Abstract
Loss of apical-basal polarity and activation of Epithelial-Mesenchymal Transition (EMT) both contribute to carcinoma progression and metastasis. Here, we report that apical-basal polarity inhibits EMT to suppress metastatic dissemination. Using mouse and human epithelial 3D organoid cultures, we show that the PAR/atypical protein kinase C (aPKC) polarity complex inhibits EMT and invasion by promoting degradation of SNAIL family protein SNAI1. Under intact apical-basal polarity, aPKC kinases phosphorylate SNAI1 on S249, leading to protein degradation. Loss of apical-basal polarity prevents aPKC-mediated SNAI1 phosphorylation and stabilizes SNAI1 protein to promote EMT and invasion. In human breast tumour xenografts, inhibition of the PAR complex-mediated SNAI1 degradation mechanism promotes tumour invasion and metastasis. Analyses of human breast tissue samples reveal negative correlations between PAR3 and SNAI1 protein levels. Our results demonstrate that apical-basal polarity functions as a critical checkpoint of EMT to precisely control epithelial-mesenchymal plasticity during tumour metastasis.
INTRODUCTION
The majority of human carcinomas show loss of epithelial apical-basal polarity during the progression from benign to invasive carcinoma. Apical-basal polarity is often regarded as a gatekeeper against tumour development and metastasis1, 2. Apical-basal polarity is regulated by three major polarity complexes, the PAR, Crumbs, and Scribble complexes3–5. The core PAR complex consists of Par3, Par6, and atypical PKCs (aPKCs) and is essential to define the apical domain of an epithelial cell6–8. Binding between Par6 and aPKCs, PKCζ or PKCι, promotes aPKCs to adopt an active and signalling-competent conformation to phosphorylate their substrates9. Loss of PAR3 results in dissociation of the PAR complex and loss of epithelial polarity10–12. PAR3 expression is frequently downregulated in primary tumour of various carcinoma types1, 2, 13.
Epithelial-Mesenchymal Transition (EMT) provides stationary carcinoma cells the ability to invade and disseminate during metastasis14–16. During EMT, epithelial cells first lose apical-basal polarity before weakening cell-cell junctions and rearranging cytoskeleton to acquire invasive capacities. This complex cellular program is orchestrated via coordinated activation of transcription factors, including the SNAIL, TWIST, and ZEB families17–20. SNAI1 is a key factor to suppress E-cadherin expression and to promote loss of epithelial characteristics, including epithelial polarity and junctions21–23.
Recent studies show that carcinoma cells undergo EMT to disseminate, while turning off EMT is needed for metastatic outgrowth24–26. While microenvironmental cues, including inflammation, hypoxia, and matrix stiffness are implicated in regulating epithelial mesenchymal plasticity27, 28, very little is known whether any intrinsic cellular machineries, such as apical-basal polarity and cell-cell junctions, could directly impinge on the EMT transcription factors to control EMT progression. Here, we employ three-dimensional (3D) epithelial organoid cultures that retain the integrity of apical-basal polarity in epithelial tissues to explore molecular links between epithelial cell polarity and EMT transcription factors and to define the impact of this feedback mechanism linking the PAR polarity complex and the SNAI1 transcription factor in tumour invasion and metastasis.
RESULTS
SNAI1 protein is destabilized and fails to induce EMT in 3D primary mouse mammary epithelial organoids
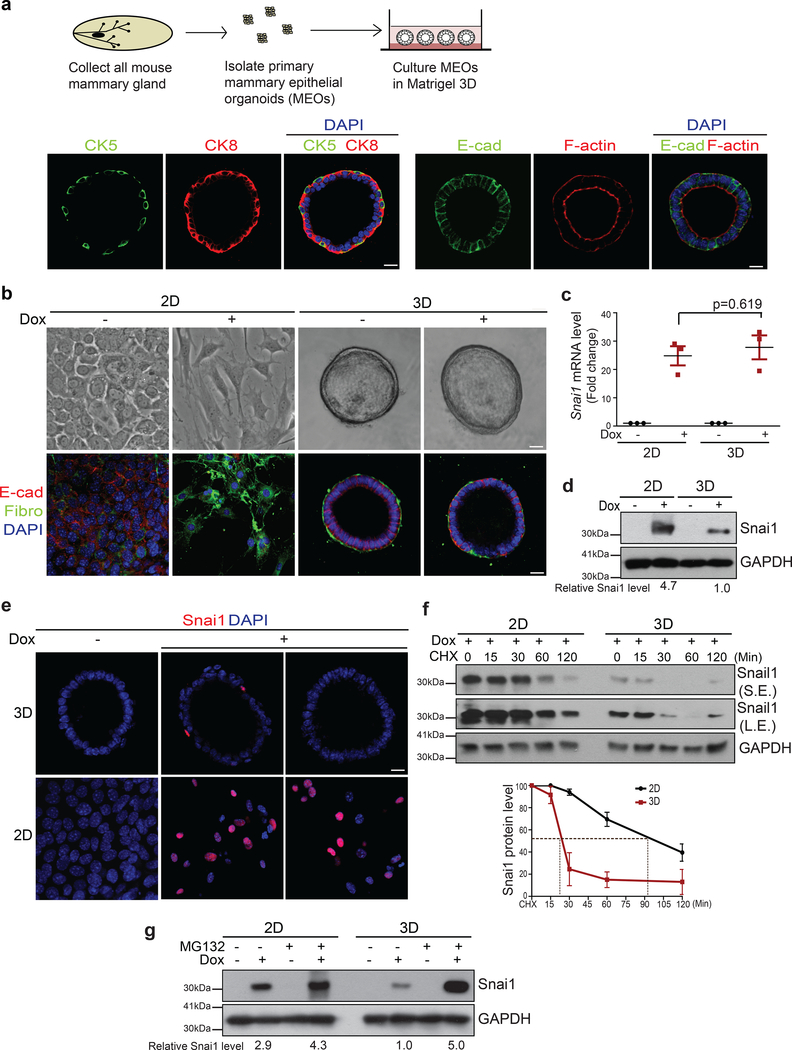
To determine the impact of apical-basal polarity on EMT progression, we employed a 3D organoids culture in which freshly isolated primary mouse mammary epithelial organoids (MEOs) form a bilayer ductal structure composed of the luminal and basal cell layers with a centrally localized, hollow lumen, membranous E-cadherin and cortical F-actin at the apical side29 (Fig. 1a). We isolated primary MEOs from the Tetracycline inducible SNAI1 (TetON-SNAI1) mice we generated. In 2D cultures, TetON-SNAI1 mammary epithelial cells (MECs) formed a monolayer with intact E-cadherin-mediated adherens junctions. Upon induction of SNAI1, these cells switched to a spindle-like cell shape and displayed many EMT features including reduced E-cadherin-mediated adherens junctions and increased Fibronectin expression within 4 days (Fig. 1b). In 3D cultures, MEOs formed hollow lumen and complete apical-basal polarity (Fig. 1b). To our surprise, these MEOs retained their acinar structure with intact adherens junctions upon SNAI1 induction (Fig. 1b). Consistent with these phenotypes, SNAI1 induction only resulted in E-cadherin mRNA suppression and vimentin mRNA expression in 2D, but not 3D cultures (Supplementary Fig. 1a). As a positive control, we also isolated primary MEOs from the TetON-TWIST1 mice we previously published25. Induction of TWIST1 in 3D MEOs and 2D MECs isolated from these TetON-TWIST1 mice both resulted in EMT and single cell invasion (Supplementary Fig. 1b–d), consistent with published data30. Together, these data show that in contrast to TWIST1, SNAI1 induction fails to promote EMT specifically in 3D polarized organoids.
Figure 1.
SNAI1 protein is unstable and fails to induce EMT in MEOs with intact apical-basal polarity. (a) The experimental scheme for isolating and culturing primary mouse epithelial organoids (MEOs). Immunofluorescence images for CK8 (cytokeratin 8), CK5 (cytokeratin 5), E-cadherin (E-cad), and F-actin in MEOs after 4 days of cultures. Scale bar, 25μm. (b) Bright-field images (top) and immunofluorescence images for E-cadherin and Fibronectin (Fibro) (bottom) in TetON-SNAI1 3D MEOs and 2D MECs cultures before and after doxycycline treatment for 4 days. Scale bar, 25μm. (c) qPCR analysis of relative SNAI1 mRNA levels normalized to GAPDH in TetON-SNAI1 MECs and MEOs before and after doxycycline induction. n=3 independent experiments, error bars represent standard deviation; unpaired two-tailed t-test with Welch’s correction. (d) Immunoblots for SNAI1 and GAPDH in lysate from TetON-SNAI1 MECs and MEOs before and after 4 days of doxycycline treatment. The values indicate relative signal intensities of SNAI1/GAPDH. (e) Immunofluorescence images for SNAI1 in TetON-SNAI1 MEOs and MECs before and after 4 days of doxycycline induction. Scale bar, 25μm. (f) TetON-SNAI1 MECs and MEOs were treated with doxycycline for 4 days and then treated with 10μM cycloheximide (CHX) following the time points indicated and analysed for SNAI1 and GAPDH by immunoblotting. S.E. indicates short exposure, L.E. indicates long exposure. The graph represents the quantification of relative SNAI1 proteins levels vs. GAPDH. n=3 independent experiments, error bars represent standard deviation. (g) TetON-SNAI1 MECs and MEOs were treated with doxycycline for 4 days and then treated with 10μM MG132 for 4hrs and analysed for SNAI1 and GAPDH by immunoblotting. The values indicate the relative signal intensity of SNAI1/GAPDH. All immunofluorescence images and Western Blots shown represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3 and unprocessed blots in Supplementary Figure 7.
