradices is regulated in response to phosphate in the environment. Mol Plant Microbe Interact 14: 1140–1148. 10.1094/MPMI.2001.14.10.1140 [DOI] [PubMed] [Google Scholar]
- Margulis L. 1991. Symbiogenesis and symbionticism. In Symbiosis as a source of evolutionary innovation: Speciation and morphogenesis (ed. Margulis L, Fester R), pp. 1–14. MIT Press, Cambridge, MA. [PubMed] [Google Scholar]
- Marschner P. 2011. Marschner's mineral nutrition of higher plants, 3rd ed Academic, London. [Google Scholar]
- Martin F, Kohler A, Murat C, Veneault-Fourrey C, Hibbett DS. 2016. Unearthing the roots of ectomycorrhizal symbioses. Nat Rev Microbiol 14: 760–773. 10.1038/nrmicro.2016.149 [DOI] [PubMed] [Google Scholar]
- Martín-Robles N, Lehmann A, Seco E, Aroca R, Rillig MC, Milla R. 2018. Impacts of domestication on the arbuscular mycorrhizal symbiosis of 27 crop species. New Phytol 218: 322–334. 10.1111/nph.14962 [DOI] [PubMed] [Google Scholar]
- Marx C, Dexheimer J, Gianinazzi-Pearson V, Gianinazzi S. 1982. Enzymatic studies on the metabolism of vesicular-arbuscular mycorrhizas. IV: Ultracytoenzymological evidence (ATPase) for active transfer processes in the host-arbuscule interface. New Phytol 90: 37–43. 10.1111/j.1469-8137.1982.tb03238.x [DOI] [Google Scholar]
- Mathieu S, Cusant L, Roux C, Corradi N. 2018. Arbuscular mycorrhizal fungi: Intraspecific diversity and pangenomes. New Phytol 220: 1129–1134. 10.1111/nph.15275 [DOI] [PubMed] [Google Scholar]
- Mereschkowsky C. 1905. Über natur und ursprung der chromatophoren im pflanzenreiche. Biol Zent Bl 25: 593–604. [Google Scholar]
- Merrild MP, Ambus P, Rosendahl S, Jakobsen I. 2013. Common arbuscular mycorrhizal networks amplify competition for phosphorus between seedlings and established plants. New Phytol 200: 229–240. 10.1111/nph.12351 [DOI] [PubMed] [Google Scholar]
- Mills BJW, Batterman SA, Field KJ. 2018. Nutrient acquisition by symbiotic fungi governs Palaeozoic climate transition. Philos Trans R Soc B Biol Sci 373: 20160503 10.1098/rstb.2016.0503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud M-C, Bechtold N, Raghothama K, Nussaume L. 2004. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741. 10.1007/s11103-004-1965-5 [DOI] [PubMed] [Google Scholar]
- Mondo SJ, Toomer KH, Morton JB, Lekberg Y, Pawlowska TE. 2012. Evolutionary stability in a 400-million-year-old heritable facultative mutualism. Evolution 66: 2564–2576. [DOI] [PubMed] [Google Scholar]
- Morel C, Plenchette C. 1994. Is the isotopically exchangeable phosphate of a loamy soil the plant-available P? Plant Soil 158: 287–297. 10.1007/BF00009502 [DOI] [Google Scholar]
- Mosse B. 1973. Plant growth responses to vesicular-arbuscular mycorrhiza. New Phytol 72: 127–136. 10.1111/j.1469-8137.1973.tb02017.x [DOI] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. 2002. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–53. 10.1046/j.1365-313X.2002.01356.x [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Ané JM. 2011. Germinating spore exudates from arbuscular mycorrhizal fungi: Molecular and developmental responses in plants and their regulation by ethylene. Mol Plant Microbe Interact 24: 260–270. 10.1094/MPMI-06-10-0146 [DOI] [PubMed] [Google Scholar]
- Murray JD, Muni RRD, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, et al. 2011. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65: 244–252. 10.1111/j.1365-313X.2010.04415.x [DOI] [PubMed] [Google Scholar]
- Nadal M, Sawers R, Naseem S, Bassin B, Kulicke C, Sharman A, An G, An K, Ahern KR, Romag A, et al. 2017. An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat Plants 3: 17073 10.1038/nplants.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD, Ferhatoglu Y. 2010. Functional categories of root exudate compounds and their relevance to AM fungal growth. In Arbuscular mycorrhizas: Physiology and function (ed. Koltai H, Kapulnik Y), pp. 33–56. Springer, Dordrecht, Netherlands. [Google Scholar]
- Nagy R, Karandashov V, Chague V, Kalinkevich K, Tamasloukht M, Xu G, Jakobsen I, Levy AA, Amrhein N, Bucher M. 2005. The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J 42: 236–250. 10.1111/j.1365-313X.2005.02364.x [DOI] [PubMed] [Google Scholar]
