ABSTRACT
We previously showed that the rdn1 and sunn supernodulation mutants of Medicago truncatula respond differentially to overexpression of the rhizobial CLAVAT3/EMBRYO SURROUNDING REGION (CLE) signaling peptides MtCLE12p and MtCLE13p, allowing the order of action of the genes to be determined in the autoregulation of nodulation (AON) signal transduction pathway. We tested the same gene constructs that lead to the production of proteolytically processed peptides (indicated by a p after the name) in plants mutant for two other proteins that control nodule number (CRN and CRA2) and were able to determine that CRN is involved in the same signaling pathway as MtCLE12p and MtCLE13p, while regulation in CRA2 mutants responds normally to the peptides, suggesting CRA2 likely signals separately from SUNN, RDN1, and CRN. Based on the analysis of the double mutant of cra2-2 and sunn-4, we also confirm recent findings that CRA2 acts independently of SUNN in nodule number regulation.
KEYWORDS: Medicago truncatula, autoregulation of nodulation, CRN, CRA2, CLE12, CLE13, nitrate demand signaling
Introduction
CLE peptides are involved in many signal transduction pathways affecting plant root growth and development (for review see1,2). A subset of CLE peptides in legumes have been shown to be involved in the autoregulation of nodulation (AON). In soybean, Lotus japonicus and Medicago truncatula, some CLE peptides negatively regulate nodule development3-7 and evidence supports a model in which the peptides genes are upregulated in the nodule meristem and peptides transported to receptors in the shoot of the plant.6,7 A subsequent signal back to the root results in a halt to further nodule development, and recent findings implicate cytokinins8 and a microRNA9 in this signal.
We showed that overexpression of the M. truncatula nodulation regulatory peptides MtCLE12p and MtCLE13p in rdn1-2 hypernodulation mutants yielded genetic evidence that the hydroxyproline O-arabinosyltransferase enzyme encoded by RDN1 modifies MtCLE12p. Further, that modification was necessary for regulatory signaling by MtCLE12p but not signaling by MtCLE13p.10 The receptor kinase SUNN, mutation of which also cause hypernodulation,11 was shown genetically to be the receptor for both MtCLE12p and MtCLE13p and sunn-4 plants were used as a negative control unresponsive to the peptides.10 In M. truncatula, SUNN has the highest homology to CLV1 in Arabidopsis.12
Two other molecules are predicted to associate with SUNN. The pseudokinase CRN has been shown by bimolecular fluorescence complementation to associate with SUNN, and mutations in CRN co-segregate with an increased nodule phenotype.13 Likewise, CLV2 associates with both SUNN and CRN13 and mutations in CLV2 cause an increased nodule phenotype.14 Evidence from Arabidopsis also indicates that CLV1, CLV2 and CRN associate with each other and signal together in some pathways.15–18
In contrast, mutation of the CRA2 receptor kinase gene in M. truncatula has the opposite effect of CRN gene mutations, reducing nodule number in a systemic manner.19 Genetic evidence suggests the CRA2 is the receptor for the CEP1 peptide.20 The CEP peptides are part of a signaling system for nitrogen demand.21 Since high nitrate reduces or eliminates nodulation, the authors of the work above postulated that CRA2 might be involved in SUNN nodule regulatory signaling as well.20
Based on this data, we hypothesized that constitutive expression of MtCLE12 and MtCLE13 could require CRN to regulate nodule number. If CRA2 is involved in SUNN regulatory signaling, we postulated constitutive expression of MtCLE12 and MtCLE13 would affect systemic nodule number signaling in CRA2 mutants as well. We performed the same experiments in10 in a M. truncatula crn mutant and the cra2-2 mutant identified in.19 Before beginning, we confirmed that we could rescue the hypernodulation phenotype of the crn mutant used in13 with the CRN message, proof that the Tnt1 insertion in the crn mutant is the cause of the phenotype. Testing both mutants allowed us to determine that cross talk between the nodule regulatory pathway and the nitrogen demand signaling pathway does not involve the M. truncatula rhizobia-induced CLEs, despite the fact that both sunn and rdn1 mutants have altered phenotypic plasticity in response to nitrate in the absence of rhizobia.22 Phenotypic analysis of the sunn-5;cra2-2 double mutant further confirmation this finding of independence.