We then examined SNAI1 mRNA and protein levels in 2D and 3D cultures. Doxycycline treatment induced SNAI1 mRNA to similar levels in 2D and 3D cultures (Fig. 1c). However, a significantly lower level of SNAI1 protein was detected in MEOs compared to MECs (Fig. 1d). Consistent with this, SNAI1 protein was detectable in only very few epithelial cells in MEOs, whereas the majority of MECs expressed SNAI1 protein after doxycycline treatment (Fig. 1e). In contrast, TWIST1 protein and mRNA could be easily detected in TetON-TWIST1 MEOs as well as in TetON-TWIST1 MECs (Supplementary Fig. 1e and 1f). These results show that despite potent induction of SNAI1 mRNA, SNAI1 protein level failed to increase in 3D polarized MEOs.
We examined SNAI1 protein degradation rates in 2D vs. 3D culture in the presence of cycloheximide. Interestingly, SNAI1 protein was remarkably unstable in MEOs with a half-life of around 20 min vs. over 100 min in MECs (Fig. 1f). Proteasome inhibitor treatment increased the SNAI1 protein level in MEOs to a similar level as in MECs (Fig. 1g). Together, these results show that SNAI1 protein stability is significantly reduced in 3D polarized MEOs, which might be responsible for the failure of SNAI1 to promote EMT.
Loss of apical-basal polarity increases SNAI1 stability in mammary epithelial organoids
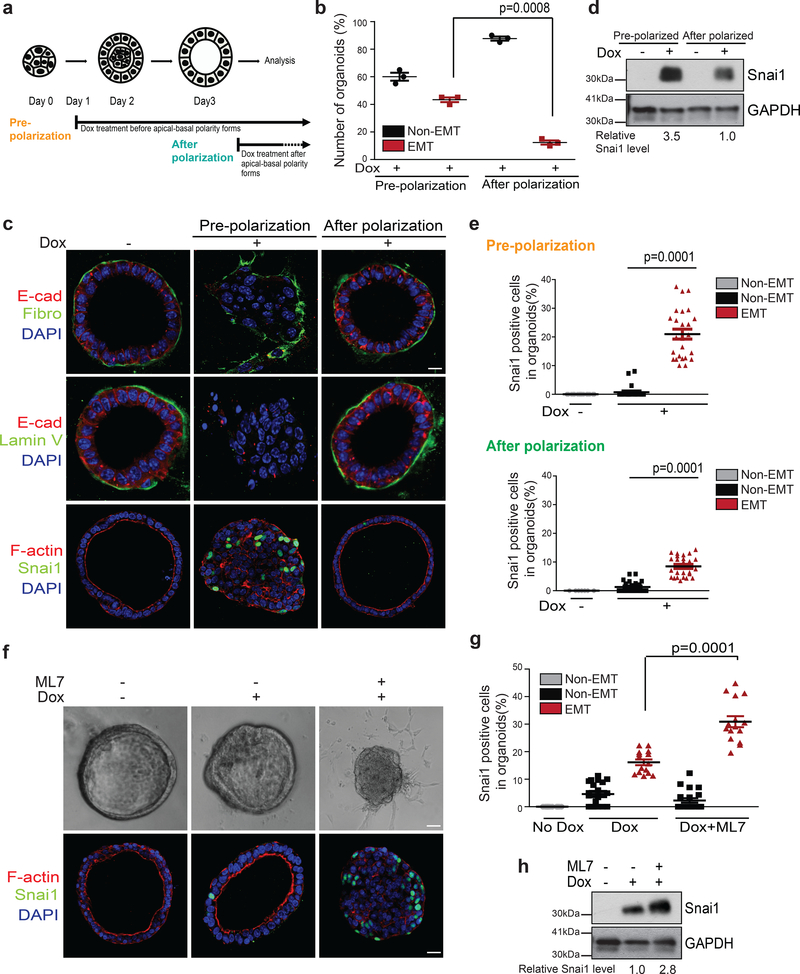
One major difference between 3D MEOs and 2D MECs is the apical-basal polarity. We therefore reasoned that polarity might directly regulate SNAI1 protein stability. Primary MEOs took 3 days to establish hollow lumen and apical-basal polarity, as indicated by the presence of cortical F-actin and the PAR complex components, Par3 and PKCζ at the apical surface. In contrast, after 1 day of the culture, the majority of MEOs are not polarized with filled lumen (Fig. 2a and Supplementary Fig. 2a). Induction of SNAI1 in these pre-polarized MEOs resulted in significantly higher percentage of organoids undergoing EMT (Fig. 2b), as judged by reduced E-cadherin-mediated adherens junctions, loss of laminin V-positive basement membrane, and increased fibronectin expression, compared to induction of SNAI1 in polarized MEOs (Fig. 2c). Furthermore, SNAI1 protein level increased in pre-polarized MEOs compared with polarized MEOs after doxycycline treatment (Fig. 2c–e), supporting a role of apical-basal polarity in regulating SNAI1 protein level.
Figure 2.
Disruption of apical-basal polarity increases SNAI1 protein stability in MEOs. (a) The experimental scheme for inducing SNAI1 in organoids before polarization and after polarization. (b) Quantification of the percentage of MEOs undergoing EMT under pre-polarization and after polarization conditions. n=3 independent experiments with 50 organoids/condition in each experiment, unpaired two-tailed t-test with Welch’s correction. Error bars represent standard deviation. (c) Immunofluorescence images for E-cadherin, Fibronectin, Laminin V (Lamin V), SNAI1 and F-actin in TetON-SNAI1 MEOs under pre-polarization and after polarization conditions as indicated. Scale bar, 25μm. (d) Immunoblot for SNAI1 and GAPDH from TetON-SNAI1 MEOs under pre-polarization and after polarization conditions as indicated. Values indicate relative signal intensities of SNAI1/GAPDH. (e) Quantification of the percentages of SNAI1-positive cells in TetON-SNAI1 MEOs under pre-polarization (Dox-; n=20 cells, Dox+; n=25 cells) and after polarization (Dox-; n= 20 cells, Dox+ non-EMT; n=25 cells, Dox+ EMT; n=20 cells), conditions as indicated. Data represent one out of three independent experiments. Unpaired two-tailed t-test with Welch’s correction. (f) Bright-field images (top) and immunofluorescence images for SNAI1 and F-actin (bottom) of TetON-SNAI1 MEOs treated with doxycycline and 10μM ML7 for 4 days. Scale bar, 25μm. (g) Quantification of the percentages of SNAI1-positive cells in non-EMT and EMT TetON-SNAI1 MEOs under different treatment conditions as indicated. (Dox-; n=20 cells, Dox+ non-EMT; n=20 cells, Dox+EMT; n=15 cells, Dox+ML7 non-EMT; n=20 cells, Dox+ML7 EMT; n=15 cells). Data represent one out of three independent experiments. Unpaired two-tailed t-test with Welch’s correction. (h) Immunoblot for SNAI1 and GAPDH from TetON-SNAI1 MEOs under different treatment conditions as indicated. The values indicate relative signal intensities of SNAI1/GAPDH. All immunofluorescence images and Western Blots shown represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3 and unprocessed blots in Supplementary Figure 7.
We also used a selective myosin light chain kinase inhibitor (ML7) to disrupt apical-basal polarity in fully polarized MEOs, as previously reported31, 32. Notably, ML7 treatment disrupted apical-basal polarity and remarkably increased the percentage of SNAI1-positive cells in individual MEOs (Fig. 2f–g) and increased total SNAI1 protein level (Fig. 2h). Together, our results strongly indicate that apical-basal polarity negatively regulates SNAI1 protein stability.
aPKC-mediated SNAI1 phosphorylation promotes SNAI1 protein degradation
Previous studies reported that SNAI1 stability is regulated by phosphorylation of SNAI1 protein by GSK3β33, 34. Treatment with a selective GSK3 inhibitor CHIR-99021 increased SNAI1 protein levels in 2D MECs and in 3D MEOs; however, the fold increases of SNAI1 protein levels were similar in 2D MECs vs. 3D MEOs and the SNAI1 protein level remained significantly lower in 3D MEOs compared to that in 2D MECs upon GSK3β inhibition (Supplementary Fig. 3a). Together, these data suggest that GSK3β is not responsible for apical-basal polarity-induced SNAI1 degradation.
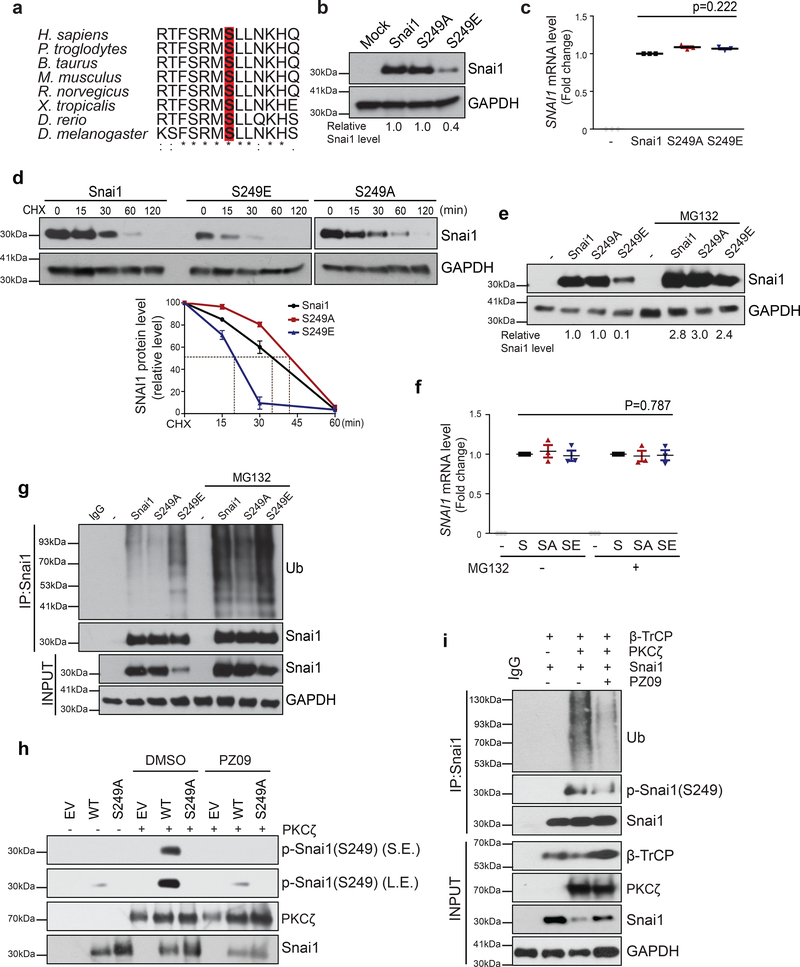
To uncover unknown mechanisms regulating SNAI1 protein stability, we used protein modification prediction softwares to identify potential conserved phosphorylation sites on SNAI1. These analyses identified S249 as a candidate aPKC phosphorylation site (Supplementary Fig. 3b–c). Sequence alignment show that the potential aPKC-phosphorylation motif, RMSLL is highly conserved from human to Drosophila (Fig. 3a). aPKCs serve as core components of the PAR polarity complex that is essential for apical-basal polarity35. Therefore we generated phospho-deficient S249A and phospho-mimetic S249E SNAI1 mutants to determine whether phosphorylation of S249 affects SNAI1 protein stability. Indeed, the steady-state levels of the S249E SNAI1 mutant protein were significantly lower than those of the wild-type or the S249A mutant SNAI1 (Fig. 3b); in contrast, their mRNA levels were similar in 2D cultures of MCF10A cells without apical-basal polarity (Fig. 3c). Consistently, S249E SNAI1 protein displayed a shorter half-life compared with WT or S249A SNAI1 proteins (Fig. 3d). Proteasome inhibitor MG132 stabilized the S249E SNAI1 protein without affecting its mRNA level (Fig. 3e,f). Furthermore, compared to wild-type SNAI1 and S249A mutant protein, the S249E mutant protein was highly ubiquitinated in the absence or presence of MG132 (Fig. 3g), further supporting the notion that phosphorylation of SNAI1 on S249 promotes SNAI1 ubiquitination and proteasome-mediated protein degradation.