- Nagy R, Vasconcelos MJV, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M. 2006. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol 8: 186–197. 10.1055/s-2005-873052 [DOI] [PubMed] [Google Scholar]
- Naito M, Morton JB, Pawlowska TE. 2015. Minimal genomes of mycoplasma-related endobacteria are plastic and contain host-derived genes for sustained life within Glomeromycota. Proc Natl Acad Sci 112: 7791–7796. 10.1073/pnas.1501676112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Schüßler A, Bonfante P. 2010. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J 4: 862–871. 10.1038/ismej.2010.21 [DOI] [PubMed] [Google Scholar]
- Newsham KK, Fitter AH, Watkinson AR. 1995. Arbuscular mycorrhiza protect an annual grass from root pathogenic fungi in the field. J Ecol 83: 991 10.2307/2261180 [DOI] [Google Scholar]
- Noë R, Hammerstein P. 1995. Biological markets. Trends Ecol Evol 10: 336–339. 10.1016/S0169-5347(00)89123-5 [DOI] [PubMed] [Google Scholar]
- Noë R, Kiers ET. 2018. Mycorrhizal markets, firms, and co-ops. Trends Ecol Evol 33: 777–789. 10.1016/j.tree.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. 2011. Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci 2: 83 10.3389/fpls.2011.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláh B, Brière C, Bécard G, Dénarié J, Gough C. 2005. Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J 44: 195–207. 10.1111/j.1365-313X.2005.02522.x [DOI] [PubMed] [Google Scholar]
- Oulhen N, Schulz BJ, Carrier TJ. 2016. English translation of Heinrich Anton de Bary's 1878 speech, “Die Erscheinung der Symbiose” (“De la symbiose”). Symbiosis 69: 131–139. 10.1007/s13199-016-0409-8 [DOI] [Google Scholar]
- Pan H, Oztas O, Zhang X, Wu X, Stonoha C, Wang E, Wang B, Wang D. 2016. A symbiotic SNARE protein generated by alternative termination of transcription. Nat Plants 2: 15197 10.1038/nplants.2015.197 [DOI] [PubMed] [Google Scholar]
- Park HJ, Floss DS, Levesque-Tremblay V, Bravo A, Harrison MJ. 2015. Hyphal branching during Arbuscule development requires reduced arbuscular mycorrhiza1. Plant Physiol 169: 2774–2788. 10.1104/pp.15.01155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M. 2008. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775. 10.1038/nrmicro1987 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. 2002. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 99: 13324–13329. 10.1073/pnas.202474599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowska TE, Gaspar ML, Lastovetsky OA, Mondo SJ, Real-Ramirez I, Shakya E, Bonfante P. 2018. Biology of fungi and their bacterial endosymbionts. Annu Rev Phytopathol 56: 289–309. 10.1146/annurev-phyto-080417-045914 [DOI] [PubMed] [Google Scholar]
- Pearson JN, Jakobsen I. 1993. The relative contribution of hyphae and roots to phosphorus uptake by arbuscular mycorrhizal plants, measured by dual labelling with 32P and 33P. New Phytol 124: 489–494. 10.1111/j.1469-8137.1993.tb03840.x [DOI] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L. 2014. Root architecture responses: In search of phosphate. Plant Physiol 166: 1713–1723. 10.1104/pp.114.244541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. 2016. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol 90: 635–644. 10.1007/s11103-015-0337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, Massicotte HB. 2004. Exploring structural definitions of mycorrhizas, with emphasis on nutrient-exchange interfaces. Can J Bot 82: 1074–1088. 10.1139/b04-071 [DOI] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: The microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799. 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- Pimprikar P, Gutjahr C. 2018. Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol 59: 673–690. 10.1093/pcp/pcy024 [DOI] [PubMed] [Google Scholar]
- Pimprikar P, Carbonnel S, Paries M, Katzer K, Klingl V, Bohmer MJ, Karl L, Floss DS, Harrison MJ, Parniske M, et al. 2016. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol 26: 987–998. 10.1016/j.cub.2016.01.069 [DOI] [PubMed] [Google Scholar]