Results
The CRN coding sequence rescues the CRN mutation
The CRN coding sequence used in13 was cloned and transformed by tissue culture into the crn mutant as described in23 and multiple T0 whole plant transgenics carrying the construct were obtained. Two independent transgenic lines carrying the construct (T1) were selected for analysis because they segregated only plants carrying the construct. T2 plants from these lines were grown in an aeroponic chamber in nodulation medium and inoculated as in.10 The crn mutant plants carrying the CRN construct displayed wild type nodule numbers when compared to the R108 wild type (no statistical difference, Student’s t-test) and different from the parental crn mutant used for transformation (Student's t-test, p > 0.001, Figure 1(a)). The results confirm the suggestion in13 that the lesion in the CRN gene is responsible for the hypernodulation phenotype of crn mutants.
Figure 1.
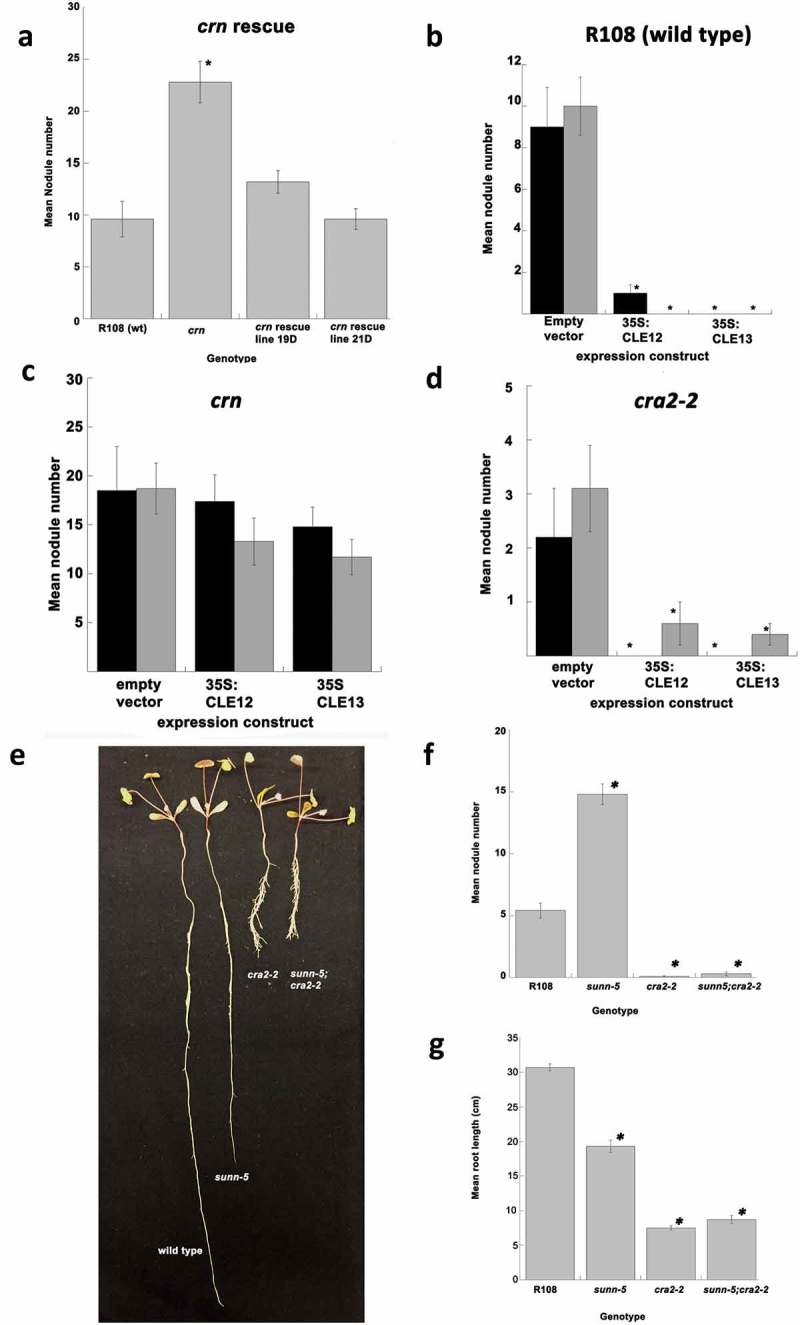
MtCLE12 and MtCLE13 are dependent on CRN but not CRA2 for AON signaling. (a) Expression of wild type CRN in crn plants rescues the mutant phenotype. Nodule number in wild type R108 and the mutant crn line compared to progeny of each of two lines from independent transformations of crn mutants expressing wild type CRN under the CaMV 35S promoter (21D and 19D). (n = 10–12 per genotype). * indicates significant difference from wild type as determined by Student's t-test, p < 0.001. (b and c) MtCLE12 and MtCLE13 overexpression effects depend on CRN. Data are mean number of nodules 14 days post inoculation with S. medicae in transgenic hairy roots constitutively expressing MtCLE12 or MtCLE13 under the CaMV 35S promoter. Grey and white bars indicate the results of two independent experiments. (b) Wildtype R108, n = 8–10 plants per construct per experiment (c) crn n = 4–8 plants per construct per experiment (d) MtCLE12 and MtCLE13 overexpression effects do not depend on CRA2. cra2-2 n = 5–9 plants per construct per experiment. Error bars indicate standard error of mean, * indicates significance of group from empty vector control, Student's t-test, p < 0.01. (e) The cra2-2;sunn-5 double mutant displays the cra2-2 phenotype. Photo left to right is of wild type, sunn-5, cra2-2 and sunn-5;cra2-2 double mutant plants. (f) Nodule phenotypes of F2 progeny of cra2-2 crossed to sunn-5. Error bars indicate standard error of mean, * indicates significance of group from wild type as determined by Student's t-test, p < 0.001. (n = 10 (wt) 15 (sunn-5) 14 (cra2-2) and 16 (sunn-5;cra2-2). (g) Root length phenotypes of the same genotypes, uninoculated. (n = 10–16). Error bars indicate standard error of mean, * indicates significance of group from wild type as determined by Student's t-test, p < 0.001.
