ABSTRACT
The TARGET OF RAPAMYCIN-SNF1-RELATED PROTEIN KINASE 1 (TOR-SnRK1) arms race is a key regulator of plant growth in response to energy fluctuations and stress. Recently, we have identified that two members of the FCS-LIKE ZINC FINGER (FLZ) protein family, FLZ6 and 10, repress SnRK1 signaling and thereby involved in the activation of the TARGET OF RAPAMYCIN (TOR) signaling. In this study, we demonstrate that FLZ6 and 10 are also involved in the regulation of osmotic stress responses. Downregulation of FLZ6 and 10 results in enhanced expression of stress-responsive genes and better resilience towards osmotic stress at the seedling stage. These results indicate that FLZ6 and 10 are involved in the regulation of stress mitigation in plants through directly affecting SnRK1 signaling.
KEYWORDS: FCS-Like Zinc finger, SNF1-related protein kinase 1, Target of Rapamycin, osmotic stress, abscisic acid
Organisms constantly monitor the energy and nutrient status to optimize growth according to nutrient/energy availability. The TARGET OF RAPAMYCIN- SNF1-RELATED PROTEIN KINASE 1 (TOR-SnRK1) arms race in response to energy status lies in the centre of nutrition-dependent growth in plants.1,2 In response to energy and nutrient sufficiency, TOR is activated which accelerates growth through the promotion of protein synthesis, translation reinitiation, cell cycle progression etc.3-5 Upon nutrient deficiency, SnRK1 inhibits TOR pathway and through other phosphorylation events prepare the cell to survive under nutrient deficiency2 Energy and stress signaling are highly interconnected. Plants balance abiotic stress and growth response through the interaction of TOR-SnRK1 and ABA signaling. The TOR and ABA signaling were found to be reciprocally regulated in response to stress and nutrient sufficiency signals6 During normal growth conditions, TOR represses ABA signaling through inhibiting the activity of PYRABACTIN RESISTANCE 1-LIKE (PYL) receptors. In response to stress, the SNF1-RELATED PROTEIN KINASE 2 (SnRK2), which is activated through PYL, phosphorylates the regulatory component of TOR, REGULATORY-ASSOCIATED PROTEIN OF TOR (RAPTOR) which culminates in the inhibition of TOR activity6 ABA positively regulates SnRK1 signaling by inhibiting clade A type 2C protein phosphatases (PP2Cs), which negatively regulate SnRK1, SnRK2, and SnRK3 family members7 Intriguingly, the upstream activating kinases of SnRK1, SnRK1-ACTIVATING KINASE 1 and 2 (SnAK1 and 2) can phosphorylate and activate SnRK3 family members8 Collectively, these studies indicate that the balance between TOR, ABA, and SnRKs is necessary for optimizing growth and abiotic stress response.
The FCS-LIKE ZINC FINGER (FLZ) proteins are a class of land-plant specific C2-C2 zinc finger proteins which promiscuously interact with the subunits of SnRK1.9-12 In Arabidopsis, the expression of this multigene family is highly responsive to sugar and energy status and various abiotic stresses and are proposed to work as adaptors of SnRK1 complex.11-14 Recently, we found that two starvation-induced FLZ genes, FLZ6 and FLZ10, act as negative regulators of SnRK1 signaling15 Mutants of these genes accumulated more SnRK1α1, which culminated in enhanced SnRK1 activity and attenuated growth even under favourable growth conditions. We found that the inhibition of TOR signaling due to the enhanced SnRK1 activity is the major reason for this growth suppression. Taken together, our results suggest that FLZ6 and 10 are involved in the regulation of TOR-SnRK1 dynamics and energy-dependent growth in plants. Similar to SnRK1α1 overexpression lines, mutant lines of FLZ6 and 10 were ABA hypersensitive indicating that these genes may have a role to play in ABA and stress responses15 The role of TOR-SnRK1 dynamics in controlling stress responses and the ABA hypersensitivity of flz6 and flz10 mutants prompted us to analyze whether FLZ6 and 10 are also involved in regulating stress response in plants. We focused our study on osmotic stress responses because TOR-SnRK1 dynamics is implicated in osmotic stress mitigation. Osmotic stress represses TOR signaling and understandably, the mutant lines of TOR and RAPTOR show altered sensitivity towards osmotic stress.6,16 SnRK1α1 was found to be essential for the induction of autophagy in response to mannitol treatment17
In order to test the role of FLZ6 and 10 in osmotic stress at the physiological level, we used previously characterized mutant lines (flz6.1 and flz10.1) with enhanced SnRK1 activity for the physiological assays15 Stratified seeds were grown for 5 days in 0.5X MS medium. Five days after germination (DAG) seedlings were transferred to 0.5X MS medium supplemented with different concentrations (0, 100, 200 and 300 mM) of mannitol and phenotype was compared with control seedlings after 5 days (Figure 1A). Mannitol treatment caused a dose-dependent reduction in the primary root length and fresh weight in all lines (Figure 1B). As observed previously,15 the mutant lines showed a significant reduction in the growth parameters in control conditions. However, growth inhibition in response to stress treatment in the mutants was significantly attenuated at many concentrations. The flz6.1 line showed stronger resistance towards mannitol treatment with significantly long primary root growth at severe osmotic stress condition (300 mM mannitol). The flz10.1 line also showed resistance towards osmotic stress treatment especially at severe osmotic stress condition (Figure 1B).