- Poulsen KH, Nagy R, Gao LL, Smith SE, Bucher M, Smith FA, Jakobsen I. 2005. Physiological and molecular evidence for Pi uptake via the symbiotic pathway in a reduced mycorrhizal colonization mutant in tomato associated with a compatible fungus. New Phytol 168: 445–454. 10.1111/j.1469-8137.2005.01523.x [DOI] [PubMed] [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. 2010. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 61: 482–494. 10.1111/j.1365-313X.2009.04072.x [DOI] [PubMed] [Google Scholar]
- Pumplin N, Zhang X, Noar RD, Harrison MJ. 2012. Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci 109: E665–E672. 10.1073/pnas.1110215109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693. 10.1146/annurev.arplant.50.1.665 [DOI] [PubMed] [Google Scholar]
- Ramos AC, Martins MA, Okorokova-Façanha AL, Olivares FL, Okorokov LA, Sepúlveda N, Feijó JA, Façanha AR. 2009. Arbuscular mycorrhizal fungi induce differential activation of the plasma membrane and vacuolar H+ pumps in maize roots. Mycorrhiza 19: 69–80. 10.1007/s00572-008-0204-3 [DOI] [PubMed] [Google Scholar]
- Rasmussen N, Lloyd DC, Ratcliffe RG, Hansen PE, Jakobsen I. 2000. 31P NMR for the study of P metabolism and translocation in arbuscular mycorrhizal fungi. Plant Soil 226: 245–253. 10.1023/A:1026411801081 [DOI] [Google Scholar]
- Rausch C, Bucher M. 2002. Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37. 10.1007/s00425-002-0921-3 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M. 2001. A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–465. 10.1038/35106601 [DOI] [PubMed] [Google Scholar]
- Raven JA, Edwards D. 2001. Roots: Evolutionary origins and biogeochemical significance. J Exp Bot 52: 381–401. 10.1093/jxb/52.suppl_1.381 [DOI] [PubMed] [Google Scholar]
- Ravnskov S, Jakobsen I. 1995. Functional compatibility in arbuscular mycorrhizas measured as hyphal P transport to the plant. New Phytol 129: 611–618. 10.1111/j.1469-8137.1995.tb03029.x [DOI] [Google Scholar]
- Redecker D, Kodner R, Graham LE. 2000. Glomalean fungi from the Ordovician. Science 289: 1920–1921. [DOI] [PubMed] [Google Scholar]
- Remy W, Taylort TN, Hass H, Kerp H. 1994. Four hundred-million-year-old vesicular arbuscular mycorrhizae (Endomycorrhiae/symbiosis/fossil ft/mut). Proc Natl Acad Sci 91: 11841–11843. 10.1073/pnas.91.25.11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack GJ. 1997. Early forest soils and their role in Devonian global change. Science 276: 583–585. [DOI] [PubMed] [Google Scholar]
- Rich MK, Schorderet M, Bapaume L, Falquet L, Morel P, Vandenbussche M, Reinhardt D. 2015. The petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol 168: 788–797. 10.1104/pp.15.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich MK, Courty PE, Roux C, Reinhardt D. 2017. Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genomics 18: 589 10.1186/s12864-017-3988-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth R, Paszkowski U. 2017. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr Opin Plant Biol 39: 50–56. 10.1016/j.pbi.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Roth R, Chiapello M, Montero H, Gehrig P, Grossmann J, O'Holleran K, Hartken D, Walters F, Yang SY, Hillmer S, et al. 2018. A rice serine/threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat Commun 9: 4677 10.1038/s41467-018-06865-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth R, Hillmer S, Funaya C, Chiapello M, Schumacher K, Lo Presti L, Kahmann R, Paszkowski U. 2019. Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat Plants 5: 204–211. [DOI] [PubMed] [Google Scholar]
- Salvioli A, Ghignone S, Novero M, Navazio L, Venice F, Bagnaresi P, Bonfante P. 2016. Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J 10: 130–144. 10.1038/ismej.2015.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders IR. 1999. No sex please, we're fungi. Nature 399: 737–738. 10.1038/21544 [DOI] [PubMed] [Google Scholar]
- Sanders IR, Croll D. 2010. Arbuscular mycorrhiza: The challenge to understand the genetics of the fungal partner. Annu Rev Genet 44: 271–292. 10.1146/annurev-genet-102108-134239 [DOI] [PubMed] [Google Scholar]