Figure 3.
Phosphorylation of Serine 249 on SNAI1 promotes SNAI1 degradation. (a) Alignment of the putative aPKC phosphorylation site in SNAI1 homologs. The alignment was generated by Clustal Omega. (b) Immunoblot for SNAI1 and GAPDH in MCF10A cells expressing wild-type, phospho-deficient S249A SNAI1 mutant, and phospho-mimetic S249E SNAI1 mutant. Values indicate relative signal intensity of SNAI1/GAPDH. Three independent experiments. (c) qPCR analysis of relative SNAI1 mRNA levels normalized to GAPDH in MCF10A cells expressing wild-type, phospho-deficient S249A SNAI1 mutant, and phospho-mimetic S249E SNAI1 mutant. n=3 independent experiments, error bars represent standard deviation, unpaired two-tailed t-test with Welch’s correction. (d) MCF10A cells expressing indicated SNAI1 constructs were treated with 10μM cycloheximide (CHX) following the time points indicated and analysed for SNAI1 and GAPDH by immunoblotting. The graph represents the quantification of relative SNAI1 proteins levels. n=3 independent experiments, error bars represent standard deviation. (e) MCF10A cells expressing indicated SNAI1 constructs were treated with 10μM MG132 for 4hrs and analysed for SNAI1 protein levels by immunoblotting. Values indicate relative signal intensities of SNAI1/GAPDH. (f) qPCR analysis of relative SNAI1 mRNA levels normalized to GAPDH in MCF10A cells expressing indicated SNAI1 constructs in the presence of MG132. n=3 independent experiments, error bars represent standard deviation, paired two-tailed t-test. (g) SNAI1 proteins were immunoprecipitated from MCF10A cells expressing indicated SNAI1 constructs with or without MG132 treatment and probed for SNAI1 and Ubiquitin. (h) in vitro kinase assay for phospho-SNAI1(S249) with purified WT and S249A SNAI1, and aPKC with or without PZ09 treatment. (i) 293T cells overexpressing SNAI1, aPKC and β-TrCP in the indicated combination were treated with 5uM PZ09 or DMSO. SNAI1 proteins were immunoprecipitated from 293T cells and probed for Ub, phospho-SNAI1(S249), SNAI1, and GAPDH. All immunofluorescence images and Western Blots shown represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3 and unprocessed blots in Supplementary Figure 7.
To directly test whether aPKCs could phosphorylate S249 on SNAI1 protein, we performed an in vitro kinase assay and found that purified PKCζ could phosphorylate purified SNAI1 WT protein on S249, as recognized by a pS249-specific SNAI1 antibody we generated. Importantly, S249 phosphorylation was inhibited in the presence of an aPKC specific inhibitor PZ0936 (Fig. 3h). We next co-expressed SNAI1 and PKCζ together with β-TrCP, the E3 ligase that is predicted to bind to the S249 region. SNAI1 ubiquitination was detected specifically in cells co-expressing PKCζ and β-TrCP and was reversed by PZ09 treatment (Fig. 3i). Furthermore, we tested the SNAI1–6AS mutant in which all GSK3β-phosphorylation sites are abolished 34 and found that this mutant is responsive to aPKC-mediated SNAI1 degradation just as well as the WT SNAI1 protein (Supplementary Fig. 3d). These results show that phosphorylation of SNAI1 on S249 by aPKCs reduces SNAI1 protein stability, which is independent of GSK3-mediated SNAI1 phosphorylation.
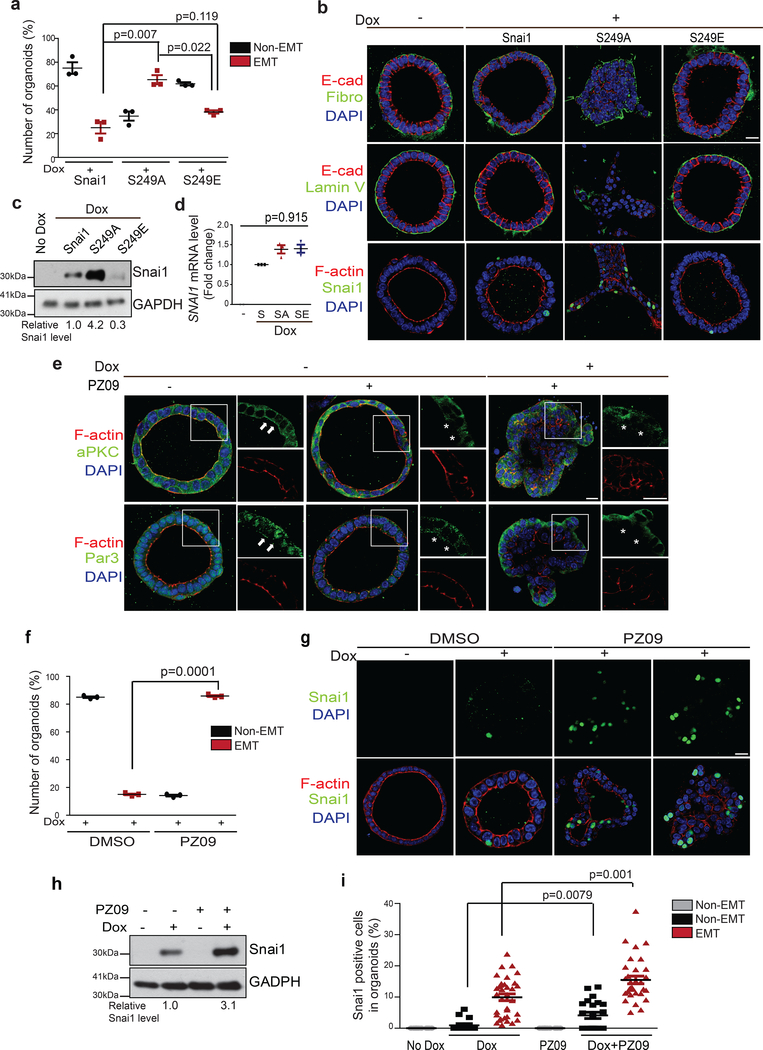
We next expressed Tet-inducible SNAI1 WT and S249 mutants in polarized MEOs by lentiviral infection. Using a lentiviral construct carrying GFP, we determined that our primary organoids infection reached about 35% cells in individual organoids (Supplementary Fig. 2b). Similar to what we observed in TetON-SNAI1 MEOs, the majority of organoids overexpressing WT SNAI1 and the S249E mutant failed to undergo EMT and invasion upon doxycycline induction; in contrast, the S249A mutant was able to induce EMT and invasion in over 65% polarized organoids (Fig. 4a,b). Consistently, the S249A mutant, but not the WT and S249E SNAI1, shows higher protein expression in MEOs without affecting SNAI1 mRNA levels (Fig. 4c,d). Together, these data suggest that apical-basal polarity regulates S249 phosphorylation to impact SNAI1 protein degradation and EMT.
Figure 4.
Inhibition of aPKC promotes SNAI1 protein stability and synergizes with SNAI1 induction to promote EMT in MEOs. (a) Quantification of the percentage of non-EMT and EMT MEOs expressing TetON-SNAI1 WT and mutants. n=3 independent experiments with 50 organoids/condition in each experiment, unpaired two-tailed t-test with Welch’s correction. Error bars represent standard deviation. (b) Immunofluorescence images for E-cadherin, Fibronectin, Laminin V, SNAI1 and F-actin in MEOs expressing TetON-SNAI1 WT and mutants. Scale bars, 25μm. (c) Immunoblot for SNAI1 and GAPDH in MEOs expressing TetON-SNAI1 WT and mutants. Values indicate relative signal intensities of SNAI1/GAPDH. (d) qPCR analysis of relative SNAI1 mRNA levels normalized to GAPDH in MEOs expressing TetON-SNAI1 WT and mutants. unpaired two-tailed t-test with Welch’s correction. Error bars represent standard deviation. (e) Immunofluorescence images for aPKC, Par3 and F-actin in TetON-SNAI1 MEOs under different treatment conditions as indicated. Arrowheads point to the presence of aPKC and F-actin at the apical membrane and Par3 at the apical/lateral region. Stars mark the loss of these proteins at the corresponding areas. Scale bars, 25μm and 50μm. (f) Quantification of the percentage of non-ENT and EMT TetON-SNAI1 MEOs in response to PZ09 treatment. n=3 independent experiments with 50 organoids/condition in each experiment, unpaired two-tailed t-test with Welch’s correction. Error bars represent standard deviation. (g) Immunofluorescence images for SNAI1 and F-actin in TetON-SNAI1 MEOs in response to PZ09 treatment. Scale bar, 25μm. (h) Immunoblot for SNAI1 and GAPDH in TetON-SNAI1 MEOs in response to PZ09 treatment. Values indicate relative signal intensities of SNAI1/GAPDH. (i) Quantification of the percentage of SNAI1-positive cells in non-EMT and EMT TetON-SNAI1 MEOs in response to PZ09 treatment. Dox-; n=20 cells, Dox+ non-EMT; n=15 cells,, PZ09; n=20 cells, Dox+ PZ09 non-EMT; n=22 cells, Dox+PZ09 EMT; n=32 cells. Error bars represent standard deviation. Unpaired two-tailed t-test with Welch’s correction. All immunofluorescence images and Western Blots shown represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3 and unprocessed blots in Supplementary Figure 7.