Figure 1.
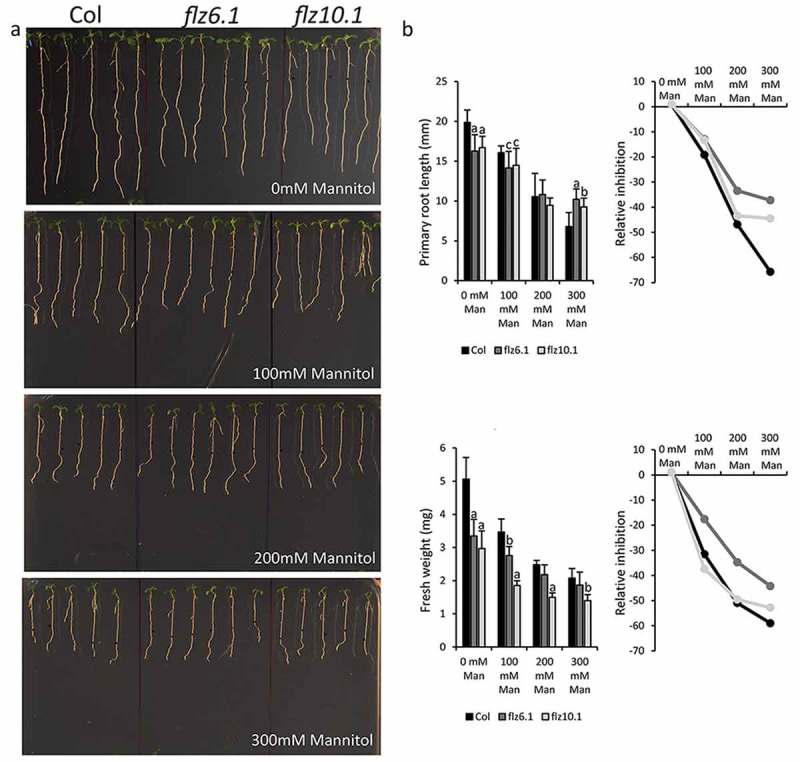
Osmotic stress sensitivity of flz6 and flz10 mutants. 5DAG seedlings were transferred to 0.5X MS medium supplemented with different concentrations of mannitol and sensitivity was assayed after the 5th day of transfer. (A) The phenotype of WT and flz6.1 and flz10.1 lines with and without mannitol treatment. (B) Primary root length and fresh weight of WT and flz mutant lines under different mannitol treatment. The bar graph represents the absolute values and the line graph represents the reduction in the studied parameter due to mannitol treatment relative to the control experiment. The experiment was repeated three times yielding similar results. At least 10 seedlings were used for each treatment. The letters above the bars indicate the statistical difference in the studied parameter in the mutant in comparison with the WT grown under the same condition (Two-tailed Student’s t-test; a = p ≤ 0.001, b = p ≤ 0.01, c = p ≤ 0.05).
Overexpression of SnRK1α1 cause induction of many stress-responsive genes.18,19 We tested the level of five stress-responsive genes in 5DAG seedlings by qRT-PCR using the primers listed in Table S1. We observed a modest increase in the expression of many of these genes in both mutant lines (Figure 2A). Remarkably the expression was more pronounced in the flz6.1 in comparison with flz10.1 which correlates with the stress response of these lines in the physiological assays. The expression of most of these genes were also high in the publically available microarray data in which SnRK1α1 is overexpressed in mesophyll protoplasts (Figure 2B)19
Figure 2.