- Sato T, Ezawa T, Cheng W, Tawaraya K. 2015. Release of acid phosphatase from extraradical hyphae of arbuscular mycorrhizal fungus Rhizophagus clarus. Soil Sci Plant Nutr 61: 269–274. 10.1080/00380768.2014.993298 [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Gutjahr C, Paszkowski U. 2008. Cereal mycorrhiza: An ancient symbiosis in modern agriculture. Trends Plant Sci 13: 93–97. 10.1016/j.tplants.2007.11.006 [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Svane SF, Quan C, Grønlund M, Wozniak B, Gebreselassie M-N, González-Muñoz E, Chávez Montes RA, Baxter I, Goudet J, et al. 2017. Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol 214: 632–643. 10.1111/nph.14403 [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Ramírez-Flores MR, Olalde-Portugal V, Paszkowski U. 2018. The impact of domestication and crop improvement on arbuscular mycorrhizal symbiosis in cereals: Insights from genetics and genomics. New Phytol 220: 1135–1140. 10.1111/nph.15152 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. 1998. Uptake phosphorus uptake by plants: From soil to cell. Plant Physiol 116: 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf A, Roose T, Schweiger P. 2008. Impact of growth and uptake patterns of arbuscular mycorrhizal fungi on plant phosphorus uptake—A modelling study. Plant Soil 312: 85–99. 10.1007/s11104-008-9749-3 [DOI] [Google Scholar]
- Schott S, Valdebenito B, Bustos D, Gomez-Porras JL, Sharma T, Dreyer I. 2016. Cooperation through competition-dynamics and microeconomics of a minimal nutrient trade system in arbuscular mycorrhizal symbiosis. Front Plant Sci 7: 912 10.3389/fpls.2016.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger PF, Jakobsen I. 1999. Direct measurement of arbuscular mycorrhizal phosphorus uptake into field-grown winter wheat. Agron J 91: 998–1002. 10.2134/agronj1999.916998x [DOI] [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG. 1995. Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol 108: 7–15. 10.1104/pp.108.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Yano K. 2003. Phosphorus acquisition from non-labile sources in peanut and pigeonpea with mycorrhizal interaction. Appl Soil Ecol 24: 133–141. 10.1016/S0929-1393(03)00093-3 [DOI] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. 2004. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642. 10.1111/j.1365-313X.2004.02161.x [DOI] [PubMed] [Google Scholar]
- Smith SE. 2003. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol 133: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis. Academic, London. [Google Scholar]
- Smith SE, Smith FA, Jakobsen I. 2003. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol 133: 16–20. 10.1104/pp.103.024380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Smith FA, Jakobsen I. 2004. Functional diversity in arbuscular mycorrhizal (AM) symbioses: The contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol 162: 511–524. 10.1111/j.1469-8137.2004.01039.x [DOI] [Google Scholar]
- Smith SE, Jakobsen I, Grønlund M, Smith FA. 2011. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: Interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol 156: 1050–1057. 10.1104/pp.111.174581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaiman MZ, Saito M. 1997. Use of sugars by intraradical hyphae of arbuscular mycorrhizal fungi revealed by radiorespirometry. New Phytol 136: 533–538. 10.1046/j.1469-8137.1997.00757.x [DOI] [PubMed] [Google Scholar]
- Solaiman MZ, Ezawa T, Kojima T, Saito M. 1999. Polyphosphates in intraradical and extraradical hyphae of an arbuscular mycorrhizal fungus, Gigaspora margarita. Appl Environ Microbiol 65: 5604–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MES, Wood AJ, Cameron DD, Brockhurst MA. 2016. Metabolic constraints for a novel symbiosis. R Soc Open Sci 3 10.1098/rsos.150708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y. 2011. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J 66: 689–699. 10.1111/j.1365-313X.2011.04532.x [DOI] [PubMed] [Google Scholar]