The PAR3/aPKC complex is required for apical-basal polarity-induced SNAI1 protein degradation
To determine whether the aPKC kinase activity is required for SNAI1 protein degradation in polarized MEOs, we treated the TetON-SNAI1 MEOs with PZ09, which inhibits both PKCζ and PKCι. In MEOs with intact apical-basal polarity, PKCζ and Par3 could be detected at the apical region, as marked by strong cortical F-actin signals (Fig. 4e). Importantly, induction of SNAI1 in the presence of PZ09 effectively promoted EMT in over 80% MEOs, compared to activation of EMT in less than 15% MEOs in the control group (Fig. 4e,f). Furthermore, PZ09 treatment drastically increased SNAI1 protein levels (Fig. 4h). Consistently, PZ09 treatment of MEOs resulted in a significant increase in the percentage of SNAI1-positive cells in individual organoids (Fig. 4g–i). Our results suggest that the aPKC kinase activity is required for SNAI1 phosphorylation and degradation in polarized MEOs.
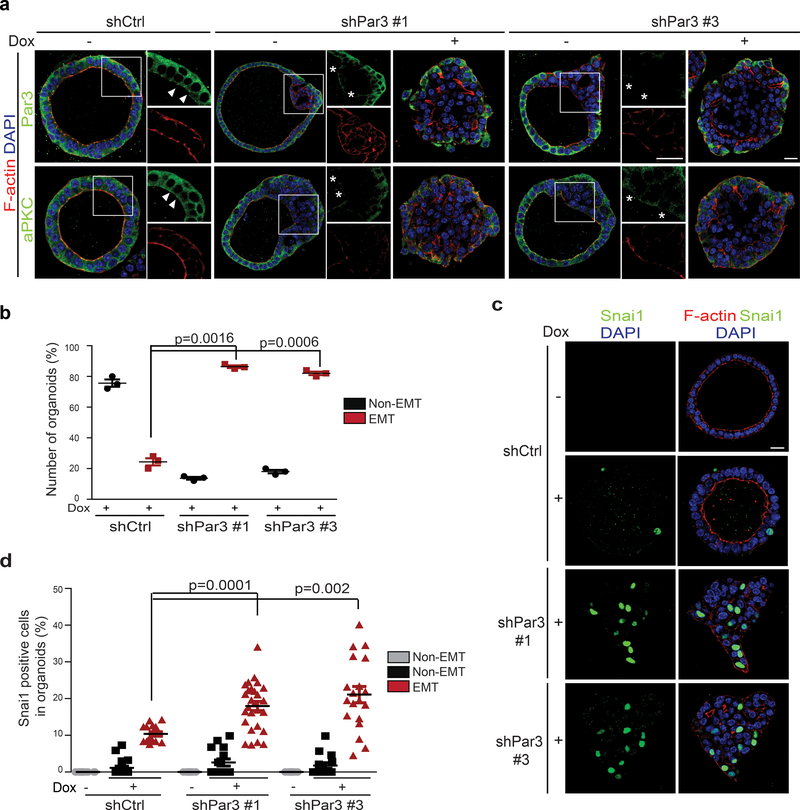
We next expressed short hairpin RNAs (shRNA) against Par3 via lentiviral infection of primary MEOs isolated from the TetON-SNAI1 mice. Partial knockdown of Par3 caused loss of apical-basal polarity in certain areas of MEOs and mislocalization of PKCζ from the apical area (Fig. 5a). Functionally, knockdown of Par3 synergized with induction of SNAI1 to promote EMT in MEOs (Fig. 5b). Also importantly, knockdown of Par3 increased the percentage of SNAI1-positive cells in individual MEOs (Fig. 5c,d). Together, these data demonstrate that the PAR complex is required for aPKC-regulated SNAI1 destabilization to prevent EMT in polarized MEOs.
Figure 5.
Knockdown of Par3 promotes SNAI1 protein stability and induces EMT in MEOs.
(a) Representative immunofluorescence images for Par3, aPKC, and F-actin in TetON-SNAI1 MEOs expressing control shRFP or shPar3#1 and #3. Arrowheads point to the presence of aPKC at the apical membrane and Par3 at the apical/lateral region. Stars mark the loss of these proteins at the corresponding areas. Scale bars, 25μm and 50μm. Three independent experiments. (b) Quantification of the percentage of non-EMT and EMT TetON-SNAI1 MEOs expressing shRFP or shPar3#1 and #3. n=3 independent experiments with 50 organoids/condition in each experiment, unpaired two-tailed t-test with Welch’s correction. Error bars represent standard deviation. (c) Immunofluorescence images for SNAI1 and F-actin in TetON-SNAI1 MEOs expressing shRFP or shPar3#1 and #3 before and after doxycycline treatment. Scale bars, 25μm. Three independent experiments. (d) Quantification of the percentage of SNAI1-positive cells in non-EMT and EMT TetON-SNAI1 MEOs expressing shRFP or shPar3#1 and #3 before and after doxycycline treatment. n values from left to right: 20, 20, 15, 20, 15, 27, 20, 15 and 19 cells, respectively. Data represent one out of three independent experiments Unpaired two-tailed t-test with Welch’s correction. Error bars represent standard deviation. All immunofluorescence images represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3.
The PAR3-aPKC complex destabilizes endogenous SNAI1 protein to prevent EMT
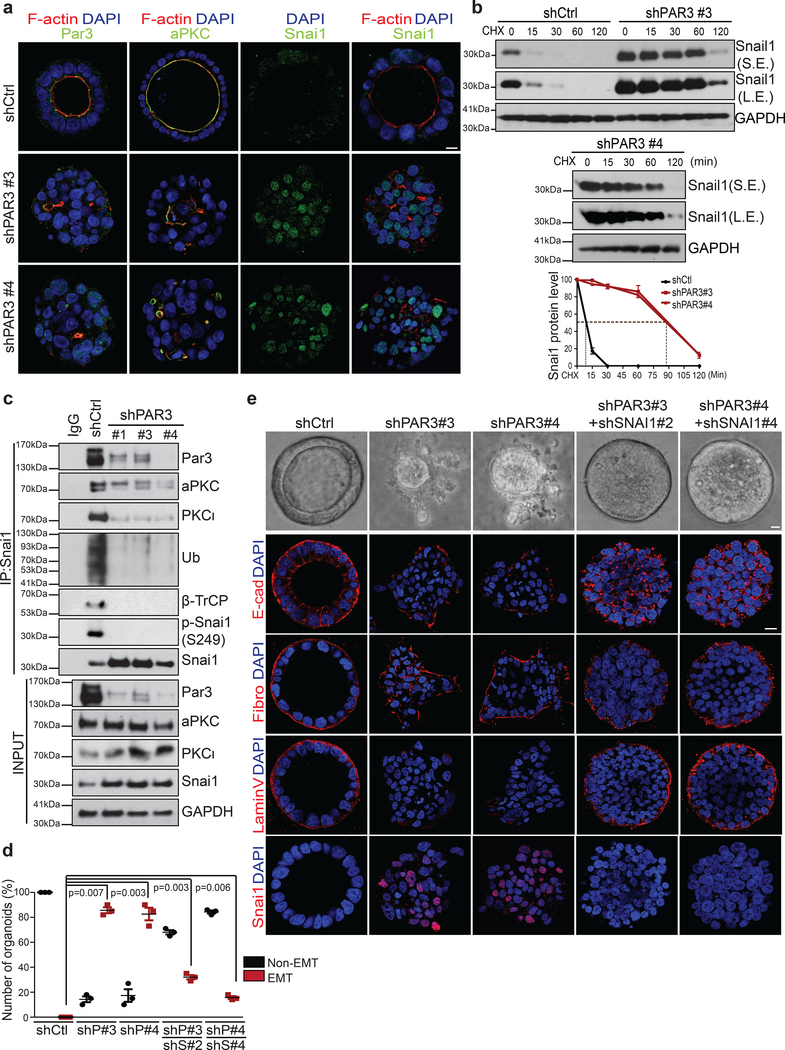
We next investigated endogenous human SNAI1 regulation by polarity using Caco2 human intestinal epithelial cells that express endogenous SNAI1 mRNA and develop well-established apical-basal polarity in 3D37. Consistently, endogenous SNAI1 protein level was significantly reduced in 3D polarized organoids compared to sparse 2D culture without polarity. Immunoprecipitated endogenous SNAI1 protein is specifically phosphorylated on Ser249 and ubiquitinated and interacts with β-TrCP only in polarized 3D organoids, but not in 2D cultures (Supplementary Fig. 4a). PAR3 and aPKC proteins were specifically localized at the apical membrane in control organoids, whereas knockdown of PAR3 completely disrupted apical-basal polarity and caused mislocalization of aPKCs (Fig. 6a). In control organoids, SNAI1 protein was largely undetectable; upon knockdown of PAR3, most cells in individual organoids became positive for endogenous SNAI1 protein (Fig. 6a). SNAI1 protein rapidly degraded with a half-life of < 10 min in control organoids, while knocking down Par3 elongated SNAI1 half-life to over 80min (Fig. 6b). Consistently, PAR3 knockdown increased endogenous SNAI1 levels without affecting SNAI1 mRNA levels (Fig. 6c and Supplementary Fig. 4b). SNAI1 immunoprecipitated from Caco2 organoids is phosphorylated on S249 and interacts with both PAR3 and β-TrCP. Interestingly, both aPKC members, PKCζ and PKCι, are associated with SNAI1 in the control organoids. Importantly, knockdown of PAR3 abolished S249 phosphorylation on SNAI1 and significantly reduced the interaction between SNAI1 and aPKCs or with β-TrCP (Fig. 6c). Consequently, over 80% organoids in 3D culture underwent EMT upon knockdown of PAR3, as immunostaining showed reduced E-cadherin-mediated junctions, loss of Laminin V and increased fibronectin expression (Fig. 6d,e). Importantly, PAR3 loss-induced EMT and invasion through basement membrane could be significantly blocked upon deletion of SNAI1 despite polarity loss caused by PAR3 knockdown (Fig. 6d,e, and Supplementary Fig. 4c). These data strongly support a critical role of the PAR complex in reducing endogenous SNAI1 protein stability and blocking SNAI1-induced EMT.