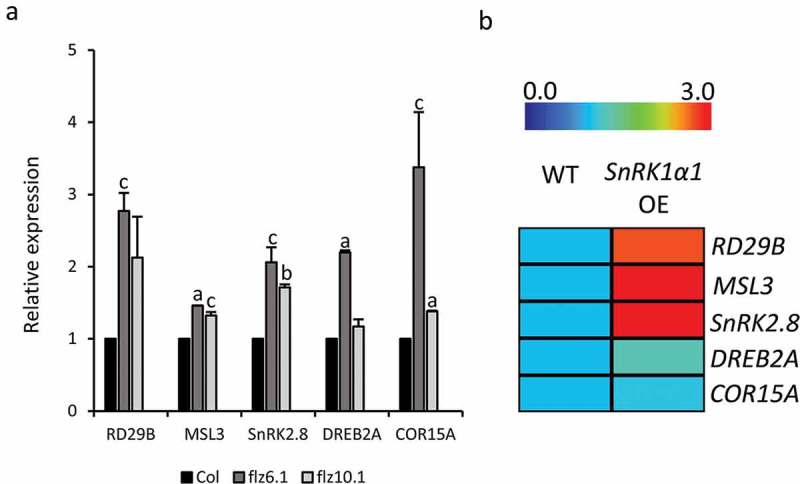
Expression of stress-regulated genes in flz6 and flz10 mutants. (A) Expression of stress-regulated genes in 5DAG flz6.1 and flz10.1 seedlings in comparison with WT. 18S rRNA was used as the endogenous control. Data shown is the average of two biological replicates with 3 technical replicates each. The letters above the bars indicate the statistical difference in the expression in the mutant in comparison with the WT (Two-tailed Student’s t-test; a = p ≤ 0.001, b = p ≤ 0.01, c = p ≤ 0.05. (B) The heat map showing the expression of stress-regulated genes in the protoplasts overexpressed with SnRK1α1 (NCBI GEO accession: GSE8257).
Along with its central role in controlling adaptive responses during energy deficiency, the role of SnRK1 in promoting tolerance towards abiotic stresses such as drought and submergence has recently been appreciated.20-22 Our physiological and gene expression assays using mutant lines indicate that FLZ6 and 10 are involved in stress responses possibly through their effect on SnRK1α1 stability. The enhanced tolerance observed in the mutant lines of FLZ6 and 10 could be partly due to constitutive downregulation of TOR activity in these lines because TOR activity was found to be rapidly inhibited in response to osmotic stress.6,15,16 Further, SnRK1 is known to activate the transcription of a wide variety of general and specific stress-responsive genes19 Indeed, a significant overlap was observed in the transcriptome of SnRK1 and ABA indicating synergism of both pathways7 However, molecular nodes of SnRK1 and stress interaction is still elusive. It would be critical to analyze whether SnRK1 can phosphorylate and activate transcription factors and key signaling proteins involved in the stress tolerance pathways. FLZ proteins might be involved in these events by working as an adaptor which can modulate the recruitment of target proteins to the kinase complex. Future experiments will be directed at testing these hypotheses at the molecular level. A deeper understanding of the intricacies of SnRK1-FLZ signaling and its interaction with stress pathways will help us in improving crop plants. Taken together, our recent report15 and the present study highlights FLZ6 and FLZ10 as the land-plant specific regulatory modules of SnRK1 and this regulation has wide influence in controlling the growth and stress resilience in plants.
Funding Statement
This work was supported by National Institute of Plant Genome Research, Department of Biotechnology, Ministry of Science and Technology (DBT) (Core Grant) and Department of Biotechnology, Ministry of Science and Technology (DBT) (Project Grant: BT/PR8001/BRB/10/1211/2013). DS and MS acknowledge the University Grant Commission, Government of India for the research fellowship. MS acknowledges Department of Biotechnology for the research fellowship. MJK and CTM acknowledge National Institute of Plant Genome Research, Department of Biotechnology, Ministry of Science and Technology (DBT) for the research fellowship. SJ acknowledges the research fellowship from the Project Grant from the Department of Biotechnology, Ministry of Science and Technology (DBT) (Project Grant: BT/PR12855/BPA/118/87/2015). MJK acknowledges research grant and a fellowship from Department of Science and Technology, Ministry of Science and Technology (DST) (INSPIRE Faculty Programme, Grant no. IFA18-LSPA110).