- Steffen W, Richardson K, Rockström J, Cornell SE, Fetzer I, Bennett EM, Biggs R, Carpenter SR, de Vries W, de Wit CA, et al. 2015. Planetary boundaries: Guiding human development on a changing planet. Science 347: 1259855 10.1126/science.1259855 [DOI] [PubMed] [Google Scholar]
- Steidinger BS, Bever JD. 2016. Host discrimination in modular mutualisms: A theoretical framework for meta-populations of mutualists and exploiters. Proc R Soc B Biol Sci 283: 20152428 10.1098/rspb.2015.2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner S, Lendzemo V, Langer I, Schweiger P, Khaosaad T, Toussaint JP, Vierheilig H, Steinkellner S, Lendzemo V, Langer I, et al. 2007. Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant–fungus interactions. Molecules 12: 1290–1306. 10.3390/12071290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strullu-Derrien C. 2018. Fossil filamentous microorganisms associated with plants in early terrestrial environments. Curr Opin Plant Biol 44: 122–128. 10.1016/j.pbi.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Strullu-Derrien C, Selosse MA, Kenrick P, Martin FM. 2018. The origin and evolution of mycorrhizal symbioses: From palaeomycology to phylogenomics. New Phytol 220: 1012–1030. 10.1111/nph.15076 [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Saito K. 2017. Transcriptional profiling of arbuscular mycorrhizal roots exposed to high levels of phosphate reveals the repression of cell cycle-related genes and secreted protein genes in Rhizophagus irregularis. Mycorrhiza 27: 139–146. 10.1007/s00572-016-0735-y [DOI] [PubMed] [Google Scholar]
- Sun J, Miller JB, Granqvist E, Wiley-Kalil A, Gobbato E, Maillet F, Cottaz S, Samain E, Venkateshwaran M, Fort S, et al. 2015a. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 27: 823–838. 10.1105/tpc.114.131326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Bonfante P, Tang M. 2015b. Effect of volatiles versus exudates released by germinating spores of Gigaspora margarita on lateral root formation. Plant Physiol Biochem 97: 1–10. 10.1016/j.plaphy.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Sun X, Chen W, Ivanov S, MacLean AM, Wight H, Ramaraj T, Mudge J, Harrison MJ, Fei Z. 2018a. Genome and evolution of the arbuscular mycorrhizal fungus Diversispora epigaea (formerly Glomus versiforme) and its bacterial endosymbionts. New Phytol 221: 1556–1573. 10.1111/nph.15472 [DOI] [PubMed] [Google Scholar]
- Sun Z, Song J, Xin X, Xie X, Zhao B. 2018b. Arbuscular mycorrhizal fungal 14-3-3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Front Microbiol 9: 91 10.3389/fmicb.2018.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen NB, Watts-Williams SJ, Joner EJ, Battini F, Efthymiou A, Cruz-Paredes C, Nybroe O, Jakobsen I. 2018. Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J 12: 1296–1307. 10.1038/s41396-018-0059-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Kobae Y, Toyotaka M, Hata S. 2012. Identification and expression analysis of arbuscular mycorrhiza-inducible phosphate transporter genes of soybean. Biotechnol Biochem 76: 309–313. 10.1271/bbb.110684 [DOI] [PubMed] [Google Scholar]
- Tang N, San Clemente H, Roy S, Bécard G, Zhao B, Roux C. 2016. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Front Microbiol 7: 233 10.3389/fmicb.2016.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani C, Ohtomo R, Osaki M, Kuga Y, Ezawa T. 2009. ATP-dependent but proton gradient-independent polyphosphate-synthesizing activity in extraradical hyphae of an arbuscular mycorrhizal fungus. Appl Environ Microbiol 75: 7044–7050. 10.1128/AEM.01519-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TN, Remy W, Hass H, Kerp H. 1995. Fossil arbuscular mycorrhizae from the early Devonian. Mycologia 87: 560 10.1080/00275514.1995.12026569 [DOI] [Google Scholar]
- Tisserant E, Kohler A, Dozolme-Seddas P, Balestrini R, Benabdellah K, Colard A, Croll D, Da Silva C, Gomez SK, Koul R, et al. 2012. The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol 193: 755–769. 10.1111/j.1469-8137.2011.03948.x [DOI] [PubMed] [Google Scholar]
- Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frei dit Frey N, Gianinazzi-Pearson V, et al. 2013. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci 110: 20117–20122. 10.1073/pnas.1313452110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, et al. 2018. Core microbiomes for sustainable agroecosystems. Nat Plants 4: 247–257. 10.1038/s41477-018-0139-4 [DOI] [PubMed] [Google Scholar]
- Toro M, Azcon R, Barea J. 1997. Improvement of arbuscular mycorrhiza development by inoculation of soil with phosphate-solubilizing rhizobacteria to improve rock phosphate bioavailability 32P and nutrient cycling. Appl Environ Microbiol 63: 4408–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cortés G, Ghignone S, Bonfante P, Schüßler A. 2015. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: Transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc Natl Acad Sci 112: 7785–7790. 10.1073/pnas.1501540112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Miller RM. 1984. Dynamics of arbuscule development and degeneration in a Zea mays mycorrhiza. Am J Bot 71: 449–460. 10.1002/j.1537-2197.1984.tb12530.x [DOI] [Google Scholar]
- Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H, Schmitt-Kopplin P, Eberl L, Nomura N. 2017. Membrane vesicle-mediated bacterial communication. ISME J 11: 1504–1509. 10.1038/ismej.2017.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki S, Handa Y, Takeda N, Kawaguchi M. 2016. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol Plant Microbe Interact 29: 277–286. 10.1094/MPMI-10-15-0234-R [DOI] [PubMed] [Google Scholar]
- Valat L, Deglène-Benbrahim L, Kendel M, Hussenet R, Le Jeune C, Schellenbaum P, Maillot P. 2018. Transcriptional induction of two phosphate transporter 1 genes and enhanced root branching in grape plants inoculated with Funneliformis mosseae. Mycorrhiza 28: 179–185. [DOI] [PubMed] [Google Scholar]
- van Aarle IM, Olsson PA, Soderstrom B. 2001. Microscopic detection of phosphatase activity of saprophytic and arbuscular mycorrhizal fungi using a fluorogenic substrate. Mycologia 93: 17 10.2307/3761601 [DOI] [Google Scholar]
- van Aarle IM, Rouhier H, Saito M. 2002. Phosphatase activities of arbuscular mycorrhizal intraradical and extraradical mycelium, and their relation to phosphorus availability. Mycol Res 106: 1224–1229. 10.1017/S0953756202006470 [DOI] [Google Scholar]
- Vannini C, Carpentieri A, Salvioli A, Novero M, Marsoni M, Testa L, de Pinto MC, Amoresano A, Ortolani F, Bracale M, et al. 2016. An interdomain network: The endobacterium of a mycorrhizal fungus promotes antioxidative responses in both fungal and plant hosts. New Phytol 211: 265–275. 10.1111/nph.13895 [DOI] [PubMed] [Google Scholar]
- Vejchasarn P, Lynch JP, Brown KM. 2016. Genetic variability in phosphorus responses of rice root phenotypes. Rice 9: 29 10.1186/s12284-016-0102-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venice F, de Pinto MC, Novero M, Ghignone S, Salvioli A, Bonfante P. 2017. Gigaspora margarita with and without its endobacterium shows adaptive responses to oxidative stress. Mycorrhiza 27: 747–759. 10.1007/s00572-017-0790-z [DOI] [PubMed] [Google Scholar]
- Viereck N, Hansen PE, Jakobsen I. 2004. Phosphate pool dynamics in the arbuscular mycorrhizal fungus Glomus intraradices studied by in vivo31P NMR spectroscopy. New Phytol 162: 783–794. 10.1111/j.1469-8137.2004.01048.x [DOI] [PubMed] [Google Scholar]
- Volpe V, Giovannetti M, Sun X-G, Fiorilli V, Bonfante P. 2016. The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non mycorrhizal roots. Plant Cell Environ 39: 660–671. [DOI] [PubMed] [Google Scholar]
- Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10: 828–840. 10.1038/nrmicro2910 [DOI] [PubMed] [Google Scholar]
- Voß S, Betz R, Heidt S, Corradi N, Requena N. 2018. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front Microbiol 9: 2068 10.3389/fmicb.2018.02068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG. 2008. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises l-glutamate-induced changes in root architecture. Plant J 54: 820–828. 10.1111/j.1365-313X.2008.03443.x [DOI] [PubMed] [Google Scholar]