Figure 6.
Loss of PAR3 stabilizes endogenous SNAI1 protein and induced EMT in 3D Caco2 organoids. (a) Immunofluorescence images for PAR3, aPKC, SNAI1 and F-actin in Caco2 organoids expressing shRFP or shPAR3#3 and #4. Scale bar, 25μm. (b) Caco2 organoids expressing shRFP, shPAR3#3 and #4 were treated with 10μM cycloheximide (CHX) following the time points indicated and analysed for SNAI1 and GAPDH by immunoblotting. S.E. indicates short exposure, L.E. indicates long exposure. The graph represents the quantification of relative SNAI1 proteins levels vs. GAPDH. n=3 independent experiments, error bars represent standard deviation. (c) Endogenous SNAI1 proteins were immunoprecipitated from Caco2 organoids expressing shRFP or shPAR3#1, #3, and #4 and probed for PAR3, aPKC, PKCι, Ubiquitin (Ub), β-TrCP, phospho-SNAI1 (S249) and SNAI1. Immunoblot for PAR3, aPKC, PKCι, SNAI1, and GAPDH in Caco2 organoids expressing shRFP or shPAR3#1, #3, and #4. (d) Quantification of the percentage of non-EMT and EMT TetON-SNAI1 MEOs expressing indicated shRNA constructs. n=50 organoids/condition in each experiment, data from three independent experiments are represented, paired two-tailed t-test, Error bars represent standard deviation. (e) Representative bright-field images (top) and immunofluorescence images for GFP, E-cadherin, Fibronectin, Laminin V, and SNAI1 (bottom) of Caco2 organoids expressing the indicated shRNA constructs. Scale bar, 25μm.
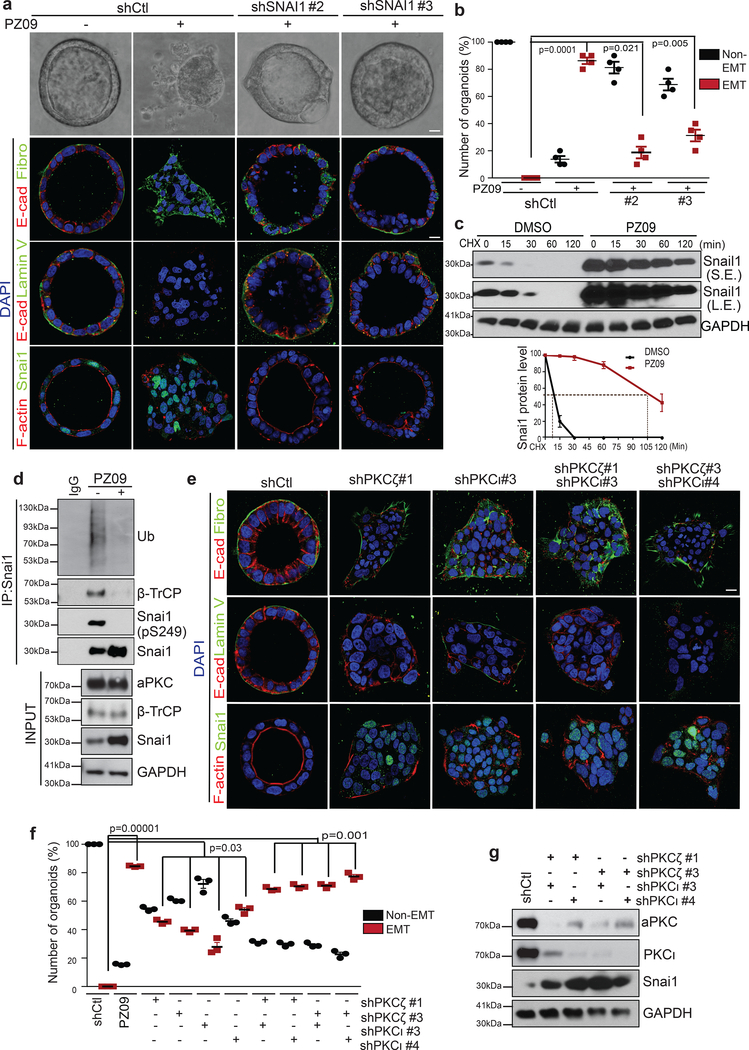
We next investigated whether inactivation of aPKCs affects endogenous SNAI1 phosphorylation on S249 and subsequent degradation in Caco2 3D organoids. PZ09 effectively inhibited the aPKC kinase activity in Caco2 3D organoids as assessed using an aPKC-specific FRET reporter38 (Supplementary Fig. 5a). Morphologically, upon PZ09 treatment, over 90% of Caco2 organoids lost apical-basal polarity, E-cadherin-mediated adherens junctions, and basement membrane integrity and increased fibronectin expression, which is consistent with increases of SNAI1-positive cells (Fig. 7a,b and Supplementary Fig. 5c). Inhibition of the aPKC kinase activity also increased SNAI1 protein level and led the majority of cells in individual organoids to express SNAI1 protein without affecting SNAI1 mRNA levels (Supplementary Fig. 5b–d). The half-life of SNAI1 protein was increased from <10 min to >100 min upon inhibition of aPKCs (Fig. 7c). Notably, we found that endogenous SNAI1 protein is phosphorylated on S249, ubiquitinated, interacts with β-TrCP and all these events were inhibited by PZ09 treatment (Fig. 7d). We next investigated whether endogenous SNAI1 protein is required for induction of EMT upon aPKC inhibition in Caco2 organoids. Knockdown of SNAI1 significantly blocked EMT induction in response to PZ09 treatment (Fig. 7a,b and Supplementary Fig. 5d), suggesting that inhibition of the aPKC kinase activity promotes EMT in a SNAI1-dependent manner.
Figure 7.
Suppression of aPKCs increases endogenous SNAI1 stability and promotes EMT in 3D Caco2 organoids. (a) Representative immunofluorescence images for E-cadherin, Fibronectin, Laminin V, SNAI1 and F-actin in Caco2 organoids expressing shRFP or shSNAI1 #2, and shSNAI1 #3 with or without PZ09 treatment. Scale bar, 25μm. Three independent experiments. (b) Quantification of the percentage of non-ENT and EMT Caco2 organoids expressing shRFP or shSNAI1 #2 and #3 with or without PZ09 treatment. n=3 independent experiments with 50 organoids/condition in each experiment paired two-tailed t-test Error bars represent standard deviation. (c) Caco2 organoids were treated with DMSO or PZ09 and then treated with 10μM cycloheximide (CHX) following the time points indicated and analysed for SNAI1 and GAPDH by immunoblotting. S.E. indicates short exposure, L.E. indicates long exposure. The graph represents the quantification of relative SNAI1 proteins levels vs. GAPDH. n=3 independent experiments, error bars represent standard deviation. (d) Endogenous SNAI1 proteins were immunoprecipitated from Caco2 organoids with or without PZ09 treatment and probed for Ubiquitin (Ub), β-TrCP, phospho-SNAI1(S249) and SNAI1. Immunoblot for aPKC, β-TrCP, SNAI1, and GAPDH in Caco2 organoids with or without PZ09 treatment. (e) Representative immunofluorescence images for E-cadherin, Fibronectin, Laminin V, SNAI1 and F-actin in Caco2 organoids expressing the indicated shRNA constructs. Scale bar, 25μm. (f) Quantification of the percentage of non-EMT and EMT Caco2 organoids expressing the indicated shRNA constructs. n=3 independent experiments with50 organoids/condition in each experiment, paired two-tailed t-test, error bars represent standard deviation. (g) Immunoblot for aPKC, PKCι, SNAI1 and GAPDH in lysate from Caco2 organoids expressing the indicated shRNA constructs. All immunofluorescence images and Western Blots shown represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3 and unprocessed blots in Supplementary Figure 7.
We next examined individual contributions of PKCζ and PKCι. Although single knockdown of PKCζ or PKCι could partially stabilize SNAI1 and promote EMT in some organoids, only double knockdown of both PKCζ and PKCι could fully recapitulate the EMT and invasion phenotype induced by PZ09 treatment (Fig. 7e–g and Supplementary Fig. 5e,f), suggesting that PKCζ and PKCι have redundant roles in the PAR complex to phosphorylate SNAI1 on S249. These data strongly indicate that both aPKC members in the PAR complex are responsible for S249 phosphorylation of endogenous SNAI1 protein and subsequent ubiquitination and degradation of the SNAI1 protein. Taken together, these findings demonstrate that apical-basal polarity promotes endogenous SNAI1 protein phosphorylation and degradation via active aPKCs in the PAR complex to prevent EMT.