Acknowledgments
The authors acknowledges DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Disclosure of Conflicts of Interest
The authors declare no conflict of interest.
Supplementary material
Supplementary data for this article can be accessed here.
References
- 1.Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C.. TOR signaling and nutrient sensing. Annu Rev Plant Biol. 2016;67:261–285. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- 2.Broeckx T, Hulsmans S, Rolland F.. The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot. 2016;67:6215–6252. doi: 10.1093/jxb/erw416. [DOI] [PubMed] [Google Scholar]
- 3.Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell. 2012;24:4850–4874. doi: 10.1105/tpc.112.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. Embo J. 2017;36:886–903. doi: 10.15252/embj.201694816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Zhao Y, Li Z, Hsu -C-C, Liu X, Fu L, Hou Y-J, Du Y, Xie S, Zhang C, et al. Reciprocal regulation of the tor kinase and ABA receptor balances plant growth and stress response. Mol Cell. 2018;69:100–112.e6. doi: 10.1016/j.molcel.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, Elias A, Rabissi A, Lumbreras V, Gonzalez-Guzman M, et al. ABI1 and PP2CA phosphatases are negative regulators of snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell. 2013;25:3871–3884. doi: 10.1105/tpc.113.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barajas-Lopez JDD, Moreno JR, Gamez-Arjona FM, Pardo JM, Punkkinen M, Zhu J-K, Quintero FJ, Fujii H. Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 2018;93:107–118. doi: 10.1111/tpj.13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamsheer KM, Laxmi A. DUF581 is plant specific FCS-Like Zinc Finger involved in protein-protein interaction. PLoS One. 2014;9:e99074. doi: 10.1371/journal.pone.0099074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamsheer KM, Mannully CT, Gopan N, Laxmi A. Comprehensive evolutionary and expression analysis of FCS-Like Zinc finger gene family yields insights into their origin, expansion and divergence. PLoS One. 2015;10:e0134328. doi: 10.1371/journal.pone.0134328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nietzsche M, Schießl I, Börnke F. The complex becomes more complex: protein-protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell- and stimulus type-specific SnRK1 signaling in plants. Front Plant Sci. 2014;5:54. doi: 10.3389/fpls.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamsheer KM, Shukla BN, Jindal S, Gopan N, Mannully CT, Laxmi A. The FCS-like zinc finger scaffold of the kinase SnRK1 is formed by the coordinated actions of the FLZ domain and intrinsically disordered regions. J Biol Chem. 2018;293(34):13134–13150. doi: 10.1074/jbc.RA118.002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamsheer KM, Laxmi A. Expression of Arabidopsis FCS-Like Zinc Finger genes is differentially regulated by sugars, cellular energy level, and abiotic stress. Front Plant Sci. 2015;6:746. doi: 10.3389/fpls.2015.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nietzsche M, Landgraf R, Tohge T, Börnke F. A protein-protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in Arabidopsis thaliana. Curr Plant Biol. 2016;5:36–44. doi: 10.1016/j.cpb.2015.10.004. [DOI] [Google Scholar]
- 15.Jamsheer KM, Sharma M, Singh D, Mannully CT, Jindal S, Shukla BN, Laxmi A. FCS-like zinc finger 6 and 10 repress SnRK1 signalling in Arabidopsis. Plant J. 2018;94:232–245. doi: 10.1111/tpj.13854. [DOI] [PubMed] [Google Scholar]
- 16.Mahfouz MM, Kim S, Delauney AJ, Verma DPS. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soto-Burgos J, Bassham DC. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One. 2017;12:e0182591. doi: 10.1371/journal.pone.0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baena-González E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Su -Z-Z, Huang L, Xia F-N, Qi H, Xie L-J, Xiao S, Chen Q-F. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front Plant Sci. 2017;8:1201. doi: 10.3389/fpls.2017.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho H-Y, Wen T-N, Wang Y-T, Shih M-C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J Exp Bot. 2016;67:2745–2760. doi: 10.1093/jxb/erw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho Y-H, Hong J-W, Kim E-C, Yoo S-D. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012;158:1955–1964. doi: 10.1104/pp.111.189829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