- Walder F, van der Heijden MGA. 2015. Regulation of resource exchange in the arbuscular mycorrhizal symbiosis. Nat Plants 1: 15159 10.1038/nplants.2015.159 [DOI] [PubMed] [Google Scholar]
- Walder F, Niemann H, Natarajan M, Lehmann MF, Boller T, Wiemken A. 2012. Mycorrhizal networks: Common goods of plants shared under unequal terms of trade. Plant Physiol 159: 789–797. 10.1104/pp.112.195727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder F, Brulé D, Koegel S, Wiemken A, Boller T, Courty P. 2015. Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol 205: 1632–1645. 10.1111/nph.13292 [DOI] [PubMed] [Google Scholar]
- Wang B, Qiu YL. 2006. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16: 299–363. [DOI] [PubMed] [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GED. 2012. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol 22: 2242–2246. 10.1016/j.cub.2012.09.043 [DOI] [PubMed] [Google Scholar]
- Wang E, Yu N, Bano SA, Liu C, Miller AJ, Cousins D, Zhang X, Ratet P, Tadege M, Mysore KS, et al. 2014. A H+-ATPase that energizes nutrient uptake during mycorrhizal symbioses in rice and Medicago truncatula. Plant Cell 26: 1818–1830. 10.1105/tpc.113.120527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yue W, Ying Y, Wang S, Secco D, Liu Y, Whelan J, Tyerman SD, Shou H. 2015. Rice SPX-major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol 169: 2822–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts-Williams SJ, Jakobsen I, Cavagnaro TR, Grønlund M. 2015. Local and distal effects of arbuscular mycorrhizal colonization on direct pathway Pi uptake and root growth in Medicago truncatula. J Exp Bot 66: 4061–4073. 10.1093/jxb/erv202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts-Williams SJ, Emmett BD, Levesque-Tremblay V, MacLean AM, Sun X, Satterlee JW, Fei Z, Harrison MJ. 2018. Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ 10.1111/pce.13509 [DOI] [PubMed] [Google Scholar]
- Wegmüller S, Svistoonoff S, Reinhardt D, Stuurman J, Amrhein N, Bucher M. 2008. A transgenic dTph1 insertional mutagenesis system for forward genetics in mycorrhizal phosphate transport of Petunia. Plant J 54: 1115–1127. 10.1111/j.1365-313X.2008.03474.x [DOI] [PubMed] [Google Scholar]
- Weremijewicz J, Janos DP. 2013. Common mycorrhizal networks amplify size inequality in Andropogon gerardii monocultures. New Phytol 198: 203–213. 10.1111/nph.12125 [DOI] [PubMed] [Google Scholar]
- Weremijewicz J, Sternberg Lda SLO, Janos DP. 2016. Common mycorrhizal networks amplify competition by preferential mineral nutrient allocation to large host plants. New Phytol 212: 461–471. 10.1111/nph.14041 [DOI] [PubMed] [Google Scholar]
- Werner GDA, Strassmann JE, Ivens ABF, Engelmoer DJP, Verbruggen E, Queller DC, Noë R, Johnson NC, Hammerstein P, Kiers ET. 2014. Evolution of microbial markets. Proc Natl Acad Sci 111: 1237–1244. 10.1073/pnas.1315980111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A. 2007. Evolutionary explanations for cooperation. Curr Biol 17: R661–R672. 10.1016/j.cub.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Westheimer FH. 1987. Why nature chose phosphates. Science 235: 1173–1178. 10.1126/science.2434996 [DOI] [PubMed] [Google Scholar]
- Wewer V, Brands M, Dörmann P. 2014. Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus Rhizophagus irregularis during mycorrhization of Lotus japonicus. Plant J 79: 398–412. 10.1111/tpj.12566 [DOI] [PubMed] [Google Scholar]
- Willmann M, Gerlach N, Buer B, Polatajko A, Nagy R, Koebke E, Jansa J, Flisch R, Bucher M. 2013. Mycorrhizal phosphate uptake pathway in maize: Vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front Plant Sci 4: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ACC, Duncan RP. 2015. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci 112: 10255–10261. 10.1073/pnas.1423305112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL, et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol 4: e188 10.1371/journal.pbio.0040188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Huang W, Liu F, Tang N, Liu Y, Lin H, Zhao B. 2013. Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol 198: 836–852. 10.1111/nph.12188 [DOI] [PubMed] [Google Scholar]