Loss of the PAR complex-mediated polarity promotes invasion and metastasis via SNAI1 in vivo
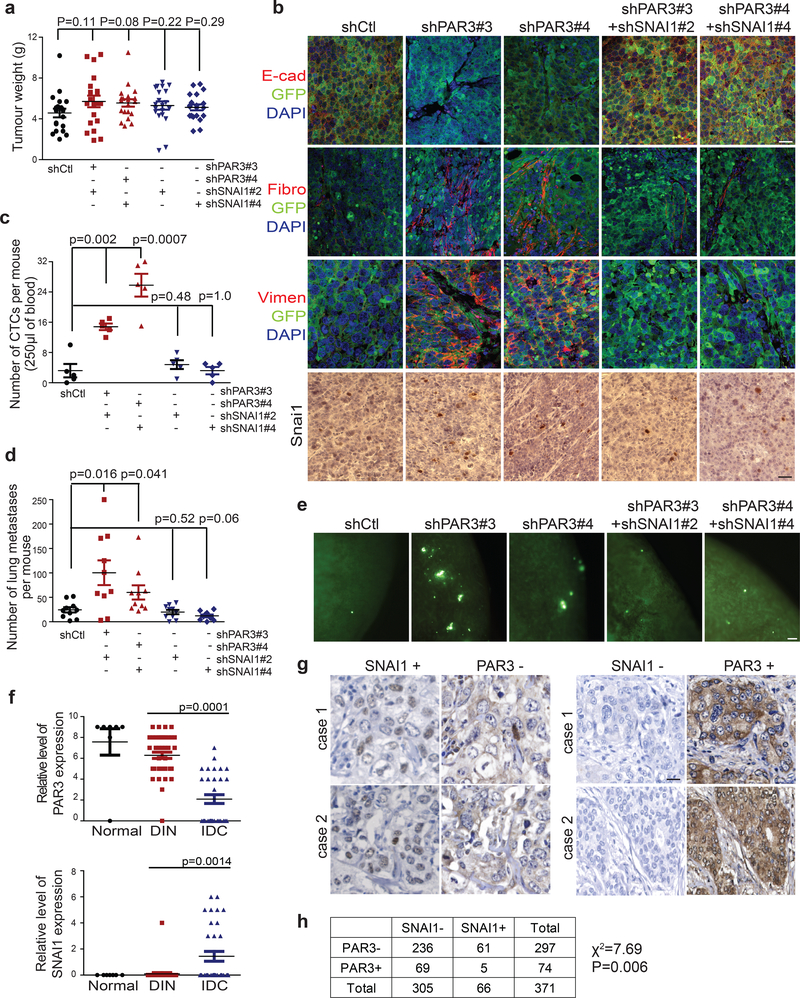
We implanted GFP-labelled Caco2 cells expressing control shRNA, shRNAs against PAR3, and shRNAs against PAR3 and SNAI1 in mice to follow primary tumour development and metastasis. Control Caco2 cells developed primary tumours that presented an epithelial morphology. In contrast, loss of PAR3 resulted in mesenchymal primary tumours with very low membranous E-cadherin, high human Vimentin and high SNAI1 expression. More importantly, knockdown of SNAI1 reversed the mesenchymal phenotype in Caco2 tumours expressing shRNAs against PAR3 (Fig. 8a,b and Supplementary Fig. 6a). Consistent with results in Fig. 6, Caco2 control tumours were largely negative for SNAI1 immunostaining, while knockdown of PAR3 increased SNAI1 protein (Fig 8b). Moreover, mice carrying PAR3-knockdown tumours presented significantly more circulating tumour cells (CTCs) compared to mice carrying the control Caco2 tumours, and knockdown of SNAI1 significantly reduced CTC numbers to the control level (Fig. 8c). Furthermore, we observed a significant increase in GFP-positive disseminated tumour cells in the lung of mice carrying PAR3-knockdown tumours compared to control, and this increase in lung dissemination was completely blocked by SNAI1 knockdown (Fig. 8d,e). These in vivo results strongly support our cellular and biochemical data showing that the PAR complex-mediated SNAI1 protein degradation inhibits EMT, tumour invasion, and metastasis in vivo.
Figure 8.
The epithelial polarity-regulated SNAI1 degradation mechanism impacts tumour invasion and metastasis in vivo and is associated with human breast cancer progression. (a) Primary tumour weight of GFP-tagged Caco2 xenograft tumours expressing shRFP, shPAR3, and shPAR3-shSNAI1 after subcutaneous injection for 6 weeks. n=20 tumours per group, unpaired two-tailed t-test with Welch’s correction, Error bars represent standard deviation. (b) Immunofluorescence images for GFP, E-cadherin, and human Vimentin (Vim), and immunohistochemistry images for SNAI1 in Caco2 xenograft tumours expressing shRFP, shPAR3 and shPAR3-shSNAI1. Scale bars, 50μm for SNAI1 staining and 25μm for all other markers. (c) Quantification of the number of circulating tumour cells (CTCs) isolated from mice carrying Caco2 tumours expressing shRFP, shPAR3 and shPAR3-shSNAI1. n=10 mice per group, unpaired two-tailed t-test with Welch’s correction, Error bars represent standard deviation. (d) Quantification of GFP-positive metastatic events in the lung of mice carrying Caco2 tumours expressing shRFP, shPAR3 and shPAR3-shSNAI1. n=10 mice per group, unpaired two-tailed t-test with Welch’s correction Error bars represent standard deviation (e) Fluorescent images of GFP-positive metastatic lesions in the lung of mice carrying Caco2 tumours expressing shRFP, shPAR3 and shPAR3-shSNAI1. GFP signal indicates disseminated tumour cells in the lung. Scale bars, 100μm. (f) Quantification of the relative PAR3, and SNAI1 signals in human breast tumour tissue samples (n=48 patients, two sections from each patient were analysed). Unpaired two-tailed t-test with Welch’s correction, Error bars represent standard deviation. (g) Representative immunohisto-chemistry images for SNAI1 and PAR3 in human breast tumour samples. Scale bars, 25μm. (h) Correlation analysis of SNAI1 and PAR3 in human stage-2 breast tumour tissue microarray. All data panels represent one out of three independent experiments. Source data for graphs can be found in Supplementary Table 3.
PAR3 and SNAI1 expression are negatively correlated in human breast tissue samples
We next evaluated the involvement of the PAR3-SNAI1 regulatory mechanism in human breast cancer initiation and progression by immunostaining analysis for PAR3 and SNAI1 expression in human breast tissue microarray (TMA). PAR3 protein was detected at the apical membrane in normal mammary ducts and in the majority of ductal intraepithelial neoplasia (DIN) samples, while these proteins expression were clearly decreased in invasive ductal carcinoma (IDC) samples. In reverse, SNAI1 protein was largely not detectable in normal and DIN samples, and was only visible in IDC samples (Fig. 8f and Supplementary Fig. 6b). We next analysed PAR3 and SNAI1 expression in a cohort of 371 Stage-2 breast tumours from the NCI Cancer Diagnosis Program. Consistent with a suppressive function of the PAR epithelial polarity complexes on SNAI1 stability, 92% of SNAI1-positive tumours were negative for PAR3 (Fig. 8g,h). It is important to note that SNAI1 expression is regulated at the transcriptional and posttranslational levels. This is consistent with Fig. 8h showing that among 236 Par3-negative tumours, 61 showed SNAI1 positivity, while the remaining 80% are still negative for SNAI1, possibly due to the lack of SNAI1 mRNA expression. Our results suggest that the molecular machinery that links the PAR3 complex with SNAI1 protein stability may also contribute to human breast tumour progression.
DISCUSSION
We demonstrate that apical-basal polarity directly impinges on the EMT program to prevent EMT and invasion, thus inhibiting metastasis. Mechanistically, we show that under intact polarity, active aPKCs in the PAR complex phosphorylate SNAI1 on S249, which in turn promotes ubiquitination and degradation of SNAI1 protein. Inactivation of the PAR3-aPKC polarity machinery stabilizes SNAI1 protein, thus promoting SNAI1-induced EMT and invasion. In vivo, aPKC-mediated phosphorylation of SNAI1 hampers the ability of SNAI1 to promote breast tumour cells invasion, disseminate and metastasize to distant organs. Downregulation of PAR3 and increase of the SNAI1 protein expression are associated with breast tumour progression in human breast cancer patients. Our findings reveal a direct molecular link between apical-basal polarity and the core EMT transcription machinery in tumour invasion and metastasis (Supplementary Fig. 6c).
Previous studies of EMT regulation of epithelial cell properties focused on cell-cell junctions, such as tight junctions and adherens junctions22. Although loss of cell polarity is an early step during the EMT process, polarity is largely thought to be a passive recipient of the EMT-inducing signals to decrease epithelial characteristics. Results from this study show that apical-basal polarity actively inhibits the core EMT transcriptional program by destabilizing SNAI1 protein. Given the key role of SNAI1 in suppressing proteins associated with cell-cell junctions33, 39, our results bring up the concept that apical-basal polarity is actively involved in maintaining epithelial cell-cell junctions via suppressing the EMT program. Also importantly, our study demonstrates that polarity is a critical checkpoint during EMT. A number of key EMT transcription factors, especially SNAI1 and ZEB1, are shown to suppress transcription of several key polarity genes, including Crb3, PATj, and LGL2, thus disrupting apical-basal polarity22, 40–42. Consequently, loss of epithelial polarity sends a positive feedback signal to further stabilize SNAI1 and move the EMT process forward to promote tumour progression (Supplemental Figure 6c). The majority of human carcinomas show loss of apical-basal polarity during the progression from intraepithelial neoplasia to invasive carcinoma43–45. Our work and several previous studies links epithelial cell polarity proteins such as PAR3 and aPKCs to metastasis suppression2. Loss of PAR3 promoted cancer metastasis through activation of Tiam1-mediated Rac-GTP pathway and inhibition of E-cadherin junction stability 12. In addition, loss of PAR3 accelerated breast cancer formation and metastasis through aPKC-mediated Jak/Stat3 signalling10. This study adds SNAI1 as a key target of the PAR/aPKC polarity complex in suppressing tumour invasion and metastasis. Together, these findings support the notion that epithelial cell polarity proteins functions as major metastasis suppressors via several pathways to maintain epithelial characteristics in carcinomas.