CRN is part of the SUNN/CLE signaling pathway, but CRA2 is not
The MtCLE12 and MtCLE13 peptide genes as well as an empty vector control were constitutively expressed in composite hairy roots of R108 ecotype wild type plants as in.10 The construct carries a DS-Red marker which allows identification of transformed roots by microscopy and only these roots were used in the analysis. Inoculation with rhizobia resulted in a normal number of nodules in the empty vector control. Significantly reduced nodulation was observed in plants constitutively expressing either the MtCLE12 or the MtCLE13 gene (Figure 1(b) p < 0.001, Student's t-test), in agreement with previous findings for this experiment performed in wild type plants of the A17 ecotype.4,10 However, expression of both the empty vector control and either the MtCLE12 or the MtCLE13 gene in composite hairy roots had no effect on the increased nodule number observed in crn mutant plants (Figure 1(c)). This is the same result we and other observed for the experiment done in a sunn mutant background4,10 and indicates that both CLE peptides signal through a pathway that involves the CRN pseudokinase. While it is possible CRN is a receptor for the CLEs, it is more likely based on experiments on the interactions of CRN with other molecules in Arabidopsis and M. truncatula that this is evidence of a downstream effect. Since CRN and SUNN have been shown to physically interact13 it may be that CRN responds to the binding of the CLEs to the SUNN kinase with which it associates, and this response does not occur in a crn mutant.
In contrast, when the MtCLE12 and MtCLE13 peptide genes were constitutively expressed in cra2-2 mutant plants in the R108 background, the low nodule phenotype of these plants was significantly lower than mutants expressing the empty vector (Figure 1(d), p < 0.01, Student's t-test), similar to the effect of expression of these genes in wild type plants. From this, we conclude that MtCLE12p and MtCLE13p are not involved in the CRA2 nodule regulatory pathway. Further evidence of independence is the phenotype of plants carrying mutations in both sunn-513 and cra2-2 .20 The sunn-5 allele in the R108 background contains a Tnt1 transposon insertion in an exon near the end of the extracellular domain of the receptor, resulting in a three-fold increase in nodule number over the corresponding wild type (Figure 1(f)). The sunn-5;cra2-2 double mutant, identified by PCR among F2 plants from a cra2-2 cross to sunn-5, was analyzed as in Figure 1(a) for nodule number and the compact root architecture phenotype of cra2-2. Plants homozygous for both mutations displayed the cra2-2 compact root with many laterals and low nodule number phenotypes (Figure 1(e–g)). The presence of the cra2-2 allele drastically reduced the nodule number in plants carrying the sunn-5 allele (Student's t-test, p < .001), however, there was no statistical difference in nodule number between the double mutant and plants containing only the cra2-2 allele. Recent work with an allelic series of cra2 mutants in the A17 background revealed an intermediate nodule phenotype close to wild type nodulation in their sunn;cra2 double mutants, 24 but the less severe reduction in nodulation is likely due to the alleles and ecotype used to create the double mutant. The cra2-2 allele in the R108 genotype has a severe reduction in nodule number (many plants don‘t make a nodule at 14 days post inoculation) in our aeroponic system compared the cra2 alleles in the A17 genotype in their pouch system (3–5 nodules per plant at 14 days post inoculation) and the sunn-5 allele in R108 is not as strong (3x increase) as the sunn-4 allele in A17 (10x increase) in our system. However, our results support their conclusion that the CEP/CRA2 and CLE/SUNN systemic pathways act independently from the shoots to regulate nodule number, 24 and implicate CRN in the CLE/SUNN/CRN systemic pathway.