- Xie X, Lin H, Peng X, Xu C, Sun Z, Jiang K, Huang A, Wu X, Tang N, Salvioli A, et al. 2016. Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Mol Plant 9: 1583–1608. 10.1016/j.molp.2016.08.011 [DOI] [PubMed] [Google Scholar]
- Xu GH, Chague V, Melamed-Bessudo C, Kapulnik Y, Jain A, Raghothama KG, Levy AA, Silber A. 2007. Functional characterization of LePT4: A phosphate transporter in tomato with mycorrhiza-enhanced expression. J Exp Bot 58: 2491–2501. 10.1093/jxb/erm096 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao H, Wan R, Liu Y, Xu Z, Tian W, Ruan W, Wang F, Deng M, Wang J, et al. 2019. Identification of vacuolar phosphate efflux transporters in land plants. Nat Plants 5: 84–94. [DOI] [PubMed] [Google Scholar]
- Xue L, Cui H, Buer B, Vijayakumar V, Delaux PM, Junkermann S, Bucher M. 2015. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol 167: 854–871. 10.1104/pp.114.255430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Klinnawee L, Zhou Y, Saridis G, Vijayakumar V, Brands M, Dörmann P, Gigolashvili T, Turck F, Bucher M. 2018. AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc Natl Acad Sci 115: E9239–E9246. 10.1073/pnas.1812275115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Paszkowski U. 2011. Phosphate import at the arbuscule: Just a nutrient? Mol Plant Microbe Interact 24: 1296–1299. 10.1094/MPMI-06-11-0151 [DOI] [PubMed] [Google Scholar]
- Yang SY, Grønlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Kumar CS, Sundaresan V, Salamin N, et al. 2012. Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell 24: 4236–4251. 10.1105/tpc.112.104901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Li X, Feng G, Christie P. 2001. Influence of extramatrical hyphae on mycorrhizal dependency of wheat genotypes. Commun Soil Sci Plant Anal 32: 3307–3317. 10.1081/CSS-120001122 [DOI] [Google Scholar]
- Yu N, Luo D, Zhang X, Liu J, Wang W, Jin Y, Dong W, Liu J, Liu H, Yang W, et al. 2014. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res 24: 130–133. 10.1038/cr.2013.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Wang C, Baldauf JA, Tai H, Gutjahr C, Hochholdinger F, Li C. 2018. Root type and soil phosphate determine the taxonomic landscape of colonizing fungi and the transcriptome of field-grown maize roots. New Phytol 217: 1240–1253. 10.1111/nph.14893 [DOI] [PubMed] [Google Scholar]
- Zeng T, Holmer R, Hontelez J, te Lintel-Hekkert B, Marufu L, de Zeeuw T, Wu F, Schijlen E, Bisseling T, Limpens E. 2018. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J 94: 411–425. 10.1111/tpj.13908 [DOI] [PubMed] [Google Scholar]
- Zhang X, Pumplin N, Ivanov S, Harrison MJ. 2015. EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Curr Biol 25: 2189–2195. 10.1016/j.cub.2015.06.075 [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu M, Liu Y, Zhang F, Hodge A, Feng G. 2016. Carbon and phosphorus exchange may enable cooperation between an arbuscular mycorrhizal fungus and a phosphate-solubilizing bacterium. New Phytol 210: 1022–1032. 10.1111/nph.13838. [DOI] [PubMed] [Google Scholar]
- Zhang L, Feng G, Declerck S. 2018. Signal beyond nutrient, fructose, exuded by an arbuscular mycorrhizal fungus triggers phytate mineralization by a phosphate solubilizing bacterium. ISME J 12: 2339–2351. 10.1038/s41396-018-0171-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lynch JP. 2004. The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays) seedlings. Funct Plant Biol 31: 949 10.1071/FP04046 [DOI] [PubMed] [Google Scholar]
- Zhu YG, Smith AF, Smith SE. 2003. Phosphorus efficiencies and responses of barley (Hordeum vulgare L.) to arbuscular mycorrhizal fungi grown in highly calcareous soil. Mycorrhiza 13: 93–100. 10.1007/s00572-002-0205-6 [DOI] [PubMed] [Google Scholar]
- Zientz E, Dandekar T, Gross R. 2004. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev 68: 745–770. 10.1128/MMBR.68.4.745-770.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]