SNAI1 stability is also regulated by phosphorylation of SNAI1 protein by GSK3β33, 34. Our study shows that inhibiting GSK3 under both polarized and unpolarized conditions stabilized Snai1 protein to the same degree. These data show that GSK3 inhibition cannot rescue polarity-mediated SNAI1 degradation and that GSK3 promotes Snail degradation regardless of the polarity status. Therefore, SNAI1 could be regulated by diverse upstream signals independently to regulate its stability to impact EMT.
Our data identify an unexpected role for aPKCs in suppressing EMT, thus inhibiting tumor invasion and metastasis. Previous studies suggest that aPKCs may promote tumour proliferation and progression10, 46–49. However, these studies were largely performed in cells lacking apical-basal polarity and may thus reflect polarity-independent functions of aPKCs. For example, in addition to function in the PAR complex, aPKCs also bind to another scaffold protein p62 to function in response to TNFα, LPS, and IL150. Therefore, it is critical to analyse aPKCs under specific physiological conditions to reveal the diverse roles of this unique PKC subfamily in tumour progression and metastasis.
ONLINE METHODS
Reagents, antibodies and DNA constructs
Primary antibodies include anti-SNAI1 (CST, 3879, 1:200 for immunostaining, 1:1000 for immunoblotting), mouse anti-SNAI1 EC3 monoclonal antibody (a kind gift from Dr. A. Garcia de Herreros, Institut Hospital del Mar d’Investigacions Mèdiques, Barcelona, Spain, 1:25 for immunostaining), anti-Twist1 (Santa Cruz, sc-81417, 1:50 for immunostaining, 1:1000 for immunoblotting), anti-E-cadherin (BD, 610182, 1:200 for immunostaining), anti-Fibronectin (Sigma, F3648, 1:200 for immunostaining), anti-integrin α5 (Millipore, AB1928, 1:100 for immunostaining), anti-laminin V (a kind gift from Dr. M. Aumailley, University of Cologne, Germany, 1:1000 for immunostaining), anti-PAR3 (Millipore, 07–330, 1:100 for immunostaining, 1:4000 for immunoblotting), anti-phospho-PKCζ (T560) (Abcam, ab62372, 1:100 for immunostaining, 1:2000 for immunoblotting), anti-Vimentin, V9 (Santa Cruz, sc-6260, 1:50 for immunostaining), anti-β-TrCP (Santa Cruz, sc-390629, sc-166492, 1:500 for immunoblotting), anti-PKCι (Genetex, GTX102900, 1:1000 for immunoblotting), anti-CK8 (cytokeratin-8: TROMA-I, DSHB, 1:10 for immunostaining), anti-CK5 (cytokeratin-5, Covance, PRB-160P, 1:200 for immunostaining), anti-aPKC (Santa Cruz, sc-216, 1:100 for immunostaining, 1:5000 for immunoblotting), anti-CD45 (BD, 550539, 1:100 for immunostaining), anti-pan-cytokeratin (Abcam, ab9377, 1:200 for immunostaining), anti-phospho-SNAI1(S249) (rabbit polyclonal antibody generated by Abgent, 1:500 for immunoblotting), and anti-GAPDH (Genetex, GTX100118, 1:20000 for immunoblotting). Secondary fluorescent antibodies used include anti-mouse, anti-rabbit, anti-rat conjugated with Alexa Fluor 488, 546 and 647 (Life Technologies). Secondary horseradish peroxidase (HRP)-conjugated antibodies used include anti-mouse and anti-rabbit (Jackson Immunoresearch). Doxycycline and ML7 were purchased from Sigma. PZ09 was synthesized by Reagency (Australia). All Plasmids such as pWZL-Blast (WB)-SNAI1 and pFASTBac-SNAI1, pLKO-Tet-On SNAI1 were generated from cDNA via PCR. Mutants were constructed using site-directed mutagenesis kit (Agilent Technologies). All shRNA constructs were purchased from Sigma and their sequences are list in Supplementary Table 1.
Cell culture
MCF10DCIS cells were grown in DMEM/F12 media supplemented with 5% horse serum, 20ng/ml human EGF, 10μg/ml insulin, 0.5μg/ml hydrocortisone, penicillin, streptomycin and 100ng/ml cholera toxin (Sigma-Aldrich). Caco2 cells were cultured in MEM media supplemented with 20% foetal bovine serum, penicillin, and streptomycin. Sf-9 cells were grown in Sf-900 II SFM media (Gibco) in shaking cultures at 27°C. To generate stable gene knockdown cells, shRNA lentiviruses (human PAR3, mouse Par3, and human SNAI1 MISSION shRNA, Sigma) were transduced into target cells with 6μg/ml protamine sulphate. To generate stable gene overexpressed cells, retroviruses (pWZL-Blast-SNAI1, SNAI1 S249A, and SNAI1 S249E mutant) were infected into target cells with 6μg/ml protamine sulphate. Infected cells were then selected for with 2μg/ml puromycin or blasticidin.
Three-dimensional (3D) cell culture
Caco2 cells were grown in 3D cell culture as previously described51. Briefly, Caco2 cells were seeded on Matrigel (Growth Factor Reduced Matrigel, BD Biosciences) in 2% Matrigel MEM media supplemented with 20% foetal bovine serum, penicillin and streptomycin.
Inducible SNAI1 and TWIST1 transgenic mice
All animal care and experiments were performed in accordance with the animal protocol approved by the Institutional Animal Care and Use Committee of the University of California, San Diego. This study is compliant with all relevant ethical regulations regarding animal research. TetON SNAI1 and TWIST1 mice were generated using a site-specific single copy integration strategy to insert a single copy of transgene in the Col1A locus52. Inducible SNAI1 and Twist1 mice were generated by crossing TetON-SNAI1 and TetON-Twist1 mice with ROSA-rtTA mice. SNAI1 and TWIST1 expression was induced by doxycycline (1μg/ml) treatment.
Isolation and 3D cultures of primary mammary epithelial organoids (MEOs)
MEOs 3D cultures were prepared as previously described53. Briefly, all mammary glands were harvested from ROSA-rtTA/TetON-SNAI1 or ROSA/rtTA/TetON-TWIST1 transgenic mice, minced and digested with 25ml of collagenase solution in DMEM/F12 media with 2% BSA, 25μg/ml gentamicin, 5μg/ml insulin, 0.1g collagenase A (Roche), and 0.2% trypsin in the flask for 30min at 37°C. MEOs were isolated by differential centrifugations (pulse up to 1,500 rpm in 10ml DMEM/F12 media). MEOs were resuspended with DMEM/F12 media. For 3D cultures, MEOs were embedded in 50% Matrigel (BD) and cultured in 3D growth media: DMEM/F12 media supplemented with 1% ITS (sigma), 25ng/ml EGF, 50ng/ml NRG (R&D system), penicillin and streptomycin in 8-well chamber slide (Nunc Lab-Tek). For 2D culture, MEOs were seeded in regular culture plate and grown in growth media with 2% fetal calf serum.
Immunostaining of mammary epithelial organoids (MEOs)
We used a protocol adapted from the method previously described54. In brief, MEOs were fixed in 4% PFA for 20min at room temperature and permeabilized with PBS-0.2% Triton X100 and 0.1% BSA for 10min at 4°C, washed three times with PBS-100mM glycine, and then incubated in PBS-5mM EDTA, 1mM NaVO4, 1.5mM NaF for 1–2 day at 4°C to remove Matrigel. Samples were blocked with 10% goat serum in Immunobuffer (PBS-0.1% Tween20 and 1% BSA) and incubated with primary antibodies in Immunobuffer for overnight, washed three times with Immunobuffer, and incubated with secondary antibodies for 1 hr, washed three times with Immunobuffer and mounted with DAPI-containing mounting medium (Vector). Images were acquired using an Olympus FV1000 confocal microscope and analysed using ImageJ software.
Immunoblotting and Immunoprecipitation
MEOs were isolated in cell recovery solution (Corning) to extract protein from 3D cultures following the manufacturer’s protocol. MEOs were lysed with RIPA buffer (50mM Tris-HCl, 1% NP-40, 0.2% SDS, 0.5% sodium deoxycholate, 150mM NaCl, and 10mM NaF). Immunoprecipitation were prepared as previously described55. Briefly, MCF10A, Caco2 cells and organoids were lysed with IP lysis buffer (20mM Tris-HCl, 1% Triton X-100, 10mM MgCl2, 10mM KCl, 2mM EDTA, 1mM NaF, 1mM sodium orthovanadate, 2.5mM β−6-glycerophosphate, 10% glycerol, pH 7.5). SNAI1 and PKCζ antibodies were conjugated to protein G beads (Invitrogen), crosslinked using disuccinimidyl suberate (Thermo Scientific Pierce) as per the manufacturer’s protocol, incubated with lysates overnight at 4°C, washed five times with IP lysis buffer supplemented with 200mM NaCl, and eluted using 50mM DTT LDS sample buffer at 70°C for 10 min.
Real-time PCR
RNA was extracted from cells and MEOs using the Trizol reagent (Thermo Fisher Scientific). cDNA was generated using random hexamer primers and a cDNA Reverse Transcription Kit (Applied Biosystems). All primer sequences for qPCR are listed in Supplementary Table 2. Expression values were generated using ddCt values normalized to GAPDH and HPRT. Experiments were performed in both biological and technical triplicate using 7500 Fast (Applied Biosystems) and CFX Connect (Bio-Rad) real-time PCR detection systems. For data analysis in each comparison, unpaired two-tailed Student’s t-tests with Welch’s correction were used to determine statistical significance.