Funding Statement
This work was supported by the National Science Foundation USA [IOS #1444461].
Acknowledgments
We thank Florian Frugier for the gift of the cra2-2 mutant and Sofie Goormachtig for the gift of the MtCLE12 and MtCLE13 constructs. This work is supported by NSF IOS #1444461 to J.F.
References
- 1.Oh E, Seo PJ, Kim J.. Signaling peptides and receptors coordinating plant root development. Trends Plant Sci. 2018;23(4):337–351. doi: 10.1016/j.tplants.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Kucukoglu M, Nilsson O.. CLE peptide signaling in plants-the power of moving around. Physiol Plant. 2015;155(1):74–87. doi: 10.1111/ppl.2015.155.issue-1. [DOI] [PubMed] [Google Scholar]
- 3.Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’haeseleer K, Holsters M, Goormachtig S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153(1):222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 2012;70(3):367–376. doi: 10.1111/tpj.2012.70.issue-3. [DOI] [PubMed] [Google Scholar]
- 5.Osipova MA, Mortier V, Demchenko KN, Tsyganov VE, Tikhonovich IA, Lutova LA, Dolgikh EA, Goormachtig S. Wuschel-related homeobox5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 2012;158(3):1329–1341. doi: 10.1104/pp.111.189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- 7.Hastwell AH, Gresshoff PM, Ferguson BJ. The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Funct Plant Biol. 2015;42(3):229–238. doi: 10.1071/FP14222. [DOI] [PubMed] [Google Scholar]
- 8.Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M. Nodule inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci U S A. 2014;111(40):14607–14612. doi: 10.1073/pnas.1412716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsikou D, Yan Z, Holt DB, Abel NB, Reid DE, Madsen LH, Bhasin H, Sexauer M, Stougaard J, Markmann K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science. 2018;362(6411):233–236. doi: 10.1126/science.aat6907. [DOI] [PubMed] [Google Scholar]
- 10.Kassaw T, Nowak S, Schnabel E, Frugoli J. ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiol. 2017;174(4):2445–2456. doi: 10.1104/pp.16.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR. Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 2003;131:998–1008. doi: 10.1104/pp.015677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnabel E, Journet E-P, de Carvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN Gene encoding a CLV1-like leucine-rich repeat receptor kinase regulates nodule number and root length. Plant Mol Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- 13.Crook AD, Schnabel EL, Frugoli JA. The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J. 2016;88(1):108–119. doi: 10.1111/tpj.13234. [DOI] [PubMed] [Google Scholar]
- 14.Krusell L, Sato N, Fukuhara I, Koch BEV, Grossmann C, Okamoto S, Oka-Kira E, Otsubo Y, Aubert G, Nakagawa T, et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011;65(6):861–871. doi: 10.1111/tpj.2011.65.issue-6. [DOI] [PubMed] [Google Scholar]
- 15.Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20(4):934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, Jin JB, Liu C-M, Lin J. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61(2):223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]
- 17.Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell. 2011;23(3):851–854. doi: 10.1105/tpc.110.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011;21(5):345–352. doi: 10.1016/j.cub.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F. Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet. 2014;10(12):e1004891. doi: 10.1371/journal.pgen.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid D, Stougaard J, Frugier F, Imin N, Djordjevic M. Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol. 2016;171(4):2536–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabata R, Sumida K, Yoshii T, Kentaro O, Shinohara H, Matsubayashi Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science. 2014;346(6207):343–346. doi: 10.1126/science.1255826. [DOI] [PubMed] [Google Scholar]
- 22.Goh CH, Nicotra AB, Mathesius U. Genes controlling legume nodule numbers affect phenotypic plasticity responses to nitrogen in the presence and absence of rhizobia. Plant Cell Environ. 2018online ahead of print. doi: 10.1111/pce.13498. [DOI] [PubMed] [Google Scholar]
- 23.Wen L, Chen Y, Schnabel E, Crook A, Frugoli J. Comparison of efficiency and time to regeneration of Agrobacterium-mediated transformation methods in Medicago truncatula. Plant Methods. 2019;15(20). doi: 10.1186/s13007-018-0385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laffont C, Huault E, Gautrat P, Endre G, Kalo P, Bourion V, Duc G, Frugier F. Independent regulation of symbiotic nodulation by the SUNN naegative and cra2 positive system pathways. Plant Physiol. 2019; 01588.2018 online ahead of print. doi: 10.1104/pp.18.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]