In vitro kinase assay
Human Myc-His6-SNAI11 WT and S249A were expressed and pulled down from Sf9 insect cells using Ni-NTA agarose resin. Human PKCζ was expressed and purified from Sf9 insect cells. Phosphorylation reactions were carried out using 1ug of purified SNAI1, 200ng of purified PKCζ in 20mM HEPES (pH 7.4), 5mM MgCl2 and 500uM ATP at 30°C for 1 hour, in the presence of 20uM PZ09 or vehicle control (DMSO).
aPKC kinase activity FRET Imaging
Caco2 cells co-transfected with the PKC activity reporter CKAR and mCherry-tagged PKMζ versus mCherry-Vec control for 2 days were plated in 3D organoids culture on 35-mm imaging dishes, and imaged in HanK’s balanced salt solutions supplemented with 1mM CaCl2. To analyse PKCζ kinase activity, cells were treated with 5μM PZ09 and the change in FRET resulting from phosphorylation of the reporter was measured using methods previously described38.
Caco2 xenograft mouse model
shControl (5.0 × 106), shPAR3 (0.5 × 106), shPAR3 and shSNAI1 (1.0 × 106) Caco2 overexpressing GFP-labelled cells suspended in 100 μl Matrigel (BD Biosciences) were injected bilaterally into the flanks of 7-week-old female BALB/c nude mice. Mice were euthanized and tumour burden was analysed at 6 weeks after tumour cell injection. Mice were dissected and tumour invasion was assessed in situ using a fluorescent dissection scope (Leica Microsystems).
Isolation and analysis of circulating tumour cells
Peripheral blood was obtained from tumour-bearing mice via intracardiac puncture at the termination of the experiment. Red blood cells (RBCs) were removed by dextran solution, RBC lysis buffer, and CD45 magnetic beads (Invitrogen). Remaining cells were fixed in 2% PFA. Cells were then spun onto slides using a cytospin and stained with rat anti-CD45 and rabbit anti-pan-cytokeratin (Abcam) antibodies, followed by DAPI nuclear stain. Images were acquired using an Olympus FV1000 confocal microscope and analysed using ImageJ software.
Tumour tissue microarrays
Tissue microarrays (TMAs) were purchased from US Biomax Inc (BB08015, BR246a, T087b). A total of 48 duplicates TMA samples included 6 normal breast, 21 ductal intraepithelial neoplasia and 21 invasive ductal carcinoma. For retrospective analysis, 371 Stage-2 TMA were also purchased from National Cancer Institute Cancer Diagnosis Program. The TMA contains human tissues obtained with informed consent according to US federal law and are exempt from Institutional Review Board review by the University of California, San Diego Human Research Protections Program. Immunostaining for SNAI1 and PAR3 was performed as described above. TMAs were concurrently imaged by confocal microscopy. All samples were scored blindly according to the following methods. Score = qp + qs. For proportion of immune-positive (qp), 1 for 0–4%, 2 for 5–19%, 3 for 20–39%, 4 for 40–59%, 5 for 60–79%, and 6 for 80–100%. For strength of staining (qs), 0 for negative, 1 for weak expression, 2 for intermediate expression, 3 for strong expression55, 56. Immunohistochemistry for SNAI1 and PAR3 were performed.
Statistics and Reproducibility
Statistical analysis was performed using GraphPad Prism Software. All P values were derived from Student’s t-test comparing two groups using unpaired two-tailed analysis with Welch’s correction or paired two-tailed t-test, unless otherwise noted. Error bars denote standard deviation unless otherwise noted. Statistical significance was defined as P < 0.05, with regard to the null hypothesis. All qualitative data shown using representative data were repeated in at least 3 independent biological replicates. Correlation of SNAI1 and PAR3 levels in human breast cancer patients was analysed using a χ2 test.
Data Availability
Source data for figures 1–8 and supplementary figures 1–6 are provided in Supplementary Table 3. All other data are available from the authors on reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Yang lab for helpful discussions, especially Tiffany Lee for mouse colony maintenance, Julia Callender for technical help, and Kay Yeung for constructive comments. We thank Antonio Garcia de Herreros for SNAI1 EC3 antibody, Monique Aumailley for anti-laminin V antibody, Andrew Ewald for advice on mouse primary mammary organoid isolation, Ian Macara for the pLV-shPar3-Venus construct used in pilot experiments. We thank UCSD Shared Microscope Facility and UCSD Cancer Centre Support Grant P30 CA23100 from NCI. This work was supported by grants from ACS (RSG-09-282-01-CSM), NCI (1RO1CA168689, 1R01CA174869, 1R01CA206880, and 1R21CA191442) to J.Y. and NIH R35 GM122523 and NIH P01 DK054441 to A. C. N.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no financial and non-financial competing interests.
REFERENCE
- 1.Royer C & Lu X Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ 18, 1470–1477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zen K et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene 28, 2910–2918 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A & Massey-Harroche D Polarity complex proteins. Biochim Biophys Acta 1778, 614–630 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lee M & Vasioukhin V Cell polarity and cancer--cell and tissue polarity as a non-canonical tumor suppressor. J Cell Sci 121, 1141–1150 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Ngok SP, Lin WH & Anastasiadis PZ Establishment of epithelial polarity--GEF who’s minding the GAP? J Cell Sci 127, 3205–3215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horikoshi Y et al. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J Cell Sci 122, 1595–1606 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Goldstein B & Macara IG The PAR proteins: fundamental players in animal cell polarization. Dev Cell 13, 609–622 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A & Ohno S Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells 7, 1161–1171 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Graybill C, Wee B, Atwood SX & Prehoda KE Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J Biol Chem 287, 21003–21011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaffrey LM, Montalbano J, Mihai C & Macara IG Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell 22, 601–614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCaffrey LM & Macara IG Signaling pathways in cell polarity. Cold Spring Harb Perspect Biol 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue B, Krishnamurthy K, Allred DC & Muthuswamy SK Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat Cell Biol 15, 189–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iden S et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell 22, 389–403 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Brabletz T To differentiate or not--routes towards metastasis. Nat Rev Cancer 12, 425–436 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Tsai JH & Yang J Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 27, 2192–2206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieto MA, Huang RY, Jackson RA & Thiery JP Emt: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Kalluri R & Weinberg RA The basics of epithelial-mesenchymal transition. J Clin Invest 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamouille S, Xu J & Derynck R Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Craene B & Berx G Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13, 97–110 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Yang J & Weinberg RA Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Barrallo-Gimeno A & Nieto MA The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132, 3151–3161 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Moreno-Bueno G, Portillo F & Cano A Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 27, 6958–6969 (2008). [DOI] [PubMed] [Google Scholar]
- 23.de Herreros AG, Peiro S, Nassour M & Savagner P Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol Neoplasia 15, 135–147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ocana OH et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Tsai JH, Donaher JL, Murphy DA, Chau S & Yang J Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran HD et al. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res 74, 6330–6340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung HY, Fattet L & Yang J Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res 21, 962–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei SC et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol 17, 678–688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewald AJ Isolation of mouse mammary organoids for long-term time-lapse imaging. Cold Spring Harb Protoc 2013, 130–133 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Shamir ER et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J Cell Biol 204, 839–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewald AJ, Brenot A, Duong M, Chan BS & Werb Z Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell 14, 570–581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou SS, Diz-Munoz A, Weiner OD, Fletcher DA & Theriot JA Myosin light chain kinase regulates cell polarization independently of membrane tension or Rho kinase. J Cell Biol 209, 275–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peinado H, Olmeda D & Cano A Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7, 415–428 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Zhou BP et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6, 931–940 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Joberty G, Petersen C, Gao L & Macara IG The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2, 531–539 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Tobias IS et al. Protein kinase Czeta exhibits constitutive phosphorylation and phosphatidylinositol-3,4,5-triphosphate-independent regulation. Biochem J 473, 509–523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durgan J, Kaji N, Jin D & Hall A Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J Biol Chem 286, 12461–12474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobias IS & Newton AC Protein Scaffolds Control Localized Protein Kinase Czeta Activity. J Biol Chem 291, 13809–13822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Estrada OM et al. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J 394, 449–457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aigner K et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26, 6979–6988 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaderna S et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 68, 537–544 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Whiteman EL, Liu CJ, Fearon ER & Margolis B The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene 27, 3875–3879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothenberg SM et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res 70, 2158–2164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee SJ & McCaffrey L Emerging role of cell polarity proteins in breast cancer progression and metastasis. Breast Cancer (Dove Med Press) 6, 15–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muthuswamy SK & Xue B Cell polarity as a regulator of cancer cell behavior plasticity. Annu Rev Cell Dev Biol 28, 599–625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker PJ, Justilien V, Riou P, Linch M & Fields AP Atypical protein kinase Ciota as a human oncogene and therapeutic target. Biochem Pharmacol 88, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunaratne A, Thai BL & Di Guglielmo GM Atypical protein kinase C phosphorylates Par6 and facilitates transforming growth factor beta-induced epithelial-to-mesenchymal transition. Mol Cell Biol 33, 874–886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guyer RA & Macara IG Loss of the polarity protein PAR3 activates STAT3 signaling via an atypical protein kinase C (aPKC)/NF-kappaB/interleukin-6 (IL-6) axis in mouse mammary cells. J Biol Chem 290, 8457–8468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosse C et al. Control of MT1-MMP transport by atypical PKC during breast-cancer progression. Proc Natl Acad Sci U S A 111, E1872–1879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diaz-Meco MT & Moscat J The atypical PKCs in inflammation: NF-kappaB and beyond. Immunol Rev 246, 154–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debnath J, Muthuswamy SK & Brugge JS Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Beard C, Hochedlinger K, Plath K, Wutz A & Jaenisch R Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Fata JE et al. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol 306, 193–207 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee GY, Kenny PA, Lee EH & Bissell MJ Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4, 359–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin J et al. Association of PKCzeta expression with clinicopathological characteristics of breast cancer. PLoS One 9, e90811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amaral T, McKenna SJ, Robertson K & Thompson A Classification and immunohistochemical scoring of breast tissue microarray spots. IEEE Trans Biomed Eng 60, 2806–2814 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figures 1–8 and supplementary figures 1–6 are provided in Supplementary Table 3. All other data are available from the authors on reasonable request.